In this study, Pérez-Calero et al. set out to investigate the role of UAP56/DDX39B in R-loop removal. Using in vitro and in vivo approaches, the authors demonstrate that UAP56 functions as a DNA–RNA helicase required to eliminate harmful cotranscriptional RNA structures that otherwise would block transcription and replication.
Keywords: double-strand breaks, genome instability, R loops, RNA–DNA helicase, RNA–DNA hybrids, replication fork stalling, UAP56/DDX39B
Abstract
Nonscheduled R loops represent a major source of DNA damage and replication stress. Cells have different ways to prevent R-loop accumulation. One mechanism relies on the conserved THO complex in association with cotranscriptional RNA processing factors including the RNA-dependent ATPase UAP56/DDX39B and histone modifiers such as the SIN3 deacetylase in humans. We investigated the function of UAP56/DDX39B in R-loop removal. We show that UAP56 depletion causes R-loop accumulation, R-loop-mediated genome instability, and replication fork stalling. We demonstrate an RNA–DNA helicase activity in UAP56 and show that its overexpression suppresses R loops and genome instability induced by depleting five different unrelated factors. UAP56/DDX39B localizes to active chromatin and prevents the accumulation of RNA–DNA hybrids over the entire genome. We propose that, in addition to its RNA processing role, UAP56/DDX39B is a key helicase required to eliminate harmful cotranscriptional RNA structures that otherwise would block transcription and replication.
R loops play a central role in many physiological processes like immunoglobulin class-switch recombination, mitochondrial DNA replication, telomere homeostasis or even regulatory roles at some regions of the genome (Ginno et al. 2012; Chan et al. 2014; Wahba et al. 2016), but they are also an important source of genome instability (Aguilera 2002; García-Muse and Aguilera 2019). Cotranscriptional R loops constitute a major barrier for replication fork (RF) progression leading to transcription-replication conflicts that can result in DNA breaks and subsequently to genome instability and chromosome fragility (Hamperl et al. 2017; García-Rubio et al. 2018a). Evidence indicating that R loops are a source of genome instability was first provided in yeast cells lacking specific RNA biogenesis and processing factors such as the THO complex (Huertas and Aguilera 2003), a result supported later on with the identification of other RNA biogenesis factors that also contribute to prevent harmful R-loop accumulation (Paulsen et al. 2009).
Under wild-type conditions, RNA–DNA hybrids would form sporadically all over the genome with a preference for highly expressed genes, high GC skew genes or specific DNA regions among others, but presumably at a low frequency, although data about this parameter are not available yet. Strikingly, THO inactivation in yeast and human cells causes R-loop accumulation and R-loop-dependent genome instability that are accompanied by disturbances in RF progression (Wellinger et al. 2006; Domínguez-Sánchez et al. 2011). Interestingly, the analysis of different histone mutants in yeast demonstrated that R loops per se do not necessarily compromise genome integrity, unless chromatin modifications occur (García-Pichardo et al. 2017). In human cells, it has been previously shown that THOC1 physically and functionally interacts with the histone deacetylase SIN3 corepressor complex to prevent R-loop accumulation (Salas-Armenteros et al. 2017).
Different studies in human cells provided evidence for the involvement of a number of other RNA processing factors in preventing R-loop accumulation, including SRSF1, SETX/Sen1, or AQR (Li and Manley 2005; Paulsen et al. 2009; Skourti-Stathaki et al. 2011), consistent with the model that a proper assembly of the messenger ribonucleoprotein particle (mRNP) prevents the RNA from hybridizing back with the template DNA (Santos-Pereira and Aguilera 2015). However, RNA–DNA hybrids might also be facilitated by changes in the DNA template such as accumulation of an excess of local negative supercoiling (Drolet 2006; Tuduri et al. 2009; El Hage et al. 2010) or the occurrence of single-strand DNA breaks (SSBs) or double-strand DNA breaks (DSBs) (Britton et al. 2014; Ohle et al. 2016), which would release the topological constriction that would limit the capacity of rotation of the DNA strand to hybridize with the nascent RNA during transcription (Aguilera and Gómez-González 2017).
Apart from specific mRNP assembly factors, cells have two other mechanisms to prevent harmful R-loop accumulation. One mechanism based on the action of nucleases such as RNase H, which degrades the RNA moiety of the RNA–DNA hybrid, or a number of RNA helicases such as DDX19 or DXH9 that in vitro have RNA–DNA unwinding activity (Hodroj et al. 2017; Cristini et al. 2018). The third mechanism would rely on DNA replication-associated repair as accredited by the observation that BRCA1, BRCA2, Fanconi anemia factors, or the FACT chromatin reorganizing complex involved in RF progression help prevent R-loop accumulation and R-loop-mediated instability at transcription-replication conflicts (Bhatia et al. 2014; Herrera-Moyano et al. 2014; García-Rubio et al. 2015; Hatchi et al. 2015).
The heteromeric human THO complex associates with a number of RNA-binding proteins including the DEAD-box RNA helicase UAP56/DDX39B (yeast Sub2) or the mRNA export adaptor protein ALY/REF (yeast Yra1) in a supramolecular structure termed TREX, which plays a central role in the packaging and export of nascent RNAs (Sträßer et al. 2002). However, UAP56/Sub2 is not an integral part of THO (Chavez et al. 2000; Peña et al. 2012). In yeast it has been shown that Sub2 seems to play a chaperone role in the process of assembly of the mRNP (Saguez et al. 2013). Mutations of SUB2 lead to hyper-recombination phenotype, whereas Sub2 overexpression partially suppresses the growth-defect and hyper-recombination associated to the hpr1Δ mutants of the THO complex (Jimeno et al. 2002). In human cells, UAP56 depletion leads to a strong genomic instability phenotype (Domínguez-Sánchez et al. 2011).
Altogether, these findings suggest a double role of UAP56/DDX39B in mRNP biogenesis/export and the maintenance of genome integrity, but whether they are unrelated or linked via a putative functional connection with the human THO complex has not been established yet. Here we show that depletion of UAP56 in human cells not only leads to a strong genome instability, but this is specifically dependent on transcription and R loops. After showing that cells depleted of UAP56 significantly increase R loops, we showed that UAP56 is recruited to the majority of the transcribed DNA regions and its depletion causes R-loop accumulation genome-wide. Importantly, we show that UAP56 has a strong in vitro RNA–DNA helicase activity, more efficient than its dsRNA helicase activity, and that overexpression of UAP56 in cells depleted of unrelated factors such as DDX23, SETX, AQR, THOC1, or FANCD2 suppressed their phenotypes of R-loop accumulation and genome instability. Our study shows that UAP56/DDX39B is an essential R-loop-unwinding factor required to prevent and eliminate harmful RNA cotranscriptional structures that otherwise would become a major source of RF blockage and genome instability.
Results
UAP56 prevents R-loop accumulation and R-loop-mediated genome instability
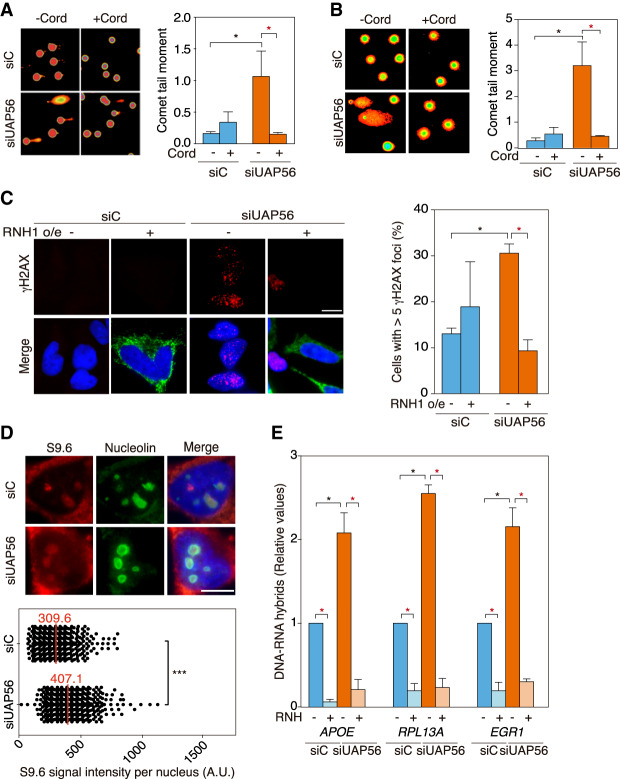
As previously reported (Domínguez-Sánchez et al. 2011), transient depletion of UAP56 in HeLa cells (Supplemental Fig. S1A,B) leads to an accumulation of DNA breaks as detected by single-cell electrophoresis (comet assay) and γH2AX foci monitored by immunofluorescence (IF) in comparison with siC control cells (Fig. 1A–C). To determine whether SSBs and DSBs are transcription-dependent, we used 3′ deoxyadenosine (cordycepin), a specific inhibitor of RNA chain elongation. Cordycepin fully suppressed the increase in DNA breaks as detected by alkaline and neutral comet assay (Fig 1A,B). We next assayed whether this increase was also dependent on RNA–DNA hybrids. For this purpose, we overexpressed RNase H1 and found that γH2AX foci were significantly reduced in siUAP56 cells (Fig. 1C), indicating that DNA damage was R-loop-dependent. Consequently, we explored whether this was due to an increased accumulation of R loops. We first assayed R loops by IF using the S9.6 monoclonal antibody and observed a significant increase in the S9.6 nuclear signal in siUAP56 cells (Fig. 1D). To confirm this result and to discard dsRNA signal that the S9.6 might detect in IF experiments (Hartono et al. 2018; Silva et al. 2018), we analyzed hybrid accumulation by DRIP-qPCR in a set of genes (APOE, RPL13A, and EGR1) that have been previously validated for this purpose (Salas-Armenteros et al. 2017). RNA–DNA hybrids increased in UAP56-depleted cells up to twofold above the siC control levels (Fig. 1E). Consistently, the RNA–DNA hybrid signals were completely removed by in vitro RNase H treatment as a confirmation of the specificity of the assay. Importantly, this increase in RNA–DNA hybrids was not due to an increase in transcription, since a decrease in mRNA levels of those genes, detected by RT-qPCR, was observed (Supplemental Fig. S1C), similar to previously described phenotypes for cells depleted in mRNP assembly factors like THO (Domínguez-Sánchez et al. 2011). Altogether, the data indicate that UAP56 depletion leads to a significant increase of R loops and R-loop-mediated genome instability. Furthermore, accumulation of γH2AX foci and S9.6 signal after UAP56 depletion was rescued by transfection with a plasmid overexpressing UAP56, indicating that the DNA damage and R-loop accumulation phenotypes of UAP56-depleted cells were due to UAP56 silencing and not to off-target effects (Supplemental Fig. S1D,E).
