Highlights
-
•
A visualized LAMP rapid detection method for PPV infection was established.
-
•
The limit of detection (LOD) for PPV by LAMP is 10 copies.
-
•
This method is specific, sensitive and suitable for PPV detection in the field.
Abbreviations: LAM, Ploop-mediatedisothermal amplification; ORF, openreading frame; PPV, porcineparvovirus; PCV2, porcinecircovirus type 2; PRRSV, porcinereproductive and respiratory syndrome virus; PRV, porcinepseudo rabies virus; CSFV, classicalswine fever virus; LOD, limit of detection; NTC, no template control
Keywords: Porcine parvovirus, Virus detection, LAMP
Abstract
Porcine parvovirus (PPV) is one of the major causes of reproductive pig disease. Due to its serious nature, wide spread and consequent great damage to the swine industry, an effective, rapid and convenient method for its detection is needed. A loop-mediated isothermal amplification (LAMP) assay was established to detect PPV infection. Two pairs of primers were specifically designed to recognize the six different sequences of open reading frame1 (ORF1) gene. The optimized LAMP program was as follows: 50 min at 59 °C followed by 3 min at 80 °C.The amplified products were analyzed both by visual inspection after staining with SYBR Green I dye and by conventional agarose gel electrophoresis. Both methods showed the same sensitivity. The limit of detection (LOD) for PPV by LAMP was 10 copies, which is 100-fold lower than conventional PCR. Our LAMP assay did not cross-react with other viruses. We used the established LAMP system to test 1100 field samples and detected 660 positives. The LAMP detection method for PPV represents a visual, sensitive and rapid assay which can detect the virus in the field, offering an attractive alternative for the PPV detection methods currently in use.
1. Introduction
Porcine parvovirus (PPV), a virus belonging to the Parvoviridae family, causes maternal reproductive failure of swine known as porcine reproductive system disease which is a serious problem in the pig breeding industry. The characteristics of PPV infection in infected sows (especially primiparous sows) are stillbirth, fetal malformation and mummification, but the infection also can cause neonatal death and piglet disease including diarrhea and dermatitis (Yin and Liu, 1997). All kinds of pigs can be infected by PPV, such as domestic pigs, wild boar, newborn piglets, finishing pigs and SPF pigs. However, the pregnant sow itself and some infected pigs do not have evident clinical symptoms (Kennedy et al., 2000; Ellis et al., 2000).
PPV has caused huge losses to the pig industry. Therefore, an effective method is necessary to detect the PPV infection. Currently, conventional PCR is used to detect and identify the virus (Caprioli et al., 2006; Huang et al., 2004; Jiang et al., 2010). But its amplification efficiency is affected by many disturbing inhibitors (Wilson, 1997; Abu and Rådström, 1998). Enzyme linked immunosorbent assay (ELISA) is also a common way to detect PPV (Jenkins, 1992). However, infected swine are difficult to diagnose by this method because they are prone to false-positive results during the analytical process (Westenbrink et al., 1989).Though the common PCR, ELISA and Real-time PCR methods (Zheng et al., 2013; Pérez et al., 2012; Chen et al., 2009a) are also suitable for the qualitative and quantitative analysis of PPV, it requires highly skilled laboratory technicians. Therefore, an alternative quick, accurate and simple method is still needed to detect PPV.
Some years ago, a novel nucleic acids amplification technique was introduced which was called loop-mediated isothermal amplification (LAMP) (Notomi et al., 2000). The method is simple and extremely specific to the target sequence, since the four primers can identify the six target sequences and amplify it (Mori et al., 2001; Zhang et al., 2010). Compared with other detection methods, the LAMP method has many advantages, in particular specificity, sensitivity and rapidity. The LAMP products have a typical ladder-like pattern and can be detected by adding SYBR Green I dye (Zhang et al., 2010; Iwamoto et al., 2003). The LAMP amplification solution can be visually turned to green in the presence of a dye SYBR Green I, while the LAMP solution remains orange in the absence of amplification (Iwamoto et al., 2003). The LAMP method has become a useful assay for the fast detection of food borne pathogenic microorganisms and infectious diseases (He et al., 2016). Other examples are the detection of heat-labile I and heat-stable I enterotoxin genes of enterotoxigenic Escherichia coli by LAMP (Yano et al., 2007).
In this study, a detection method based on the LAMP technology is described which is suitable for the clinical detection of porcine parvovirus. The diagnostic kit was developed, tested and applied. The present study offers the necessary technological basis for the prevention and control of porcine parvovirus infection.
