Abstract
The European Commission asked EFSA for a scientific opinion on chronic wasting disease in two parts. Part one, on surveillance, animal health risk‐based measures and public health risks, was published in January 2017. This opinion (part two) addresses the remaining Terms of Reference, namely, ‘are the conclusions and recommendations in the EFSA opinion of June 2004 on diagnostic methods for chronic wasting disease still valid? If not, an update should be provided’, and ‘update the conclusions of the 2010 EFSA opinion on the results of the European Union survey on chronic wasting disease in cervids, as regards its occurrence in the cervid population in the European Union’. Data on the performance of authorised rapid tests in North America are not comprehensive, and are more limited than those available for the tests approved for statutory transmissible spongiform encephalopathies surveillance applications in cattle and sheep. There are no data directly comparing available rapid test performances in cervids. The experience in Norway shows that the Bio‐Rad TeSeE™ SAP test, immunohistochemistry and western blotting have detected reindeer, moose and red deer cases. It was shown that testing both brainstem and lymphoid tissue from each animal increases the surveillance sensitivity. Shortcomings in the previous EU survey limited the reliability of inferences that could be made about the potential disease occurrence in Europe. Subsequently, testing activity in Europe was low, until the detection of the disease in Norway, triggering substantial testing efforts in that country. Available data neither support nor refute the conclusion that chronic wasting disease does not occur widely in the EU and do not preclude the possibility that the disease was present in Europe before the survey was conducted. It appears plausible that chronic wasting disease could have become established in Norway more than a decade ago.
Keywords: chronic, wasting, cervids, diagnostic, occurrence
Summary
In 2016, the European Food Safety Authority (EFSA) was asked by the European Commission to deliver a scientific opinion on three Terms of Reference (ToRs): (1) surveillance, (2) public health and (3) (animal health risk‐based measures) by 31 December 2016. On 18 January 2017, EFSA published a scientific opinion on chronic wasting disease (CWD) in cervids addressing these three ToRs (EFSA BIOHAZ Panel, 2017a). Within the same mandate, EFSA was asked to deliver by 31 December 2017 a scientific opinion on the following ToR: (4) are the conclusions and recommendations in the EFSA opinion of June 2004 on diagnostic methods for CWD still valid? If not, an update should be provided, and (5) update the conclusions of the 2010 EFSA opinion on the results of the European Union (EU) survey on CWD in cervids, as regards the occurrence of CWD in the cervid population in the EU.
No formal validation of test performance equivalent to the existing EU requirements for tests used for statutory surveillance in cattle and sheep has been undertaken for cervid material. A qualitative evaluation of the suitability of the Bio‐Rad and the IDEXX rapid tests (RT) commercially available for the diagnosis of CWD was carried out by means of literature review (both an ad hoc literature review on the diagnosis of CWD and the references retrieved by the search conducted for the 2017 Opinion (EFSA BIOHAZ Panel, 2017a)), the data provided by the manufacturers, and the knowledge and expertise of the Working Group (WG) members.
A review of the available approaches to the diagnosis of CWD including the considerations underpinning the selection of animals, tissues and diagnostic tests has been conducted, as well as a review of the different diagnostic methods applied for the detection of CWD, both in the context of large‐scale surveillance and for research purposes. Screening tests and confirmatory diagnostic methods have been reviewed along with methods for classification of isolates based on data from confirmatory testing, bioassay in potential natural host species and bioassay in rodent models. Requirements for the validation of new diagnostic tests, in particular, the steps and different pathways as defined by the International Organization of Animal Health (OIE) for validation of tests for wild populations, were considered. A review of all the validation exercises of RT for the detection of bovine spongiform encephalopathy (BSE) and for the diagnosis of transmissible spongiform encephalopathies (TSE) in small ruminants conducted in the EU has been included for comparison with the data current available for the rapid tests presently used for the detection of CWD in North America.
Sensitive amplification methods, such as protein misfolding cyclic amplification (PMCA) and real‐time quaking‐induced conversion (rtQuiC) that are currently under development for in vivo screening, or for the detection of environmental contamination, are also considered, but they are not yet at a point in their development where they could be applied in a statutory surveillance context.
To demonstrate how the potential for patchy CWD distribution could complicate surveillance in a heterogeneous geographic area the size of Europe, historical and contemporary maps of CWD distribution in the 28 contiguous US states east of the Mississippi River, spanning ~ 2.5 M km2, were used. This area approximates the EU (28 Member States (MS): > 4.4 M km2) with respect to several ecological, epidemiological and jurisdictional features relevant to CWD surveillance in the context of ToR 5.
Data on surveillance in Europe in 2015, 2016 and 2017 were extracted from annual reports submitted by the MS, and from the background information provided by the European Commission, and included in the mandate and the European Commission database. Surveillance data from Norway for the period 1 January 2017–27 November 2017 have been provided by the Norwegian Veterinary Institute, upon request. These data were used together with historical surveillance data from five Colorado mule deer herds collected over 15–21 years to provide a temporal reference of the estimated prevalence in new incursions of CWD and potential time lags in ‘epidemic’ emergence. Data from North America were used to generate a composite epidemic curve, and data from a published model were graphed for comparison with the observed data. The point estimate of comparable survey data from Norwegian reindeer (Nordfjella 1 region) was also calculated.
The experience in Norway so far shows that the Bio‐Rad RT (TeSeE™ SAP) has detected cases of CWD in reindeer, moose and red deer. It has also been shown that antibodies raised against the core or C‐terminal parts of the prion protein used for immunohistochemistry (IHC) and western blot (WB) were able to detect these cases.
Developments in immunoblotting techniques have resulted in the ability to discriminate experimental BSE from CWD in red deer. However, there is only limited information on the biological and molecular characteristics that define different strains in the North American cervid population against which the EU isolates could be compared and classified.
The conclusions (1, 2, 3, 4) and recommendations (3, 5, 6, 7) of the 2004 EFSA opinion on diagnostic methods for CWD remain valid. The available formal data on the performance of authorised RT for the detection of CWD in cervids in North America are not comprehensive and are much more limited than those available for the detection of BSE in cattle and scrapie in sheep. The lack of sufficient positive reference samples Europe, and a current lack of information on the strain(s) that might be circulating, make the estimation of the diagnostic sensitivity (DSe) of any test unfeasible for cervid samples, and preclude the development of alternative tests for use in European TSE surveillance in cervids. No direct comparison of test performance (i.e. parallel testing on the same panel of samples) can be made from the data available so there is no possibility to identify any differences between the two RT available on the market.
The generation of positive control material for European CWD strain/s, as recommended in both the 2004 and 2010 EFSA opinions (EFSA, 2004a,b; EFSA BIOHAZ Panel, 2010), for example, by experimental inoculation of a range of cervid species would be useful but is very difficult to perform, and would raise a number of practical and welfare issues. It would require the maintenance of experimentally infected individuals from non‐domesticated species in high biosafety facilities for a long period of time. In the absence of the specific pathogenesis data that such studies would provide and in the light of the results from the Norwegian surveillance, both brainstem and lymphoid tissue should be tested from each animal to improve sensitivity possible from collected material. The added sensitivity conferred by the testing of lymphoid tissues in addition to the brainstem is further corroborated by the experience from the testing conducted in Norway; three out of the eight positive reindeer were positive on lymphoid tissue only and five were positive in both brainstem and lymphoid tissue. Similarly, the Norwegian experience indicates that there was no detectable lymphoid involvement in the moose and red deer cases.
The tissue distribution of infectivity in some CWD‐infected cervids is now known to extend beyond the central nervous system and lymphoid tissues. While the removal of these specific tissues from the food chain, as recommended in the 2004 Opinion, would reduce human dietary exposure to infectivity, exclusion from the food chain of the whole carcass of any infected animal would be required to eliminate human dietary exposure.
The conclusions (1, 2, 4, 5, 6) and all recommendations (1, 2, 3, 4) of the 2010 EFSA opinion remain valid. Shortcomings in the 2006–2010 EU CWD survey design and subsequent implementation limited the reliability of inferences that could be made about the potential occurrence of CWD in Europe. Despite the lack of substantial surveillance in the EU since that time, cases of CWD have now been detected in wild Norwegian reindeer, moose and red deer, confirming the long‐held suspicion that at least some European cervid species are susceptible. Since the implementation of the 2006–2010 EU survey, testing activity has been low in Europe until the detection of CWD in Norway triggered a substantial testing effort in this country in 2016 and 2017. The surveillance programme proposed in the 2017 EFSA opinion supersedes the specifications of the EU‐wide survey that was implemented following the recommendations of the 2004 EFSA opinion.
