Abstract
Immunoaffinity based EV isolation technologies use antibodies targeting surface markers on EVs to provide higher isolation specificity and purity compared to existing approaches. One standing challenge for researchers is how to release captured EVs from the substrate to increase downstream and biological studies. The strong binding between the antibody and antigen or the antibody and substrate is commonly unbreakable without operating at conditions outside of the critical physiological range, making the release of EVs problematic. Additionally, immuno-affinity approaches are usually low-throughput due to their low flow velocity to ensure adequate time for antibody-antigen binding. To overcome these limitations, we modified the OncoBean chip, a previously reported circulating tumor cell isolation microfluidic device. The OncoBean chip is a radial flow microfluidic device with bean-shape microposts functionalized with biotin-conjugated EPCAM antibody through biotin-avidin link chemistry. It was demonstrated that the high surface area and varying shear rate provided by the bean-shaped posts and the radial flow design in the chip, enabled efficient capture of CTCs at high flow rate. We replace the anti-EPCAM with antibodies that recognize common EV surface markers to achieve high-throughput EV isolation. Moreover, by incorporating desthiobiotin-conjugated antibodies, EVs can be released from the device after capture, which offers a significant improvement over the existing isolation. The released EVs were found to be functional by confirming their uptake by cells using flow cytometry and fluorescent microscopy. We believe the proposed technology can facilitate both the study of EVs as cell-to-cell communicators and the further identification of EV markers.
Introduction
Extracellular vesicles (EVs) are a group of heterogeneous membrane-bound vesicles that include exosomes, microvesicles and apoptotic bodies, which are actively secreted by almost all cell types into extracellular spaces.1,2 These vesicles have been widely investigated and are considered to be powerful mediators of cell-to-cell communications. They can travel across great distances within the human body through the circulation and release their cargos upon internalization by recipient cells.2 Emerging evidence has shown that genetic information carried by these nanovesicles supports various biological functions including activating anti-apoptosis, enhancing blood vessel formation, and regulating immune response.2–5 Moreover, they have been shown to carry and transfer proteins and nucleic acids that are reflective of their originated cells.
The role of EVs and their cargo in promoting pathological processes in various disease, such as cancer and neurodegenerative diseases is becoming clearer, with many studies linking specific EV cargo to disease progression and outlook.6–11 As such, proteomic and genomic analysis of EVs can potentially provide a valuable biomarker for the detection, characterization, and monitoring of disease progression. For instance, miRNA dysregulation in EVs have been detected in various types of cancer, such as brain and lung cancer.12–14 The miRNAs carried by EVs released from the tumor or the tumor microenvironment have been shown to deeply influence tumorigenesis and therapeutic response.12,15 For example, miRNAs found in EVs secreted by lung cancer cells were shown to be promote tumor growth and metastasis through alteration of the immune response.16 Furthermore, dysregulated miRNAs in EVs have also been considered as a diagnostic tool for many cancer types.17,18 In addition to cancers, recent studies have shown that the cargos shuttled by EVs can be biomarkers for neurodegenerative disease, such as Alzheimer’s and Parkinson’s disease.19,20 Thus, isolating and analyzing the contents of EVs can provide researchers and clinicians valuable information about a patient’s diseases status, potentially even informing future diagnostic or prognostic tests.
Despite the valuable information housed in EVs, the lack of efficient isolation methods is still a major limitation for the study of EVs. The extremely small and heterogeneous size of EVs within a sample, 30–2000 nm, makes isolation challenging. The current standard isolation method is differential ultracentrifugation (UC), which is used to isolate EVs from various sample types including cell culture supernatant, blood, urine, and cerebral fluid.21–23 Using UC, samples are processed through serial centrifugation steps with increasing speeds to remove cells and cellular debris before pelleting the target population of vesicles. Critical drawbacks such as lengthy processing time and inefficient yields make it challenging for EV studies where the sample volume is low or the target EVs are low in number.24,25 Furthermore, several studies have shown that the high centrifugation force damages the membrane integrity of EVs and promotes EV rupture and coagulation, hindering potential downstream analysis. A recent push to move away from ultracentrifugation, reduce EV loss, increase purity, and preserve sample integrity has led the development of new isolation technologies to replace ultracentrifugation.26–28
Besides ultracentrifugation, techniques such as ultrafiltration and precipitation have been developed for EV isolation. These methods often exploit the different physical properties among EVs, including size, density, and solubility, to isolate EVs from various sample types. These techniques are often limited by contamination with non-vesicular proteins or off-target extracellular vesicles, as summarized in Table S1. One of the most promising alternative EV isolation techniques, immunoaffinity-based capture, use antibodies to target surface markers on EVs. Common targets of immunoaffinity based capture are tetraspanins, specifically CD9, CD63, and CD81. These are a group of proteins that are broadly accepted EV markers.26,29 For example, Koliha et al have demonstrated EV capture using magnetic beads coated with antibodies against CD9, CD63, and CD81.30 Boriachek et al. functionalized gold-loaded ferric oxide nanocubes with CD9 and CD63 antibodies to isolate EVs from culture media.31 Since the molecular composition of EVs is also dependent on their parent cells, novel markers such as epithelial cellular adhesion molecule (EPCAM) and epidermal growth factor receptor (EGFR) have also been used for capturing specific EV populations. For instance, magnetic beads coated with chondroitin sulphate peptidoglycan 4 antibody has been reported to isolate EVs from plasma to study melanoma cancer.32 Zhou et al. designed a microfabricated chip with multiplexed gold sensors that capture EVs with EPCAM antibody.33
Among the types of immunoaffinity isolation technologies, microfluidic platforms with antibody-coated surfaces have become a promising alternative EV isolation strategy. These devices have small dimensions which facilitate minimized reagent volumes, isolation times, and procedural costs while enhancing the product purity and sensitivity.24,34,35 For instance, Kanwar et al. developed a microfluidic platform, Exochip, that isolates EVs from plasma from pancreatic cancer patient using anti-CD63.36 The device enables rapid EV quantification and facilitates EV protein and miRNA characterizations. Zhang et al. designed a device with graphene oxide nanoposts coated with CD81 antibody to detect and isolate EVs at low detection limit.37 Vaidyanathan et al. fabricated a microfluidic device with functionalized gold electrodes coated with CD9, human epidermal growth factor receptor 2 (HER2) and prostate specific antigen (PSA) antibodies.38 The capture of EVs is enhanced by nanoshearing between the electrodes induced by an alternating electric current. Zhang et al. devised a 3D-nanopatterned microfluidic chip with a porous herringbone mixer fabricated by assembled silica colloids to capture EVs from serum samples in ovarian cancer.39 They have shown that their device is compatible with downstream analysis including ELISA, western blotting and digital PCR.
