Abstract
Direct lineage reprogramming of abundant and accessible cells into therapeutically useful cell types holds tremendous potential in regenerative medicine. To date, a number of different cell types have been generated by lineage reprogramming methods, including cells from the neural, cardiac, hepatic, and pancreatic lineages. The success of this strategy relies on developmental biology and the knowledge of cell-fate-defining transcriptional networks. Hepatocytes represent a prime target for β cell conversion for numerous reasons, including close developmental origin, accessibility, and regenerative potential. We present here an overview of pancreatic and hepatic development, with a particular focus on the mechanisms underlying the divergence between the two cell lineages. Additionally, we discuss to what extent this lineage relationship can be exploited in efforts to reprogram one cell type into the other and whether such an approach may provide a suitable strategy for regenerative therapies of diabetes.
Cell identity is imparted by temporal and spatial integration of extrinsic signals and intrinsic determinants. It is now well established that cell differentiation is not only a unidirectional process (Gurdon 2006; Shi et al. 2017). The differentiated state can be unlocked and cells can be forced to change identity, either to revert to a pluripotent state or to adopt another differentiated identity, so-called lineage reprogramming (Gurdon 2006; Shi et al. 2017). Defining the mechanisms that allow mature cells to switch identities is not only leading to an improved understanding of developmental processes and disease pathogenesis but it is also opening doors to cellular replacement therapies (Heinrich et al. 2015; Tanabe et al. 2015; Xu et al. 2015). For instance, such ability to adopt an alternative fate can be harnessed therapeutically to generate damaged or lost cells for regenerative medicine.
Lineage reprogramming is conceptually attractive in regenerative medicine, providing an alternative approach with a relatively lower risk of tumorigenesis when compared to the use of pluripotent stem cells (Cohen and Melton 2011; Xu et al. 2015). It also provides the opportunity to directly convert cells in situ, which is relevant in certain regenerative strategies (Heinrich et al. 2015). Finally, lineage reprogramming strategies can be designed to use abundant and easily accessible autologous patient-derived cell types as a source, like skin or liver biopsies, which would tremendously facilitate the clinical application.
Successful experimental strategies for reprogramming of one specialized cell type into another are inspired by embryonic development (Heinrich et al. 2015; Tanabe et al. 2015; Xu et al. 2015). These approaches are typically based on the ectopic expression of defined transcription factors (TFs), which are necessary for the acquisition of the desired cell fate. Also, an embryological origin that is common to both the cellular source and desired cell types might facilitate the fate switch (Heinrich et al. 2015; Tanabe et al. 2015; Xu et al. 2015).
Particularly relevant in this context are the so-called pioneer TFs, which act as master regulators of cell fate during normal development via their interaction with chromatin and in cooperation with lineage-specific TFs, including FoxA, GATA4, C/ebpα, Ascl1 (Morris 2016). Often, reprogramming strategies are based on the combination of multiple TFs to superimpose the program of the desired cell type; such combinations can either facilitate stepwise conversion from a progenitor to a mature state, for example with one TF being involved in the initial fate specification and the other one in subsequent maturation, or alternatively one of the TFs might act as a repressor to erase the original cellular identity (Heinrich et al. 2015; Tanabe et al. 2015; Xu et al. 2015). Finally, developmental factors acting at the branchpoint between alternate lineages appear also particularly suitable for lineage reprogramming (Tanabe et al. 2015; Cerdá-Esteban et al. 2017).
The liver represents an ideal cellular source for generating pancreatic cells through lineage reprogramming because of the close developmental origin between the two organs but also because of its regenerative ability and accessibility (Si-Tayeb et al. 2010; Zaret 2016; Michalopoulos 2017). It might be potentially clinically feasible to reprogram ex vivo human hepatic biopsies and transplant them back into the same patient, allowing the diabetic patient to be the donor of her/his own healthy tissue. Besides a common developmental origin, the two organs share many features also in adult life, including a common set of TFs and metabolic properties (Zaret and Grompe 2008). Moreover, as a result of these similarities, they represent a remarkable example of intercellular plasticity persistent throughout adulthood (Shen et al. 2003; Zaret and Grompe 2008). For instance, occurrence of hepatic foci has been reported in the pancreas of rodents upon injury, in response to special dietary regimens (e.g., copper depletion-diet), carcinogens, or extracellular cues, as well as in rare human pancreatic cancers, such as hepatoid carcinomas (Rao et al. 1989; Krakowski et al. 1999; Grompe 2003; Shen et al. 2003). Similarly, the emergence of cells displaying a pancreatic phenotype in the liver, with expression of pancreatic gene markers, has been reported in experimental animal models (Rao et al. 1986; Shen et al. 2003; Shanmukhappa et al. 2005) or human liver cancers, particularly cholangiocarcinoma (Banales et al. 2016).
We review here the molecular and cellular mechanisms underlying the acquisition of pancreatic and hepatic cell identities from endoderm progenitors, focusing in particular on the divergence between the two lineages. We also discuss how this knowledge has been applied in the context of lineage reprogramming and summarize past and current efforts to induce the generation of insulin-producing β cells from liver cells. We draw mainly from findings in the mouse model that are also conserved in other model organisms, including in humans.
THE LIVER AND PANCREAS: STRUCTURE AND FUNCTIONS
The liver and pancreas are essential regulators of the systemic metabolism in vertebrates. The liver performs a wide range of metabolic functions, such as nutrient processing, maintenance of blood metabolites and protein concentrations, as well as life-saving detoxification processes (Abdel-Misih and Bloomston 2010). Several cell types of different embryological origin, including hepatocytes, biliary epithelial cells, stellate cells, Kupffer cells, and liver sinusoidal endothelial cells, compose the liver. The hepatocytes are the major component of the organ and operate in anatomical units termed lobules that are radially polarized by blood flow and morphogens (Abdel-Misih and Bloomston 2010). Within this microenvironment, hepatocytes subspecialize based on their position along the porto-central axis of the liver lobule into either “periportal” or “perivenous” hepatocytes, in a phenomenon called “liver zonation” (Ben-Moshe and Itzkovitz 2019). For example, hepatocytes in the region adjacent to the portal veins are metabolically active and involved in cholesterol synthesis, fatty acid oxidation, and bile acid production, while the hepatocytes in the region around the central vein are the major effector of glycolysis and xenobiotic metabolism (Ben-Moshe and Itzkovitz 2019).
The adult pancreas is a gland composed of exocrine and endocrine tissues with divergent metabolic functions (Klatt and Kumar 2015). The exocrine pancreas produces and secretes digestive enzymes, which are transported to the duodenum via a network of pancreatic ducts, while the endocrine cells produce the hormones regulating glucose metabolism and homeostasis, including glucagon and insulin (Klatt and Kumar 2015). Hormone-secreting endocrine cells cluster together to form the pancreatic islets, also known as islets of Langerhans, whereas in the exocrine compartment acinar cells are organized in functional units along the ductal network (Spagnoli 2007). The endocrine islets consist of mostly insulin-producing β cells (∼60% in humans) and glucagon-producing α cells (∼30% in humans), with the remaining 10% made up of δ cells, γ or pancreatic polypeptide (PP) cells, and ε cells (Spagnoli 2007; Romer and Sussel 2015).
The importance of liver and pancreas for human physiology and health is reflected in the severity of the diseases associated with the two organs, including chronic liver disease, cancer, and diabetes mellitus. End-stage liver diseases as well as diabetes are incurable conditions, often life threatening, and represent huge socioeconomic burdens, stressing the need for novel regenerative therapies to treat them (Abdel-Misih and Bloomston 2010; Stanger and Hebrok 2013; ADA 2014; Michalopoulos 2017; Sneddon et al. 2018).
Because of the close embryonic origin and intercellular plasticity between the two cell types, targeting liver cells to generate insulin-producing cells through lineage reprogramming strategies has emerged as a valuable approach. Moreover, the high regenerative ability and accessibility of the adult liver (Michalopoulos 2017) make it an ideal renewable source of new β cells, whereas the opposite is not conceivable, being the pancreas in an inaccessible anatomical location and characterized by a very limited capacity for regeneration (Stanger and Hebrok 2013; Klatt and Kumar 2015).
THE LIVER AND PANCREAS ARE DEVELOPMENTALLY RELATED
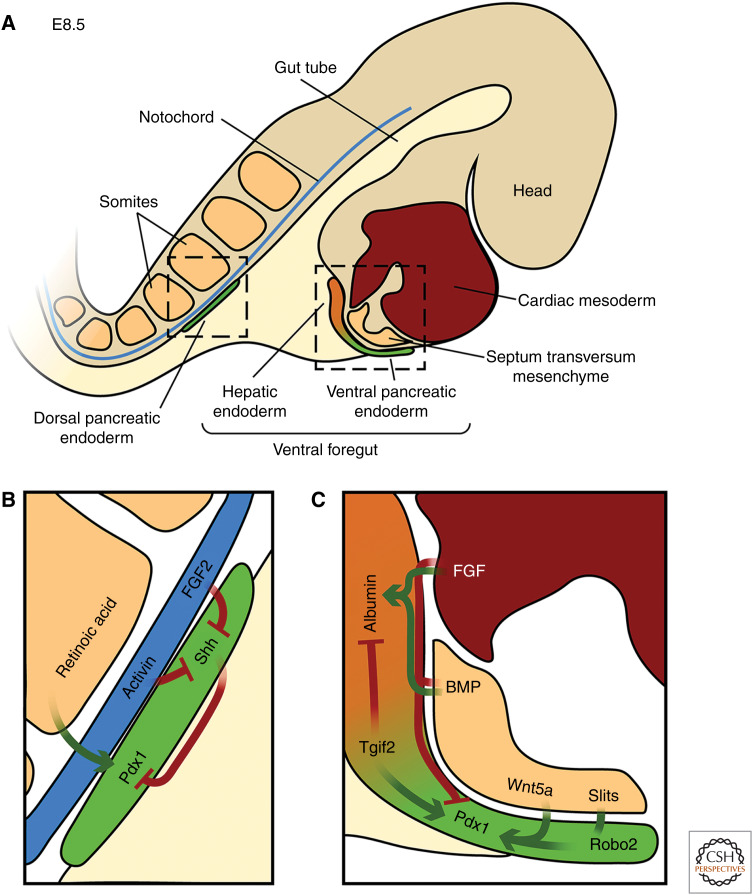
Many excellent and comprehensive reviews have been written on the cellular and molecular mechanisms underlying pancreas and liver development separately (Duncan 2003; Gittes 2009; Si-Tayeb et al. 2010; Pan and Wright 2011; Shih et al. 2013; Romer and Sussel 2015; Zaret 2016), here we mostly focus on the mechanisms underlying the divergence between the two lineages. Fate mapping and lineage tracing experiments in different vertebrate models have shown that liver and pancreas are both derivatives of the endodermal germ layer and arise from cells of the posterior foregut (Deutsch et al. 2001; Tremblay and Zaret 2005; Zorn and Wells 2009; Angelo et al. 2012). In the mouse, hepatic and pancreatic organ domains become specified around embryonic (E) day 8.5 from two distinct domains in the posterior foregut, one located dorsally and the other ventrally (Zorn and Wells 2009). The dorsal foregut endoderm will give rise to pancreatic tissue only, while the ventral foregut endoderm harbors progenitors of the liver, pancreas, gallbladder, and bile ducts (Fig. 1; Zorn and Wells 2009).