2. Material and methods
2.1. Viral materials
The viral strains used for the LAMP assays were obtained from the Institute of Animal Husbandry and Veterinary Science, Shanghai Academy of Agricultural Sciences. Porcine Parvoviruse (PPV), Classical swine fever virus (CSFV), Porcine circovirus type 2(PCV2), porcine pseudorabies virus (PRV) and porcine reproductive and respiratory syndrome virus (PRRSV) were included. The geographical origin, and year of isolation of these viruses were summarized in Table 1 . Pig sera were gathered from a slaughterhouse in Shanghai (China) and used as clinical samples for the detection of PPV by LAMP.
Table 1.
Origins of virus strains tested in this study.
| Virus name | Strain name | Country of isolation | Year of isolation |
|---|---|---|---|
| PPV | S-1 | China | 1983 (Pan et al., 1983) |
| PPV | S-2 | China | 1985 (Pan et al., 1985) |
| PPV | NJ | China | 2012 (Zhang et al., 2012) |
| CSFV | C | China | 1955−1956(Wang et al., 2000) |
| PCV2 | SH | China | 2006 (Guo et al., 2010) |
| PRV | Bartha K61 | Hungary | 1961 (Yuan et al., 1983) |
| PRRSV | ATCC VR2332 | USA | 1992 (Collins et al., 1992) |
2.2. DNA and RNA extraction and purification
DNA was extracted from PPV, PCV2 and PRV by the Blood Viral DNA/RNA kit (BIOMIGA Inc, San Diego, CA). The DNA from PPV obtained in the previous step was used as template to optimize the test reaction temperature.
RNA from PRRSV and CSFV was extracted following the same method as the DNA. The process from RNA to cDNA was achieved by reverse transcription (Takara Corp., Japan). These cDNA templates were used for the next specific experiment.
2.3. Primer design
Two pairs of primers were designed by Primer Explorer 3 based on the PPV VP1 gene (capsid protein 1) gene of PPV genome (https://www.ncbi.nlm.nih.gov/ nuccore KF913351.1). They were called FIP, BIP, F3 and B3 and the information of them were shown in Table 2 . The F3 and B3 primers were used in the PCR reaction and the target sequence was 201 bp.
Table 2.
VP1 gene-derived primers for LAMP detection of PPV.
| Primers | Sequences | Location |
|---|---|---|
| PPV-F3 | 5′- ACGGAGGTAAAATTGGACA -3′ | 2586−2604 |
| PPV-B3 | 5′- TCCCTTTAGCTTTTTTTTTAGC-3′ | 2765−2786 |
| PPV-FIP | 5′- GAGATGTAGTTGGTGAGTCTGTTTTTTTTTCTTCAGAGCAAAGCGTG -3′ | 2649−2672+TTTT + 2609−2627 |
| PPV-BIP | 5′- CAACCAGAGGTAAGAAGATCGCTTTTAAAAATATGTCTTGGAGCAGGT -3′ | 2675−2696+TTTT +2734−2755 |
2.4. Conventional PCR assay and plasmid construction
PCR assays were performed in 25 μL reaction volumes containing 2.5 μL 10×buffer (TaKaRa), 0.2 mM dNTPs, 0.4 μM each of F3 and B3, 2.5 U Taq DNA polymerase (TaKaRa biotechnology Co., Ltd, Dalian, China) and 1 μL template DNA. The program consisted of an initial denaturation at 95 °C for 5 min, followed by 35 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 5 min in an Applied Biosystem 2720 thermal cycler (Applied Biosystem., US). The PCR products were sequenced and analyzed.
Meanwhile, the 201bp PCR products were purified using the QIAquick PCR purification kit (Qiagen, Germany),following the manufacturer's instructions. Then the fragments were cloned into pEASY®-T3 Cloning Vector and transformed into Trans1-T1 competent cell using pEASY®-T3 Cloning Kit (TransGen Biotech Co., Ltd., Beijing, China). The positive plasmid were obtained according to blue/white selection and identified by colony PCR and sequencing. The resulting positive plasmid containing VP1 gene fragment of PPV was extracted for further experiments.
2.5. Reaction protocol of LAMP
LAMP reactions were performed in volumes of 25 μL, which contained 2.5 μL 10×buffer (-Mg2+)(TaKaRa), 0.48 mM Mg2+, 0.4 mM dNTPs, 0.4 μM each of FIP and BIP, 0.8 μM each of F3 and B3, 1 M betaine (Sigma), and 9.6 U Bst DNA polymerase (Vazyme Biotech Co.,Ltd). After adding 1 μL template DNA, the mixture was incubated for 5 min at 95 °C and cooled on ice for 5 min, after which the Bst polymerase was added. The LAMP reaction was performed in a conventional heating block.