Although current immunoaffinity isolation technologies provide more specific EV enrichment and isolation, these technologies suffer from limitations of low throughput as high flow rates would hinder the antigen-antibody interaction. Additionally, surface-antibody-EV binding is tight, making it challenging to release and retrieve viable EVs post isolation. Being unable to recover intact EVs is a critical drawback for studying the interaction between cells and EVs, hindering both in-vitro and in-vivo studies. Despite advances in the field, a need exists to further improve microfluidics platforms to isolate and harvest EVs.
Many immunoaffinity-based technologies for EV capture rely on the high-affinity binding of biotin-avidin to immobilize the capture antibodies on the micro/nanostructured surface or beads. Owing to the simple procedure and stable binding, biotin-conjugated antibodies have been used for antibody-based capture not just for EVs but also for other targets including cells, lipids, and enzymes.40,41 However, the irreversibility of the biotin-avidin binding limits the use of captured EVs as the EVs would need to be lysed in some way for removal. Strategies to break biotin-avidin binding frequently operate at conditions outside of the physiological range, including extreme pH and high temperatures, which potentially reduce the integrity of the captured samples.42,43
The use of desthiobiotin, an analogue of biotin, has been reported to be an effective alternative to standard biotin.42 Desthiobiotin has a lower binding affinity to avidin than biotin. Hence, samples captured using desthiobiotin-conjugated antibodies can be eluted or released from an avidin-coated surface using a biotin solution. This elution leads to the replacement of desthiobiotin with biotin, effectively releasing the sample from the capture surface and allowing for downstream applications.44 This capture-release method has been used for the isolation and release cells and proteins.45 In the present study, we incorporated this desthiobiotin release method to achieve the release of intact EVs using a high throughput immunoaffinity based microfluidic device.
We have previously developed the OncoBean chip, a radial flow microfluidic device with bean-shape microposts functionalized with biotin-conjugated anti-epithelial cell adhesion molecule (EpCAM) antibody through biotin-avidin link chemistry.46 It was demonstrated that the high surface area and stable shear rate provided by the bean-shaped posts, as well as the radial flow design in the chip, enabled efficient capture of circulating tumor cells (CTCs) at high flow rates.46,47 Here, we apply this chip with antibodies against common EV surface markers, CD9, CD63, and CD81, to achieve high throughput EV isolation. Our results showed that the device is well suited to process large volumes of samples, which is ideal for obtaining high concentrations of EVs for downstream analysis. Furthermore, by using desthiobiotin-conjugated antibodies instead of biotinylated ones, EVs can be released and harvested from the device after capture. The released EVs were shown to be functional by demonstrating their internalization by cells based on flow cytometry. One exciting application of releasing exosomes is to investigate the uptake of exosomes into distant cells. We performed initial experiments to first test the integrity of the EVs following release. To do so we performed immunofluorescent labelling of EVs for common EV proteins (CD81 and CD63) before demonstrating a successful staining using flow cytometry. We then used fluorescently dyed EVs to perform an initial uptake experiment and demonstrated the uptake of released EVs by cells. We believe that this novel strategy can be used for a high throughput EV enrichment which will enable both systematic analysis of EV cargo and investigations into their interactions with recipient cells.
Experimental
OncoBean fabrication and functionalization
The OncoBean Chip, a previously reported microfluidic device for isolating circulating tumor cells, was utilized for EV capture in the present study.40 The device is made of a polydimethylsiloxane (PDMS) top bonded to a glass slide. The mold for the polydimethylsiloxane (PDMS) device is fabricated using a negative photoresist, SU-8 100, patterned silicon wafer using standard soft lithography techniques. Briefly, negative photoresist SU-8 100 (MicroChem Corp) was spin coated onto a silicon wafer at 2350 rpm before performing UV exposure, post exposure baking, developing, and feature measuring. Well-mixed PDMS and curing agent were poured onto a SU-8 mold at a ratio of 10:1 and degassed in a desiccator for 30 minutes to remove all bubbles. The polymer was cured at 65 °C overnight in the oven. After baking, the polymer was peeled off from the mold and cut for surface functionalization. The PDMS chamber was bonded to a standard sized glass slide via plasma surface activation of oxygen.
The completed device was processed with 3-mercaptopropyltrimethoxysilane (Gelest) in ethanol by syringe injection and incubated for an hour. This device was then washed with ethanol and treated with N-gamma-Maleimidobutyryloxy-Succinimide (GMBS) (ThermoScientific) for 30 min. This was followed by rinsing with ethanol and adding NeutrAvidin before incubating. The device was then stored at 4 °C for future use.
Sample Preparation and EV isolation
The sample collection and experiments were approved by University of Michigan Institutional Review Board (IRB). Informed consents were obtained from all participants of this clinical study and the blood samples from all patients and healthy donors were obtained after approval of the institution review board (IRB) at the University of Michigan. All experimental protocols were performed in accordance with the approved guidelines and regulations by the ethics committee at the University of Michigan. Whole blood samples from healthy donors were drawn into EDTA tubes and were subsequently centrifuged at 2000 x g for 15 minutes to collect the top layer of plasma. The isolated plasma samples were kept at −80°C until use.