Current available data do not preclude the possibility that CWD was present in Norway and perhaps elsewhere in Europe before the 2006–2010 EU CWD survey was conducted, whether in epidemic form or not. Comparing the point estimates of CWD prevalence among ‘adult’ (> 1 year old) reindeer harvested in Nordfjella 1 in 2016 (0.97%, 95% C.I.: 0.2–2.8%) and for the period 1 January–27 November 2017 (0.68%, 95% CI: 0.22–1.6%) to the epidemic curve for mule deer in investigated herds in the US, it appears plausible that CWD could have become established in Norway more than a decade ago.
Adhering to contemporary surveillance recommendations (EFSA BIOHAZ Panel, 2017a), especially with respect to focusing sampling on high‐risk individuals and developing a biologically meaningful spatial sampling framework relevant to the populations being monitored, with the aim of achieving set target sample sizes at the primary sampling unit level, should improve the reliability and value of data arising from renewed CWD surveillance efforts by some MS in coming years. The finding of the first case of CWD in red deer in Norway means that the surveillance scheme as in Reg. 999/2001, as amended, does not cover geographically all the MS in which red deer are present.
Further recommendations have been made, among them, the incorporation of sampling and testing for CWD into any wildlife health surveillance programmes, and the increase of awareness and dissemination of information about CWD in appropriate forums in the EU in order to improve the reporting of suspect cases. In addition, it is recommended to use only trained personnel for sample collection, and to avoid any test or detection method that uses antibodies for which the epitope is known to be polymorphic in cervids, unless successful binding in positive animals with those polymorphisms can be demonstrated. Residual samples, including relevant metadata, should be retained from all positive animals, and from as many tissues as possible, for isolate classification, future test evaluation, epidemiology or research purposes. Complementary studies should be conducted to identify any relevant differences influencing the epidemiology of the disease and to investigate the presence and frequency of potentially resistant alleles in the European cervid population. Finally, it is recommended to keep the performance of all currently applied tests, including those still being developed, under review and revise and update statutory testing protocols as new data become available.
1. Introduction
1.1. Background and Terms of Reference as provided by the requestor
The former Scientific Steering Committee (SSC)1 of the European Commission (EC) adopted on 6–7 March 2003 an opinion on chronic wasting disease (CWD) and tissues that might carry a risk for human and animal feed chains’. In summary, it highlighted that a risk of prion transmissions to humans consuming products of CWD‐affected cervids could not be excluded.
In its scientific opinion of 3 June 2004 on a surveillance programme for CWD in the European Union (EU) (EFSA, 2004b), the European Food Safety Authority (EFSA) stressed ‘a potential risk to consumers if a transmissible spongiform encephalopathy (TSE) would be present in European cervids’. EFSA further highlighted that ‘it might be prudent considering appropriate measures to reduce such a risk, e.g. excluding tissues such as central nervous system (CNS) and lymphoid tissues from the human food chain, which would greatly reduce any potential risk for consumers. However, it is stressed that currently, no data regarding a risk of TSE infections from cervid products for humans are available’.
In its 2011 scientific opinion on possible associations between TSE in animals and humans EFSA BIOHAZ Panel (2011) concluded regarding CWD that, although CWD agents have failed to induce disease in transgenic mice expressing human prion protein (PrP), experimental transmission to certain non‐human primate species has been reported. EFSA also mentioned ongoing experiments to assess the zoonotic potential of CWD strains in primate models.
The SSC Opinion of 6–7 March 2003 (SSC, 2003) also recommended the instigation of a surveillance programme for TSE in cervids in the EU. As a result, the EC asked EFSA for recommendations concerning such surveillance, and EFSA recommended in its opinion of June 2004 to initiate an EU‐wide experimental screening, targeting at‐risk groups of animals.
On that basis, a survey on CWD in the EU was launched by Commission Decision 2007/182/EC2 and implemented between 2007 and 2010. In this framework, more than 13,000 samples were collected from 21 Member States (MS) and Norway, mainly from red deer and white‐tailed deer (the survey also included 74 samples from reindeer), without any sample found positive to TSE. Therefore, EFSA concluded in 2010 that, while occurrences of cases of TSE in cervids in the EU could not be excluded, especially in remote and presently unsampled geographical areas, there was no cervid TSE epidemic in the EU.
In mid‐March 2016, a sick animal was observed during an exercise of identification and registration of wild reindeer (Rangifer tarandus) by the Norwegian Institute for Nature Research, in the locality of Laerdal. The animal subsequently died and its carcass was sent to the Norwegian Veterinary Institute for necropsy. The necropsy included testing for TSE. On 4 April 2016, the Norwegian National Reference Laboratory (NRL) for TSE confirmed the presence of TSE by enzyme‐linked immunosorbent assay (ELISA), western blot (WB) and immunohistochemistry (IHC). On 7 April 2016, the European Reference laboratory (EURL) for TSE confirmed that the samples received were strongly positive for TSE and were presumptive for CWD.
On 27 April 2016, the International Organization of Animal Health (OIE) reference laboratory for CWD in Canada (Canadian Food Inspection Agency (CFIA)) confirmed the CWD‐positive diagnosis, noting that the sample was consistent with CWD in farmed and wild cervids in Canada, and reindeer experimentally infected with CWD by the oral route.
On 25 May 2016, a second case of CWD was confirmed in Norway, this time in a wild moose (Alces alces) in the locality of Selbu in mid Norway. The moose was a 13 years old pregnant female that was euthanized due to abnormal behaviour. The animal was dehydrated, cachectic and had increased urination. It was found in the Selbu municipality. The Norwegian NRL for TSE performed ELISA, IHC and WB, which were all positive.
1.2. Terms of Reference
EFSA is requested by the European Commission to provide a scientific opinion on the following questions:
EFSA is asked to provide recommendations on surveillance of the cervid populations at the country level aimed at detecting CWD and/or estimating the prevalence of CWD in Norway, Sweden, Finland, Iceland, Estonia, Latvia and Poland, which are the EU and European Economic Area (EEA) countries with reindeer and/or moose populations, depending on the level of prevalence which is wished to be detected.
Has new evidence become available with regard to possible public health risks due to the occurrence of CWD in cervids since the publication of the 2011 joint EFSA/ECDC opinion? Does the natural exposure of consumers to cervid products originating from regions where CWD cases are detected represent a risk for public health?
EFSA is asked to recommend, if necessary, additional animal health risk‐based measures to prevent the introduction of CWD into the EU cervid populations and to prevent its spread within the EU?
Are the conclusions and recommendations in the EFSA opinion of June 2004 on diagnostic methods for CWD still valid? If not, an update should be provided.
EFSA is asked to update the conclusions of the 2010 EFSA opinion on the results of the EU survey on CWD in cervids, as regards the occurrence of CWD in the cervid population in the EU.
1.3. Interpretation of the Background and Terms of Reference
In the light of the sensitivity of this emerging issue, EFSA was asked by the European Commission to deliver its scientific opinion as soon as possible and according to the following schedule:
EFSA is asked to provide its scientific opinion on the Terms of Reference (ToR) No 1 (surveillance), 2 (public health) and 3 (risk mitigation measures) by 31 December 2016;
EFSA is asked to provide its scientific opinion on the Terms of Reference No 4 (diagnosis of CWD) and 5 (review of 2010 EFSA opinion) by 31 December 2017.
On 18 January 2017, EFSA published a scientific opinion on CWD in cervids addressing the Terms of Reference 1, 2 and 3 (EFSA BIOHAZ Panel, 2017a). The current scientific opinion provides scientific opinion on the Terms of Reference 4 and 5.
As described in the EFSA 2017 opinion (EFSA BIOHAZ Panel, 2017a), cervids belong to the taxonomic family Cervidae. The family Cervidae has two subfamilies: Cervinae and Capreolinae (or Odocoileinae). Since there may be confusion and misunderstanding regarding the common names of cervid species (e.g. ‘Eurasian elk’ is the moose in Europe (Alces alces alces) but ‘North American elk’ (wapiti; Cervus elaphus nelsoni) are more equivalent to European ‘red deer’), it is necessary to refer to their Latin names. The cervid species below are referenced by common names hereafter. Unless preceded by a descriptor (e.g. ‘red’, ‘mule’) or otherwise denoted, the term ‘deer’ refers generically to animals of North American species in the genus Odocoileus.
The following species or subspecies in Europe are referred to in this report:
-
Subfamily Capreolinae:
-
–
Eurasian tundra reindeer (Rangifer tarandus tarandus)
-
–
Finnish (Eurasian) forest reindeer (Rangifer tarandus fennicus)
-
–
Moose (or Eurasian/European elk) (Alces alces alces)
-
–
Roe deer (Capreolus capreolus)
-
–
White‐tailed deer (Odocoileus virginianus)
-
–
-
Subfamily Cervinae:
-
–
Red deer (Cervus elaphus)
-
–
Fallow deer (Dama dama).
