Abstract
Accumulation of protoporphyrin IX (PPIX) and Zn-PPIX, are the clinical hallmarks of protoporphyria. Phenotypic expression of protoporphyria is due to decreased activity of ferrochelatase (FECH) or to increased activity of aminolevulinic acid synthase (ALAS) in red blood cells. Other genetic defects have been shown to contribute to disease severity including loss of function mutations in the mitochondrial AAA-ATPase, CLPX and mutations in the Iron-responsive element binding protein 2 (IRP2), in mice. It is clear that multiple paths lead to a common phenotype of excess plasma PPIX that causes a phototoxic reaction on sun exposed areas. In this study we examined the association between mitochondrial iron acquisition and utilization with activity of FECH. Our data show that there is a metabolic link between the activity FECH and levels of MFRN1 mRNA. We examined the correlation between FECH activity and MFRN1 mRNA in cell lines established from patients with the classical protoporphyria, porphyria due to defects in ALAS2 mutations. Our data confirm MFRN1 message levels positively correlated with FECH enzymatic activity in all cell types.
1. Introduction
The nuclear encoded mitochondrial enzyme ferrochelatase (FECH) catalyzes the insertion of ferrous iron into protoporphyrin IX in the last step of heme biosynthesis. The active form of human FECH is a homodimer, each half containing a 2Fe-2S cluster that is essential for FECH enzymatic activity and stability [1,2]. FECH is part of an oligomeric complex in mitochondria that includes Mitoferrin-1 (MFRN1) and ABCB10 [3,4]. Mitoferrin-1, located in the inner mitochondrial membrane, transports ferrous iron in mitochondria for formation of the 2Fe-2S clusters, and is the source of the ferrous iron used in the final step of heme biosynthesis [5,6]. ABCB10 enhances MFRN1 stability and function through physical interaction, by binding on the N-terminus of MFRN1 as a MFRN1-ABCB10 complex [7].
Enzymatic activity of FECH is deficient in patients with Erythropoietic Protoporphyria (EPP) [8,9]. The most common genotype in patients with EPP is a mutation in one FECH allele that severely alters enzyme structure and/or function, and an intronic polymorphism in the other FECH allele (IVS3-48 T > C) that causes alternative splicing, resulting in low FECH expression allele (LEA) and decreased enzymatic activity [10-13]. The diminished FECH activity causes excess accumulation of Protoporphyrin IX (PPIX), primarily in the bone marrow, with an increase of free PPIX in circulating erythrocytes, and increased hepatobiliary excretion of PPIX. The major clinical features that result are lifelong skin photosensitivity from sunlight exposure in nearly all cases [14], and a variable degree of hepatobiliary injury in approximately 20% of patients due to the toxic effects of hepatic PPIX [15,16], which can lead to liver failure necessitating liver transplantation in 1 to 5% of cases [17].
Biochemical and clinical manifestations like those in EPP also occur in patients who have no genetic deficiency of FECH enzymatic activity, but have C-terminal truncation mutations of the ALAS2 gene expressed in the developing erythroid population of the bone marrow [18]. These mutations cause a 2–3 fold increase in enzymatic activity of ALAS2 [19], the rate limiting step of heme biosynthesis, resulting in the excess production and accumulation of PPIX, a condition designated X-Linked Protoporphyria (XLP) [18,20].
A pilot study, published in 2011, examined the relationship between FECH enzymatic activity and the MFRN1 mRNA level in cultured lymphoblasts of five normal individuals, one individual with classical EPP, and four individuals with XLP (3 members of the same pedigree), and two individuals with positive biochemical studies for an EPP phenotype, but without identifiable mutations of FECH or ALAS2 enzymes [21]. This study showed a positive relationship between FECH activity and MFRN1 mRNA level, with correlation coefficient of 0.75. However, there were insufficient sample numbers from each well characterized erythropoietic porphyria variant (FECH nonsense/LEA, FECH missense/LEA, XLP), were not available to draw statistically significant conclusions. The present study was performed on increased sample numbers to determine if this relationship between MFRN1 message levels positively correlated with FECH enzymatic activity.