Cell culture
Patu8988t pancreatic cancer cell line was cultured in DMEM medium with 10% FBS. Cultures were incubated at 37°C in a humidified 5% CO2 incubator (Thermo Fisher Scientific, Waltham, MA). Cells were grown until they reached 70–80% confluence, at which time, they were subjected to experimentation.
EV Capture and release
Before experiments, the device was functionalized with the capture antibody by injecting biotin or desthio-biotin conjugated antibody in 1% bovine serum albumin (BSA) (Sigma Aldrich) in phosphate buffered saline (PBS) and incubating for 60 minutes. After antibody immobilization and washing with 1mL of PBS at a flowrate of 50 μl/min, 3% BSA was processed at a flow rate of 50 μl/min for 10 min to block the excess reaction sites and prevent non-specific binding. Human plasma or cell culture medium were processed through the OncoBean Chip for EV capture, followed by washing with PBS at a flow rate of 50 μl/min for 20 min. The EVs captured by the desthiobiotin antiCD63 conjugated device were released from the device using a 0.5 mM biotin solution. The biotin solution was incubated for 1 hour in the device, followed by a wash with biotin solution and the collection of released EVs. The released EVs were either characterized by nanoparticle tracking analysis, NTA, using the NanoSight NS300 (Malvern Instruments, UK) to determine the size and concentrations, or proceeded to functional studies. The EVs immobilized by the device using biotinylated antibodies were lysed for RNA and protein extraction.
Electron Microscope (EM) analysis of captured Evs
A small portion of the PDMS top of the device after EV isolation was cut out using a biopsy punch. Each punched PDMS specimen was fixed in 2.5% glutaraldehyde in PBS at room temperature for one hour and then rinsed with PBS, followed by sequential dehydration with ethanol at concentrations of 50%, 70%, 90%, 95%, and 100% for 10 min each. The specimen was then immersed for 10 min in solution of 1:1 ethanol: hexamethyldisilazane (HMDS) and then transferred to 100% HMDS, followed by overnight air drying in the hood. The dehydrated specimen was then attached to carbon double sided tape and was mounted on a SEM stub before coating with conductive a layer. The EVs were examined by FEI Nova 200 Nanolab Dualbeam FIB scanning electron microscope under low beam energies (2.0–5.0 kV) at the Michigan Center for Materials Characterization (MC2) at University of Michigan.
Western blotting and protein quantification
To harvest the protein of the EVs captured, the device chamber was injected with RIPA buffer (Sigma) with 1% Halt protease inhibitor (Thermo) at a flowrate of 50 μl/min for 2 min, followed by a 10 min incubation on ice. This was followed by an injection of 70μL per device at the same rate and the effluent was collected. After another 10 min incubation, the remaining effluent in the devices was pushed out by pumping air manually. The collected samples were stored at −80 °C until used.
Total protein was measured by standard micro-BCA analysis according to the manufacturer’s instructions (Thermo). Lysed protein was mixed with 4x Laemmli buffer with 2-mercaptoethanol at a ratio of 4:1 and heated at 95° for 7 minutes. The protein samples were then loaded on a 10-lane 4–20% SDS gradient gel (BioRad) and run for 47 minutes at 120V in Tris-Glycine-SDS buffer (BioRad). The gel was then transferred to a methanol-activated PVFD membrane at 120V for 1hr using BioRad’s Minigel wet transfer system. Following transfer, the membrane was rinsed with DI water before drying for 1 hour. The membrane was then reactivated with methanol, rinsed with DI water and submerged in tris-buffered saline with 1% Tween 20 (TBST) before blocking in 5% non-fat milk in TBST for 1 hour at room temperature on a rocker. Primary antibodies then incubated overnight in 4° on a rocker at a concentration of 1:1000 for both CD9 (Cell Signaling) and Beta-Actin (Cell Signaling). Primary antibodies were rinsed 6 times, 3 quick rinses and 3× 5 minutes on a rocker, in TBST before applying anti-rabbit HRP secondary at a concentration of 1:1500 (Cell Signaling) for 1.5 hrs on a rocker at room temperature. Secondary antibody was rinsed as previously described and Thermo’s SuperSignal PICO Reagant was applied for 5 minutes before imaging using BioRad’s ChemiDoc Imager.
RNA preparation, RT, and real-time qPCR
Total RNA from EVs was extracted using Qiazol (QIAGEN). After the EVs were immobilized within the device, 120μl of Qiazol was flowed through the device at 50 μl/min, followed by 10 min incubation. The device was then processed with another 150 μl at the same flow rate while the effluent was collected. After another 10 min incubation, the remaining effluent in the devices was pushed out by pumping air manually. The collected samples were stored at −80 °C until used.
SYBR Green-based real-time qPCR technique was performed for detection of miRNAs, as previously described.42 Total RNAs were purified from isolated EVs using a Single Cell RNA Extraction Kit (Norgen). Purified RNA amount was measured by NanoDrop Lite Spectrophotometer (Thermo Fisher Scientific). Reverse Transcription Kit (Thermo Fisher Scientific) was used to generate single-stranded cDNA from an equal amount of purified RNAs.
Flow Cytometry
For directly exosome surface staining, EVs were incubated with preconjugated FITC mouse anti-human CD63 (BioLegend) and APC Anti-Human CD81 (BioLegend) for 30 min at room temperature. After incubation, the samples were washed with PBS, ultracentrifuged and then resuspended in PBS buffer. Stained samples were analyzed using ZE5 (Bio-Rad) flow cytometer and FlowJo software (Treestar).
