Escherichia coli is the favored workhorse of biochemists for heterologous protein overexpression. Despite the deceptively straightforward approaches for expressing foreign proteins at high levels in E. coli, poor expression and misfolding are two common roadblocks that cripple production of recombinant proteins. Tackling these issues is essential to enable structure-function studies of proteins and scale-up efforts of biotherapeutics. Here, we highlight an advance1 that could be leveraged for predictably ramping protein expression in vivo and in vitro.
The rates of initiation and early elongation, the pacemakers of translation, are determined by various factors, including mRNA secondary structure, tRNA abundance, and codon usage. There is a growing body of evidence that this list should include the nucleotide sequence immediately downstream from the start codon. For example, Voges et al.2 highlighted the importance of this factor using 756 green fluorescent protein (GFP) fusion constructs, each with a different 39-bp sequence starting at the second codon to mirror its respective native context in a naturally occurring open reading frame (ORF). They observed that GFP expression anticorrelated with the GC content and predicted base-pairing likelihood, especially between codons 2 and 7. Goodman et al.3 reached somewhat similar conclusions using their library of 14,234 superfolder GFP genes, each with a different combination of promoters, ribosome binding sites, and N-terminal sequences that were taken from the first 33 bp of 137 E. coli essential genes. Finally, Han et al.4 used ribosome profiling to demonstrate ribosomal pausing at codon 5 of translating ORFs in several mammalian cell lines, a finding further cemented during their reanalysis of yeast, worm, and zebrafish Ribo-Seq data. Therefore, translation efficiency (i.e., number of full-length polypeptides produced per mRNA per unit time) is influenced by the nucleotides around +10 of an ORF2,3 and by an obligatory checkpoint around codon 5 that signals elongation commitment.4
Inspired by the insights described above, Verma et al.1 sought to dissect the mechanistic basis for how codons 3–5 engender productive translation. First, they constructed a library of ~259,000 eGFP variants in which nucleotides +7 to +15 were randomized. Second, they employed a sort-and-sequence approach to parse E. coli cells into five bins based on the level of expressed eGFP, using the relative fluorescence units that ranged from 20 to 12,000 as a proxy for expression. Third, they assigned a GFP score to each sequence based on its weighted distribution over all five bins, an outcome attributable to stochastic variation in recombinant protein expression. This approach led to a score of 2.03 for wild-type eGFP, bookended by low-scoring (1) to high-scoring (5) variants.
Analysis of variants with a GFP score >4 revealed an AU-content bias (6–9 A/Us at nucleotides +7 to +15). Enriched among these high-scoring sequences were two motifs: AAD UAU (D = A, G, or U; encoding KY or NY) and AAV AUU (V = A, C, or G; encoding KI or NI) at codons 3 and 4 or codons 4 and 5 (Figure 1). Additionally, all eGFP variants with these dipeptides in positions 3–5, regardless of the codons used, displayed elevated levels of eGFP expression as inferred from their high median GFP score of 4.31. Shifting the high-scoring sequence to codons 9–11 by insertion of a His6 tag at codons 3–8 eliminated payoffs in expression, underscoring the strict positional dependence for these motifs. The broad applicability of these favorable sequences was amply illustrated by in vitro translation experiments and E. coli expression studies that employed different promoters, upstream sequences, and three test-candidate ORFs (coral mEOS2, human Gai, and human RGS2).1
Figure 1.
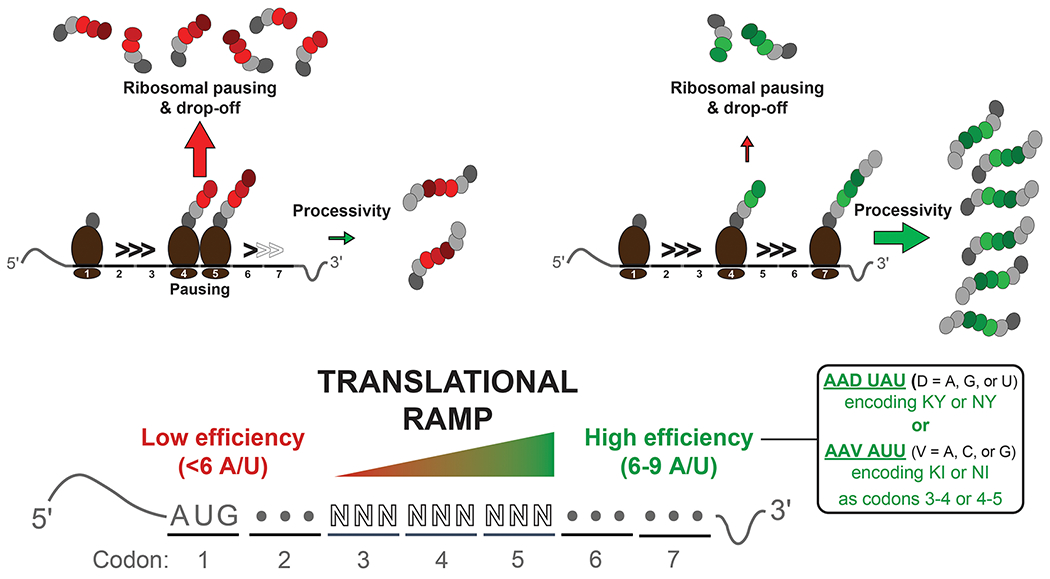
Codons 3–5 and their amino acids act as a translational ramp in a sequence-dependent manner. Low-efficiency sequences are associated with ribosomal pausing at codon 4 or 5 and termination of protein synthesis. In contrast, AU-rich sequences and the specified dipeptides (inset) favor high efficiency, leading to processive translation through codons 3–5 and an increased level of full-length polypeptide synthesis.