Figure 1.
UAP56 prevents R-loop accumulation and R-loop-mediated genome instability. (A) Representative images and quantification of SSBs and DSBs measured by alkaline single-cell gel electrophoresis (comet assay) in siC (control) and siUAP56 transfected HeLa cells untreated or treated for 4 h with 50 µM cordycepin. More than 100 cells were counted in each experiment, from which a median value was obtained. Data are plotted as mean of the medians + SEM (n = 3). (*) P < 0.05, Student's t-test, two tailed. (B) Representative images and quantification of DSBs measured by neutral comet assay in siC (control) and siUAP56 transfected HeLa cells untreated or treated for 4 h with 50 µM cordycepin. More than 100 cells were counted in each experiment, from which a median value was obtained. Data are plotted as mean of the medians + SEM (n = 3). (*) P < 0.05, Student's t-test, two tailed. (C) Representative images and quantification of γH2AX foci by IF in siC and siUAP56 HeLa cells transfected with pcDNA3 (−RNH1) or pcDNA3-RNaseH1 (+RNH1) for RNase H1 overexpression. Immunostaining with anti-γH2AX antibody (red), anti-RNase H1 antibody (green), and DAPI (blue) are shown. The graph shows the quantification of cells containing more than five γH2AX foci. More than 70 cells per condition were considered in each experiment. Data are plotted as mean + SEM (n = 3). Scale bar, 25 μm. (*) P < 0.05, Student's t-test, two-tailed. (D) Representative images and quantification of nuclear RNA–DNA hybrid accumulation in UAP56-depleted HeLa cells. Immunostaining with S9.6 monoclonal antibody (red), anti-nucleolin antibody (green), and DAPI (blue) in siC and siUAP56 transfected HeLa cells. More than 100 cells per condition were counted in each of the three experiments. The median of the S9.6 signal intensity per nucleus after nucleolar signal removal is shown (n = 3). Scale bar, 10 μm. (***) P < 0.001, Mann-Whitney U-test, two-tailed. (E) DRIP-qPCR using the anti-RNA–DNA hybrids S9.6 monoclonal antibody in siC and siUAP56 transfected HeLa cells in APOE, RPL13A, and EGR1 genes. Signal values normalized with respect to the siC control are plotted as mean + SEM (n = 3). (*) P < 0.05, paired Student's t-test, two tailed. Data information: Black stars denote significant increases, whereas red stars denote significant decreases.
UAP56 depletion causes R-loop-dependent replication fork stalling
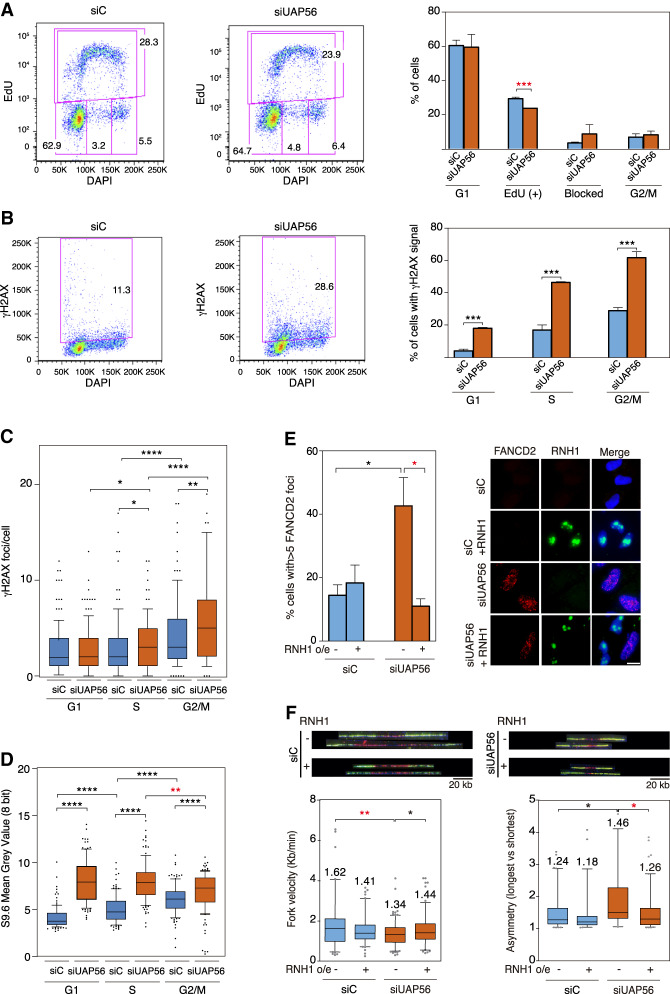
Given the accumulated evidence indicating that RF stalling is a major cause of R-loop-dependent DNA damage (Hamperl et al. 2017; Gómez-González and Aguilera 2019), next we determined the impact of UAP56 depletion on replication in HeLa cells. Analysis of the ability of UAP56-depleted cells to incorporate EdU in combination with γH2AX staining by flow cytometry revealed a decrease in the percentage of EdU-positive cells that is accompanied by an increase in the percentage of cells blocked in S phase, suggesting replication defects (Fig. 2A). However, UAP56 depletion led to a significant increase in the percentage of damaged cells in all phases of the cell cycle, as determined by nuclear γH2AX intensity (Fig. 2B). To specifically determine DSBs we examined the number of γH2AX foci per cell using IF high-throughput analysis after classifying cells according to cell cycle by its DNA content. The validation of this methodology to quantify cells in S phase by EdU incorporation and cells in G2/M phase using H3S10P as a marker are shown in Supplemental Figure S2A,B. γH2AX foci per cell were increased from G1 to S and from S to G2 (Fig. 2C). However, while the increase in γH2AX foci was not observed in G1 in UAP56-depleted cells, it was significant at S and G2/M phases with respect to control cells. This suggests that DSBs could be formed after RF collisions with R loops in UAP56-depleted cells. Consequently, we analyzed RNA–DNA hybrid accumulation by S9.6 IF intensity throughout cell cycle phases after transient depletion of UAP56. As previously reported (Barroso et al. 2019), a significant increase from G1 to S and S to G2 phases was observed in control cells (Fig. 2D). Instead, an increase of S9.6 signal in UAP56-depleted cells versus control cells was observed at all cell cycle stages. Importantly, however, the major increase in hybrids caused by UAP56-depletion occurred in G1 cells, in which it was reached the highest level (Fig. 2D; Supplemental Fig. S2C).
Figure 2.
