Abstract
Sparse arrays reduce the number of active channels that effectively increases the inter-element spacing. Large inter-element spacing results in grating lobe artifacts degrading the ultrasound image quality and reducing the contrast-to-noise ratio. A deep learning-based custom algorithm is proposed to estimate inactive channel data in periodic sparse arrays. The algorithm uses data from multiple active channels to estimate inactive channels. The estimated inactive channel data effectively reduces the inter-element spacing for beamforming, thus suppressing the grating lobes. Estimated inactive element channel data was combined with active element channel data resulting in a pseudo fully sampled array. The channel data was beamformed using a simple delay-and-sum method and compared with the sparse array and fully sampled array. The performance of the algorithm was validated using a wire target in a water tank, multi-purpose tissue-mimicking phantom, and in-vivo carotid data. Grating lobes suppression up to 15.25 dB was observed with an increase in contrast-to-noise (CNR) for the pseudo fully sampled array. Hypoechoic regions showed more improvement in CNR than hyperechoic regions. Root-mean-square error for unwrapped phase between fully sampled array and the pseudo fully sampled array was low, making the estimated data suitable for Doppler and elastography applications. Speckle pattern was also preserved; thus, the estimated data can also be used for quantitative ultrasound applications. The algorithm can improve the quality of sparse array images and has applications in small scale ultrasound devices and 2D arrays.
Keywords: convolutional neural networks, deep learning, gap-filling, sparse array, ultrasound imaging
1. INTRODUCTION
The use of sparse array (SA) technique in uniform linear arrays and two dimensional (2D) matrix probes has been extensively explored [1], [2]. A fully sampled array (FSA) allows each element in the transducer to be continuously driven by using the ultrasound system, which implies that the number of channels in the ultrasound system and the number of transducer elements should be the same. However, a high number of system channels come at an increased cost and form factor. SA works by driving only a limited number of transducer elements, referred to as active elements, thus reducing the active element count and the active channel count in an ultrasound imaging system. However, reducing the number of active elements comes at the cost of increased inter-element spacing. Inter-element spacing larger than half of a wavelength (λ/2) introduces grating lobes that degrades the image quality. Typically, uniform linear arrays have an inter-element spacing of λ, except phase arrays, which typically have an inter-element spacing of λ/2. Different techniques have been proposed to suppress these grating lobes artifacts by optimally designing or experimentally finding the minimal-redundancy array. These techniques include non-uniform weighted periodic sparse array [3], randomized active arrays [4], minimally redundant 2D array designs [5], optimized sparse periodic linear arrays [6], multi-depth radiation optimization using simulated annealing and spiral-array inspired energy functions [7], optimal 2D non-grid sparse arrays [8], circular ring arrays for IVUS imaging [9], and thinning and weighting of large planar arrays by simulated annealing [10]. However, the above-mentioned methods suffer from low signal-to-noise ratio (SNR).
A technique termed gap-filling (GF) or gap-interpolation (GI) has been proposed previously [11], which generates the unknown and unacquired RF-echo data by using estimation and prediction. In GF, neighboring RF-echo channel data are used to virtually generate new RF-echo channels data. The technique is similar to sonogram gap-filling used for interpolating spectral Doppler signal [12], [13]. By creating a virtual signal for the inactive elements, the inter-element spacing can be effectively reduced, resulting in suppression of grating lobes. Similarly, virtual sub-wavelengths receiving elements have shown to suppress the grating and side lobes while retaining the mainlobe signal [14], [15]. However, simple interpolation techniques, like averaging, bi-cubic interpolation, and copying the signal, would result in only a limited reduction of the grating lobes. The limited success of such techniques can be explained due to the low correlation between the neighboring channel data lines (pre-beamformed RF data). A delay of λ/4 between the neighboring channel lines can result in zero correlation between the two lines. Thus, interpolation techniques fail as the wave patterns in both axial and lateral direction needs to be accounted for.
Previous works have shown that low-rank interpolation problems of the RF data can be solved using a convolutional neural network (CNN) approach [16], [17]. A CNN approach is used in this paper to demonstrate that missing channel data from inactive elements can be estimated, resulting in decreased inter-element spacing and, thus, suppression of the grating lobes. Here, a periodic sparse array is considered in which the even elements are inactive. A new method is proposed to suppress the grating lobe artifacts while reducing the active channel count by accounting for the periodicity of the wave signal in both axial and lateral directions.
The paper introduces a custom CNN algorithm for estimating the inactive channel data in a periodic sparse array. The algorithm is trained on data gathered from tissue-mimicking phantoms. The performance of the algorithm is evaluated by analyzing the point spread function of a wire target, another tissue-mimicking phantom, and in-vivo carotid data from two different users. Quantitative parameters, such as resolution, contrast-to-noise ratio, speckle quality, and unwrapped phase, are used to access the quality of reconstruction in comparison to those of SA and FSA.
II. MATERIALS AND METHODS
A. THEORY
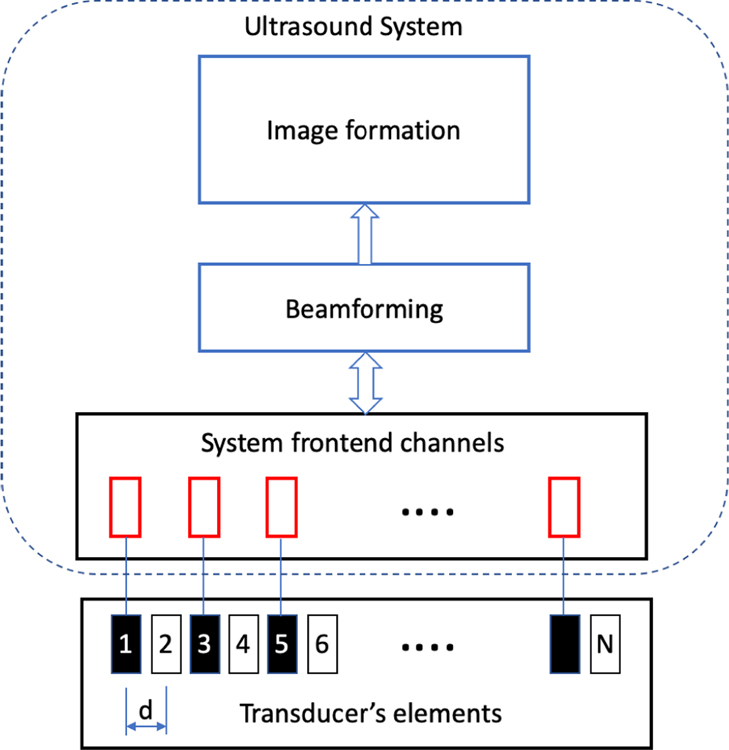
Fig. 1 represents a periodic SA with N total elements compromising of N/2 active elements, represented in black, and N/2 inactive elements, represented in white. The inter-element spacing is d. The channel data received at jth channel can be represented as xj(n), where n represents the nth sample of the data. The inactive channel can be estimated using a non-linear combination of the active elements which can be represented as
| (1) |
For a periodic SA, the inter-element spacing becomes 2d, which is typically greater than λ/2, resulting in the grating lobes being unavoidable. Estimating the data for the inactive elements, effectively reduces the inter-element spacing to d, thus shifting the grating lobes further away from the main lobe.
Fig. 1.
Periodic sparse array with active channels in black and inactive channels in white. Inactive channel data can be estimated by using the information from the active channel data. The estimated channel data is referred to as pseudo channel data.
