In this study, Irizarry et al. investigated whether the role of graded transcription factors changes over time during embryonic patterning. They used a light-regulated protein degradation system to assay temporal dependence of the transcription factor Dorsal in dorsal–ventral axis patterning of Drosophila embryos, and found that continuous input is not required for some Dorsal targets, and downstream responses, such as twist, function as molecular ratchets.
Keywords: Drosophila melanogaster, BLID, optogenetics, Dorsal Rel transcription factor, Snail, Twist, morphogen, cis-regulatory module, light-inducible degron
Abstract
Graded transcription factors are pivotal regulators of embryonic patterning, but whether their role changes over time is unclear. A light-regulated protein degradation system was used to assay temporal dependence of the transcription factor Dorsal in dorsal–ventral axis patterning of Drosophila embryos. Surprisingly, the high-threshold target gene snail only requires Dorsal input early but not late when Dorsal levels peak. Instead, late snail expression can be supported by action of the Twist transcription factor, specifically, through one enhancer, sna.distal. This study demonstrates that continuous input is not required for some Dorsal targets and downstream responses, such as twist, function as molecular ratchets.
The maternally deposited transcription factor Dorsal (Dl) is considered a morphogen as it forms a nuclear gradient that specifies distinct cell fates along the dorsal–ventral (DV) axis of Drosophila embryos (for review, see Reeves and Stathopoulos 2009). How Dorsal nuclear concentration, which varies along the DV axis, impacts target gene expression has been studied, but few studies have focused on the temporal action of this transcription factor on its targets (Liberman et al. 2009; Reeves et al. 2012; Rushlow and Shvartsman 2012). Several recent studies have used optogenetic approaches to study the temporal contributions of other maternal transcription factors, Bicoid (Bcd) and Zelda, finding that they are continuously required in the early embryo to support expression of target genes (Huang et al. 2017; McDaniel et al. 2019). In particular, high-threshold targets of Bcd require continuous input early and late—both preceding and concurrent with cellularization. As Dl levels steadily increase over time, in contrast to Bcd levels, which remain relatively constant (for review, see Sandler and Stathopoulos 2016b), we hypothesized that target gene dependency on Dl may also be dynamic. In this study, we investigated whether Dl input to target genes is required continuously, as for Bcd, or whether instead Dl input is only required at particular time points.
Results
An optogenetic approach was used to examine the temporal action of Dl in supporting target gene expression, initially focusing analysis on the target gene snail (sna). sna is expressed in ventral regions of the embryo in cells that ultimately contain the highest levels of nuclear Dl (Kosman et al. 1991) and is therefore considered a high-threshold target. However, while Dl levels peak in ventral regions of the embryo during nuclear cycle (nc) 14, studies have shown that sna is expressed within ventral regions at nc13, suggesting that lower levels of Dl are in fact sufficient for this high-threshold target (Reeves et al. 2012).
To assay the temporal dependence on Dl for expression of target genes, including the high-threshold response gene sna as well as low-threshold responses including genes short gastrulation (sog) and decapentaplegic (dpp) (for review, see Reeves and Stathopoulos 2009), an optogenetic Blue Light-Inducible Degron (BLID) sequence was fused to Dl in-frame at the C terminus through modification of the endogenous gene locus using CRISPR/Cas9 technology (Fig. 1A). BLID consists of a LOV2 domain and a degron sequence, such that in the dark, when the α-helix of the LOV2 domain interacts with the LOV core domain, the degron is inaccessible; but upon illumination with blue light (∼400–500 nm), the helix dissociates from the LOV core domain, the degron is exposed, and the entire fusion protein, Dl-BLID in this case, is degraded (Bonger et al. 2014).
Figure 1.
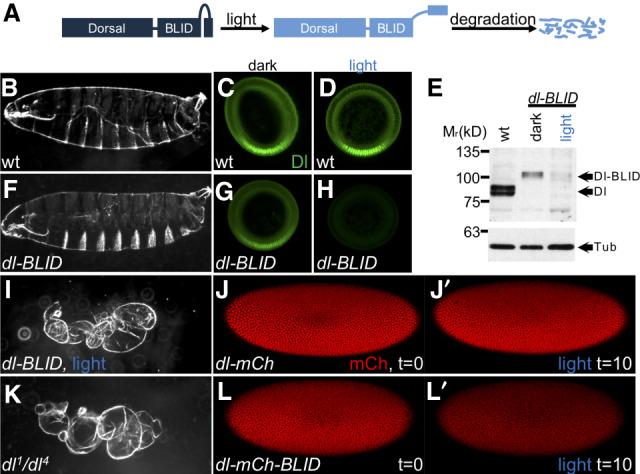
Illumination with blue light induces degradation of Dl-BLID fusion proteins. (A) The Dl-BLID construct. Blue-light illumination causes a degradation sequence to be exposed, resulting in the degradation of the entire fusion protein. (B,F,I,K) Cuticle preparations of embryos derived from wild-type mothers without illumination (B; n = 181/190), dl-BLID mothers without illumination (F; n = 147/310), dl-BLID mothers with 4 h of blue LED illumination (I; n = 31/36), and dl null mutant (dl1/dl4) mothers without illumination (K; 142/142). (C,D,G,H) Manually crossed sectioned embryos stained with anti-Dl antibody (green) derived from wild-type mothers without illumination (C; n = 5/5), wild-type mothers with 1 h of blue LED illumination (D; n = 5/5), dl-BLID mothers without illumination (G; n = 5/5), and dl-BLID mothers with 1 h of blue LED illumination (H; n = 4/5). All embryos in C,D,G,H were imaged at the same settings, demonstrating a clear decrease in Dl levels in H. (E) Western blot of wild type (lane 1), dl-BLID without illumination (lane 2), and dl-BLID with 30-min blue LED illumination (lane 3). Top blot is probed with anti-Dl antibody. Bottom blot is probed with anti-Tubulin antibody to serve as a loading control. Arrows indicate the approximate locations of Dl, Dl-BLID, and Tubulin bands. (J,J′,L,L′) Snapshots from live imaging movies of dl-mCherry (n = 1) and dl-mCherry-BLID (n = 3) at the start (J,L; t = 0) and after 10 min of 40% power blue laser illumination (J′,L′; t = 10). All embryos/larval cuticles are oriented with anterior to left and dorsal up, except cross-sections, which are oriented with the ventral side at the bottom and the dorsal side at the top.
