Abstract
Background
Middle East respiratory syndrome coronavirus (MERS-CoV) has continued to cause sporadic outbreaks of severe respiratory tract infection over the last 8 years.
Methods
Complete genome sequencing using next-generation sequencing was performed for MERS-CoV isolates from cases that occurred in Riyadh between 2015 and 2019. Phylogenetic analysis and molecular mutational analysis were carried out to investigate disease severity.
Results
A total of eight MERS-CoV isolates were subjected to complete genome sequencing. Phylogenetic analysis resulted in the assembly of 7/8 sequences within lineage 3 and one sequence within lineage 4 showing complex genomic recombination. The isolates contained a variety of unique amino acid substitutions in ORF1ab (41), the N protein (10), the S protein (9) and ORF4b (5).
Conclusion
Our study shows that MERS-CoV is evolving. The emergence of new variants carries the potential for increased virulence and could impose a challenge to the global health system. We recommend the sequencing every new MERS-CoV isolate to observe the changes in the virus and relate them to clinical outcomes.
Keywords: MERS-CoV, SARS-CoV, Substitutions, Zoonosis, Saudi Arabia
Introduction
The 21st century is witnessing a 3rd outbreak of zoonosis [1] associated with a novel coronavirus, ‘SARS-CoV-2’ in this case. Similar to its predecessors, the severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV) [2,3], infection with SARS-CoV-2 (officially named COVID-19 by the World Health Organization on February 11, 2020) can manifest as bronchitis, pneumonia, or a severe respiratory illness.
Interestingly, these novel human coronaviruses (hCoVs) emerged 8–10 years apart. SARS-CoV emerged in China in November 2003, where the pandemic infected more than 8096 people, resulting in 774 deaths (WHO 2002). The pandemic warning was removed by the World Health Organization (WHO) in 2004, and there was no evidence of new cases of infection in humans thereafter. Approximately eight years later in 2012, a new coronavirus, ‘MERS-CoV’, surfaced in Saudi Arabia [3], and periodic outbreaks of MERS-CoV are still a matter of immense concern for global health security. A total of 2494 laboratory-confirmed cases have been documented to date, resulting in 858 deaths spanning 27 countries [4]. Overall, Saudi Arabia has led in the number of infections, with 2102 confirmed cases and 780 deaths [5].
Similar to many other CoVs, MERS-CoV has a very large RNA genome, of approximately 30 kb including the 5′ and 3′ UTRs, which encodes three broad protein classes [6,7]. Several structural and non-structural proteins are encoded within the intact virion. The structural proteins include the spike (S) protein, nucleocapsid (N) protein, envelope (E) protein, and membrane (M) protein. The two main polyproteins (pp1ab and pp1a) encoded by ORF1ab are cleaved into 15/16 different non-structural proteins [6,7]. Together, they form the replicase complex, which is essential for polyprotein processing, and efficient virus replication. MERS-CoV encodes accessory proteins including ORF 3, ORF 4a, ORF4b, and ORF 5 [6,7], similar to many other coronaviruses. The exact roles and precise locations of the accessory proteins of MERS-CoV are not quite clear. However, it has been suggested that these proteins influence viral replication, pathogenesis, and disease outcomes, as observed in SARS-CoV and other coronaviruses [8].
The entry of MERS-CoV into human cells is facilitated by a heavily glycosylated type I transmembrane protein (S) that binds to the dipeptidyl peptidase 4 (DPP4) protein, a receptor on host cells [9]. There is a strong consensus that dromedary camels (Camelus dromedarius) are the primary source of the transmission of MERS-CoV to humans [10]. Whether camels serve as mixing vessels for the emergence of variant MERS-CoV strains warrants further investigation [11]. A low mutation rate has been demonstrated in MERS strains isolated from humans, which is attributed to the low level of immunological pressure exerted on this coronavirus in humans [12]. A high frequency of mutations and recombinant events is observed in MERS-CoV strains isolated from camels [13]. Sporadic cases of MERS-CoV are in continuous circulation in Saudi Arabia, and the currently available epidemiological data cannot explain the changing epidemiology of the virus.
