Highlights
-
•
Linear B-cell epitopes in RBD of S protein predicted against SARS-CoV-2.
-
•
Discontinuous B-cell epitopes from S protein predicted against SARS-CoV-2.
-
•
T-cell epitopes from S, M and N protein predicted against SARS-CoV-2.
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; HLA, human leukocyte antigen; MHC, major histocompatibility complex; WHO, world health organization; PHEIC, public health emergency of international concern; S, spike; RBD, receptor-binding domain; NCBI, national center of biotechnology information; PDB, protein data bank; GRAVY, grand average of hydropathicity; IEDB, immune-epitope-database and analysis-resource
Keywords: SARS-CoV-2, Spike protein, T-cell epitope, B-cell epitope, Immune-informatics, Vaccine design
Abstract
The outbreak of the 2019 novel coronavirus (SARS-CoV-2) has infected millions of people with a large number of deaths across the globe. The existing therapies are limited in dealing with SARS-CoV-2 due to the sudden appearance of the virus. Therefore, vaccines and antiviral medicines are in desperate need. We took immune-informatics approaches to identify B- and T-cell epitopes for surface glycoprotein (S), membrane glycoprotein (M) and nucleocapsid protein (N) of SARS-CoV-2, followed by estimating their antigenicity and interactions with the human leukocyte antigen (HLA) alleles. Allergenicity, toxicity, physiochemical properties analysis and stability were examined to confirm the specificity and selectivity of the epitope candidates. We identified a total of five B cell epitopes in RBD of S protein, seven MHC class-I, and 18 MHC class-II binding T-cell epitopes from S, M and N protein which showed non-allergenic, non-toxic and highly antigenic features and non-mutated in 55,179 SARS-CoV-2 virus strains until June 25, 2020. The epitopes identified here can be a potentially good candidate repertoire for vaccine development.
1. Introduction
The SARS-CoV-2 (coronavirus disease 2019; previously 2019-nCoV) has recently emerged as a human pathogen leading to millions of confirmed cases globally and more than 100,000 deaths (Wrapp et al., 2020). The SARS-CoV-2 virus is an enveloped, positive single-stranded RNA coronavirus with a genome size of approximately 29.9 kb. SARS-CoV-2 is closely related to several bat coronaviruses and the SARS-CoV virus (Lu et al., 2020; Wu et al., 2020), and all belong to the B lineage of the beta-coronaviruses (Zhou et al., 2020). The transmission of SARS-CoV-2 appears to contain the way from human to human and from contact with infected surfaces and objects, causing WHO to declare a Public Health Emergency of International Concern (PHEIC) on January 30th, 2020 (Chan et al., 2020; Chen et al., 2020a; Li et al., 2020).
Structural proteins are important targets for vaccine and anti-viral drug development due to their indispensable function to fuse and enter into the host cell (Lindenbach and Rice, 2003). SARS-CoV-2 utilizes glycosylated spike (S) protein to gain entry into host cells. The S protein is a trimeric class I fusion protein and exists in a metastable prefusion conformation that undergoes a dramatic structural rearrangement to fuse the viral membrane with the host cell membrane (Wrapp et al., 2020; Bosch et al., 2003; Li, 2016). The S protein includes the receptor binding S1-subunit and the membrane fusion S2-subunit. The S1 subunit receptor-binding domain (RBD) is specifically recognized by the host receptor. When the S1 subunit binds to a host-cell receptor, the prefusion trimer is destabilized, resulting in the shedding of the S1 subunit, and the state transition of the S2 subunit to a stable postfusion conformation (Walls et al., 2017). The critical function of the S protein can be a breakthrough in vaccine design and development.
The SARS-CoV-2 coronavirus membrane glycoprotein (M Protein) is a 222 aa structural protein that is the most abundant in coronavirus, and it is normally highly conserved as a candidate antigen for developing the SARS-CoV-2 vaccine (Neuman et al., 2011). Immunization with the full length of M protein was reported to be able to elicit neutralizing antibodies in SARS patients (Pang et al., 2004). The SARS-CoV-2 coronavirus nucleocapsid phosphoprotein (N Protein) is a 419 aa structural protein highly conserved with multiple functions including the formation of nucleocapsids, signal transduction virus budding, RNA replication, and mRNA transcription (McBride et al., 2014). N protein is highly antigenic, 89 % of patients who developed SARS, produced antibodies to this antigen (Leung et al., 2004). The immunogenicity of E protein is limited, owing to that it consists of 76–109 aa in different coronaviruses with channel activity (Zhang et al., 2020), therefore it is not suitable for use as an immunogen.
Great efforts are being made for the discovery of antiviral drugs, even so, there are no licensed therapeutic or vaccine for the treatment of SARS-CoV-2 infection available in the market. Developing an effective treatment for SARS-CoV-2 is, therefore, a research priority. It is time-consuming and expensive to design novel vaccines against viruses by the use of kits and related antibodies (Tahir Ul Qamar et al., 2018; Chen et al., 2020b). Previously, numerous methods including the whole virus, DNA, subunit, and virus-like particles were used in developing vaccines for SARS and MERS (Song et al., 2019; Yong et al., 2019; Schindewolf and Menachery, 2019; Prompetchara et al., 2020). There were epitopes screened to develop vaccine targets for SARS-CoV (Liu et al., 2017) and MERS-CoV (Shi et al., 2015), respectively. These epitopes can be prepared by chemical synthesis techniques and are easier in quality control, but structural modifications, delivery systems, and adjuvants are additionally required in the formulation due to the low immunogenicity caused by their low molecular weight and structural complexity (Azmi et al., 2014). Currently, a set of B and T cell epitopes highly conserved in SARS-CoV-2 were identified from S and N proteins of SARS-CoV that may help develop SARS-CoV-2 vaccines (Ahmed et al., 2020). Among those that can be analyzed, B-cell can recognize and activate defense responses against viral infection, T-cell, and antibody reactions that may recover extreme respiratory infection. Thus, we chose the method of immune-informatics, which is more efficient and more applicable for deep analysis of viral antigens, B- and T- cell linear epitope prediction, and evaluation of immunogenicity and virulence of pathogens.
In this manuscript, we applied immuno-informatics approaches to identify potential B- and T-cell epitopes based on the S protein of SARS-CoV-2. The antigenicity of all the epitopes was estimated and the interactions with the human leukocyte antigen (HLA) alleles were evaluated for MHC class-I epitopes. Allergenicity, toxicity, stability, and physiochemical properties were also investigated for exploring the antigenicity, stability, and safety of the identified epitopes. The conservation of all B- and T- cell epitopes were examined across all isolates from different locations. Some of these identified epitopes could be used as promising vaccine candidates.