-
–
For purposes of discussion in this opinion, the term ‘chronic wasting disease’ is used generically to include TSE – caused by prion infection – occurring naturally in any cervid species. Whether CWD in Norwegian moose, reindeer and red deer are caused by a common prion strain has not been determined, but early indications suggest that there are differences in the expression of disease in reindeer as compared to moose and red deer. Moreover, whether CWD cases in Norway or other European countries are caused by the same prion strain(s) that occur in North America has not been determined. The prion strain(s) associated with CWD in North America is/are infectious among susceptible hosts. Based on reported initial similarities, the current opinion assumes that the epidemiology of CWD in Norwegian reindeer will resemble that described for CWD in North American cervids.
Following the cases in 2016, Norway has expanded its surveillance of cervids for TSE in 2017. Norway's objective is to test those cervids found sick or that died but were not slaughtered for human consumption. In addition, the Norwegian authorities encourage hunters in the concerned regions to bring heads of animals killed during the hunting season to control points in view of TSE sampling and testing.
In 2016, Norway tested a total of 10,064 cervids (see Appendix A, Table A.1) and the total number of cases confirmed in 2016 was five: three in wild Eurasian tundra reindeer out of 842 tested (0.35%) and two in wild moose out of 4,403 tested (0.04%).
Table A.1.
Results of the TSE surveillance in cervids in Norway in the period 1 January 2015–27 November 2017
| Wild/semi‐domesticated | Captive/farmed | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Year | Wild Eurasiatundra reindeer | Semi‐domesticated Eurasian tundra reindeer | Finnish (Eurasian) forest reindeer | Moose | Roe deer | White‐tailed deer | Red deer | Fallow deer | Eurasian tundra reindeer | Finnish (Eurasian} forest reindeer) | Moose | Roe deer | White‐tailed deer | Red deer | Fallow deer | Other | Unknown |
| 2015 | 4 | 7 | 1 | 3 | |||||||||||||
| 2016 | 842 (3) | 1,738 | 4,403 (2) | 484 | 2,454 | 143 | 88 | ||||||||||
| 2017 a | 2,491 (5) | 6,982 | 5,252 (1) | 1,641 | 2,740 (1) | 389 | |||||||||||
1/1/17–27/11/17 only. Number of positive cases in brackets (Hopp, 2017). Empty cell means no testing.
Up to the 27 November, a total of 19,786 cervids had been tested in 2017, and the total number of cases seven: five in wild Eurasian tundra reindeer out of 2,491 tested (0.2%), one moose out of 5,252 tested (0.019%) and one red deer out of 2,740 tested wild red deer (0.036%).
At the time of writing this scientific opinion, (29 November 2017), a total of 12 cases of CWD have been confirmed in Norway: eight in wild reindeer, three in moose and one in red deer. A surveillance programme for the 2017–2018 hunting season is being implemented in Norway as well as the cull of the wild reindeer population in Nordfjella, where all cases of CWD in wild reindeer have been detected to date.
2. Data and methodologies
The data used in Section 3.4 have been sourced via a literature search, looking at new evidence on the diagnosis of CWD. Additional data have been extracted from scientific papers that were out of the scope of the search due to the specific subject.
Data on the performance of two rapid tests (RT) for the detection of CWD were provided by Bio‐Rad and IDEXX upon request. When presented in the opinion, these will be referred to as manufacturers’ data. Additional data have been retrieved from the scientific literature and documents provided by the manufacturers or from the outputs of the literature search.
According to Part I.A, Chapter B.I Annex III of Regulation (EC) 999/20013, the information to be presented by the MS in their annual report should include animals other than bovine, ovine and caprine, and the number of samples and confirmed TSE cases per species. Surveillance data in Europe in 2015 have been extracted from the above‐mentioned annual reports submitted by the MS and from the background information provided by the European Commission and included in the mandate. TSE surveillance data in cervids for 2016 have been extracted from the European Commission database as submitted by reporting countries (EU MS and Iceland, Norway and Switzerland). TSE surveillance data in cervids for the period 1 January 2017–27 November 2017 in Norway have been provided by the Norwegian Veterinary Institute (NVI), upon request.
Data from North America were used to address the spatial and temporal aspects of CWD dynamics relevant to interpreting European surveillance data. Historical (ca 2005 and 2010) and contemporary (ca 2017) maps of CWD distribution prepared by the US Geological Survey National Wildlife Health Center4 summarising historical CWD surveillance data (ca 2002–2012) provided by the US Department of Agriculture, Animal and Plant Health Inspection Service, Veterinary Services, were used to illustrate the spatio‐temporal distributions. Mule deer harvest surveillance data from Colorado, US (Miller et al., 2000; Miller and Conner, 2005; Conner et al., 2007a), and from Nordfjella 1 reindeer area in Norway as well as data from a published model (Miller et al., 2000) were used to illustrate points related to temporal considerations and potential time lags in ‘epidemic’ emergence.
2.1. Methodologies
2.1.1. Literature review
To retrieve data on the diagnosis of CWD, a literature search in the PubMed databases was undertaken. Neither the time of publication nor the language of publication was restricted. The following search string was used: (*deer* OR cervid* OR moose OR elk* OR wapiti* OR carib* OR muntjac*) AND (“chronic wasting disease” OR CWD OR TSE* OR spongiform encephalopa* OR prion* OR prp* OR PK) AND (diagnos* OR test* OR detect* OR assay* OR confirm* OR method* OR technique* OR generat* OR misfold* OR conversion OR “in vitro” OR “in‐vitro” OR “in vivo” OR “in‐vivo” OR amplification OR passage* OR cycl* OR substrate OR *quic OR “asa” OR pmca OR quaking OR western OR blot* OR WB OR elisa OR immuno* OR *histo* OR microscop* OR typ* OR strain* OR fibril* OR inocul* OR rapid OR ante‐mortem OR post‐mortem OR marker* OR replic*). These terms were searched in all types of publications.
A total of 434 references were retrieved and a double screening looking for potentially relevant references was conducted. Two pairs of reviewers conducted the screening: each pair independently screened 217 references. Discrepancies were discussed by the two pairs of reviewers until a final shortlist of references was agreed for each of the two lots. A subset of 120 relevant references was selected and considered in this assessment by reviewing the full papers. The references retrieved by the search conducted for the 2017 Opinion (EFSA BIOHAZ Panel, 2017a) were also double screened for any additional references that might be relevant to the ToR of the current opinion.
No formal evaluation of test performance equivalent to the existing EU requirements for cattle and sheep has been undertaken for cervid material, so it was agreed to carry out a qualitative evaluation for the diagnostics of CWD by means of literature review, the data provided by the manufacturers and the knowledge and expertise of the Working Group (WG) members. In addition to the shortlist of references as in the literature review, the experts in the WG selected other relevant references starting from scientific papers, including review papers, book chapters, non‐peer‐review papers known by the experts themselves or retrieved through non‐systematic searches, until the information of the subject was considered sufficient to undertake the assessment by the WG.
2.1.2. Examples of spatial and temporal CWD dynamics
Historical and contemporary maps of CWD distribution and historical surveillance data were used to demonstrate how the potential for patchy CWD distribution could complicate surveillance in a Europe‐sized geographic area. The 28 contiguous US states east of the Mississippi River that span ~ 2.5 M km2 were selected as a relevant example for illustrating CWD surveillance concepts and challenges in the context of ToR 5.
Harvest surveillance data from five Colorado mule deer herds collected over 15–21 years (Miller et al., 2000; Miller and Conner, 2005; Conner et al., 2007a) were graphed. Because outbreaks were asynchronous, prevalence of individual herd data segments were aligned by point times rather than by calendar year. Values were then averaged across each generic ‘year’ to generate a composite epidemic curve. Data from a published model (Miller et al., 2000) were also graphed for comparison. The point estimate of comparable harvest survey data from Nordfjella reindeer was also calculated to provide a temporal reference used in addressing ToR 5.
3. Assessment
3.1. Historical background of CWD diagnosis
Reports in the late 1960s and 1970s described CWD as a fatal wasting syndrome of mule deer (Odocoileus hemionus) and black‐tailed deer (Odocoileus hemionus columbianus) held in captivity in several wildlife facilities in northern Colorado and southern Wyoming (US) (Williams and Young, 1980). Classification of CWD as a TSE required histopathological examination of brains from diseased animals (Williams and Young, 1980). These and subsequent initial analyses (Williams and Young, 1993) were limited to microscopic evaluation of the CNS to detect neuropathological features typical of, but not necessarily exclusive to, prion diseases including neuronal vacuolation, and attendant spongiform degeneration of the neuropil, reactive astrocytic gliosis and florid amyloid plaques. Such lesions vary in intensity, anatomic location and with stage of disease. As such, diagnosis by histopathology may be ambiguous in naturally affected wild animals with early stage disease, and impossible if the integrity of brain tissue is compromised. The development of appropriate antibodies (Abs) capable of recognising PrP in formalin‐fixed, paraffin‐embedded sections after hydrated autoclaving was a valuable development in diagnostic and pathogenesis studies of CWD, and led to reports of disease‐specific PrP detection by IHC and WB in the brains and lymphoid tissues of CWD affected deer and wapiti.