2. Methods
2.1. Study population
Studies were done on a) ten patients with confirmed ALAS2 mutations, b) ten patients with a FECH C411G mutation (C411, one of the four cysteine residues coordinating 2Fe-2S cluster binding, located on C-terminal domain is substituted with glycine), that prevents the 2Fe-2S complex from binding to FECH, and c) ten patients with FECH nonsense or splicing null mutations. All individuals with FECH mutations (groups b and c) had the low expression allele (IVS3-48c > t) in trans, which was not present in any of the individuals with ALAS2 mutations. The sample size of 10 was chosen because it is the smallest number that can provide a correlation coefficient of > 0.6 with p = 0.05 and study power of 80%. Studies were also done on 21 healthy individuals with no personal or family history of porphyria. The control population tested negative for FECH or ALAS2 mutations and the low expression allele. The study was approved by the Institutional Review Boards at the University of Alabama at Birmingham, Mount Sinai School of Medicine in New York, University of Utah in Salt Lake City, University of California at San Francisco, University of Texas Medical Branch at Galveston, Wake Forest University at Winston Salem, and Children's Hospital of Boston.
2.2. Lymphoblast generation and culture
After written informed consent from participants, peripheral blood samples were obtained to establish Epstein-Barr virus-transformed lymphoblasts (EBVTL), using methods previously described [22,23]. EBVTL were cultured in RPMI 1640 containing penicillin/streptomycin sulfate (Cellgro) and 20% heat-inactivated fetal bovine serum (Biomedia) until growing well, followed by culture in the same medium containing 15% fetal bovine serum for three to four days to reach the log phase of growth. They were then harvested for measurement of FECH enzymatic activity, MFRN1 mRNA, and mitochondrial iron levels. The iron concentration in fetal bovine serum used in the cultures ranged from 279 to 309 μg/dL.
2.3. Measurements of FECH activity and MFRN1 mRNA in cultured lymphoblasts
FECH enzymatic activity was assessed in sonicates of EBVTL, using a fluorometric assay that measures the formation of zinc-deuteroporphyrin, and is expressed as nmol/mg protein/h [22]. Quantitative levels of MFRN1 mRNA were measured in total mRNA isolated from EBVTL; qRT-PCR using the Quanti Tect™ Custom Assay (Qiagen) as previously described [21]. Primers and probes for MFRN1 were designed by Qiagen Quantiprobe Design Software (Qiagen). Real-time PCR was performed on an ABI Prism 7700 instrument (Applied Biosystems).
Mitochondrial isolation from EBVTL was done with a commercial kit (Thermo Scientific Cat # 89874), followed by traditional Dounce homogenization to separate mitochondria from cytosolic components [24]. The iron level in the mitochondria was determined by measurement of bathophenanthroline sulfonate Fe (BPS) 3 formation after incubation with nitric acid at 100 °C overnight [25,26].
2.4. Western blot
Total protein was extracted from cultured cells, 80 μg protein were subjected to SDS-PAGE followed by Western blotting onto PVDF membranes. Western transfers were immunoblotted with anti-FECH (1:1000), anti-MFRN1 (1:1000), anti-GAPDH (1:2000) antibodies. Western blot assay was performed to evaluate levels of FECH protein and MFRN1 protein in mitochondrial fractions extracted from EBVTL [22]. Polyclonal Anti-FECH (SC-99138) obtained from Santa Cruz Biotechnology, polyclonal anti-MFRN1 (SLC25A37/PAS-26720) from Thermo Scientific, and Polyclonal anti-GAPDH (ab9485) purchased from Abcam Inc. In all experiments, membranes were subsequently incubated with a secondary horseradish peroxidase (HRP)-labelled goat anti-rabbit secondary antibody (1:3000) for 1 h at room temperature. The ECL detection reagent (Pierce Chemical) was prepared according to the manufacturer's instructions.
3. Results
3.1. EBVTL measurements
The levels of FECH enzymatic activity in EBVTL classic EPP (FECH nonsense/LEA) were significantly reduced compared to the normal controls (Table 1). There was also a reduction in FECH enzymatic activity in the group with ALAS2 mutations (XLP) compared to normal controls. This was accompanied by decreased formation of normal MFRN1 mRNA and reduced FECH enzymatic activity compared to normal lines.