In this study, we obtained complete genome sequences of MERS-CoV from eight individuals from the Riyadh region in Saudi Arabia. We carried out molecular epidemiology analysis to investigate their epidemiological connection with one another and with past strains. An in-depth genetic analysis was performed to construct an interface linking genomic variations and disease severity.
Materials and methods
Patients and samples
The patients included in this study (n = 8) presented with severe upper and lower respiratory tract infections and were admitted to the Intensive Care Unit (ICU) of the King Abdulaziz Medical City, Ministry of National Guard – Health Affairs (MNG-HA), Riyadh. Sputum, nasopharyngeal swab, endotracheal aspirate, or bronchial lavage samples were obtained. The supernatants were carefully separated from the clinical samples by centrifugation at 2000 rpm for 10 min and stored at −80 °C until processing.
RNA extraction and cDNA preparation
Viral RNA was extracted from 1 mL of the stored samples using a QIAamp Viral RNA Mini Kit (Qiagen, Valencia CA, USA). The samples were processed in multiple aliquot at 200 μL/turn and passed through a single column to obtain a high concentration of RNA. The RNA was eluted in 60 μl of water and stored at −80°C until further processing. The RNA concentration was adjusted to a minimum of 100 ng/μl in a Qubit 3.0 instrument using the Qubit RNA BR assay kit (Thermo Fisher Scientific, USA). The RNA extracted from each sample was reverse transcribed into cDNA using the superscript VILO IV cDNA kit (Thermo Fisher, USA) and stored at −80°C. Screening for MERS-CoV was performed using the Coronavirus MERS-Cov RT-PCR kit (TIB Molbion/Roche, Germany), targeting the upstream region of the envelope gene (upE) and open reading frame 1a (ORF1a). The amplicons were subjected to complete genome sequencing using Ion Torrent Ampliseq technology sequencing.
Complete genome sequencing
Viral cDNA was subjected to Ampliseq library preparation according to the manufacturer’s protocol using MERS Ampliseq Panels (Thermo Fisher Scientific, USA) with a mean insert size of 200 bps. These panels were designed on the basis of data from 233 reference strains retrieved from NCBI databases. A total of 33,108 amplicons with an average length of 200 bp were included in the two primer pools. Each library was assigned a distinct barcode using the Ion Xpress Barcode Adapters 1–16 kit and purified using Agencourt AMpure Xp beads (Beckman Coulter, Brea, CA, USA).
Each purified library was efficiently quantified in a StepOnePlus Real-Time PCR system using the TaqMan Library quantification kit (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol and normalized to 25 pM. Template-positive Ion Sphere particles were obtained by pooling all 10 normalized libraries and clonally amplified in a OneTouch 2 system using the Ion PGM HiQ OT2 200 kit (Thermo Fisher Scientific, USA.). Template-positive ISPs were loaded onto Ion 318 chips and sequenced in an Ion PGM instrument using the Ion PGM HiQ Sequencing kit (Thermo Fisher Scientific, USA).
Gap-filling PCR and Sanger sequencing
Fourteen bidirectional primer pairs were designed (Supplement Table 1) for MERS-CoV genome gaps using Primer Express software v3.0 (Applied Biosystems, Foster City, CA, USA). Gap-filling PCR was performed, and the amplicons were sequenced by Sanger sequencing in a 3730xl Genetic Analyser using the BigDye Terminator v 3.1 Cycle Sequencing kit (Thermo Fisher Scientific, USA) following the manufacturer’s protocol.
Table 1.