B. TRAINING ON SMALL PARTS PHANTOM
Deep learning algorithms require a training dataset to train the model. To fulfill this requirement, data was gathered from a tissue-mimicking phantom rather than simulating ultrasound data as the simulation are computationally intensive. Tissue-mimicking phantoms are constructed such that they encompass the majority of the sonographic features observed in in vivo tissue. A tissue-mimicking phantom, Sono404 small parts phantom (SPP) (Gammex, Middleton, Wisconsin, USA), was used for gathering the training dataset. The phantom had an attenuation of 0.7dB/cm-MHz. A programmable ultrasound research system from Verasonics (Vantage, Verasonics Inc., Kirkland, Washington, USA) was used to collect the data. A linear array transducer, L11–4v, (Verasonics Inc., Kirkland, Washington, USA) with a center frequency of 6.25 MHz, pitch of 0.3 mm, and 128 elements was used. A custom Verasonics script was developed to save channel data from all 128 elements. Plane-wave imaging, with a pulse duration of 1 cycle and no compounding (angle 0o), was used to capture the channel data. Kaiser apodization was used for the transmit waveform, whereas the received waveform had no apodization. Channel data collected with all 128 elements in transmit and receive is referred to as FSA in the manuscript. For sparse array channel data, 128 elements were used in transmission, and 64 elements (odd-numbered elements) were used in receiving. The number of elements used in transmission was kept constant at 128 to maintain the same insonification power for both FSA and SA. A simple delay-and-sum beamformer with dynamic aperture control was used to beamform the channel data. The acquisition depth was kept constant at 63.08 mm. The transducer was excited at a peak-to-peak excitation value of 100 volts.
C. ALGORITHM
1). MODEL:
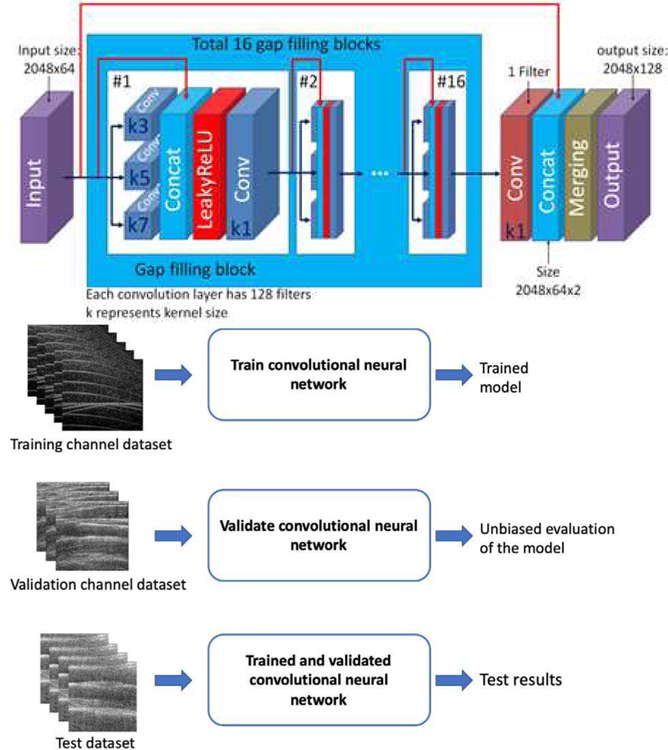
Fig. 2 illustrates the convolutional neural network model used to predict the channel data. The model consists of an input layer with a default size of 2048 rows and 64 channels. The input data provided to the algorithm is the SA channel data, Is, from 64 channels. An example of a gap-filling block is shown in Fig. 2, which consists of three different convolutional layers with a kernel size of 3, 5, and 7. The output of the convolutional layers is concatenated along with the input to the layer and a non-linear activation function, leaky rectified linear units is applied, followed by a convolutional layer with a kernel size of 1. In total, the model includes 16 gap-filling blocks. After the data had passed through the 16 blocks, the convolution layer of kernel size 1 was used to select the biggest activation and then concatenated with the original input image. The concatenated images were aligned alternately to create channel data of size 2048 rows and 128 channels.
Fig. 2.
Convolutional neural network model for estimating inactive channel data. Training and validation are performed using channel datasets from phantoms. Testing is done using phantom and in-vivo data.
2). LOSS FUNCTION:
Structural dissimilarity metric was used as the loss function to be minimized by the algorithm as shown in equation 2
| (2) |
where Is is the reconstructed pseudo-FSA channel data, Ir is the reference FSA channel data, µ is the mean operator, σ is the variance operator, c1 = (k1L)2, c2 = (k2L)2, k1 = 0.01, k2 = 0.03 and L = 2#bits per pixel −1. The Structural dissimilarity metric was based on equally weighted measurements of luminance, contrast, and structure. These metrics treated the pre-beamformed data as an image and calculated the amplitude-based information. Phase information was not taken into account. The algorithm was developed using Python (version 3.6.1, Python Software Foundation) and open-source Keras Python library (version 2.2.1) with TensorFlow ([18] backend (version 1.10.0, Google Inc., Mountain View, California). A hyperparameter optimization was performed on the kernel initializer, the number of filters, optimizer, and the loss function. The parameters reported in Table 1 provided the best results.
TABLE I.
Parameters for convolutional neural network algorithm.
| Parameter | Value |
|---|---|
| input size | 2048 × 64 |
| output size | 2048 × 128 |
| optimizer, learning rate | Adam, 1×10−4 |
| decay rate after 3 epochs | 90% |
| loss function | Structural dissimilarity |
| kernel initializer | Lecun normal |
| # filters | 128 |
| # gap filling blocks | 16 |
3). PROCESSING STEPS:
(A) Training: The input data provided to the algorithm is the SA channel data, Is, from 64 channels. The FSA channel data, Ir, was also provided to the algorithm as the reference for training purposes. The training dataset consisted of 1640 channel data images. (B) Validation: 183 validation images were used from the small parts phantom. The algorithm met the stop criterion after 28 epochs with training and validation DSSIM scores of 0.013 and 0.013, respectively. (C) Testing: Channel data from different phantoms and in-vivo experiments were used for testing. The performance of the model was evaluated in quantitative terms in each case, as described below.
D. POINT SPREAD FUNCTION ANALYSIS
The reduction in the grating lobe can be analyzed by studying the beam profile of a wire target in a water tank. Raw channel data was collected from a wire target phantom immersed in a water tank using the same transducer. The raw channel data was not used for training and only for testing the characteristics of the beam profile. Sixteen channel images were collected. Similar to the previously reported study [19], the contrast ratio was calculated by equation 3
| (3) |
where x represents the lateral points, z represents the axial points, R is a circle of diameter 5λ concentric with the wire target, and IQ is the in-phase/quadrature data.
E. SMALL PARTS PHANTOM (SPP)
An additional dataset of 425 channel data was taken with the SPP to evaluate the performance of the algorithm. The channel data was beamformed offline and compared with SA and FSA. Quantitative metrics were used to compare the hypoechoic and hyperechoic regions from a sample image.
F. MULTI-PURPOSE PHANTOM (MPP)
The performance of the algorithm was tested using another tissue-mimicking phantom, multi-purpose multi-tissue ultrasound phantom (MPP) model 040GSE (CIRS, Norfolk, Virginia, USA). The ultrasound transducer was linearly swept over the transducer with an attenuation of 0.5dB/cm-MHz. A set of 585 raw channel data images were collected. The channel data was tested using the proposed algorithm, and pseudo-FSA channel data was reconstructed. The channel data was then beamformed offline for comparison with SA and FSA. Quantitative metrics were used to compare the hypoechoic and hyperechoic regions from a sample image.
G. IN-VIVO CAROTID DATA
To demonstrate the performance of the algorithm in clinical applications, in-vivo data was collected from longitudinal and transverse orientations of the carotid artery from two different users. Approximately 1000 raw channel data images were collected for each orientation of each user. The raw channel data was only used for testing by reconstructing the pseudo-FSA channel data. The channel data was beamformed offline and was compared with SA and FSA. Quantitative metrics were used to compare the carotid region from a sample image.
H. QUANTITATIVE METRICS TO EVALUATE PERFORMANCE
Quantitative metrics were defined to compare the results from SA, FSA, and pseudo-FSA. The contrast-to-noise ratio (CNR) was calculated using equation 4
| (4) |
where µ is the mean of intensity values, σ is the standard deviation of the intensity values, and ROI is the region of interest.
Speckle statistics were calculated as it is an important parameter representing the intrinsic characteristic of ultrasound images. A fully developed speckle pattern follows a Rayleigh distribution. The Kolmogorov-Smirnov (KS) test was used to access the speckle quality [20]. If a significance level of 0.05 was obtained, the speckle pattern was considered to follow the Rayleigh distribution.