Table 1.
Ferrochelatase Enzymatic Activity and Mitoferrin-1 mRNA levels in EBVTL.
| Genotype | Wild type N = 21 |
FECH C411G/ LEA N = 10 |
FECH Nonsense/ LEA N = 10 |
ALAS2 Mutations N = 10 |
|---|---|---|---|---|
| FECH activitya | ||||
| Mean(SD) | 19.7(3.8) | 5.2 (1.3)* | 4.4(1.6)* | 13.1(3.0)* |
| Mitoferrin-1 mRNAb | ||||
| Mean(SD) | 1728.7(372.3) | 488.5(139.5)* | 318.5(60.7)* | 1495.3(316.1)** |
p value < .001.
p value = .09.
nmol zinc-deuteroporphyrin/mg protein/h.
copies/μg total RNA.
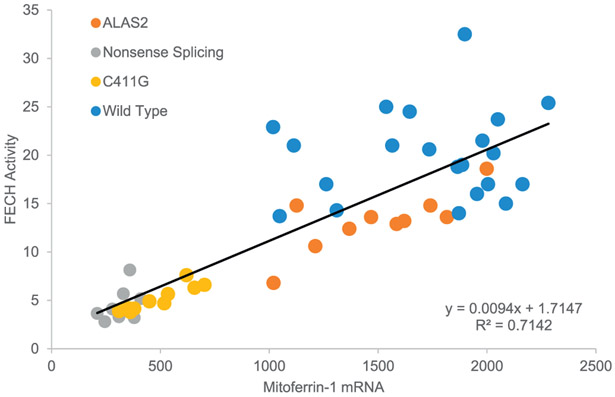
The correlation coefficient between FECH enzymatic activity and MFRN1 mRNA level in this study confirms the presence of a positive relationship between these two mitochondrial entities. The correlation coefficient is 0.94 when the three types of mutant cell lines are combined, and remains high at 0.85 when the normal cell lines are also included (Fig. 1 and 2). Iron levels, in mitochondria isolated from each of the four EBVTL groups, were highest in the normal EBVTL group, and lowest in the EBVTL group with FECH Nonsense/Splicing mutations (p-value 0.0015 between the two groups). (Table 2).
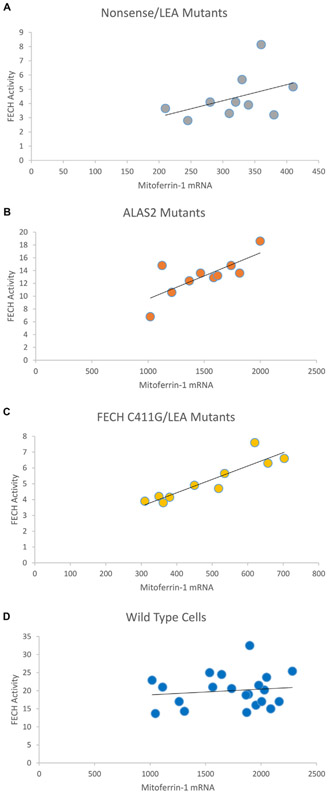
Fig. 1.
Correlation between FECH activity (nmol/mg protein/h) and Mitoferrin-1 mRNA (relative to total mRNA level) in patients with protoporphyria due to A) FECH Nonsense/LEA alleles, B) ALAS2 mutants, and C) FECH C411G/LES alleles. Panel D is on healthy controls with wild-type lymphoblasts.
Fig. 2.
Correlation between FECH activity (nmol/mg protein/h) and Mitoferrin-1 mRNA (relative to total mRNA level) in patients with protoporphyria and healthy controls.
Table 2.
Mitochondrial Iron in EBVTL (μg/g protein).
| Genotype | Wild type N = 8 |
FECH C411G N = 7 |
FECH Nonsense/ Splicing N = 10 |
ALAS2 Mutations N = 8 |
|---|---|---|---|---|
| Mean(SD) | 154(58) | 112(84) | 66(28) | 149(60) |
| *p | – | 0.15 | 0.0015 | 0.428 |
p value – compared with normal group.