In Europe, the initial diagnosis of TSE in all species was based on passive surveillance (i.e. clinical presentation) and assessment via conventional histopathology and subsequently IHC, both of which are time‐consuming, technically demanding, and expensive. Large‐scale active surveillance programmes for TSE in cattle and small ruminants were facilitated by the commercial development of rapid immunologically based screening tests (RT). These were originally developed for the detection of bovine spongiform encephalopathy (BSE) in cattle brain, and extensive formal test evaluation of a wide range of these RT was undertaken at EU level (EFSA, 2007b). It is important to note that BSE in cattle represented a single‐species population, with one epidemic agent strain, and a host population without relevant polymorphisms in the prion protein (PRNP) gene.
When surveillance programmes were extended to include small ruminants, the tests approved for use in cattle were initially applied without species‐specific evaluation or validation. Following the emergence of atypical scrapie in some countries, which were all using the same screening test, a specific evaluation of tests against ovine samples representing different strains resulted in the refining of the approved test lists (EFSA BIOHAZ Panel, 2009), and the delisting of some tests for sheep.
Similar evaluations have never been carried out for either caprine or cervid samples within Europe, and the tests used are those validated and approved for cattle and sheep. In a previous cervid surveillance exercise in the EU (EFSA BIOHAZ Panel, 2010), 15 different tests/test formats were used on brain samples from a range of species. All test results were negative and only diagnostic specificity (DSp) could be inferred from these data.
In North America, CWD diagnosis was similarly reliant initially upon clinical ascertainment and histopathological confirmation (Williams and Young, 1980, 1982, 1992; Spraker et al., 1997), but came to rely on IHC for large‐scale surveillance (Miller et al., 2000; Miller and Williams, 2002). As surveillance efforts intensified, the commercially available tests for BSE were increasingly applied to cervids, and often used lymphoid tissue rather than brain as the test substrate (Hibler et al., 2003). Full formal validation of these tests has never been undertaken for their use in cervids, or for testing lymphoid tissue, but some assessments have been conducted (see Section 3.3.5).
3.2. Designing a diagnostic protocol for surveillance
When designing a testing protocol for surveillance, there are many factors that can influence both the selection of samples and the range of tests applied to them, depending on the intended purpose of the surveillance (e.g. case or strain detection, prevalence estimates), that will determine the resulting case definition. Ideally, the best test/tissue combination for surveillance should be determined at a species level, also taking into account any knowledge on agent strain and host genotype variables within each species. Risk managers may then have to factor in the feasibility and costs of particular sampling and testing regimes, considering trade‐offs between tissues to be sampled and tested, sample size, selected tests, per‐sample collection and testing costs, and the impacts of all these factors on the detection probability. The schematic in Figure 1 summarises the relationships between each stage of the decision‐making process.
Figure 1.
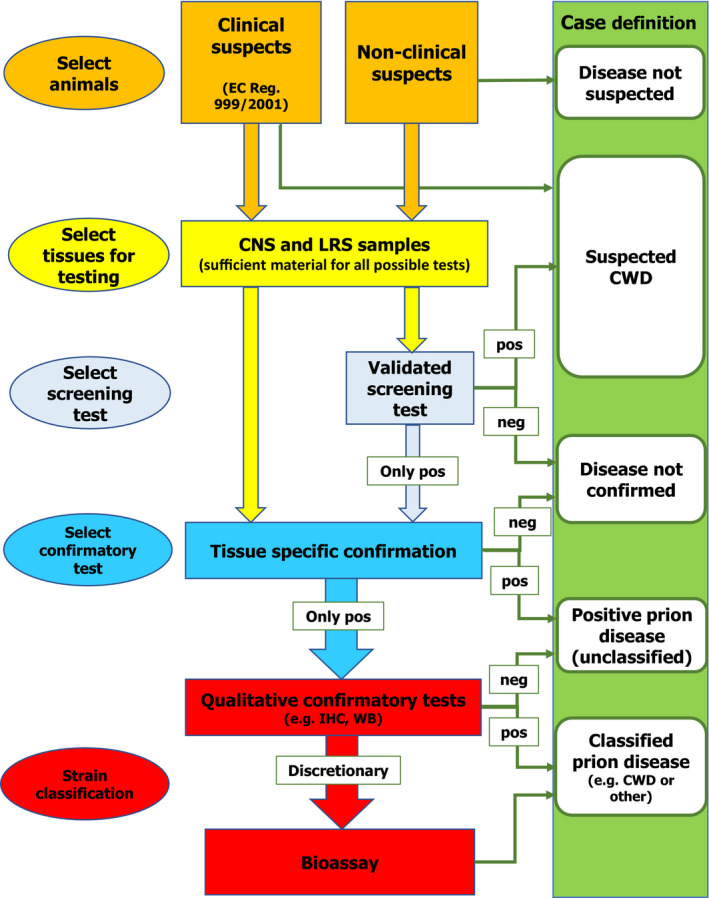
Flow chart of a proposed diagnostic protocol and case definition outcomes for TSE in cervids
- CNS: central nervous system; LRS: lymphoreticular system; IHC: immunohistochemistry; WB: western blot; CWD: chronic wasting disease; pos: positive; neg: negative.
3.2.1. Selection of animals
The importance of selecting the most relevant animals for a robust surveillance programme – able to detect CWD – should be emphasised and is discussed in detail in the 2017 EFSA Opinion (EFSA BIOHAZ Panel, 2017a). In summary, testing high‐risk animals (e.g. found dead, hunted or slaughtered animals considered not fit for human consumption, vehicle/predator kills and animals killed because they are sick or in poor body condition and not fit for human consumption) of any susceptible cervid species will maximise detection probabilities. Because there is no prior knowledge of differences in species susceptibility (with the exception of fallow deer which appear likely to have reduced natural susceptibility to CWD in North America), and multiple species coexist within most habitats, all species could contribute. However, a subset of these species may be selected or emphasised based on local or national risk assessments (EFSA BIOHAZ Panel, 2017a), or expand as new data on susceptibility emerges.
In addition, the OIE has affirmed that ‘surveillance of wildlife diseases must be considered equally as important as surveillance and control of diseases in domestic animals’ (OIE, 2017a). The OIE Terrestrial Animal Health Code defines wildlife health surveillance as ‘the systematic ongoing collection, collation and analysis of information related to wildlife health and the timely dissemination of information so that action can be taken’. Wildlife health (or disease) surveillance can be classified into general surveillance (also referred to as passive surveillance), i.e. the further investigation of animals found dead or ill, and targeted surveillance (also referred to as active surveillance), which implies testing of animals for specific pathogens (OIE, 2009). The OIE has developed an organisation of national focal points for wildlife in its member countries, a major component being the reinforcement and support to conduct national wildlife health programmes, which includes the testing of wild, feral and semi‐domesticated animals (OIE, 2010).
Every year, at least 18,000 wild animals, including mammals and birds, are examined within general surveillance programmes and over 50,000 wild animals are tested within various targeted surveillance programmes implemented at national level in Europe (Kuiken et al., 2011). The number of cervids examined in these surveillance programmes in Europe is not known. However, the specific testing for CWD in both surveillance categories in Europe has been limited even though testing for TSE in conjunction with monitoring programmes for cervids in EU Member States has been recommended (EFSA BIOHAZ Panel, 2010, 2017a), with the exception of the survey set up by the European Commission between 2006 and 2010 (EFSA BIOHAZ Panel, 2010). Furthermore, according to Regulation EC No 999/2001, the suspected presence of any TSE in any animal should be notified to the competent authority, which should take all appropriate measures, including the investigation for TSE, with no exception for the reporting and testing of suspected cases of CWD in farmed, semi‐domesticated and wild cervids.
Therefore, it is recommended that the specific sampling and testing for CWD are incorporated into the general wildlife surveillance programmes implemented at national level in Europe in addition to, but not in place of, the targeted surveillance previously proposed in the CWD opinion. The risk category of cervids found dead or ill gives the highest likelihood of detecting cases of CWD (Miller et al., 2000; Samuel et al., 2003; Walsh and Miller, 2010; EFSA BIOHAZ Panel, 2017a).