UAP56 depletion causes R-loop-dependent replication fork stalling. (A) FACS analysis of HeLa cells depleted of UAP56 (asynchronous culture). DNA synthesis and cell cycle were evaluated with EdU and DAPI staining, respectively. Percentage of cells in each phase of the cell cycle is represented. Data are plotted as mean and SD (n = 3). (***) P < 0.005, Student's t-test, two-tailed. (B) Quantification of DNA damage analysis by FACS in siUAP56 HeLa cells. Cells were separated based on their EdU signal and DNA content (DAPI) (gates coming from the top panel). DNA damage was evaluated using an anti-γH2AX antibody. Percentage of damaged cells (γH2AX positive) in each phase of the cell cycle is represented. Data are plotted as mean and SD (n = 3). (***) P < 0.005, Student's t-test, two-tailed. (C) Quantification of γH2AX foci by IF in siC control and UAP56-depleted HeLa cells according to cell cycle. The graph shows the number of γH2AX foci per cell. More than 150 total cells per condition were analyzed. The median values of each population are shown. Boxes and whiskers indicate 10–90 percentiles (n = 3). (*) P < 0.05; (**) P < 0.01; (****) P < 0.000, Mann-Whitney U-test, two-tailed. (D) Quantification of nuclear RNA–DNA hybrid accumulation in UAP56-depleted HeLa cells according to cell cycle. The median of the S9.6 signal intensity per nucleus after nucleolar signal removal is shown. More than 150 total cells per condition were analyzed. Boxes and whiskers indicate 10–90 percentiles (n = 3). (**) P < 0.01; (****) P < 0.0001, Mann-Whitney U-test, two-tailed) (E) Representative images and quantification of FANCD2 IF in siC and siUAP56 HeLa cells with or without RNase H1 overexpression. The graph shows the quantification of the percentage of cells containing more than five FANCD2 foci. More than 130 total cells per condition were considered. Data are plotted as mean + SEM (n = 3). Scale bar, 25 μm. (*) P < 0.05, Student's t-test, two-tailed. (F) Representative pictures of DNA fibers labeled by IdU and CIdU for single DNA molecule analysis. Profiles of RF velocity and asymmetry of siC and siUAP56 cells transfected with the empty vector pcDNA3 or pcDNA3-RNaseH1 (+RNH1) for RNase H1 overexpression are shown. Median values are indicated. Boxes and whiskers indicate 5–95 percentiles. For fork velocity, >90 tracks were considered. For fork asymmetry, from 30 to 100 measurements per condition were considered. (*) P < 0.05; (**) P < 0.01, Mann-Whitney U-test, two-tailed. Data information: Black stars denote significant increases, whereas red stars denote significant decreases.
These results suggest that UAP56 controls hybrid homeostasis all over the cell cycle but has a major impact in G1 cells. Since the major accumulation of DNA damage is, instead, observed in S–G2 cells we wonder whether this was caused by replication fork (RF) stalling on hybrids produced both in G1 and S cells. To assay RF stalling in UAP56-deprived cells, we analyzed the levels of FANCD2 foci by IF, provided that Fanconi anemia factors accumulate at sites of putative RF blockages caused by R loops (García-Rubio et al. 2015). A significant increase of FANCD2 foci was observed in UAP56-depleted cells as compared with the siC control (Fig. 2E). Importantly, overexpression of RNase H1 in these cells drastically reduced this increase of FANCD2 foci, suggesting that R-loop accumulation in UAP56-depleted cells leads to RF blockage that require the action of Fanconi anemia for its processing.
To confirm this, we analyzed RF progression directly by single-molecule DNA combing assays. The experiments revealed a significantly slower RF progression in siUAP56 cells than in the siC control cells (Fig. 2F). This reduction in RF velocity was paralleled by an increase in the frequency of RF stalling as measured by RF asymmetry (Fig. 2F). Importantly, overexpression of RNase H1 restored the wild-type levels of fork velocity and asymmetry in UAP56-depleted cells (Fig 2F). Therefore, our results indicate that silencing of UAP56 promotes slower RF progression and RF stalling that are mediated by R loops, which are the primary cause of the increased DNA damage. We discarded that the recovery caused by RNase H1 overexpression was mediated by changes in the cell cycle, since it does not induce major changes in cell cycle profile of UAP56-depleted cells (Supplemental Fig. S3A).
Finally, even though our results suggest that R loops caused by UAP56-depletion leads to DNA damage by inducing replication fork stalling in S phase, we assayed whether nuclease action on the displaced ssDNA strand could also be an important source of damage, as suggested previously (Sollier et al. 2014; Kim et al. 2019b). For this we assayed the effect of the inactivation of XPG and TOP2 by siXPG depletion and by treatment with the TOP2 inhibitor dexrazoxane in siUAP56-depleted cells (Supplemental Fig. S3B,C). Whereas TOP2 inactivation even increased DNA damage, double XPG and UAP56 depletion caused a slight but nonsignificant decrease when compared with siUAP56 cells. Therefore, neither of those mechanisms seem to detectably contribute to the generation of DNA breaks in UAP56-deficient cells.
UAP56 is an RNA–DNA helicase and R-loop resolvase
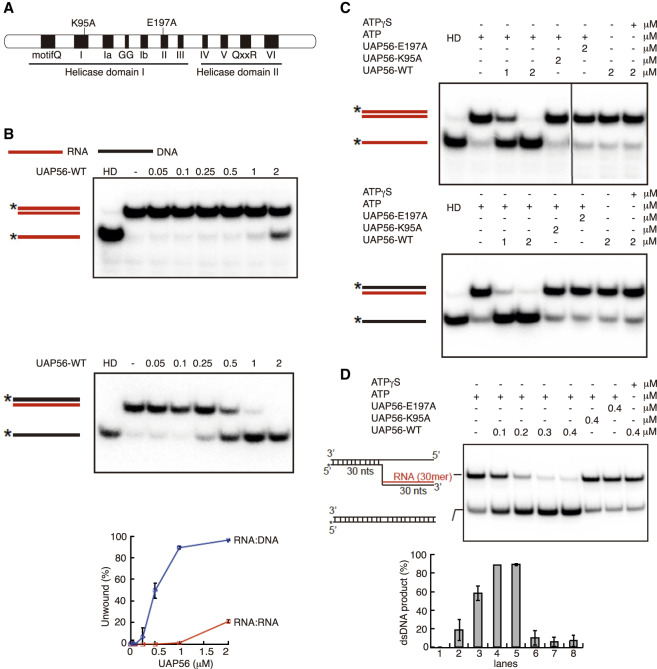
Given that UAP56 is a DEAD-box RNA dependent ATPase (Shen et al. 2007), we wondered whether it possesses RNA–DNA unwinding activity that could explain the strong R-loop accumulation of siUAP56 cells. For this we purified to homogeneity UAP56 and two mutants, UAP56-K95A and -E197A, that have been previously reported to be deficient for the ATPase activity and, therefore, for its helicase activity (Fig. 3A; Supplemental Fig. S4A; Shen et al. 2007). Using a blunt-ended RNA duplex (dsRNA) of 13 base pairs, we observed that UAP56 could unwind this substrate in a protein concentration-dependent manner, consistent with previous results (Fig. 3B; Shen et al. 2007). Similar results were obtained using other dsRNA substrates with either a 5′ or 3′ overhang (Supplemental Fig. S4B). Next, we tested RNA–DNA hybrids with blunt and 5′ or 3′ RNA overhang substrates. UAP56 could unwind all substrates in a protein concentration and ATP-dependent manner (Fig 3B; Supplemental Fig. S4C). In contrast, UAP56 was unable to unwind dsDNA (Supplemental Fig. S4D). Importantly, the percentage of unwound product with the RNA–DNA hybrids was up to fourfold of that obtained for dsRNA (Fig. 3B). Therefore, UAP56 is more adept at unwinding RNA–DNA hybrids than dsRNA. To confirm the molecular identity of this unwinding activity, we proved that neither UAP56-K95A nor UAP56-E197A could unwind dsRNA or RNA–DNA substrates (Fig. 3C; Supplemental Fig. S4B,C). Finally, we tested UAP56 with a 5′ RNA–DNA flap structure that resembles a branch migratable R loop (Schwab et al. 2015). As shown in Figure 3D, UAP56, but not the UAP56-K95A or UAP56-E197A mutant, could dissociate the flap structure to yield a dsDNA product. Therefore, our results indicate that UAP56 resolves RNA–DNA hybrids as well as R-loop-mimicking structures.
Figure 3.
UAP56 is an RNA–DNA helicase and R-loop resolvase. (A) Schematic representation of mutations in the helicase core of UAP56 analyzed in this study. (B) RNA-unwinding assay using same amount of dsRNA or RNA–DNA duplex with a serial dilution of UAP56-WT protein (micromolar). The positions of duplex substrate and unwound products are indicated at the left, where the stars show the position of the radiolabel. Gels were dried and subject to phosphorimaging analysis. Graph shows the percentage of unwound product respect to the UAP56 concentration-dependent manner. (HD) Heat-denatured substrate. (C) RNA-unwinding assay with UAP56-WT, UAP56-E197A, and UAP56-K95A using a dsRNA or a RNA–DNA duplex as a substrate. Other details as in B. (D) RNA–DNA unwinding assay with UAP56-WT, UAP56-E197A, and UAP56-K95A using RNA–DNA flap structures mimicking R loops as substrates. Other details as in B. Graph shows the percentage of dsDNA product recovered after the reaction. Concentrations of UAP56-WT of 0.3–0.4 μM exhibit almost 90% of dsDNA recovery (R-loop resolution).
