Abstract
Genomic analyses have revolutionized our understanding of the biology of B-progenitor acute lymphoblastic leukemia (ALL). Studies of thousands of cases across the age spectrum have revised the taxonomy of B-ALL by identifying multiple new subgroups with diverse sequence and structural initiating events that vary substantially by age at diagnosis and prognostic significance. There is a growing appreciation of the role of inherited genetic variation in predisposition to ALL and drug responsiveness and of the nature of genetic variegation and clonal evolution that may be targeted for improved diagnostic, risk stratification, disease monitoring, and therapeutic intervention. This review provides an overview of the current state of knowledge of the genetic basis of B-ALL, with an emphasis on recent discoveries that have changed our approach to diagnosis and monitoring.
B-progenitor acute lymphoblastic leukemia (B-ALL) is the most common childhood cancer, with cure rates exceeding 90% in most developed countries (Hunger and Mullighan 2015). However, the prognosis for ALL declines with increasing age, with historic cure rates of just 30%–40% in adults (age ≥ 40 yr) (Frey and Luger 2015). B-progenitor acute lymphoblastic leukemia (B-ALL) comprises multiple subtypes characterized by recurrent disease-initiating genetic alterations that are important for risk stratification. These include aneuploidy (gain or loss of whole chromosomes) or chromosomal translocations that deregulate genes through the formation of chimeric fusions or by juxtaposition to strong enhancers and commonly involve hematopoietic transcription factors, epigenetic modifiers, cytokine receptors, and tyrosine kinases (Iacobucci and Mullighan 2017). Cooperating genetic events that contribute to leukemogenesis include copy number alterations and sequence mutations that perturb multiple cellular pathways. In recent years, the rapid development and implementation of next-generation sequencing techniques has revolutionized our understanding of the genomic landscape of ALL by identifying genomic alterations that were previously cryptic and by enabling comprehensive characterization of both germline and somatic alterations that define each subtype across the age spectrum, as well as characterizing the nature of clonal variegation, genetic heterogeneity, and disease progression (Mullighan et al. 2008b; Ma et al. 2015; Tzoneva et al. 2018). In addition to refining risk stratification, these studies have also identified new therapeutic targets that guide precision medicine approaches intended to improve the cure rate while reducing adverse treatment effects.
Here, we will review the genomic landscape of B-ALL with particular emphasis on new subtypes and prognosis and discuss both somatic and inherited variants that contribute to leukemogenesis. The role of the interaction between leukemic cells and the bone marrow microenvironment in disease development and response to treatment will also be discussed.
RECURRENT CHROMOSOMAL ALTERATIONS AND PROGNOSIS
The frequency of subtype-defining alterations varies with age (Table 1; Fig. 1). Secondary genetic alterations may be acquired or enriched during disease progression (Mullighan et al. 2007, 2008b; Moorman et al. 2012). Common targets include lymphoid transcription factors (IKZF1, PAX5, EBF1, ETV6), cell cycle regulators and tumor suppressors (CDKN2A/B, TP53, RB1), regulators of lymphoid signaling (BTLA and CD200), Ras pathway signaling (NRAS, KRAS, PTPN11), and chromatin modifiers (CREBBP, SETD2, WHSC1) (Kuiper et al. 2007; Mullighan et al. 2007). The prevalence, gene, and type of alteration vary between subtypes and have different prognostic relevance. Current risk stratification and treatment algorithms incorporate age, sex, presentation white blood cell count, established cytogenetic alterations, and response to initial therapy as measured by levels of minimal residual disease (MRD). Genomic alterations including composite copy number alterations have recently been proposed as important factors for determining prognosis (Hamadeh et al. 2019). Because MRD is such a central component of risk stratification, future clinical trials should aim to integrate new genomic information with response to therapy to develop a comprehensive relapse prediction model (O'Connor et al. 2018).
Table 1.
Prevalence and prognosis of subtypes in B-ALL
| ALL subtype | Category | Median age (yr) | Prevalence | Genomic alterations | Clinical features | Reference(s) |
|---|---|---|---|---|---|---|
| Hyperdiploid (>50 chromosomes) | Aneuploid | 4 | High in children (25%) | Ras pathway Epigenetic modifiers | Excellent prognosis | Paulsson et al. 2015 |
| Low-hypodiploid (31–39 chromosomes) | Aneuploid | 47 | High in adults (10%–15%) | IKZF2 del, TP53 mut (commonly inherited) | Poor prognosis | Holmfeldt et al. 2013 |
| Near-haploid (24–30 chromosomes) | Aneuploid | 5.4 | <3% in all ages | Ras pathway, IKZF3 del | Intermediate prognosis | Holmfeldt et al. 2013 |
| iAMP21 | Copy number gain | 10 | ∼3% in children and AYA | Complex structural alterations of chromosome 21 | Good prognosis with intensive therapy, low WBC | Harrison 2015 |
| ETV6-RUNX1 t(12;21)(p13;q22) | TF rearrangement | 4 | High in children (25%) | PAX5 del, WHSC1 mut | Excellent prognosis | Mullighan et al. 2007; Jaffe et al. 2013 |
| ETV6-RUNX1-like | TF rearrangement | 3 | ∼3% in children | ETV6 fusions and del, IKZF1 fusions and del | Unknown | Lilljebjörn et al. 2016; Zaliova et al. 2017 |
| DUX4-rearranged | TF rearrangement | 14.3 | Peak in AYA (∼8%) | ERG del, IKZF1 del, Ras pathway | Excellent prognosis | Lilljebjörn et al. 2016; Yasuda et al. 2016; Zhang et al. 2016 |
| KMT2A-rearranged | TF rearrangement | 40 | High in infants (∼90%) and adults (∼15%) | Ras pathway (commonly subclonal) | Poor prognosis, sensitive to bortezomib or DOT1L inhibition | Andersson et al. 2015 |
| TCF3-PBX1 t(1;19)(q23;p13) | TF rearrangement | 8 | ∼5% in children, rarely in adults | Good prognosis, CNS relapse | Barber et al. 2007; Burmeister et al. 2010 | |
| ZNF384-rearranged | TF rearrangement | 15 | Peak in AYA (∼5%) | Epigenetic modifiers, Ras pathway | Intermediate prognosis | Liu et al. 2016; Shago et al. 2016; Yasuda et al. 2016 |
| MEF2D-rearranged | TF rearrangement | 14 | Peak in AYA (∼7%) | Ras pathway | Intermediate prognosis, sensitive to HDAC inhibition | Gu et al. 2016; Suzuki et al. 2016 |
| NUTM1-rearranged | TF rearrangement | 3 | Exclusively in children (1%) | Unknown | Excellent prognosis | Li et al. 2018; Gu et al. 2019 |
| TCF3-HLF t(17;19)(q22;p13) | TF rearrangement | 15 | Very rare in all ages (<1%) | TCF3 mut, PAX5 del, Ras pathway | Very poor prognosis, sensitive to Bcl2 inhibition | Fischer et al. 2015 |
| PAX5alt | Other TF-driven | 10 | Highest in children (∼11%) | PAX5 fusion, mut, amp | Intermediate prognosis | Li et al. 2018; Gu et al. 2019 |
| PAX5 P80R | Other TF-driven | 22 | Highest in adults (∼4%) | Ras pathway | Intermediate prognosis | Li et al. 2018; Gu et al. 2019 |
| IKZF1 N159Y | Other TF-driven | Very rare in all ages (<1%) | Unknown | Unknown | Li et al. 2018; Gu et al. 2019 | |
| BCL2/MYC-rearranged | Other TF-driven | 48 | Almost exclusively in AYA and adults (∼3%) | Unknown | Poor prognosis | Gu et al. 2019 |
| Ph-like | Kinase-driven | 21 | Peaks in AYA (25%–30%) | Multiple kinase alterations, IKZF1 del and mut, CDKN2A/B del | Poor prognosis, amenable to TKI therapy | Roberts et al. 2014a, 2017a |
| BCR-ABL1 t(9;22)(q34;q11.2) | Kinase-driven | 40–45 | 5% in children, highest in adults (40%–50%) | IKZF1 del and mut, CDKN2A/B del | Historically poor prognosis, improved with TKI | Mullighan et al. 2008a; Roberts et al. 2014a, 2017a |
| Other | 16 | ∼5% in children, ∼10% in AYA and adults | Unknown | Intermediate prognosis |
(AYA) Adolescent and young adult, (amp) amplification, (B-ALL) B-progenitor acute lymphoblastic leukemia, (CNS) central nervous system, (del) deletion, (HDAC) histone deacetylase, (mut) sequence mutation, (TF) transcription factor, (TKI) tyrosine kinase inhibitor.
Figure 1.
(A) tSNE plot showing B-progenitor acute lymphoblastic leukemia (B-ALL) subtypes based on RNA-seq gene expression profiling of 1988 cases. (B) Distribution of B-ALL subtypes within each age group. Subtypes are grouped as gross chromosomal abnormalities (aneuploidy or copy number gain), transcription factor (TF) rearrangement, other TF-driven, kinase-driven, and all others (Gu et al. 2019).
Gross Chromosomal Abnormalities
High hyperdiploidy (nonrandom gain of at least five chromosomes) is present in ∼25% of childhood ALL patients, but accounts for <5% of adolescents and young adults (16–39 yr old; AYA) and adults, and is associated with a favorable outcome. The patterns of chromosomal gain are nonrandom, and most commonly involve chromosomes 4, 10, 14, and 21 and the X chromosome. Mutations involving the Ras pathway (KRAS, NRAS, PTPN11) and epigenetic modifiers are frequent genetic events in hyperdiploid patients (Paulsson et al. 2015). Hypodiploid ALL with less than 44 chromosomes comprises two principal subtypes with distinct transcriptional profiles and genetic alterations. Historically, hypodiploid ALL has been associated with an unfavorable prognosis (Harrison et al. 2004); however, the outcome is improved with contemporary studies utilizing MRD risk-stratified regimens, and transplantation provides no additional survival benefit compared to chemotherapy alone in MRD-negative patients (Mullighan et al. 2015; Pui et al. 2019). Patients with low hypodiploidy (31–39 chromosomes) commonly harbor deletion of IKZF2 and sequence mutations of TP53 that are frequently inherited (Holmfeldt et al. 2013). This subtype is rare in children (<1%), but increases with age, accounting for >10% of adults, and is associated with a very poor outcome (Moorman et al. 2007; Gu et al. 2019). Patients with near-haploid ALL (24–30 chromosomes) present at a younger age, accounting for ∼2% of childhood ALL (Holmfeldt et al. 2013). Frequent secondary alterations in this subtype include Ras-activating mutations and deletions of IKZF3 (Holmfeldt et al. 2013; Gu et al. 2019). Doubling of the hypodiploid clone (known as masked hypodiploidy) is common in both near-haploid and low-hypodiploid ALL and results in a modal chromosome number in the hyperdiploid range. Given the markedly differing prognoses of hypodiploid and hyperdiploid ALL, it is important to distinguish masked hypodiploidy (which typically shows four copies of multiple chromosomes in the doubled clone, and copy-neutral loss of heterozygosity of multiple chromosomes) from hyperdiploidy (which typically has multiple trisomic chromosomes).