Because GFP scores did not correlate with the abundance of tRNAs (for decoding codons 3–5), distinctions in expression of eGFP variants could not be ascribed to differences in ribosome dwell times caused by decoding delays due to rare tRNAs. To establish a molecular basis for the sequence-dependent variability in translation efficiency, single-molecule FRET assays were used to monitor ribosome conformational changes and tRNA binding during translation of seven-codon mRNAs containing a low- or high-scoring motif at codons 3–5.1 Despite equivalent formation of the translation initiation complexes with these mRNAs, only 27% of ribosomes completed translation with the low-scoring mRNA compared to 84% with the high-scoring mRNA (Figure 1). Ribosomes that did not complete translation stalled at codon 4 or 5 and eventually dissociated from the mRNA. This translation arrest was rationalized1 based on an intrinsic dynamic feature of the ribosome: each round of elongation entails an intersubunit rotation that reflects the translocation of tRNAs to their adjacent sites. FRET assays used to monitor this rotation revealed that most arrested ribosomes are in a decoding-incompetent state despite having an empty A site. Persisting in this state undergirds the “processivity barrier” for translation during early elongation.1 Additional studies are required to better understand how codons 3–5 act to decrease the probability of the nascent polypeptide making it to the mandatory exit tunnel-mediated commitment to elongation.4
Interestingly, native E. coli (and Saccharomyces cerevisiae) ORFs are enriched in the high-scoring sequences at codons 3–5 from the eGFP library.1 Based on a computational analysis, Moreira et al.5 found that two other enterobacteria (Klebsiella oxytoca and Enterobacter asburiae) are also enriched for high-scoring sequences at codons 3–5, indicating a strong positive selection for particular sequences in this region. The idea that codons 3–5 function as key translation success factors is bolstered by another observation: native E. coli ORFs with high eGFP-scoring sequences1 are associated with slightly higher translation efficiencies and protein abundances as determined from ribosome profiling and proteomic inventory data, respectively.5 Why was a stronger correlation not observed between the codon 3–5 sequences and native protein synthesis rates? Bacterial translation is choreographed by a complex suite of determinants, making it difficult to unmask the exclusive contribution of the N-terminal sequence to systemwide expression of endogenous proteins without fully parsing potential counterbalancing determinants (e.g., mRNA-specific differences in coupling of transcription and translation, variable length of 5′-UTRs5).
By identifying and characterizing a sequence-specific translational ramp at codons 3–5, Djuranovic and co-workers1 have uncovered a redeployable “translation rheostat” that could tune recombinant protein expression in vivo and in vitro. As proof of concept, they demonstrated a higher level of expression for three different ORFs when each was engineered to have a codon 3–5 sequence with a GFP score >4. Of equal appeal is the testable hypothesis that use of a translation-dampening, low-scoring sequence could help overcome challenges associated with proteins that are prone to misfolding or forming insoluble aggregates. Exploiting a scalable translational ramp1 either alone or in conjunction with chaperone fusions (e.g., SUMO) should help realize the full potential of heterologous protein expression in bacteria.
ACKNOWLEDGMENTS
The authors thank Kurt Fredrick (OSU) for valuable discussion.
Funding
The authors are grateful for funding from the National Institutes of Health (GM-120582, AI-116119, and AI-140541 to V.G. and T32-GM086252 to B.E.S.) and an OSU Pelotonia Postdoctoral Fellowship (to W.J.Z.).
Footnotes
The authors declare no competing financial interest.
Contributor Information
Walter J. Zahurancik, Department of Chemistry & Biochemistry and Center for RNA Biology, The Ohio State University, Columbus, Ohio 43210, United States
Blake E. Szkoda, Department of Chemistry & Biochemistry and Ohio State Biochemistry Program, The Ohio State University, Columbus, Ohio 43210, United States
Lien B. Lai, Department of Chemistry & Biochemistry and Center for RNA Biology, The Ohio State University, Columbus, Ohio 43210, United States
Venkat Gopalan, Department of Chemistry & Biochemistry, Center for RNA Biology, and Ohio State Biochemistry Program, The Ohio State University, Columbus, Ohio 43210, United States.
REFERENCES
- (1).Verma M, Choi J, Cottrell KA, Lavagnino Z, Thomas EN, Pavlovic-Djuranovic S, Szczesny P, Piston DW, Zaher HS, Puglisi JD, and Djuranovic S (2019) A short translational ramp determines the efficiency of protein synthesis. Nat. Commun. 10, 5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Voges D, Watzele M, Nemetz C, Wizemann S, and Buchberger B (2004) Analyzing and enhancing mRNA translational efficiency in an Escherichia coli in vitro expression system. Biochem. Biophys. Res. Commun 318, 601–614. [DOI] [PubMed] [Google Scholar]
- (3).Goodman DB, Church GM, and Kosuri S (2013) Causes and effects of N-terminal codon bias in bacterial genes. Science 342, 475–479. [DOI] [PubMed] [Google Scholar]
- (4).Han Y, Gao X, Liu B, Wan J, Zhang X, and Qian S (2014) Ribosome profiling reveals sequence-independent post-initiation pausing as a signature of translation. Cell Res. 24, 842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Moreira MH, Barros GC, Requiao RD, Rossetto S, Domitrovic T, and Palhano FL (2019) From reporters to endogenous genes: the impact of the first five codons on translation efficiency in Escherichia coli. RNA Biol. 16, 1806–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]