2.6. Temperature optimization
To optimize the reaction temperature, LAMP was carried at 54 ℃, 54.5 ℃, 55.2 ℃, 56.1 ℃, 57.5 ℃, 58.7 ℃, 59.6 ℃, 60.9 ℃, 62.1 ℃, 63.1 ℃ 63.7 ℃ and 64 ℃ for 50 min and terminated at 80℃ for 3 min, respectively. The reaction system was the same as method 2.5 mentioned above, and the template was plasmid containing PPV VP1 gene fragment with the concentration 104 copies per μL. Two reactions with and without inner set of primers were carried out at 59.0 ℃ as control.
2.7. Specificity of the LAMP method
To verify the specificity of PPV detection by LAMP, the DNA (PPV, PCV2, PRV) and cDNA samples (PRRSV, CSFV) were amplified as sample templates by the LAMP reaction at 59.0 ℃ for 50 min and terminated at 80 ℃ for 3 min, respectively. As a positive control we used the PPV VP1 plasmid; as a no template control (NTC) water was used. All reactions were repeated in duplicates.
2.8. The sensitivity of LAMP and PCR
Ten-fold serial dilutions of the PPV plasmid were made to obtain a gradient of 107 to 1 copy per μL. These plasmids of different concentration were prepared to define the limit of detection (LOD) of PPV DNA by LAMP and PCR assays. The amplification products were checked on agarose gel electrophoresis. In addition, the LAMP products were also checked by adding a dye SYBR Green I.
2.9. Analysis of LAMP and PCR products
LAMP and PCR amplified products were detected by 2 % (w/v) agarose gel electrophoresis and observed by staining with Goldview (SBS Genetech Co., Ltd., Shanghai). The LAMP products were also directly observed depending on their color by mixing each sample with 2 μL 1:10-diluted SYBR Green I (Thermo Fisher Scientific, USA). The positive sample will turn green while the negative sample will still remain orange.
2.10. Detection of PPV in clinical samples
1100 whole blood samples were collected as random from "duroc × landrace × Yorkshire" pigs with the weight of 105−120 kg and 170–175 days old. These blood samples were placed at 4 ℃ for 6 h, then the sera were separated by centrifugation at 3000 r/min for 5 min. 1100 serum samples were tested using the LAMP and PCR method for the presence of PPV to determine if their source were infected with PPV.
We also compared the LAMP results of straightly using the serum samples with the DNA extraction by the Blood Viral DNA/RNA kit. 3 samples of serum were used as template in LAMP reactions, meanwhile the genomic DNA of these 3 serum samples were extracted as template to perform this experiment. All reactions were carried out at 59.0 ℃ for 50 min and terminated at 80 ℃ for 3 min, respectively.
3. Results
3.1. Temperature optimization of the PPV LAMP method
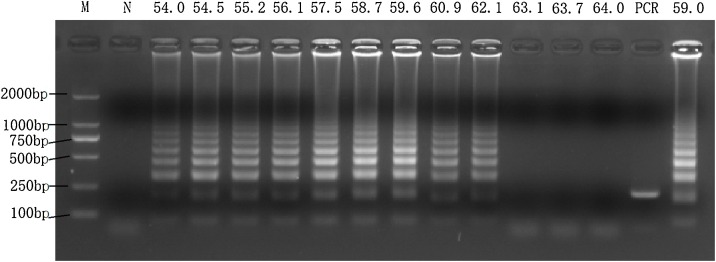
The optimum temperature for the LAMP reaction was determined to be ±59 °C (Fig. 1 ). At this temperature, the typical ladder-like pattern was the brightest and clearest, corresponding to the highest amount of product. The comparison between reactions with and without inner set primers confirmed the feasibility of this LAMP protocol.
Fig. 1.
Optimal temperature of the LAMP assay for detecting PPV.
M: DL2000 DNA marker; N: no template control; LAMP reactions were carried out at 54 ℃, 54.5 ℃, 55.2 ℃, 56.1 ℃, 57.5 ℃, 58.7 ℃, 59.6 ℃, 60.9 ℃, 62.1 ℃, 63.1 ℃, 63.7 ℃ and 64 ℃, respectively. The other 2 reactions with and without inner set of primers were carried out at 59.0 ℃ as control. Plasmids containing PPV VP1 gene fragment with the concentration 104 copies per μL were as template. All reactions were incubated at above temperature for 50 min and terminated at 80 ℃ for 3 min. The amplification products were detected by 2 % (w/v) agarose gel electrophoresis staining with Goldview.