UAP56 overexpression rescues R-loop and R-loop-mediated genome instability in a helicase-dependent manner
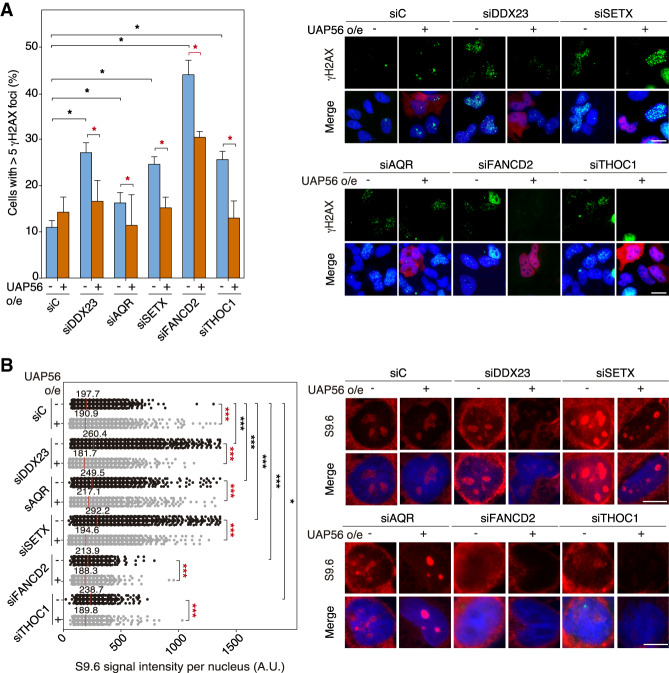
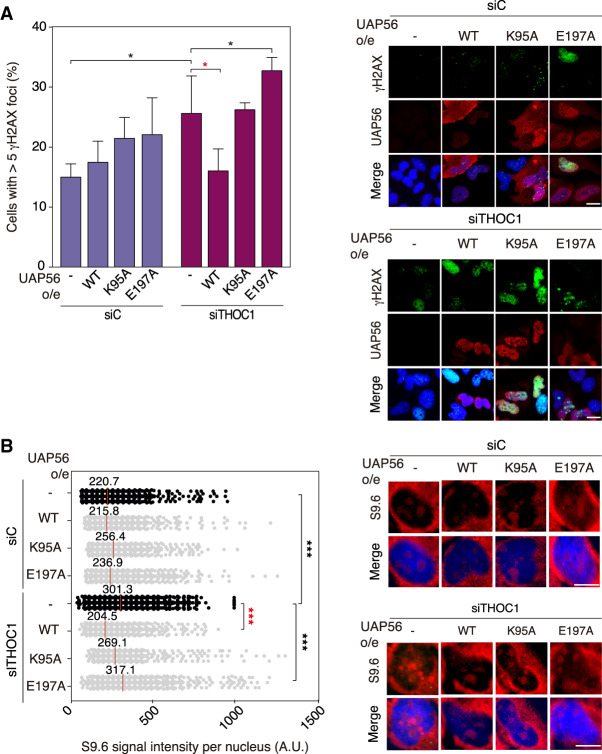
In the last years a number of known RNA helicases like DHX9 or DDX1 have been shown to have RNA–DNA unwinding activity in vitro (Li et al. 2008; Chakraborty and Grosse 2011), but evidence for this activity being relevant in vivo is scarce or absent. According to the way of action of DEAD-box RNA helicases (Yang et al. 2007) it may not be surprising that any RNA helicase so far tested is able to unwind in vitro the RNA strand, regardless of whether paired with an RNA or a DNA strand. For this reason, we assayed whether overexpression of WT and helicase-dead mutants of UAP56 suppressed the R-loop accumulation and R-loop-mediated genome instability of a number of unrelated conditions, such as those created by depletion of three RNA helicases DDX23, SETX, and AQR and two different factors FANCD2 and THOC1, all of which accumulate R loops by different mechanisms (Domínguez-Sánchez et al. 2011; Skourti-Stathaki et al. 2011; Sollier et al. 2014; García-Rubio et al. 2015; Sridhara et al. 2017). First, we confirmed the absence of any effect of overexpressing UAP56 in cell cycle by FACS (Supplemental Fig. S5A). Then, we assayed DNA damage by γH2AX foci and R-loop accumulation via IF. Depletion of all these factors via siRNA led to an increase in γH2AX foci (Fig. 4A) and S9.6 signal (Fig. 4B), as expected. Notably, wild-type UAP56 overexpression rescued not only RNA–DNA hybrid accumulation but also the R-loop-dependent genome instability associated with depletion of such factors (Fig. 4A,B). Next, we overexpressed the two UAP56 helicase-dead mutants UAP56-K95A and UAP56-E197A in siTHOC1-depleted cells to assay whether the in vivo ability to suppress R loops resided in the active sites. The two mutant forms were unable to suppress the increased levels of DNA damage, as detected by γH2AX foci, or the high levels of R-loop accumulation in THOC1-depleted cells (Fig. 5A,B) consistent with the helicase activity being responsible for R-loop control. It is worth noting that overexpression of the UAP56-K95A mutant in UAP56-depleted cells partially reduced the R-loop accumulation (Supplemental Fig. S5B). This could be explained by reasons unrelated with the direct role of UAP56 in R-loop resolution, but linked to the role of UAP56 in prevention of R-loop formation. For example, the helicase dead-protein would stick to the nascent RNA, thus reducing transcription and impeding it to hybridize back with the DNA template. Thus, it may rescue the RNA protection function of UAP56 but not the unwinding activity of the protein. In this sense, it has been previously shown that overexpression of another helicase-dead mutant (URH49), for instance, also rescues other phenotypes caused by siURH49 (Yoo and Chung 2011). Altogether, our results indicate that UAP56 has the in vivo ability to use its RNA–DNA helicase and R-loop-resolving activity to remove R loops regardless of the origin of its accumulation.
Figure 4.
UAP56 overexpression rescues R-loop and R-loop-mediated genome instability. (A) Representative images and detection of γH2AX foci by IF in the indicated HeLa-depleted cells transfected with pFlag (−UAP56) or pFlag-UAP56 (+UAP56) overexpressing UAP56. Immunostaining with anti-γH2AX antibody (green), anti-Flag antibody (red) to detect UAP56 overexpression, and DAPI (blue) are shown. The graph shows the percentage of cells containing more than five γH2AX foci. More than 50 cells per condition were considered in each experiment. Data are plotted as mean + SEM (n ≥ 5). Scale bar, 25 μm. (*) P < 0.05, Student's t-test, two-tailed. (B) Representative images and quantification of S9.6 immunofluorescence signal in the indicated HeLa-depleted cells transfected with the empty vector pFlag (−UAP56) or pFlag-UAP56 (+UAP56) for UAP56 overexpression. Immunostaining shows S9.6 monoclonal antibody (red) and DAPI (blue). More than 100 cells per condition were counted in each experiment. The graph shows the median of the S9.6 signal intensity per nucleus (n = 3). Scale bar, 10 μm. (*) P < 0.05; (***) P < 0.001, Mann-Whitney U-test, two-tailed. Data information: Black stars denote significant increases, whereas red stars denote significant decreases.
Figure 5.
UAP56 overexpression rescues R-loop and R-loop-mediated genome instability in a helicase-dependent manner. (A) Representative images and detection of γH2AX foci by IF in siC and siTHOC1 HeLa cells transfected with pFlag (−UAP56), pFlag-UAP56 (+UAP56) for UAP56 overexpression, and with pFlag-UAP56-K95A (+UAP56-K95A) and pFlag-UAP56-E197A (+UAP56-E197A) for helicase-dead UAP56 overexpression. Other details as in Figure 4A. More than 70 cells per condition were considered in each experiment. Data are plotted as mean + SEM (n = 4). Scale bar, 25 μm. (*) P < 0.05, Student's t-test, two-tailed. (B) Representative images and quantification of S9.6 immunofluorescence signal siC and siTHOC1 HeLa cells transfected with pFlag (−UAP56), pFlag-UAP56 (+UAP56) for UAP56 overexpression, pFlag-UAP56-K95A (+UAP56-K95A) and pFlag-UAP56-E197A (+UAP56-E197A) for helicase-dead UAP56 overexpression. More than 100 cells per condition were counted in each experiment. The graph shows the median of the S9.6 signal intensity per nucleus (n ≥ 3). Other details as in Figure 4B. Scale bar, 10 μm. (***) P < 0.001, Mann-Whitney U-test, two-tailed. Data information: Black stars denote significant increases, whereas red stars denote significant decreases.