3.2. Western blot results
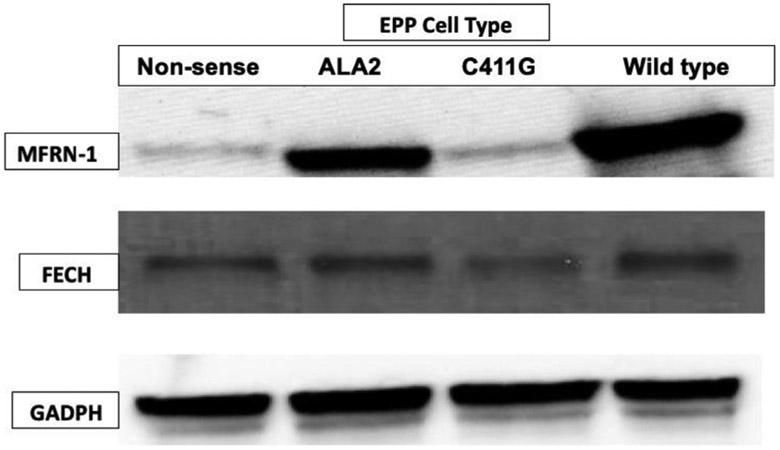
Western Blot evaluations of FECH protein and MFRN1 protein (Fig. 3) levels in mitochondrial fractions of the four EBVTL groups show that the highest protein levels were in normal cells, followed by ALAS2 cells (XLP), FECH C411G/LEA cells, and FECH Nonsense/LEA cells. The levels of FECH are normal in the ALAS2 cells and approximately 35% of wild type in cells with the C411G and the non-sense FECH alleles. FECH activity was 25.4 in wild type, 4.2 in C411G 11.7 in ALAS2 and 4.1 nmol/mg protein/h and correlated highest with the levels of mitoferrin 1 protein (Fig. 3).
Fig. 3.
Western Blot of MFRN1/FECH/GAPDH in patients with protoporphyria due to mutations in either FECH (non-sense or C411G) or gain-of-function mutations in ALAS2. Total protein, 80 μg, from EBVTL of patients were subjected to SDS-PAGE then transferred to PVDF membranes. Blots were developed with anti-FECH (1:1000), anti-MFRN1 (1:1000), anti-GAPDH (1:2000) antibodies followed by a secondary horseradish peroxidase (HRP)-labelled goat anti-rabbit secondary antibody (1:3000).
4. Discussion
The main findings of our study are: a) reduction in FECH enzymatic activity in patients with EPP and XLP, b) reduction in MFRN1 mRNA levels in both EPP and XLP patients, c) excellent correlation between MFRN1 mRNA and FECH activity, and d) reduction in mitochondrial iron levels among patients with EPP with FECH nonsense/LEA mutations.
Reduction in FECH activity positively correlating with MFRN1 mRNA levels was previously shown in a pilot study [21]. In the present study, these findings were confirmed with better correlation between FECH activity and MFRN1 mRNA levels, with a positive correlation coefficient of 0.94. Further, results in the current study extend these findings to all cause of increased PPIX accumulation, mutations in FECH or ALAS2. However, mitochondrial iron level reduction was seen among only one subgroup of EPP patients, those with FECH nonsense/LEA alleles, and not in subjects with FECH C411G/LEA alleles or in XLP patients. Reasons for these discordant findings on mitochondrial iron levels remain unclear and need further investigation. It would also be interesting to examine the effects of exogenous ferrous iron administration in EPP and XLP patients by measuring the changes in the FECH activity, MFRN1 mRNA, and mitochondrial iron levels.