Figure 1.
Specification of the hepatic and pancreatic organ domains. (A) Sagittal view of a mouse embryo at E8.5. The endoderm (light beige) receives inductive signals from neighboring mesodermal tissues (septum transversum mesenchyme, cardiac mesoderm, somites, notochord). Consequently, hepatic (orange) and ventral pancreatic (green) organ domains form in the ventral foregut endoderm and a dorsal pancreatic (green) progenitor domain in the dorsal foregut. (B) Schematics of the factors controlling fate specification of the dorsal pancreatic endoderm. Activin and FGF2 secreted by the notochord repress expression of Sonic hedgehog (Shh) in the presumptive dorsal pancreatic endoderm, which in turn allows Pdx1 expression (Hebrok 2003). Pdx1 expression is further promoted by retinoic acid from the somites (Gittes 2009). (C) Schematic overview of the factors controlling the fate specification of hepatic and ventral pancreatic endoderm. Fibroblast growth factors (FGFs) secreted from the cardiac mesoderm and bone morphogenetic proteins (BMPs) produced by the septum transversum mesenchyme promote hepatic fate in the anterior ventral foregut, while suppressing ventral pancreatic identity (Zaret 2016). In the posterior ventral foregut, extrinsic signaling cues, such as Wnt5a (Rodríguez-Seguel et al. 2013), and cell-intrinsic transcriptional regulators, such as Tgif2 (Cerdá-Esteban et al. 2017), establish ventral pancreatic identity while suppressing hepatic fate. Subsequently, specified organ domains turn on hepatic (Albumin) or pancreatic marker genes (Pdx1). Slit ligands from the overlying mesenchyme bind to Robo2 receptors expressed in the ventral pancreatic endoderm and facilitate the maintenance of pancreatic identity (Escot et al. 2018).
Although pancreatic progenitors arising from the dorsal and ventral foregut are exposed to different signaling environments and receive different instructive cues, they form two organ rudiments, which fuse during later development to form a single organ, and give rise to functionally indistinguishable pancreatic cell types (Spagnoli 2007; Gittes 2009). To date, very little differences in the cellular composition of pancreatic tissues of dorsal or ventral origin have been identified (Spagnoli 2007).
In the posterior ventral foregut, hepatic and pancreatic progenitors develop in close proximity to one another and express a set of common TFs, such as Prox1, Hhex, and members of the FoxA and Gata families (Sosa-Pineda et al. 2000; Bort et al. 2004; Watt et al. 2007; Zorn and Wells 2009; Zaret 2016). Fate mapping and lineage tracing experiments in mouse and fish, respectively, have supported the notion that both hepatic and pancreatic progenitors arise from a common bipotent progenitor domain in the ventral foregut and gradually segregate into lineage restricted progenitor populations as development progresses (Deutsch et al. 2001; Chung et al. 2008; Angelo et al. 2012). Yet, defining the precise nature of this population as well as the temporal dynamics of the segregation require more comprehensive in vivo analyses.
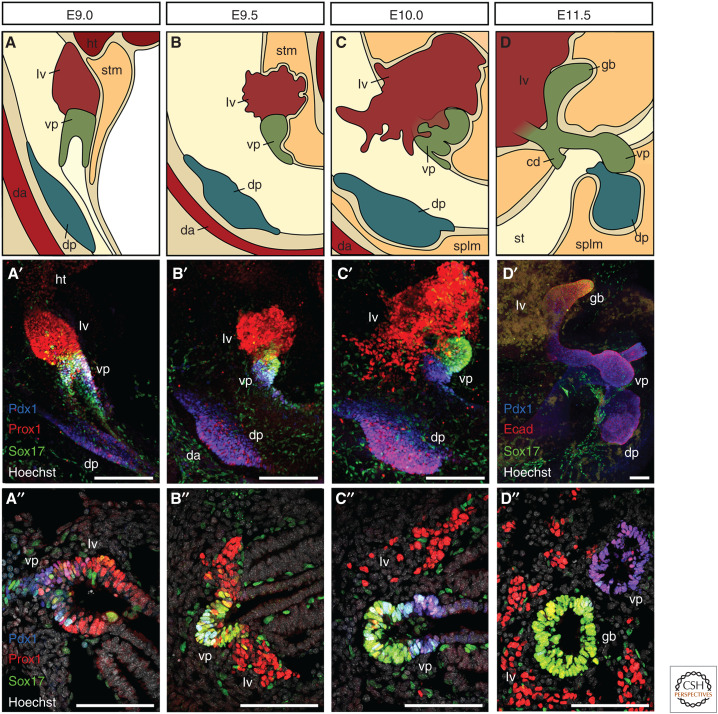
The first identifiable ventral pancreatic progenitors in the mouse emerge at the posterior end of the ventral foregut, a region called the foregut lip, while hepatic progenitors become specified more anteriorly in close proximity of the developing heart and septum transversum mesenchyme (Deutsch et al. 2001; Angelo et al. 2012). Within the next 24 hours of development, dorsal and ventral hepatopancreatic organ domains form morphologically distinct organ buds and grow into the surrounding mesenchymal tissue (Figs. 1 and 2; Spagnoli 2007). Between E9.5 and E10.5, the ventral pancreatic organ domain undergoes further morphological changes and separates into two distinct buds, which subsequently give rise to pancreatic and biliary tissues, respectively (Figs. 1 and 2; Spence et al. 2009; Uemura et al. 2010).
Figure 2.
Organogenesis of the liver and pancreas. (A–D) Schematic representation of the developing hepatopancreatic organ systems in the mouse embryos at the indicated stages. The liver grows dramatically during this developmental time window, forms characteristic hepatic chords, and invades the surrounding septum transversum mesenchyme. Dorsal and ventral pancreatic organ domains form epithelial buds and expand into the surrounding mesenchyme. The ventral pancreatic organ domain (A–C) eventually gives rise to gallbladder, common duct, and ventral pancreas (D). (A′–D′, A″–D″) Representative immunofluorescence stainings of the hepatopancreatic organ system at E9.0 (A′, A″), E9.5 (B′, B″), E10.0 (C′, C″), or E11.5 (D′, D″) either in wholemount (A′–D′) or on cryosections (A″–D″). Prox1 (red) marks all hepatopancreatic tissues, Pdx1 (blue) marks ventral and dorsal pancreas, and Sox17 (green) marks ventral pancreas, gallbladder, and endothelial cells. Staining for Ecad (red, D′) defines all epithelial tissues. (cd) common duct, (da) dorsal aorta, (dp) dorsal pancreas, (gb) gallbladder, (ht) heart, (lv) liver, (splm) splanchnic mesoderm, (stm) septum transversum mesenchyme, (vp) ventral pancreas. Scale bars, 100 µm.
Studies over the last two decades have begun to uncover the mechanisms by which a putative homogenous progenitor population within the ventral foregut endoderm segregates into distinct hepatic and pancreatic organ domains and how cells acquire lineage-specific fates. Experiments done using explant cultures of E8.0–8.5 mouse endoderm identified fibroblast growth factor (FGF) signaling molecules released from the cardiac mesoderm and bone morphogenetic proteins (BMPs) from the septum transversum mesenchyme as necessary for hepatic fate specification (Deutsch et al. 2001; Rossi et al. 2001; Wandzioch and Zaret 2009). In the absence of these prohepatic signals, ventral foregut cells appeared to undertake a pancreatic differentiation program, referred to as the “default fate” of this foregut region (Deutsch et al. 2001; Zaret 2016).
Studies in different vertebrate models highlighted a role for Hedgehog signaling in restricting pancreatic progenitor population size and location, but interestingly this pathway is not modulated in the ventral foregut and does not seem to antagonize ventral pancreatic fate there (Hebrok 2003). FGF10 signaling has been well characterized for its roles in growth and differentiation of pancreatic progenitors in the mouse (Bhushan et al. 2001) and was also reported for being essential in lineage segregation between hepatic, pancreatic, and biliary tissues in zebrafish (Dong et al. 2007). Notch signaling has also been involved in this fate decision; specifically, its downstream effector Hes-1 is required for gallbladder formation and Hes1 genetic ablation promotes the conversion of biliary tissue into pancreatic fate in the mouse embryo (Sumazaki et al. 2004; Fukuda et al. 2006).