ALL with intrachromosomal amplification of chromosome 21 (iAMP21) defines a subtype of ALL that is more common in older children (median age 10 yr), and is rarely observed in patients older than 30 yr (Harrison et al. 2014). The role of iAMP21 in leukemogenesis is unclear, but a common region of amplification includes ERG and DYRK1A with gain of at least two copies of RUNX1 (Li et al. 2014). Secondary events include the P2RY8-CRLF2 fusion and genetic alterations in kinase signaling, including IL7R and FLT3. Improved risk stratification and treatment with intensive therapy can rescue the poor outcome of these patients when treated as standard-risk (Moorman et al. 2013).
Translocations
ETV6-RUNX1, encoded by the t(12;21)(p13;q22) translocation, is another favorable cytogenetic alteration with a high frequency in childhood ALL (25%) and <5% in AYAs and adults. Secondary DNA copy number alterations, notably PAX5 deletion, and mutation of WHSC1 are frequent genetic events in patients harboring ETV6-RUNX1 (Mullighan et al. 2007; Jaffe et al. 2013; Papaemmanuil et al. 2014). KMT2A (MLL) rearrangements are a hallmark of infant ALL (age < 1 yr). They also account for a significant proportion of adults with ALL (∼15%) and are associated with a poor prognosis in all ages (Hunger and Mullighan 2015). The reasons underlying the biphasic distribution in age are not well understood. In infant cases, KMT2A rearrangement is frequently acquired in utero (Ford et al. 1993), and patients harbor very few cooperating lesions, suggesting the rearrangement itself is sufficient to induce leukemia (Andersson et al. 2015). The most commonly perturbed pathways include PI3K and Ras signaling, with the majority of mutations present at a low tumor burden (Driessen et al. 2013; Andersson et al. 2015; Agraz-Doblas et al. 2019). Subclonal activating mutations of FLT3 were recently shown to accelerate disease onset in a mouse model of KMT2A-rearranged leukemia, suggesting these alterations can influence the rate of leukemogenesis even at low levels (Hyrenius-Wittsten et al. 2018).
TCF3-PBX1, encoded by the t(1;19)(q23;p13) translocation, is present in ∼5% of children and less in AYAs and adults. Previously considered a high-risk subtype with a propensity to central nervous system relapse, it is now associated with a favorable outcome on contemporary ALL therapies (Barber et al. 2007; Burmeister et al. 2010). By contrast the t(17;19)(q22;p13) translocation, encoding the TCF3-HLF fusion gene, defines a rare subtype of ALL (<1% in all ages) with a distinct transcriptional profile that is typically associated with an overall survival of <2 yr from diagnosis (Inaba et al. 1992; Hunger 1996). Interestingly, primary leukemic cells harboring TCF3-HLF show sensitivity to the BCL2 inhibitor venetoclax (ABT-199), identifying a new therapeutic option for this fatal subtype (Fischer et al. 2015).
BCR-ABL1 ALL is uncommon in children (2%–5% of patients), but accounts for at least 25% of adults (Roberts et al. 2014a, 2017a). The addition of ABL1 tyrosine kinase inhibitors (TKIs) to chemotherapeutic regimens in both children and adults has significantly improved the survival of BCR-ABL1-positive patients (Ravandi et al. 2010; Schultz et al. 2014; Slayton et al. 2018). IKZF1 alterations (deletion or mutation) are a hallmark of kinase-driven ALL (Ph+ and Ph-like) and are associated with treatment failure and relapse, even in the era of TKI therapy (Mullighan et al. 2008a; Martinelli et al. 2009; Roberts et al. 2014a; Slayton et al. 2018). The co-occurrence of IKZF1 deletions with CDKN2A/B, PAX5, or PAR1 deletions in the absence of ERG deletions (termed IKZF1plus) detected by multiplex ligation probe amplification (MLPA) in childhood ALL confers a worse prognosis compared to patients with IKZF1 deletion who do not fulfill the criteria for IKZF1plus (Stanulla et al. 2018). Although technically straightforward, identification of IKZF1plus as a biomarker of poor outcome is limited by the inability of the MLPA approach to identify the full spectrum of IKZF1 alterations, cases with high-risk ALL that do not have IKZF1 alterations, and the lack of ERG deletion in approximately one-third of DUX4 cases that commonly have IKZF1 alterations and favorable outcome.
NEW SUBTYPES IN B-ALL
The application of comprehensive sequencing and integrative analyses continues to refine the genomic landscape of ALL, resulting in the identification of new entities with prognostic and therapeutic significance. Rearrangements in these new subtypes involve a diverse range of partners that converge on a single gene (e.g., MEF2D and ZNF384-rearranged ALL) or are cryptic by cytogenetic analysis (e.g., DUX4-rearranged ALL). Other subtypes may harbor alteration of a range of driver genes by diverse mechanisms (e.g., Ph-like ALL) or are initiated by sequence mutations (e.g., PAX5 P80R and IKZF1 N159Y). Additional groups have similar gene expression profiles to known subtypes with different genetic alterations (Ph-like and ETV6-RUNX1-like ALL).
Ph-like ALL: An Opportunity for Targeted Therapies
Philadelphia chromosome like (Ph-like or BCR-ABL1-like) ALL was incorporated as a provisional entity to the revision of the World Health Organization (WHO) classification of acute leukemia in 2016 (Arber et al. 2016). Leukemic cells from patients with Ph-like ALL have similar transcriptional profiles to Ph+ ALL but lack the BCR-ABL1 fusion gene (Den Boer et al. 2009; Mullighan et al. 2009). Similar to with Ph+ ALL, the incidence of Ph-like ALL increases with age, comprising 10%–15% of childhood ALL cases, >20% of adults, and peaking at 25%–30% in AYAs (Loh et al. 2013; Roberts et al. 2014a, 2017a, 2018; Jain et al. 2017a; Reshmi et al. 2017; Tasian et al. 2017a). In all ages, Ph-like ALL is associated with elevated MRD levels and/or higher rates of treatment failure compared to non-Ph-like ALL patients (Roberts et al. 2014a, 2017a; Tasian et al. 2017b). Thus, the inferior treatment outcomes in AYA and adults may be partly explained by the high prevalence of Ph-like ALL. In Children's Oncology Group (COG) cohorts of National Cancer Institute (NCI) standard-risk (SR) ALL, Ph-like ALL is less common and confers a better prognosis compared to children with high-risk (HR) ALL (Roberts et al. 2018). Furthermore, children with Ph-like ALL treated on St. Jude Total XV studies had a favorable outcome with MRD risk-directed therapy intensification (Roberts et al. 2014b).
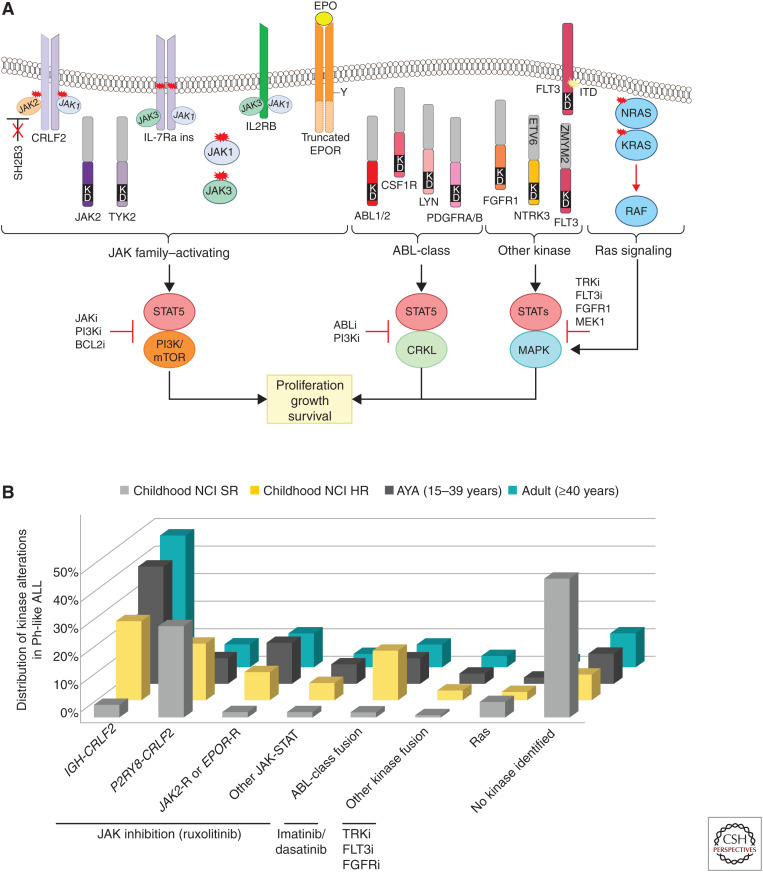
Ph-like ALL is genetically heterogeneous and is characterized by rearrangements, copy number alterations, and sequence mutations that activate tyrosine kinase or cytokine receptor signaling. Despite this complexity, most alterations can be divided into a limited number of distinct subgroups based on the activated kinase and signaling pathways. These include rearrangements or, less commonly, sequence mutations of CRLF2 (IGH-CRLF2, P2RY8-CRLF2), fusions involving ABL-class genes (ABL1, ABL2, CSF1R, LYN, PDGFRA, PDGFRB), alterations activating JAK-STAT signaling (including rearrangements of JAK2, EPOR or TYK2) and mutations/deletions of IL7R, SH2B3, JAK1, JAK3, TYK2, IL2RB), Ras signaling pathways (NRAS, KRAS, PTPN11), and less common fusions (FLT3, FGFR1, NTRK3, PTK2B) (Fig. 2A; Roberts et al. 2014a, 2017a; Reshmi et al. 2017). The frequency of each kinase subgroup varies with age, particularly with respect to CRLF2-rearrangements, in which IGH-CRLF2 accounts for almost 50% of Ph-like ALL in AYAs and adults but is less common in children. ABL-class fusions are most prevalent in children with HR ALL (Fig. 2B). Fewer kinase alterations are identified in Ph-like ALL patients with SR ALL (Roberts et al. 2018). A small subset of children harboring rearrangement of CRLF2—most commonly P2RY8-CRLF2 and with Down syndrome ALL —lack the Ph-like ALL gene expression signature (Gu et al. 2019).
Figure 2.
(A) Kinase alterations and signaling pathways dysregulated in Philadelphia chromosome-like (Ph-like) ALL. The majority of kinase and cytokine receptor alterations converge on two pathways that activate JAK-family member signaling or ABL signaling. Alterations that activate JAK-STAT signaling can be targeted with JAK and PI3K inhibitors. ABL-class alterations can be targeted with ABL-inhibitors such as dasatinib. Other kinase alterations and those that activate Ras signaling can be targeted with specific inhibitors including those that inactivate TRK, FLT3, FGFR1, and MEK for the MAPK pathway. (B) Distribution of kinase subtypes in Ph-like ALL within each age group (Roberts et al. 2014a, 2017a, 2018; Reshmi et al. 2017). Combined prevalence of Ph-like ALL subtypes in childhood National Cancer Institute (NCI) standard-risk (SR; age 1–9.99 yr and WBC < 50,000/µL), NCI high-risk (HR; age 10–15 yr or WBC ≥ 50,000/µL), adolescent and young adults (1639 yr), and adults (≥40 yr). Genomic subtypes include IGH-CRLF2, P2RY8-CRLF2, and ABL-class fusions (ABL1, ABL2, CSF1R, LYN, PDGFRA, and PDGFRB); JAK2 and EPOR rearrangements and other mutations in JAK–STAT signaling (JAK1/3, IL7R, SH2B3, TYK2, and IL2RB); and other kinase alterations (FLT3, FGFR1, NTRK3), Ras mutations (KRAS, NRAS, NF1, PTPN11, BRAF, and CBL), and unknown alterations.