To assay the degradation efficiency of Dl-BLID, embryos laid by homozygous dl-BLID mothers were collected and illuminated with blue light for 4 h (see the Materials and Methods). Larval cuticles were examined as (1) a proxy for changes to Dl levels that manifest as DV patterning defects (Roth et al. 1989), and (2) to assay for any phenotypes induced indirectly by blue-light treatment. The majority of dl-BLID embryos illuminated for 4 h with blue light exhibit cuticles similar in phenotype to dorsalized embryos laid by dl null (dl1/dl4) mothers (Fig. 1, cf. I and 1K) suggesting that Dl-BLID is successfully degraded upon blue light illumination. However, while half of the dl-BLID embryos that were not subjected to blue light exhibited normal cuticles (Fig. 1B,F), the remaining half exhibited a range of subtle defects including a small number with the more severe, dorsalized cuticle phenotype (Supplemental Fig. S1B–D). In contrast, wild-type and dl null mutant embryos appear unaltered when exposed to blue light for 4 h (Supplemental Fig. S1A,E), supporting the view that differences in dl-BLID cuticles, in the light versus dark, result from light-induced degradation and not indirect effects of blue-light exposure. These results suggest that blue light degrades Dl, but that the degradation process is likely leaky, occurring to some degree even in the dark.
To directly test whether Dl is degraded upon illumination, we stained embryos with anti-Dl antibody and imaged cross-sections to assay for changes to the nuclear concentration gradient. As expected, we found that levels of Dl in wild-type embryos containing an unmodified, native dl gene are unaltered both for embryos kept in the dark as well as those exposed to blue light for 1 h (Fig. 1C,D). In the dark, the Dl gradient signal associated with dl-BLID embryos appears qualitatively lower compared with wild type (Fig. 1C,G). However, when dl-BLID embryos were exposed to blue light for 1 h, almost all of the signal, especially the nuclear gradient, is lost (Fig. 1H). Taken together, Dl-BLID appears to support a relatively normal Dorsal nuclear gradient that is efficiently degradable with blue-light illumination but exhibits increased variability in levels/shape compared with wild type, even in the dark. We used this finding to our advantage, as lower levels of Dorsal initially are likely to be more easily manipulated by short light exposures.
To further confirm that Dl-BLID is being degraded, Dl protein levels in embryos were examined by Western blot using anti-Dl antibodies (Fig. 1E). After 30 min in the blue light, Dl-BLID protein levels were indeed reduced to barely detectable levels (Fig. 1E). For embryos that were kept in the dark, Dl-BLID proteins levels were lower compared with wild-type embryos, possibly due to leaky degradation of the degron (Fig. 1E). This lower Dl level may contribute to the broad range of cuticle phenotypes observed in dl-BLID embryos kept in the dark (Supplemental Fig. S1B). The results from the cuticle preparation, Dl antibody staining, and Western suggested that controlling Dl levels using blue light illumination with temporal resolution is feasible.
To directly observe Dl-BLID degradation by blue light, we created and assayed Dl-BLID fluorescent protein fusions. While we found that Dl-mCherry-BLID fusions do not retain Dl function, this fusion does permit visualization of the kinetics of blue-light-induced degradation. Embryos expressing Dl-mCherry and Dl-mCherry-BLID were imaged live using confocal microscopy. When control embryos are exposed to a high power (40%) blue laser of 488-nm wavelength for 10 min, Dl-mCherry embryos (lacking BLID sequence) exhibit little to no decrease in Dl signal (Fig. 1J,J′; Supplemental Movie S1). On the other hand, Dl-mCherry-BLID embryos undergo a dramatic decrease in Dl signal (Fig. 1L,L′; Supplemental Movie S1) indicating that Dl-BLID degradation is occurring in embryos, with appreciable degradation observable within minutes rather than hours observed in other systems (Baaske et al. 2018). Taken together, these results warrant use of the Dl-BLID system to finely assay temporal dependence of target genes on Dl over time during early embryonic development.
To determine whether high levels of Dl are required continuously throughout early embryonic development, we utilized confocal microscopy to illuminate individual embryos with blue light for either 20 min starting at nc14a (laser early, LE) or 20 min starting at nc14c (laser late [LL]) (Fig. 2A). In addition, a triple fluorescent protein (FP) reporter system (H2A-BFP, MCP-GFP, and PCP-mCherry) (Bothma et al. 2015) was introduced by genetic crosses into the dl-BLID background in order to monitor embryonic development and gene expression responses. The H2A-BFP fusion identifies nuclei, which is useful for monitoring all cells in the developing embryos, whereas the MCP-GFP and PCP-mCherry fusions bind to particular RNA stem–loops, which can be used to monitor nascent transcription.
Figure 2.