Recent RNA-seq analysis of hepatic and pancreatic progenitors isolated from mouse embryos at the time of their lineage divergence has provided further insights into intrinsic and extrinsic factors regulating the fate decision process (Rodríguez-Seguel et al. 2013). Many unique progenitor receptor–ligand signatures have been unveiled from these data sets, including noncanonical Wnts and Slit/Robo signaling (Rodríguez-Seguel et al. 2013; Escot et al. 2018). Specifically, noncanonical Wnt ligands and receptors have been found enriched in foregut and pancreatic progenitors, while absent in hepatoblasts, and to control the lineage segregation by favoring the acquisition of pancreatic fate within the ventral foregut (Rodríguez-Seguel et al. 2013). More recently, the Slit/Robo guidance pathway has been shown to establish a propancreatic niche to preserve pancreatic identity in cells of the mouse ventral foregut (Escot et al. 2018). Consistently, in the absence of the Robo1/2 receptors, progenitor cells with hepatic features were found within the ventral pancreas (Escot et al. 2018).
Overall, the experimental manipulation of the developmental signals identified so far results in a shift in the balance between hepatic and pancreatic progenitor domains, underlining the important cellular plasticity in this embryonic territory. Further studies are needed to better define the source of these signaling factors and whether the surrounding mesenchyme adjacent to the pancreatic endoderm is the same as the one next to the hepatic endoderm or whether a separate mesenchyme migrates to surround and guide pancreatic differentiation.
Besides the fine tuning of multiple signaling pathways, mouse mutant studies indicated the relevance of a subset of transcriptional regulators in ventral foregut cells (Zorn and Wells 2009). For instance, genetic ablation of Hhex, Onecut1 (also known as Hnf6), or Hnf1β in the mouse embryo resulted in concurrent phenotypes affecting ventral foregut derivatives, including malformations or developmental delay in liver, biliary, and ventral pancreas organogenesis (Clotman et al. 2002; Bort et al. 2004; Haumaitre et al. 2005; Hunter et al. 2007).
Recent work on the TF Sox9 has established a first connection between the intrinsic transcriptional program and extrinsic signaling molecules. Specifically, Seymour et al. showed that the deletion of Sox9 in pancreatic progenitors leads to the loss of pancreatic cell identity and gain of the hepatic one (Seymour et al. 2012). This is because of the down-regulation of Fgfr2b expression, which in turn is required for transducing FGF10 mesenchymal signals (Seymour et al. 2012). Likewise, genetic ablation of the TALE-homeodomain transcriptional regulator TGIF2 in the mouse endoderm resulted in ventral pancreas hypoplasia and a concurrent increase in liver bud size (Cerdá-Esteban et al. 2017). TGIF2 acts as a transcriptional corepressor for TGFβ/BMP-activated Smads and is specifically expressed in ventral pancreatic progenitors at early developmental stages, possibly defining a cell-autonomous mechanism to locally repress prohepatic BMP signaling (Cerdá-Esteban et al. 2017). Additionally, genetic ablation of Sox17 in the mouse foregut endoderm resulted in gallbladder agenesis and the emergence of ectopic Pdx1-positive pancreatic cells in the liver bud and common duct (Spence et al. 2009; Uemura et al. 2010).
Although the precise hierarchy of signaling events leading to the separation of the ventral foregut endoderm into hepatic, biliary, and pancreatic tissues is not yet established, the aforementioned studies underscore the intricate relationship between cells of the ventral foregut. Such cellular plasticity within the ventral foregut has also been highlighted by recent single-cell (sc)-RNA-seq analyses, pinpointing to intermediate progenitor states with the potential to give rise to the pancreas as well as to the hepatobiliary tract (Li et al. 2018). Future genetic lineage tracing experiments combined to further single-cell transcriptomics are required to establish the exact lineage relationship existing between the two cell types and the extent of plasticity.
Importantly, human genetic syndromes, such as the Martínez-Frías syndrome (Martínez-Frías et al. 1992), Mitchell–Riley syndrome (Mitchell et al. 2004), as well as a number of congenital human defects featuring multiorgan phenotypes (Ashraf et al. 2005; Galán-Gómez et al. 2007; Chappell et al. 2008), suggest an intertwined development of ventral foregut derivatives in humans, like in the mouse. Most of these human genetic disorders are caused by autosomal recessive genetic defects of unknown etiology and usually present a combination of gallbladder agenesis, intestinal atresia, duodenal malrotation, and pancreatic hypoplasia. In some cases, including the Mitchell–Riley syndrome, patients also suffer from neonatal diabetes. It is possible that gene defects affecting proper lineage segregation of common ventral foregut progenitors toward hepatic, pancreatic, and biliary fates underlie these congenital malformations. Conservation of these developmental processes in the human embryo is also supported by recent transcriptome analyses, which revealed many gene expression signatures in common between human and mouse pancreatic and hepatic progenitors (Jennings et al. 2017; Petersen et al. 2018; Ramond et al. 2018).
In sum, the mechanistic and temporal events underlying hepatic and pancreatic fate acquisition within the ventral foregut remain incompletely understood. A comprehensive molecular and cellular characterization of the bipotent potential of ventral foregut cells in vivo at the single-cell level is still missing as well as the identity of its surrounding microenvironment is elusive.
REGENERATIVE THERAPEUTIC APPROACHES TO TREAT DIABETES
Diabetes is a chronic, metabolic disease that affects over 420 million people worldwide (ADA 2014). Type 2 diabetes (T2D) is the most common form of the disease, it develops during adulthood and is characterized by β cell dysfunction, insulin resistance, and metabolic stress, which eventually lead to a β cell mass reduction (Ashcroft and Rorsman 2012; Weir and Bonner-Weir 2013). Type 1 diabetes (T1D) is an autoimmune disorder characterized by the specific immune destruction of pancreatic β cells, which leads to insufficient production of insulin and hyperglycemia (Weir and Bonner-Weir 2013; Pociot and Lernmark 2016). Loss (T1D) or dysfunction (T2D) of insulin-producing cells result in impaired control of glycemia, which leads to long-term complications, such as retinopathies, kidney failure, and heart attacks, that ultimately reduce life expectancy (Weir and Bonner-Weir 2013).
Despite the availability of insulin as a treatment to temporarily restore the glucostasis in diabetic patients, this remedy is unable to avoid either the acute dangers of hypoglycemia or the long-term complications of hyperglycemia. Current efforts in regenerative medicine aim at restoring glycemic control in diabetic patients either by replacing β cells or by preserving or enhancing the remnant endogenous β cell mass (Fig. 3; Ellis et al. 2017; Sneddon et al. 2018). So far, among these different approaches, the replacement of β cells by whole-organ or pancreatic islets transplantation have been successful in achieving glycemic control in diabetic patients (Cogger and Nostro 2015; Shapiro et al. 2017). Islet transplantation when combined with proper immunomodulatory regimens leads to insulin independence for 5 years in at least 25% of patients in certain specialized centers (Cogger and Nostro 2015; Shapiro et al. 2017; Sneddon et al. 2018). Thus, the proof-of-concept for cell replacement therapy in T1D has been laid down. However, this approach is extremely limited by the scarcity of cadaveric donor tissue and the high risks linked to immunosuppressive regimens. Establishing a renewable source of insulin-producing cells that can be used in transplantation would eliminate the reliance on cadavers and permit the broad application of these therapies. Moreover, if the cellular source could be autologous in origin (from the same patient), this would help to address the adverse consequences of the immune response normally triggered by the transplanted tissue (Sneddon et al. 2018).
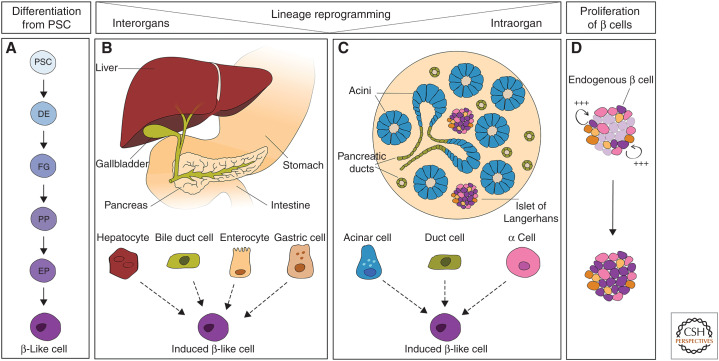
Figure 3.
Cell-replacement strategies for treating diabetes. Schematic representation of different approaches to generate de novo β cells to restore the glycemic function in the context of diabetes. A β-like cell state can be induced through (A) differentiation from pluripotent stem cells, (B) direct lineage reprogramming of endoderm-derived cell types (liver, gallbladder, intestine, and stomach), or (C) other pancreatic cell types (acinar cells, duct cells, endocrine α cells). Alternatively, the proliferative capacity of the endogenous β cells could be reawakened to replenish the pool of insulin-secreting cells. (PSC) pluripotent stem cells, (DE) definitive endoderm, (FG) foregut, (PP) pancreatic progenitor, (EP) endocrine progenitor.
Efforts into establishing an insulin-producing cell supply from human pluripotent stem cells or different somatic cell types have seen promising advances over the past decade. Also, a first clinical trial has been launched by the company Viacyte based on human ESC-derived pancreatic endoderm cells (#NCT03163511). Moreover, significant efforts are directed toward defining strategies on how to protect transplanted cells from the body's immune response either by improving immune modulation or by encapsulation devices. Excellent reviews have been written on the current state-of-the-art in β cell replacement relying on stem-cell-derived cells (Cogger and Nostro 2015; Johnson 2016; Ellis et al. 2017; Sneddon et al. 2018); here, we review instead efforts aimed at generating β-like cells through direct lineage reprogramming, especially from liver cells.