The resolution was also calculated by using full width at half maximum (FWHM) for both axial and lateral direction.
Unwrapped phase information was calculated by using equation 5
| (5) |
where tan−1 is the inverse tangent, IQ is the in-phase quadrature data, and k is an integer. The root-mean-square error between pseudo-FSA and FSA for unwrapped phase information was also calculated.
3. RESULTS
A. POINT SPREAD FUNCTION
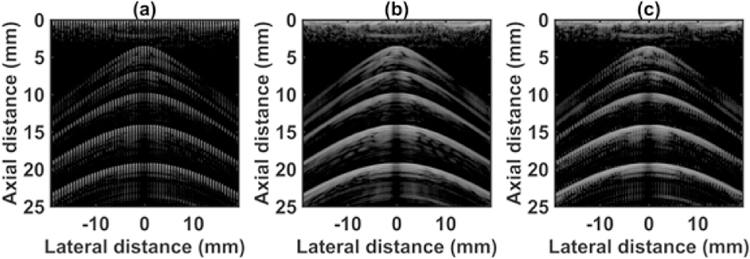
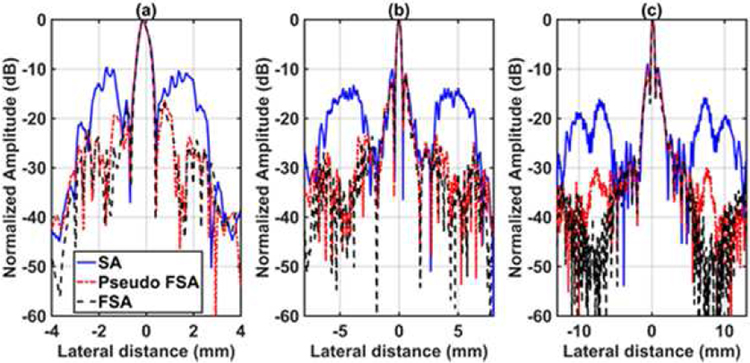
The quantitative results for testing with the wire target in a water tank are shown in Table 2. Fig. 3 shows the channel data for the SA, FSA and pseudo-FSA. The corresponding beamformed images are illustrated in Fig. 4 with a dynamic range of 60 dB. The grating lobes in SA were clearly visible. The FSA exhibits very small grating lobes. The grating lobes were reduced in the pseudo-FSA, however not completely eliminated. Figs. 5(a), 5(b) and 5(c) show the beam profile for the first, third, and fifth wire target. Table 3 summarizes the results for the ratio of grating lobe energy to the total energy, as defined in equation 4. The grating lobe energy was lowest for FSA, followed by pseudo-FSA and SA. The grating lobe energy in pseudo-FSA was suppressed by a mean value of 2.12 dB compared to SA. However, the grating lobe energy for pseudo-FSA was lower than the FSA. No difference was observed in axial or lateral resolution. Speckle quality was not evaluated due to the small size of the target.
TABLE II.
Mean error between Pseudo-FSA and FSA.
| Metric | WT | SPP | MPP | cTu1 | cTu2 | cLu1 | cLu2 |
|---|---|---|---|---|---|---|---|
| # Images |
16 | 425 | 585 | 1050 | 1030 | 1055 | 1045 |
| RMSE | 128 | 38 | 73 | 297 | 261 | 231 | 217 |
| MAE | 36 | 19 | 35 | 101 | 88 | 76 | 71 |
| DSSIM | 0.04 | 0.01 | 0.04 | 0.10 | 0.09 | 0.07 | 0.07 |
RMSE is the root mean square error rounded to the nearest integer
MAE is the mean absolute error rounded to the nearest integer
DSSIM is the structural dissimilarity error rounded to two significant digits
WT is wire target
SPP is the small parts phantom
MPP is the multi-purpose multi-tissue phantom
cTu1 is carotid in transverse orientation for volunteer1
cTu2 is carotid in transverse orientation for volunteer2
cLu1 is carotid in longitudinal orientation for volunteer1
cLu2 is carotid in longitudinal orientation for volunteer2
Fig. 3.
Channel data from wire target in water tank using (a) 64 channel RX sparse array, (b) 128 channel RX fully sampled array, (c) 64 channel RX pseudo fully sampled array. Data is displayed at a dynamic range of 60 dB.
Fig. 4.
Beamformed RF data from wire target in water tank using (a) 64 channel RX sparse array, (b) 128 channel RX fully sampled array, (c) 64 channel RX pseudo fully sampled array. Data is displayed at a dynamic range of 60 dB.
Fig. 5.
Lateral beam profile for 64 channel RX sparse array, 128 channel RX fully sampled array, and 64 channel RX pseudo fully sampled array at a depth of (a) 3.6 mm, (b) 10.5 mm, (c) 20 mm.
TABLE III.
Ratio of grating lobe energy to total energy for different receive configurations in wire target experiment.