The majority of Ph-like alterations can be targeted effectively in preclinical models using a combinatorial approach of chemotherapy with ABL1 (e.g., dasatinib) or JAK inhibition (e.g., ruxolitinib) (Roberts et al. 2017b), and a number of case reports demonstrate efficacy of ABL1 TKI treatment in Ph-like ALL patients with refractory disease (Lengline et al. 2013; Weston et al. 2013; Kobayashi et al. 2015; Schwab et al. 2016). This approach is currently being tested in frontline studies of patients treated at St. Jude Children's Research Hospital (Total XVII, NCT03117751) (Inaba et al. 2017) and on COG protocols (AALL1131, NCT01406756; AALL1521, NCT02723994) (Tasian et al. 2017b).
ETV6-RUNX1-Like ALL
Analogous to Ph-like ALL, ETV6-RUNX1-like ALL is defined by having a gene expression profile and immunophenotype (CD27 positive, CD44 low to negative) similar to ETV6-RUNX1 ALL, but lacking the ETV6-RUNX1 fusion (Lilljebjörn et al. 2016; Zaliova et al. 2017). Unsurprisingly, like ETV6-RUNX1 ALL, ETV6-RUNX1-like ALL is almost exclusively identified in children (∼3%) and confers a favorable prognosis. This subtype is associated with alternate lesions (gene fusions or copy number alterations) in ETV6, IKZF1, or TCF3, suggesting global deregulation of lymphoid development is a hallmark of this transcriptional signature (Gu et al. 2019).
DUX4-rearranged ALL
An interesting subtype of B-ALL with a very distinctive gene expression profile and immunophenotype (CD2 and CD371 positive) is characterized by genetic alterations and deregulation of the transcription factor genes DUX4 (double homeobox 4) and ERG (ETS-related gene) (Yeoh et al. 2002; Harvey et al. 2010; Lilljebjörn et al. 2016; Yasuda et al. 2016; Zhang et al. 2016; Schinnerl et al. 2019). DUX4 is located in microsatellite D4Z4 repeat domains in the subtelomeric region of chromosome 4 that is duplicated on chromosome 10q and is normally exclusively expressed in germinal tissues (Gatica and Rosa 2016). In DUX4-rearranged ALL, translocation or insertion of DUX4 to IGH is the initiating event that results in overexpression of a 3′ truncated isoform of DUX4 not normally expressed in B cells. The aberrantly expressed DUX4 binds to an intragenic region of ERG, resulting in gross transcriptional deregulation of ERG, and, commonly, expression of ERGalt, a transcript that utilizes a noncanonical first exon that encodes a truncated carboxy-terminal ERG protein. ERGalt retains the DNA-binding and transactivating domain of ERG, inhibits the transcriptional activity of wild-type ERG, and is transforming in mouse models of B-ALL (Zhang et al. 2016). This subtype accounts for 5%–10% of B-ALL, with a slight peak in AYAs. Of clinical relevance, DUX4-rearranged ALL is associated with an excellent prognosis in both children and adults (Gu et al. 2019), even despite the presence of secondary genetic alterations otherwise associated with poor outcome, such as IKZF1 deletions, which are present in ∼40% of DUX4-rearranged ALL (Zhang et al. 2016).
New Transcription Factors: MEF2D and ZNF384
Recurrent rearrangements of MEF2D and ZNF384 account for ∼4% and 5% of children and up to 7% and 10% in AYA patients, respectively. Accordingly, both subtypes are associated with older age of onset (median age 14 and 15 yr) (Gu et al. 2016; Liu et al. 2016; Suzuki et al. 2016).
Multiple 3′ partners have been identified for MEF2D (encoding myocyte enhancer factor 2D), including BCL9, CSF1R, DAZAP1, FOXJ2, HNRNPUL1, HNRNPH1, and SS18 (Gu et al. 2016; Ohki et al. 2019). All fusions preserve the MEF2D MADS-box domain that mediates DNA binding, resulting in enhanced transcriptional activity and deregulation of MEF2D targets (Gu et al. 2016). An exception is MEF2D-CFS1R, which displays the Ph-like gene expression profile (Roberts et al. 2014a). MEF2D-rearranged ALL is associated with an aberrant immunophenotype (CD10 negative, CD38 positive) and an intermediate to poor outcome (Gu et al. 2016; Suzuki et al. 2016; Ohki et al. 2019). Alterations of PHF6, recurrently mutated in T-cell ALL, were the most frequent cooperating lesions identified by targeted sequencing (Ohki et al. 2019). Deregulation of MEF2D also results in the overexpression of HDAC9 (histone deacetylase 9), which can be targeted therapeutically using HDAC inhibitors (Gu et al. 2016).
Rearrangements of ZNF384 (encoding zinc finger 384) define a subtype of acute leukemia that transcends immunophenotypic classification and may manifest as classical pre-B ALL without lineage aberrancy, B-ALL with expression of the myeloid markers (CD13/33), or B/myeloid mixed phenotype acute leukemia (Alexander et al. 2018). To date, 11 different 5′ fusion partners, usually involving a transcriptional regulator or chromatin modifier, have been identified for ZNF384: ARIDIB, BMP2K, CLTC, CREBBP, EP300, EWSR1, NIPBL, SMARCA2, SYNRG, TAF15, and TCF3 (Liu et al. 2016; Shago et al. 2016; Yasuda et al. 2016; Hirabayashi et al. 2017). An intermediate prognosis has been described in small pediatric cohorts (Liu et al. 2016). The rearrangements are also distinctive, usually involving the entire coding region of ZNF384, resulting in the expression of wild-type ZNF384 in a lineage inappropriate manner, as well as the chimeric fusion protein. Studies of hematopoietic progenitor cells from primary leukemia samples, as well as xenografting of immunophenotypically multiclonal populations, has shown that ZNF384 rearrangements are acquired in a subset of hematopoietic stem cells and prime leukemic cells for lineage plasticity (Alexander et al. 2018). More recently, cases harboring rearrangement of the zinc finger ZNF362 to SMARCA2 and TAF15 were shown to cluster with ZNF384-rearranged ALL, indicating deregulation of similar downstream targets (Li et al. 2018).
REDEFINING “OTHER” B-ALL
Despite the advances made in refining the classification of B-ALL, until recently, almost one-quarter of cases across the age spectrum lacked a subtype defining lesion and were collectively known as “Other.” These cases were excluded from risk stratification, commonly relapsed, and lacked targeted therapeutic approaches. To systematically define the frequency and prognostic significance of subtypes across the age spectrum, two groups recently performed an integrated large scale genomic analysis of 1223 and 1988 B-ALL cases, respectively, using transcriptional profiling to refine subtype classification (Li et al. 2018; Gu et al. 2019). In addition to known groups, including those defined by aneuploidy, up to five new subtypes were identified with distinct gene expression signatures, accounting for an additional 15% of B-ALL. As such, >90% of ALL cases may be classified into distinct genetic subtypes using these algorithms.
PAX5-Driven Subtypes
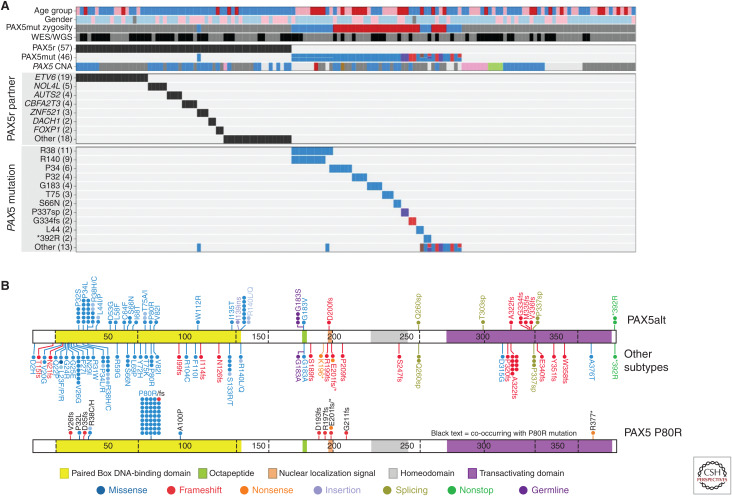
PAX5 is largely considered to function as a haploinsufficient tumor suppressor in ALL, with secondary heterozygous deletions and loss-of-function mutations present in one-third of all patients with B-ALL across a range of subtypes (Kuiper et al. 2007; Mullighan et al. 2007). In mouse models, Pax5 heterozygosity cooperates with constitutive activation of the JAK-STAT pathway to promote B-ALL development, supporting its role as a tumor suppressor (Dang et al. 2015). PAX5 translocations are reported in 2%–3% of B-ALL (Nebral et al. 2009; Coyaud et al. 2010). Recent analyses identified two PAX5 subtypes defined by distinct transcriptional profiles and genetic alterations. The first subtype, referred to as PAX5-altered (PAX5alt), comprises cases with diverse PAX5 rearrangements (most commonly to ETV6 or NOL4L), sequence mutations or intragenic amplification (Schwab et al. 2017), with the highest prevalence observed in children and AYA (10% each vs. 7% in adults) (Gu et al. 2019). The second group of PAX5-driven ALL is defined by the presence of the PAX5 P80R mutation, which is homozygous in almost all cases because of deletion or frameshift mutation of the wild-type PAX5 allele, suggesting that loss of both PAX5 alleles drives the unique gene expression profile of this subtype (Fig. 3; Li et al. 2018; Gu et al. 2019; Passet et al. 2019). The prevalence of PAX5 P80R increases with age, accounting for almost 5% of adults. This subtype confers an intermediate to favorable prognosis in both children and adults (Bastian et al. 2019; Gu et al. 2019; Passet et al. 2019; Zaliova et al. 2019). Cooperating lesions identified in PAX5 P80R patients include a high frequency of signaling mutations, particularly in the Ras, JAK-STAT, and other kinase signaling pathways (FLT3, PIK3CA), highlighting the potential for targeted therapies (Gu et al. 2019; Passet et al. 2019). Notably, heterozygous Pax5P80R/+ or homozygous Pax5P80R/P80R knock-in mice develop B-progenitor ALL that is transplantable, and tumors that arise in Pax5P80R/+ mice genetically inactivate the wild-type Pax5 allele by deletion or truncation, recapitulating the loss of wild-type PAX5 observed in human ALL (Gu et al. 2019). In a mouse model of B-ALL, PAX5-ETV6 activated distinct transcriptional pathways including pre-B cell receptor signaling and migration/adhesion, confirming its role as an oncoprotein rather than simply acting as a competitive inhibitor of the wild-type PAX5 protein (Smeenk et al. 2017). The identification of PAX5 subtypes as distinct entities highlights the importance of this gene in regulating B-cell differentiation, and confirms PAX5 alterations as founding lesions in B-lymphoid leukemogenesis as opposed to secondary cooperating events as previously thought.
Figure 3.