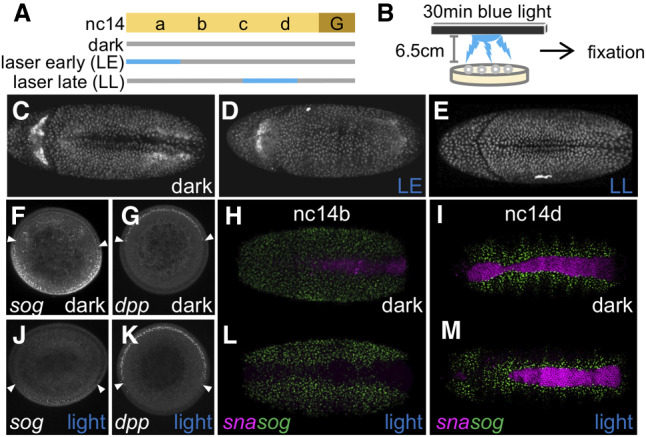
High levels of Dl at late stages are not required to support sna or gastrulation. (A) Scheme of 20 min 40% blue laser illumination on single embryos using a confocal microscope. (Gray bar) Low-power (0.8%) 405-nm laser to image H2A.BFP; (blue bar) Low-power (0.8%) 405-nm laser and 20-min high-power (40%) 488-nm laser. (B) Scheme of 30-min blue LED illumination on a batch of embryos, which was followed by immediate fixation. (C–E) dl-BLID embryos at stage 6 illuminated using a laser (see A). Embryos kept in the dark (“dark,” n = 2) (C), with 20-min blue laser early illumination (“LE,” n = 3) (D), and with 20-min blue laser late illumination (“LL,” n = 3) (E). (F,G,J,K) Manually cross-sectioned nc14a embryos stained for sog (F,J) or dpp (G,K) transcripts kept in the dark (F,G), or with 30-min blue LED illumination (“light,” J,K). White arrowheads mark the expression boundaries. (H,I,L,M) sna (purple) and sog (green) transcript expressions were assayed in dl-BLID embryos kept in the dark (H [n = 4], I [n = 7]) or illuminated with a blue LED (see B) for 30 min (L [n = 6], M [n = 6]). The stages of embryos at fixation were nc14b (H,L) and nc14d (I,M). All whole-mount images are a ventral view with anterior to the left. Cross-sectioned embryos are aligned with the ventral side at the bottom and the dorsal side at the top.
To start, H2A-BFP signal was used to assay whether blue-light illumination affects developmental progression of embryos by observing gastrulation, which involves invagination of the presumptive mesoderm. dl-BLID embryos invaginate ventrally and proceed through gastrulation even when illuminated at the low power (0.8%) blue laser needed to image H2A-BFP, despite some low-level degradation of Dl-BLID (“dark”) (Fig. 2C; Supplemental Movie S2). Alternatively, when additionally subjected to high power (40%) blue laser illumination during an early time window (“LE”) (Fig. 2A), dl-BLID embryos fail to ventrally invaginate, and therefore do not gastrulate (Fig. 2D; Supplemental Movie S3). Embryos obtained from females lacking nuclear Dl also fail to undergo gastrulation (Leptin and Grunewald 1990) supporting the idea that the failure of dl-BLID embryos illuminated early to gastrulation is due to decrease in Dl levels. In contrast, dl-BLID embryos illuminated during a late time window (“LL”) (Fig. 2A), surprisingly, are able to invaginate (Fig. 2E; Supplemental Movie S4). These differences in developmental progression between embryos illuminated early or late suggest that high levels of Dl achieved by late nc14 are not necessary for embryos to proceed through gastrulation.
To test how Dl target gene expression is altered by lower Dl levels, we performed fluorescent in situ hybridization (FISH) using riboprobes to monitor expressions of the genes dpp, sog, and sna, which span the DV axis (Reeves et al. 2012) comparing expression in the dark to that after illumination. In order to collect enough embryos to carry out FISH experiments, we illuminated embryos en masse on plates as opposed to using confocal microscope laser illumination (Fig. 2B; see the Materials and Methods). dl-BLID embryos kept in the dark were analyzed by FISH and show dorsal dpp expression at nc14a-b (Fig. 2G) but a narrower sna expression domain with increased variability at the anterior (Fig. 2H; Supplemental Fig. S2). In addition, sog expression is repressed in this more narrow domain encompassed by its repressor, Sna (Fig. 2F,H,I; Cowden and Levine 2002). Narrowing of the sna domain is likely due to lower levels of total Dl present in the dl-BLID background, even in the dark (Fig. 1E,G).
Embryos illuminated for 30 min before being fixed at nc14a (likely illuminated between nc13-nc14a) exhibit ventrally expanded dpp (Fig. 2K) but retracted sog (Fig. 2J). As dpp and sog expression share a boundary, where dpp is repressed by Dl and sog expression is supported by Dl, these genes likely share the same threshold response but with opposite effect (for review, see Reeves and Stathopoulos 2009). Furthermore, sna expression is lost in embryos fixed at nc14b (likely illuminated between nc14a-b), but sog expression appears unaltered (Fig. 2L; see Discussion). sog transcription is absent from ventral-most regions, presumably due to the presence and action of Sna protein despite the lack of sna transcripts. As sna transcripts have a half-life of ∼13 min (Boettiger and Levine 2013), Sna protein made before blue-light illumination may perdure and continue to repress sog (at least partially) in ventral regions (Bothma et al. 2011).
In contrast, embryos illuminated for 30 min before being fixed at nc14d (likely illuminated later between nc14c-d), express both sna and sog similar to embryos kept in the dark (Fig. 2I,M). These results support the view that the decrease in Dl levels upon illumination affects multiple target genes, but in a temporally dependent manner. Collectively, these results suggest that embryos exposed to light late (i.e., nc14c to nc14d) can still gastrulate because of maintained expression of target genes including sna, a critical regulator of gastrulation (Leptin and Grunewald 1990), whereas embryos exposed to light early (i.e., nc14a to nc14b) fail to gastrulate due to loss of sna.
To distinguish whether maintenance of sna expression at the late time point relates to retention of transcripts made earlier or to an ability to produce new transcripts late, even when Dl is degraded, we turned to live imaging. The sna transcripts identified by in situ hybridization within fixed embryos comprise both mature and nascent transcripts; it is difficult to distinguish nascent sna transcripts in part because this gene is expressed at high levels and transcripts accumulate. Instead, the MS2-MCP system was used to monitor nascent transcription in vivo. Combining the MS2-MCP system with dl-BLID allows nascent transcription to be assayed under different illumination schemes. Specifically, transgenic lines containing a previously defined sna MS2-based reporter were used to assay sna transcriptional activity (Bothma et al. 2015). In these constructs, ∼20 kb spanning the sna locus is used as a reporter in which sna is replaced with the yellow gene sequence including intronic MS2 RNA stem loop sequences (Fig. 3A; Bothma et al. 2015). When this reporter is actively transcribed, MCP-GFP fusion proteins bind to the stem loops and produce visible nuclear puncta, allowing live monitoring of sna expression.