| EML/EBK | SA | FSA | Pseudo-FSA |
|---|---|---|---|
| First wire target | −1.17 | −3.96 | −3.52 |
| Third wire target | −0.75 | −3.40 | −2.44 |
| Fifth wire target | −1.10 | −3.87 | −3.42 |
EML is the energy associated with the main lobe
EBK is the energy outside 5λ
SA is the sparse array
FSA is the fully sampled array
Pseudo-FSA is the virtually created FSA with the proposed method
B. SMALL PARTS PHANTOM
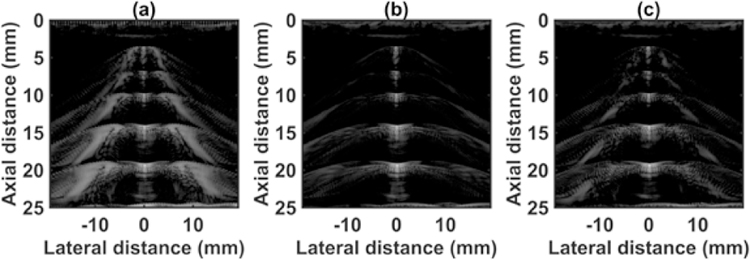
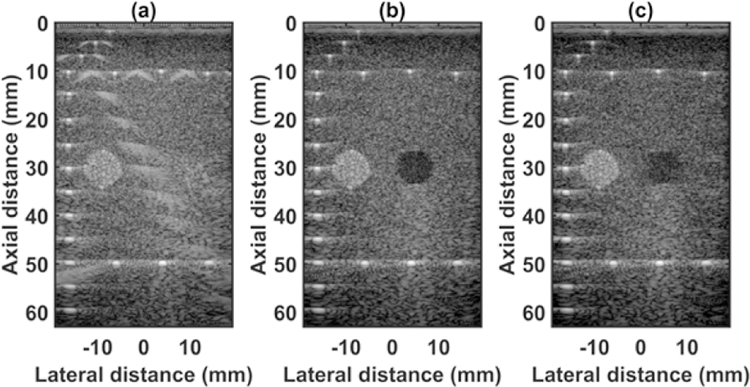
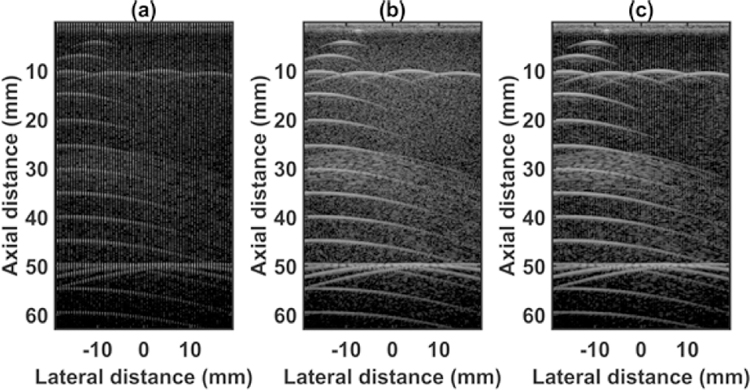
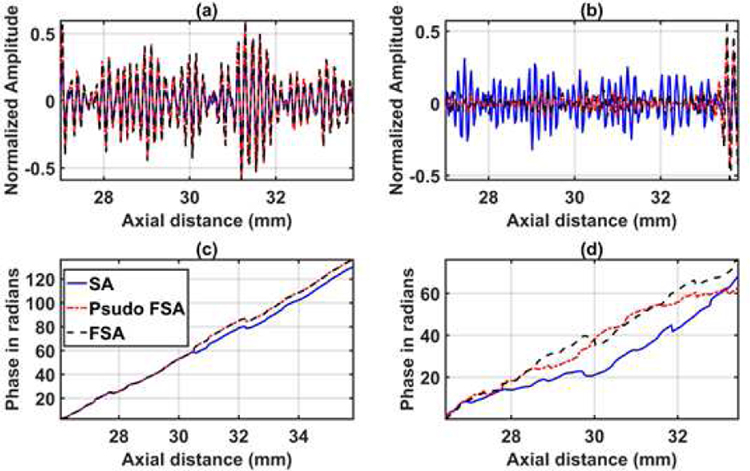
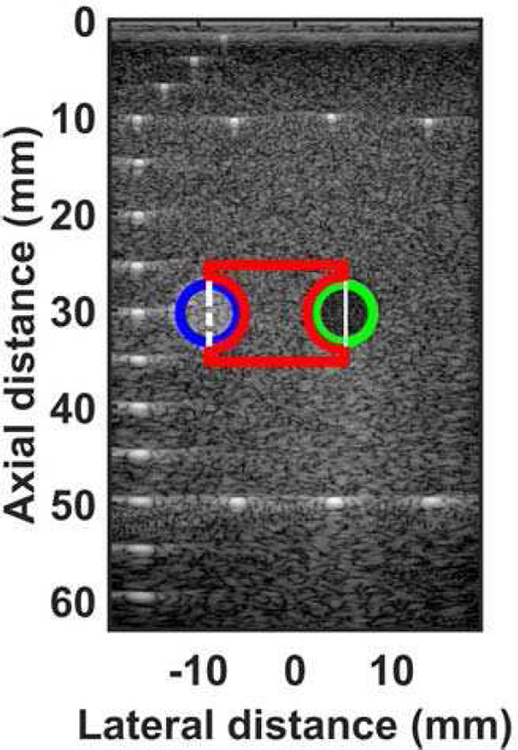
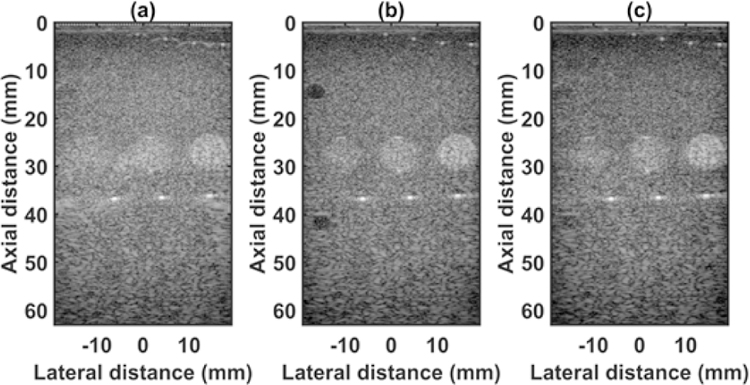
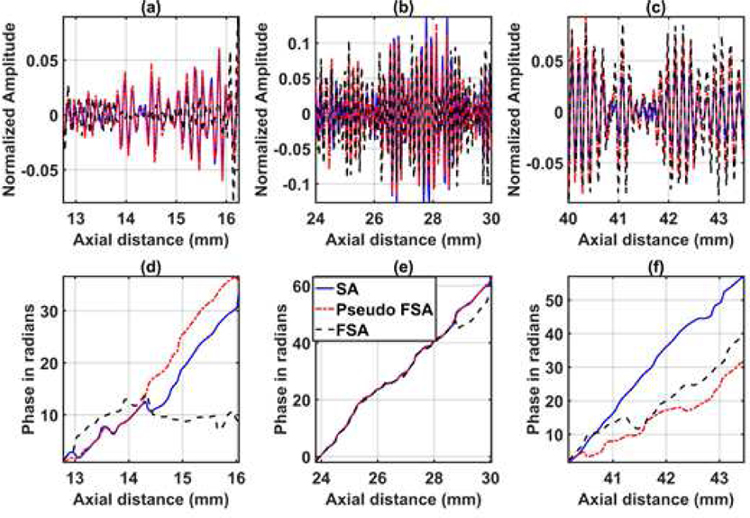
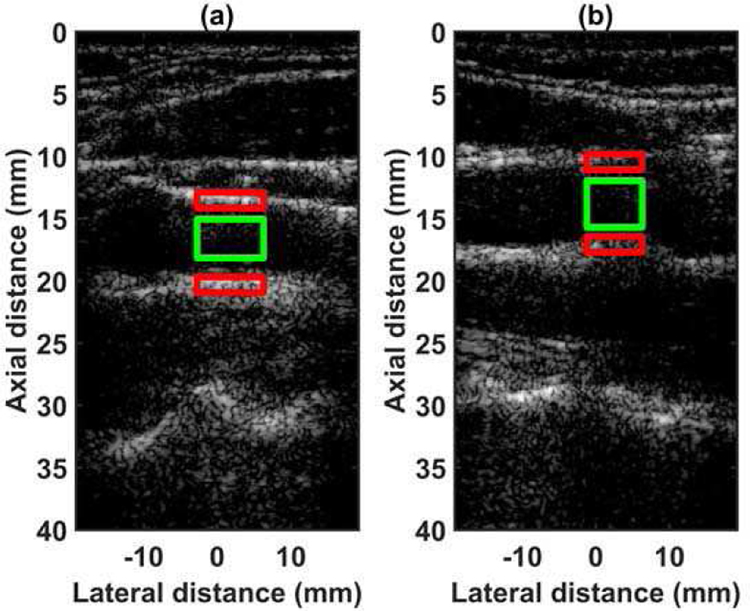
The quantified estimation error results for SPP data using different metrics are summarized in Table 2. Fig. 7 shows the beamformed images for the SA, FSA, and pseudo-FSA displayed with a dynamic range of 60dB. Fig. 6 shows a sample channel data image for SA, FSA and pseudo-FSA. The corresponding beamformed data can be observed in Fig. 7. The sample image contains wire targets that are spread laterally at the depths of 1 cm and 5 cm, one hyperechoic region, and one hypoechoic region. Wire targets are also present at different depths on the left side of the image. In the case of SA, the grating lobes from wire targets at a depth of 3 cm completely obfuscate the hypoechoic region. The pseudo-FSA was able to reconstruct the hypoechoic region, but some geometrical distortion is observed after reconstruction. Table 4 summarizes the CNR for the hyperechoic and the hypoechoic regions. Pseudo-FSA had higher CNR than SA for both regions and had higher CNR than FSA for the hyperechoic region. The beamformed RF A-line passing through the center of the hyperechoic region and hypoechoic region is shown in Fig. 8(a), and 8(b). For the hyperechoic region, the difference between the SA, FSA, and pseudo-FSA is not clear. However, for the hypoechoic region, the SA had a much higher amplitude, whereas the pseudo-FSA closely follows the FSA. Fig. 8(c), and 8(d) shows the unwrapped phase values for the hyperechoic and hypoechoic RF A-line. Phase information for pseudo-FSA from the hyperechoic region was precisely reconstructed and matched the FSA. However, the phase information for pseudo-FSA from the hypoechoic region had some errors but performed better than SA. The hyperechoic region has a root-mean-square unwrapped phase error of 4.54 rad between SA and FSA, and 0.15 rad between pseudo-FSA and FSA. The hypoechoic region has the root-mean-square unwrapped phase error of 11.86 rad between SA and FSA, and 4.71 rad between pseudo-FSA and FSA. The mask used to calculate CNR and the lines from which the RF A-line was selected is shown in Fig. 9. Axial and lateral resolution for SA, FSA, and pseudo-FSA were the same. The homogenous background region in SPP followed Rayleigh distribution for FSA and pseudo-FSA but not for SA. Similarly, for hypoechoic region FSA and pseudo-FSA followed Rayleigh distribution, whereas SA did not. However, for the hyperechoic region, none of them followed Rayleigh distribution.