Uptake of Evs
Following release from the capture device, EVs were labeled by PKH26 Red Fluorescent Cell Linker Kit (Sigma), and Exosome Spin Columns (MW 3000) (Thermo Fisher Scientific) were used to remove excess dye as previously described.14 Patu8988t cells were seeded into a 6 well plate and allowed to settle and adhere for 48 hours before the dyed EVs were added for a 12 h incubation in a 37° cell culture incubator as previously described. Flow cytometry was performed to measure mean fluorescence intensity (MFI) of cells.
Results and discussion
Evaluation of EV capture with OncoBean chip
While previous work assessed the ability of OncoBean to isolate circulating tumor cells from blood samples, the present study explored the OncoBean’s capture potential and throughput for isolating extracellular vesicles. Fig. 1 schematically illustrates EVs being captured on the OncoBean’s antibody-coated bean-shaped microposts. The increasing cross-sectional area in radial flow design provides a decreasing flow velocity from inlet to outlet, which allows for higher flow rate compared to linear flow devices. The high surface area provided by the bean-shaped microposts also increases the contact of EVs with capture antibodies, thus enabling high capture at high flow rates.
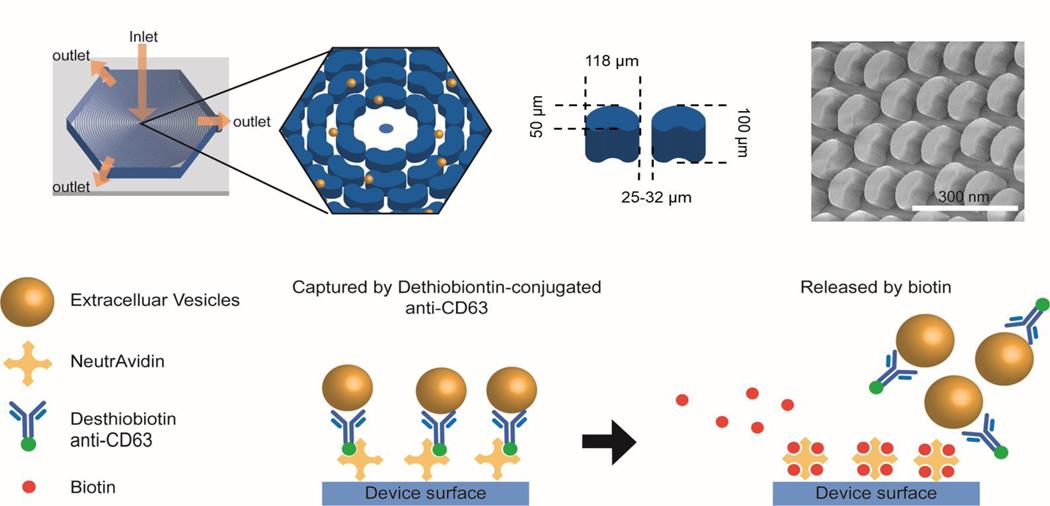
Fig 1.
Schematic illustration of OncoBean Chip and EV isolation. The design of the device can be seen in the previous report. In general, the bean posts are 50 μm in width, 118 μm along the longest axis, and 100 μm in height. The posts were placed 25–32 μm apart from each other. The device surface functionalized with Neutravidin can be coated with desthiobiotin-conjugated antibody that recognize surface markers of EVs. The EVs are captured by flowing samples through the chip. Compared to biotin, desthiobiotin has a lower binding affinity to avidin, thus facilitating a release mechanism for the EVs. Though the introduction of the biotin, the desthiobiotin-antibody-EV complex releases from the Neutravidin coated surface, allowing for EV collection.
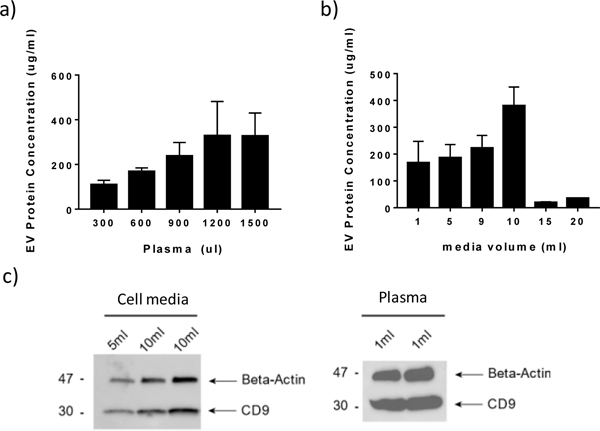
To demonstrate the ability of the OncoBean chip to perform high-throughput isolation of EVs, we used the two most common sources of EVs: plasma and cell culture media. First, we used biotinylated CD63 and CD9 antibodies as capture antibodies to optimize the flowrate and sample volume for EV isolation. With the goal of minimizing the processing time, we fixed the processing time at 1 hour and tested the capture using varying sample volumes to determine the capture capacity of the device. Protein was extracted by flowing RIPA extraction buffer through the device post EV capture. We used the protein concentration measured by microBCA kit to quantify the EVs captured. As can be seen in Fig 2a, when the volume of plasma was increased to 1200 μl, the protein concentration reached the maximum value and further increasing the plasma volume did not increase the protein concentration. For cell culture media, the chip was capable of processing up to 10 mL of culture media in an hour, demonstrating high throughput EV isolation (Fig 2b). However, the protein concentration decreased greatly at flow rates higher than 10 ml/hr. This could be caused by the limited contact time between the EVs and antibodies. Further, we performed Western blot analysis on the protein extracted from the chip to verify the EV capture, as shown in Fig 2c. The presence of CD9, a specific exosome marker, was confirmed in both culture media and plasma samples, and β-actin was used as a loading control.
Fig 2.