2. Methods
2.1. Data retrieval and structural analysis
The primary sequence of SARS-CoV-2 protein was retrieved from the NCBI database using accession number MN908947.3 (Wu et al., 2020). Experimentally known 3D structure of SARS-CoV-2 S protein (PDB ID: 6VSB) and N protein (PDB ID: 6VYO) were retrieved from Protein Data Bank (Wrapp et al., 2020). There is no 3D structure of M protein available yet. The predicted interaction conformation between RBD of SARS-CoV-2 S protein and human ACE-2 was retrieved from a very recent report (Fast and Chen, 2020; Tai et al., 2020; Shang et al., 2020; Lan et al., 2020; Qiu et al., 2020; Ortega et al., 2020). The protein sequence was analyzed for its chemicals and physical properties including GRAVY (Grand average of hydropathicity), half-life, molecular weight, stability index, and amino acid atomic composition via an online tool Protparam (Gasteiger et al., 2003). TMHMM v2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was applied to examine the transmembrane topology of S and M protein. The secondary structure of the SARS-CoV-2 S, M and N protein was analyzed by PSIPRED (Buchan et al., 2013). The existence of disulfide-bonds was examined through an online tool DIANNA v1.1 which uses a trained neural system to make predictions (Ferre and Clote, 2006). Antigenicity of full-length S, M and N protein were evaluated by VaxiJen v2.0 (Doytchinova and Flower, 2007).
2.2. B-cell epitope prediction
IEDB (Immune-Epitope-Database And Analysis-Resource) (Peters et al., 2005) were used to predict linear B-cell epitopes using Bepipred and Bepipred2.0 with default parameter settings, Kolaskar and Tongaonkar antigenicity, Parker hydrophilicity, Chou and Fasman beta-turn, and Karplus and Schulz flexibility. BcePred (Saha and Raghava, 2004) was also used to predict linear B-cell epitopes using accessibility, antigenic propensity, exposed surface, flexibility, hydrophilicity, polarity, and turns. Predicted linear B-cell epitopes by IEDB and BcePred were combined to the linear B-cell epitope candidate list. Based on the transmembrane topology of S and M protein predicted by TMHMM v2.0, only epitopes on the outer surface remained, and other intracellular epitopes were eliminated. VaxiJen 2.0 (Doytchinova and Flower, 2007) was applied to evaluate the antigenicity of the remained epitopes. A stringent criterion was used to have epitopes with an antigenicity score of 0.9 viewed adequate to initiate a defensive immune reaction. A B-cell discontinuous epitope forms the antigen-binding interface through fragments scattered along the protein sequence. DiscoTope2.0 (Kringelum et al., 2012) with a discotope score threshold of -3.7 was used to predict discontinuous epitopes. As the 3D structure of M protein is not available, open-source Pymol was used to examine the positions of selected linear and discontinuous epitopes on the 3D structure of SARS-CoV-2 S protein or the interacting conformation of S protein RBD and human ACE-2(Tai et al., 2020; Shang et al., 2020; Lan et al., 2020).
2.3. T-cell epitope prediction
Cytotoxic T-lymphocyte epitopes are important in developing vaccines. Peptide_binding_to_MHC_class_I_molecules tool of IEDB and HLA class I set (Weiskopf et al., 2013) was utilized to predict MHC class I binding T-cell epitopes. Peptide_binding_to_MHC_class_II_molecules tool of IEDB and HLA class II set (Greenbaum et al., 2011) was utilized to predict T-cell epitopes. Percentile rank with a threshold of 1% for MHC class I binding epitopes and 10 % for MHC class II binding epitopes were used to filter out peptide-allele with weak binding affinity. The antigenicity score of each epitope was calculated by VaxiJen v2.0. A high stringent standard was used to filter peptides with antigenicity score larger than or equal to 1, the number of binding alleles larger than or equal to 3 for MHC class I binding epitopes and 5 for MHC class II binding epitopes.
2.4. Characterization of selected B-cell and T-cell epitopes
All selected linear B-cell and MHC class I and II binding T cell epitopes were examined for their allergenicity, hydro and physiochemical features, toxicity, and digestion. Allergenicity of linear B-cell and T-cell epitopes were assessed by Allergen FP 1.0 (http://ddg-pharmfac.net/AllergenFP/). Toxicity of linear B-cell and T-cell epitopes along with hydrophobicity, hydropathicity, hydrophilicity, and charge were evaluated by ToxinPred (https://webs.iiitd.edu.in/raghava/toxinpred/index.html). The peptides that can be digested by several enzymes are usually non-stable, while the peptides digested by fewer enzymes are more stable, making them more favorable candidate vaccines (Tahir Ul Qamar et al., 2019). Examined by protein digest server (http://db.systemsbiology.net:8080/proteomicsToolkit/proteinDigest.html), the digestion of linear B- and T-cell epitopes by 13 enzymes including Trypsin, Chymotrpsin, Clostripain, Cyanogen Bromide, IodosoBenzoate, Proline Endopept, Staph Protease, Trypsin K, Trypsin R, AspN, Chymotrypsin (modified), Elastase, and Elastase/Trypsin/Chymotryp.
2.5. Protein-epitope interaction evaluation
The 3D structure of human HLA-B35:01(PDB ID: 1A9E) at a resolution of 2.5 Å, HLA-B*51:01 (PDB ID: 1E27) at a resolution of 2.2 Å, HLA-B*53:01 (PDB ID: 1A1O) at a resolution of 2.3 Å, HLA-B*57:01 (PDB ID: 3 × 11) at a resolution of 2.15 Å, HLA-B*58:01 (PDB ID: 5IM7) at a resolution of 2.50 Å, HLA-A*01:01 (PDB ID: 6AT9) at a resolution of 2.95 Å, HLA-A*68:01 (PDB ID: 6PBH) at a resolution of 1.89 Å, HLA-A*11:01 (PDB ID: 5GRG) at a resolution of 1.94 Å, HLA-A*03:01 (PDB ID: 6O9B) at a resolution of 2.20 Å, and HLA-B*15:01 (PDB ID: 5TXS) at a resolution of 1.70 Å were downloaded from protein databank (RCSB PDB) and used for evaluating their interactions with selected epitopes. Protein-peptide interactions were performed by PepSite (Trabuco et al., 2012) with the top prediction chosen from a total of 10 epitope-protein interaction reports. pepATTRACT (de Vries et al., 2017) was adopted to estimate the Docking score of each peptide with the corresponding HLA allele.
2.6. Conservation analysis of selected B- and T-cell epitopes
ConSurf (Ashkenazy et al., 2010) was used to examine the conservation status for each residue of SARS-CoV-2 by analyzing the amino acid sequences of S, M, and N protein from seven known coronaviruses including SARS-CoV-2 (YP_009724390.1), SARS-CoV (NP_828851.1), MERS-CoV (YP_009047204.1), alpha coronavirus 229E (NP_073551.1), alpha coronavirus NL63 (AFV53148.1), beta coronavirus OC43 (YP_009555241.1) and beta coronavirus HKU1 (AAT98580.1). The S, M, and N protein sequences of different SARS-CoV-2 virus strains were taken from an open-access database NGDC (https://bigd.big.ac.cn/ncov/), where 55,179 SARS-CoV-2 virus strains’ sequences were documented with 11,813 mutations reported in the virus genome until June 25, 2020.