Patient clinical details.
| GenBank accession | Age (year) | Gender | History | Admission diagnosis | Specimen date | # Stay (days) |
|---|---|---|---|---|---|---|
| MH013216 | 57 | Male | HTN, DM | Pulmonary Edema; Carotid Artery Stenosis | Mar 2014 | 38 |
| MH454272 | 65 | Male | CPD | Fever | Jul 2015 | 168 |
| MH306207 | 31 | Female | SLE, HTN | DVT | Aug2015 | 37 |
| MH371127 | 36 | Male | HTN, DM, IHD | Pneumonia | Nov 2016 | 10 |
| MH359139 | 50 | Female | HTN, DM | Pneumonia | Nov 2016 | 30 |
| MH432120 | 68 | Male | HTN, BA | Exacerbation | Feb 2017 | 35 |
| MN120513 | 69 | Male | HTN, BPH, DM | Pneumonia | Mar 2019 | 91 |
| MN120514 | 58 | Male | HTN, RD | Pneumonia | Mar 2019 | 7 |
*BA, Bronchial Asthma; BPH, Benign Prostatic hyperplasia; CPD, Chronic Pulmonary Disease; DM, Diabetes Mellitus; HTN, Hypertension; RD, Renal Disease; SLE, Systemic Lupus Erythematosus; DVE, Deep Vein Thrombosis.
MERS-CoV genome annotation assembly
The next-generation sequencing (NGS) and Sanger sequencing data were preprocessed (base calling, base quality recalibration, alignment, and consensus sequence assembly) using Torrent Suite Server v 5.6 and Ion Torrent software. The nucleotide genome sequences obtained in this study were deposited in GenBank under the accession numbers MH013216, MN120513, MN120514, MH306207, MH359139, MH371127, MH432120 and MH454272.
Phylogenetic analyses of MERS-CoV
The complete genome sequences of MERS-CoVs obtained from the clinical isolates (n = 8) were aligned with the MERS-CoV sequences (both partial and complete) available in GenBank using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) program included in the MEGA v7 software package (www.megasoftware.net). Homologous basic local alignment search tool (BLAST) searches were carried out for each strain to determine the most closely related sequences.
To generate a composite phylogenetic tree, we randomly removed sequences to create a dataset proportional to these newly isolated sequences, keeping in mind that the homologous sequences remain in the final alignment. The final alignment contained 113 sequences from the complete genome phylogeny, including 79 from human cases and 34 of animal origin. The best-fit substitution model was selected using jModelTest [14] and employed in the maximum-likelihood analysis.
The maximum likelihood (ML) phylogenetic tree for the near-complete genomes (>30,000 nt in length) was inferred using the ML procedure in the RaxML version 8.2.10 package [15], employing the GTR + G nucleotide substitution model and 100 bootstrap replicates. Phylogenetic analyses of RF1a, ORF1ab and S gene segments were performed to determine whether any of these sequences exhibited recombination within different lineages.
Analysis of genomic variations
The nucleotide and amino acid sequences of each protein 8 MERS-CoV isolates were aligned with the corresponding human MERS-CoV and camel MERS-CoV protein sequences via multiple alignment in Molecular Evolutionary Genetics Analysis (MEGA) software. Amino acid changes were calculated using BioEdit (www.bioedit.com) with reference to Saudi isolate JX869059 (EMC/2012).
Recombination analysis
To detect possible recombination, a bootscan analysis was performed using Simplot version 3.5.1. Individual sequences from this study were aligned with reference MERS-CoV sequences representing different clades. The analysis was conducted using a sliding window of 200 nucleotides, a 20-bp step size and 100 bootstrap replicates through gap-stripped alignments and neighbour-joining analysis. Possible sites of recombination suggested by the boot scan analysis were confirmed through multiple sequence alignments.
Results
Baseline clinical characteristics of patients
Table 1 presents the baseline and clinical characteristics of the 8 studied patients infected with MERS-CoV. The mean age of the patients was 57 years, and they included 6 males and 2 females. Most of the patients had a history of hypertension, and 4 of the 8 had diabetes. All these patients were admitted to the intensive care unit (ICU) of KAMC, MNG-HA, and 4 of the patients died.