(A) Genetic alterations of PAX5, including gene rearrangements (PAX5r), sequence mutations (PAX5mut). and focal intragenic amplifications (PAX5amp, pink in PAX5 CNA) observed in the PAX5alt cohort. (B) Protein domain plot of PAX5 showing the mutations detected in PAX5alt and other B-ALL subtypes (top panel) and in the PAX5 P80R subtype (bottom panel). (CNA) Copy number alteration.
IKZF1 N159Y
Another uncommon subtype (accounting for <1% of ALL) defined by a single mutation in a lymphoid transcription factor includes cases harboring a heterozygous N159Y missense mutation in IKZF1 (Li et al. 2018; Gu et al. 2019). In contrast to PAX5 P80R ALL, the nonmutated wild-type allele of IKZF1 is retained in patients with IKZF1 N159Y ALL. The N159 residue is located within the DNA-binding domain of IKZF1. Mutation of this residue results in nuclear mislocalization and enhanced intercellular adhesion that is characteristic of perturbed IKZF1 function (Churchman et al. 2015). Such cases exhibit a distinct gene expression profile compared to other IKZF1-altered cases, with increased expression of genes involved in oncogenesis (YAP1), chromatin remodeling (SALL1), and JAK-STAT signaling (Li et al. 2018; Gu et al. 2019).
IGH Rearrangements
Rearrangements of the IGH locus to a range of partners—including CRLF2, CEBP family members (CCAAT/enhancer binding protein), and ID4—are frequent in AYA and adult ALL (∼10%) and generally confer a poor prognosis (Russell et al. 2014). In addition to these partners, we identified a subset of cases with pre-B immunophenotype and a unique transcriptional signature characterized by rearrangement of IGH to BCL2, MYC, and/or BCL6 (BCL2/MYC) (Gu et al. 2019) that resemble those observed in “double-hit” lymphoma and are rarely identified in ALL (Moorman et al. 2012; Russell et al. 2014; Uchida et al. 2017; Wagener et al. 2018). This subtype is predominantly identified in adults (median age 48.5 yr) and is associated with an extremely unfavorable outcome.
NUTM1 Rearrangements
An additional subtype present exclusively in 1% of childhood ALL (median age 3 yr) involves fusion of almost all the coding region of NUTM1 (nuclear protein in testis midline carcinoma family 1) to six different 5′ partners—ACIN1, BRD9, CUX1, IKZF1, SLC12A6, and ZNF618—resulting in increased expression of NUTM1 (Li et al. 2018; Gu et al. 2019). NUTM1 is normally expressed in the testis and acts as a chromatin modifier by recruiting EP300 (p300) to increase local histone acetylation (Alekseyenko et al. 2015). Fusions of NUTM1 (commonly BRD4-NUTM1) are a hallmark of NUT midline carcinoma (NMC), an aggressive and fatal subtype of squamous cell carcinoma that also arises frequently in children (French 2014). BRD4-NUTM1 acts to repress differentiation in NMC by recruiting histone acetyltransferases and other transcriptional cofactors to regions of chromatin that are actively transcribing pro-proliferative and antidifferentiation genes, including MYC (French 2014). Thus, fusions such as BRD9-NUTM1 in ALL may have a similar mechanism of action, although experimental studies are required to elucidate the role of NUTM1 in leukemogenesis. In contrast to NMC, ALL patients with NUTM1 rearrangements have an excellent prognosis. Given the involvement of BRD9, bromodomain or HDAC inhibitors would be a logical targeted therapeutic approach for these patients.
MIXED PHENOTYPE ALL
Mixed phenotype acute leukemia (MPAL) is characterized by expression of cell surface proteins characteristic of multiple lineages, most commonly B and myeloid (B/M MPAL) or T and myeloid (T/M MPAL) markers, either in a single (biphenotypic) or multiple (bilineal) immunophenotypic subpopulations. Prior studies had identified rearrangements of KMT2A (MLL) or the BCR-ABL1 fusion in a minority of cases, but until recently the genetics of MPAL had been poorly understood. However, this is of great interest given the phenotypic plasticity and poor prognosis of this form of leukemia. Genomic analyses have shown that T/M and B/M are genetically distinct, with T/M leukemia characterized by founder mutations or rearrangements in transcription factors and chromatin modifiers (WT1, ETV6, RUNX1, CEBPA) and the majority of B/M cases to harbor rearrangements of ZNF384 (Alexander et al. 2018; Takahashi et al. 2018; Xiao et al. 2018). The phenotypic plasticity and characteristic of MPAL (that has bedeviled the selection of appropriate therapy) is largely independent of genetic variegation and, rather, is due to the acquisition of founding lesions in very early hematopoietic progenitors. Thus, MPAL forms part of a spectrum of immature/stem cell leukemias (for T/M MPAL, like early T cell precursor ALL) (Zhang et al. 2012), and future studies are integrating ALL-directed therapy and genomic analysis to further refine optimal diagnostic and classification approaches (Hrusak et al. 2018).
INHERITED VARIANTS IN ALL
Genome-wide association studies (GWASs) have identified risk loci with common genetic polymorphisms that are associated with a modest increase in ALL susceptibility, including IKZF1 (7p12.2), CDKN2A/CDKN2B (9p21), PIP4K2A (10p12.2), GATA3 (10p14), ARID5B (10q21.2), CEBPE, and ERG (14q11.2) (Moriyama et al. 2015b; Qian et al. 2019). Associations with several of these loci exhibit a degree of ALL subtype specificity—for example, GATA3 with Ph-like ALL (Perez-Andreu et al. 2013; Jain et al. 2017b) and ERG with TCF3-PBX1—suggesting an interplay of germline and somatic alterations in leukemogenesis. More recently, studies of families with multiple individuals with ALL and complementary examinations of large cohorts of patients with presumed sporadic ALL have identified deleterious germline variants in genes that are also targets of somatic mutation in ALL, including PAX5, ETV6, IKZF1, TP53, and ERG.
A role for PAX5 in autosomal dominant predisposition to B-ALL was identified by the description of three unrelated families who harbored a germline PAX5 c547G > A mutation in the octapeptide domain (PAX5 G183S) that resulted in moderate attenuation of transcriptional activity in vitro (Shah et al. 2013; Auer et al. 2014). Notably, all affected individuals had somatic loss of the wild-type allele, suggesting that biallelic inactivation of PAX5 is also important for B-cell leukemogenesis in this context.
Deleterious germline variants within the DNA-binding domain of ETV6 are present in 1% of sporadic B-ALL and affect transcriptional repression either by abrogating binding to ETS-containing DNA sequences or through altered intracellular localization (Moriyama et al. 2015a; Noetzli et al. 2015; Topka et al. 2015; Zhang et al. 2015). Multiple subsequent reports suggest that ETV6 sequence mutations may be the most common germline alterations predisposing to ALL (Feurstein and Godley 2017; Hock and Shimamura 2017; Duployez et al. 2018). Moreover, a focal germline ETV6 splice site deletion resulting in exon skipping and protein truncation has been reported in a highly penetrant family (Rampersaud et al. 2019). Another report identified a constitutional translocation disrupting ETV6 (Jarviaho et al. 2019). These studies indicate that careful analysis of germline structural variants is required to describe the full repertoire of deleterious germline alterations in ALL.
Churchman et al. reported inherited germline variants in IKZF1 that impair its function in a similar manner to somatic mutations. In contrast to somatic IKZF1 alterations that are most commonly deletions or mutations in the amino-terminal (DNA-binding) or carboxy-terminal (dimerizing) zinc fingers (Churchman et al. 2015), the germline variants are scattered throughout the gene in regions of poorly characterized function and were not predicted to be deleterious by in silico analyses, but were highly deleterious in more sophisticated cellular assays including subcellular mislocalization, cell–cell adhesion, and cell stromal adhesion in vivo (Churchman et al. 2018).
TP53 alterations are a hallmark of low-hypodiploid ALL, with almost half occurring in the germline, suggesting that low-hypodiploid ALL is another manifestation of Li–Fraumeni syndrome (Holmfeldt et al. 2013). In a large cohort of childhood ALL, 49 nonsilent rare TP53 coding variants were identified in 77 patients, of which 22 variants were classified as pathogenic (Qian et al. 2018). Children with TP53 pathogenic variants presented at an older age, had inferior outcomes to children with wild-type TP53, and were more likely to develop second malignancies. This study also confirmed the association of inherited TP53 variants with hypodiploid ALL (Qian et al. 2018). A recent GWAS identified novel susceptibility variants at the ERG locus that were enriched in Hispanics (Qian et al. 2019), providing additional insight into the relationship of germline genetic variation in racial occurrence and outcomes in ALL (Yang et al. 2011; Karol et al. 2017). Together, these studies highlight the importance of these genes in both de novo and familial ALL.
RELAPSED ALL
Relapsed ALL remains a leading cause of childhood cancer death (Curtin et al. 2016) and is associated with high rates of treatment failure and death in older individuals (Fielding et al. 2007; Stock 2010; Frey and Luger 2015). The main curative approach for adults is an allogenic stem cell transplant; however, survival rates for relapsed ALL are improving with the implementation of new immunotherapeutic approaches including blinatumomab (CD19/CD3 bispecific T-cell engager), inotuzumab ozagamicin (anti-CD22 antibody conjugated to calicheamicin), and CAR T cells (chimeric antigen receptor) (Davila et al. 2014; Kantarjian et al. 2016, 2017; Maude et al. 2018; Park et al. 2018).
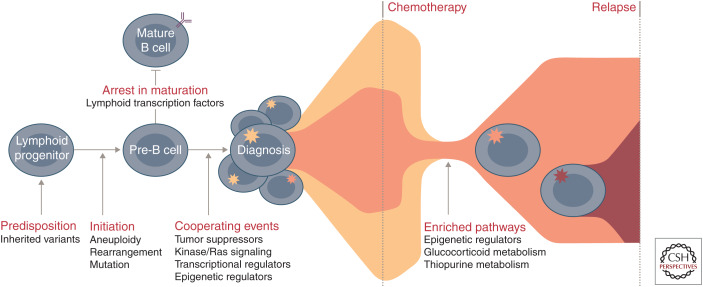
Genomic studies in childhood ALL show that predominant clones at diagnosis are often eradicated, and relapse arises from a minor clone that already harbors and/or acquires additional genomic alterations that drive resistance in a drug-specific or -agnostic manner (Fig. 4; Mullighan et al. 2008b, 2011; Li et al. 2015; Ma et al. 2015; Oshima et al. 2016; Tzoneva et al. 2018). Mutations in genes encoding epigenetic regulators and chromatin modifiers are recurrent events in relapsed ALL and can directly influence response to treatment (Mullighan et al. 2011; Mar et al. 2014; Ma et al. 2015). In particular, mutations in the transcriptional coactivator and acetyl transferase CREBBP occur in up to 20% of relapsed ALL and impair sensitivity to glucocorticoid therapy (Mullighan et al. 2011). Mutations in NT5C2 (5′-nucleotidase catalytic enzyme II) confer increased resistance to purine analogs at the cost of impaired leukemia cell growth and leukemia-initiating cell activity (Meyer et al. 2013; Tzoneva et al. 2018). In addition, loss of MSH6, a major component of the mismatch repair (MMR) system, results in intrinsic chemoresistance to thiopurines because of an inability to recognize thioguanine nucleotide mismatching and failure to initiate MMR. Thus, cells defective for MSH6 do not undergo cell cycle arrest or apoptosis and continue to proliferate in the presence of thiopurine (Evensen et al. 2018). Other recurrent somatic alterations in relapsed ALL include deletions of the glucocorticoid receptor NR3C1 and mutations in the H3K36 trimethyltransferase SETD2, the lysine-specific demethylase KDM6A, and the epigenetic regulator MLL2 (Mar et al. 2014; Ma et al. 2015). Enhancing our knowledge of relapse-enriched or acquired alterations is important for initial risk stratification and has implications for molecular monitoring given the increasingly widespread application of deep sequencing approaches to identify low levels of MRD.