An example of the value of this approach was the detection of the first cases of CWD in Europe when visibly ‘diseased’ reindeer and moose were tested for CWD during general surveillance of wildlife animals in Norway in 2016 (Benestad et al., 2016). Targeted surveillance programmes that involve sampling cervids, such as those for tuberculosis, paratuberculosis and bluetongue, could also incorporate sampling and testing for CWD to augment, but not supplant, surveillance explicitly focused on CWD detection since animals showing clinical signs characteristic for other diseases may instead, or also, be CWD‐infected.
An additional and important component of wildlife health surveillance is the ‘timely dissemination of information so that action can be taken’. It is recommended to disseminate information about CWD in appropriate forums in the EU to increase awareness of CWD and improve the detection and reporting of suspect cases.
3.2.2. Selection of the tissues
TSE present very specific diagnostic challenges because of their strain variation, their very long incubation period and the lack of pathognomonic clinical signs. Moreover, the disease pathogenesis can vary both among and within species due to the influence of strain, host genotype, or a combination of the two.
Based on the knowledge currently available from experimental challenges of a range of species with TSE by a range of routes, early TSE infection is silent, and cannot be detected in any tissue by any test. Infectivity (as detected by rodent bioassay) precedes detectable accumulation of the abnormal form of PrP (PrPSc) using the existing in vitro detection methods, but whether this reflects true pathogenesis, or just the limitations of test sensitivity, is open to argument. It does however mean that with the currently available methods, animals in the very early stages of infection will not be detected in any surveillance programme. However, abnormal PrP can be detected in some tissues (from within a few weeks–months of first infection through to death (Sigurdson et al., 1999; Fox et al., 2006)) and thus the accepted diagnostic tests would be able to detect the majority of infected animals encountered in the course of routine surveillance provided the appropriate tissues are sampled and tested and especially where surveillance targets ‘high‐risk’ case material.
Experimental pathogenesis studies in sheep, goats, cattle and deer indicate that TSE is present with two main patterns of pathogenesis (Sisó et al., 2010). In the first, PrPSc accumulates in the peripheral lymphoid tissues where it is detectable from a few weeks post‐infection, although the lymphoid tissue with the earliest detection sensitivity can vary with route of challenge. Detectable neuroinvasion does not occur until several months post‐infection. This is the most common pattern seen in all four of the North American cervid species naturally infected with CWD. In the second pattern, lymphoid accumulation is either minimal (e.g. most cattle) or not detectable (e.g. sheep of certain genotypes; atypical forms of disease in sheep and cattle), so the first detectable PrPSc is in the brain following neuroinvasion. It has also been proposed, for atypical disease in cattle and small ruminants, that disease may arise from some form of spontaneous neurodegeneration (Baron et al., 2007), in which case the brain would be the first tissue to be affected.
Sampling protocols for large‐scale TSE surveillance of cattle and small ruminants in the EU have focussed on the brainstem as the tissue of choice. This has been driven by a number of factors. Before the development of immunodetection methods, the brain was the only tissue with microscopically visible lesions (spongiform change) in cattle clinically suspected of having BSE, and was subsequently shown to be positive consistently using immunodetection methods in both clinically affected and preclinical animals (Arnold et al., 2007). While some lymphoreticular involvement is detectable in preclinically affected cattle (Terry et al., 2003; Stack et al., 2011), this is neither a prominent nor a consistent feature, and would be a poor diagnostic target. In scrapie‐infected small ruminants, widespread accumulations of PrPSc in gut‐associated lymphoid tissues can be detected a few months after infection, preceding neuroinvasion by several months. This has been used to offer a live screening option using either tonsil or rectoanal mucosa‐associated lymphoid tissue (RAMALT) biopsies (Gonzalez et al., 2005; González et al., 2008). More critically, it has been shown that both host genotype and prion strain can substantially affect the extent to which the lymphoreticular system (LRS) is visibly involved, e.g. sheep carrying the ARR allele, or animals with atypical Nor98 scrapie (Benestad et al., 2003). As a consequence of this variability, a positive LRS result is very informative, while a negative result is less so. To date, CWD cases in Norwegian reindeer appear consistent with the early lymphoid propagation pattern (Benestad et al., 2016), whereas the CWD cases in Norwegian moose and red deer showed no lymphoid involvement as analysed with currently available diagnostic methods (Benestad, 2017a).
This variation and the limited experience thus far in Europe mean that there is no tissue that can be proposed as the most sensitive for surveillance in all circumstances. Sampling only brainstem could reduce the DSe for strain–host combinations that are characterised by early lymphoid PrPSc accumulation. Indeed, analyses of a large number of both retropharyngeal lymph node (RPLN) and obex samples of CWD‐infected white‐tailed deer (WTD) have shown that a substantial proportion of animals (20.4%) had deposits of PrPSc only in lymphoid tissue (Keane et al., 2008a). Similar patterns were described in mule deer (Miller and Williams, 2002; Hibler et al., 2003). Alternatively, sampling only lymphoid tissue could also reduce DSe, e.g. screening of captive wapiti suggests 7–12% of infected individuals may have detectable PrPSc in brain tissue only (Spraker et al., 2004; Haley et al., 2016b) or, in cases like the Norwegian moose and red deer, could preclude diagnosis altogether.
Knowledge of the pathogenesis and the related distribution of tissue infectivity from experimental studies should drive a fit‐for‐purpose selection of tissues for sampling. Despite the current lack of such studies in Europe, the Norwegian outbreak is providing helpful information, e.g. in that abnormal PrP was detectable in the lymphoid tissues of all confirmed Norwegian reindeer cases, even in those cases (37.5%) in which neuroinvasion had not yet occurred. In the moose cases, as mentioned in the recent EFSA Opinion (EFSA BIOHAZ Panel, 2017a), the lymph nodes (available from only one of the first two affected animals) were negative. The analysis of the third positive moose, and the one case identified in red deer, confirmed the absence of detectable PrPSc in the RPLN. This differs from natural cases in hunted moose from Colorado where both obex and a RPLN showed the accumulation of PrPCWD (Baeten et al., 2007). This indicates that in order to maximise the sensitivity of surveillance there is a need to target and test both obex and RPLN. This would also optimise the monitoring of EU species other than reindeer, moose or red deer for which no experimental or field data currently exists. Therefore, while the results from pathogenesis studies would be of great help, the lack of such data does not prevent the design and the implementation of effective large‐scale surveillance programmes. In North America, much of the surveillance screening focusses on lymphoid tissue as the primary tissue for testing. However, it is acknowledged that, as is the case for small ruminants, there may be affected animals that do not have widespread lymphoid involvement and will therefore be missed by the surveillance screening (Spraker et al., 2004; Haley et al., 2016a). This in turn might lead to an incomplete understanding of the strain variation present in wild populations.
Consequently, both lymphoid tissue (preferably the RPLN, which is consistently affected early in the disease or the tonsil) and brainstem at the level of the obex should be tested to maximise the DSe in any surveillance programme. Trained personnel should be used to ensure the accurate sampling of the target tissues. However, collection of samples from (wild) cervids in real life is not always made under optimal conditions. Field experience in Norway and North America shows that samples collected by trained personnel result in fewer samples of low quality. Experience is especially critical when the tissues are autolysed, which can make it difficult or impossible to be sure of the precise neuroanatomy of brainstem samples, or even to identify specific lymph nodes reliably.
3.2.3. Selection of diagnostic tests
The interlinked effects of age, sex, genotype and disease progression on the DSe of different tests has been evaluated on cervid material, but not comprehensively, and it is not possible to individually assess the impact of each combination on test performance from the data available. All the current diagnostic tests for CWD rely on the immunodetection of prion protein in tissues, tissue homogenates/extracts or body fluids, either directly or following some amplification step.
The PrP is a normal host protein, highly conserved across mammalian species, so it is difficult to raise antibodies against, and no antibodies are specific for the disease‐associated isoforms of the protein (PrPSc) or for the host‐encoded prion protein (PrPC) of any one species. However, over the years a number of monoclonal antibodies (mAb) have been produced against various linear or conformational epitopes using PrPSc from a variety of host species and recombinant protein fragments. Not every antibody recognises every species/strain combination, so again this might affect the performance of individual tests, depending on whether the antibodies used are able to detect the host/strain isoform under investigation. Positive controls from field samples should be used whenever possible, but these may not exist (if an uncharacterised population is being screened, for example, the EU cervid population), or may not be available in the quantity required. Moreover, kit positive samples tend to contain recombinant protein, which does not control for sample extraction/preparation steps, or autolysis. It also means that the interpretation of test results from an ‘unknown’ or ‘undefined’ population need to be approached with caution. Polymorphisms in the prion protein gene (PRNP) occur in species known to be affected by TSE, but in general the codons that are polymorphic are different for each species (Table 2) as detailed in the 2017 EFSA opinion (EFSA BIOHAZ Panel, 2017a). Particular care should be taken with test interpretation if a polymorphism occurs within the epitope relevant to a test mAb. For example, CWD‐infected mule deer homozygous for phenylalanine at PRNP codon 225 (225FF) showed relatively poor staining when immunolabelled using mAb F99/97.6.1 (Wolfe et al., 2014), an antibody widely used for detection of CWD that has a binding epitope that aligns with PRNP codons 220–225 (Dassanayake et al., 2013; Madsen‐Bouterse et al., 2015).