Phylogenetic analysis
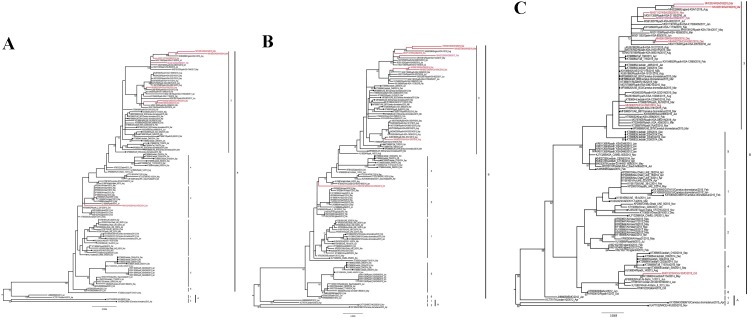
The evolutionary relationships of these newly sequenced Saudi MERS-CoV isolates were assessed in combination with the globally isolated sequences available in the GenBank database (Figs. 1,2 ). Seven of these isolates were assembled with lineage B3, while one strain belonged to the lineage B4 sequences in the complete genome phylogenetic analysis (Fig. 1). All the studied isolates were closely related to previous strains from Riyadh isolated during 2015−2017. Two of the 2019 strains and one 2017 strain specifically exhibited longer branch lengths due to a large number of substitutions. The only lineage B4 strain (MH013216) appeared in the complete genome tree together with a recombinant strain from Jeddah isolated from a camel. In the subgenomic phylogenetic analysis, all B3 strains remained in the same clade (Fig. 2A, 2B, 2C), whereas the B4 strains resembled the B2 strains more closely in the ORF1a gene phylogeny (Fig. 2A). Further analysis using the Simplot program [16] confirmed that MH013216 is a complex recombinant strain with contributions from multiple lineages of MERS-CoV (Fig. 3 ). The close similarity of MH013216 with B4 sequences was mainly located between nucleotides 16,500 and 24,000. In ORF1, however, the strain sequence matched parts of A2, B1, B2, and B3. The assembly of the strain within B2 occurred because of similar nucleotides at certain sites within lineages and an exclusive B2-like arrangement between nucleotides 12,000 and 12,500 (Fig. 3).
Fig. 1.
A near-complete phylogenetic analysis of Middle East respiratory syndrome coronavirus (MERS-CoV) genomes. This maximum likelihood tree was estimated from sequences >30,000 nt in length (n = 111), including those of six newly sequenced isolates from Saudi Arabia, using RAxML. The names of viruses sequenced in this work are shown in red. Camel viruses are indicated by bold circles, while all other samples are from human infections. Branch lengths reflect the number of nucleotide substitutions per site. Intra- and inter-lineage genetic distances (p-distance) are shown in the inset.
Fig. 2.
Phylogenetic analysis of the Middle East respiratory syndrome coronavirus (MERS-CoV) based on partial ORF1 (A), ORF2 (B), and S gene (C) regions. These maximum likelihood trees were estimated with the inclusion of six newly sequenced isolates from Saudi Arabia using RAxML. The names of viruses sequenced in this work are shown in red. Camel viruses are indicated by bold circles, while all other samples are from human infections. Branch lengths reflect the number of nucleotide substitutions per site. Intra- and inter-lineage genetic distances (p-distance) are shown in the inset.
Fig. 3.
Bootscan analysis demonstrating the complex recombination of MH013216. Mers-CoV strains representing lineages 1-6 of group B along with strains from group A were subjected to bootscan analysis across the complete genome using the Simplot program with a 200-bp window size, 20-bp step size, and 100 bootstrap replicates using gap-stripped alignments and neighbour-joining analysis. Vertical lines show the common estimated breakpoints of recombination in the analysed strains vs MH013216.
Mutational analysis
We compared the deduced amino acid sequences of the studied isolates with the reference JX869059 (EMC/2012) and searched for amino acid substitutions in MERS-CoV proteins that are probably associated with disease severity or transmissibility in humans. We were astounded to note a variety of unique mutations not detailed previously in MERS-CoV.
The majority of these mutations were found in ORF1ab (41 unique mutations), followed by the N protein (10 mutations), the S protein (9 mutations) and ORF4b (5 mutations) (supplement Table 2A and supplement Table 2B). In the ORF1ab replicase, mutations were quite evident in nsp3 (n = 17) (putative papain-like protease (PLpro)), nsp12 (n = 9) (putative RNA-dependent RNA polymerase (RdRp)), nsp13 (n = 6) (putative helicase (Hel)) and nsp14 (n = 6) (putative exonuclease (Exon)). Random mutations were also found in nsp4 and nsp6 (putative hydrophobic regions), nsp16 (putative S-adenosylmethionine-dependent 2′-O-ribose methyltransferase), and nsp1 and nsp2 (proteins of unknown function).