Fig. 7.
Beamformed RF data from small parts phantom using (a) 64 channel RX sparse array, (b) 128 channel RX fully sampled array, (c) 64 channel RX pseudo fully sampled array. Data is displayed at a dynamic range of 60 dB.
Fig. 6.
Channel data from small parts phantom using (a) 64 channel RX sparse array, (b) 128 channel RX fully sampled array, (c) 64 channel RX pseudo fully sampled array. Data is displayed at a dynamic range of 60 dB.
TABLE IV.
Contrast to noise ratio for Hyperechoic and Hypoechoic regions of small parts phantom.
| CNR | SA | FSA | Pseudo-FSA |
|---|---|---|---|
| ROI 1 | −4.48 | 1.63 | 2.70 |
| ROI 2 | −15.25 | 2.41 | 0.00 |
ROI I is the hyperechoic region
ROI 2 is the hypoechoic region
Fig. 8.
RF A-line data for 64 channel RX sparse array, 128 channel RX fully sampled array, and 64 channel RX pseudo fully sampled array acquired from small parts phantom in (a) hyperechoic region of ROI 1, (b) hypoechoic region of ROI 2. Unwrapped phase information for 64 channel RX sparse array, 128 channel RX fully sampled array, and 64 channel RX pseudo fully sampled array acquired from small parts phantom in (c) hyperechoic region of ROI 1 (d) hypoechoic region of ROI 2.
Fig. 9.
Mask showing regions for calculating contrast-to-noise ratio and RF A-line for small parts phantom. The blue circle shows the ROI 1 for hyperechoic region, the green circle shows the ROI 2 for hypoechoic region. The red area shows the region used as background for noise calculation. The dashed white line shows the RF A-line associated with hyperechoic region. The solid white line shows the RF A-line associated with hypoechoic region.
C. MULTI-PURPOSE PHANTOM
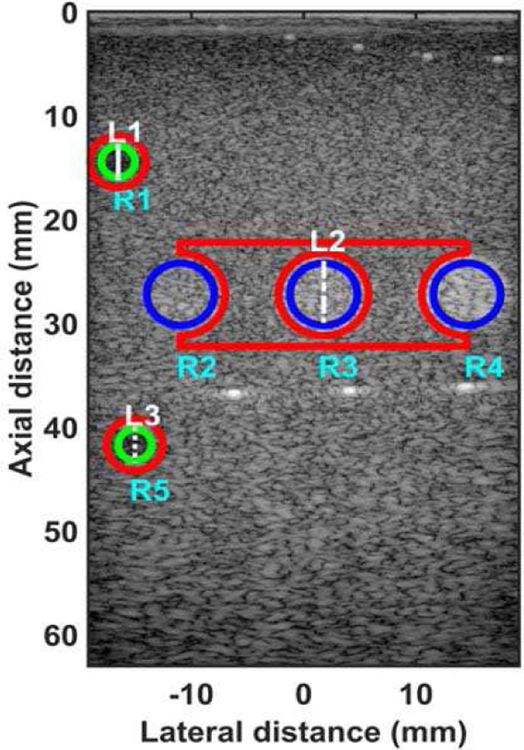
The quantified estimation error for MPP data using different metrics is summarized in Table 2. Fig. 10 shows the beamformed images for the SA, FSA, and pseudo-FSA displayed with a dynamic range of 60dB. The sample image contains three hyperechoic regions of varying echogenicity at a depth of 25 mm, two hypoechoic regions on the left side of the image, wire targets at a depth of 5 mm, and wire targets at a depth of 36 mm. For SA, grating lobes can be observed for the wire targets, and the hyperechoic regions are geometrically distorted. The hypoechoic region was completely obfuscated by the grating lobes. The pseudo-FSA was able to suppress the grating lobes associated with the wire targets. Pseudo-FSA partially reconstructed the hypoechoic regions. The geometrical shape of the hyperechoic region from pseudo-FSA is similar to FSA. Table 5 summarizes the CNR for the 3 hyperechoic and 2 hypoechoic regions. Pseudo-FSA had higher CNR for the hypoechoic region compared to SA. Pseudo-FSA showed improvement in CNR over SA for the hyperechoic regions. The beamformed RF A-line for the top hypoechoic region, central hyperechoic region, and the bottom hypoechoic region is shown in Fig. 11(a), 11(b), and 11(c), respectively. Pseudo-FSA has better reconstruction for RF A-line of the hyperechoic region than hypoechoic regions. Fig. 11(d), 11(e), and 11(f) show the unwrapped phase values for the top hypoechoic region, central hyperechoic region and bottom hypoechoic region. The top hypoechoic region has a root-mean-square unwrapped phase error of 9.99 rad between SA and FSA, and 13.91 rad between pseudo-FSA and FSA. The middle hyperechoic region has a root-mean-square unwrapped phase error of 3.90 rad between SA and FSA, and 3.81 rad between pseudo-FSA and FSA. The bottom hypoechoic region has a root-mean-square unwrapped phase error of 13.72 rad between SA and FSA, and 5.33 rad between pseudo-FSA and FSA. The mask used to calculate CNR, and the line from which the RF A-line was selected is shown in Fig. 12. Axial and lateral resolution for SA, FSA, and pseudo-FSA was the same. The homogenous background region in MPP followed Rayleigh distribution for FSA, pseudo-FSA, and SA. The top hypoechoic region and mid hyperechoic region follow Rayleigh distribution only for SA. The lower hypoechoic region does not follow Rayleigh distribution for any technique.
Fig. 10.
Beamformed RF data from multi-purpose phantom using (a) 64 channel RX sparse array, (b) 128 channel RX fully sampled array, (c) 64 channel RX pseudo fully sampled array. Data is displayed at a dynamic range of 60 dB.
TABLE V.
Contrast to noise ratio for Hyperechoic and Hypoechoic regions of multi-purpose phantom.
| CNR | SA | FSA | Pseudo-FSA |
|---|---|---|---|
| ROI 1 | −7.81 | 1.28 | −6 81 |
| ROI 2 | −9.26 | −7.39 | −5.70 |
| ROI 3 | −2.89 | −1.73 | −1 54 |
| ROI 4 | −1.89 | 1.08 | 1.52 |
| ROI 5 | −13.73 | 1.96 | −2.80 |
ROI I is the top hypoechoic region
ROI 2 is the left hyperechoic region
ROI 3 is the middle hyperechoic region
ROI 4 is the right hyperechoic region
ROI 5 is the bottom hypoechoic region
Fig. 11.
RF A-line data for 64 channel RX sparse array, 128 channel RX fully sampled array, and 64 channel RX pseudo fully sampled array acquired from multi-purpose phantom in (a) hypoechoic region of ROI 1, (b) hyperechoic region of ROI 3, (c) hypoechoic region of ROI 5. Unwrapped phase information for 64 channel RX sparse array, 128 channel RX fully sampled array, and 64 channel RX pseudo fully sampled array acquired from multi-purpose phantom in (d) hypoechoic region of ROI 1, (e) hyperechoic region of ROI 3, (f) hypoechoic region of ROI 5.
Fig. 12.
Mask showing regions for calculating contrast-to-noise ratio and RF A-line for multi-purpose phantom. The green circles show the ROI 1 (labelled R1) and ROI 5 (labelled R5) associated with the two hypoechoic regions. The blue circles show the ROI 2 (labelled R2), ROI 3 (labelled R3), and ROI 4 (labelled R4) for the three hyperechoic regions. The red area shows the region used as background for noise calculation. The solid white line (labelled L1) shows the RF A-line associated with ROI 1 hypoechoic region. The dashed white line (labelled L2) shows the RF A-line associated with ROI 3 hyperechoic region. The dashed white line (labelled L3) shows the RF A-line associated with ROI 5 hypoechoic region.