Figure 3.
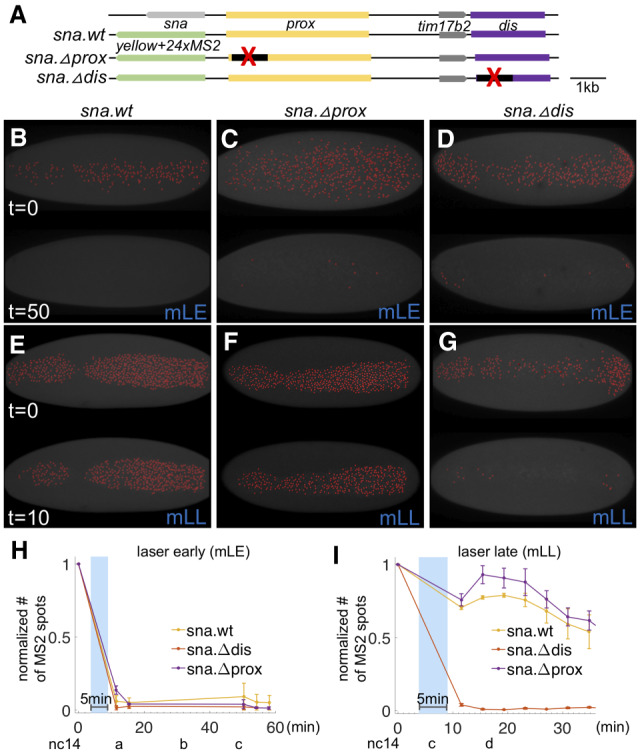
High levels of Dl are required for sna activativation only at early stages, but not at late stages, in which sna expression is predominantly supported by the sna distal enhancer. (A) Scheme of large reporter constructs used to assay sna transcriptional activities by MS2-MCP system (Bothma et al. 2015). (B–G) MCP.GFP signals associated with the sna MS2 reporter were imaged (false-colored red dots) in dl-BLID with early (B–D) or late (E–G) blue laser illumination that is MS2-MCP imaging compatible (“mLE” and “mLL,” respectively) (see also Supplemental Fig. S3) in various sna regulatory conditions including wildtype (sna.wt; B,E), proximal enhancer deletion (sna.Δprox; C,F), and distal enhancer deletion (sna.Δdis; D,G). Images are snapshots from movies, before illumination (top) and after illumination (bottom) of each panel. Three movies were taken for each condition. Ventral views of embryos are shown with anterior oriented to the left. (H,I) Quantitative analysis of the number of MCP.GFP dots associated with the sna MS2 reporter in dl-BLID embryos with sna.wt, sna.Δprox, or sna.Δdis sna regulatory condition. Number of MS2-MCP.GFP spots are counted in each time frame, and the values are normalized to the initial value detected in the first frame (before 5 min blue-laser illumination with 15% laser power) with early laser (H) or late laser (I) illumination. Blue shade indicates a time frame of 5-min 15% blue-laser illumination. Error bars represent standard error of the mean. For individual traces, see Supplemental Figure S5. For details for detection of sna.MS2-MCP.GFP and blue-laser illumination, see Supplemental Figure S3.
An intermediate power laser setting (5%) was used to image the MS2-MCP signal, while the high-power setting was used to degrade Dl-BLID (Supplemental Fig. S3A; see the Materials and Methods). Again, under these imaging conditions, illumination of dl-BLID embryos with high power at the early time point (i.e., nc14a-nc14b, “mLE”) leads to gastrulation failure, whereas illumination of embryos later (i.e., nc14c, “mLL”) has no effect on gastrulation despite the use of intermediate laser power to image the MS2-MCP signal for an extended period of time (Supplemental Fig S3B). We used this scheme, in which MS2-MCP imaging and Dl-BLID degradation are compatible, to determine how sna transcription is affected by temporal changes in Dl levels.
We found that wild-type sna MS2-MCP signal (sna.wt) was retained when embryos were illuminated with high-power laser late (mLL) (Fig. 3E,I; Supplemental Movie S7), but was diminished when embryos were illuminated early (mLE) (Fig. 3B,H; Supplemental Movie S5). Two enhancers are known to support early sna expression during embryogenesis, one proximal (sna.prox) and one distal (sna.dis) (Ip et al. 1992; Perry et al. 2010; Dunipace et al. 2011; Ozdemir et al. 2011). In order to understand which cis-regulatory sequences drive sna gene expression even when Dl is degraded at the late time point, we also assayed two reporter variants in which portions of these two early embryonic enhancers had been deleted, constructed in a previous study (Fig. 3A; Bothma et al. 2015). The sna.Δprox reporter behaves as the sna.wt reporter: Embryos illuminated early lose signal, whereas those illuminated late retain it (Fig. 3C,F,H,I; Supplemental Movies S6, S8). In contrast, the sna.Δdis reporter loses expression when illuminated at either time point (Fig. 3D,G–I; Supplemental Movies S6, S8). Thus, the distal enhancer is necessary for late sna expression when no or very little Dl is present, while the proximal enhancer cannot support late sna expression in the absence of Dl. This suggests that the proximal sna enhancer likely requires high Dl levels for activity. Taken together, these results support a model in which Dl acts through either enhancer (directly or indirectly) early, but that an additional input is required to sustain late sna expression through the sna.dis enhancer, specifically.
Another Dl target gene encoding a bHLH transcription factor, twist (twi), is expressed in ventral regions, and also provides input to sna (for review, see Reeves and Stathopoulos 2009). sna expression is either lost or greatly diminished in dl and twi mutants, respectively (Ip et al. 1992). twi transcript levels increase rapidly at the onset of nc14 and activation of mesodermal genes follows (Sandler and Stathopoulos 2016a), suggesting that Twi may be an important input into these target genes. Furthermore, peak Dl levels are not required to support sna expression as ectopic Twi gradients can support its expression even in conditions of low, but not completely absent, Dl (Stathopoulos and Levine 2002). These previous studies had suggested that Twi may suffice to support sna activation at the late time point, even in the absence of Dl. However, it was previously not possible to remove Dl but retain Twi as twi gene expression is Dl-dependent.