Table 2.
Variability of genotypes in selected codons of the PRNP identified in investigated cervids of Europe
| PRNP type | 59 | 98 | 109 | 168 | 176 | 209 | 225 | 226 | Citation | |
|---|---|---|---|---|---|---|---|---|---|---|
| Consensus amino acid sequence | No. animals | G | T | K | P | N | M | S | Q | |
| Red deer | 323a |
G/G (322) G/S (1) |
T/T(279) T/A (38) A/A(2) |
P/P (322) P/S (1) |
Q/Q (123) Q/E(112) E/E(88) |
Peletto et al. (2009) | ||||
| Fallow deer | 11 | K/K | N/N | M/M | S/S | E/E | Wik et al. (2012) | |||
| European Moose | 15 |
K/K (6) K/Q (6) Q/Q (3) |
N/N | S/S | Q/Q | Wik et al. (2012) | ||||
| Roe deer | 11 | K/K | N/N | M/M | S/S | Q/Q | Wik et al. (2012) | |||
| Reindeer | 9 | K/K |
N/N (8) D/D (1) |
M/M |
S/S (5) S/Y(2)Y/Y(2) |
Q/Q | Wik et al. (2012) |
Numbers of animals for each specific change in the consensus amino acid sequence are reported in brackets.
There has been no systematic evaluation of antibody binding capacity in relation to cervid genotype. While it is not possible to assess the analytical sensitivity (ASe) of the currently available RT against known positive examples of each genotype (assuming that all genotypes are susceptible), with the available knowledge provided by the manufacturers, there is no reason to conclude that known test/genotype combinations might adversely affect test performance. The Abs used to investigate the Norwegian CWD cases in reindeer, moose and red deer and their binding epitopes in the PrP protein are displayed in Figure 2. A number of Abs have successfully detected CWD in cervids of different species and genotypes in the world, within various immunodetection test formats. These are listed in Table 1.
Figure 2.
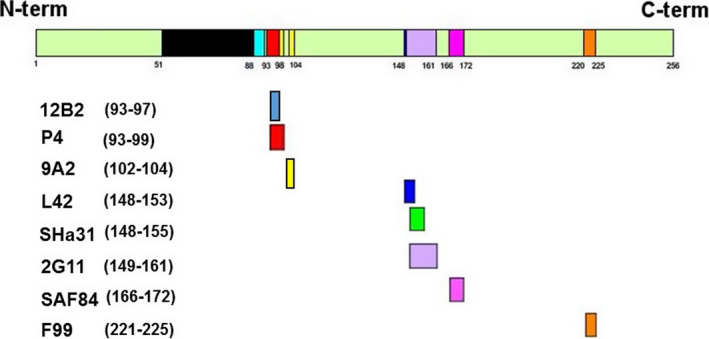
Antibodies used to investigate the Norwegian CWD cases in reindeer, moose and red deer and their binding epitopes in the PrP protein (Benestad, 2017b)
Table 1.
Test characteristics in experimental challenges and detected natural cases of TSE in cervids
| Test format | Antibody/test | Species | Genotype (when reported) | References |
|---|---|---|---|---|
| ELISA | Bio‐Rad (proprietary) | Elk/Wapiti | Hibler et al. (2003) | |
| Bio‐Rad | Moose | 209MM | Baeten et al. (2007) | |
| Bio‐Rad | Mule deer | 225SF, 225SS, 225FF | Hibler et al. (2003), Jewell et al. (2005), Wolfe et al. (2007), Race et al. (2009a), Wolfe et al. (2014) | |
| Bio‐Rad | Red deer | 226E/Q, 132MM, 96GG | Balachandran et al. (2010) | |
| Bio‐Rad | Reindeer | 2VV, 129GG, 138SS, 169VV | Mitchell et al. (2012), Benestad et al. (2016) | |
| Bio‐Rad | White‐tailed deer (WTD) | 96GG, GS | Hibler et al. (2003), Wolfe et al. (2007), Keane et al. (2008b), Masujin et al. (2007), Thomsen et al. (2012) | |
| IDEXX (proprietary) | Elk/Wapiti | 132MM 132 ML | Yang et al. (2011) | |
| IDEXX | WTD | 96GG | Keane et al. (2008b) | |
| IDEXX | Mule deer | Miller (2017a) | ||
| IDEXX | Moose | Miller (2017a) | ||
| Fujirebio | WTD | Masujin et al. (2007) | ||
| IHC | BAR224 | Red deer | 132MM | Dagleish et al. (2008)a, Martin et al. (2009)a, Dagleish et al. (2015) |
| BAR24 | Muntjac | 95QQ, 96GG, 98SS,132MM, 225SS | Elder et al. (2013), Nalls et al. (2013) | |
| BAR24 | WTD | 96GG, 96GS | Mathiason et al. (2009), Haley et al. (2012), Elder et al. (2013) | |
| B103 | WTD | Masujin et al. (2007) | ||
| F89 | Mule deer | Spraker et al. (2002)e, Baszler et al. (2006) | ||
| F89 | Norwegian moose | Benestad (2017c) | ||
| Pirisinu et al. (2017) | ||||
| F89 | WTD | Baszler et al. (2006) | ||
| F89/99 cocktail | Elk/Wapiti | 132MM, 132ML | Hamir et al. (2003)b, Hamir et al. (2004)b, Huang et al. (2005), Hamir et al. (2006) | |
| F89/2G11 cocktail | Norwegian reindeer | Benestad et al. (2016) | ||
| F99 | Elk/Wapiti | 132MM, 132 ML | Peters et al. (2000), Hibler et al. (2003), Hamir et al. (2004)b, Spraker et al. (2006, 2009), Yang et al. (2011), Nichols et al. (2012), Monello et al. (2013), Selariu et al. (2015), Wyckoff et al. (2015), Haley et al. (2016b) | |
| F99 | Moose | 209MM | Baeten et al. (2007) | |
| F99 | Mule deer | 225SS, 225SF | Spraker et al. (2002), Sigurdson et al. (2002), Wild et al. (2002), Miller and Williams (2002), Hibler et al. (2003), Jewell et al. (2005), Baszler et al. (2006), Fox et al. (2006), Race et al. (2009a), Wolfe et al. (2007) | |
| F99 | Norwegian moose | Benestad (2017c) | ||
| F99 | Norwegian red deer | Benestad (2017c) | ||
| F99 | Norwegian reindeer | Benestad (2017c) | ||
| F99 | Red deer | 226E/Q, 132MM, 96GG | Dagleish et al. (2008)a, Balachandran et al. (2010), Dagleish et al. (2015) | |
| F99 | Reindeer | 2VV, 129GG, 138 SS, 169VV | Mitchell et al. (2012) | |
| F99 | WTD | 96GG, 96GS, 96SS | Wild et al. (2002), Hibler et al. (2003), Baszler et al. (2006), Wolfe et al. (2007), Keane et al. (2008a,b, 2009), Greenlee et al. (2011)b, Thomsen et al. (2012), Henderson et al. (2013) | |
| F99 | WTD | 95Q/H, 96G/S, 116A, 226Q/K | Haley et al. (2016a) | |
| F99 | 225FF | Wolfe et al. (2014)d | ||
| L42 | Norwegian moose | Benestad (2017c)c | ||
| L42 | Norwegian red deer | Benestad (2017c)c | ||
| L42 | Norwegian reindeer | 2VV, 129GG, 138SS, 169VV | Benestad et al. (2016) | |
| P4 | Elk/Wapiti | Wyckoff et al. (2015) | ||
| P4 | Norwegian moose | Benestad (2017c) | ||
| P4 | Norwegian red deer | Benestad (2017c) | ||
| P4 | Norwegian reindeer | Benestad (2017c) | ||
| R145 | Norwegian reindeer | 2VV, 129GG, 138SS, 169VV | Benestad (2017c) | |
| Sha31 | Norwegian reindeer | 2VV, 129GG, 138SS, 169VV | Benestad et al. (2016), Pirisinu et al. (2017) | |
| Sha31 | Norwegian moose | Benestad (2017c) | ||
| T2 | WTD | Masujin et al. (2007) | ||
| 12B2 | Red deer | Martin et al. (2009)a | ||
| 12B2 | Norwegian moose | Benestad (2017c)c | ||
| 12B2 | Norwegian red deer | Benestad (2017c)c | ||
| 12B2 | Norwegian reindeer | 2VV, 129GG, 138SS, 169VV | Benestad et al. (2016) | |
| 2G11 | Norwegian moose | Benestad (2017c) | ||
| 2G11 | Norwegian red deer | Benestad (2017c) | ||
| 2G11 | Norwegian reindeer | Benestad (2017c) | ||
| 44B1 | WTD | Masujin et al. (2007) | ||
| 6H4 | Elk/wapiti | Angers et al. (2009) | ||
| 6H4 | Mule deer | Sigurdson et al. (2002) | ||
| 9A2 | Norwegian moose | Benestad (2017c)c | ||
| 9A2 | Norwegian red deer | Benestad (2017c)c | ||
| 9A2 | Norwegian reindeer | Benestad (2017c) | ||
| WB | Bio‐Rad Western | Red deer | 226E/Q, 132 MM, 96GG | Balachandran et al. (2010) |
| Bio‐Rad Western | Reindeer | 2VV, 129GG, 138SS, 169VV | Mitchell et al. (2012), Benestad et al. (2016) | |
| Bio‐Rad Western | WTD | 96GG, 96GS | Thomsen et al. (2012) | |
| B103 | WTD | Masujin et al. (2007) | ||
| BAR221 | WTD | 96GS | Henderson et al. (2015b) | |