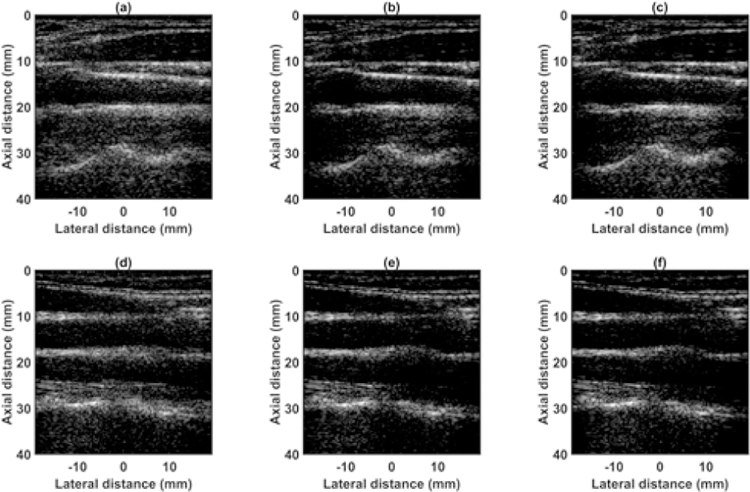
D. IN-VIVO CAROTID
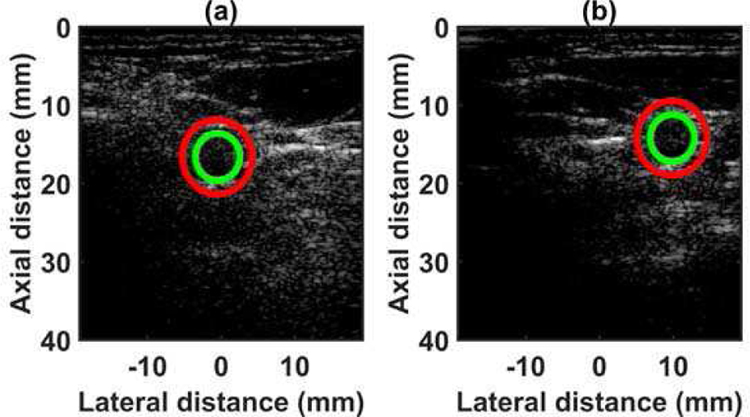
The quantified estimation error for in vivo carotid data from two volunteers using different metrics is summarized in Table 2. Fig. 13 shows the beamformed images for the SA, FSA, and pseudo-FSA displayed with a dynamic range of 40dB. The carotid artery was observed between depths of 10 mm and 20 mm. For SA, the hypoechoic region inside the carotid artery is affected by the grating lobe artifact. The pseudo-FSA is able to partially reconstruct the carotid artery. The CNR values for the transverse orientation of the carotid artery are summarized in Table 6, and the mask used to calculate CNR is displayed in Fig. 15. Axial and lateral resolution for SA, FSA, and pseudo-FSA was the same. Speckle pattern was not accessed for in-vivo data as fully developed speckle pattern was hard to observe close to the carotid artery. The root-mean-square unwrapped phase error for transverse orientation of volunteer 1 between SA and FSA was 12.86 rad, and between pseudo-FSA and FSA was 10.60 rad. The root-mean-square unwrapped phase error for transverse orientation of volunteer 2 between SA and FSA was 3.08 rad, and between pseudo-FSA and FSA was 4.76 rad.
Fig. 13.
Beamformed RF data from the carotid artery of volunteer 1 with the probe in transverse orientation acquired using (a) 64 channel RX sparse array, (b) 128 channel RX fully sampled array, (c) 64 channel RX pseudo fully sampled array. Channel data from the carotid artery of volunteer 2 with the probe in transverse orientation acquired using (d) 64 channel RX sparse array, (e) 128 channel RX fully sampled array, (f) 64 channel RX pseudo fully sampled array. Data is displayed at a dynamic range of 40 dB.
TABLE VI.
Contrast to noise ratio for carotid artery.
| CNR | SA | FSA | Psuedo FSA |
|---|---|---|---|
| cTv 1 | −19.79 | −3.82 | −5.10 |
| cTv 2 | −14.32 | −1.13 | −5 72 |
| cLv 1 | 6.40 | 6.68 | 7.07 |
| cLv 2 | 1.83 | 2.39 | 1.91 |
cTV1 is carotid in transverse orientation for volunteer 1
cTv2 is carotid in transverse orientation for volunteer 2
cLv1 is carotid in longitudinal orientation for volunteer 1
cLv2 is carotid in longitudinal orientation for volunteer 2
Fig. 15.
Beamformed RF data from carotid artery of volunteer 1 with probe in longitudinal orientation acquired using (a) 64 channel RX sparse array, (b) 128 channel RX fully sampled array, (c) 64 channel RX pseudo fully sampled array. Channel data from carotid artery of volunteer 2 with probe in longitudinal orientation acquired using (d) 64 channel RX sparse array, (e) 128 channel RX fully sampled array, (f) 64 channel RX pseudo fully sampled array. Data is displayed at a dynamic range of 40 dB.
Fig. 14 shows the beamformed images for the SA, FSA, and pseudo-FSA displayed with a dynamic range of 40dB. The carotid artery was observed between depths of 10 mm and 20 mm. For SA, the hypoechoic region inside the carotid artery had some artifacts. The pseudo-FSA was able to reduce these artifacts. Furthermore, the anatomical structures are well defined in pseudo-FSA compared to SA. The CNR values for the longitudinal orientation of the carotid artery are summarized in Table 6, and the mask used to calculate CNR is displayed in Fig. 16. The axial and lateral resolutions for the SA, FSA, and pseudo-FSA are similar. Speckle pattern was not accessed for in-vivo data as fully developed speckle pattern was hard to observe close to the carotid artery. The root-mean-square unwrapped phase error for longitudinal orientation of volunteer 1 between SA and FSA was 3.60 rad, and between pseudo-FSA and FSA was 3.32 rad. The root-mean-square unwrapped phase error for longitudinal orientation of volunteer 2 between SA and FSA was 13.32 rad, and between pseudo-FSA and FSA was 14.51 rad.
Fig. 14.
Mask showing regions for calculating contrast-to-noise ratio from the carotid artery in the transverse orientation of (a) volunteer 1, (b) volunteer 2. The green circle shows the carotid artery, and the circle shows the region used for background noise.
Fig. 16.
Mask showing regions for calculating contrast-to-noise ratio from the carotid artery in the transverse orientation of (a) volunteer 1, (b) volunteer 2. The green region shows the carotid artery, and the red area shows the region used for background noise.
IV. DISCUSSION
A CNN algorithm was proposed to estimate the inactive channel data in periodic sparse arrays from neighboring active channels. The proposed algorithm was trained on an SPP and was validated using wire target data, MPP, and in-vivo carotid data. The best performance of the algorithm was observed when the phantom used for training, SPP, was also used for testing. The high prediction errors observed for the in-vivo data are due to the sonographic features associated with in-vivo data, which are not emulated in tissue-mimicking phantoms, e.g., varying attenuation, heterogeneity of the tissue, varying speed of sound, and phase aberration. The performance of the algorithm can be improved for in-vivo data by training it on in-vivo data from multiple volunteers. Although the quantitative performance of in-vivo data is lower compared to that of the tissue-mimicking phantom, the pseudo-FSA performed better than SA in both qualitative and quantitative terms. No change was observed in the lateral resolution as the main lobe widths for the SA, FSA, and pseudo-FSA were the same. The main lobe width has no dependence on inter-element spacing and only depends on the F-number as shown in equation 6
| (6) |
where n is a constant dependent on the measurement technique, F# is the F-number, and λ is the wavelength. The axial resolution depends only on the center frequency of the transmitted signal, and thus, no change was observed in the axial resolution as the center frequency for the three techniques was the same.
The uniform linear array used in this study had inter-element spacing, d, of 0.3 mm, whereas the wavelength used for acquiring the data was 0.25 mm, thus rendering the grating lobes unavoidable even in FSA. The inter-element spacing for the SA was doubled, bringing the grating lobes even closer to the main lobe. The increased inter-element spacing resulted in the degradation of the CNR for SA. To recover the CNR, grating lobes had to be moved away from the main lobe. By estimating the inactive channel data using the proposed method, the inter-element spacing for beamforming was reduced; thus, the grating lobes were moved further away from the main lobe to improve the CNR. The proposed method could be used to further reduce the inter-element spacing by predicting the sub-wavelength virtual channel, which could further suppress the grating lobes and improve the CNR [14], [15].