Genome-wide chromatin-associated action of UAP56
Since UAP56 interacts with a number of cotranscriptional RNA-binding factors, such as THO and RNA export factors (Sträßer et al. 2002), we assayed whether it also functioned in association with chromatin and chromatin remodelers, as previously shown for THO (Salas-Armenteros et al. 2017). Provided that THO physically interacts with the SIN3 histone deacetylase complex, we first tested whether this was also the case for UAP56. Western analysis showed that UAP56 coimmunoprecipitates (co-IP) with the Sin3A component of SIN3 complex (Supplemental Fig. S6A). We confirmed in situ this colocalization by proximity ligation assay (PLA). Importantly, this physical proximity is detected only in the nucleus, where both proteins are known to function (Supplemental Fig. S6B). Next, we assayed whether these factors work together in R-loop prevention. Interestingly, double depletion of UAP56 and Sin3A conferred a significant increase in R loops, determined by S9.6 IF assays, compared with control cells, which is lower when compared with cells depleted of each factor individually, suggesting an epistatic relationship between UAP56 and SIN3 in R-loop homeostasis (Supplemental Fig. S6C). The result is consistent with the idea that UAP56 as THO, and likely other RNA-binding factors participating in the assembly of the nascent mRNP, form a high-order cotranscriptional structure in the nucleus.
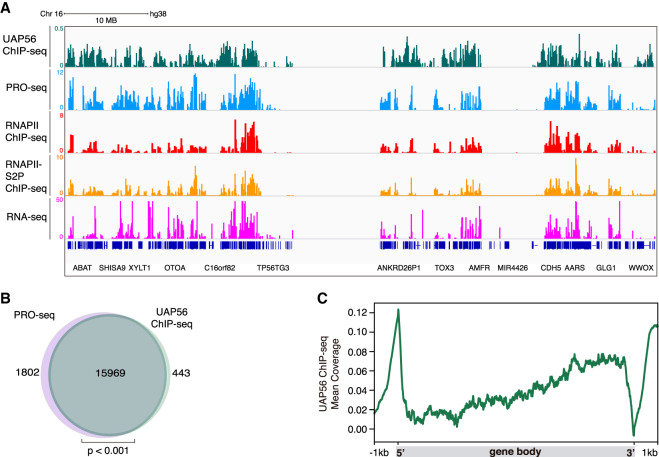
Next, we asked whether UAP56 was present all over the genome, as would be expected by a general cotranscriptional RNA-binding and processing factor. For this, we performed ChIP-seq and RNA-seq analysis in the K562 cell line and obtained reproducible results (Supplemental Fig. S7A,B). This line is regularly used in genome-wide occupancy analysis of different transcription factors, secondary DNA structures, and chromatin modifications, so that there is an increasing set of data to compare with any genome-wide analysis (Sloan et al. 2016). We compared UAP56 ChIP-seq results with transcription-related data available at ENCODE and GEO databases. We crossed data from different data sets such as the precision nuclear run-on sequencing (PRO-seq), which maps active RNAPIIs, RNAPII ChIP-seq, and RNAPII-S2P ChIP-seq, which marks transcription elongation regions and our RNA-seq data from siC control cells. The majority of UAP56 clusters coincided with those from PRO-seq and RNAPII ChIP-seq (Fig. 6A). These data are in concordance with an UAP56 global cotranscriptional function all over the transcribed genome. Indeed, UAP56 could be found associated with chromatin in the majority of RNAPII active sites (97%), according to the overlap between UAP56 ChIP-seq and PRO-seq data (Fig. 6B). Furthermore, to analyze the distribution of UAP56 along the length of all genes, we performed metagenomic analysis along all of these gene bodies and their flanking regions. The binding profile revealed that UAP56 peaks at promoter and termination regions. Inside the gene body, UAP56 is increased in a gradient manner toward the 3′ end of genes (Fig. 6C). The presence of UAP56 at promoters coincided with the mapping of nascent RNAs at these regions corresponding to antisense RNA, as detected by metaplot analysis of PRO-seq data (Supplemental Fig. S7C). These findings support the physiological role of UAP56 in removing cotranscriptional R loops and are consistent with the reported genome-wide occupancy of the yeast ortholog Sub2, where it is recruited in a continuous and increasing manner toward the 3′ end of transcribed genes (Gomez-Gonzalez et al. 2011). Thus, we can conclude that UAP56 is a helicase that is present in active chromatin all over the genome (Fig. 6A,B).
Figure 6.
Genome-wide chromatin-associated action of UAP56. (A) Representative screenshot of a genomic region in which UAP56 ChIP-seq (green), PRO-seq (light blue), RNAPII ChIP-seq (red), RNAPII-S5P ChIP-seq (orange), and RNA-seq (magenta) in K562 cells is shown. (B) Venn diagram showing the overlap between protein-coding genes transcribed in K562 cells detected by PRO-seq (light purple) and UAP56 chromatin-bound protein-coding genes detected by ChIP-seq (green). P < 0.001, hypergeometric test. (C) Metaplot analysis of the distribution of the UAP56 ChIP-seq signal (IP-Input) along a model gene body.
Genome-wide accumulation of RNA–DNA hybrids in UAP56-depleted cells
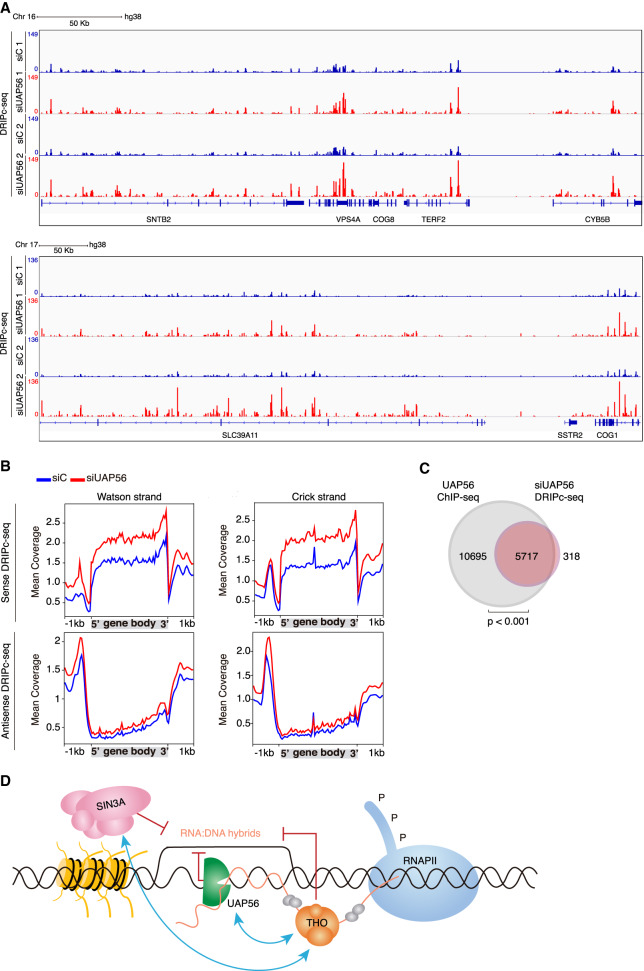
Next, we investigated the global impact of UAP56 depletion on R-loop accumulation by determining the RNA–DNA hybrid distribution along the entire genome in UAP56-depleted cells. We performed DRIPc-seq (DRIP followed by cDNA conversion coupled to high-throughput sequencing) in siC (control) and siUAP56 silenced K562 cells. This technique allows reproducible high-resolution and strand-specific R-loop detection genome-wide (Sanz et al. 2016). First, we confirmed that our S9.6 immunoprecipitation experiments specifically recover RNA–DNA hybrids by performing DRIP-seq in K562 cells with and without RNase H treatment. DRIP-seq signal was highly RNase H-sensitive; that is, strongly reduced in RNase H-treated DRIP-seq (Supplemental Fig. S7D). These results were validated by DRIP-qPCR in different loci and by regression analysis of the genome-wide data (Supplemental Fig. S7E,F). Our DRIPc-seq analysis, a technique exhibiting superior accuracy to that of DRIP-seq, less background and strand specificity, showed also high sensitiveness to RNase H treatment. Regression analysis of DRIP-seq and DRIPc-seq data of untreated and RNase H-treated cells gave comparable results (Supplemental Fig. S7F–H), establishing that the immunoprecipitated material corresponded to RNA–DNA hybrids and the sequences obtained mapped faithfully to DNA–RNA hybrids, regardless of whether the DNA or the RNA strand was used for sequencing. Indeed, both DRIP and DRIPc experiments gave similar profiles (Supplemental Fig. S7D), supporting the reliability and reproducibility of the experiments.
The R-loop peak distribution analysis is shown for a representative genomic region in Figure 7A. In siUAP56 cells, R loops were observed at 73,473 reproducible peaks upon UAP56 depletion. In 31,909 the signal was significantly higher (R-loop gain) and in 42,383 was either similar or slightly lower (R-loop no gain) than that of the siC control (Supplemental Fig. S8A). The R-loop gain peaks included peaks undetected in siC controls (de novo R loops) and peaks with a signal significantly increased over the control (increased R loops) (Supplemental Fig. S8B). Consistently, metaplot analysis of those peaks revealed an increased DRIPc-seq signal in siUAP56 cells when compared with the control, being higher in the case of increased R-loop peaks. Both kinds of R-loop gain peaks covered similar genomic features, detectable mainly at protein-coding gene bodies and ncRNAs (Supplemental Fig. S8C,D). Interestingly, R-loop gain genes were significantly longer than the median of the genome and expressed at higher levels when compared with R-loop no gain genes (Supplemental Fig. S8E,F), consistent with the idea that longer and highly expressed genes are more prone to R-loop accumulation.