Figure 4.
Commonly altered pathways and stepwise progression of B-progenitor acute lymphoblastic leukemia (B-ALL). Common genetic polymorphisms (IKZF1, CDKN2A/B, PIP4K2A, GATA3, ARID5B, CEBPE, and ERG) and deleterious nonsilent inherited variants (PAX5, ETV6, IKZF1, TP53, and ERG) increase the risk of ALL susceptibility. Driving or founding lesions of ALL define genomic subtypes: aneuploidy and other chromosomal abnormalities (hyperdiploid, low-hypodiploid, near-haploid, iAMP21), rearrangements deregulating transcription factors (ETV6-RUNX1, ETV6-RUNX1-like, KMT2A, TCF3-PBX1, DUX4, ZNF384, MEF2D, NUTM1, TCF3-HLF, PAX5, BCL2/MYC) or kinase genes (Ph-like, BCR-ABL1), and specific mutations in lymphoid transcription factors (PAX5 P80R, IKZF1, N195Y). Deletion and loss of lymphoid transcription factors (e.g., IKZF1, PAX5, EBF1) coupled with the alteration of tumor suppressors and cell cycle regulators (CDKN2A/B, TP53), kinase signaling pathway genes (e.g., NRAS, KRAS, FLT3), other transcriptional regulators (e.g., ETV6, ERG), or epigenetic regulators (e.g., CREBBP, WHSC1, CTCF) result in the accumulation of immature lymphoid blasts and presentation at diagnosis. During treatment, the predominant diagnosis clone is commonly eradicated and relapse arises from a minor clone that already harbors and/or acquires additional genetic alterations that drive resistance. Pathways that are enriched at relapse include those involving epigenetic regulators (e.g., CREBBP, SETD2, KDM6A), the glucocorticoid response (e.g., CREBBP, NR3C1), and thiopurine metabolism (e.g., NT5C2, MSH6).
ROLE OF THE MICROENVIRONMENT IN ALL
Most studies of mechanisms of leukemogenesis and treatment response have focused on leukemic cell-intrinsic features, but it is increasingly apparent that tumor cell-extrinsic factors, including the nature of nonleukemic hematopoietic cells, and the interaction of leukemic cells with the bone marrow microenvironment, are important determinants of response to therapy and may also be directly influenced by genetic alterations of the leukemic cell. This is exemplified by the finding that alterations of IKZF1 (Ikaros) in kinase-driven (Ph+ and Ph-like) ALL drive high-risk disease by derepressing expression of adhesion molecules that result in acquisition of a hematopoietic stem cell like phenotype and aberrant leukemic intercellular and cell-stromal adhesion (Joshi et al. 2014; Churchman et al. 2015). This leads to perturbed bone marrow mislocalization and resistance to therapy that may be circumvented, at least in this context, by rexinoids (that bind to retinoid X receptor α, which is also derepressed by loss of Ikaros) that result in differentiation and up-regulation of wild-type Ikaros. Another approach is focal adhesion kinase (FAK) inhibitors, which inhibit FAK signaling downstream of integrin activation (Churchman et al. 2016), an approach that is entering the clinic for the treatment of solid tumors (Lee et al. 2015) and in conjunction with immunotherapy (Jiang et al. 2016).
Although there is extensive evidence that remodeling of, and interaction with, the bone marrow hematopoietic niche has an important role in the survival of acute myeloid leukemia cells (Tabe and Konopleva 2014), the nature and importance of the ALL cell microenvironment interaction is less well studied, but is likely important in light of findings that disruption of CXCR4-CXCL12-mediated interaction can improve drug responsiveness in experimental models of B-ALL and T-cell acute lymphoblastic leukemia (T-ALL) (Pitt et al. 2015; Randhawa et al. 2016). Our recent data indicate that interaction of leukemic cells with bone marrow stromal cells results in profound deregulation of adhesion, signaling cascades, and epithelial to mesenchymal transition–like phenotype in ALL cells and accompanying drug resistance that can potentially be exploited therapeutically (Yoshihara et al. 2018).
CONCLUSIONS
Within the last decade, integrated genomic analyses of large cohorts of childhood ALL, and more recently AYA and adult ALL, has revolutionized our understanding of the genetic basis of ALL by identifying new subtypes, dysregulated pathways, and therapeutic targets that have led to improved risk stratification and treatment strategies. Despite these advances, a proportion of ALL cases cannot be categorized into any of the currently established subtypes, and ongoing discovery studies are required to fully define the genomic landscape. Recent discoveries have already had substantial impact on diagnosis and management of the disease. For example, targeted approaches are being tested in multiple trials of Ph-like ALL, and the appreciation that accurate classification and risk stratification requires genomic approaches that detect complex structural events in addition to sequence alterations has led to the increasingly widespread adoption of RNA sequencing and, in some centers, whole-genome sequencing. It is envisaged that genomic sequencing will become the clinical standard of care, and the field will continue to explore novel and sensitive approaches to detect and monitor disease, including cell-free technology and mutation-directed measurement of measurable residual disease.
ACKNOWLEDGMENTS
The authors thank colleagues at St. Jude, the Children's Oncology Group, and the multiple centers and leukemia cooperative study groups that have contributed samples and expertise to many of the studies described in this review, including Joshua Stokes from Biomedical Communications at St. Jude. The authors are supported by a National Institutes of Health Outstanding Investigator Award, a St. Baldrick's Foundation Robert J. Arceci Innovation Award, and the Henry Schueler 41&9 Foundation (to C.G.M.).
Footnotes
Editors: Michael G. Kharas, Ross L. Levine, and Ari M. Melnick
Additional Perspectives on Leukemia and Lymphoma: Molecular and Therapeutic Insights available at www.perspectivesinmedicine.org
REFERENCES
- Agraz Doblas A, Bueno C, Bashford-Rogers R, Roy A, Schneider P, Bardini M, Ballerini P, Cazzaniga G, Moreno T, Revilla C, et al. 2019. Unraveling the cellular origin and clinical prognostic markers of infant B-cell acute lymphoblastic leukemia using genome-wide analysis. Haematologica 104: 1176–1188. 10.3324/haematol.2018.206375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Walsh EM, Wang X, Grayson AR, Hsi PT, Kharchenko PV, Kuroda MI, French CA. 2015. The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev 29: 1507–1523. 10.1101/gad.267583.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander TB, Gu Z, Iacobucci I, Dickerson K, Choi JK, Xu B, Payne-Turner D, Yoshihara H, Loh ML, Horan J, et al. 2018. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature 562: 373–379. 10.1038/s41586-018-0436-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, Nakitandwe J, Holmfeldt L, Parker M, Easton J, et al. 2015. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet 47: 330–337. 10.1038/ng.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. 2016. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127: 2391–2405. 10.1182/blood-2016-03-643544 [DOI] [PubMed] [Google Scholar]
- Auer F, Rüschendorf F, Gombert M, Husemann P, Ginzel S, Izraeli S, Harit M, Weintraub M, Weinstein OY, Lerer I, et al. 2014. Inherited susceptibility to pre B-ALL caused by germline transmission of PAX5 c.547G>A. Leukemia 28: 1136–1138. 10.1038/leu.2013.363 [DOI] [PubMed] [Google Scholar]
- Barber KE, Harrison CJ, Broadfield ZJ, Stewart AR, Wright SL, Martineau M, Strefford JC, Moorman AV. 2007. Molecular cytogenetic characterization of TCF3 (E2A)/19p13.3 rearrangements in B-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer 46: 478–486. 10.1002/gcc.20431 [DOI] [PubMed] [Google Scholar]
- Bastian L, Schroeder MP, Eckert C, Schlee C, Tanchez JO, Kampf S, Wagner DL, Schulze V, Isaakidis K, Lazaro-Navarro J, et al. 2019. PAX5 biallelic genomic alterations define a novel subgroup of B-cell precursor acute lymphoblastic leukemia. Leukemia 33: 1895–1909. 10.1038/s41375-019-0430-z [DOI] [PubMed] [Google Scholar]
- Burmeister T, Gokbuget N, Schwartz S, Fischer L, Hubert D, Sindram A, Hoelzer D, Thiel E. 2010. Clinical features and prognostic implications of TCF3-PBX1 and ETV6-RUNX1 in adult acute lymphoblastic leukemia. Haematologica 95: 241–246. 10.3324/haematol.2009.011346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, Low J, Qu C, Paietta EM, Kasper LH, Chang Y, Payne-Turner D, Althoff MJ, Song G, Chen SC, et al. 2015. Efficacy of retinoids in IKZF1-mutated BCR-ABL1 acute lymphoblastic leukemia. Cancer Cell 28: 343–356. 10.1016/j.ccell.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, Evans K, Richmond J, Robbins A, Jones L, Shapiro IM, Pachter JA, Weaver DT, Houghton PJ, Smith MA, et al. 2016. Synergism of FAK and tyrosine kinase inhibition in Ph+ B-ALL. JCI Insight 1: e86082 10.1172/jci.insight.86082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, Qian M, Te Kronnie G, Zhang R, Yang W, Zhang H, Lana T, Tedrick P, Baskin R, Verbist K, et al. 2018. Germline genetic IKZF1 variation and predisposition to childhood acute lymphoblastic leukemia. Cancer Cell 33: 937–948.e8. 10.1016/j.ccell.2018.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyaud E, Struski S, Prade N, Familiades J, Eichner R, Quelen C, Bousquet M, Mugneret F, Talmant P, Pages MP, et al. 2010. Wide diversity of PAX5 alterations in B-ALL: A groupe francophone de cytogénétique hématologique study. Blood 115: 3089–3097. 10.1182/blood-2009-07-234229 [DOI] [PubMed] [Google Scholar]
- Curtin SC, Miniño AM, Adnderson RN. 2016. Declines in cancer death rates among children and adolescents in the United States, 1999–2014, NCHS data brief, no 257 National Center for Health Statistics, Hyattsville, MD. [PubMed] [Google Scholar]
- Dang J, Wei L, de Ridder J, Su X, Rust AG, Roberts KG, Payne-Turner D, Cheng J, Ma J, Qu C, et al. 2015. PAX5 is a tumor suppressor in mouse mutagenesis models of acute lymphoblastic leukemia. Blood 125: 3609–3617. 10.1182/blood-2015-02-626127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila ML, Riviere I, Wang X, Bartido S, Park J, Curran K, Chung SS, Stefanski J, Borquez-Ojeda O, Olszewska M, et al. 2014. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med 6: 224ra25 10.1126/scitranslmed.3008226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven LJ, Beverloo HB, Van der Spek PJ, Escherich G, et al. 2009. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: A genome-wide classification study. Lancet Oncol 10: 125–134. 10.1016/S1470-2045(08)70339-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen EM, van Roon EH, Spijkers-Hagelstein JA, Schneider P, de Lorenzo P, Valsecchi MG, Pieters R, Stam RW. 2013. Frequencies and prognostic impact of RAS mutations in MLL-rearranged acute lymphoblastic leukemia in infants. Haematologica 98: 937–944. 10.3324/haematol.2012.067983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duployez N, Abou Chahla W, Lejeune S, Marceau-Renaut A, Letizia G, Boyer T, Geffroy S, Peyrouze P, Grardel N, Nelken B, et al. 2018. Detection of a new heterozygous germline ETV6 mutation in a case with hyperdiploid acute lymphoblastic leukemia. Eur J Haematol 100: 104–107. 10.1111/ejh.12981 [DOI] [PubMed] [Google Scholar]