3. Results
3.1. Structural analysis of SARS-CoV-2 S protein, m protein, and N protein
S protein is an important target for vaccine development because of its important function in entering the host cell. M protein and N protein of coronavirus have also been reported to generate immunogenic epitopes. Therefore, the physiochemical properties of SARS-CoV-2 S protein, M protein, and N protein were first examined by Protparam, demonstrating their amino acids (aa) and molecular weight (1273 aa and 141.18 kDa for S protein, 222 aa and 25.15 kDa for M protein, and 419 aa and 45.63 kDa for N protein) (Supplementary Table 1). 110 aa and 103 aa were found as negatively and positively charged for S protein, respectively. There were 13 aa negatively and 21 aa positively charged for M protein, and 36 aa negatively and 60 aa positively charged for N protein. The theoretical iso-electric point (PI) of the S, M, and N protein were 6.24, 9.51, and 10.07, respectively. The instability-index (II) was computed to be 33.01 for S protein, 39.14 for M protein, and 55.09 for N protein, which categories the S protein as stable but not for M and N protein. Aliphatic-index was 84.67 with GRAVY (grand average of hydropathicity) value of -0.079 for S protein (120.86 and 0.446 for M protein, 52.53, and -0.971 for N protein). The number of Carbon (C), Oxygen (O), Nitrogen (N), Hydrogen (H), and Sulfur (S) of a total of 19,710 atoms were formulated as C6336H9770N1656O1984S54 for S protein with M and N protein formulated as C1165H1823N303O301S8 and C1971H3137N607O629S7, respectively. The details of the physiochemical properties of SARS-CoV-2 S, M, and N protein can be seen in Supplementary Table 1.
The secondary structure of S, M, and N protein were generated by PSIPRED (Buchan et al., 2013), showing that the Beta strand (26.3 % for S, 24.3 % for M, 12.2 % for N), Helixes (24.4 % for S, 40.1 % for M, 21 % for N), and coil (49.3 % for S, 35.6 % for M, 66.8 % for N) are present in structure for S, M, and N protein respectively (Supplementary Fig. 1–3 for S, M and N protein, respectively). 20, and 6 disulfide (S—S) bond positions were identified by DiANNA for S and M protein without S—S bond positions found for N protein (Supplementary Table 2). 40 cysteine residues were identified by DiANNA in the full-length of the S protein sequence, which made 20 disulfide (S—S) bonds at the following positions (15–1240, 131–391, 136–662, 166–1236, 291–671, 301–336, 361–488, 379–743, 432–1235, 480–1248, 525–1247, 538–1043, 590–617, 649–1241, 738–1243, 749–1126, 760–1250, 840–1032, 851–1254, 1082–1253) (Supplementary Table 2). 20 cystein residues were identified in the M protein sequence, generating 6 disulfide (S—S) bonds at positions of 33–64, 33–86, 33–159, 64–86, 64–159, and 86–159 (Supplementary Table 2). Antigenicity analysis of the full-length protein by Vaxijen confirmed that they were expected antigens with an antigenicity score of 0.4646 for S protein, 0.5102 for M protein, and 0.5059 for N protein. As S and M proteins are transmembrane protein, the transmembrane protein topologies were therefore predicted by TMHMM for S and M protein, respectively. The residues from 1 to 1213 were exposed on the surface, residues from 1214 to 1236 were inside transmembrane-region and residues from 1237 to 1273 were within the core-region of the S protein (Supplementary Fig. 4A). 1–19 residues were exposed on the surface, with 20–99 residues inside transmembrane-region and 100–222 residues within the core-region of the M protein (Supplementary Fig.5A). B-cell epitopes can bind to antigen receptors on the surface of B cells, but N protein is inside the virus. Considering both S and M protein are transmembrane proteins, we attempted to predict B-cell epitopes only for S and M protein (even there is no neutralization activity well known for M protein) in the downstream analysis. We predicted T-cell epitopes for S, M, and N protein.
3.2. Identification of linear B-cell epitopes from S and M protein
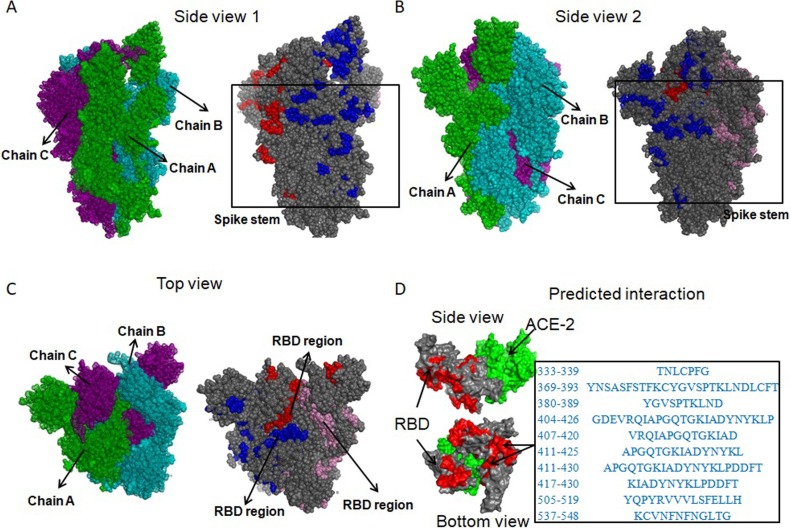
B-cell epitopes can guide B-cell to recognize and activate defense responses against viral infection. Recognition of B-cell epitopes depended on predictions of linear epitopes, antigenicity, hydrophilicity, accessibility of surface, beta-turn, and flexibility (Fieser et al., 1987). B-cell epitopes of S and M protein were predicted by methods with default settings provided in IEDB (Peters et al., 2005) including Bepipred, Bepipred2.0, Kolaskar, and Tongaonkar antigenicity scale, Parker hydrophilicity, Emini surface accessibility, Chou and Fasman beta-turn and Karplus and Schulz flexibility. A total of 262 and 33 linear epitopes were identified based on the combination of the results for S and M protein, respectively (Supplementary Table 3A, Supplementary Fig. 4B-F for S, Supplementary Fig 5B-F for M). BcePred (Saha and Raghava, 2004) was used to predict B-cell epitopes using