We hypothesized that Twi is responsible for the late expression of sna, essentially taking over for Dl. To test this idea, embryos were fixed after 30-min blue LED illumination (Fig. 2B) and assayed for Dl and Twi proteins using antibodies, and for sna transcripts by FISH. Embryos exposed to light early or late exhibited low levels or no Dl as expected but, surprisingly, retained Twi expression (Supplemental Fig. S4) demonstrating that even low levels of Dl in nc14 are sufficient to support low levels of Twi expression. sna expression is also retained when embryos are illuminated late (Fig. 4A–B′′), but not early (Supplemental Fig. 4A–B′′) suggesting early nc14 sna expression is Dl-dependent. However, when the twi mutant is recombined with dl-BLID, even when embryos are kept in the dark and high levels of Dl are present, sna expression is lost if Twi is absent (Fig. 4C–C′′). Taken together, these results suggested that Twi is a pivotal input for sna activation, particularly at late stages when sna expression is independent of high Dl levels.
Figure 4.
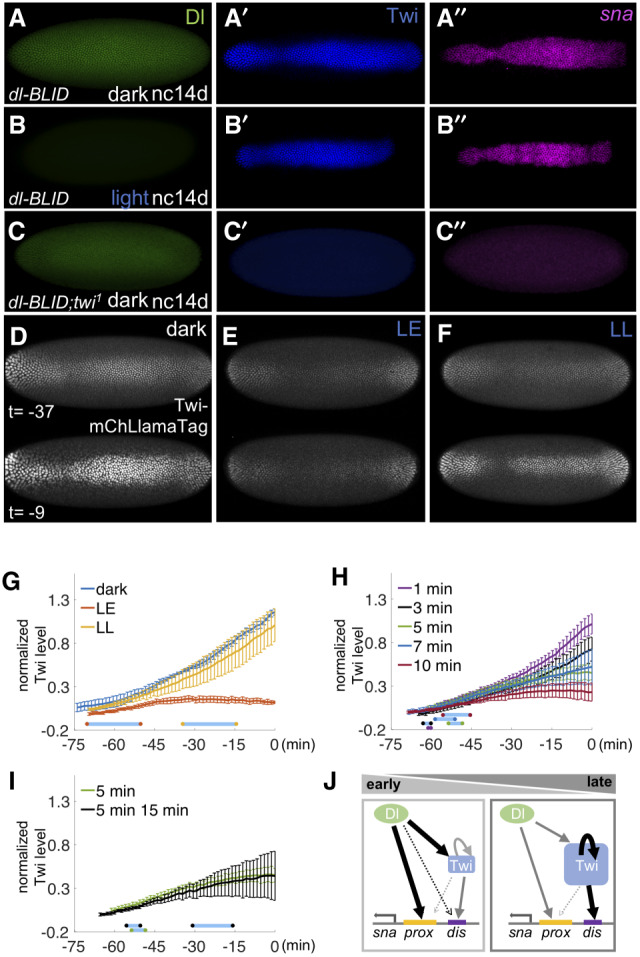
Twi suffices to support sna expression at late stages in the absence of high levels of Dl. (A–C) Expression of Dl proteins (green), Twi proteins (blue), and sna transcripts (pink) were examined in dl-BLID embryos with twi wild-type (A,B) or twi1 mutant background at nc14d without (A,C) or with (B; n = 11) 30-min blue LED illumination. (D–F) Snapshots from movies showing mCherry signal associated with the Twi-mCherryLlamaTag (Twi-mChLlamaTag) (Bothma et al. 2018) under various confocal 40% blue-laser illumination conditions: no illumination (dark) (D), 20-min early illumination at nc14a (LE) (E), and 20-min late illumination at nc14c (LL) (F). Time indicates the time length preceding the germband extension. (G–I) Quantitative analysis of the levels of mCherry associated by Twi-mChLlamaTag with varying 40% blue-laser illumination conditions: no illumination (dark, blue), 20-min early illumination at nc14a (LE, red), and 20-min late illumination at nc14c (LL, yellow) (G); illumination at nc14b for 1 min (purple), 3 min (black), 5 min (green), 7 min (blue), and 10 min (red) (H); and the 5-min (green) data replotted from H to compare with 5 min at nc14b followed by additional 15-min illumination at nc14c (black) (I). Three movies were taken for each condition. For the individual traces, see Supplemental Figure S6. (J) A model of regulatory shift, such as from high level of Dl to high level of Twi-dependent regulatory states, to support sna expression throughout early embryonic development. Dl proteins (green circle), Twi proteins (blue square), sna proximal enhancer (yellow bar), and sna distal enhancer (purple bar). All embryo images are ventral views with anterior to the left. Blue bars in G–I represent the average time window of confocal blue laser illumination to their respective curves. Error bars represent standard error of the mean.
In order to understand the temporal relationship between Dl and Twi transcription factor dynamics, we assayed Twi dynamics with fine time resolution in combination with temporally controlled Dl-BLID levels. Twi levels were detected in dl-BLID embryos using a previously described Twi-mCherryLlamaTag fusion protein, which allows early zygotic proteins to be visualized without having to wait for fluorescence maturation (Bothma et al. 2018). When embryos are kept in the dark, mCherry signal intensifies throughout nc14, suggesting exponential production of Twi protein (Fig. 4D,G; Supplemental Movie S9). However, in embryos exposed to high-power blue laser illumination at nc14a, no increase in Twi levels is observed (“LE”) (Fig. 4E,G; Supplemental Movie S10). In contrast, for embryos illuminated at nc14c, Twi levels increase (“LL”) (Fig. 4F,G; Supplemental Movie S10) similarly to embryos without illumination (“dark”) (Fig. 4D,G; Supplemental Movie S9). These results support the view that Twi is only responsive to Dl levels early, but is able to maintain its expression late even if Dl levels fall.