3.2. Specificity of the LAMP method
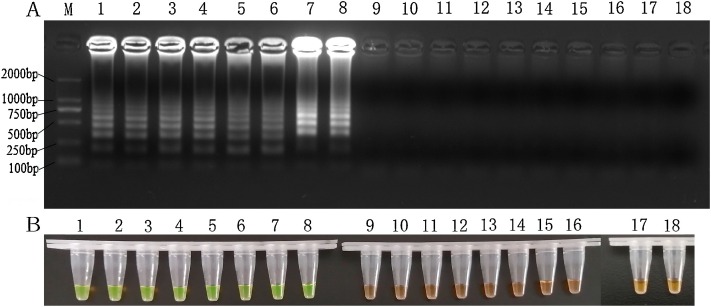
In this assay, only the genomic DNA samples of 3 PPV strains and PPV plasmid (positive control) used as template gave the typical ladder-like bands and a green color, while the DNA/cDNA samples obtained for other virus species as well as the NTC had no bands and showed an orange color (Fig.2 A and B). The results indicated the primers could only amplify PPV nucleic acid. Consistently, the length of the amplified product was 201 bp as predicted. Its sequence was confirmed to be 100 % identical to the corresponding sequence in the PPV VP1 gene.
Fig. 2.
Specificity of the PPV LAMP Assay.
M: DL2000 DNA marker; Lane1 and 2: PPV S-1 strain; Lane 3 and 4: PPV S-2 strain; Lane 5 and 6: PPV NJ strain; Lane 7 and 8: positive control; Lane 9 and 10: PCV2; Lane 11and 12: PRRSV; Lane 13 and 14: PRV; Lane 15 and 16: CSFV; Lane 17 and 18: no template control. All reactions were carried out at 59.0 ℃ for 50 min and terminated at 80 ℃ for 3 min. The concentration of DNA/RNA template of virus samples is 10 ng per μL. Plasmids containing PPV VP1 gene fragment with the concentration 104 copies per μL were as positive control.
Results on 2 % (w/v) agarose gel electrophoresis staining with Goldview.
Visual results by adding SYBR Green I.
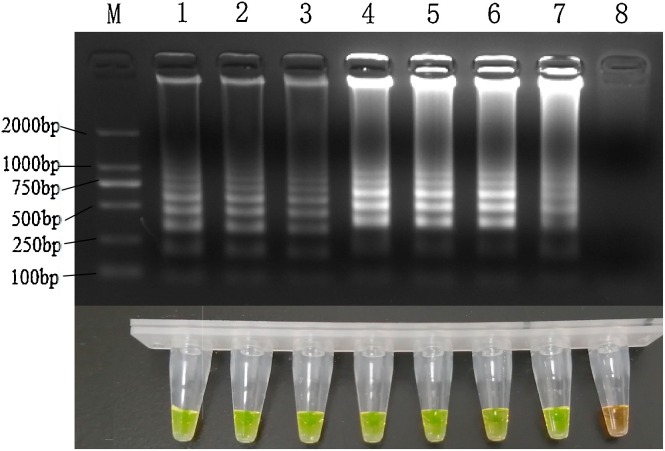
3.3. Sensitivity of LAMP and PCR
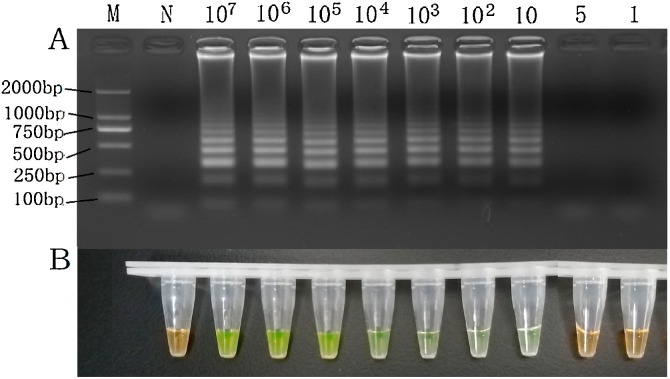
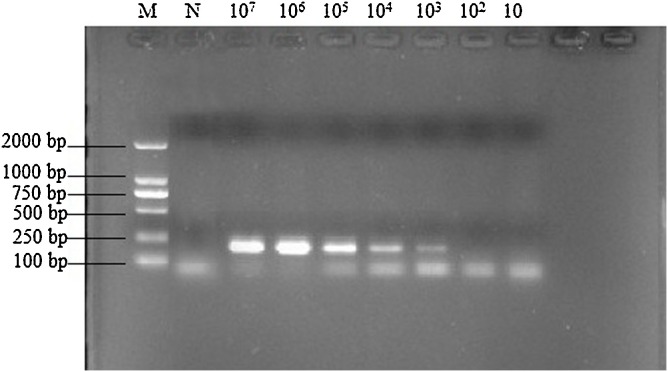
The lower LOD of PPV by LAMP was found to be 101 copies based on the results of the agarose gel electrophoresis analysis (Fig. 3 A) and visual observation (Fig. 3B). In contrast, the LOD for the PCR was 103copies (Fig. 4 ). These results indicate that the LAMP method is about 100 times more sensitive than the conventional PCR assay and this sensitivity is in line with the daily testing requirements.