Reprogramming of Somatic Cells to a Pancreatic β Cell State
Lineage reprogramming potentially offers a cure for diabetes (Cohen and Melton 2011; Heinrich et al. 2015). The discovery of a high degree of cellular plasticity in the adult pancreas has indeed reinvigorated this field, and several studies have pointed to pancreas-resident cells as potential sources for new β cells (Fig. 3; Cavelti-Weder et al. 2015; Heinrich et al. 2015). Seminal work from Melton and colleagues provided the first evidence that pancreatic acinar cells can be directed to acquire a β cell phenotype upon in vivo forced expression of the TFs Pdx1, MafA, and Ngn3 (hereafter referred to as PMN), which are key developmental regulators of β cell fate (Zhou et al. 2008). Since then, the same triplet of genetic factors has been applied to convert multiple cell types into β-like fate, including pancreatic ducts (Wang et al. 2018), as well as nonpancreatic cell types, enteroendocrine cells (Chen et al. 2014; Ariyachet et al. 2016), liver cells (Banga et al. 2012), and gallbladder cells (Wang et al. 2016; Galivo et al. 2017). Given the close origin and common endocrine functions (Pan and Wright 2011; Romer and Sussel 2015), fate interconversion between pancreatic islet cells has also been extensively explored by various groups (Bramswig et al. 2013; Courtney et al. 2013; Wilcox et al. 2013; van der Meulen and Huising 2015; Chera and Herrera 2016; Chakravarthy et al. 2017; Furuyama et al. 2019). Nevertheless, the pancreas resides in an anatomically inaccessible location, making both exocrine and islet cells a poor cellular source for ex vivo reprogramming followed by cell-replacement approaches. Thus, an alternative approach with more immediate therapeutic applications might be to use other cellular sources, which are more easily accessible outside of the pancreas (referred here to as interorgans reprogramming) (Fig. 3).
An in vivo reprogramming screen based on a transgenic mouse model, which allows doxycycline-regulated expression of the PMN TFs in all tissues, identified the enteroendocrine cells of the gut as a putative source of insulin-producing cells (Chen et al. 2014). More recently, Zhou and colleagues showed that the enteroendocrine cells resident in the stomach undergo conversion into insulin-positive cells upon PMN overexpression more efficiently than the intestinal ones (Ariyachet et al. 2016). Moreover, the enteroendocrine cells in the intestinal crypts have been shown to respond to Foxo1 ablation in vivo in the mouse by inducing ectopic insulin expression, even though they retain intestinal properties (Talchai et al. 2012).
Grompe's group reported that the gallbladder, which is part of the extrahepatic biliary system and shares common development with the ventral pancreatic rudiment, retains a certain potential to reprogram into the pancreatic lineage. Specifically, adenoviral-mediated overexpression of the PMN transgene, along with inhibition of the Hedgehog and BMP pathways, induce endocrine molecular features both in mouse (Wang et al. 2016) and human (Galivo et al. 2017) gallbladder cells. Nevertheless, the obtained cell products do not inactivate completely the gallbladder program and display multihormonal properties.
Reprogramming of Hepatocytes into Pancreatic Cell Types
The similarities between hepatic and pancreatic cells together with the high regenerative ability of the adult liver and its accessibility make the liver a very attractive cellular source of new β cells in the context of interorgan lineage reprogramming (Fig. 3).
To date, most of the liver-to-pancreas reprogramming efforts have relied on forcing the expression of pancreatic TFs in hepatic cells (Table 1). Experiments in the laboratory of Sarah Ferber pioneered the use of an adenoviral-mediated expression of Pdx1 in hepatopancreatic transdifferentiation (Ferber et al. 2000). This work was then expanded by her group and others by combining Pdx1 with other pancreatic TFs (e.g., Ngn3, NeuroD1, Nkx6.1, MafA, Pax6) and/or by supplementing the culture medium with various small molecules and growth factors in mouse and human liver cell cultures (Table 1; Ber et al. 2003; Kojima et al. 2003; Gefen-Halevi et al. 2010; Ham et al. 2013; Nagasaki et al. 2014). Quite commonly these studies showed that adult liver cells transduced with Pdx1-adenovirus in vitro are able to produce insulin to some extent and ameliorate the glycemic status of diabetic mice upon transplantation (Ferber et al. 2000; Ber et al. 2003; Sapir et al. 2005).
Table 1.
List of liver-to-pancreas reprogramming studies
| Species | Cell source | System | Reprogramming strategy | In vivo functional outcome | References |
|---|---|---|---|---|---|
| Mouse | Adult liver cells | In vivo | Pdx1 (Ad) | Amelioration of hyperglycemia | Ferber et al. 2000 |
| Mouse | Adult liver cells | In vivo | Pdx1 (Ad) | Amelioration of hyperglycemia | Ber et al. 2003 |
| Mouse | Adult liver cells | In vivo | NeuroD1 (HDAd), betacellulin | Amelioration of hyperglycemia | Kojima et al. 2003 |
| Mouse | Adult liver cells | In vivo | Pdx1 (OE in Alb-Cre mice) | n.d. | Miyatsuka et al. 2003 |
| Mouse | Adult liver cells | In vivo | Pdx1, Ngn3 (AAV8) | No amelioration of hyperglycemia | Wang et al. 2007 |
| Mouse | Adult liver: hepatocytes and oval cells | In vivo | Ngn3 (HDAd), betacellulin | Transient expression of insulin | Yechoor et al. 2009 |
| Mouse | Hepatic ducts | In vivo | Pdx1, Ngn3, MafA (Ad) | Long-term reversal of hyperglycemia | Banga et al. 2012 |
| Mouse | Adult liver cells | In vivo | Pdx1, NeuroD1, MafA (Ad) | Short-term reversal of hyperglycemia | Nagasaki et al. 2014 |
| Mouse | Adult liver cells | In vivo | Ngn3 (HDAd), betacellulin, Socs1 | Long-term reversal of hyperglycemia | Li et al. 2015 |
| Xenopus and human | Immature liver of transgenic tadpoles (TTR-Xlhbox8) and HepG2 | In vivo and in vitro | Pdx1–VP16 | n.d. | Horb et al. 2003 |
| Mouse | Adult primary hepatocytes and BAML | In vivo and in vitro | Tgif2 (Lv and AAV8) | Amelioration of hyperglycemia | Cerdá-Esteban et al. 2017 |
| Mouse | Hepatic oval cell line | In vitro | Pdx1–VP16, exposure to high glucose | Amelioration of hyperglycemia | Cao et al. 2004 |
| Mouse | Adult primary hepatocytes | In vitro | NeuroD, Ngn3, Pax4 (Ad), HGF, betacellulin, dexamethasone (-) | n.d. | Yatoh et al. 2007 |
| Mouse | Embryonic liver cells | In vitro | Pdx1, Ngn3, MafA (Ad) | n.d. | Yang et al. 2013 |
| Rat | Liver stem cells (oval) | In vitro | High glucose | Amelioration of hyperglycemia | Yang et al. 2002 |
| Rat | Liver epithelial WB cells (stem-cell like) | In vitro | Pdx1–VP16, Pax4 (Lv) | Amelioration of hyperglycemia | Tang et al. 2006 |
| Rat | Liver epithelial WB cells (stem-cell like) | In vitro | 5-AZA, TSA, RA, insulin, transferrin, and selenite (ITS) and nicotinamide | Amelioration of hyperglycemia | Liu et al. 2013 |
| Human | Fetal liver cells | In vitro | hTERT, Pdx1 (Lv) | Amelioration of hyperglycemia | Zalzman et al. 2003 |
| Human | Adult and fetal liver cells | In vitro | Pdx1 (Ad), nicotinamide, EGF | Amelioration of hyperglycemia | Sapir et al. 2005 |
| Human | Adult liver cells | In vitro | Pdx1, Nkx6.1 (Ad) | n.d. | Gefen-Halevi et al. 2010 |
| Human | Adult liver cells | In vitro | Pdx1 (Lv) | n.d. | Meivar-Levy et al. 2011 |
| Human | Hepatocellular carcinoma cells (Huh7) | In vitro | Pdx1, Pdx1–VP16, Ngn3 (Lv) | n.d. | Donelan et al. 2015 |
| Pig | Neonatal liver cells | In vitro | Pdx1–VP16, Beta2/NeuroD1, MafA (Ad) | Amelioration of hyperglycemia | Ham et al. 2013 |
List of published strategies used for direct lineage reprogramming of hepatic cells into pancreatic cells.
(Lv) lentivirus, (AAV) adeno-associated virus, (Ad) adenovirus, (HDAd) helper-dependant adenoviral vectors, (OE) overexpression, (n.d.) not determined.
Despite the well-established and fundamental role of Pdx1 in pancreatic development, little is known about its downstream transcriptional cascade and how this TF might work in the liver-to-pancreas conversion. Pdx1 has been shown to promote liver-to-pancreas conversion more efficiently in Xenopus and mouse when modified and fused to a VP16 transcriptional activation domain (Horb et al. 2003; Kaneto et al. 2005). These findings suggest that Pdx1 works together with an activator, in the context of pancreatic progenitors, and perhaps the identification of such a factor might help Pdx1 “reprogramming” activity, for example, in a hepatocyte cellular context. More recent ChIP-Seq analyses from the Dunn's group showed that PDX1 occupies and binds at hepatic gene loci, not only in differentiating human ESCs but also in HepG2 hepatoma cells (Teo et al. 2015). Consistently, Pdx1 overexpression in hepatoma lines suppresses the expression of a subset of endogenous liver genes, such as Albumin (Teo et al. 2015) as well as Hnf1a and Hnf4a (Donelan et al. 2015). Thus, Pdx1 might act as a context-dependent transcriptional repressor and activator within the same cell type, possibly explaining its activity when overexpressed in liver cells.
In lineage reprogramming experiments, Pdx1 is often combined with Ngn3 and MafA (Table 1; Fig. 3). Adenoviral vectors have been used by various groups to induce abundant expression of the PMN transgene in vivo in the liver as well as in vitro in hepatic cultures (Table 1). Slack and colleagues showed that PMN overexpression in vivo in the liver of NOD-SCID mice, rendered diabetic by treatment with streptozotocin, results in the induction of insulin-positive cells; these cells showed a mixed phenotype, displaying some typical features of duct cells and some β cell properties (Banga et al. 2012). Moreover, the induced “insulin-positive duct-like” cells were shown to arise from a Sox9-positive cell population, which are probably cells of the small bile ducts (Banga et al. 2012). The PMN transgene induces a similar partial reprogramming also in fetal liver cultures; but in this context, the hepatoblasts seem more prone to undergo reprogramming than the Sox9-positive cells (Yang et al. 2013). Thus, PMN-mediated reprogramming in the liver context results in an immature and hybrid cell type, yet to be fully characterized.