In addition to being a substrate for FECH in heme biosynthesis, iron has other important roles in heme metabolism. Experiments with K562 erythroleukemia cells and Cos7 cells expressing human FECH have shown that formation of the 2Fe-2S cluster bound to the C-terminal region FECH is regulated by availability of intracellular iron [23]. When intracellular iron levels are low, rapid degradation of FECH protein occurs, indicating that the post-translational stability of FECH depends on iron availability and formation of the structurally important 2Fe-2S cluster. Thus, MFRN1 is an essential component of heme synthesis, as it transports ferrous iron for the final step in heme biosynthesis in the mitochondria, as part of the FECH metabolon [27]. The multi-protein complex is thought to sequester iron, preventing tissue damage that could occur if ferrous iron were free in solution. When MFRN1 expression is low, there is a decrease in the levels of 2Fe-2S clusters being produced by the mitochondrial machinery, leading to decreased FECH activity and potentially disrupting overall iron homeostasis leading to alterations in translation of mRNAs controlled by the Iron Responsive Element/Iron Responsive Element-Binding Protein (IRE/IRE-BP) system [28]. These observations have been made in context to ferritin and other components responsible for iron homeostasis [5]. While the mechanism tying mRNA levels for MFRN1 to activity of FECH are not clear, the evolution of a system to prevent excess free iron and to prevent excess PPIX seems clear [29].
Independent regulatory systems to prevent the toxicity associated with “free iron” and/or “free PPIX” have evolved in multiple cells types. Excess PPIX is pumped out of erythrocytes by ABCG2, a transporter that has a role in regulating PPIX levels during erythroid differentiation, and therefore, could be a genetic determinant of EPP [30]. The plasma membrane bound heme transporter FLVCR has also been shown to export potentially toxic PPIX from the erythron during development [31]. The liver transferrin pathway has a role in the orchestration of iron distribution between peripheral iron stores, the spleen, and the bone marrow [32]. In FECH-deficient protoporphyria patients, ALAS2 expression is enhanced, and the erythrocytic PPIX concentration correlates with iron availability [33].
Other proteins that could also impact phenotypic expression in EPP are the mitochondrial protein that co-localizes with FECH in mitochondria of mouse erythroleukemia cells [34], heme-regulated elF2-kinase (HRI) that is essential for translational regulation of alpha and beta globulins [35], and the iron-regulatory protein 2 (IRP2) that post-transcriptionally regulates iron-responsive proteins [36]. It is notable that patients with EPP often have a mild anemia, with variable evidence of iron deficiency that includes microcytosis, low serum iron levels, and low serum ferritin levels. Iron availability has been shown to modulate aberrant splicing of FECH through the iron-and 2-oxoglutarate dependent dioxygenase Jmjd6 and U2AF65 [33]. However, the role of iron replacement in EPP remains controversial, with both clinical improvement and exacerbation of photosensitivity having been reported [37]. The ultimate goal of these studies is to understand how the mitochondria communicates to the nucleus to coordinate proteins levels where a single member of a multi-subunit complex is deficient. In this specific case, tying the ability to import iron, that can be toxic in excess, to the capacity to produce an essential cofactor- heme.
Acknowledgements
This research was supported in part by the Porphyrias Consortium (U54 DK083909), which is a part of the NCATS Rare Diseases Clinical Research Network (RDCRN). RDCRN is an initiative of the Office of Rare Diseases Research (ORDR), NCATS, and is funded through a collaboration between NCATS and the NIDDK. MB is supported in part by the NIH Career Development Award (K23 DK095946). BW is supported in part by the American Porphyria Foundation’s “Protect the Future Program”.