- Evensen NA, Madhusoodhan PP, Meyer J, Saliba J, Chowdhury A, Araten DJ, Nersting J, Bhatla T, Vincent TL, Teachey D, et al. 2018. MSH6 haploinsufficiency at relapse contributes to the development of thiopurine resistance in pediatric B-lymphoblastic leukemia. Haematologica 103: 830–839. 10.3324/haematol.2017.176362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurstein S, Godley LA. 2017. Germline ETV6 mutations and predisposition to hematological malignancies. Int J Hematol 106: 189–195. 10.1007/s12185-017-2259-4 [DOI] [PubMed] [Google Scholar]
- Fielding AK, Richards SM, Chopra R, Lazarus HM, Litzow MR, Buck G, Durrant IJ, Luger SM, Marks DI, Franklin IM, et al. 2007. Outcome of 609 adults after relapse of acute lymphoblastic leukemia (ALL); an MRC UKALL12/ECOG 2993 study. Blood 109: 944–950. 10.1182/blood-2006-05-018192 [DOI] [PubMed] [Google Scholar]
- Fischer U, Forster M, Rinaldi A, Risch T, Sungalee S, Warnatz HJ, Bornhauser B, Gombert M, Kratsch C, Stütz AM, et al. 2015. Genomics and drug profiling of fatal TCF3-HLF–positive acute lymphoblastic leukemia identifies recurrent mutation patterns and therapeutic options. Nat Genet 47: 1020–1029. 10.1038/ng.3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford AM, Ridge SA, Cabrera ME, Mahmoud H, Steel CM, Chan LC, Greaves M. 1993. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature 363: 358–360. 10.1038/363358a0 [DOI] [PubMed] [Google Scholar]
- French C. 2014. NUT midline carcinoma. Nat Rev Cancer 14: 149–150. 10.1038/nrc3659 [DOI] [PubMed] [Google Scholar]
- Frey NV, Luger SM. 2015. How I treat adults with relapsed or refractory Philadelphia chromosome-negative acute lymphoblastic leukemia. Blood 126: 589–596. 10.1182/blood-2014-09-551937 [DOI] [PubMed] [Google Scholar]
- Gatica LV, Rosa AL. 2016. A complex interplay of genetic and epigenetic events leads to abnormal expression of the DUX4 gene in facioscapulohumeral muscular dystrophy. Neuromuscul Disord 26: 844–852. 10.1016/j.nmd.2016.09.015 [DOI] [PubMed] [Google Scholar]
- Gu Z, Churchman M, Roberts K, Li Y, Liu Y, Harvey RC, McCastlain K, Reshmi SC, Payne-Turner D, Iacobucci I, et al. 2016. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun 7: 13331 10.1038/ncomms13331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, Hagiwara K, Pelletier S, Gingras S, Berns H, et al. 2019. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet 51: 296–307. 10.1038/s41588-018-0315-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamadeh L, Enshaei A, Schwab C, Alonso CN, Attarbaschi A, Barbany G, den Boer ML, Boer JM, Braun M, Dalla Pozza L, et al. 2019. Validation of the United Kingdom copy-number alteration classifier in 3239 children with B-cell precursor ALL. Blood Adv 3: 148–157. 10.1182/bloodadvances.2018025718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CJ. 2015. Blood spotlight on iAMP21 acute lymphoblastic leukemia (ALL), a high-risk pediatric disease. Blood 125: 1383–1386. 10.1182/blood-2014-08-569228 [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Moorman AV, Broadfield ZJ, Cheung KL, Harris RL, Reza Jalali G, Robinson HM, Barber KE, Richards SM, Mitchell CD, et al. 2004. Three distinct subgroups of hypodiploidy in acute lymphoblastic leukemia. Br J Haematol 125: 552–559. 10.1111/j.1365-2141.2004.04948.x [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Moorman AV, Schwab C, Carroll AJ, Raetz EA, Devidas M, Strehl S, Nebral K, Harbott J, Teigler-Schlegel A, et al. 2014. An international study of intrachromosomal amplification of chromosome 21 (iAMP21): cytogenetic characterization and outcome. Leukemia 28: 1015–1021. 10.1038/leu.2013.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ, Chen IM, Atlas SR, Kang H, Ar K, et al. 2010. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: Correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood 116: 4874–4884. 10.1182/blood-2009-08-239681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi S, Ohki K, Nakabayashi K, Ichikawa H, Momozawa Y, Okamura K, Yaguchi A, Terada K, Saito Y, Yoshimi A, et al. 2017. ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica 102: 118–129. 10.3324/haematol.2016.151035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H, Shimamura A. 2017. ETV6 in hematopoiesis and leukemia predisposition. Semin Hematol 54: 98–104. 10.1053/j.seminhematol.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, Payne-Turner D, Churchman M, Andersson A, Chen SC, et al. 2013. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet 45: 242–252. 10.1038/ng.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrusak O, de Haas V, Stancikova J, Vakrmanova B, Janotova I, Mejstrikova E, Capek V, Trka J, Zaliova M, Luks A, et al. 2018. International cooperative study identifies treatment strategy in childhood ambiguous lineage leukemia. Blood 132: 264–276. 10.1182/blood-2017-12-821363 [DOI] [PubMed] [Google Scholar]
- Hunger SP. 1996. Chromosomal translocations involving the E2A gene in acute lymphoblastic leukemia: Clinical features and molecular pathogenesis. Blood 87: 1211–1224. [PubMed] [Google Scholar]
- Hunger SP, Mullighan CG. 2015. Acute lymphoblastic leukemia in children. N Engl J Med 373: 1541–1552. 10.1056/NEJMra1400972 [DOI] [PubMed] [Google Scholar]
- Hyrenius-Wittsten A, Pilheden M, Sturesson H, Hansson J, Walsh MP, Song G, Kazi JU, Liu J, Ramakrishan R, Garcia-Ruiz C, et al. 2018. De novo activating mutations drive clonal evolution and enhance clonal fitness in KMT2A-rearranged leukemia. Nat Commun 9: 1770 10.1038/s41467-018-04180-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci I, Mullighan CG. 2017. Genetic basis of acute lymphoblastic leukemia. J Clin Oncol 35: 975–983. 10.1200/JCO.2016.70.7836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T, Roberts WM, Shapiro LH, Jolly KW, Raimondi SC, Smith SD, Look AT. 1992. Fusion of the leucine zipper gene HLF to the E2A gene in human acute B-lineage leukemia. Science 257: 531–534. 10.1126/science.1386162 [DOI] [PubMed] [Google Scholar]
- Inaba H, Azzato EM, Mullighan CG. 2017. Integration of next-generation sequencing to treat acute lymphoblastic leukemia with targetable lesions: The St. Jude Children's Research Hospital approach. Front Pediatr 5: 258 10.3389/fped.2017.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe JD, Wang Y, Chan HM, Zhang J, Huether R, Kryukov GV, Bhang HE, Taylor JE, Hu M, Englund NP, et al. 2013. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet 45: 1386–1391. 10.1038/ng.2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Roberts KG, Jabbour E, Patel K, Eterovic AK, Chen K, Zweidler-McKay P, Lu X, Fawcett G, Wang SA, et al. 2017a. Ph-like acute lymphoblastic leukemia: A high-risk subtype in adults. Blood 129: 572–581. 10.1182/blood-2016-07-726588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Zhang H, Roberts KG, Qian MX, Yang WJ, Jabbour EJ, Kantarjian HM, Mullighan CG, Yang JJ, Konopleva M. 2017b. GATA3 rs3824662A allele is overrepresented in adult patients with Ph-like ALL, especially in patients with CRLF2 abnormalities. Blood 130: 1430 10.1182/blood-2017-03-771576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarviaho T, Bang B, Zachariadis V, Taylan F, Moilanen J, Mottonen M, Smith CIE, Harila-Saari A, Niinimaki R, Nordgren A. 2019. Predisposition to childhood acute lymphoblastic leukemia caused by a constitutional translocation disrupting ETV6. Blood Adv 24: 2722–2731. 10.1182/bloodadvances.2018028795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Hegde S, Knolhoff BL, Zhu Y, Herndon JM, Meyer MA, Nywening TM, Hawkins WG, Shapiro IM, Weaver DT, et al. 2016. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat Med 22: 851–860. 10.1038/nm.4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi I, Yoshida T, Jena N, Qi X, Zhang J, Van Etten RA, Georgopoulos K. 2014. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat Immunol 15: 294–304. 10.1038/ni.2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian HM, DeAngelo DJ, Stelljes M, Martinelli G, Liedtke M, Stock W, Gokbuget N, O'Brien S, Wang K, Wang T, et al. 2016. Inotuzumab ozogamicin versus standard therapy for acute lymphoblastic leukemia. N Engl J Med 375: 740–753. 10.1056/NEJMoa1509277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H, Stein A, Gökbuget N, Fielding AK, Schuh AC, Ribera JM, Wei A, Dombret H, Foà R, Bassan R, et al. 2017. Blinatumomab versus chemotherapy for advanced acute lymphoblastic leukemia. N Engl J Med 376: 836–847. 10.1056/NEJMoa1609783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karol SE, Larsen E, Cheng C, Cao X, Yang W, Ramsey LB, Fernandez CA, McCorkle JR, Paugh SW, Autry RJ, et al. 2017. Genetics of ancestry-specific risk for relapse in acute lymphoblastic leukemia. Leukemia 31: 1325–1332. 10.1038/leu.2017.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Miyagawa N, Mitsui K, Matsuoka M, Kojima Y, Takahashi H, Ootsubo K, Nagai J, Ueno H, Ishibashi T, et al. 2015. TKI dasatinib monotherapy for a patient with Ph-like ALL bearing ATF7IP/PDGFRB translocation. Pediatr Blood Cancer 62: 1058–1060. 10.1002/pbc.25327 [DOI] [PubMed] [Google Scholar]
- Kuiper RP, Schoenmakers EF, van Reijmersdal SV, Hehir-Kwa JY, van Kessel AG, van Leeuwen FN, Hoogerbrugge PM. 2007. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia 21: 1258–1266. 10.1038/sj.leu.2404691 [DOI] [PubMed] [Google Scholar]