Figure 7.
Genome-wide accumulation of RNA–DNA hybrids in UAP56-depleted cells. (A) Representative screenshot of different genomic regions showing the DRIPc-seq signal profiles for siC (blue) and siUAP56 (red) (n = 2). (B) Distribution of antisense and sense DRIPc-seq signal (mean coverage) along a gene metaplot for siC control and siUAP56 cells over Watson (left) and Crick strand (right). (C) Venn diagram showing the overlap between R-loop gain genes (red) and genes to which UAP56 binds in normal K562 cells detected by ChIP-seq (gray). P < 0.001, hypergeometric test. (D) Model to explain the role of UAP56 in R-loop resolution during transcription. In the model, THO contributes to the proper assembly of the mRNP and talks to the Sin3A histone deacetylase complex to presumably transiently close the chromatin. The RNA–RNA and RNA–DNA helicase UAP56/DDX39B, apart from its role in RNA processing, ensures that unscheduled hybrids formed behind the transcription machinery are removed cotranscriptionally, thus potentially releasing the intact nascent RNA molecule for further processing and export.
Gene metaplot analysis of DRIPc signals along the entire genome shows that R loops are found at promoter regions that correspond to antisense RNA and throughout the gene body and transcription termination regions (Fig. 7B). The results are in consonance with previous studies in normal K562 cells (Sanz et al. 2016). Importantly, DRIPc-seq results and conclusions were validated by DRIP-qPCR at representative test loci (Supplemental Fig. S9A) and regression analysis (Supplemental Fig. S9B,C). In addition, we show that the increase in R-loop signal is not caused by higher transcription levels as can be seen globally by RNA-seq and at specific loci (Supplemental Fig. S9D,E).
Finally, the comparative analysis of DRIPc-seq data of siUAP56 cells with ChIP-seq data of the normal K562 cells highlights that the vast majority (94.7%) of genes increasing R loops upon UAP56 depletion coincide with genes where UAP56 is present (Fig. 7C), a conclusion confirmed by the significant enrichment of UAP56 in K562 normal cells at genes that accumulate R loops in UAP56-depleted cells (Supplemental Fig. S9F). Therefore, our results support a global role in R-loop protection caused by UAP56 as well as a global action of this protein during transcription of RNAPII genes.
Discussion
Here we show that UAP56/DDX39B, a conserved RNA-dependent ATPase involved in RNA splicing and export, is an RNA–DNA helicase crucial to prevent the accumulation of unscheduled R loops during transcription, such R loops being an important source of DNA breaks promoted by R-loop-mediated transcription-replication conflicts. Overexpression of UAP56 in different genetic backgrounds that accumulate high R-loop levels suppresses them and the R-loop-mediated genome instability, confirming the relevance of its RNA–DNA unwinding activity in vivo. This activity is able to remove R loops accumulated as a consequence of the inactivation of RNA helicases such as DDX23, SETX, or AQR, the FANCD2 Fanconi anemia repair factor or the mRNP biogenesis factor THOC1, indicating that the ability of UAP56/DDX39B to eliminate hybrids is additional to its role in mRNA processing and export. These conclusions are supported by the fact that helicase-dead mutant proteins are unable to unwind RNA–DNA hybrids in vitro and failed to recover R-loop accumulation and genome instability as efficiently as UAP56-WT does in such genetic background.
The action of UAP56 is extended all over the genome and not restricted to specific regions or to a subset of genes according to function or structure. UAP56 is located in the majority of the transcribed regions of genes from 5′ to 3′ regions (Fig. 6). This is consistent with the expected function of UAP56 in mRNP biogenesis and export, which are intimately linked to transcription. Our proximity ligation assay and co-IP analysis showing colocalization and epistatic interaction between UAP56/DDX39B and the histone deacetylase complex SIN3, the same as previously shown for THOC1 (Salas-Armenteros et al. 2017), together with UAP56 ChIP-seq data suggest a crosstalk of UAP56 with chromatin during transcription. Importantly, the vast majority of these hybrid-prone genes in the absence of UAP56 coincides with those genes in which UAP56 is chromatin-bound in K562 cells, confirming the protective role of UAP56 against the cotranscriptional accumulation of nonscheduled R loops genome-wide. Interestingly, this protective role is more evident at longer and highly transcribed genes, consistent with the idea that these genes are more prone to R-loop accumulation (García-Muse and Aguilera 2019).
This whole body of results lights up a scenario in which during transcription, a specific number of proteins are recruited to active chromatin in the form of a multiprotein structure necessary for processing and exporting the nascent RNA, but at the same time protecting it from forming RNA–DNA hybrids. The latter function would be provided by UAP56/DDX39B, which would act cotranscriptionally unwinding unscheduled RNA–DNA hybrids (Fig. 7D). As we and others have shown, a major mechanism by which hybrids lead to genome instability is by blocking the progression of the replication fork. Consistently, we show in this study that UAP56/DDX39B depletion causes replication fork stalling as determined by DNA combing (Fig. 2F). Interestingly, the increase in R-loop accumulation after UAP56 depletion is observed at all stages of the cell cycle, consistent with a general role of this factor in R-loop prevention, with the largest increase observed in G1 (Fig. 2D). Instead, even though cells with DNA damage after UAP56 depletion accumulate at all stages of the cell cycle (Fig. 2B), DNA breaks are preferentially increased in S and G2 cells as determined by the number of γH2AX foci per cell (Fig. 2C). This, together with the increase in RF stalling as determined by FANCD2 foci and DNA combing and the lack of any reduction in damage produced by inactivation of XPG and TOP2, two nucleases previously suggested to mediate R-loop-mediated damage (Sollier et al. 2014; Kim et al. 2019b), indicated that R loops accumulated upon depletion of UAP56 lead to DNA damage by promoting transcription-replication conflicts.
In recent years different reports have provided evidence for an in vitro RNA–DNA unwinding activity of an increasing number of known RNA helicases. These include Senataxin (Sen1/SETX), the RNA helicases Dbp2/DDX5, DHX9, Dbp5/DDX19, DDX21, DDX23, AQR, Mph1/FANCM, or DDX1 (Sollier et al. 2014; Hatchi et al. 2015; Schwab et al. 2015; Li et al. 2016; Hodroj et al. 2017; Song et al. 2017; Sridhara et al. 2017; Cristini et al. 2018; Mersaoui et al. 2019). IF studies have revealed that silencing of the corresponding genes causes an increase in S9.6 signal. However, a comparative in vitro analysis of RNA–RNA versus RNA–DNA unwinding or a demonstration of the in vivo RNA–DNA unwinding activity is lacking. Because RNA helicases of the DEAD-box family could act efficiently on short RNA duplexes or even RNA–DNA heteroduplexes (Linder and Jankowsky 2011) and considering that these proteins generally work during transcription in association with other RNA-binding proteins and processing factors, it is highly difficult to separate its RNA–DNA unwinding activity from its potential function as RNA chaperons. These considerations would imply that different DDX proteins may not have redundant RNA–DNA unwinding abilities. Thus, SETX has been shown to localize and resolve R loops at termination sites or double-strand breaks (Hatchi et al. 2015; Cohen et al. 2018) and to be expressed in S–G2 phase of the cell cycle in yeast (Mischo et al. 2018). Alternatively, there are other RNA helicases such as DDX21 or DDX23 that seem to be recruited to RNAPII stalled at R loops to promote transcription elongation (Song et al. 2017; Sridhara et al. 2017), while DDX19 is proposed to act on R loops formed upon replication stress and DNA damage (Hodroj et al. 2017). It would certainly be possible that each DDX protein specifically works on a subset of R loops whether located in different DNA regions or nuclear structures (nucleolus, nuclear pore, away from pores) or whether formed by failures of different nuclear processes (transcription, splicing, or export), among other possibilities. Since these proteins could act at a particular phase of the cell cycle or could act preferentially at particular locations or RNA sequences or could interact differentially with other mRNP components, we need further evidence to consider them master cotranscriptional RNA–DNA unwinding factors.
However, our results clearly show that UAP56/DDX39B is able to efficiently remove R loops accumulated under different conditions in vivo. Given its genome-wide distribution all over the vast majority of transcribed DNA sequences in which UAP56 is found associated with chromatin, as detected by ChIP-seq, UAP56/DDX39B emerges as the major RNA–DNA helicase removing unscheduled R loops formed during transcription. This has a key physiological relevance, because RNA–DNA hybrids need to be cotranscriptionally removed not only to avoid RF blockage and replication stress, but also to prevent transcription elongation impairment and premature transcription termination. It is unlikely that cells rely on RNases H to eliminate occasional cotranscriptional hybrids, because this would result in nascent RNA degradation, highly costly to cells. Instead, a master transcription-coupled RNA–RNA and DNA–RNA helicase such as UAP56/DDX39B would release the nascent RNA from the DNA giving it a second chance to be properly coated into a full export-competent RNA-protein particle. We thus propose that in addition to its role as an RNA processing and mRNP biogenesis factor, UAP56/DDX39b represents an essential transcription-associated R-loop-unwinding factor that removes occasional hybrids that otherwise would block transcription and replication, providing new insights into our understanding on how cells prevent and eliminate harmful RNA cotranscriptional structures.