References
- [1].Dailey HA, Dailey TA, Wu CK, Medlock AE, Wang KF, Rose JP, Wang BC, Ferrochelatase at the millennium: structures, mechanisms and [2Fe-2S] clusters, Cell. Mol. Life Sci 57 (2000) 1909–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wu CK, Dailey HA, Rose JP, Burden A, Sellers VM, Wang BC, The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis, Nat. Struct. Biol 8 (2001) 156–160. [DOI] [PubMed] [Google Scholar]
- [3].Chen W, Dailey HA, Paw BH, Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis, Blood 116 (2010) 628–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Piel RB III, Shiferaw MT, Vashisht AA, Marcero JR, Praissman JL, Phillips JD, Wohlschlegel JA, et al. , A novel role for Progesterone Receptor Membrane Component 1 (PGRMC1): a partner and regulator of ferrochelatase, Biochemistry 55 (2016) 5204–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, Gwynn B, et al. , Mitoferrin is essential for erythroid iron assimilation, Nature 440 (2006) 96–100. [DOI] [PubMed] [Google Scholar]
- [6].Medlock AE, Najahi-Missaoui W, Ross TA, Dailey TA, Burch J, O'Brien JR, Lanzilotta WN, et al. , Identification and characterization of solvent-filled channels in human ferrochelatase, Biochemistry 51 (2012) 5422–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen W, Paradkar PN, Li L, Pierce EL, Langer NB, Takahashi-Makise N, Hyde BB, et al. , Abcb10 physically interacts with mitoferrin-1 (Slc25a37) to enhance its stability and function in the erythroid mitochondria, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 16263–16268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bottomley SS, Tanaka M, Everett MA, Diminished erythroid ferrochelatase activity in protoporphyria, J. Lab. Clin. Med 86 (1975) 126–131. [PubMed] [Google Scholar]
- [9].Bonkowsky HL, Bloomer JR, Ebert PS, Mahoney MJ, Heme synthetase deficiency in human protoporphyria. Demonstration of the defect in liver and cultured skin fibroblasts, J. Clin. Invest 56 (1975) 1139–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang X, Poh-Fitzpatrick M, Taketani S, Chen T, Piomelli S, Screening for ferrochelatase mutations: molecular heterogeneity of erythropoietic protoporphyria, Biochim. Biophys. Acta 1225 (1994) 187–190. [DOI] [PubMed] [Google Scholar]
- [11].Sellers VM, Dailey TA, Dailey HA, Examination of ferrochelatase mutations that cause erythropoietic protoporphyria, Blood 91 (1998) 3980–3985. [PubMed] [Google Scholar]
- [12].Gouya L, Puy H, Robreau AM, Bourgeois M, Lamoril J, Da Silva V, Grandchamp B, et al. , The penetrance of dominant erythropoietic protoporphyria is modulated by expression of wildtype FECH, Nat. Genet 30 (2002) 27–28. [DOI] [PubMed] [Google Scholar]
- [13].Risheg H, Chen FP, Bloomer JR, Genotypic determinants of phenotype in north American patients with erythropoietic protoporphyria, Mol. Genet. Metab 80 (2003)196–206. [DOI] [PubMed] [Google Scholar]
- [14].Poh-Fitzpatrick MB, Molecular and cellular mechanisms of porphyrin photosensitization, Photo-Dermatology 3 (1986) 148–157. [PubMed] [Google Scholar]
- [15].Bloomer JR, Phillips MJ, Davidson DL, Klatskin G, Bloomer, Hepatic disease in erythropoietic protoporphyria, Am. J. Med 58 (1975) 869–882. [DOI] [PubMed] [Google Scholar]
- [16].Doss MO, Frank M, Hepatobiliary implications and complications in protoporphyria, a 20-year study, Clin. Biochem 22 (1989) 223–229. [DOI] [PubMed] [Google Scholar]
- [17].McGuire BM, Bonkovsky HL, Carithers RL Jr., Chung RT, Goldstein LI, Lake JR, Lok AS, et al. , Liver transplantation for erythropoietic protoporphyria liver disease, Liver Transpl. 11 (2005) 1590–1596. [DOI] [PubMed] [Google Scholar]