To examine how responsive Twi is to Dl levels early, we manipulated Dl levels using various durations of blue laser illumination at nc14b and measured the effect live using the Twi-mCherryLlamaTag as a proxy for Twi levels (see the Materials and Methods). Short time window illuminations with blue laser (<5 min) early presumably lead to small or negligible changes in Dl-BLID levels and therefore had little or no effect on Twi levels throughout nc14 (Fig. 4H). However, with incremental increase in duration of blue laser illumination (5, 7, or 10 min), Twi levels also fail to increase, with severity corresponding to the duration of illumination. The rate of change for Twi levels decreases substantially, most apparent with the 10-min exposure (Fig. 4H), instead of undergoing the exponential increase observed in dark, laser late, or short time window illuminations of 1 and 3 min (Fig. 4G,H). These results suggest that Twi levels are reflective of the underlying Dl levels early, and that levels of Dl early impact levels of Twi present later.
Interestingly, intermediate exposure of Dl-BLID to blue laser (e.g., 5–10 min) results in loss of the late nc14 exponential increase in Twi levels that is normally observed in control embryos (e.g., dark) (Fig. 4G) as the rate of change in levels decreases, but it initially remained unclear why (Fig. 4H). In order to explain why Twi levels do not grow exponentially after intermediate duration Dl degradation, we hypothesized that low levels of Dl are retained that continue to support low levels of Twi. In this scenario, a second blue-light illumination to knock down the remaining Dl would be expected to further decrease Twi levels. However, we found that exposure to a second illumination (e.g., 15 min at nc14c) has no effect; Twi is maintained at levels similar to embryos exposed to a single 5-min illumination at nc14b (Fig. 4I). This observation suggests that in late nc14 activation of Twi shifts to a gene regulatory state that is independent of Dl levels. Collectively, these data support the view that a Twi-dependent threshold exists above which Twi can activate its own expression independently of Dl at this late stage and supports a model where levels of Dl in early nc14 determine twi expression, but during late nc14, twi expression is Dl independent.
Discussion
In this study, we examined whether Dl is continuously required to activate target genes in the early embryo by utilizing a Dl-BLID fusion. Dl is required early for the initiation of expression of the sna target gene in ventral regions, but surprisingly is not needed late to maintain its expression. Like sna, expression of htl, mes3 and netA are sustained in dl-BLID embryos illuminated with blue LED light late (Supplemental Fig. S7) suggesting that other target genes are similarly regulated. In contrast, we found that the lateral gene sog is still expressed no matter when Dl degradation occurs during nc14. This unexpected result appears to contradict the model where the sog dorsal boundary is formed by limiting levels of nuclear Dorsal. One possible explanation that is consistent with this model is that low levels of Dorsal remain after illumination and are enough to activate sog. However, this should result in either a narrow sog expression domain or requires asymmetrical degradation of Dorsal, neither of which was observed (Fig. 1H; Supplemental Fig. S2; Supplemental Movie S1). Another explanation is that once the sog domain is established by lower levels of Dl, sog does not require Dl to remain active because another factor acts to retain its expression. A simpler explanation is that sog transcripts are long and the detected signal could be from sog transcripts that were initiated at an earlier time point, when Dorsal was present. These possible explanations for how sog transcription fails to respond to Dorsal degradation upon illumination are not mutually exclusive. Addressing how sog transcription becomes Dl independent in future studies will be an important step forward in our understanding of how the sog dorsal boundary is set.
Our results also provide insight into how a transcriptional network may buffer changes in levels of a maternal patterning morphogen. In the case of sna, high levels of Dl are required early to activate sna gene expression. Dl acts both directly, and indirectly by controlling twi expression, as Twi is also an input to sna (Fig. 4J, left). In contrast, Dl is dispensable for sna activation at later time points. When Dl levels are reduced, sna expression remains (Fig. 4J, right), likely maintained by Twi once Twi is expressed. The ability to retain expression of a morphogen target gene despite a decrease in morphogen levels has been termed a “ratchet response,” and was demonstrated for targets of the activin morphogen in Xenopus (Gurdon et al. 1995). Twi can maintain its own expression through autoregulation (Kosman et al. 1991; Crews and Pearson 2009), and we propose this autoregulatory feedback serves to support this ratchet response that is able to buffer against decreases in Dl levels. However, simple Twi autoregulatory feedback would predict a single steady-state concentration for Twi. Instead, we observed that Twi levels increase exponentially or reach intermediate levels of Twi when varying the length of illumination. While this result would not support simple autoregulatory feedback as a mechanism for maintaining Twi expression in the absence of high Dl, it requires Twi levels to be at steady state. It is possible that the observed responses have not reached steady-state, and if given enough time they might all converge to the same steady-state concentration (i.e., a single response supported by autoregulation). It is likely that other factors contribute to twi regulation; however, these results support the model that Dl activates twi, and Twi is able to maintain its own expression through autoregulation.
Taken together, we propose that once Twi reaches sufficient levels to support its own auto-activation, Dl is no longer required to support sna expression (Fig. 4J, right). This is in sharp contrast to the Bcd morphogen, which patterns the anterior–posterior (AP) axis and to the early maternal pioneer factor Zelda (Huang et al. 2017; McDaniel et al. 2019). Both Bcd and Zelda have been found to be required continuously; perturbations at any stage cause loss of gene expression. Alternatively, the DV gene regulatory network shifts from a state of high Dl dependence to a state of Dl independence for several target genes expressed in the presumptive mesoderm. It is possible that this ratchet response relates to the ability of twi gene expression to buffer changes in Dl concentration, and allows the DV patterning network to respond only to increasing Dl levels. Taken together, ratchet-like responses are crucial steps during animal development not only because they support morphogen-dependent patterning, but also they may serve to buffer expression of target genes against fluctuations in morphogen levels due to genetic and environmental changes.