It should be noted that the aforementioned studies used adenoviral vectors for mediating the expression of pancreatic TFs (Table 1). Actually, when the same TFs (e.g., Pdx1 and Ngn3) were delivered to the liver of diabetic mice using adeno-associated virus (AAV) vectors, which have a high transduction efficiency but lower immunogenic potential compared to adenoviruses, the mice remained hyperglycemic (Wang et al. 2007). This study went further to show that unspecific elements of the adenovirus capsid are required together with the pancreatic TFs for inducing some insulin expression (Wang et al. 2007). It is therefore likely that adenoviral vectors may establish an inflammatory microenvironment that is more conducive to reprogramming, as previously suggested in other contexts (Lee et al. 2012).
Hence, it seems that the most appropriate route for reprogramming hepatocytes into pancreatic β cells has yet to be defined. To define additional reprogramming factors and successful strategies for converting hepatocytes into pancreatic cells, we and others carried out thorough analyses of developmental regulators controlling the fate decision between the two lineages (Rodríguez-Seguel et al. 2013; Escot et al. 2018; Li et al. 2018). From these studies, we found for example that TGIF2 acts as a regulator of the cell fate binary choice between liver and pancreas (Cerdá-Esteban et al. 2017). In the embryo, TGIF2 promotes the establishment of the pancreatic identity at the expenses of the hepatic one (Cerdá-Esteban et al. 2017), being therefore a suitable candidate to induce cell conversion. Consistently, when ectopically expressed in mouse hepatocytes in vivo and in vitro as well as in hepatic cell lines, Tgif2 triggers repression of the hepatic phenotype and induces activation of a pancreatic progenitor program (Cerdá-Esteban et al. 2017). The obtained reprogrammed cells can be further differentiated along the pancreatic cell lineage and produce some insulin upon transplantation in diabetic mouse models (Cerdá-Esteban et al. 2017). This strategy represents therefore a starting point for achieving full conversion into mature β cells upon combination of TGIF2 with additional TFs or modified culture conditions.
Another open question is whether all hepatocytes are equally competent to undergo reprogramming. Alternatively, which cells in the liver undergo reprogramming? Only few of the liver-to-pancreas reprogramming studies performed lineage tracing to answer this question or used genetic tools to specifically target adult hepatocytes (Yechoor et al. 2009; Banga et al. 2012; Cerdá-Esteban et al. 2017). Banga et al. (2012) used lineage tracing to show that the cells in the liver that respond to the PMN transgene in their experimental set-up and acquire some β cell features are Sox9-descendant cells. In vivo studies based on the adenoviral-mediated expression of Ngn3 in the liver suggested that putative progenitor-like cells (also known as oval cells) localized in the periportal region can undergo fate conversion and become insulin-positive cells (Yechoor et al. 2009; Li et al. 2015). In Cerdá-Esteban et al., Tgif2 expression was specifically targeted in vivo to the hepatocytes by the virtue of the AAV serotype and the use of the hepatocyte-specific thyroid-binding globulin promoter (Cerdá-Esteban et al. 2017). Nevertheless, further investigations are required to determine whether a subset of hepatocytes is more prone to undergo reprogramming and whether a more plastic population could be targeted to increase the reprogramming efficiency.
CONCLUDING REMARKS
Direct lineage reprogramming technologies are opening new doors in regenerative medicine, whereby the patient's own tissue might constitute a therapeutic source either by in situ targeting or by in vitro genetic modification and subsequent transplantation. However, translating these findings into clinical applications still presents significant challenges.
First, all findings obtained in the mouse or other models need to be validated and transposed onto human model systems, which often require important modifications of the experimental strategies. Human primary cells of good quality are not always accessible. For instance, even though methods for hepatocyte isolation have improved resulting into higher yields of viable cells, to culture them in vitro and for long-term remains very challenging (Guguen-Guillouzo and Guillouzo 2010). Moreover, high-quality primary hepatocytes are primarily reserved to the clinics for transplantation and less available for research. At present, 3D organoid cultures in combination with human pluripotent stem cells are helping to improve long-term culture and expansion of adult human liver cells and also to develop human liver tissue models (Shinozawa et al. 2016). These models will move forward the ex vivo reprogramming efforts in human cells or organ-like contexts. Moreover, humanized models will serve to perform in vivo lineage reprogramming of human cells but also to assess the functionality of reprogrammed cells before any reprogramming strategy can be tested in patients.
Comprehensive and standardized assays need to be put in place to more rigorously characterize the cells obtained through reprogramming. Also, reprogrammed cells need to be always compared to their in vivo counterparts to assess how similar and interchangeable they are. Specifically, surrogate pancreatic β cells induced by lineage reprogramming should undergo deep phenotyping at the transcriptome, epigenome, and functional levels (e.g., insulin content, secretion, and dynamic response to blood glucose) (Johnson 2016). The gold standard assay commonly used for testing induced β-like cells is their ability to counteract hyperglycemia upon transplantation into diabetic mouse models (Table 1); however, the field is reconsidering the value of a comprehensive in vitro testing of the properties of induced β-like cells. Moreover, it is now well accepted that not all insulin-secreting β cells are equal (Tritschler et al. 2017). For example, phenotyping platforms should become available for single-cell functional analysis of the generated β-like cells, including measuring metabolic parameters and electrophysiological properties of the cells. Importantly, publicly available data sets obtained from deep-phenotyping of human islets should be used for benchmarking the β-like cells obtained in a laboratory.
Full understanding of the basic mechanisms behind lineage reprogramming is required for improving the efficiency and specificity of the process. Most of the studies reviewed here reported incomplete conversion of liver cells into pancreatic cells, generating hybrid states that often express only a subset of pancreatic genes and fail to silence the original hepatic program (Ferber et al. 2000; Ber et al. 2003; Cao et al. 2004; Yatoh et al. 2007; Yechoor et al. 2009; Gefen-Halevi et al. 2010; Yang et al. 2013). The activation or repression of certain genes might be impeded by their chromatin configuration. Thus, including in the “reprogramming factors cocktail” pioneer TFs or epigenetic modifiers that have the ability to trigger remodeling of chromatin domains and recruit specific reprogramming factors to the target sites (Iwafuchi-Doi and Zaret 2014) might help to enhance a full cell identity switch. In line with this, the pioneer factors FoxA1/2 and GATA4 have been used for the reprogramming of fibroblasts into hepatic cells (Huang et al. 2011; Sekiya and Suzuki 2011). Likewise, the standard pancreatic reprogramming cocktail could benefit from the addition of lineage-specific pioneer factors, yet to be identified.
Single-cell technologies are offering unprecedented insight into cell heterogeneity, revealing the behavior of rare cell populations that are typically masked in bulk population analyses (Natarajan et al. 2017; Kester and van Oudenaarden 2018). Therefore, sc-RNA-seq experiments will allow full appreciation of (1) cellular heterogeneity during reprogramming of hepatocytes into pancreatic cells, (2) discovery of cell-state-specific genes, and (3) alternative “off-target” programs emerging during reprogramming. New experimental techniques that combine single-cell transcriptome sequencing with genetic lineage labels or viral barcode approaches have been developed, by which it is possible to reconstruct lineage trajectories during reprogramming and identify barriers or predict elements that could enhance the full conversion (Biddy et al. 2018). Thus, insights from sc-RNA-seq will help to increase efficiency and precision in reprogramming approaches toward the generation of functional pancreatic β cells.
Finally, the next step will be to combine lineage reprogramming with tissue engineering approaches to move from the “one-cell scale” to the “tissue scale.” Often, cells derived from ESC programming or lineage reprogramming need three-dimensional environments and proper cell–cell interactions to complete their maturation. Pancreatic organogenesis relies on the continuous interplay between pancreatic progenitors and the surrounding tissues (Sakhneny et al. 2019; Seymour and Serup 2019). Moreover, each organ is composed of multiple cell types within a well-organized structure, which represents an additional challenge to reproduce. For instance, the pancreatic tissue displays a completely different cellular organization compared to the liver. Thus, the next challenge in lineage reprogramming is to take into account the tissue architecture and reproduce the functional pancreatic tissue or islet units along with their respective organ niche(s). This combination will have transformative impact on regenerative medicine.
ACKNOWLEDGMENTS
Work in the Spagnoli laboratory related to the findings described here has been supported by ERC Proof-of-Concept (TheLiRep 641036), FET Open (Pan3DP 800981), and JDRF (1-INO-2018-634-A).