| BAR224 | WTD | 96GG, 96GS | Mathiason et al. (2009), Haley et al. (2011), Henderson et al. (2013) | |
| F99 | Elk/Wapiti | Davidowitz et al. (2005), Huang et al. (2005) | ||
| F99 | Elk/Wapiti | 132MM, 132 ML, 132 LL | O'rourke et al. (2007) | |
| F99 | Red deer | Martin et al. (2009)a | ||
| ICSM18 | Elk/Wapiti | 132MM, 132 ML | Yang et al. (2011) | |
| ICSM18 | WTD | Daus et al. (2011) | ||
| L42 | Deer | Race et al. (2007) | ||
| L42 | Elk/Wapiti | Race et al. (2007) | ||
| L42 | Mule deer | Race et al. (2009a) | ||
| L42 | Norwegian moose | Benestad (2017c), Pirisinu et al. (2017) | ||
| L42 | Red deer | 132MM | Dagleish et al. (2015)a | |
| L42 | Norwegian reindeer | 2VV, 129GG, 138SS, 169VV | Benestad et al. (2016), Pirisinu et al. (2017) | |
| Prionics Check | Elk/Wapiti | Hamir et al. (2003)b | ||
| P4 | Elk/Wapiti | 132MM, 132ML | O'rourke et al. (2007) | |
| P4 | Norwegian moose | Benestad (2017c)c | ||
| P4 | Norwegian red deer | Benestad (2017c)c | ||
| P4 | Norwegian reindeer | 2VV, 129GG, 138SS, 169VV | Benestad (2017c), Pirisinu et al. (2017) | |
| P4 | Red deer | Martin et al. (2009)a, Dagleish et al. (2015)a / f | ||
| R35 | Deer | Race et al. (2007) | ||
| R35 | Elk/Wapiti | Race et al. (2007) | ||
| SAF70 | Elk/Wapiti | Davidowitz et al. (2005) | ||
| SAF84 | Norwegian moose | Benestad (2017c), Pirisinu et al. (2017) | ||
| SAF84 | Norwegian reindeer | 2VV, 129GG, 138SS, 169VV | Benestad et al. (2016), Pirisinu et al. (2017) | |
| Sha31 | Norwegian moose | Benestad (2017c) | ||
| Sha31 | Red deer | Martin et al. (2009)a | ||
| Sha31 | Norwegian reindeer | 2VV, 129GG, 138SS, 169VV | Benestad et al. (2016), Pirisinu et al. (2017) | |
| T2 | WTD | Masujin et al. (2007) | ||
| 11F12 | WTD | Rubenstein et al. (2010, 2011), Chang et al. (2008) | ||
| 12B2 | Norwegian moose | Benestad (2017c)c, Pirisinu et al. (2017) | ||
| 12B2 | Norwegian red deer | Benestad (2017c)c | ||
| 12B2 | Norwegian reindeer | Benestad (2017c), Pirisinu et al. (2017) | ||
| 12B2 | Red deer | Martin et al. (2009)a | ||
| 2A11 | ‘Deer’ | Brun et al. (2004) | ||
| 44B1 | WTD | Masujin et al. (2007) | ||
| 5D6 | WTD | Rubenstein et al. (2010, 2011) | ||
| 5D6 | WTD | Chang et al. (2008) | ||
| 6H4 | Elk/Wapiti | Davidowitz et al. (2005), Angers et al. (2009) | ||
| 6H4 | Mule deer | Race et al. (2009a) | ||
| 6H4 | Red deer | Martin et al. (2009)a | ||
| 6H4 | WTD | 96GG, 96GS, 96SS | Greenlee et al. (2011)b | |
| 8E9 | WTD | Chang et al. (2008), Rubenstein et al. (2010, 2011) | ||
| 8G8 | Elk/Wapiti | 132MM, 132 ML, 132 LL | O'rourke et al. (2007) | |
| 9A2 | Norwegian moose | Benestad (2017c)c | ||
| 9A2 | Norwegian red deer | Benestad (2017c)c | ||
| 9A2 | Norwegian reindeer | Benestad (2017c)c |
BSE source of infection.
Scrapie source of infection.
Poor or no signal.
Poor signal in this genotype.
Specificity using LRS questionable.
No signal with BSE.
The impact of genotype on DSe has been studied for WTD with a common polymorphism (glycine (G) or serine (S)) at codon 96 PRNP, in which prion genotype was strongly linked to the temporal progression of prion accumulation in the obex, and hence the ability to detect it (Keane et al., 2008b). Thomsen et al. (2012) found that the DSe of IHC in rectal biopsy samples of WTD was also dependent on disease progression, linked to the genotype at codon 96. DSe was 76% (95% CL: 49–91%) for WTD that were homozygous for the G polymorphism (96GG), but only 42% (95% CL: 13–79%) for WTD that were heterozygous (96GS). As might be predicted from knowledge of disease pathogenesis, DSe was only 36% for deer in the earliest stage of disease but was 100% for deer in the last stages of preclinical disease. The authors applied the Bio‐Rad TeSeE™ ELISA to all samples either in parallel with IHC and/or WB, or in series, but no data were shown on the performance of the RT. Similar findings were reported by Keane et al. (2008b). Differences have been observed in the detection of cases using serial protein misfolding cyclic amplification assay (sPMCA) and IHC in 96GG compared with 96GS WTD (Wolfe et al., 2007; Haley et al., 2012). The PRNP of wapiti is polymorphic at codon 132, encoding either methionine (M) or leucine (L). The 132ML polymorphism in wapiti also influences the DSe of tests when applied to peripheral lymphoid tissues (Haley et al., 2016b).
The effect of species variation on the detection of PrPSc also has been studied, but this may be a reflection of the different polymorphisms in the different species. For example, the accumulation of PrPres in tonsils and RPLN was shown to be greater in deer than in wapiti using immunoblotting (Race et al., 2007), but these data do not indicate whether the variation in levels of detected PrPres was due to the number of follicles affected, or a difference in the amount of PrPres in single follicles.
Very few studies have addressed the genetic diversity of PRNP in cervid populations of Europe. It appears that the extent of genetic diversity is linked to the cervid species, with reindeer and red deer being more diverse than others. However, the limited number of animals and geographical sources studied to date precludes any conclusion on the presence and frequency of polymorphic alleles in PRNP of European cervids. Table 2 shows the available data on genotypes of selected codons identified in PRNP of cervid species in Europe. Consequently, the recommendations of the 2017 EFSA Opinion on CWD (EFSA BIOHAZ Panel, 2017a) remain valid. Genotyping all cervids tested positive by surveillance and a representative subset of cervids tested negative by surveillance would help generate information on the PRNP gene in European cervid populations. The collation of these data also would help inform on the probable susceptibility or resistance of these species to CWD.
3.3. Validation of diagnostic tests
Validation is a procedure that determines the fitness of a diagnostic test which has been developed, optimised, and standardised for a defined intended purpose. Validation comprises estimates of the analytical and diagnostic performance characteristics of a test. The international guidelines for validation of diagnostic tests for infectious diseases in animals, including for specific application in wildlife populations, are described in the OIE Terrestrial Manual, chapter 1.1.6. (OIE, 2017b), and summarised in Appendix B. The tests should be validated for the species in which they will be applied. Diagnostic tests can be classified into direct and indirect identification methods. Direct diagnostic techniques detect/identify the presence of pathogens while indirect methods detect immunological cellular or antibody response of the animal host.