accessibility, antigenic propensity, exposed surface, flexibility, hydrophilicity, polarity, and turns. Overall, we obtained a total of 129 and 24 linear B-cell epitopes for S and M protein respectively (Supplementary Table 3B). VaxiJen v2.0 was further used to estimate the antigenicity of all linear B-cell epitopes, resulting in a total of 80 and 4 epitopes for S and M protein with the antigenicity score larger than or equal to 0.9, respectively (Supplementary Table 3C). Based on the transmembrane topology of S and M protein predicted by TMHMM v2.0, intracellular epitopes were further eliminated. As a result, 78 linear B-cell epitopes from S protein were retained as candidates (Supplementary Table 3C). Allergenicity and toxicity of the 78 linear B-cell epitopes was further assessed by Allergen FP 1.0 and ToxinPhred, respectively, leading to the result that a total of 34 linear B-cell epitopes were retained as neither allergen nor toxin (Supplementary Table 3C). The 34 linear B-cell epitopes were mapped to the 3D structure of the SARS-CoV-2 S protein (PDB ID: 6VSB), showing that 24 epitopes were in spike stem region (Fig. 1 A-B)(Supplementary Table 3C) and ‘TNLCPFG’, ‘YNSASFSTFKCYGVSPTKLNDLCFT’, ‘YGVSPTKLND’, ‘GDEVRQIAPGQTGKIADYNYKLP’, ‘VRQIAPGQTGKIAD’, ‘APGQTGKIADYNYKL’, ‘APGQTGKIADYNYKLPDDFT’, ‘KIADYNYKLPDDFT’, ‘YQPYRVVVLSFELLH’, and ‘KCVNFNFNGLTG’ located in the RBD region of the spike head, which is the most exposed region (Table 1 ) (Fig. 1C). Based on the predicted interacting conformation between RBD domain of SARS-CoV-2 S protein and ACE-2 (Fast and Chen, 2020; Tai et al., 2020; Shang et al., 2020; Lan et al., 2020), the ten linear B-cell epitopes in the spike head substantially overlaps with the interacting surface where ACE-2 binds to RBD(Tai et al., 2020; Tai et al., 2020; Shang et al., 2020; Lan et al., 2020), demonstrating that an antibody binding to this surface may block viral entry into cells (Fig. 1D). After examining the antigenicity of recently reported B-cell epitopes(Grifoni et al., 2020; Srivastava et al., 2020), we discovered that all except for one epitope from Orf3a (antigenicity score of QGEIKDATPSDF: 1.1542) (Supplementary Table 3D) have much less antigenicity score than the ten linear B-cell epitopes we identified from the most exposed region in spike protein (antigenicity scores ranging from 0.9567 to 1.6969) (Table 1).
Fig. 1.
The locations of the 34 non-allergenic and non-toxic linear B-cell epitopes in the 3D structure of SARS-CoV-2 S protein (PDB ID 6VSB). (A-B) The locations of the 24 non-allergenic and non-toxic B-cell epitopes in the spike stem region; (C) The locations of the ten non-allergenic and non-toxic B-cell epitopes in the spike head which is the most exposed region. (D) The locations of the ten linear non-allergenic and non-toxic B-cell epitopes mapped to the predicted interacting conformation between the RBD domain of SARS-CoV-2 S protein and ACE-2. From A to C, Green, cyan and purple are chain A, chain B and chain C, respectively; Blue, red and pink are the locations of the 34 non-allergenic and non-toxic linear B-cell epitopes in chain A, chain B, and chain C, respectively. In D, light green is the ACE-2; Grey is the RBD of S protein; Light red is the locations of the ten non-allergenic and non-toxic B-cell epitopes in RBD.
Table 1.
Predicted linear B-cell epitopes with antigenicity score.
| start | end | peptide | Antigenicity | method | |
|---|---|---|---|---|---|
| 333 | 339 | TNLCPFG | 1.1812 | Kolaskar and Tongaonkar antigenicity | |
| 369 | 393 | YNSASFSTFKCYGVSPTKLNDLCFT | 1.4031 | Bepipred2.0 | |
| 380 | 389 | YGVSPTKLND | 1.4531 | Chou and Fasman beta turn | |
| 404 | 426 | GDEVRQIAPGQTGKIADYNYKLP | 1.1017 | Bepipred2.0 | |
| 407 | 420 | VRQIAPGQTGKIAD | 1.2606 | Bepipred | |
| 411 | 425 | APGQTGKIADYNYKL | 1.4441 | Parker hydrophilicity | |
| 411 | 430 | APGQTGKIADYNYKLPDDFT | 1.0425 | Chou and Fasman beta turn | |
| 417 | 430 | KIADYNYKLPDDFT | 0.9567 | Accessibility | |
| 505 | 519 | YQPYRVVVLSFELLH | 0.9711 | Antigenic_Propensity | |
| 537 | 548 | KCVNFNFNGLTG | 1.6969 | Chou and Fasman beta turn | |
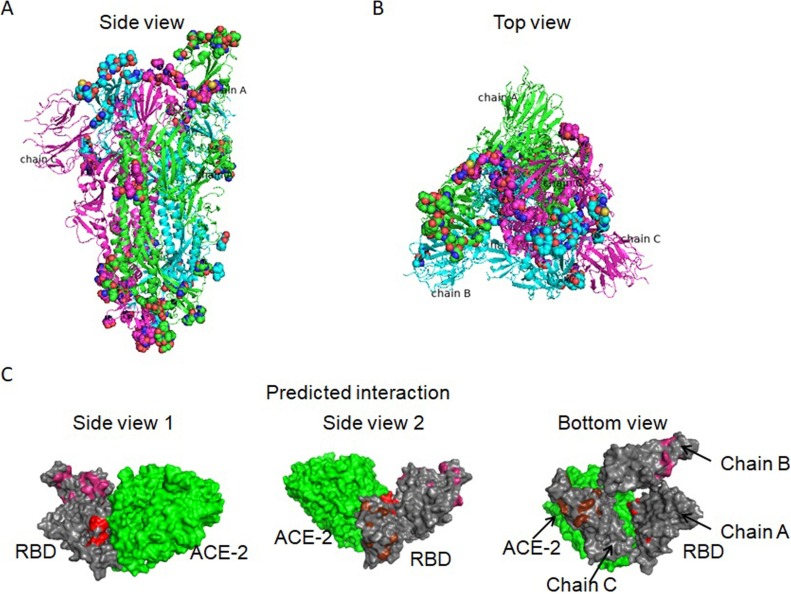
As there is no 3D structure of M protein available, discontinuous B-cell epitopes were predicted for S protein by Discotope 2.0 using A, B, and C chain of the 3D structure of S protein (PDB ID: 6VSB). The positions of discontinuous epitopes were mapped on the surface of the 3D structure of S protein (Fig. 2 A, Supplementary Fig. 6). Most discontinuous B-cell epitopes were mapped on the fully-exposed ‘spike head’ region (Fig. 2B) (Supplementary Table 4) and exposed ‘spike stem’ region, while a few located in the ‘spike root’ region (Supplementary Table 3E). The main discontinuous B-cell epitopes on the ‘spike head’ region overlapped with the interacting surface of ACE-2 binding to S protein (Fig. 2C), suggesting their roles in blocking virus’ fusion with cells.
Fig. 2.