Footnotes
Editors: Cristina Lo Celso, Kristy Red-Horse, and Fiona M. Watt
Additional Perspectives on Stem Cells: From Biological Principles to Regenerative Medicine available at www.cshperspectives.org
REFERENCES
- Abdel-Misih SRZ, Bloomston M. 2010. Liver anatomy. Surg Clin North Am 90: 643–653. 10.1016/j.suc.2010.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ADA. 2014. Diagnosis and classification of diabetes mellitus. Diabetes Care 37(Suppl 1): S81–S90. [DOI] [PubMed] [Google Scholar]
- Angelo JR, Guerrero-Zayas MI, Tremblay KD. 2012. A fate map of the murine pancreas buds reveals a multipotent ventral foregut organ progenitor. PLoS ONE 7: e40707 10.1371/journal.pone.0040707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyachet C, Tovaglieri A, Xiang G, Lu J, Shah MS, Richmond CA, Verbeke C, Melton DA, Stanger BZ, Mooney D, et al. 2016. Reprogrammed stomach tissue as a renewable source of functional β cells for blood glucose regulation. Cell Stem Cell 18: 410–421. 10.1016/j.stem.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft F, Rorsman P. 2012. Diabetes mellitus and the β cell: the last ten years. Cell 148: 1160–1171. 10.1016/j.cell.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf A, Abdullatif H, Hardin W, Moates JM. 2005. Unusual case of neonatal diabetes mellitus due to congenital pancreas agenesis. Pediatr Diabetes 6: 239–243. 10.1111/j.1399-543X.2005.00114.x [DOI] [PubMed] [Google Scholar]
- Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, et al. 2016. Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol 13: 261–280. 10.1038/nrgastro.2016.51 [DOI] [PubMed] [Google Scholar]
- Banga A, Akinci E, Greder LV, Dutton JR, Slack JM. 2012. In vivo reprogramming of Sox9+ cells in the liver to insulin-secreting ducts. Proc Natl Acad Sci 109: 15336–15341. 10.1073/pnas.1201701109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Moshe S, Itzkovitz S. 2019. Spatial heterogeneity in the mammalian liver. Nat Rev Gastroenterol Hepatol 16: 395–410. 10.1038/s41575-019-0134-x [DOI] [PubMed] [Google Scholar]
- Ber I, Shternhall K, Perl S, Ohanuna Z, Goldberg I, Barshack I, Benvenisti-Zarum L, Meivar-Levy I, Ferber S. 2003. Functional, persistent, and extended liver to pancreas transdifferentiation. J Biol Chem 278: 31950–31957. 10.1074/jbc.M303127200 [DOI] [PubMed] [Google Scholar]
- Bhushan A, Itoh N, Kato S, Thiery JP, Czernichow P, Bellusci S, Scharfmann R. 2001. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development 128: 5109–5117. [DOI] [PubMed] [Google Scholar]
- Biddy BA, Kong W, Kamimoto K, Guo C, Waye SE, Sun T, Morris SA. 2018. Single-cell mapping of lineage and identity in direct reprogramming. Nature 564: 219–224. 10.1038/s41586-018-0744-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bort R, Martinez-Barbera JP, Beddington RSP, Zaret KS. 2004. Hex homeobox gene-dependent tissue positioning is required for organogenesis of the ventral pancreas. Development 131: 797–806. 10.1242/dev.00965 [DOI] [PubMed] [Google Scholar]
- Bramswig NC, Everett LJ, Schug J, Dorrell C, Liu C, Luo Y, Streeter PR, Naji A, Grompe M, Kaestner KH. 2013. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J Clin Invest 123: 1275–1284. 10.1172/JCI66514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao LZ, Tang DQ, Horb ME, Li SW, Yang LJ. 2004. High glucose is necessary for complete maturation of Pdx1-VP16-expressing hepatic cells into functional insulin-producing cells. Diabetes 53: 3168–3178. 10.2337/diabetes.53.12.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavelti-Weder C, Li W, Zumsteg A, Stemann M, Yamada T, Bonner-Weir S, Weir G, Zhou Q. 2015. Direct reprogramming for pancreatic β-cells using key developmental genes. Curr Pathobiol Rep 3: 57–65. 10.1007/s40139-015-0068-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdá-Esteban N, Naumann H, Ruzittu S, Mah N, Pongrac IM, Cozzitorto C, Hommel A, Andrade-Navarro MA, Bonifacio E, Spagnoli FM. 2017. Stepwise reprogramming of liver cells to a pancreas progenitor state by the transcriptional regulator Tgif2. Nat Commun 8: 14127 10.1038/ncomms14127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy H, Gu X, Enge M, Dai X, Wang Y, Damond N, Downie C, Liu K, Wang J, Xing Y, et al. 2017. Converting adult pancreatic islet α cells into β cells by targeting both Dnmt1 and Arx. Cell Metab 25: 622–634. 10.1016/j.cmet.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell L, Gorman S, Campbell F, Ellard S, Rice G, Dobbie A, Crow Y. 2008. A further example of a distinctive autosomal recessive syndrome comprising neonatal diabetes mellitus, intestinal atresias and gall bladder agenesis. Am J Med Genet A 146A: 1713–1717. 10.1002/ajmg.a.32304 [DOI] [PubMed] [Google Scholar]
- Chen YJ, Finkbeiner SR, Weinblatt D, Emmett MJ, Tameire F, Yousefi M, Yang C, Maehr R, Zhou Q, Shemer R, et al. 2014. De novo formation of insulin-producing “neo-β cell islets” from intestinal crypts. Cell Rep 6: 1046–1058. 10.1016/j.celrep.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chera S, Herrera PL. 2016. Regeneration of pancreatic insulin-producing cells by in situ adaptive cell conversion. Curr Opin Genet Dev 40: 1–10. 10.1016/j.gde.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Shin CH, Stainier DYR. 2008. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev Cell 15: 738–748. 10.1016/j.devcel.2008.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotman F, Lannoy VJ, Reber M, Cereghini S, Cassiman D, Jacquemin P, Roskams T, Rousseau GG, Lemaigre FP. 2002. The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development 129: 1819–1828. [DOI] [PubMed] [Google Scholar]
- Cogger K, Nostro MC. 2015. Recent advances in cell replacement therapies for the treatment of type 1 diabetes. Endocrinology 156: 8–15. 10.1210/en.2014-1691 [DOI] [PubMed] [Google Scholar]
- Cohen DE, Melton D. 2011. Turning straw into gold: directing cell fate for regenerative medicine. Nat Rev Genet 12: 243–252. 10.1038/nrg2938 [DOI] [PubMed] [Google Scholar]
- Courtney M, Gjernes E, Druelle N, Ravaud C, Vieira A, Ben-Othman N, Pfeifer A, Avolio F, Leuckx G, Lacas-Gervais S, et al. 2013. The inactivation of Arx in pancreatic α-cells triggers their neogenesis and conversion into functional β-like cells. PLoS Genet 9: e1003934 10.1371/journal.pgen.1003934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch G, Jung J, Zheng M, Lóra J, Zaret KS. 2001. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development 128: 871–881. [DOI] [PubMed] [Google Scholar]
- Donelan W, Li S, Wang H, Lu S, Xie C, Tang D, Chang LJ, Yang LJ. 2015. Pancreatic and duodenal homeobox gene 1 (Pdx1) down-regulates hepatic transcription factor 1α (HNF1α) expression during reprogramming of human hepatic cells into insulin-producing cells. Am J Transl Res 7: 995–1008. [PMC free article] [PubMed] [Google Scholar]
- Dong PDS, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, Neumann CJ, Stainier DYR. 2007. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet 39: 397–402. 10.1038/ng1961 [DOI] [PubMed] [Google Scholar]
- Duncan SA. 2003. Mechanisms controlling early development of the liver. Mech Dev 120: 19–33. 10.1016/S0925-4773(02)00328-3 [DOI] [PubMed] [Google Scholar]
- Ellis C, Ramzy A, Kieffer TJ. 2017. Regenerative medicine and cell-based approaches to restore pancreatic function. Nat Rev Gastroenterol Hepatol 14: 612–628. 10.1038/nrgastro.2017.93 [DOI] [PubMed] [Google Scholar]
- Escot S, Willnow D, Naumann H, Di Francescantonio S, Spagnoli FM. 2018. Robo signalling controls pancreatic progenitor identity by regulating Tead transcription factors. Nat Commun 9: 5082 10.1038/s41467-018-07474-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber S, Halkin A, Cohen H, Ber I, Einav Y, Goldberg I, Barshack I, Seijffers R, Kopolovic J, Kaiser N, et al. 2000. Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat Med 6: 568–572. 10.1038/75050 [DOI] [PubMed] [Google Scholar]
- Fukuda A, Kawaguchi Y, Furuyama K, Kodama S, Horiguchi M, Kuhara T, Koizumi M, Boyer DF, Fujimoto K, Doi R, et al. 2006. Ectopic pancreas formation in Hes1-knockout mice reveals plasticity of endodermal progenitors of the gut, bile duct, and pancreas. J Clin Invest 116: 1484–1493. 10.1172/JCI27704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama K, Chera S, van Gurp L, Oropeza D, Ghila L, Damond N, Vethe H, Paulo JA, Joosten AM, Berney T, et al. 2019. Diabetes relief in mice by glucose-sensing insulin-secreting human α-cells. Nature 567: 43–48. 10.1038/s41586-019-0942-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán-Gómez E, Sánchez EB, Arias-Castro S, Cardesa-García JJ. 2007. Intrauterine growth retardation, duodenal and extrahepatic biliary atresia, hypoplastic pancreas and other intestinal anomalies: further evidence of the Martínez-Frías syndrome. Eur J Med Genet 50: 144–148. 10.1016/j.ejmg.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Galivo F, Benedetti E, Wang Y, Pelz C, Schug J, Kaestner KH, Grompe M. 2017. Reprogramming human gallbladder cells into insulin-producing β-like cells. PLoS ONE 12: e0181812 10.1371/journal.pone.0181812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefen-Halevi S, Rachmut IH, Molakandov K, Berneman D, Mor E, Meivar-Levy I, Ferber S. 2010. NKX6.1 promotes PDX-1-induced liver to pancreatic β-cells reprogramming. Cell Reprogram 12: 655–664. 10.1089/cell.2010.0030 [DOI] [PubMed] [Google Scholar]