Materials and methods
Fly stocks/husbandry and plasmids
All flies were kept at 18°C, unless otherwise noted. yw was used as wild type. Fly stocks used were as follows: dl4/CyO (Bloomington Drosophila Stock Center [BDSC] 7096), dl1/CyO (#3236, BDSC), twi1/CyO (BDSC 2381), nos > MCP-GFP, nos > mCherry-PCP, His2Av-eBFP2 (from Michael Levine, Princeton University) (Lim et al. 2018), snailBAC > MS2 with both proximal and distal enhancers (WT, sna.wt), proximal deletion (NoPrimary, sna.Δprox), or distal deletion (NoShadow, sna.Δprox) (from Michael Levine, Princeton University) (Bothma et al. 2015), vasa-mCherry and Twi-mCherryLlamaTag (from Hernan Garcia, University of California at Berkeley) (Bothma et al. 2018). For details regarding fly crosses, see the Supplemental Material.
Genome editing
CRISPR was performed as described previously (Gratz et al. 2014; Port et al. 2014). Briefly, the gRNA fly line (targeting before the C-term and after the 3′UTR of Dorsal) (see the Supplemental Materials; Supplemental Table S1) and the Cas9 line Sp/CyO, P{nos-Cas9}2A, (NIG-FLY, CAS-0004) were mated. Embryos were collected and the homology-directed repair (HDR) template containing the C-term of dl fused to BLID (see the Supplemental Material) was injected into these embryos. Flies were screened for DsRed. The integration was confirmed by PCR and sequencing.
Blue-light illumination
Embryos were collected for 1 h at 18°C followed by 4-h incubation for aging and illuminated with blue light using either a set of LEDs (2501BU blue 225 LED 13.8-W square grow light panel 110) or the 488-nm laser on a Zeiss LSM 800 confocal microscope. For blue LED light illumination, embryos on agar plates were placed 6.5 cm below the LED light panel and illuminated for appropriate time lengths. After blue light exposure, the embryos were fixed. For 488-nm blue laser illumination, the embryos were dechorionated and mounted on a heptane glued slide. The embryos were immersed in water, and a blue laser was applied using a 25× water immersion objective. All the embryos were prepared under red filtered light to avoid possible Dl-BLID degradation by light coming from microscopes or other ambient sources.
Cuticle preparations
Embryos were collected for 2 h at 18°C, aged 1.5 h in the dark, and illuminated with blue LEDs for 4 h. Subsequently, embryos were aged for an additional 36–40 h in the dark and then processed by standard cuticle preparation using lactic acid.
Western blot analysis
Aged embryos were dechorionated and mounted in Halocarbon 27 oil (Sigma-Aldrich). Embryos at nc14b were manually selected and illuminated for 30 min with LED blue light. After light exposure, embryos at nc14c were prepared for standard Western blot.
Immunostaining and fluorescent in situ hybridization (FISH)
Immunostaining and FISH protocols were followed as previously described (Kosman et al. 2004). Sheep antidigoxigenin (Life Technology PA185378), or rabbit anti-FITC (Invitrogen A889), mouse anti-Dl (1:10; Developmental Studies Hybridoma Bank 7A4) or guinea pig anti-Twi (1:200) (Trisnadi and Stathopoulos 2015) were used together with Alexa conjugate secondaries (1:400; Thermo Fisher). DAPI staining (1:10,000; Molecular Probes) was used to mark nuclei.
Live imaging and quantification
To test efficiency of Dl-BLID degradation upon blue laser illumination, 488 nm of blue laser with 40% laser power (high power) was applied to the embryos heterozygous for either dl-mCherry or dl-mCherry-BLID, while also applying 555-nm laser to monitor mCherry signal.
To examine the continuous requirement of high level of Dl at blastoderm stage, dl-BLID embryos were illuminated by 488 nm blue laser with 40% laser power (high power) for 20 min, starting at the appropriate developmental stage. Early embryonic development was examined by live imaging of H2A-BFP (i.e., His2Av.eBFP2) using 405 nm blue laser with 0.8% laser power (low power) (Lim et al. 2018).
To test sna transcriptional activities, the MS2-MCP system was used (Bothma et al. 2015) in combination with dl-BLID to optogenetically manipulate Dl levels and assay target gene expression live. To both detect sna.MS2-MCP.GFP signals and degrade Dl-BLID, a 488-nm blue laser was used for both purposes but using different laser power: 5% (intermediate level) and 15% (high power), respectively. To distinguish this MS2-MCP imaging scheme from standard approach (i.e., Fig. 2A), we refer to MS2-MCP imaging laser early and laser late as “mLE” and “mLL” with exact conditions outlined in Supplemental Fig. S3A.
To image Twi protein dynamics, Twi-mCherryLlamaTag system, which recognizes maternally deposited mature mCherry fluorescent protein, was utilized (Bothma et al. 2018). mCherry was imaged live from the onset of nc14a to gastrulation while Dl-BLID was degraded by 488 nm blue laser at 40% laser power (high power) with varying lengths of time at appropriate developmental stages. All images were taken using a 25× water immersion objective. For additional details regarding imaging and quantification, see the Supplemental Material.
Supplementary Material
Acknowledgments
We are grateful to Mike Levine and Hernan Garcia for providing fly stocks, Leslie Dunipace for injections and guidance, Theodora Koromila for help with imaging strategy, and Susan Newcomb for edits to manuscript. This study was supported by funding from National Institutes of Health grants R21HD095639 and R35GM118146 to A.S. and R21OD017964 to D.S.
Author contributions: A.S., D.S., J.I., and J.M. conceived the project and planned the experimental approach. A.S. directed the project. J.I. performed all imaging. J.M. performed all quantitative analysis of the imaging data, Crispr/Cas9 genomic engineering, Western analysis, and viability studies. J.I. and J.M. performed stainings. D.S. and G.K. validated the use of BLID for studies in Drosophila and helped to develop protocols for its use. J.I., J.M., and D.S. developed the protocol for fixed embryo analysis, whereas J.I. and J.M. developed protocols for BLID live imaging. Data were analyzed by J.I., J.M., and A.S. The manuscript was written by J.I., J.M., and A.S. with edits provided by D.S.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.338194.120.