The positions of discontinuous B-cell epitopes were on the 3D structure of S protein (PDB ID 6VSB). (A-B) The side view and top view displayed the epitopes (dots mode) in the S protein (cartoon mode). Green, cyan and magenta represent chain A, chain B and chain C, respectively. The spheres represents the discontinuous epitopes. (C) The main discontinuous B-cell epitopes on the ‘spike head’ region overlapped with the interacting surface of ACE-2 binding to S protein. Green is ACE-2, and grey is RBD of S protein. Red, magenta and brown are the locations of the discontinuous B-cell epitopes on chain A, chain B and chain C.
3.3. Hydro and physiochemical property and stability analysis of the ten linear B-cell epitopes in RBD
As the ten linear B-cell epitopes in RBD of the S protein were predicted to be of both non-allergen and non-toxin, we further examined their hydrophobicity, hydropathicity, hydrophilicity, and charge by a support vector machine (SVM) based method, ToxinPred (Supplementary Table 5A). The stability of the ten linear B-cell epitopes was evaluated by the number of peptide-digesting enzymes through the protein digest server (http://db.systemsbiology.net:8080/proteomicsToolkit/proteinDigest.html). More non-digesting enzymes predicted for an epitope suggests its potentially higher stability. All the ten linear B-cell epitopes were found to have multiple non-digesting enzymes varying from 2 to 8 enzymes (Supplementary Table 5B).
3.4. Identification of T-cell epitopes from S, m and N protein
Peptide_binding_to_MHC_class_I_molecules tool of IEDB and HLA class I set (Weiskopf et al., 2013) was utilized to predict T-cell epitopes for S protein. Percentile rank with a threshold of 1% was used to filter out peptide-allele with weak binding affinity. The antigenicity score of each peptide was calculated by VaxiJen v2.0 to evaluate its antigenicity. A peptide having both high antigenicity score and capacity to bind with a larger number of alleles is considered to have high potentials to initiate a strong defense response. High stringent criteria were used to filter peptides with antigenicity score larger than or equal to 1 and the number of binding alleles larger than or equal to 3. Utilizing the evaluating method above, we obtained a total of 27 MHC class-I allele binding peptides from S, M, and N protein (Supplementary Table 6A for S, 6B for M and 6C for N protein). The peptide ‘IPFAMQMAYR’ binding with A*68:01, B*35:01, and A*33:01 (antigenicity: 1.5145) from S protein, ‘RTRSMWSF’ binding with B*57:01, A*30:01, A*32:01 and B*58:01 (antigenicity: 1.4716) from M protein and ‘KLDDKDPNF’ binding with A*32:01, A*02:06, A*02:01, A*01:01, A*30:02, and B*15:01 (antigenicity: 2.6591) from N protein have the highest antigenicity scores among MHC class-I binding epitopes derived from S, M or N protein, respectively. The peptide ‘FAMQMAYRF’ binding with six alleles from S protein, ‘EQWNLVIGF’ binding with six alleles from M protein, and ‘KMKDLSPRW’ binding with eight alleles from N protein have the highest number of binding MHC class-I alleles with strong antigenicity score of 1.0278, 1.3869, and 1.7462 among MHC class-I binding epitopes derived from S, M and N protein, respectively.
Peptide_binding_to_MHC_class_II_molecules tool of IEDB and HLA class II set (Greenbaum et al., 2011) was utilized to predict T-cell epitopes for S protein. Percentile rank with a threshold of 10 % was used to filter out peptide-allele with weak binding affinity. The antigenicity score of each peptide was calculated by VaxiJen v2.0 to evaluate its antigenicity. A high stringent standard was used to filter peptides with antigenicity score larger than or equal to 1 and the number of binding alleles larger than or equal to 5. As a result, we obtained a total of 26 MHC class-II allele binding peptides from S and M protein (Supplementary Table 7A for S, 7B for M protein). No MHC class-II allele binding peptides were identified for N protein. The peptide ‘VGYQPYRVVVLSFEL’ from S protein binding with six alleles and ‘WNLVIGFLFLTWICL’ from M protein binding with six alleles have the highest antigenicity score of 1.3858 and 1.4689 among epitopes derived from S or M protein respectively. The peptides ‘GVVFLHVTYVPAQEK’ and ‘GYQPYRVVVLSFELL’ binding with 11 alleles from S protein and ‘LACFVLAAVYRINWI’ binding with 12 alleles from M protein have the highest number of binding MHC class-II alleles with strong antigenicity score of 1.1043, 1.074 and 1.2905 among epitopes derived from S or M protein, respectively.
3.5. Allergenicity, toxicity and stability analysis of T-cell epitopes from S, M and N protein
Allergenicity of T-cell epitopes were assessed by Allergen FP 1.0. Results showed that two of nine, three of nine, two of nine MHC class-I binding peptides from S, M, and N protein were probably non-allergen, respectively (Supplementary Table 8A-C). Nine of thirteen and nine of thirteen MHC class-II binding peptides from S and M protein were predicted to be non-allergen, respectively (Supplementary Table 8A-C). Toxicity of T-cell epitopes along with hydrophobicity, hydropathicity, hydrophilicity, and charge was evaluated by ToxinPred. All but two T-cell epitopes were predicted to be non-toxin (Supplementary Table 8A-C). The stability of T-cell epitopes was evaluated through the number of peptides digesting enzymes by the protein digest server. All T-cell epitopes but ‘KMKDLSPRWY’ were found to have multiple non-digesting enzymes varying from 3 to 11 enzymes (Supplementary Table 9A-C). We compared the selected 25 T-cell epitopes (11, 12 and 2 epitopes from S, M, and N protein, respectively), determined as both non-allergen and non-toxin (including two MHC-I and nine MHC-II binding T-cell epitopes) with the five recently reported SARS-CoV-2 S protein epitopes (‘SYGFQPTNGVGYQPY’, ‘SQSIIAYTMSLGAEN’, ‘IPTNFTISVTTEILP’, ‘AAAYYVGYLQPRTFL’, and ‘APHGVVFLHVTYVPA’) (Fast and Chen, 2020). We found that four of 11 T-cell epitopes from S protein substantially overlapped with two of the five T-cell epitopes reported in the literature (‘PTNFTISVTTEILPV’, ‘TNFTISVTTEILPVS’, ‘TNFTISVTTEILPVS’ overlapped with ‘IPTNFTISVTTEILP’; ‘VVFLHVTYVPAQEKN’ overlapped with ‘APHGVVFLHVTYVPA’).