Characterization of EVs captured using OncoBean chip. a-b) Protein levels in EVs isolated from a) Serum and b) cell culture medium. c) a representative western blot analysis of the proteins from EVs isolated from OncoBean using anti-CD81 and anti-CD63 and characterized for CD9 expression levels in plasma and cell culture medium.
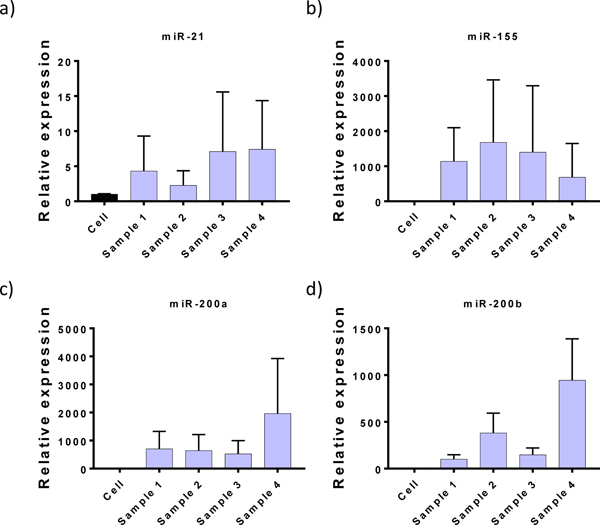
In addition to the protein analysis, we also evaluated our device for its applications in performing miRNA analysis of EVs. A variety of different types of RNA molecules have been identified in EVs. Messenger RNAs (mRNAs), long non-coding RNAs (lncRNAs), ribosomal RNA (rRNA), microRNAs (miRNAs) and the fragments of these intact RNA molecules have all been identified.49 MiR-21, miR-155 and miR-200 have been shown to be enriched in EVs and their expression levels have been correlated with poor prognosis in pancreatic cancer.50 To test the capabilities of our device to capture EVs for miRNA analysis, we performed RT-PCR to measure the level of miR-21, miR-155 and miR-200 in the EVs. Similar to the previously reported pancreatic cancer EVs, we observed enrichment of miR-21, miR-155 and miR-200 in the device isolated EVs (Fig 3). The characterization of EV protein and miRNA demonstrates the utility of the OncoBean Chip for downstream EV analysis after high throughput capture of both low and high sample volumes.
Fig 3.
a) miR-21, b) miR-155, c) miR-200a, and d) miR-200b expressions from device isolated EVs. The RNA was obtained by lysing the captured EVs in the device and quantified using the real-time quantitative PCR (RT-qPCR). Individual miRNA expression levels are shown in bar graphs.
Release and harvest EVs from the device
A critical disadvantage of using biotin-conjugated antibodies for EV capture is the irreversible binding between biotin and avidin, hindering the harvest of intact EVs from the device. To overcome this challenge, we used desthiobiotin-conjugated anti-CD63 instead of biotinylated anti-CD63 to capture EVs. Compared to biotin-avidin binding, desthiobiotin-avidin binding can be reversed because of the lower binding affinity between desthiobiotin and avidin. Desthiobiotin binds with avidin with a lower affinity than biotin and can be released from avidin through displacement by a biotin molecule. When we used desthiobiotin-conjugated antibodies instead of biotinylated antibodies, immobilized EVs captured on the surface of the device were released through exchange reactions by the replacement, as illustrated in Fig 1. The stronger affinity between the biotin and Neutravidin causes the replacement of desthiobiotin with biotin, and thus enabling the release of the EVs from the neutravidin-coated surface.
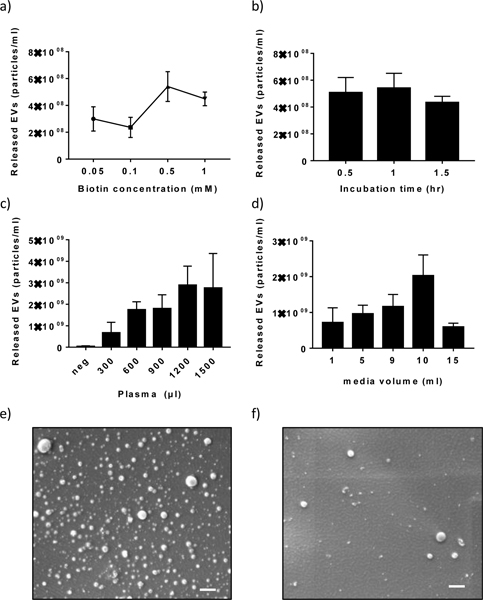
Optimization studies were conducted in order to determine a reasonable biotin concentration that allowed for effective release of EVs from the device. Pre-purified EVs from heathy plasma (System Biosciences) were captured using desthiobiotin-conjugated anti-CD63 on the OncoBean chip and the released EVs were then quantified with NTA analysis. As shown in Fig 4a, the concentration of released EVs increased along with increasing concentrations of biotin and reached a maximum at a concentration of 0.5 mM. This result led us to use 0.5 mM for all further experiments to achieve effective release of EVs. In addition to the biotin concentration, we also evaluated the influence of biotin solution incubation time. However, as illustrated in Fig 4b, the incubation time was not a significant factor for EV release. Fig. 4c and 4d shows the concentration of EVs collected after release from the outlet at different sample volumes using the 0.5mM biotin solution. These graphs showed that our device can harvest EVs from up to 1.2 ml of plasma or 10 ml of cell culture medium. The lower optimal flow rate for plasma was expected due to the higher viscosity. According to the previous report, the adhesion of antigen to antibody can be drop rapidly with increasing shear stress.51 Therefore, a lower flow rate is required to compensate the higher shear stress experienced by EVs in samples with higher viscosities. To the best of our knowledge, this is the highest throughput achieved on a microfluidic platform for EV isolation from culture medium reported to date. Furthermore, the strength of the technology was evaluated by testing clinical specimens from cancer patients. Plasma samples from stage III non-small cell lung cancer patients and healthy donors were processed using the OncoBean chip to harvest EVs for NTA measurement (Fig. S1). Patient demographics are provided in Table S2. We observed no significant difference in total EV concentration between cancer patients and healthy donors, which corroborates previous reports.35,52 These results demonstrated the potential clinical utility of the OncoBean chip for EV capture and analysis.