Fig. 3.
Sensitivity of the PPV LAMP Assay.
M: DL2000 DNA marker; N: no template control; LAMP was carried out with 107 to 1 copy of plasmids of PPV as template, respectively.
All reactions were carried out at 59.0 ℃ for 50 min and terminated at 80 ℃ for 3 min.
Results on 2 % (w/v) agarose gel electrophoresis staining with Goldview.
Visual results by adding SYBR Green I.
Fig. 4.
Sensitivity of the PPV PCR Assay.
M: DL2000 DNA marker; N: no template control; PCR was carried out at 107 to 101 copies of plasmids of PPV as template, respectively. The amplification products were detected by 2 % (w/v) agarose gel electrophoresis staining with Goldview.
3.4. PPV recognition in clinical samples
A total of 1100 serum samples were tested using the LAMP and PCR methods to determine whether their sources were infected by PPV. Among them, 660 clinical samples were found to be positive by LAMP and 623 samples were positive by PCR (Table 3 ). In summary, 620 serum samples were detected positive and 437 samples were negative by both LAMP and PCR. The coincidence rate of these two methods was 96.1 % (1057/1100) for clinical samples detection. The comparison between serum template and genomic DNA template showed that DNA extraction is not necessary and can be omitted. Using the serum samples straightly as template doesn’t affect the LAMP results (Fig. 5 ).
Table 3.
Detection results of 1100 serum samples by LAMP and PCR.
| LAMP | PCR | Coincidence rate | ||
|---|---|---|---|---|
| + | – | |||
| + | 660 | 620 | 40 | 96.1 % (1057/1100) |
| – | 440 | 3 | 437 | |
+ Positive; − Negative.
Fig. 5.
Comparison between samples of genomic DNA and serum for the LAMP assay.
M: DL2000 DNA marker; Lane 1, 2 and 3: LAMP reactions were carried out with 3 samples of serum, respectively; Lane 4, 5 and 6: LAMP reactions were carried out with genomic DNA of the same 3 samples, rspectively; Lane 7: positive control; Lane 8: no template control.
All reactions were carried out at 59.0 ℃ for 50 min and terminated at 80 ℃ for 3 min. The concentration of genomic DNA templates of serum samples is 10 ng per μL. Plasmids containing PPV VP1 gene fragment with the concentration 104 copies per μL were as positive control.
Upper: Results on 2 % (w/v) agarose gel electrophoresis staining with Goldview.
Lower: Visual results by adding SYBR Green I.
4. Discussion
In this study, we developed a method for the visual and rapid detection of PPV using an optimized LAMP technique. LAMP has a number of advantages when compared to PCR, particularly its high sensitivity, easy manipulation in addition to its visual and time saving detection.
We investigated the optimized PPV LAMP method and observed its high specificity for PPV, showing no amplification products for any of the other viruses tested. Importantly, two pairs of primers were used to identify the target gene by LAMP (Nagamine et al., 2002), while only one pair of primers was used in conventional PCR. Our LAMP method has a high specificity because it targets the conserved region of PPV ORF1 gene in the design of the two pairs of primers.
The LAMP reaction can be carried out under isothermal conditions in a relatively short time, without specific equipment like a PCR thermo cycler. The LAMP reaction took only 50 min total time and did not need either intricate pretreatment or an expensive apparatus. The reaction was more quickly than the enzyme linked immunosorbent assay and the real-time PCR. Hence, the LAMP method we developed has the advantages of easy manipulation and easy popularization.