References
- Baaske J, Gonschorek P, Engesser R, Dominguez-Monedero A, Raute K, Fischbach P, Müller K, Cachat E, Schamel WWA, Minguet S, et al. 2018. Dual-controlled optogenetic system for the rapid down-regulation of protein levels in mammalian cells. Sci Rep 8: 15024 10.1038/s41598-018-32929-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. 2013. Rapid transcription fosters coordinate snail expression in the Drosophila embryo. Cell Rep 3: 8–15. 10.1016/j.celrep.2012.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonger KM, Rakhit R, Payumo AY, Chen JK, Wandless TJ. 2014. General method for regulating protein stability with light. ACS Chem Biol 9: 111–115. 10.1021/cb400755b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothma JP, Magliocco J, Levine M. 2011. The snail repressor inhibits release, not elongation, of paused Pol II in the Drosophila embryo. Curr Biol 21: 1571–1577. 10.1016/j.cub.2011.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothma JP, Garcia HG, Ng S, Perry MW, Gregor T, Levine M. 2015. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. Elife 4: e07956 10.7554/eLife.07956 10.7554/eLife.07956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothma JP, Norstad MR, Alamos S, Garcia HG. 2018. LlamaTags: a versatile tool to image transcription factor dynamics in live embryos. Cell 173: 1810–1822.e16. 10.1016/j.cell.2018.03.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowden J, Levine M. 2002. The snail repressor positions notch signaling in the Drosophila embryo. Development 129: 1785–1793. [DOI] [PubMed] [Google Scholar]
- Crews ST, Pearson JC. 2009. Transcriptional autoregulation in development. Curr Biol 19: R241–R246. 10.1016/j.cub.2009.01.015 10.1016/j.cub.2009.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunipace L, Ozdemir A, Stathopoulos A. 2011. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development 138: 4075–4084. 10.1242/dev.069146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O'Connor-Giles KM. 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961–971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Mitchell A, Mahony D. 1995. Direct and continuous assessment by cells of their position in a morphogen gradient. Nature 376: 520–521. 10.1038/376520a0 [DOI] [PubMed] [Google Scholar]
- Huang A, Amourda C, Zhang S, Tolwinski NS, Saunders TE. 2017. Decoding temporal interpretation of the morphogen Bicoid in the early embryo. Elife 6: e26258 10.7554/eLife.26258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M. 1992. dorsal–twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev 6: 1518–1530. 10.1101/gad.6.8.1518 [DOI] [PubMed] [Google Scholar]
- Kosman D, Ip YT, Levine M, Arora K. 1991. Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science 254: 118–122. 10.1126/science.1925551 [DOI] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. 2004. Multiplex detection of RNA expression in Drosophila embryos. Science 305: 846 10.1126/science.1099247 [DOI] [PubMed] [Google Scholar]
- Leptin M, Grunewald B. 1990. Cell shape changes during gastrulation in Drosophila. Development 110: 73–84. [DOI] [PubMed] [Google Scholar]
- Liberman LM, Reeves GT, Stathopoulos A. 2009. Quantitative imaging of the Dorsal nuclear gradient reveals limitations to threshold-dependent patterning in Drosophila. Proc Natl Acad Sci U S A 106: 22317–22322. 10.1073/pnas.0906227106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Heist T, Levine M, Fukaya T. 2018. Visualization of transvection in living Drosophila embryos. Mol Cell 70: 287–296.e6. 10.1016/j.molcel.2018.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel SL, Gibson TJ, Schulz KN, Fernandez Garcia M, Nevil M, Jain SU, Lewis PW, Zaret KS, Harrison MM. 2019. Continued activity of the pioneer factor zelda is required to drive zygotic genome activation. Mol Cell 74: 185–195.e4. 10.1016/j.molcel.2019.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir A, Fisher-Aylor KI, Pepke S, Samanta M, Dunipace L, McCue K, Zeng L, Ogawa N, Wold BJ, Stathopoulos A. 2011. High resolution mapping of Twist to DNA in Drosophila embryos: efficient functional analysis and evolutionary conservation. Genome Res 21: 566–577. 10.1101/gr.104018.109 10.1101/gr.104018.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. 2010. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol 20: 1562–1567. 10.1016/j.cub.2010.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Chen H-M, Lee T, Bullock SL. 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A 111: E2967–E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves GT, Stathopoulos A. 2009. Graded dorsal and differential gene regulation in the Drosophila embryo. Cold Spring Harb Perspect Biol 1: a000836 10.1101/cshperspect.a000836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves GT, Trisnadi N, Truong TV, Nahmad M, Katz S, Stathopoulos A. 2012. Dorsal–ventral gene expression in the Drosophila embryo reflects the dynamics and precision of the dorsal nuclear gradient. Dev Cell 22: 544–557. 10.1016/j.devcel.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Stein D, Nüsslein-Volhard C. 1989. A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell 59: 1189–1202. 10.1016/0092-8674(89)90774-5 [DOI] [PubMed] [Google Scholar]
- Rushlow CA, Shvartsman SY. 2012. Temporal dynamics, spatial range, and transcriptional interpretation of the Dorsal morphogen gradient. Curr Opin Genet Dev 22: 542–546. 10.1016/j.gde.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Sandler JE, Stathopoulos A. 2016a. Quantitative single-embryo profile of Drosophila genome activation and the dorsal–ventral patterning network. Genetics 202: 1575–1584. 10.1534/genetics.116.186783 10.1534/genetics.116.186783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler JE, Stathopoulos A. 2016b. Stepwise progression of embryonic patterning. Trends Genet 32: 432–443. 10.1016/j.tig.2016.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. 2002. Linear signaling in the Toll-Dorsal pathway of Drosophila: activated Pelle kinase specifies all threshold outputs of gene expression while the bHLH protein Twist specifies a subset. Development 129: 3411–3419. [DOI] [PubMed] [Google Scholar]
- Trisnadi N, Stathopoulos A. 2015. Ectopic expression screen identifies genes affecting Drosophila mesoderm development including the HSPG Trol. G3 5: 301–313. 10.1534/g3.114.015891 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