- Lee BY, Timpson P, Horvath LG, Daly RJ. 2015. FAK signaling in human cancer as a target for therapeutics. Pharmacol Ther 146: 132–149. 10.1016/j.pharmthera.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Lengline E, Beldjord K, Dombret H, Soulier J, Boissel N, Clappier E. 2013. Successful tyrosine kinase inhibitor therapy in a refractory B-cell precursor acute lymphoblastic leukemia with EBF1-PDGFRB fusion. Haematologica 98: e146–e148. 10.3324/haematol.2013.095372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Schwab C, Ryan SL, Papaemmanuil E, Robinson HM, Jacobs P, Moorman AV, Dyer S, Borrow J, Griffiths M, et al. 2014. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature 508: 98–102. 10.1038/nature13115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Li H, Bai Y, Kirschner-Schwabe R, Yang JJ, Chen Y, Lu G, Tzoneva G, Ma X, Wu T, et al. 2015. Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL. Nat Med 21: 563–571. 10.1038/nm.3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Dai YT, Lilljebjörn H, Shen SH, Cui BW, Bai L, Liu YF, Qian MX, Kubota Y, Kiyoi H, et al. 2018. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci 115: E11711–E11720. 10.1073/pnas.1814397115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilljebjörn H, Henningsson R, Hyrenius-Wittsten A, Olsson L, Orsmark-Pietras C, von Palffy S, Askmyr M, Rissler M, Schrappe M, Cario G, et al. 2016. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun 7: 11790 10.1038/ncomms11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YF, Wang BY, Zhang WN, Huang JY, Li BS, Zhang M, Jiang L, Li JF, Wang MJ, Dai YJ, et al. 2016. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine 8: 173–183. 10.1016/j.ebiom.2016.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh ML, Zhang J, Harvey RC, Roberts K, Payne-Turner D, Kang H, Wu G, Chen X, Becksfort J, Edmonson M, et al. 2013. Tyrosine kinome sequencing of pediatric acute lymphoblastic leukemia: A report from the children's oncology group TARGET project. Blood 121: 485–488. 10.1182/blood-2012-04-422691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, Song G, Easton J, Harvey RC, Wheeler DA, et al. 2015. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun 6: 6604 10.1038/ncomms7604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K, Neuberg DS, Sinha AU, Sallan SE, Silverman LB, et al. 2014. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun 5: 3469 10.1038/ncomms4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli G, Iacobucci I, Storlazzi CT, Vignetti M, Paoloni F, Cilloni D, Soverini S, Vitale A, Chiaretti S, Cimino G, et al. 2009. IKZF1 (Ikaros) deletions in BCR-ABL1–positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: A GIMEMA AL WP report. J Clin Oncol 27: 5202–5207. 10.1200/JCO.2008.21.6408 [DOI] [PubMed] [Google Scholar]
- Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, Bader P, Verneris MR, Stefanski HE, Myers GD, et al. 2018. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med 378: 439–448. 10.1056/NEJMoa1709866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JA, Wang J, Hogan LE, Yang JJ, Dandekar S, Patel JP, Tang Z, Zumbo P, Li S, Zavadil J, et al. 2013. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet 45: 290–294. 10.1038/ng.2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman AV, Harrison CJ, Buck GA, Richards SM, Secker-Walker LM, Martineau M, Vance GH, Cherry AM, Higgins RR, Fielding AK, et al. 2007. Karyotype is an independent prognostic factor in adult acute lymphoblastic leukemia (ALL): Analysis of cytogenetic data from patients treated on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial. Blood 109: 3189–3197. 10.1182/blood-2006-10-051912 [DOI] [PubMed] [Google Scholar]
- Moorman AV, Schwab C, Ensor HM, Russell LJ, Morrison H, Jones L, Masic D, Patel B, Rowe JM, Tallman M, et al. 2012. IGH@ translocations, CRLF2 deregulation, and microdeletions in adolescents and adults with acute lymphoblastic leukemia. J Clin Oncol 30: 3100–3108. 10.1200/JCO.2011.40.3907 [DOI] [PubMed] [Google Scholar]
- Moorman AV, Robinson H, Schwab C, Richards SM, Hancock J, Mitchell CD, Goulden N, Vora A, Harrison CJ. 2013. Risk-directed treatment intensification significantly reduces the risk of relapse among children and adolescents with acute lymphoblastic leukemia and intrachromosomal amplification of chromosome 21: a comparison of the MRC ALL97/99 and UKALL2003 trials. J Clin Oncol 31: 3389–3396. 10.1200/JCO.2013.48.9377 [DOI] [PubMed] [Google Scholar]
- Moriyama T, Metzger ML, Wu G, Nishii R, Qian M, Devidas M, Yang W, Cheng C, Cao X, Quinn E, et al. 2015a. Germline genetic variation in ETV6 and risk of childhood acute lymphoblastic leukaemia: A systematic genetic study. Lancet Oncol 16: 1659–1666. 10.1016/S1470-2045(15)00369-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama T, Relling MV, Yang JJ. 2015b. Inherited genetic variation in childhood acute lymphoblastic leukemia. Blood 125: 3988–3995. 10.1182/blood-2014-12-580001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446: 758–764. 10.1038/nature05690 [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, et al. 2008a. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453: 110–114. 10.1038/nature06866 [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, Downing JR. 2008b. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 322: 1377–1380. 10.1126/science.1164266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, et al. 2009. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 360: 470–480. 10.1056/NEJMoa0808253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, Heatley SL, Holmfeldt L, Collins-Underwood JR, Ma J, et al. 2011. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 471: 235–239. 10.1038/nature09727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Jeha S, Pei D, Payne-Turner D, Coustan-Smith E, Roberts KG, Waanders E, Choi JK, Ma X, Raimondi SC, et al. 2015. Outcome of children with hypodiploid ALL treated with risk-directed therapy based on MRD levels. Blood 126: 2896–2899. 10.1182/blood-2015-09-671131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebral K, Denk D, Attarbaschi A, König M, Mann G, Haas OA, Strehl S. 2009. Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia 23: 134–143. 10.1038/leu.2008.306 [DOI] [PubMed] [Google Scholar]
- Noetzli L, Lo RW, Lee-Sherick AB, Callaghan M, Noris P, Savoia A, Rajpurkar M, Jones K, Gowan K, Balduini CL, et al. 2015. Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia. Nat Genet 47: 535–538. 10.1038/ng.3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor D, Enshaei A, Bartram J, Hancock J, Harrison CJ, Hough R, Samarasinghe S, Schwab C, Vora A, Wade R, et al. 2018. Genotype-specific minimal residual disease interpretation improves stratification in pediatric acute lymphoblastic leukemia. J Clin Oncol 36: 34–43. 10.1200/JCO.2017.74.0449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki K, Kiyokawa N, Saito Y, Hirabayashi S, Nakabayashi K, Ichikawa H, Momozawa Y, Okamura K, Yoshimi A, Ogata-Kawata H, et al. 2019. Clinical and molecular characteristics of MEF2D fusion-positive B-cell precursor acute lymphoblastic leukemia in childhood, including a novel translocation resulting in MEF2D-HNRNPH1 gene fusion. Haematologica 104: 128–137. 10.3324/haematol.2017.186320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K, Khiabanian H, da Silva-Almeida AC, Tzoneva G, Abate F, Ambesi-Impiombato A, Sanchez-Martin M, Carpenter Z, Penson A, Perez-Garcia A, et al. 2016. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci 113: 11306–11311. 10.1073/pnas.1608420113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Rapado I, Li Y, Potter NE, Wedge DC, Tubio J, Alexandrov LB, Van Loo P, Cooke SL, Marshall J, et al. 2014. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet 46: 116–125. 10.1038/ng.2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Rivière I, Gonen M, Wang X, Sénéchal B, Curran KJ, Sauter C, Wang Y, Santomasso B, Mead E, et al. 2018. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N Engl J Med 378: 449–459. 10.1056/NEJMoa1709919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passet M, Boissel N, Sigaux F, Saillard C, Bargetzi M, Ba I, Thomas X, Graux C, Chalandon Y, Leguay T, et al. 2019. PAX5 P80R mutation identifies a novel subtype of B-cell precursor acute lymphoblastic leukemia with favorable outcome. Blood 133: 280–284. 10.1182/blood-2018-10-882142 [DOI] [PubMed] [Google Scholar]
- Paulsson K, Lilljebjörn H, Biloglav A, Olsson L, Rissler M, Castor A, Barbany G, Fogelstrand L, Nordgren A, Sjogren H, et al. 2015. The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat Genet 47: 672–676. 10.1038/ng.3301 [DOI] [PubMed] [Google Scholar]
- Perez-Andreu V, Roberts KG, Harvey RC, Yang W, Cheng C, Pei D, Xu H, Gastier-Foster J ES, Lim JY, et al. 2013. Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat Genet 45: 1494–1498. 10.1038/ng.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt LA, Tikhonova AN, Hu H, Trimarchi T, King B, Gong Y, Sanchez-Martin M, Tsirigos A, Littman DR, Ferrando AA, et al. 2015. CXCL12-producing vascular endothelial niches control acute T cell leukemia maintenance. Cancer Cell 27: 755–768. 10.1016/j.ccell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH, Rebora P, Schrappe M, Attarbaschi A, Baruchel A, Basso G, Cave H, Elitzur S, Koh K, Liu HC, et al. 2019. Outcome of children with hypodiploid acute lymphoblastic leukemia: A retrospective multinational study. J Clin Oncol 37: 770–779. 10.1200/JCO.18.00822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian M, Cao X, Devidas M, Yang W, Cheng C, Dai Y, Carroll A, Heerema NA, Zhang H, Moriyama T, et al. 2018. TP53 germline variations influence the predisposition and prognosis of B-cell acute lymphoblastic leukemia in children. J Clin Oncol 36: 591–599. 10.1200/JCO.2017.75.5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian M, Xu H, Perez-Andreu V, Roberts KG, Zhang H, Yang W, Zhang S, Zhao X, Smith C, Devidas M, et al. 2019. Novel susceptibility variants at the ERG locus for childhood acute lymphoblastic leukemia in Hispanics. Blood 133: 724–729. 10.1182/blood-2018-07-862946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampersaud E, Ziegler DS, Iacobucci I, Payne-Turner D, Churchman ML, Schrader KA, Vijai J, Offit K, Tucker K, Sutton R, et al. 2019. Germline deletion of ETV6 in familial acute lymphoblastic leukemia. Blood Adv 3: 1039–1046. 10.1182/bloodadvances.2018030635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa S, Cho BS, Ghosh D, Sivina M, Koehrer S, Müschen M, Peled A, Davis RE, Konopleva M, Burger JA. 2016. Effects of pharmacological and genetic disruption of CXCR4 chemokine receptor function in B-cell acute lymphoblastic leukaemia. Br J Haematol 174: 425–436. 10.1111/bjh.14075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravandi F, O'Brien S, Thomas D, Faderl S, Jones D, Garris R, Dara S, Jorgensen J, Kebriaei P, Champlin R, et al. 2010. First report of phase 2 study of dasatinib with hyper-CVAD for the frontline treatment of patients with Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia. Blood 116: 2070–2077. 