3.6. Interaction of MHC class I binding T-cell epitopes with HLA alleles
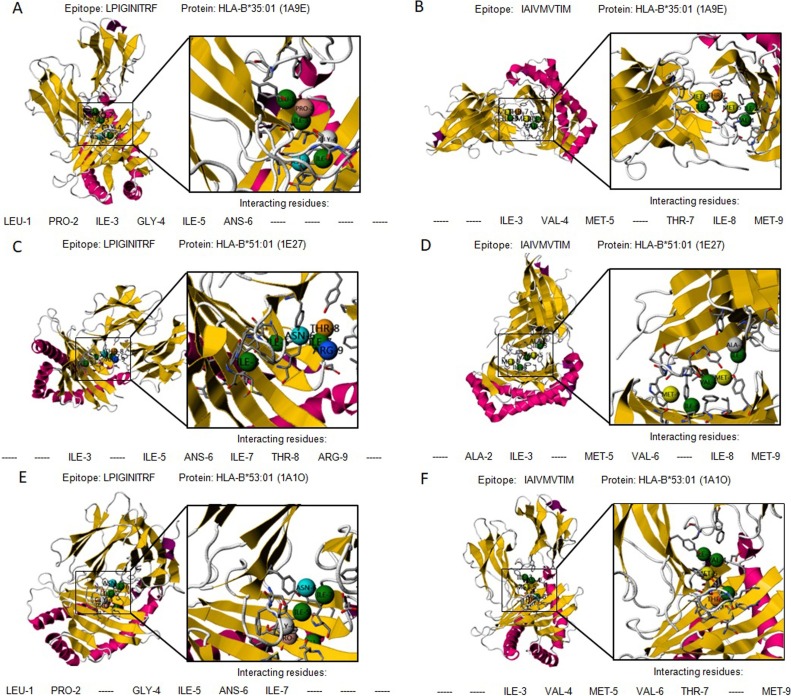
Protein-peptide interactions are critical in cellular signaling pathways. Seven MHC class-I binding epitopes from S, M and N protein (‘LPIGINITRF’ and ‘IAIVMVTIM’ for S, ‘EQWNLVIGF’, ‘LVIGAVILR’, and ‘DSGFAAYSRY’ for M, and ‘GKMKDLSPRW’ and ‘SSRSRNSSR’ for N protein), were predicted to be non-allergic and non-toxic. The peptides ‘LPIGINITRF’ and ‘IAIVMVTIM’ from S protein were predicted to bind to HLA-B35:01, HLA-B*51:01, and HLA-B*53:01. The 3D structure of human HLA-B35:01(PDB ID: 1A9E) (Menssen et al., 1999), HLA-B*51:01 (PDB ID: 1E27) (Maenaka et al., 2000) and HLA-B*53:01 (PDB ID: 1A1O) (Smith et al., 1996) protein were accessible with co-crystallized peptide in PDB database. Protein-peptide interactions were performed by PepSite (Trabuco et al., 2012). 10 epitope-protein interactions were reported and the top prediction was chosen. HLA-B*35:01 (1A9E) is of a hetero 2mer structure with 386 residues. Epitope ‘LPIGINITRF’ having a docking score of -13.9943 kcal/mol was predicted to significantly bind on the surface of HLA-B35:01(PDB ID: 1A9E) through six hydrogen bonds with Leu-1, Pro-2, Ile-3, Gly-4, Ile-5, and Ans-6 (Fig. 3 A). Epitope ‘IAIVMVTIM’ having a docking score of -17.4708 kcal/mol moderately significantly bond to HLA-B35:01(PDB ID: 1A9E) via six hydrogen bonds with Ile-3, Val-4, Met-5, Thr-7, Ile-8, and Met-9 (Fig. 3B). Similarly, both epitope ‘LPIGINITRF’ and ‘IAIVMVTIM’ show strong and stable bonding with HLA-B*51:01 (1E27) residues (Fig. 3C-D) (docking score of -14.4615 kcal/mol for ‘LPIGINITRF’ and -18.3599 kcal/mol for ‘IAIVMVTIM’), and HLA-B*53:01 (PDB ID: 1A1O) residues (Fig. 3D-F) (docking score of -13.9565 kcal/mol for ‘LPIGINITRF’ and -14.1208 kcal/mol for ‘IAIVMVTIM’), respectively.
Fig. 3.
The graphical presentation of predicted interactions between MHC class I binding T-cell epitopes from S protein and HLA alleles. Two epitopes ‘LPIGINITRF’ and ‘IAIVMVTIM’ with HLA-B:*35:01(PDB ID 1A9E) (A-B), HLA-B*51:01 (PDB ID 1E27) (C-D), and HLA-B*53:01(PDB ID 1A1O) (E-F), respectively.
Among the HLA alleles bond by the peptides ‘EQWNLVIGF’, ‘LVIGAVILR’, and ‘DSGFAAYSRY’ from M, and ‘GKMKDLSPRW’ and ‘SSRSRNSSR’ from N protein, the 3D structure of HLA-B*15:01 (PDB ID: 5TXS), HLA-A*68:01 (PDB ID: 6PBH), HLA-A*03:01 (PDB ID: 6O9B, 5GRG), HLA-A*01:01 (PDB ID: 6AT9), HLA-B*57:01 (PDB ID: 3 × 11) and HLA-B*58:01 (PDB ID: 5IM7) were available with co-crystallized peptide in the PDB database. ‘EQWNLVIGF’ binding with HLA-B*15:01, ‘LVIGAVILR’ binding with HLA-A*68:01 and HLA-A*03:01, and ‘DSGFAAYSRY’ binding with HLA-A*01:01 were confirmed (Supplementary Fig. 7A-E) (docking score of -16.1239 kcal/mol for ‘EQWNLVIGF’ binding with HLA-B*15:01′, -16.5349 kcal/mol for ‘LVIGAVILR’ with HLA-A*68:01, -13.6659 kcal/mol with HLA-A*03:01 (PDB ID 5GRG) and -12.367 with HLA-A*03:01 (PDB ID 6O9B), and -14.9894 kcal/mol for ‘DSGFAAYSRY’ with HLA-A*01:01). ‘GKMKDLSPRW’ from N protein were confirmed binding with HLA-B*57:01 (-14.2185 kcal/mol) and HLA-B*58:01 (-13.3366 kcal/mol) (Supplementary Fig. 8A-B).