- Gittes GK. 2009. Developmental biology of the pancreas: a comprehensive review. Dev Biol 326: 4–35. 10.1016/j.ydbio.2008.10.024 [DOI] [PubMed] [Google Scholar]
- Grompe M. 2003. Pancreatic–hepatic switches in vivo. Mech Dev 120: 99–106. 10.1016/S0925-4773(02)00336-2 [DOI] [PubMed] [Google Scholar]
- Guguen-Guillouzo C, Guillouzo A. 2010. General review on in vitro hepatocyte models and their applications. Methods Mol Biol 640: 1–40. 10.1007/978-1-60761-688-7_1 [DOI] [PubMed] [Google Scholar]
- Gurdon J. 2006. From nuclear transfer to nuclear reprogramming: the reversal of cell differentiation. Annu Rev Cell Dev Biol 22: 1–22. 10.1146/annurev.cellbio.22.090805.140144 [DOI] [PubMed] [Google Scholar]
- Ham DS, Shin J, Kim JW, Park HS, Cho JH, Yoon KH. 2013. Generation of functional insulin-producing cells from neonatal porcine liver-derived cells by PDX1/VP16, BETA2/NeuroD and MafA. PLoS ONE 8: e79076 10.1371/journal.pone.0079076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haumaitre C, Barbacci E, Jenny M, Ott MO, Gradwohl G, Cereghini S. 2005. Lack of TCF2/vHNF1 in mice leads to pancreas agenesis. Proc Natl Acad Sci 102: 1490–1495. 10.1073/pnas.0405776102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebrok M. 2003. Hedgehog signaling in pancreas development. Mech Dev 120: 45–57. 10.1016/S0925-4773(02)00331-3 [DOI] [PubMed] [Google Scholar]
- Heinrich C, Spagnoli FM, Berninger B. 2015. In vivo reprogramming for tissue repair. Nat Cell Biol 17: 204–211. 10.1038/ncb3108 [DOI] [PubMed] [Google Scholar]
- Horb ME, Shen CN, Tosh D, Slack JMW. 2003. Experimental conversion of liver to pancreas. Curr Biol 13: 105–115. 10.1016/S0960-9822(02)01434-3 [DOI] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. 2011. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475: 386–389. 10.1038/nature10116 [DOI] [PubMed] [Google Scholar]
- Hunter MP, Wilson CM, Jiang X, Cong R, Vasavada H, Kaestner KH, Bogue CW. 2007. The homeobox gene Hhex is essential for proper hepatoblast differentiation and bile duct morphogenesis. Dev Biol 308: 355–367. 10.1016/j.ydbio.2007.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret K. 2014. Pioneer transcription factors in cell reprogramming. Genes Dev 28: 2679–2692. 10.1101/gad.253443.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings RE, Berry AA, Gerrard DT, Wearne SJ, Strutt J, Withey S, Chhatriwala M, Piper Hanley K, Vallier L, Bobola N, et al. 2017. Laser capture and deep sequencing reveals the transcriptomic programmes regulating the onset of pancreas and liver differentiation in human embryos. Stem Cell Rep 9: 1387–1394. 10.1016/j.stemcr.2017.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JD. 2016. The quest to make fully functional human pancreatic β cells from embryonic stem cells: climbing a mountain in the clouds. Diabetologia 59: 2047–2057. 10.1007/s00125-016-4059-4 [DOI] [PubMed] [Google Scholar]
- Kaneto H, Nakatani Y, Miyatsuka T, Matsuoka T, Matsuhisa M, Hori M, Yamasaki Y. 2005. PDX-1/VP16 fusion protein, together with NeuroD or Ngn3, markedly induces insulin gene transcription and ameliorates glucose tolerance. Diabetes 54: 1009–1022. 10.2337/diabetes.54.4.1009 [DOI] [PubMed] [Google Scholar]
- Kester L, van Oudenaarden A. 2018. Single-cell transcriptomics meets lineage tracing. Cell Stem Cell 23: 166–179. 10.1016/j.stem.2018.04.014 [DOI] [PubMed] [Google Scholar]
- Klatt EC, Kumar V. 2015. The pancreas. In Robbins and Cotran pathologic basis of disease (ed. Kumar V, Abbas AK, Fausto NVR, Stanley L, Cotran RS), 492 pp. Elsevier/Saunders, Philadelphia. [Google Scholar]
- Kojima H, Fujimiya M, Matsumura K, Younan P, Imaeda H, Maeda M, Chan L. 2003. NeuroD-betacellulin gene therapy induces islet neogenesis in the liver and reverses diabetes in mice. Nat Med 9: 596–603. 10.1038/nm867 [DOI] [PubMed] [Google Scholar]
- Krakowski M, Kritzik M, Jones EM, Krahl T, Lee J, Arnush M, Gu D, Sarvetnick N. 1999. Pancreatic expression of keratinocyte growth factor leads to differentiation of islet hepatocytes and proliferation of duct cells. Am J Pathol 154: 683–691. 10.1016/S0002-9440(10)65315-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Pera RR, Yakubov E, Cooke JP. 2012. Activation of innate immunity is required for efficient nuclear reprogramming. Cell 151: 547–558. 10.1016/j.cell.2012.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Buras E, Lee J, Liu R, Liu V, Espiritu C, Ozer K, Thompson B, Nally L, Yuan G, et al. 2015. Gene therapy with neurogenin3, betacellulin and SOCS1 reverses diabetes in NOD mice. Gene Ther 22: 876–882. 10.1038/gt.2015.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Qiu WL, Zhang YW, Xu ZR, Xiao YN, Hou C, Lamaoqiezhong YP, Cheng X, Xu CR. 2018. Single-cell transcriptomic analyses reveal distinct dorsal/ventral pancreatic programs. EMBO Rep 19: e46148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yanmei L, Wang H, Hao H, Han Q, Shen J, Shi J, Li C, Mu Y, Han W. 2013. Direct differentiation of hepatic stem-like WB cells into insulin-producing cells using small molecules. Sci Rep 3: 1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Frías ML, Frías JL, Galán E, Domingo R, Paisán L, Blanco M. 1992. Tracheoesophageal fistula, gastrointestinal abnormalities, hypospadias, and prenatal growth deficiency. Am J Med Genet 44: 352–355. 10.1002/ajmg.1320440316 [DOI] [PubMed] [Google Scholar]
- Meivar-Levy I, Sapir T, Berneman D, Weissbach T, Polak-Charcon S, Ravassard P, Tzakis AG, Mor E, Ricordi C, Ferber S. 2011. Human liver cells expressing albumin and mesenchymal characteristics give rise to insulin-producing cells. J Transplant 2011: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK. 2017. Hepatostat: liver regeneration and normal liver tissue maintenance. Hepatology 65: 1384–1392. 10.1002/hep.28988 [DOI] [PubMed] [Google Scholar]
- Mitchell J, Punthakee Z, Lo B, Bernard C, Chong K, Newman C, Cartier L, Desilets V, Cutz E, Hansen IL, et al. 2004. Neonatal diabetes, with hypoplastic pancreas, intestinal atresia and gall bladder hypoplasia: search for the aetiology of a new autosomal recessive syndrome. Diabetologia 47: 2160–2167. 10.1007/s00125-004-1576-3 [DOI] [PubMed] [Google Scholar]
- Miyatsuka T, Kaneto H, Kajimoto Y, Hirota S, Arakawa Y, Fujitani Y, Umayahara Y, Watada H, Yamasaki Y, Magnuson MA, et al. 2003. Ectopically expressed PDX-1 in liver initiates endocrine and exocrine pancreas differentiation but causes dysmorphogenesis. Biochem Biophys Res Commun 310: 1017–1025. [DOI] [PubMed] [Google Scholar]
- Morris SA. 2016. Direct lineage reprogramming via pioneer factors; a detour through developmental gene regulatory networks. Development 143: 2696–2705. 10.1242/dev.138263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H, Katsumata T, Oishi H, Tai PH, Sekiguchi Y, Koshida R, Jung Y, Kudo T, Takahashi S. 2014. Generation of insulin-producing cells from the mouse liver using β cell-related gene transfer including Mafa and Mafb. PLoS ONE 9: e113022 10.1371/journal.pone.0113022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan KN, Teichmann SA, Kolodziejczyk AA. 2017. Single cell transcriptomics of pluripotent stem cells: reprogramming and differentiation. Curr Opin Genet Dev 46: 66–76. 10.1016/j.gde.2017.06.003 [DOI] [PubMed] [Google Scholar]
- Pan FC, Wright C. 2011. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn 240: 530–565. 10.1002/dvdy.22584 [DOI] [PubMed] [Google Scholar]
- Petersen MBK, Gonçalves CAC, Kim YH, Grapin-Botton A. 2018. Recapitulating and deciphering human pancreas development from human pluripotent stem cells in a dish. Curr Top Dev Biol 129: 143–190. 10.1016/bs.ctdb.2018.02.009 [DOI] [PubMed] [Google Scholar]
- Pociot F, Lernmark A. 2016. Genetic risk factors for type 1 diabetes. Lancet 387: 2331–2339. 10.1016/S0140-6736(16)30582-7 [DOI] [PubMed] [Google Scholar]
- Ramond C, Beydag-Tasöz BS, Azad A, van de Bunt M, Petersen MBK, Beer NL, Glaser N, Berthault C, Gloyn AL, Hansson M, et al. 2018. Understanding human fetal pancreas development using subpopulation sorting, RNA sequencing and single-cell profiling. Development 145: dev165480 10.1242/dev.165480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MS, Bendayan M, Kimbrough RD, Reddy JK. 1986. Characterization of pancreatic-type tissue in the liver of rat induced by polychlorinated biphenyls. J Histochem Cytochem 34: 197–201. 10.1177/34.2.2418098 [DOI] [PubMed] [Google Scholar]
- Rao MS, Dwivedi RS, Yeldandi AV, Subbarao V, Tan X, Usman MI, Thangada S, Nemali MR, Kumar S, Scarpelli DG, et al. 1989. Role of periductal and ductular epithelial cells of the adult rat pancreas in pancreatic hepatocyte lineage. A change in the differentiation commitment. Am J Pathol 134: 1069–1086. [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Seguel E, Mah N, Naumann H, Pongrac IM, Cerdá-Esteban N, Fontaine JF, Wang Y, Chen W, Andrade-Navarro MA, Spagnoli FM. 2013. Mutually exclusive signaling signatures define the hepatic and pancreatic progenitor cell lineage divergence. Genes Dev 27: 1932–1946. 10.1101/gad.220244.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer AI, Sussel L. 2015. Pancreatic islet cell development and regeneration. Curr Opin Endocrinol Diabetes Obes 22: 255–264. 10.1097/MED.0000000000000174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi JM, Dunn NR, Hogan BL, Zaret KS. 2001. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev 15: 1998–2009. 10.1101/gad.904601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakhneny L, Khalifa-Malka L, Landsman L. 2019. Pancreas organogenesis: approaches to elucidate the role of epithelial–mesenchymal interactions. Semin Cell Dev Biol 92: 89–96. 10.1016/j.semcdb.2018.08.012 [DOI] [PubMed] [Google Scholar]