The LAMP detection limit for PPV based on visual observation by addition of SYBR Green I and by gel electrophoresis analysis was 101 copies. The sensitivity of detection by LAMP was 100 times higher than by conventional PCR. This indicated that the visual observation method could be used to analyze the LAMP product and could reliably replace the conventional agarose gel electrophoresis (Li et al., 2013). The sensitivity of the LAMP method is in line with reports about other virus species, such as swine transmissible gastroenteritis coronavirus, H5 avian influenza virus and yellow head virus (Chen et al., 2010; Imai et al., 2006; Mekata et al., 2006).
In the analysis of clinical samples, DNA extraction and purification steps were not needed. Serum samples can be used straightly as templates for the LAMP reaction. The sensitivity of the LAMP method was less affected by the composition of the clinical samples than observed with PCR. This feature not only can decrease the time and cost of the LAMP reaction, but also can simplify many troublesome programs. For the evaluation of clinical samples, we tested randomly 1100 sera by LAMP and PCR, and verified this feature. Compared to conventional PCR, the detection rate of PPV by ELISA was 5.91 % (Jenkins, 1992). Furthermore, ELISA is known to easily cause false-positive results (Westenbrink et al., 1989). Moreover, the ELISA method is troublesome and time-consuming in contrast of the LAMP method. Although the LOD by real-time PCR is 10 times lower than by conventional PCR, the detection rate of real-time PCR for PPV is only 55.56 %–60 % (Zheng et al., 2013; Chen et al., 2009b). LAMP method can make it applicable to laboratories, small-scale hospitals, private clinics and pig industry.
For a reliable LAMP test, some precautions should be adopted to prevent the occurrence of false positive results. For example, separate work areas and aerosol-resistant pipette tips should be used. Meanwhile, the used pipette tips and reaction vessels should also be collected in airtight containers. Moreover, it is advisable to divide reagents into aliquots in order to avoid contaminations. Notably, negative control samples should be firstly finished as soon as possible when total reaction system is finished to aliquot.
A LAMP method for PPV has been developed successfully by others (Chen et al., 2009a; Liu et al., 2010; Qu et al., 2010). Chen et al. chose to amplify the VP2 gene of PPV by using a set of four primers at 62 ℃ for 45 min. In this study, we selected four different primers to amplify the VP1 gene and used the SYBR Green I dye for the detection of PPV. Although both LAMP methods were established using specific primers based on the highly conserved PPV NS1 protein gene(Qu et al., 2010), the primers we designed are in different region of NS1 from them. We had aligned many PPV genome sequences and chose the most conserved region for primer design. Furthermore, we combined with the dye SYBR Green I and realized the visual detection for PPV LAMP instead of by the conventional gel electrophoresis analysis or fluorescent detection (Li et al., 2013).
In addition, DNA extraction was omitted in order to save time. In the present study, porcine serum as sample can be used straightly in the LAMP assay without DNA extraction, as the LAMP reaction is well tolerant against biological substances. Therefore, in the LAMP assay the DNA extraction step can be ignored (Kaneko et al., 2007). Thus, the LAMP method can be used to detect PPV in the field without the need of a PCR thermocycle instrument, electrophoresis apparatus or turbidimeter. Hence, our LAMP protocol provides an attractive new method for the detection of PPV. The results of this study illustrate that LAMP detection offers a convenient visual approach to detect PPV rapidly, sensitively, specifically and simply. Above all, the LAMP method was improved from the point of high reaction efficiency and accurateness. For PPV detection, it supplements and extends the former approach; the method was developed into a diagnostic kit that is well received and applied in the field.
Funding
This study was funded by the Science and Technology Commission of Shanghai Municipality, project grant No. 18391902000and 14140900702, and the Minhang Science Commission of Shanghai Municipality,project grant no. 2014MH076.
Data availability
All relevant data are within the paper and its supporting information.
CRediT authorship contribution statement
Kai Zhao: Data curation, Formal analysis, Funding acquisition, Methodology, Supervision, Visualization, Writing - review & editing. Ruili Hu: Formal analysis, Methodology, Investigation, Visualization, Writing - original draft. Jianping Ni: Project administration, Investigation, Resources. Jieling Liang: Investigation, Visualization. Xizhong He: Investigation, Resources. Yanan Du: Investigation, Validation, Writing - original draft. Yan Xu: Methodology, Writing - review & editing. Binan Zhao: Methodology, Writing - review & editing. Qi Zhang: Formal analysis, Investigation, Validation. Chunhua Li: Conceptualization, Data curation, Funding acquisition, Methodology, Resources, Supervision, Validation, Writing - review & editing.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
We are grateful to Professor Peter J.M. Rottier for editing the manuscript.