3.7. Conservation of B- and T-cell epitopes
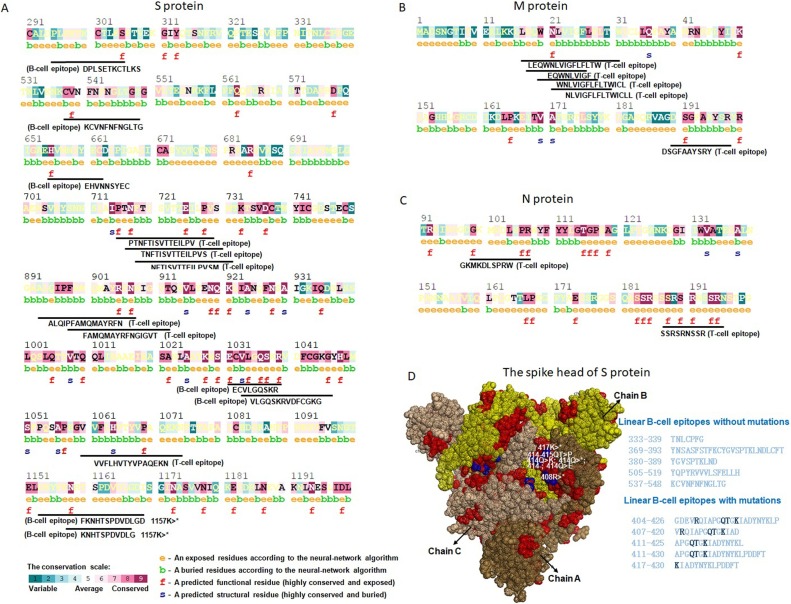
The conservation status of each residue in the selected B- and T-cell epitopes were examined by ConSurf with the use of seven known coronaviruses including SARS-CoV-2 (YP_009724390.1), SARS-CoV (NP_828851.1), MERS-CoV (YP_009047204.1), alpha coronavirus 229E (NP_073551.1), alpha coronavirus NL63 (AFV53148.1), beta coronavirus OC43 (YP_009555241.1) and beta coronavirus HKU1 (AAT98580.1). The result revealed that RBD region (from 319 to 514) of the S protein was not conserved among the seven coronaviruses (Supplementary Fig. 9). The highly conserved and exposed residues mainly located from 711 to 1221 in S protein (Supplementary Fig. 9), from 21 to 204 in. M protein (Supplementary Fig. 10), and from 18 to 311 in. N protein (Supplementary Fig. 11). Particularly, the epitopes without allergenicity and toxicity containing one functional residue (highly conserved and exposed) included B-cell epitopes of ‘DPLSETKCTLKS’, ‘KCVNFNFNGLTG’, ‘EHVNNSYEC’, ‘ECVLGQSKR’, ‘VLGQSKRVDFCGKG’, ‘FKNHTSPDVDLGD’, ‘KNHTSPDVDLG’, and T-cell epitopes of ‘PTNFTISVTTEILPV’, ‘TNFTISVTTEILPVS’, ‘NFTISVTTEILPVSM’, ‘ALQIPFAMQMAYRFN’, ‘FAMQMAYRFNGIGVT’, ‘VVFLHVTYVPAQEKN’ from S protein (Fig. 4 A). Five (‘LEQWNLVIGFLFLTW’, ‘EQWNLVIGF’, ‘WNLVIGFLFLTWICL’, ‘NLVIGFLFLTWICLL’ and ‘DSGFAAYSRY’ of 12 T-cell epitopes from M protein (Fig. 4B), and all two T-cell epitopes (‘GKMKDLSPRW’ and ‘SSRSRNSSR’) from N protein (Fig. 4C) contained at least one functional residue (highly conserved and exposed).
Fig. 4.
Conservation of B- and T-cell epitopes in SARS-CoV-2. The position of conserved epitopes in the protein sequence. (A) S protein, (B) M protein, (C) N protein. The e colored by orange is exposed residues according to the neural-network algorithm; the b colored by green is buried residues according to the neural-network algorithm; the f colored by red is predicted functional residue (highly conserved and exposed); the s colored by dark blue is predicted structural residues (highly conserved and buried). The conservation scale represents the status of conversation from variable, average to convserved. (D) The mutations observed in the ten non-allergenic and non-toxic linear B-cell epitopes in the RBD. Brown, yellow and light brown represent chain A, chain B and chain C. Red represent the locations of the ten non-allergenic and non-toxic linear B-cell epitopes in RBD region; blue represent the observed mutations. The black AAs in the epitopes are the mutated ones.
To investigate the presence of mutations in the B- and T-cell epitopes, 51,150 sequences of SARS-CoV-2 in the NGDC database were subjected to multiple sequence alignment of all selected epitopes. Four positions (408, 414, 415, 417) were observed mutated in five of ten non-allergenic and non-toxic linear B-cell epitopes in RBD of S protein (Fig. 4D) (Supplementary Table 10). Mutations occurred in nine of 24 non-allergenic and non-toxic linear B-cell epitopes in non-RBD regions (Supplementary Table 10). No mutations were observed in non-allergenic and non-toxic T-cell epitopes from S, M and N protein.
4. Discussion
The emergence of SARS-CoV-2 is a serious health threat for the whole society, thus there is an urgent need for drugs and preventative measures. The SARS-CoV-2 infection is characterized by lung infections with symptoms including fever, cough, and shortness of breath. Based on the information from CDC (Centers for Disease Control and Prevention), the symptoms can appear in as few as 2 days or as long as 14 days after exposure to the virus which can transmit from human to human or from contact with infected surfaces and objects (Chan et al., 2020; Chen et al., 2020a; Li et al., 2020).
It is essential to identify immune epitopes as quickly as possible. The S protein is crucial in the fuse and entry of the virus into host cells (Wrapp et al., 2020), therefore it is a primary target for neutralizing antibodies. The specificity of epitope-based vaccines can be enhanced by selecting parts of S protein exposed on the surface (Bakhshesh et al., 2018). Medical biotechnology is important in developing vaccines against SARS-CoV-2(Chen et al., 2020b). While computer-based immune-informatics can improve time and economic effectiveness, and therefore, it is also an essential method in immunogenic analysis and vaccine development.
In this study, we characterized the physio-chemical characteristics of the SARS-CoV-2 viral genome for epitope candidates and adopted an immune-informatics based pipeline with highly stringent criteria to identify S, M and N protein targeted B- and T-cell epitopes that may potentially promote an immune response in the host. The antigenicity, flexibility, solvent accessibility, disulfide bonds of predicted epitopes were evaluated, yielding a small repertoire of potential B-cell epitope and vaccine candidates. Allergenicity and toxicity analysis suggested the ten linear B-cell epitopes in RBD region are of non-allergen and non-toxin. Stability analysis revealed that they cannot be digested by multiple enzymes. Also, two MHC class-I and nine MHC class-II binding T-cell epitopes were predicted to interact with numerous HLA alleles and to be highly antigenic. Allergenicity, toxicity, and physiochemical properties of T-cell epitopes were analyzed to increase specificity and selectivity. The stability and safety were confirmed by digestion analysis. Conservation anlaysis of seven known coronaviruses revealed that RBD region is not conserved. Mutations generated from 51,150 sequences of SARS-CoV-2 in the NGDC database were observed in five of ten linear B-cell epitopes in RBD region. The B- and T-cell (MHC class I and II) epitopes without mutations would be considered to be vaccine candidates with full potentials of being antigenicity.