Characteristic nsp3 (PLpro) amino acid substitutions were observed at positions G981S, P1099R, and K1255R in 5/8 isolates and V1375I and M2119I in 4/8 isolates. Similarly, an nsp13 (Hel) substitution at position V5551A and annsp14 (Exon) substitution at position V6030 F were present in 5/8 isolates (supplement Table 2A).
The S protein of MERS-CoV is a heavily glycosylated type 1 transmembrane protein found on the surface of the virus, forming spikes consisting of receptor-binding subunit S1 and membrane fusion subunit S2 [7]. The noteworthy substitutions in the receptor-binding domain (RBD) of the S1 subunit were T424I, in 2/8 isolates, and S459 T, in 1/8 isolates. Another substitution, W553R, was found in the overlapping RBD/RBM (receptor binding motif) region in 2/8 isolates. The S2 subunit substitutions included S950 T in the fusion peptide, Q1009 L in hepatad repeat region 1 (HR1), and C1313S in the transmembrane (TM) region (supplement Table 2B).
N proteins are essential for the packaging of viral RNA into viable virus-like particles. The characteristic substitutions included V178A in the N-terminal domain (NTD), present in 6/8 isolates, and A300 V in the C-terminal domain (CTD), present in all studied isolates (Table 2 ).
Table 2.
Non-synonymous genomic variations observed in study isolates as compared to global sequences excluding the lineage-specific genomic variations.
| STRAIN | S | ORF (n) | M | N |
|---|---|---|---|---|
| MH013216 | P97 L, T424I, C1313S | S733R/E736 K, F1609 L, P1883S, L3785 F, M4574I, V5557A, V51Iδ | – | W293C, A300V** |
| MH454272 | W553R, R700 L, S950T | T649I, P777S, A1045 V, V1202I, E1835A, M2119I, P2742S, G3117E/V3120D | – | V178A, A300V** |
| F5011I, R5027S/D5028Y/R5029 K, G5061R, V5551A***, V6030 F, E102C, H243Cδ, H73Nδ | ||||
| MH306207 | – | C167 G, A1045 V, E1835A, M2119I, G3117E, L3785F | – | S11 F, L283 F, A300V** |
| A5362 L, V5551A*** | ||||
| MH371127 | – | V117D, G981S, P1099R, V1375I, A1462 T, P1971S, D2204A | – | V178A, A300V** |
| V5551A***, E102C | ||||
| MH359139 | W553R, R700L | T649I, P777S, A1045 V, V1202I, E1835A, M2119I, P2742S | – | V178A, A300V** |
| V5551A***, T5560S, V6030 F, E102C, H73Nδ | ||||
| MH432120 | – | T636I, G981S, P1099R, V1375I, A1462 T, P1971S, F3098 L/T3101N | T127I | G28 V, V178A, D242E, V263A, A300V** |
| F5072 L, V5551A***, I5559 M/T5560 F/G5561W/L5562 V/Y5563 V, I6668 M, V6431F | ||||
| G85D/P86 F/T87N*, E102C | ||||
| MN120513 | V26 L, S191 P, S459T | G981S, P1099R, K1255R, V1375I, A1462 T, H1714Y, P1971S, N4385 K, I4611 V, V5551A***, A5764S, M6272I, L6373R, L6958 P, V62F*, G85D/P86F*, A218S δ | P7L, V178A, G198S, A300V** | |
| MN120514 | V26 L, T424I, Q1009L | T586S, G981S, P1099R, V1375I, E1428 G, A1462 T, A1301 V, H1714Y, P1971S, S2111R, N4385 K, K5019 N, V6030 F, L6412 F, V62F*, P86L*, I147L δ, A218S δ | V178A, G198S, R292 P, A300V**, S391I, |
Other CTD amino acid substitutions observed were L293 F, present in 2/8 isolates, and V263A, R292 P, and W293C present in one isolate each. The substitutions in the linker region (LKR) of the N protein include G198S, which was present in 2/8 isolates, and D242E, which was present in one isolate. The N arm of the N protein included S11 F, which was present in 2/8 isolates, and P7L and G28 V, which were present in one isolate each. The C-tail of the N protein also contained a single amino acid substitution, S391I (supplement Table 2B).