10.1182/blood-2009-12-261586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshmi SC, Harvey RC, Roberts KG, Stonerock E, Smith A, Jenkins H, Chen IM, Valentine M, Liu Y, Li Y, et al. 2017. Targetable kinase gene fusions in high-risk B-ALL: A study from the children's oncology group. Blood 129: 3352–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, McCastlain K, Ding L, Lu C, Song G, et al. 2014a. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 371: 1005–1015. 10.1056/NEJMoa1403088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Pei D, Campana D, Payne-Turner D, Li Y, Cheng C, Sandlund JT, Jeha S, Easton J, Becksfort J, et al. 2014b. Outcomes of children with BCR-ABL1–like acute lymphoblastic leukemia treated with risk-directed therapy based on the levels of minimal residual disease. J Clin Oncol 32: 3012–3020. 10.1200/JCO.2014.55.4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Gu Z, Payne-Turner D, McCastlain K, Harvey RC, Chen IM, Pei D, Iacobucci I, Valentine M, Pounds SB, et al. 2017a. High frequency and poor outcome of Philadelphia chromosome-like acute lymphoblastic leukemia in adults. J Clin Oncol 35: 394–401. 10.1200/JCO.2016.69.0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Yang YL, Payne-Turner D, Lin W, Files JK, Dickerson K, Gu Z, Taunton J, Janke LJ, Chen T, et al. 2017b. Oncogenic role and therapeutic targeting of ABL-class and JAK-STAT activating kinase alterations in Ph-like ALL. Blood Adv 1: 1657–1671. 10.1182/bloodadvances.2017000547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Reshmi SC, Harvey RC, Chen IM, Patel K, Stonerock E, Jenkins H, Dai Y, Valentine M, Gu Z, et al. 2018. Genomic and outcome analyses of Ph-like ALL in NCI standard-risk patients: A report from the children's oncology group. Blood 132: 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell LJ, Enshaei A, Jones L, Erhorn A, Masic D, Bentley H, Laczko KS, Fielding AK, Goldstone AH, Goulden N, et al. 2014. IGH@ translocations are prevalent in teenagers and young adults with acute lymphoblastic leukemia and are associated with a poor outcome. J Clin Oncol 32: 1453–1462. 10.1200/JCO.2013.51.3242 [DOI] [PubMed] [Google Scholar]
- Schinnerl D, Mejstrikova E, Schumich A, Zaliova M, Fortschegger K, Nebral K, Attarbaschi A, Fiser K, Kauer MO, Popitsch N, et al. 2019. CD371 cell surface expression: A unique feature of DUX4-rearranged acute lymphoblastic leukemia. Haematologica 104: e352–e355. 10.3324/haematol.2018.214353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz KR, Carroll A, Heerema NA, Bowman WP, Aledo A, Slayton WB, Sather H, Devidas M, Zheng HW, Davies SM, et al. 2014. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children's oncology group study AALL0031. Leukemia 28: 1467–1471. 10.1038/leu.2014.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab C, Ryan SL, Chilton L, Elliott A, Murray J, Richardson S, Wragg C, Moppett J, Cummins M, Tunstall O, et al. 2016. EBF1-PDGFRB fusion in pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL): Genetic profile and clinical implications. Blood 127: 2214–2218. 10.1182/blood-2015-09-670166 [DOI] [PubMed] [Google Scholar]
- Schwab C, Nebral K, Chilton L, Leschi C, Waanders E, Boer JM, Žaliová M, Sutton R, Öfverholm II, Ohki K, et al. 2017. Intragenic amplification of PAX5: A novel subgroup in B-cell precursor acute lymphoblastic leukemia? Blood Adv 1: 1473–1477. 10.1182/bloodadvances.2017006734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shago M, Abla O, Hitzler J, Weitzman S, Abdelhaleem M. 2016. Frequency and outcome of pediatric acute lymphoblastic leukemia with ZNF384 gene rearrangements including a novel translocation resulting in an ARID1B/ZNF384 gene fusion. Pediatr Blood Cancer 63: 1915–1921. 10.1002/pbc.26116 [DOI] [PubMed] [Google Scholar]
- Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, Wechsler J, Yang J, Hayes J, Klein RJ, et al. 2013. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet 45: 1226–1231. 10.1038/ng.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayton WB, Schultz KR, Kairalla JA, Devidas M, Mi X, Pulsipher MA, Chang BH, Mullighan C, Iacobucci I, Silverman LB, et al. 2018. Dasatinib plus intensive chemotherapy in children, adolescents, and young adults with Philadelphia chromosome-positive acute lymphoblastic leukemia: Results of Children's Oncology Group trial AALL0622. J Clin Oncol 36: 2306–2314. 10.1200/JCO.2017.76.7228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk L, Fischer M, Jurado S, Jaritz M, Azaryan A, Werner B, Roth M, Zuber J, Stanulla M, den Boer ML, et al. 2017. Molecular role of the PAX5-ETV6 oncoprotein in promoting B-cell acute lymphoblastic leukemia. EMBO J 36: 718–735. 10.15252/embj.201695495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanulla M, Dagdan E, Zaliova M, Möricke A, Palmi C, Cazzaniga G, Eckert C, Te Kronnie G, Bourquin JP, Bornhauser B, et al. 2018. IKZF1plus defines a new minimal residual disease–dependent very-poor prognostic profile in pediatric B-cell precursor acute lymphoblastic leukemia. J Clin Oncol 36: 1240–1249. 10.1200/JCO.2017.74.3617 [DOI] [PubMed] [Google Scholar]
- Stock W. 2010. Adolescents and young adults with acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program 2010: 21–29. 10.1182/asheducation-2010.1.21 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Okuno Y, Kawashima N, Muramatsu H, Okuno T, Wang X, Kataoka S, Sekiya Y, Hamada M, Murakami N, et al. 2016. MEF2D-BCL9 fusion gene is associated with high-risk acute B-cell precursor lymphoblastic leukemia in adolescents. J Clin Oncol 34: 3451–3459. 10.1200/JCO.2016.66.5547 [DOI] [PubMed] [Google Scholar]
- Tabe Y, Konopleva M. 2014. Advances in understanding the leukaemia microenvironment. Br J Haematol 164: 767–778. 10.1111/bjh.12725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Wang F, Morita K, Yan Y, Hu P, Zhao P, Zhar AA, Wu CJ, Gumbs C, Little L, et al. 2018. Integrative genomic analysis of adult mixed phenotype acute leukemia delineates lineage associated molecular subtypes. Nat Commun 9: 2670 10.1038/s41467-018-04924-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasian SK, Hurtz C, Wertheim GB, Bailey NG, Lim MS, Harvey RC, Chen IM, Willman CL, Astles R, Zebrowski A, et al. 2017a. High incidence of Philadelphia chromosome-like acute lymphoblastic leukemia in older adults with B-ALL. Leukemia 31: 981–984. 10.1038/leu.2016.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasian SK, Loh ML, Hunger SP. 2017b. Philadelphia chromosome-like acute lymphoblastic leukemia. Blood 130: 2064–2072. 10.1182/blood-2017-06-743252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topka S, Vijai J, Walsh MF, Jacobs L, Maria A, Villano D, Gaddam P, Wu G, McGee RB, Quinn E, et al. 2015. Germline ETV6 mutations confer susceptibility to acute lymphoblastic leukemia and thrombocytopenia. PLoS Genet 11: e1005262 10.1371/journal.pgen.1005262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoneva G, Dieck CL, Oshima K, Ambesi-Impiombato A, Sánchez-Martín M, Madubata CJ, Khiabanian H, Yu J, Waanders E, Iacobucci I, et al. 2018. Clonal evolution mechanisms in NT5C2 mutant-relapsed acute lymphoblastic leukaemia. Nature 553: 511–514. 10.1038/nature25186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida A, Isobe Y, Uemura Y, Nishio Y, Sakai H, Kato M, Otsubo K, Hoshikawa M, Takagi M, Miura I. 2017. De novo acute lymphoblastic leukemia-like disease of high grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements: A case report and literature review. BMC Clin Pathol 17: 21 10.1186/s12907-017-0060-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagener R, López C, Kleinheinz K, Bausinger J, Aukema SM, Nagel I, Toprak UH, Seufert J, Altmüller J, Thiele H, et al. 2018. IG-MYC+ neoplasms with precursor B-cell phenotype are molecularly distinct from Burkitt lymphomas. Blood 132: 2280–2285. 10.1182/blood-2018-03-842088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston BW, Hayden MA, Roberts KG, Bowyer S, Hsu J, Fedoriw G, Rao KW, Mullighan CG. 2013. Tyrosine kinase inhibitor therapy induces remission in a patient with refractory EBF1-PDGFRB–positive acute lymphoblastic leukemia. J Clin Oncol 31: e413–e416. 10.1200/JCO.2012.47.6770 [DOI] [PubMed] [Google Scholar]
- Xiao W, Bharadwaj M, Levine M, Farnhoud N, Pastore F, Getta BM, Hultquist A, Famulare C, Medina JS, Patel MA, et al. 2018. PHF6 and DNMT3A mutations are enriched in distinct subgroups of mixed phenotype acute leukemia with T-lineage differentiation. Blood Adv 2: 3526–3539. 10.1182/bloodadvances.2018023531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JJ, Cheng C, Devidas M, Cao X, Fan Y, Campana D, Yang W, Neale G, Cox NJ, Scheet P, et al. 2011. Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat Genet 43: 237–241. 10.1038/ng.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Tsuzuki S, Kawazu M, Hayakawa F, Kojima S, Ueno T, Imoto N, Kohsaka S, Kunita A, Doi K, et al. 2016. Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat Genet 48: 569–574. 10.1038/ng.3535 [DOI] [PubMed] [Google Scholar]
- Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, et al. 2002. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1: 133–143. 10.1016/S1535-6108(02)00032-6 [DOI] [PubMed] [Google Scholar]
- Yoshihara H, Churchman ML, Peters JL, Finkelstein DB, Paietta EM, Litzow MR, Mullighan CG. 2018. Functional and genomic characterization of the interaction between acute lymphoblastic leukemia cells and the microenvironment identifies pathways for therapeutic intervention. Blood 132: 1550 10.1182/blood-2018-08-867010 [DOI] [PubMed] [Google Scholar]
- Zaliova M, Kotrova M, Bresolin S, Stuchly J, Stary J, Hrusak O, Te Kronnie G, Trka J, Zuna J, Vaskova M. 2017. ETV6/RUNX1-like acute lymphoblastic leukemia: A novel B-cell precursor leukemia subtype associated with the CD27/CD44 immunophenotype. Genes Chromosomes Cancer 56: 608–616. 10.1002/gcc.22464 [DOI] [PubMed] [Google Scholar]
- Zaliova M, Stuchly J, Winkowska L, Musilova A, Fiser K, Slamova M, Starkova J, Vaskova M, Hrusak O, Sramkova L, et al. 2019. Genomic landscape of pediatric B-other acute lymphoblastic leukemia in a consecutive European cohort. Haematolica 104: 1396–1406. 10.3324/haematol.2018.204974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481: 157–163. 10.1038/nature10725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MY, Churpek JE, Keel SB, Walsh T, Lee MK, Loeb KR, Gulsuner S, Pritchard CC, Sanchez-Bonilla M, Delrow JJ, et al. 2015. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet 47: 180–185. 10.1038/ng.3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, McCastlain K, Yoshihara H, Xu B, Chang Y, Churchman ML, Wu G, Li Y, Wei L, Iacobucci I, et al. 2016. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet 48: 1481–1489. 10.1038/ng.3691 [DOI] [PMC free article] [PubMed] [Google Scholar]