- Sapir T, Shternhall K, Meivar-Levy I, Blumenfeld T, Cohen H, Skutelsky E, Eventov-Friedman S, Barshack I, Goldberg I, Pri-Chen S, et al. 2005. Cell-replacement therapy for diabetes: generating functional insulin-producing tissue from adult human liver cells. Proc Natl Acad Sci 102: 7964–7969. 10.1073/pnas.0405277102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. 2011. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475: 390–393. 10.1038/nature10263 [DOI] [PubMed] [Google Scholar]
- Seymour PA, Serup P. 2019. Mesodermal induction of pancreatic fate commitment. Semin Cell Dev Biol 92: 77–88. 10.1016/j.semcdb.2018.08.008 [DOI] [PubMed] [Google Scholar]
- Seymour PA, Shih HP, Patel NA, Freude KK, Xie R, Lim CJ, Sander M. 2012. A Sox9/Fgf feed-forward loop maintains pancreatic organ identity. Development 139: 3363–3372. 10.1242/dev.078733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmukhappa K, Mourya R, Sabla GE, Degen JL, Bezerra JA. 2005. Hepatic to pancreatic switch defines a role for hemostatic factors in cellular plasticity in mice. Proc Natl Acad Sci 102: 10182–10187. 10.1073/pnas.0501691102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro AMJ, Pokrywczynska M, Ricordi C. 2017. Clinical pancreatic islet transplantation. Nat Rev Endocrinol 13: 268–277. 10.1038/nrendo.2016.178 [DOI] [PubMed] [Google Scholar]
- Shen CN, Horb ME, Slack JM, Tosh D. 2003. Transdifferentiation of pancreas to liver. Mech Dev 120: 107–116. 10.1016/S0925-4773(02)00337-4 [DOI] [PubMed] [Google Scholar]
- Shi Y, Inoue H, Wu JC, Yamanaka S. 2017. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov 16: 115–130. 10.1038/nrd.2016.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Wang A, Sander M. 2013. Pancreas organogenesis: from lineage determination to morphogenesis. Annu Rev Cell Dev Biol 29: 81–105. 10.1146/annurev-cellbio-101512-122405 [DOI] [PubMed] [Google Scholar]
- Shinozawa T, Yoshikawa HY, Takebe T. 2016. Reverse engineering liver buds through self-driven condensation and organization towards medical application. Dev Biol 420: 221–229. 10.1016/j.ydbio.2016.06.036 [DOI] [PubMed] [Google Scholar]
- Si-Tayeb K, Lemaigre F, Duncan S. 2010. Organogenesis and development of the liver. Dev Cell 18: 175–189. 10.1016/j.devcel.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Sneddon JB, Tang Q, Stock P, Bluestone JA, Roy S, Desai T, Hebrok M. 2018. Stem cell therapies for treating diabetes: progress and remaining challenges. Cell Stem Cell 22: 810–823. 10.1016/j.stem.2018.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa-Pineda B, Wigle JT, Oliver G. 2000. Hepatocyte migration during liver development requires Prox1. Nat Genet 25: 254–255. 10.1038/76996 [DOI] [PubMed] [Google Scholar]
- Spagnoli FM. 2007. From endoderm to pancreas: a multistep journey. Cell Mol Life Sci 64: 2378–2390. 10.1007/s00018-007-7184-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Lange AW, Lin SC, Kaestner KH, Lowy AM, Kim I, Whitsett JA, Wells JM. 2009. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell 17: 62–74. 10.1016/j.devcel.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger BZ, Hebrok M. 2013. Control of cell identity in pancreas development and regeneration. Gastroenterology 144: 1170–1179. 10.1053/j.gastro.2013.01.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumazaki R, Shiojiri N, Isoyama S, Masu M, Keino-Masu K, Osawa M, Nakauchi H, Kageyama R, Matsui A. 2004. Conversion of biliary system to pancreatic tissue in Hes1-deficient mice. Nat Genet 36: 83–87. 10.1038/ng1273 [DOI] [PubMed] [Google Scholar]
- Talchai C, Xuan S, Kitamura T, DePinho RA, Accili D. 2012. Generation of functional insulin-producing cells in the gut by Foxo1 ablation. Nat Genet 44: 406–412. 10.1038/ng.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K, Haag D, Wernig M. 2015. Direct somatic lineage conversion. Philos Trans R Soc Lond B Biol Sci 370: 20140368 10.1098/rstb.2014.0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Cao L, Chou W, Shun L, Farag C, Atkinson MA, Li S, Chang L, Yang L. 2006. Role of Pax4 in Pdx1-VP16-mediated liver-to-endocrine pancreas transdifferentiation. Lab Invest; J Tech Methods Pathol 86: 829–841. [DOI] [PubMed] [Google Scholar]
- Teo AKK, Tsuneyoshi N, Hoon S, Tan EK, Stanton LW, Wright CVE, Dunn NR. 2015. PDX1 binds and represses hepatic genes to ensure robust pancreatic commitment in differentiating human embryonic stem cells. Stem Cell Rep 4: 578–590. 10.1016/j.stemcr.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Zaret KS. 2005. Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Dev Biol 280: 87–99. 10.1016/j.ydbio.2005.01.003 [DOI] [PubMed] [Google Scholar]
- Tritschler S, Theis FJ, Lickert H, Böttcher A. 2017. Systematic single-cell analysis provides new insights into heterogeneity and plasticity of the pancreas. Mol Metab 6: 974–990. 10.1016/j.molmet.2017.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, Hara K, Shitara H, Ishii R, Tsunekawa N, Miura Y, Kurohmaru M, Taya C, Yonekawa H, Kanai-Azuma M, et al. 2010. Expression and function of mouse Sox17 gene in the specification of gallbladder/bile-duct progenitors during early foregut morphogenesis. Biochem Biophys Res Commun 391: 357–363. 10.1016/j.bbrc.2009.11.063 [DOI] [PubMed] [Google Scholar]
- van der Meulen T, Huising MO. 2015. Role of transcription factors in the transdifferentiation of pancreatic islet cells. J Mol Endocrinol 54: R103–R117. 10.1530/JME-14-0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandzioch E, Zaret KS. 2009. Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science 324: 1707–1710. 10.1126/science.1174497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AY, Ehrhardt A, Xu H, Kay M. 2007. Adenovirus transduction is required for the correction of diabetes using Pdx-1 or neurogenin-3 in the liver. Mol Ther 15: 255–263. 10.1038/sj.mt.6300032 [DOI] [PubMed] [Google Scholar]
- Wang Y, Galivo F, Pelz C, Haft A, Lee J, Kim SK, Grompe M. 2016. Efficient generation of pancreatic β-like cells from the mouse gallbladder. Stem Cell Res 17: 587–596. 10.1016/j.scr.2016.10.009 [DOI] [PubMed] [Google Scholar]
- Wang Y, Dorrell C, Naugler WE, Heskett M, Spellman P, Li B, Galivo F, Haft A, Wakefield L, Grompe M. 2018. Long-term correction of diabetes in mice by in vivo reprogramming of pancreatic ducts. Mol Ther 26: 1327–1342. 10.1016/j.ymthe.2018.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt AJ, Zhao R, Li J, Duncan SA. 2007. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev Biol 7: 37 10.1186/1471-213X-7-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir GC, Bonner-Weir S. 2013. Islet β cell mass in diabetes and how it relates to function, birth, and death. Ann NY Acad Sci 1281: 92–105. 10.1111/nyas.12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CL, Terry NA, Walp ER, Lee RA, May CL. 2013. Pancreatic α-cell specific deletion of mouse Arx leads to α-cell identity loss. PLoS ONE 8: e66214 10.1371/journal.pone.0066214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Du Y, Deng H. 2015. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell 16: 119–134. 10.1016/j.stem.2015.01.013 [DOI] [PubMed] [Google Scholar]
- Yang L, Li S, Hatch H, Ahrens K, Cornelius JG, Petersen BE, Peck AB. 2002. In vitro trans-differentiation of adult hepatic stem cells into pancreatic endocrine hormone-producing cells. Proc Natl Acad Sci 99: 8078–8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Akinci E, Dutton JR, Banga A, Slack JMW. 2013. Stage specific reprogramming of mouse embryo liver cells to a β cell-like phenotype. Mech Dev 130: 602–612. 10.1016/j.mod.2013.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatoh S, Akashi T, Chan PP, Kaneto H, Sharma A, Bonner-Weir S, Weir GC. 2007. NeuroD and reaggregation induce β-cell specific gene expression in cultured hepatocytes. Diabetes Metab Res Rev 23: 239–249. 10.1002/dmrr.678 [DOI] [PubMed] [Google Scholar]
- Yechoor V, Liu V, Espiritu C, Paul A, Oka K, Kojima H, Chan L. 2009. Neurogenin3 is sufficient for transdetermination of hepatic progenitor cells into neo-islets in vivo but not transdifferentiation of hepatocytes. Dev Cell 16: 358–373. 10.1016/j.devcel.2009.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalzman M, Gupta S, Giri RK, Berkovich I, Sappal BS, Karnieli O, Zern MA, Fleischer N, Efrat S. 2003. Reversal of hyperglycemia in mice by using human expandable insulin-producing cells differentiated from fetal liver progenitor cells. Proc Natl Acad Sci 100: 7253–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS. 2016. From endoderm to liver bud: paradigms of cell type specification and tissue morphogenesis. Curr Top Dev Biol 117: 647–669. 10.1016/bs.ctdb.2015.12.015 [DOI] [PubMed] [Google Scholar]
- Zaret K, Grompe M. 2008. Generation and regeneration of cells of the liver and pancreas. Science 322: 1490–1494. 10.1126/science.1161431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. 2008. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature 455: 627–632. 10.1038/nature07314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. 2009. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol 25: 221–251. 10.1146/annurev.cellbio.042308.113344 [DOI] [PMC free article] [PubMed] [Google Scholar]