We predict the B- and T-cell epitopes identified here may assist the development of potent peptide-based vaccines to address the SARS-CoV-2 challenge. Particularly, those epitopes without mutations from the conserved regions could generate immunity that is not only cross-protective across Beta coronaviruses but also relatively resistant to ongoing virus evolution (Grifoni et al., 2020). The epitopes predicted here can also potentially be used in the design of more sensitive serological assays for epidemiological or vaccine efficiency assessments. But the replication of SARS-CoV-2 must be error-prone, which is similar to SARS-CoV with a reported mutation rate of 4 × 10−4 substitutions/site/year (Huang et al., 2012). Anti-viral vaccines are necessary to be developed before the predicted epitopes are potentially obsolete. Moreover, our immune-informatics based pipeline also provides a framework to identify B- and T- cell epitopes for SARS-CoV-2, but not limited to a specific virus. At the same time, we also have to mention that there are limitations in predicting T-cell epitopes. The prerequisite that an epitope can elicit T cell response is the epitope can bind to both MHC alleles and T cell receptors. However, the binding prediction between MHC alleles and an epitope is relatively more accurate, and the binding between an epitope and T cell receptors is extremely difficult to be predicted. In short, these results here will be useful to guide the design and evaluation of efficient and specific serological assays against epitopes, as well as help prioritize vaccine target designs during this unprecedented crisis (Poh et al., 2020).
Authors’ contributions
JZ and YBF conceived and designed this study; JZ, LL, TS, YFH, and WDL performed immune-informatics analysis. JZ and YBF wrote the manuscript. JZ, YBF, LL, TS, YFH, and WDL improved and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (NSFC No. 11421202, and 11827803 to YBF), the Youth Thousand Scholar Program of China (J.Z.) and Beijing Advanced Innovation Center for Biomedical Engineering, BUAA (J.Z.)
CRediT authorship contribution statement
Lin Li: Data curation, Formal analysis, Visualization, Writing - review & editing. Ting Sun: Data curation, Methodology, Writing - review & editing. Yufei He: Data curation, Methodology, Writing - review & editing. Wendong Li: Data curation, Methodology, Writing - review & editing. Yubo Fan: Conceptualization, Supervision, Writing - review & editing. Jing Zhang: Conceptualization, Supervision, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
Acknowledgments
We thank Qiwen Gan (Postdoctoral research scientist at Columbia University Medical Center) for the discussion about the 3D structure of the RBD domain of S protein and ACE-2.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.198082.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Wrapp D. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Rice C.M. Molecular biology of flaviviruses. Adv. Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- Bosch B.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 2017;114(42):11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang H. Protective humoral responses to severe acute respiratory syndrome-associated coronavirus: implications for the design of an effective protein-based vaccine. J. Gen. Virol. 2004;85(Pt 10):3109–3113. doi: 10.1099/vir.0.80111-0. [DOI] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.T. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 2004;190(2):379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Progress and prospects on vaccine development against SARS-CoV-2. Vaccines (Basel) 2020;8(2) doi: 10.3390/vaccines8020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir Ul Qamar M. Peptide vaccine against chikungunya virus: immuno-informatics combined with molecular docking approach. J. Transl. Med. 2018;16(1):298. doi: 10.1186/s12967-018-1672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.H. The SARS-CoV-2 vaccine pipeline: an overview. Curr. Trop. Med. Rep. 2020:1–4. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1) doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong C.Y. Recent advances in the vaccine development against middle east respiratory syndrome-coronavirus. Front. Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindewolf C., Menachery V.D. Middle East respiratory syndrome vaccine candidates: cautious optimism. Viruses. 2019;11(1) doi: 10.3390/v11010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- Liu W.J. T-cell immunity of SARS-CoV: implications for vaccine development against MERS-CoV. Antiviral Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. Epitope-based vaccine target screening against highly pathogenic MERS-CoV: an in silico approach applied to emerging infectious diseases. PLoS One. 2015;10(12):e0144475. doi: 10.1371/journal.pone.0144475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi F. Recent progress in adjuvant discovery for peptide-based subunit vaccines. Hum. Vaccin. Immunother. 2014;10(3):778–796. doi: 10.4161/hv.27332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3) doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast E.C., Chen Binbin. Potential T-cell and B-cell epitopes of 2019-nCoV. BioRxiv. 2020 [Google Scholar]
- Tai W. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Qiu Y. Predicting the angiotensin converting enzyme 2 (ACE2) utilizing capability as the receptor of SARS-CoV-2. Microbes Infect. 2020 doi: 10.1016/j.micinf.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J.T. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan D.W. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013;41(Web Server issue):W349–57. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre F., Clote P. DiANNA 1.1: an extension of the DiANNA web server for ternary cysteine classification. Nucleic Acids Res. 2006;34(Web Server issue):W182–5. doi: 10.1093/nar/gkl189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3(3):e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Raghava G.P.S. Springer Berlin Heidelberg; Berlin, Heidelberg: 2004. BcePred: Prediction of Continuous B-Cell Epitopes in Antigenic Sequences Using Physico-chemical Properties. [Google Scholar]
- Kringelum J.V. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput. Biol. 2012;8(12):e1002829. doi: 10.1371/journal.pcbi.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. U.S.A. 2013;110(22):E2046–53. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63(6):325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir Ul Qamar M. Epitope-based peptide vaccine design and target site depiction against Middle East respiratory syndrome coronavirus: an immune-informatics study. J. Transl. Med. 2019;17(1):362. doi: 10.1186/s12967-019-2116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco L.G. PepSite: prediction of peptide-binding sites from protein surfaces. Nucleic Acids Res. 2012;40(Web Server issue):W423–7. doi: 10.1093/nar/gks398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S.J. The pepATTRACT web server for blind, large-scale peptide-protein docking. Nucleic Acids Res. 2017;45(W1):W361–W364. doi: 10.1093/nar/gkx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38(Web Server issue):W529–33. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieser T.M. Influence of protein flexibility and peptide conformation on reactivity of monoclonal anti-peptide antibodies with a protein alpha-helix. Proc. Natl. Acad. Sci. U.S.A. 1987;84(23):8568–8572. doi: 10.1073/pnas.84.23.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S. Structural basis to design multi-epitope vaccines against Novel Coronavirus 19 (COVID19) infection, the ongoing pandemic emergency: an in silico approach. bioRxiv. 2020 2020.04.01.019299. [Google Scholar]
- Menssen R. Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus. J. Mol. Biol. 1999;285(2):645–653. doi: 10.1006/jmbi.1998.2363. [DOI] [PubMed] [Google Scholar]
- Maenaka K. Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes. J. Immunol. 2000;165(6):3260–3267. doi: 10.4049/jimmunol.165.6.3260. [DOI] [PubMed] [Google Scholar]
- Smith K.J. Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53. Immunity. 1996;4(3):215–228. doi: 10.1016/s1074-7613(00)80430-6. [DOI] [PubMed] [Google Scholar]
- Bakhshesh M., Ranjbar M., Almasi S. Immunoinformatic analysis of glycoprotein from bovine ephemeral fever virus. Biomed. Biotechnol. Res. J. 2018;2(3):208–212. [Google Scholar]
- Huang J. Residue analysis of a CTL epitope of SARS-CoV spike protein by IFN-gamma production and bioinformatics prediction. BMC Immunol. 2012;13:50. doi: 10.1186/1471-2172-13-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh C.M. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020;11(1):2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.