Fig 4.
Characterization of EVs isolated using the OncoBean chip. a-b) NTA analysis of harvested EV with varying a) biotin concentration and b) incubation time for the release of EV using pre-purified EVs (System Biosciences). c-d) NTA measurement of EVs harvested from c) plasma and d) cell culture media. e-f) Electron microscope images show the presence of EVs immobilized e) before and f) after the biotin release step (bars = 1μm)
To confirm the successful release of EVs from the chip, SEM images were taken showing the device with and without the injection of biotin, or with and without EV release. The images clearly showed that most of the EVs captured on the chip were released after the biotin. (Fig 4e–f), and thus further validating our ability to capture and release EVs form microfluidic chip.
Characterization and cell uptake of harvested EVs
Accumulating evidence has shown that EVs can act as powerful mediators of cell-to-cell communication, facilitating various biological events. Importantly, EVs and their cargo have been shown to play important roles in disease progression. Therefore, harvesting EVs that are functional is important for in-vivo and in-vitro studies. To demonstrate that the captured and released EVs are functional and preserve their surface markers, we performed flow cytometry analysis to examine EV surface markers and cellular internalization of EVs.
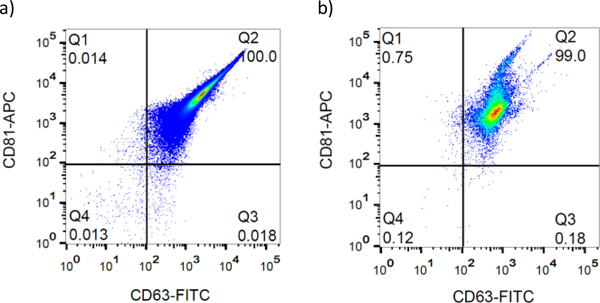
We used direct flow cytometry to examine two common EV surface markers, CD63 and CD81, on EVs isolated using the OncoBean with release and ultracentrifugation. As shown in Fig 5a, the population of positive EVs from ultracentrifugation is found in the top right demonstrating the presence of both CD63 and CD81 on each EV. Similar to the ultracentrifuged EVs, device isolated and released EVs have more than 99% CD63 and CD81 positive populations (Fig 5b). This data not only demonstrated that these vesicles represent EVs but also highlighted a sensitive method for surface marker characterization of EVs using flow cytometry. Therefore, this approach can be used to differentiate and identify heterogenous populations of EVs, thus providing insights into identifying EV surface markers.
Fig 5.
Flowcytometry analysis of a) ultracentrifuged and b) device isolated and released EVs. The anti-CD63 antibody conjugated with FITC and anti-CD81 antibody conjugated with APC were used to immunolabel the EVs.
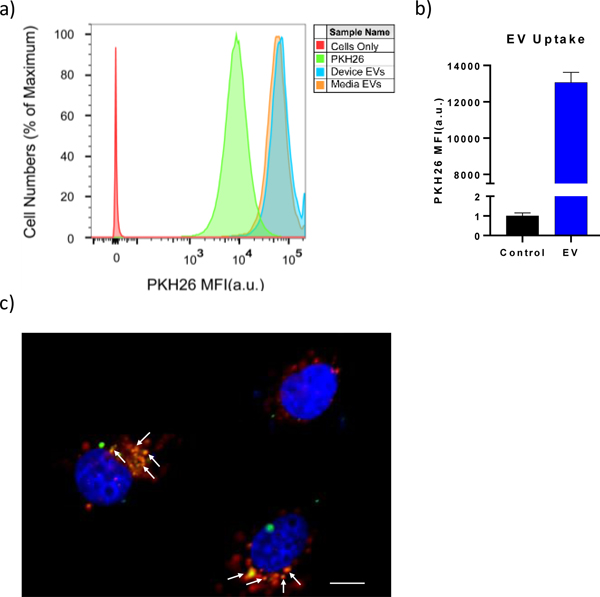
The goal of recovering viable EVs from a sample is to study the influence of EVs on the behavior of recipient cells after internalization. In order to demonstrate the biological functionality of released EVs, we demonstrate that they can be internalized by cancer cells. To do this, we compared internalization between EVs isolated from cell culture media using both the OncoBean device and ultracentrifugation. To examine whether EVs are taken up by Patu8988t cancer cells, we pre-labelled isolated EVs with PKH green dye and incubated them with Patu8988t cells for 12h and analyzed their internalization into the cancer cells using flow cytometry. As indicated by the higher green peak, the ultracentrifuged sample had similar uptake to the device-released EVs while they both had markedly more uptake into cells than the no-EV negative control using only PKH dye. This demonstrated that EVs are indeed taken up by Patu8988t cancer cells. Additionally, we found that device isolated EVs were similarly functional compared to the ultracentrifuged ones (Fig 6a and Fig 6b).
Fig 6.
a) Flow cytometry analysis shows uptake of device-derived and ultracentrifuged EVs by Patu8988t cells. The EVs were pre-labeled with PKH67 dye before the uptake. Two samples without added EVs were used as negative controls: one with PKH dye, one without PKH dye. b) The comparison of mean fluorescence intensity (MFI) of cells treated with EVs to the negative control of cells with no EVs. c) PKH67-labeled EVs (green) were trafficked to lysosomes labeled by Lysotracker (red). The traffic of PKH67-labeled EVs through lysosomes was confirmed by the colocalization of PKH67 and Lysotracker in Patu8988t cells, as indicated by arrowheads. The yellow regions indicate colocalization of green and red, or EVs and lysosomes.