Similar to other lineage C betacoronaviruses, MERS-CoV encodes five unique accessory proteins, designated 3, 4a, 4b, 5 and 8b [7]. We found a two-amino acid substitutions in ORF3, encoding a protein of unknown function, which was G85D/P86 F, in 3/8 isolates, and the single-amino acid substitutions V62 F, in 2/8 isolates, and T87 N, in one isolate. The ORF4a, 4b and M proteins have recently been found to antagonize type I INF production.
Four of the studied isolates exhibited the ORF4a substitution E102Q. The substitutions observed in ORF4b were H73 N and A218S, in 2/8 isolates, and V51I, I147 L, and H243Q, in 1 isolate each. Two of the isolates exhibited an amino acid substitution in the M protein (T127I), while one isolate exhibited a mutation in ORF5 (I98 M) (supplement Table 2B).
Discussion
Sporadic cases of MERS-CoV continue to occur in Middle Eastern regions, including Saudi Arabia. In this study, we conducted sequencing to determine the genome sequences of 8 MERS-CoV isolates. We performed evolutionary and genomic analyses to address several (among many unanswered) questions related to persistent MERS-CoV infection.
We found a number of genomic variations in the crucial regions of MERS-CoV, including the replication/transcription complex (RTC), the nucleocapsid protein N, the surface protein S, and ORF4a and ORF44b, involved in the host-mediated immune response. We identified a genomic recombinant isolate exhibiting characteristic substitutions representing almost all the group A and B MERS-CoV strains. Phylogenetic analysis resulted in the assembly of 7/8 sequences into 3 subclusters within lineage 3 of group B MERS-CoV. The longer branch lengths of these strains from the common node of their closest relatives in the phylogenetic tree were notable. New isolates were detected within a period of 6, 9, or 12 months after the detection of their closest relatives. One possible explanation for the longer branch length observed is the large number of substitutions found in these isolates, and another that we missed or could not detect were their close relatives during that period. Homology searches did not reveal any specific relatedness of MERS-CoV variants with SARS-CoV or MERS-CoV-2, in agreement with recently reported studies [1,21].
The recombination frequency in coronaviruses is quite high, and recombination is known to occur due to the exchange of functional motifs or even entire genes [17]. Some previous reports have also suggested the occurrence of recombination in association with host switching in many coronaviruses [[18], [19], [20]]. The MH013216 strain showed complex genomic recombination, including a major portion of the genome resembling strains of lineage 4; however, it harboured lineage-specific residues of both groups A and B of MERS-CoV. This strain was assembled within lineage 2 of the group B MERS-CoV strains in the subgenomic phylogenetic analysis of ORF1a.
The MERS-CoV 30 kb genome encodes structural proteins, accessory proteins, and two ORFs (ORF1a and ORF1b), from which two large polyproteins are translated and subsequently processed by the viral proteinase into nonstructural proteins (nsp) that together form RTCs [7]. The synthesis of genomic RNA (gRNA) and subgenomic messenger RNAs (sgmRNA) occurs at RTCs [8], involving interaction with the N protein [22,23]. Amino acid substitutions in the nsp proteins as well as the N protein are therefore crucial in the CoV life cycle. In fact, yeast two-hybrid (Y2H) assays of mouse hepatitis virus (MHV) and SARS-CoV revealed that nsp3 is the main RTC component that interacts with the N protein [24,25]. The direct interaction of the N protein with RTCs is considered critical, as impairment of this interaction greatly reduces CoV replication and progeny production [24].