- [18].Whatley SD, Ducamp S, Gouya L, Grandchamp B, Beaumont C, Badminton MN, Elder GH, et al. , C-terminal deletions in the ALAS2 gene lead to gain of function and cause X-linked dominant protoporphyria without anemia or iron overload, Am. J. Hum. Genet 83 (2008) 408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bishop DF, Tchaikovskii V, Nazarenko I, Desnick RJ, Molecular expression and characterization of erythroid-specific 5-aminolevulinate synthase gain-of-function mutations causing X-linked protoporphyria, Mol. Med 19 (2013) 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Balwani M, Doheny D, Bishop DF, Nazarenko I, Yasuda M, Dailey HA, Anderson KE, et al. , Loss-of-function ferrochelatase and gain-of-function erythroid-specific 5-aminolevulinate synthase mutations causing erythropoietic protoporphyria and x-linked protoporphyria in north American patients reveal novel mutations and a high prevalence of X-linked protoporphyria, Mol. Med 19 (2013) 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang Y, Langer NB, Shaw GC, Yang G, Li L, Kaplan J, Paw BH, et al. , Abnormal mitoferrin-1 expression in patients with erythropoietic protoporphyria, Exp. Hematol 39 (2011) 784–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bloomer J, Bruzzone C, Zhu L, Scarlett Y, Magness S, Brenner D, Molecular defects in ferrochelatase in patients with protoporphyria requiring liver transplantation, J. Clin. Invest 102 (1998) 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Taketani S, Adachi Y, Nakahashi Y, Regulation of the expression of human ferrochelatase by intracellular iron levels, Eur. J. Biochem 267 (2000) 4685–4692. [DOI] [PubMed] [Google Scholar]
- [24].Kesner EE, Saada-Reich A, Lorberboum-Galski H, Characteristics of mitochondrial transformation into human cells, Sci. Rep 6 (2016) 26057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Miao R, Kim H, Koppolu UM, Ellis EA, Scott RA, Lindahl PA, Biophysical characterization of the iron in mitochondria from Atm1p-depleted Saccharomyces cerevisiae, Biochemistry 48 (2009) 9556–9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tamarit J, Irazusta V, Moreno-Cermeno A, Ros J, Colorimetric assay for the quantitation of iron in yeast, Anal. Biochem 351 (2006) 149–151. [DOI] [PubMed] [Google Scholar]
- [27].Medlock AE, Shiferaw MT, Marcero JR, Vashisht AA, Wohlschlegel JA, Phillips JD, Dailey HA, Identification of the mitochondrial heme metabolism complex, PLoS One 10 (2015) e0135896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Paradkar PN, Zumbrennen KB, Paw BH, Ward DM, Kaplan J, Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2, Mol. Cell. Biol 29 (2009) 1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sachar M, Anderson KE, Ma X, Protoporphyrin IX: the good, the bad, and the ugly, J. Pharmacol. Exp. Ther 356 (2016) 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhou S, Zong Y, Ney PA, Nair G, Stewart CF, Sorrentino BP, Increased expression of the Abcg2 transporter during erythroid maturation plays a role in decreasing cellular protoporphyrin IX levels, Blood 105 (2005) 2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Quigley JG, Yang Z, Worthington MT, Phillips JD, Sabo KM, Sabath DE, Berg CL, et al. , Identification of a human heme exporter that is essential for erythropoiesis, Cell 118 (2004) 757–766. [DOI] [PubMed] [Google Scholar]
- [32].Lyoumi S, Abitbol M, Andrieu V, Henin D, Robert E, Schmitt C, Gouya L, et al. , Increased plasma transferrin, altered body iron distribution, and microcytic hypochromic anemia in ferrochelatase-deficient mice, Blood 109 (2007) 811–818. [DOI] [PubMed] [Google Scholar]
- [33].Barman-Aksozen J, Minder EI, Schubiger C, Biolcati G, Schneider-Yin X, In ferrochelatase-deficient protoporphyria patients, ALAS2 expression is enhanced and erythrocytic protoporphyrin concentration correlates with iron availability, Blood Cells Mol. Dis 54 (2015) 71–77. [DOI] [PubMed] [Google Scholar]
- [34].Taketani S, Kakimoto K, Ueta H, Masaki R, Furukawa T, Involvement of ABC7 in the biosynthesis of heme in erythroid cells: interaction of ABC7 with ferrochelatase, Blood 101 (2003) 3274–3280. [DOI] [PubMed] [Google Scholar]
- [35].Han AP, Fleming MD, Chen JJ, Heme-regulated eIF2alpha kinase modifies the phenotypic severity of murine models of erythropoietic protoporphyria and beta-thalassemia, J. Clin. Invest 115 (2005) 1562–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cooperman SS, Meyron-Holtz EG, Olivierre-Wilson H, Ghosh MC, McConnell JP, Rouault TA, Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2, Blood 106 (2005) 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wahlin S, Floderus Y, Stal P, Harper P, Erythropoietic protoporphyria in Sweden: demographic, clinical, biochemical and genetic characteristics, J. Intern. Med 269 (2011) 278–288. [DOI] [PubMed] [Google Scholar]