The lateral beam profile for wire target in the water tank, Fig 5, demonstrates that the main lobe width does not change while the grating lobes are suppressed. No change in the axial beam profile was observed.
Suppression of the grating lobes in the hypoechoic region of SPP demonstrates the capability of the proposed algorithm. The presence of hyperechoic wire targets at a depth of 10 mm creates high-intensity grating lobes, which move away from the main lobe with increasing depth, obfuscates the hypoechoic region. Hyperechoic regions are common in soft tissue, for example, in connective tissue, calcifications, tendons, nerves, and parenchymal tissues. The grating lobes from the hyperechoic regions can potentially obfuscate hypoechoic or anechoic regions present in the nearby tissues. An improvement of 15.25 dB was observed in the hypoechoic region of SPP. The CNR for the pseudo-FSA in the hyperechoic region is higher than FSA, for both SPP and MPP. This phenomenon is not due to the improvement in contrast but due to the reduction in noise variance, which stems from the inability of the proposed algorithm to predict low amplitude signals. Transverse in-vivo data shows improvement in CNR up to 14.69 dB for the hypoechoic carotid artery. However, it barely shows any improvement in the longitudinal orientation due to the absence of hyperechoic objects.
The speckle pattern is commonly used to estimate quantitative ultrasound parameters [21]–[24]. It is essential that the pseudo-FSA follows the same speckle pattern trend as the FSA so that the data generated by pseudo-FSA could be used for quantitative analysis. In all cases, the pseudo-FSA had the same speckle pattern as the FSA. However, the SA did not follow the same trend, thus rendering pseudo-FSA better suited for quantitative analysis than SA.
Phase information is another important parameter that is used in Doppler imaging [25] and shear wave elastography applications [26]–[28]. Predicting phase data can be challenging, as seen in Fig. 8(c), 8(d) and Fig. 13(d), 13(e), 13(f). The phase information changes rapidly over samples and is very specific to the local region through which the signal travels. The DSSIM loss function only considers amplitude information. In the case of a high amplitude signal, the algorithm was able to reconstruct the phase information; however, the algorithm struggled in the hypoechoic regions. Phase information prediction can be improved by modifying the loss function to consider both amplitude and phase-related information.
This study, in its current form, has some limitations in estimating the missing channel data, which could be addressed in future works. The scope of this paper was limited to introducing the algorithm, showcasing its ability to predict the channel data and suppressing the grating lobes. Only zero angle plane wave imaging was used to simplify the problem. However, in the future, different angles can be trained separately, and there is no limitation on using spatially compounded images. Here, a periodic sparse array was used to demonstrate the ability of the algorithm. However, in the future, the algorithm can be extended to non-periodic sparse arrays.
Another limitation that was observed from beamformed RF A-line data was that the algorithm performed poorly when the amplitude of the signal was low. The limitation can also be observed in Figs. 7, 9, 11, and 12 in which the reconstruction of the hypoechoic region is poor. The estimation error for the hyperechoic region was low compared to the hypoechoic region. The error associated with a high amplitude signal is large; thus, all filters are optimized to reduce errors in large-amplitude signals. This results in poor prediction for low amplitude signal regions.
A high quantitative error was observed for the in-vivo data. However, qualitatively the difference between FSA and pseudo-FSA for the longitudinal images was barely observable. The error could be further reduced in the future by training the algorithm using in-vivo datasets.
Here, only the Kaiser apodization was considered in transmit, and no apodization was considered on the receive. Future studies can explore the best apodization window in both transmit and the receive side.
V. CONCLUSION
An algorithm is proposed, which can estimate the inactive element channel data in periodic sparse arrays to reduce the inter-element spacing for beamforming, thus suppressing the grating lobes. Suppression of the grating lobe was demonstrated in both tissue-mimicking phantoms and in-vivo conditions. Considerable CNR improvement, up to 15.25 dB, was observed in the hypoechoic region while maintaining the physical attributes of the ultrasound signal. The algorithm can be used to reduce the number of active elements in arrays.
ACKNOWLEDGMENT
The authors are grateful to Kathryn Schinke-Bierly, Erin Jarrod, and Jennifer Poston for administrative support. The research reported in this publication was supported by the National Institutes of Health grants R01CA168575, R01CA174723, and R01CA195527 from the National Cancer Institute. We acknowledge the support of NVIDIA Corporation with the donation of the Titan XP GPU used for this research. Also, we acknowledge the support of Amazon web services for the donation of credits used for this research. The authors also wish to thank Dr. Desiree J. Lanzino, PT, PhD, for her editorial services and Ms. Shaheeda Adusei for formatting the manuscript.
This work was supported by the National Institute of Health under the award numbers R01CA168575, R01CA174723, R01CA195527, and R01EB17213. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Biography

Viksit Kumar, PhD received his Ph.D. degree in Mechanical Engineering from the University of Iowa in 2014. He then joined the Mayo Clinic Ultrasound Research Lab, in the Department of Physiology and Biomedical Engineering as a post-doctoral research fellow where he became a research associate in 2018. At the time of this work he was with Mayo Clinic, since 2019 he is no longer at Mayo clinic. The focus of his research is quantitative ultrasound, ultrasound shearwave elastography, Ultrasonic guidance of non-invasive devices and application of deep learning in ultrasound imaging.

Po-Yang Lee, MS received the B.S. degree in electronic engineering from the Ming Chuan University in Taiwan in 2009; and M.S. degree in electrical engineering from the Fu Jen Catholic University in Taiwan in 2014. From 2017 to 2018, he worked as Research Trainee at the department of physiology and biomedical engineering, Mayo Clinic, Rochester, MN. Currently, he is a Ph.D. candidate in the biomedical engineering, National Cheng Kung University, Taiwan. His research interests include the development of high-frequency ultrasound imaging system, point-of-care ultrasonic system and ultrasound beamforming method with deep learning.

Bae-Hyung Kim, PhD received the B.Sc., M.Sc., and Ph.D. degrees from the Department of Electronic Engineering, Sogang University, Seoul, in 2001, 2003, and 2007, respectively. From 2007 to 2010, he was a Principal Engineer with Siemens Ultrasound Systems Korea, Seoul. From 2010 to 2015, he worked as a research Staff at the Samsung Advanced Institute of Technology (SAIT), Samsung Electronics Company Ltd., Gyeonggi-do, South Korea, Member, where he led the development of volumetric ultrasound system for 2-D CMUT-on-ASIC array. After about 9 years of experience in industry, in 2015 he joined the Department of Biomedical Engineering, King’s College London, UK, as a post-doctoral research associate. At the time of this work, 2016–2018, he was with the Department of Physiology and Biomedical Engineering, Mayo Clinic College of Medicine, Rochester, MN, USA, as a Senior Research Fellow. Currently, he is the director of Vave Health Inc., San Mateo, CA, USA. He holds 43 patents filed including 13 granted patents in ultrasound imaging and system technologies. His research interests include principles of imaging system design and signal processing, development of ultrasonic instrumentation for ultrasimple front-end architecture, and estimation.

Mostafa Fatemi, PhD (S’M’77-SM’07-F’12-LF’18) received his Ph.D. degree in Electrical Engineering from Purdue University. He joined Mayo in 1994, and currently, he is a Professor of Biomedical Engineering at the Department of Physiology and Biomedical Engineering of Mayo Clinic College of Medicine in Rochester, MN. At the Mayo Clinic, he is also an active member of the Mayo Clinic Cancer Center and the Center for Clinical and Translational Science. Dr. Fatemi’s current research areas include developing novel ultrasonic methods for cancer imaging, microvasculature imaging, and bladder function evaluation. His research program is continuously supported by multiple major federal grants funded by the National Institute of Health (NIH) and Komen Breast Foundation for the Cure and Department of Defense. He is a Life Fellow of IEEE and holds Fellow membership at these institutions: American Institute of Medical and Biological Engineering (AIMBE), Acoustical Society of America (ASA), and American Institute of Ultrasound in Medicine (AIUM). Dr. Fatemi is recipient of the IEEE-UFFC Distinguished Lecturer award.