We also studied the intracellular trafficking of internalized EVs to lysosomes in Patu8988t cells to further verify the EV uptake. Lysosomes are membrane-bound organelles found in nearly all animal cells. They are spherical vesicles which contain hydrolytic enzymes that can break down many kinds of biomolecules.53 Lysosomes are an essential part of the vesicular compartment and connect the outside medium with many classes of cellular targets in the cytosol, nucleus, mitochondria, endoplasmic reticulum, and Golgi.54 EVs can reach the lysosome through endocytosis. The capture of EVs occurs through specific endocytic mechanisms according to the nature of the cargo. After uptake, EVs are routed to early endosomes. From the endosomes, the EVs can either be recycled back to the plasma membrane or sorted and targeted for lysosomal degradation.
To verify the migration of EVs into lysosomes, cells were incubated with Lysotracker (red; to label lysosomes) and cocultured with PKH67-labeled (green) EVs from human plasma and analyzed by confocal microscopy. PKH67 detected in lysosomes demonstrated the colocalization of EV and lysosomes (Fig 6c), demonstrating that the EVs from device are biologically active.
Using flow cytometry and fluorescent microscopy, we have shown the feasibility of using the immunoaffinity based OncoBean device for EV capture and release. We were then able to show the uptake of isolated and enriched EVs by cells and confirmed their surface markers using flow cytometry. We strongly believe this technology will facilitate studies into the role of EVs as cell-to-cell communicators.
Conclusions
In this study, we have successfully demonstrated the utility of the high-throughput OncoBean for EV isolation from cell culture supernatant and human plasma. The high surface area and radial flow design provided by the bean-shaped microposts facilitate capture of EVs not only at high flow rate, but also from larger volumes of media form cell culture supernatant. Furthermore, our results indicate that the OncoBean chip facilitates the analysis of EV proteins and RNAs using western blot and qPCR. Most excitingly, through desthiobiotin antibody capture and biotin elution, we were able to release functional EVs from the device for cell uptake and identification of surface markers, which were verified using flow cytometry. We believe our microfluidic device can facilitate specific enrichment of EVs enabling the study of EVs in cell-to-cell communication.
Supplementary Material
Acknowledgements
The authors acknowledge the Lurie Nanofabrication Facility at the University of Michigan. The authors acknowledge the financial support of the University of Michigan College of Engineering and NSF grant #DMR-0320740, and technical support from the Michigan Center for Materials Characterization. S.N. is supported by (NIH) U01CA210152 and R01-CA-208335–01-A1, D.N. is supported by grants from National Cancer Institute (NCI) R01CA227622, R01CA222251, & R01CA204969.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Willms E, Johansson HJ, Mager I, Lee Y, Blomberg KE, Sadik M, Alaarg A, Smith CI, Lehtio J, El Andaloussi S, Wood MJ and Vader P, Sci Rep, 2016, 6, 22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raposo G and Stoorvogel W, J Cell Biol, 2013, 200, 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boukouris S and Mathivanan S, Proteomics Clin Appl, 2015, 9, 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Balkom BW, de Jong OG, Smits M, Brummelman J, den Ouden K, de Bree PM, van Eijndhoven MA, Pegtel DM, Stoorvogel W, Wurdinger T and Verhaar MC, Blood, 2013, 121, 3997–4006, S3991–3915. [DOI] [PubMed] [Google Scholar]
- 5.Barros FM, Carneiro F, Machado JC and Melo SA, Front Immunol, 2018, 9, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katsuda T, Kosaka N and Ochiya T, Proteomics, 2014, 14, 412–425. [DOI] [PubMed] [Google Scholar]
- 7.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW and Simpson RJ, Nat Rev Clin Oncol, 2018, 15, 617–638. [DOI] [PubMed] [Google Scholar]
- 8.Candelario KM and Steindler DA, Trends Mol Med, 2014, 20, 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson AG, Gray E, Heman-Ackah SM, Mager I, Talbot K, Andaloussi SE, Wood MJ and Turner MR, Nat Rev Neurol, 2016, 12, 346–357. [DOI] [PubMed] [Google Scholar]
- 10.Boriachek K, Islam MN, Moller A, Salomon C, Nguyen NT, Hossain MSA, Yamauchi Y and Shiddiky MJA, Small, 2018, 14. [DOI] [PubMed] [Google Scholar]
- 11.Poudineh M, Sargent EH, Pantel K and Kelley SO, Nat Biomed Eng, 2018, 2, 72–84. [DOI] [PubMed] [Google Scholar]
- 12.Yang F, Ning Z, Ma L, Liu W, Shao C, Shu Y and Shen H, Mol Cancer, 2017, 16, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi R, Wang PY, Li XY, Chen JX, Li Y, Zhang XZ, Zhang CG, Jiang T, Li WB, Ding W and Cheng SJ, Oncotarget, 2015, 6, 26971–26981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN and Pass HI, J Thorac Oncol, 2013, 8, 1156–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, Marini JC, Tudawe T, Seviour EG, San Lucas FA, Alvarez H, Gupta S, Maiti SN, Cooper L, Peehl D, Ram PT, Maitra A and Nagrath D, Elife, 2016, 5, e10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D and Croce CM, Proc Natl Acad Sci U S A, 2012, 109, E2110–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita Y, Yoshioka Y and Ochiya T, Cancer Sci, 2016, 107, 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salido-Guadarrama I, Romero-Cordoba S, Peralta-Zaragoza O, Hidalgo-Miranda A and Rodriguez-Dorantes M, Onco Targets Ther, 2014, 7, 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vinaiphat A and Sze SK, Expert Rev Mol Diagn, 2019, 19, 813–824. [DOI] [PubMed] [Google Scholar]