References
- Abu A.W., Rådström P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl. Environ. Microbiol. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli A., McNeilly F., McNair I., Lagan-Tregaskis P., Ellis J., Krakowka S., McKillen J., Ostanello F., Allan G. PCR detection of porcine circovirus type 2 (PCV2) DNA in blood, tonsillar and faecal swabs from experimentally infected pigs. Res. Vet. Sci. 2006;81:287–292. doi: 10.1016/j.rvsc.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Chen H.T., Zhang J., Yang S.H., Ma L.N., Ma Y.P., Liu X.T., Cai X.P., Zhang Y.G., Liu Y.S. Rapid detection of porcine parvovirus DNA by sensitive loop-mediated isothermal amplification. J. Virol. Methods. 2009;158:100–103. doi: 10.1016/j.jviromet.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.Y., Li X.K., Cui B.A., Wei Z.Y., Li X.S., Wang Y.B., Zhao L., Wang Z.Y. A TaqMan-based real-time polymerase chain reaction for the detection of porcine parvovirus. J. Virol. Methods. 2009;156:84–88. doi: 10.1016/j.jviromet.2008.10.029. [DOI] [PubMed] [Google Scholar]
- Chen Q., Li J., Fang X.E., Xiong W. Detection of swine transmissible gastroenteritis coronavirus using loop-mediated isothermal amplification. Virol. J. 2010;7:206. doi: 10.1186/1743-422X-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J.E., Benfield D.A., Christianson W.T., Harris L., Hennings J.C., Shaw D.P., Goyal S.M., McCullough S., Morrison R.B., Joo H.S., Gorcyca D., Chladek D. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Invest. 1992;4:117–126. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- Ellis J.A., Bratanich A., Clark E.G., Allan G., Meehan B., Haines D.M., Harding J., West K.H., Krakowka S., Konoby C., Hassard L., Martin K., McNeilly F. Coinfection by porcine circoviruses and porcine parvovirus in pigs with naturally acquired postweaning multisystemic wasting syndrome. J. Vet. Diagn. Invest. 2000;12:21–27. doi: 10.1177/104063870001200104. [DOI] [PubMed] [Google Scholar]
- Guo L., Lu Y., Wei Y., Huang L., C Liu. Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol. J. 2010;7:273. doi: 10.1186/1743-422X-7-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Xue F., Xu S., Wang W. Rapid and sensitive detection of Lily symptomless virus by reverse transcription loop-mediated isothermal amplification. J. Virol. Methods. 2016;238:38–41. doi: 10.1016/j.jviromet.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Huang C., Hung J.J., Wu C.Y., Chien M.S. Multiplex PCR for rapid detection of pseudorabies virus, porcine parvovirus and porcine circoviruses. Vet. Microbiol. 2004;101:209–214. doi: 10.1016/j.vetmic.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Imai M., Ninomiya A., Minekawa H., Notomi T., Ishizaki T., Tashiro M., Odagiri T. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine. 2006;24:6679–6682. doi: 10.1016/j.vaccine.2006.05.046. [DOI] [PubMed] [Google Scholar]
- Iwamoto T., Sonobe T., Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. Intracellulare in sputum samples. J. Clin. Microbiol. 2003;41:2616–2622. doi: 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins C.E. An enzyme-linked immunosorbent assay for detection of porcine parvovirus in fetal tissues. J. Virol. Methods. 1992;39:179–184. doi: 10.1016/0166-0934(92)90136-2. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Shang H., Xu H., Zhu L., Chen W., Zhao L., Fang L. Simultaneous detection of porcine circovirus type 2, classical swine fever virus, porcine parvovirus and porcine reproductive and respiratory syndrome virus in pigs by multiplex polymerase chain reaction. Vet. J. 2010;183:172–175. doi: 10.1016/j.tvjl.2008.11.016. [DOI] [PubMed] [Google Scholar]
- Kaneko H., Kawana T., Fukushima E., Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods. 2007;70:499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Kennedy S., Moffett D., McNeilly F., Meehan B., Ellis J., Krakowka S., Allan G.M. Reproduction of lesions of post weaning multisystem wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J. Comp. Pathol. 2000;122:9–24. doi: 10.1053/jcpa.1999.0337. [DOI] [PubMed] [Google Scholar]