We found amino acid substitutions in the nsp3, helicase (nsp13, an important enzyme for viral tropism and virulence), RDRp (nsp12), and exoribonuclease (nsp14) proteins. How these proteins (either individually or together) interact with the N protein in viral replication, sgmRNA production, and translation warrants functional studies. Most of the adaptive events in MERS-CoV in humans and camels [26] are now thought to occur through nsp3 via its deubiquitination and de-esterification activity to inhibit the IFN response [27].
Irrespective of the CoV strain involved, accessory ORFs have been implicated in the context of infection, specifically in the antagonization of the host response [28,29]. MERS-CoV ORF4a suppresses IFN production by binding to dsRNA [28], and the 4b gene product is reportedly associated with the evasion of host cell IFN defence mechanisms [30,31]. Amino acid substitutions in these proteins may impact the host immune response to the disease and, ultimately, its pathogenesis. The variant strains may have acquired high pathogenicity, but intense surveillance studies will be required to elucidate how this will be reflected in viral spreading and persistence in the future.
Similar to other coronaviruses, the MERS-CoV spike protein (S) is highly exposed to the virus surface and is the first line of contact in virus infection and the host immune response. Amino acid substitutions in the RBD/RBM region that play any role in virus binding to host DPP4 need to be investigated. The heptad repeat region (HR1) has recently been reported to be a major selection target among MERS-CoVs for their interspecies jump and spread to humans. One of the studied isolates presented the HR1 substitution Q1009L, and a nearby amino acid substitution, T1015 N, has been implicated in an increased MERS-CoV infection efficiency in vitro [32].
Although camels are thought to be the primary zoonotic reservoir responsible for human transmission, there is some evidence that bats might be the ancestral reservoir host for MERS-CoV [17]. Direct contact with dromedary camels or camel products has been reported in only 55 % of primary MERS-CoV cases, and the contact history in the remaining cases is unknown [33]. This study has some limitations, as we do not have samples from camels or other domestic and wild animals to confirm the possible contact history of the disease in our patients.
We identified novel point mutations and mutation complexes, indicating a changing epidemiology of MERS-CoV, potentially raising the alarm regarding new viral outbreaks and associated morbidity and mortality. The search for intermediate hosts in MERS-CoV evolution is necessary, as are functional studies to determine the pathogenic potential of such substitutions, which may aid in the screening of patients at higher risk.
Conclusion
In conclusion, the results of this study lead to the recommendation that the complete genome sequencing of all new cases of MERS-CoV to monitor virus evolution. A clear understanding of the recombination events involved in MERS-CoV evolution would aid in developing effective tools for controlling new human coronavirus infections worldwide.
Author contributions
MAB designed and supervised the study, including contributing to the collection, analysis and interpretation of the data and writing; MD and BM contributed to the wet lab work, interpretation of the data and writing; AK, IA and UG contributed to the analysis and interpretation of the data and writing; YA, SJ and MG contributed to the data collection, analysis, and interpretation and writing; AH initiated the manuscript writing and description of the data; AA and AAJ contributed to writing the manuscript.
Funding
This work was supported by King Abdullah International Medical Research Center, Saudi Arabia (RC16/094/R).
Competing interests
The authors have no conflicts of interest to declare.
Ethical approval
This study was ethically approved by the International Review Board (IRB) committee, King Abdullah International Medical Research Center, Riyadh, Saudi Arabia.