- 20.Croese T and Furlan R, Mol Aspects Med, 2018, 60, 52–61. [DOI] [PubMed] [Google Scholar]
- 21.Momen-Heravi F, Methods Mol Biol, 2017, 1660, 25–32. [DOI] [PubMed] [Google Scholar]
- 22.Thery C, Amigorena S, Raposo G and Clayton A, Curr Protoc Cell Biol, 2006, Chapter 3, Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 23.Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Nolte-’t Hoen EN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH and Hochberg F, J Extracell Vesicles, 2013, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liga A, Vliegenthart AD, Oosthuyzen W, Dear JW and Kersaudy-Kerhoas M, Lab Chip, 2015, 15, 2388–2394. [DOI] [PubMed] [Google Scholar]
- 25.Jayachandran M, Miller VM, Heit JA and Owen WG, J Immunol Methods, 2012, 375, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M and Thery C, Proc Natl Acad Sci U S A, 2016, 113, E968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sunkara V, Kim CJ, Park J, Woo HK, Kim D, Ha HK, Kim MH, Son Y, Kim JR and Cho YK, Theranostics, 2019, 9, 1851–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang YT, Purcell E, Palacios-Rolston C, Lo TW, Ramnath N, Jolly S and Nagrath S, Small, 2019, DOI: 10.1002/smll.201903600, e1903600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreu Z and Yanez-Mo M, Front Immunol, 2014, 5, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koliha N, Wiencek Y, Heider U, Jungst C, Kladt N, Krauthauser S, Johnston IC, Bosio A, Schauss A and Wild S, J Extracell Vesicles, 2016, 5, 29975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boriachek K, Masud MK, Palma C, Phan HP, Yamauchi Y, Hossain MSA, Nguyen NT, Salomon C and Shiddiky MJA, Anal Chem, 2019, 91, 3827–3834. [DOI] [PubMed] [Google Scholar]
- 32.Sharma P, Ludwig S, Muller L, Hong CS, Kirkwood JM, Ferrone S and Whiteside TL, J Extracell Vesicles, 2018, 7, 1435138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou YG, Mohamadi RM, Poudineh M, Kermanshah L, Ahmed S, Safaei TS, Stojcic J, Nam RK, Sargent EH and Kelley SO, Small, 2016, 12, 727–732. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M and Irimia D, Lab Chip, 2010, 10, 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang YT, Purcell E, Hadlock T, Lo TW, Mutukuri A, Jolly S and Nagrath S, Analyst, 2019, 144, 5785–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanwar SS, Dunlay CJ, Simeone DM and Nagrath S, Lab Chip, 2014, 14, 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang P, He M and Zeng Y, Lab Chip, 2016, 16, 3033–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaidyanathan R, Naghibosadat M, Rauf S, Korbie D, Carrascosa LG, Shiddiky MJ and Trau M, Anal Chem, 2014, 86, 11125–11132. [DOI] [PubMed] [Google Scholar]
- 39.Zhang P, Zhou X, He M, Shang Y, Tetlow AL, Godwin AK and Zeng Y, Nat Biomed Eng, 2019, 3, 438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luka G, Ahmadi A, Najjaran H, Alocilja E, DeRosa M, Wolthers K, Malki A, Aziz H, Althani A and Hoorfar M, Sensors (Basel), 2015, 15, 30011–30031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, Lin J, Ramnath N, Wicha MS, Hayes DF, Simeone DM and Nagrath S, Nat Nanotechnol, 2013, 8, 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirsch JD, Eslamizar L, Filanoski BJ, Malekzadeh N, Haugland RP, Beechem JM and Haugland RP, Anal Biochem, 2002, 308, 343–357. [DOI] [PubMed] [Google Scholar]
- 43.Holmberg A, Blomstergren A, Nord O, Lukacs M, Lundeberg J and Uhlen M, Electrophoresis, 2005, 26, 501–510. [DOI] [PubMed] [Google Scholar]
- 44.Wan Y, Cheng G, Liu X, Hao SJ, Nisic M, Zhu CD, Xia YQ, Li WQ, Wang ZG, Zhang WL, Rice SJ, Sebastian A, Albert I, Belani CP and Zheng SY, Nat Biomed Eng, 2017, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ansari A, Lee-Montiel FT, Amos JR and Imoukhuede PI, Biotechnol Bioeng, 2015, 112, 2214–2227. [DOI] [PubMed] [Google Scholar]
- 46.Murlidhar V, Zeinali M, Grabauskiene S, Ghannad-Rezaie M, Wicha MS, Simeone DM, Ramnath N, Reddy RM and Nagrath S, Small, 2014, 10, 4895–4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murlidhar V, Reddy RM, Fouladdel S, Zhao L, Ishikawa MK, Grabauskiene S, Zhang Z, Lin J, Chang AC, Carrott P, Lynch WR, Orringer MB, Kumar-Sinha C, Palanisamy N, Beer DG, Wicha MS, Ramnath N, Azizi E and Nagrath S, Cancer Res, 2017, 77, 5194–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Z, Zhang D, Lee H, Menon AA, Wu J, Hu K and Jin Y, J Leukoc Biol, 2017, 101, 1349–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crescitelli R, Lasser C, Szabo TG, Kittel A, Eldh M, Dianzani I, Buzas EI and Lotvall J, J Extracell Vesicles, 2013, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, Jamaluddin MS, Weakley SM, Yao Q and Chen C, World J Surg, 2011, 35, 1725–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tempelman LA and Hammer DA, Biophys J, 1994, 66, 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D and Kalluri R, Nature, 2015, 523, 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mindell JA, Annu Rev Physiol, 2012, 74, 69–86. [DOI] [PubMed] [Google Scholar]
- 54.Ferri KF and Kroemer G, Nat Cell Biol, 2001, 3, E255–263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.