- Li B., Sun B., Du L.P., Mao A.H., Wen L.B., Ni Y.X., Zhang X.H., He K.W. Development of a loop-mediated isothermal amplification assay for the detection of porcine hokovirus. J. Virol. Methods. 2013;193:415–418. doi: 10.1016/j.jviromet.2013.06.040. [DOI] [PubMed] [Google Scholar]
- Liu Y.B., Zhang L., Ning Y.B., Wang Q., Fan X.Z. Establishment of porcine parvovirus detection by loop-mediated isothermal amplification assay. Scientia Agri. Sinica. 2010;43:3012–3018. [Google Scholar]
- Mekata T., Kono T., Savan R., Sakai M., Kasornchandra J., Yoshida T., Itami T. Detection of yellow head virus in shrimp by loop-mediated isothermal amplification (LAMP) J. Virol. Methods. 2006;135:151–156. doi: 10.1016/j.jviromet.2006.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Liu H., Yan Z., Tang Q. Isolation and identification of porcine parvovirus S-1 strain (in Chinese) Shanghai J. Animal. Husbandry Vet. Med. 1983;1:1–3. [Google Scholar]
- Pan X., Ye X., Yan Z., Tang Q., Liu H., Yang S., Zhang W. Isolation and identification of porcine parvovirus S-2 strain (in Chinese) Chin. J. Prev. Vet. Med. 1985;4(23–24):34. [Google Scholar]
- Pérez L.J., Perera C.L., Frías M.T., Núñez J.I., Ganges L., deArce H.D. A multiple SYBR Green I-based real-time PCR system forthe simultaneous detection of porcinecircovirus type 2, porcine parvovirus, pseudorabies virus and Torque teno sus virus 1 and 2 inpigs. J. Virol. Methods. 2012;179:233–241. doi: 10.1016/j.jviromet.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Qu G., Fu S., Shen N., Wang J., Xiao Y., Guan Y., Tang N., Chen L., Gao S., Shen Z. Rapid and sensitive diagnosis of porcine parvovirus by loop-mediated isothermal amplification (LAMP) method. J. Appl. Anim. Res. 2010;37:113–116. [Google Scholar]
- Wang Z., Lu Y., Ding M. Some multiplication properties of the lapinized chinese strain (C strain) of classical swine fever virus in primary bovine testicular cells (in Chinese) Virol. Sinica. 2000;2:170–179. [Google Scholar]
- Westenbrink F., Veldhuis M.A., Brinkhof J.M. An enzyme-linked immunosorbent assay for detection of antibodies to porcine parvovirus. J. Virol. Methods. 1989;23:169–178. doi: 10.1016/0166-0934(89)90130-4. [DOI] [PubMed] [Google Scholar]
- Wilson I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano A., Ishimaru R., Hujikata R. Rapid and sensitive detection of heat-labile I and heat-stable I enterotoxin genes of enterotoxigenic Escherichia coli by loop-mediated isothermal amplification. J. Microbiol. Methods. 2007;68:414–420. doi: 10.1016/j.mimet.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Yin Z., Liu J.H. Science Press; Beijing: 1997. Animal virology, 2nd; pp. 1145–1155. [Google Scholar]
- Yuan Q.Z., Wu Y.X., Li Y.X., Li Z.R., Nan X. The pseudorabies vaccination research. I: pseudorabies attenuated vaccine research (in Chinese) Chin. J. Prev. Vet. Med. 1983;1:1–6. [Google Scholar]
- Zhang C.F., Cui S.J., Zhu C. Loop-mediated isothermal amplification for rapid detection and differentiation of wild-type pseudorabies and gene-deleted virus vaccines. J. Virol. Methods. 2010;169:239–243. doi: 10.1016/j.jviromet.2010.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Zhang X., Tang B., Xu M., Hou J. Isolation and identification of NJ strain of porcine parvovirus(in Chinese) Jiangsu J. Agr. Sci. 2012;1:99–103. [Google Scholar]
- Zheng L.L., Wang Y.B., Li M.F., Chen H.Y., Guo X.P., Geng J.W., Wang Z.Y., Wei Z.Y., Cui B.A. Simultaneous detection of porcine parvovirus and porcine circovirus type 2 by duplex real-time PCR and amplicon melting curve analysis using SYBR Green. J. Virol. Methods. 2013;187:15–19. doi: 10.1016/j.jviromet.2012.06.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and its supporting information.