Materials and methods
siRNA transfection
Transient transfection of siRNA (50 nM) was performed using DharmaFECT 1 (Dharmacon) according to the manufacturer's instructions. All assays were performed 72 h after siRNA transfection. Different siRNAs knockdown efficiency experiments are depicted in Supplemental Figure S10.
Genome instability and replication assays
Immunofluorescence (IF), comet assay and DNA combing were essentially performed as described previously (Domínguez-Sánchez et al. 2011; Barroso et al. 2019). For further details on IF, see the Supplemental Material.
DNA:RNA immunoprecipitation (DRIP) assays
DRIP assays were performed by immunoprecipitating DNA–RNA hybrids using the S9.6 antibody from gently extracted and enzymatically digested DNA with the following cocktail of restriction enzymes: HindIII, EcoRI, XbaI, SspI, and BsrGI, treated or not with RNase H (New England Biolabs) in vitro as described (García-Rubio et al. 2018b). The relative abundance of DNA–RNA hybrid immunoprecipitated in each region was normalized to the input values and the siC control when needed (Salas-Armenteros et al. 2017). qPCR primers are listed in the Supplemental Material. DRIP assays were performed 72 h after siRNA transfection. All experiments were performed in triplicate unless otherwise indicated; mean and SEM of results are provided.
Protein purification and nucleic acid unwinding assays
UAP56-WT, UAP56-K95A, and UAP56-E197A were purified using the same procedure as described (Shen et al. 2007), as well as the RNA–RNA duplexes use for the helicase assays. RNA–DNA hybrids without and with a 5′ or 3′ overhang and DNA–DNA duplex with a 5′ overhang were prepared by annealing oligonucleotides (with one of the oligonucleotides being labeled with 32P). The 5′ RNA–DNA flap structure that resembles a branch migratable R-loop structure was constructed as described (Schwab et al. 2015).
RNA-seq
RNA was isolated from K562 cells with an RNeasy minikit (Qiagen). Then, total RNA-seq was performed after ribosomal RNA depletion applying the TruSeq stranded total RNA library and sequenced on the platform NextSEq500 (Illumina).
ChIP-seq
K562 cells were cross-linked and processed for ChIP-seq essentially as reported (Johnson et al. 2007). Immunoprecipitation was performed using 10 μg of anti-UAP56 (Proteintech 14798-1-AP) antibody. DNA was purified and used to build the libraries using the ThruPLEX DNA-seq 6S kit (Rubicon Genomics) according to manufacturer's instructions and then sequenced on the Illumina platform NextSEq500.
DRIP-seq
DRIP-seq was performed as previously described (Sanz and Chédin 2019). Basically, after the immunoprecipitation (DRIP) the DNA was sonicated and checked on a 2100 Agilent Bioanalyzer. Afterward, this DNA was used to build the libraries using the ThruPLEX DNA-seq 6S kit (Rubicon Genomics) according to manufacturer's instructions and sequenced on an Illumina NextSeq500 platform.
DRIPc-seq
DRIPc-seq was performed essentially as described (Sanz and Chédin 2019). The resulting RNA was subjected to library construction using the TruSeq stranded total RNA protocol (Illumina) from the fragmentation step.
DRIP-seq, DRIPc-seq, RNA-seq, and ChIP-seq read mapping, peak calling, and annotation
Sequenced paired-ends reads were subjected to quality control pipeline using the FASTQ Toolkit v.1.0.0 software (Illumina) and then mapped to the human reference genome hg38 canonical female using Bowtie2/BWA (Li and Durbin 2009; Langmead and Salzberg 2012) except for RNA-seq where HISAT2 (Kim et al. 2019a) was used. For DRIPc-seq, reads were separated into Watson and Crick strand using SAMTools (Li et al. 2009).
Peak calling on DRIPc-seq was performed with MACS2 package (Zhang et al. 2008) using FDR < 0.01, allowing broad region detection with a 0.1 cutoff. Next, regions covered by peaks in both replicates in both conditions were merged and fused when <5-kb distance for comparative analysis using BEDtools (Quinlan and Hall 2010). Then, the number of counts per peak was calculated using FeatureCounts and RPKM normalized. For analysis, R-loop gain peaks were established selecting those peaks whose DRIPc signal fold change was >1.25× in siUAP56 respect to the siC control cells in both replicates. Afterward, peaks were annotated to genes using ChIPseeker (Yu et al. 2015) and genes retrieved from Ensembl release 94 2018 (Zerbino et al. 2018). For our purposes, only protein-coding genes were analyzed, considering promoter as −2 kb from TSS and downstream as +2 kb from TTS.
For UAP56 ChIP-seq, peak calling was performed using MACS2 package (Zhang et al. 2008). Peak annotation was performed as described previously.
Coverage profiling of ChIP-seq, RNA-seq, DRIP-seq, and DRIPc-seq signal was obtained using deepTools2 (Ramírez et al. 2016).
For RNA-seq, counts per peak were established using FeatureCounts and RPKM-normalized.
Miscellanea
Human cells culture, plasmids, and primers are shown in the Supplemental Material. ChIP-seq, RNA-seq, DRIP-seq, and DRIPc-seq data from this study have been deposited to the Gene Expression Omnibus database under accession number GSE127979. Published ENCODE and GEO data sets used in this study are as follows: precision nuclear run-on sequencing (PRO-seq) (GSE104800), RNAPII ChIP-seq (ENCSR000BMR), and RNAPII-S2P ChIP-seq (ENCSR000EGF).
Supplementary Material
Acknowledgments
We thank Arijit Dutta for his help with the in vitro assay, and Eloisa Andújar and Mónica Pérez (CABIMER's Genomic Unit) for DNA sequencing service. WT and helicase-dead UAP56 plasmids for in vitro experiments were a gift from Rui Zhao (University of Colorado, Denver). pUBC-EGFP-UAP56 was a gift from Sonia Silva (A.A.’s laboratory). Research in A.A.’s laboratory was funded by the European Research Council (grant ERC2014 AdG669898 TARLOOP), the Spanish Ministry of Economy and Competitiveness (grants BFU2013-42918-P and BFU2016-75058-P), and the European Union (FEDER). P.S. and X.X. were supported by National Institutes of Health grants R35 CA241801 and R21 ES028792, respectively. A.B.-F. was supported by a Juan de la Cierva postdoctoral contract from the Spanish Ministry of Science and Innovation.
Author contributions: C.P.-C., X.X., S.I.B., S.M., and A.B.-F. performed the experiments. C.P.-C., X.X., A.B.-F., S.M., S.I.B., P.S., and A.A. designed the experiments. C.P.-C., A.B.-F., and V.M.G.-B. performed the bioinformatics analysis. C.P.-C., P.S., and A.A. wrote the manuscript. All authors read, discussed, and agreed with the final version of this manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.336024.119.