Acknowledgements
The authors thank the family members for their participation in this study. We are grateful to Ms. Zoe Poral for her administrative assistance in proofreading and editing the manuscript.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jiph.2020.06.030.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhanet W. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020 doi: 10.1101/2020.01.22.914952. [DOI] [Google Scholar]
- 2.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211724. https://www.who.int/csr/sars/country/table2004_04_21/en/ WHO. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. 2003; [DOI] [PubMed] [Google Scholar]
- 4.WHO . 2019. MERS situation update.http://applicationsemrowhoint/docs/EMRPUBCSR-241-2019-ENpdf?ua=1&ua=1&ua=1 [Google Scholar]
- 5.van Boheemen S., deGraaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;20:3. doi: 10.1128/mBio.00473-12. https://doi:10.1128/mBio.00473-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y.H., Chen Y.H., Wu N.P., Yao H.P., Li L.J. Molecular characteristics, functions, and related pathogenicity of MERS-CoV proteins. Engineering. 2019;5:940–947. doi: 10.1016/j.eng.2018.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McBride R., Fielding B.C. The role of severe acute respiratory syndrome (SARS) coronavirus accessory proteins in virus pathogenesis. Viruses. 2012;4:2902–2923. doi: 10.3390/v4112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau S.K., Wernery R., Wong E.Y., Joseph S., Tsang A.K., Patteril N.A. Polyphyletic origin of MERS coronaviruses and isolation of a novel clade A strain from dromedary camels in the United Arab Emirates. Emerg Microbes Infect. 2016;5:e128. doi: 10.1038/emi.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudas G., Rambaut A. MERS-CoV recombination: implications about the reservoir and potential for adaptation. Virus Evol. 2016;2:vev023. doi: 10.1093/ve/vev023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assiri A.M., Midgley C.M., Abedi G.R., Bin Saeed A., Almasri M.M., Lu X. Epidemiology of a novel recombinant middle east respiratory syndrome coronavirus in humans in Saudi Arabia. J Infect Dis. 2016;214:712–721. doi: 10.1093/infdis/jiw236. https://doi:10.1093/infdis/jiw236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alnazawi M., Altaher A., Kandeel M. Comparative genomic analysis MERS CoV isolated from humans and camels with special reference to virus encoded helicase. Biol Pharm Bull. 2017;40:1289–1298. doi: 10.1248/bpb.b17-00241. [DOI] [PubMed] [Google Scholar]
- 13.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G. Fulllength human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anthony S.J., Gilardi K., Menachery V.D., Goldstein T., Ssebide B., Mbabazi R. Further evidence for bats as the evolutionary source of middle east respiratory syndrome coronavirus. mBio. 2017;8 doi: 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du J., Yang L., Ren X., Zhang J., Dong J., Sun L. Genetic diversity of coronaviruses in Miniopterus fuliginosus bats. Sci China Life Sci. 2016;59:604–614. doi: 10.1007/s11427-016-5039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau S.K., Feng Y., Chen H., Luk H.K., Yang W.H., Li K.S. Severe acute respiratory syndrome (SARS) coronavirus ORF8 protein is acquired from SARS-Related coronavirus from greater horseshoe bats through recombination. J Virol. 2015;89:10532–10547. doi: 10.1128/JVI.01048-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood) 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 20.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verheije M.H., Hagemeijer M.C., Ulasli M., Reggiori F., Rottier P.J., Masters P.S. The coronavirus nucleocapsid protein is dynamically associated with the replicationtranscription complexes. J Virol. 2010;84:11575–11579. doi: 10.1128/JVI.0056910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol. 2019 doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurst K.R., Koetzner C.A., Masters P.S. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase transcriptase complex. J Virol. 2013;87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forni D., Cagliani R., Mozzi A., Pozzoli U., Al-Daghri N., Clerici M. Extensive positive selection drives the evolution of nonstructural proteins in lineage C betacoronaviruses. J Virol. 2016;90:3627–3639. doi: 10.1128/JVI.02988-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siu K.L., Yeung M.L., Kok K.H., Yuen K.S., Kew C., Lui P.Y. Middle east respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol. 2014;88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthews K.L., Coleman C.M., van der Meer Y., Snijder E.J., Frieman M.B. The ORF4bencoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J Gen Virol. 2014;95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y., Ye F., Zhu N. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci Rep. 2015;5:17554. doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forni D., Filippi G., Cagliani R., De Gioia L., Pozzoli U., Al-Daghri N. The heptad repeat region is a major selection target in MERS-CoV and related coronaviruses. Sci Rep. 2015;5:14480. doi: 10.1038/srep14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banerjee A., Kulcsar K., Misra V., Frieman M., Mossman K. Bats and coronaviruses. Viruses. 2019;11(1) doi: 10.3390/v11010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.