Azra Alizad, MD (M’06–SM’15) received her medical degree and fellowship from Tehran University Medical Sciences. Currently, Dr. Alizad is a professor of Radiology, Department of Radiology and Professor of Biomedical Engineering as well as an associate professor of Medicine at the Mayo Clinic College of Medicine in Rochester MN. In addition to her training in Medicine, she has a broad background and expertise in medical ultrasound research. She directs a translational research laboratory, involving development and application of novel ultrasound technologies such as microvasculature imaging, functional ultrasound, elastography, vibro-acoustic imaging for diagnosis of abnormalities in breast, thyroid, prostate, neck masses, and axillary lymph nodes, brain and bone. Dr. Alizad is the principal investigator of multiple major federal grants funded by the National Institute of Health (NIH) and recipient of grant funded by Komen Breast Foundation for the Cure. She is an elected Fellow of American Institute for Medical and Biological Engineering (AIMBE) and American Institute of Ultrasound in Medicine (AIUM) and senior member of Institute of Electrical and Electronics Engineers-UFFC. At the Mayo Clinic, she is also an active member of the Mayo Clinic Cancer Center and the Center for Clinical and Translational Science.
References
- [1].Lockwood GR and Foster FS, “Sparse array structures,” ed: Google Patents, 1996. [Google Scholar]
- [2].Schwartz JL, Jackson JI, and Plugge JS, “Diagnostic medical ultrasound system and method for using a sparse array,” ed: Google Patents, 2001. [Google Scholar]
- [3].Song T.-k. and Kim G-D, “Method for optimal design of non-uniform weighted periodic sparse arrays for ultrasound imaging system,” ed: Google Patents, 2010. [Google Scholar]
- [4].Turnbull DH and Foster FS, “Beam steering with pulsed two-dimensional transducer arrays,” IEEE transactions on ultrasonics, ferroelectrics, and frequency control, vol. 38, no. 4, pp. 320–333, 1991. [DOI] [PubMed] [Google Scholar]
- [5].Karaman M, Wygant IO, Oralkan Ö, and Khuri-Yakub BT, “Minimally redundant 2-D array designs for 3-D medical ultrasound imaging,” IEEE transactions on medical imaging, vol. 28, no. 7, pp. 1051–1061, 2009. [DOI] [PubMed] [Google Scholar]
- [6].Lockwood GR, Li P-C, O’Donnell M, and Foster FS, “Optimizing the radiation pattern of sparse periodic linear arrays,” IEEE Transactions on Ultrasonics, Ferroelectrics, and frequency control, vol. 43, no. 1, pp. 7–14, 1996. [Google Scholar]
- [7].Roux E, Ramalli A, Tortoli P, Cachard C, Robini MC, and Liebgott H, “2-D ultrasound sparse arrays multidepth radiation optimization using simulated annealing and spiral-array inspired energy functions,” IEEE transactions on ultrasonics, ferroelectrics, and frequency control, vol. 63, no. 12, pp. 2138–2149, 2016. [DOI] [PubMed] [Google Scholar]
- [8].Diarra B, Robini M, Tortoli P, Cachard C, and Liebgott H, “Design of optimal 2-D nongrid sparse arrays for medical ultrasound,” IEEE Transactions on Biomedical Engineering, vol. 60, no. 11, pp. 3093–3102, 2013. [DOI] [PubMed] [Google Scholar]
- [9].Tekes C, Karaman M, and Degertekin FL, “Optimizing circular ring arrays for forward-looking IVUS imaging,” IEEE transactions on ultrasonics, ferroelectrics, and frequency control, vol. 58, no. 12, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Trucco A, “Thinning and weighting of large planar arrays by simulated annealing,” IEEE transactions on ultrasonics, ferroelectrics, and frequency control, vol. 46, no. 2, pp. 347–355, 1999. [DOI] [PubMed] [Google Scholar]
- [11].Kim B, Kim K, and Park S, “Ultrasonic imaging apparatus, ultrasonic probe apparatus, signal processing apparatus and method of controlling ultrasonic imaging apparatus,” U.S. Pub. No. 2016/0209502 A1, 07/21/2016 2015.
- [12].Klebæk H, Jensen JA, and Hansen LK, “Neural network for sonogram gap filling,” in Ultrasonics Symposium, 1995. Proceedings., 1995 IEEE, 1995, vol. 2: IEEE, pp. 1553–1556. [Google Scholar]
- [13].Zhang Y and Li X, “Method and apparatus for filling doppler signal gaps in ultrasound diagnostic imaging,” ed: Google Patents, 2012. [Google Scholar]
- [14].Lin C-Y and Li M-L, “Plane wave imaging using virtual sub-wavelength receiving elements,” in Ultrasonics Symposium (IUS), 2016 IEEE International, 2016: IEEE, pp. 1–3. [Google Scholar]
- [15].Peng S-Y and Li M-L, “Linear array beamformation using virtual sub-wavelength receiving elements,” in Ultrasonics Symposium (IUS), 2015 IEEE International, 2015: IEEE, pp. 1–3. [Google Scholar]
- [16].Yoon YH and Ye JC, “Deep Learning for Accelerated Ultrasound Imaging,” in 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 2018: IEEE, pp. 6673–6676. [Google Scholar]
- [17].Yoon YH, Khan S, Huh J, and Ye JC, “Deep Learning in RF Sub-sampled B-mode Ultrasound Imaging,” arXiv preprint arXiv:1712.06096, 2017.
- [18].Abadi M and TensorFlow AABP, “Large-scale machine learning on heterogeneous distributed systems,” in Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI’16)( Savannah, GA, USA, 2016, pp. 265–283. [Google Scholar]
- [19].Montaldo G, Tanter M, Bercoff J, Benech N, and Fink M, “Coherent plane-wave compounding for very high frame rate ultrasonography and transient elastography,” IEEE transactions on ultrasonics, ferroelectrics, and frequency control, vol. 56, no. 3, pp. 489–506, 2009. [DOI] [PubMed] [Google Scholar]
- [20].Liebgott H, Rodriguez-Molares A, Cervenansky F, Jensen JA, and Bernard O, “Plane-wave imaging challenge in medical ultrasound,” in Ultrasonics Symposium (IUS), 2016 IEEE International, 2016: IEEE, pp. 1–4. [Google Scholar]
- [21].McFarlin BL et al. , “Development of an ultrasonic method to detect cervical remodeling in vivo in full-term pregnant women,” Ultrasound in medicine & biology, vol. 41, no. 9, pp. 2533–2539, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].McFarlin BL et al. , “Beyond cervical length: A pilot study of ultrasonic attenuation for early detection of preterm birth risk,” Ultrasound in medicine & biology, vol. 41, no. 11, pp. 3023–3029, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nam K et al. , “Cross-imaging system comparison of backscatter coefficient estimates from a tissue-mimicking material,” The Journal of the Acoustical Society of America, vol. 132, no. 3, pp. 1319–1324, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nam K et al. , “Ultrasonic attenuation and backscatter coefficient estimates of rodent-tumor-mimicking structures: comparison of results among clinical scanners,” Ultrasonic imaging, vol. 33, no. 4, pp. 233–250, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kumar V et al. , “Unambiguous Identification and Visualization of an Acoustically Active Catheter by Ultrasound Imaging in Real Time: Theory, Algorithm, and Phantom Experiments,” IEEE Transactions on Biomedical Engineering, vol. 65, no. 7, pp. 1468–1475, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bayat M et al. , “Diagnostic features of quantitative comb-push shear elastography for breast lesion differentiation,” PloS one, vol. 12, no. 3, p. e0172801, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bayat M et al. , “Correlation of ultrasound bladder vibrometry assessment of bladder compliance with urodynamic study results,” PloS one, vol. 12, no. 6, p. e0179598, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gregory A et al. , “Differentiation of Benign and Malignant Thyroid Nodules by Using Comb-push Ultrasound Shear Elastography: A Preliminary Two-plane View Study,” Academic radiology, 2018. [DOI] [PMC free article] [PubMed]