References
- Aguilera A. 2002. The connection between transcription and genomic instability. EMBO J 21: 195–201. 10.1093/emboj/21.3.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Gómez-González B. 2017. DNA–RNA hybrids: the risks of DNA breakage during transcription. Nat Struct Mol Biol 24: 439–443. 10.1038/nsmb.3395 [DOI] [PubMed] [Google Scholar]
- Barroso S, Herrera-Moyano E, Munoz S, Garcia-Rubio M, Gomez-Gonzalez B, Aguilera A. 2019. The DNA damage response acts as a safeguard against harmful DNA–RNA hybrids of different origins. EMBO Rep 20: e47250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, García-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. 2014. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511: 362–365. 10.1038/nature13374 [DOI] [PubMed] [Google Scholar]
- Britton S, Dernoncourt E, Delteil C, Froment C, Schiltz O, Salles B, Frit P, Calsou P. 2014. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res 42: 9047–9062. 10.1093/nar/gku601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P, Grosse F. 2011. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst) 10: 654–665. 10.1016/j.dnarep.2011.04.013 [DOI] [PubMed] [Google Scholar]
- Chan YA, Aristizabal MJ, Lu PY, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P. 2014. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet 10: e1004288 10.1371/journal.pgen.1004288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez S, Beilharz T, Rondón AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19: 5824–5834. 10.1093/emboj/19.21.5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Puget N, Lin YL, Clouaire T, Aguirrebengoa M, Rocher V, Pasero P, Canitrot Y, Legube G. 2018. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat Commun 9: 533 10.1038/s41467-018-02894-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristini A, Groh M, Kristiansen MS, Gromak N. 2018. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep 23: 1891–1905. 10.1016/j.celrep.2018.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Sánchez MS, Barroso S, Gómez-González B, Luna R, Aguilera A. 2011. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet 7: e1002386 10.1371/journal.pgen.1002386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet M. 2006. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol 59: 723–730. 10.1111/j.1365-2958.2005.05006.x [DOI] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, Tollervey D. 2010. Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24: 1546–1558. 10.1101/gad.573310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Muse T, Aguilera A. 2019. R loops: from physiological to pathological roles. Cell 179: 604–618. 10.1016/j.cell.2019.08.055 [DOI] [PubMed] [Google Scholar]
- García-Pichardo D, Cañas JC, García-Rubio ML, Gómez-González B, Rondón AG, Aguilera A. 2017. Histone mutants separate R loop formation from genome instability induction. Mol Cell 66: 597–609.e5. 10.1016/j.molcel.2017.05.014 [DOI] [PubMed] [Google Scholar]
- García-Rubio ML, Pérez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E, Rosado IV, Aguilera A. 2015. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet 11: e1005674 10.1371/journal.pgen.1005674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rubio M, Aguilera P, Lafuente-Barquero J, Ruiz JF, Simon MN, Geli V, Rondón AG, Aguilera A. 2018a. Yra1-bound RNA–DNA hybrids cause orientation-independent transcription-replication collisions and telomere instability. Genes Dev 32: 965–977. 10.1101/gad.311274.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rubio M, Barroso SI, Aguilera A. 2018b. Detection of DNA–RNA hybrids in vivo. Methods Mol Biol 1672: 347–361. 10.1007/978-1-4939-7306-4_24 [DOI] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. 2012. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45: 814–825. 10.1016/j.molcel.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-González B, Aguilera A. 2019. Transcription-mediated replication hindrance: a major driver of genome instability. Genes Dev 33: 1008–1026. 10.1101/gad.324517.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, García-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marín A, Foiani M, Aguilera A. 2011. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J 30: 3106–3119. 10.1038/emboj.2011.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. 2017. Transcription-replication conflict orientation modulates R-Loop levels and activates distinct DNA damage responses. Cell 170: 774–786.e19. 10.1016/j.cell.2017.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartono SR, Malapert A, Legros P, Bernard P, Chédin F, Vanoosthuyse V. 2018. The affinity of the S9.6 antibody for double-stranded RNAs impacts the accurate mapping of R-loops in fission yeast. J Mol Biol 430: 272–284. 10.1016/j.jmb.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, et al. 2015. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell 57: 636–647. 10.1016/j.molcel.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Moyano E, Mergui X, Garcia-Rubio ML, Barroso S, Aguilera A. 2014. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev 28: 735–748. 10.1101/gad.234070.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodroj D, Recolin B, Serhal K, Martinez S, Tsanov N, Abou Merhi R, Maiorano D. 2017. An ATR-dependent function for the Ddx19 RNA helicase in nuclear R-loop metabolism. EMBO J 36: 1182–1198. 10.15252/embj.201695131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Aguilera A. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721. 10.1016/j.molcel.2003.08.010 [DOI] [PubMed] [Google Scholar]
- Jimeno S, Rondón AG, Luna R, Aguilera A. 2002. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–3535. 10.1093/emboj/cdf335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. 2007. Genome-wide mapping of in vivo protein–DNA interactions. Science 316: 1497–1502. 10.1126/science.1141319 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019a. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Lee SY, Gong F, Battenhouse AM, Boutz DR, Bashyal A, Refvik ST, Chiang CM, Xhemalce B, Paull TT, et al. 2019b. Systematic bromodomain protein screens identify homologous recombination and R-loop suppression pathways involved in genome integrity. Genes Dev 33: 1751–1774. 10.1101/gad.331231.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL. 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378. 10.1016/j.cell.2005.06.008 [DOI] [PubMed] [Google Scholar]
- Li L, Monckton EA, Godbout R. 2008. A role for DEAD box 1 at DNA double-strand breaks. Mol Cell Biol 28: 6413–6425. 10.1128/MCB.01053-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Germain DR, Poon HY, Hildebrandt MR, Monckton EA, McDonald D, Hendzel MJ, Godbout R. 2016. DEAD box 1 facilitates removal of RNA and homologous recombination at DNA double-strand breaks. Mol Cell Biol 36: 2794–2810. 10.1128/MCB.00415-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. 2011. From unwinding to clamping—the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12: 505–516. 10.1038/nrm3154 [DOI] [PubMed] [Google Scholar]
- Mersaoui SY, Yu Z, Coulombe Y, Karam M, Busatto FF, Masson JY, Richard S. 2019. Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids. EMBO J 38: e100986 10.15252/embj.2018100986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Chun Y, Harlen KM, Smalec BM, Dhir S, Churchman LS, Buratowski S. 2018. Cell-cycle modulation of transcription termination factor Sen1. Mol Cell 70: 312–326.e7. 10.1016/j.molcel.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, Fischer T. 2016. Transient RNA–DNA hybrids are required for efficient double-strand break repair. Cell 167: 1001–1013.e7. 10.1016/j.cell.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239. 10.1016/j.molcel.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña A, Gewartowski K, Mroczek S, Cuéllar J, Szykowska A, Prokop A, Czarnocki-Cieciura M, Piwowarski J, Tous C, Aguilera A, et al. 2012. Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. EMBO J 31: 1605–1616. 10.1038/emboj.2012.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation Web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguez C, Gonzales FA, Schmid M, Boggild A, Latrick CM, Malagon F, Putnam A, Sanderson L, Jankowsky E, Brodersen DE, et al. 2013. Mutational analysis of the yeast RNA helicase Sub2p reveals conserved domains required for growth, mRNA export, and genomic stability. RNA 19: 1363–1371. 10.1261/rna.040048.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Armenteros I, Pérez-Calero C, Bayona-Feliu A, Tumini E, Luna R, Aguilera A. 2017. Human THO-Sin3A interaction reveals new mechanisms to prevent R-loops that cause genome instability. EMBO J 36: 3532–3547. 10.15252/embj.201797208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Pereira JM, Aguilera A. 2015. R loops: new modulators of genome dynamics and function. Nat Rev Genet 16: 583–597. 10.1038/nrg3961 [DOI] [PubMed] [Google Scholar]
- Sanz LA, Chédin F. 2019. High-resolution, strand-specific R-loop mapping via S9.6-based DNA–RNA immunoprecipitation and high-throughput sequencing. Nat Protoc 14: 1734–1755. 10.1038/s41596-019-0159-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz LA, Hartono SR, Lim YW, Steyaert S, Rajpurkar A, Ginno PA, Xu X, Chédin F. 2016. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol Cell 63: 167–178. 10.1016/j.molcel.2016.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang CC, Cohn MA, Gibbons RJ, Deans AJ, Niedzwiedz W. 2015. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol Cell 60: 351–361. 10.1016/j.molcel.2015.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zhang L, Zhao R. 2007. Biochemical characterization of the ATPase and helicase activity of UAP56, an essential pre-mRNA splicing and mRNA export factor. J Biol Chem 282: 22544–22550. 10.1074/jbc.M702304200 [DOI] [PubMed] [Google Scholar]
- Silva S, Camino LP, Aguilera A. 2018. Human mitochondrial degradosome prevents harmful mitochondrial R loops and mitochondrial genome instability. Proc Natl Acad Sci 115: 11024–11029. 10.1073/pnas.1807258115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. 2011. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42: 794–805. 10.1016/j.molcel.2011.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, Gabdank I, Narayanan AK, Ho M, Lee BT, et al. 2016. ENCODE data at the ENCODE portal. Nucleic Acids Res 44: D726–D732. 10.1093/nar/gkv1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Stork CT, García-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. 2014. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell 56: 777–785. 10.1016/j.molcel.2014.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Hotz-Wagenblatt A, Voit R, Grummt I. 2017. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev 31: 1370–1381. 10.1101/gad.300624.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhara SC, Carvalho S, Grosso AR, Gallego-Paez LM, Carmo-Fonseca M, de Almeida SF. 2017. Transcription dynamics prevent RNA-mediated genomic instability through SRPK2-dependent DDX23 phosphorylation. Cell Rep 18: 334–343. 10.1016/j.celrep.2016.12.050 [DOI] [PubMed] [Google Scholar]
- Sträßer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondón AG, Aguilera A, Struhl K, Reed R, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308. 10.1038/nature746 [DOI] [PubMed] [Google Scholar]
- Tuduri S, Crabbé L, Conti C, Tourrière H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11: 1315–1324. 10.1038/ncb1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. 2016. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev 30: 1327–1338. 10.1101/gad.280834.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A. 2006. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol Cell Biol 26: 3327–3334. 10.1128/MCB.26.8.3327-3334.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. 2007. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell 28: 253–263. 10.1016/j.molcel.2007.08.016 [DOI] [PubMed] [Google Scholar]
- Yoo HH, Chung IK. 2011. Requirement of DDX39 DEAD box RNA helicase for genome integrity and telomere protection. Aging Cell 10: 557–571. 10.1111/j.1474-9726.2011.00696.x [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, He QY. 2015. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31: 2382–2383. 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. 2018. Ensembl 2018. Nucleic Acids Res 46: D754–D761. 10.1093/nar/gkx1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.