The purpose of the test needs to be defined at early stages of test development. The purpose may be, for example, screening populations for the occurrence of infections, or confirming a positive result of a screening test. It is important to appreciate how this pre‐determined purpose affects case definition, and to exercise care in the comparison of data gathered over a long period of time, or with different tests, where case definitions may be different. A particular test may be validated for one or more intended purposes by optimising its performance characteristics for each purpose. Some tests may detect the presence of infection/disease; others may also provide qualitative data that contribute to disease characterisation and subsequent classification.
3.3.1. Previous evaluations of Rapid Tests for TSE in the EU
The nature of the prion agent is a confounding factor for test validation. Although the abnormal disease‐associated isoform of the PrP (PrPSc) of the host‐encoded prion protein PrP is widely accepted as the causal agent of prion diseases, it cannot be purified and used in assays as a ‘gold standard’. Consequently, a conventional ‘gold standard’ is not available. PrPSc also occurs as different isoforms, depending on variables such as the species and genotype of the host, and these different isoforms are thought to be the underlying mechanism for agent strain variation. This means that different ‘gold standards’ would be necessary for each combination, and, in the absence of such specific standards, extrapolation from one situation to another should be cautious.
In addition to the wealth of field data on test performance that has been built up over years of testing, the methods used for surveillance for TSE in cattle, sheep and goats in the EU have been subject to detailed laboratory trials and evaluations (see below) and ongoing scrutiny of field performance through annual proficiency testing exercises and related troubleshooting coordinated by the EURL through the EU MS NRL network. In addition, any changes to test formats made by test manufacturers have to be approved by the EURL prior to the release of amended kits.5
3.3.2. Initial test evaluation for BSE detection in cattle
The European Commission carried out a first evaluation of rapid post‐mortem BSE tests in 1999. Four tests were evaluated on brain tissue from clinical BSE cases. Three of these tests, including Bio‐Rad TeSeE™, performed satisfactorily and were later approved pursuant to Regulation (EC) No 999/2001 (European Commission, 1999).
The SSC opinion (2002a), aligning with OIE guidance, required that
Estimation of sensitivity relative to approved tests (confirmatory) using 200 known positive samples should be tested by a new rapid test (ensuring with a 95% probability that the sensitivity of the new rapid test is not below 98.5% compared with the approved test).
Estimation of specificity relative to approved tests performed using 10,000 consecutive samples from healthy slaughtered animals that tested negative using an approved test.
In 2003, an opinion of the SSC published the results of the field trial evaluation of two new rapid BSE post‐mortem tests and the following year EFSA published the EFSA Scientific Report on the Evaluation of Seven New Rapid post‐mortem BSE Tests, which included the IDEXX HerdChek BSE (EFSA, 2004b). In 2005, EFSA conducted an additional evaluation of two rapid post‐mortem BSE tests for which the field trial evaluation had not been completed. Other tests were submitted for assessment, but failed to pass the full evaluation and were not approved for statutory testing use (EFSA, 2005a). These evaluations were divided into three phases, and a test could only progress to the next phase if the previous one was completed satisfactorily, as follows:
-
1
Assessment of the application dossier (pre‐existing data from the test developer/manufacturer)
-
2
Laboratory evaluation:
DSe was assessed using 50 known positive samples (supplied frozen). Testing done under NRL supervision.
DSp was evaluated by testing 150 samples of known negative tissue (supplied frozen).
Detection limit was assessed using both positive tissue (diluted at the test site in test specific buffer) and freshly prepared homogenate dilutions of 1:5, 1:50, 1:100 and 1:200. Aliquots of each dilution were blind coded by Commission staff present on site, and were tested in duplicate on at least three different plates. This also enabled assessment of repeatability.
-
3
Field trial:
Each new test was compared to at least two reference tests for assessment of sensitivity and specificity. The maximum proportion analysed by a single reference test was ≤ 70%.
DSe was assessed relative to reference tests using 200 true positive samples that should have been well documented (origin and age of the sample, e.g. subpopulation; condition of the sample, e.g. autolysis; brain region used; storage conditions; duration of storage).
DSp was assessed using 10,000 consecutive samples from healthy slaughtered cattle that were tested negative using a reference test, and on 200 poor quality negative samples. Testing was performed with the agreement of the NRL in experienced high throughput routine laboratories, using at least two batches of test kits.
Sensitivity was assessed against pre‐existing approved rapid test performance. Increasingly these assessments included a number of autolysed samples to more accurately reflect field conditions, and also included dilution series, to explore detection limits/ASe (of increasing importance as tests were being applied to healthy slaughter populations, and amounts of PrPSc were smaller).
The field trial phase also enabled the assessment of robustness, applicability in different laboratories and the expected rate of false initial results under high‐throughput field conditions. It also provided the opportunity to adjust cut‐off values (if required) before market introduction.
One EFSA Scientific Report (EFSA, 2006) also included small subsets of negative samples to look at whether differences could be detected in animals that were clinically suspected of BSE, but not confirmed post‐mortem, and also animals for which an alternative neurological diagnosis was obtained.
Data from these evaluations were considered robust across field applications given the relative homogeneity of European cattle populations, and the fact that BSE epidemic involved a single agent strain.
3.3.3. Initial test evaluation for TSE diagnosis in small ruminants
Until 2004, no evaluation of RT for the detection of TSE on material from small ruminants had been conducted by the European Commission, although five post‐mortem BSE RT were provisionally approved by the European Commission for the TSE monitoring of small ruminants, in accordance with the TSE Regulation (EC) No 999/2001, by extrapolation from the bovine validation exercises.
The need to validate the available RT for the detection of TSE in small ruminants initiated a series of similar exercises to those conducted in the previous years for bovine testing. Overall, 12 tests were accepted for evaluation, including the Bio‐Rad TeSeE™, Bio‐Rad TeSeE™ sheep/goat and the IDEXX Herdchek (EFSA, 2005b). In addition to assessing the diagnostic capabilities of these tests in relation to a panel of 240 positive classical scrapie cases, their ability to detect ‘atypical’ scrapie (Nor 98) in sheep samples (n = 3) was evaluated. Six RT successfully completed the laboratory evaluation and were re‐evaluated against dilutions of brain homogenates from sheep experimentally infected with BSE (n = 3), in order to provide ASe for this material comparable to that previously obtained for scrapie. All tests performed satisfactorily in terms of DSe when applied to brainstem from clinical, confirmed cases of classical scrapie and BSE in sheep. Additional scrapie‐specific test formats were also subject to formal evaluation, using 250 positives and more than 1,000 negatives (EFSA BIOHAZ Panel, 2012).
No goat samples were included in these evaluations. On the basis of the limited scientific knowledge at the time it was recommended that, in terms of testing, goats should be considered equivalent to sheep. Subsequent work on PRNP polymorphisms in goats and their effects on mAb binding (Madsen‐Bouterse et al., 2015) illustrated the importance of understanding genetic variation and its potential influence on immunodiagnostics in each host species targeted for TSE screening.
Given the wider tissue distribution of PrPSc in many classical scrapie cases, one of these evaluations (EFSA, 2005c) also included a preliminary assessment of test sensitivity and specificity when applied to lymphoid tissue.
3.3.4. Additional evaluations of the analytical sensitivity of approved tests for BSE and scrapie
In 2009, EFSA conducted a scientific evaluation of the report prepared by the Community Reference Laboratory (CRL) for TSE in 2008 that assessed the ASe of all the approved TSE RT against the same sample sets for the three main types of ruminant TSE (BSE, classical scrapie and atypical scrapie), following the EFSA protocol for the evaluation of rapid post‐mortem tests to detect TSE in small ruminants (EFSA, 2007a). Due to the inability to meet requirements for all types of TSE agents on known positive samples, some tests were not recommended for use for TSE monitoring in small ruminants (EFSA BIOHAZ Panel, 2009).
3.3.5. EU Test evaluation for TSE in cervids
No test has been evaluated across a panel of samples comprehensively representing the range of species, genotypes, strains and tissue types that might be expected from our knowledge of these diseases. Similarly, a single panel of samples has not been presented to a panel of all potentially available tests. So, no direct comparison of test performance across all prospective diagnostic approaches can be made from the data available, although there are limited data on comparisons of different protocols/tests from individual manufacturers.
The detection of disease‐specific PrPSc in a range of tissues from a range of animals representing species and genotype using a test gives an indication that that test can be used for disease detection (i.e. epitopes for the antibodies used in any particular test format are present, and accessible for binding). If such test positives are confirmed by an independent method (usually one of the confirmatory methods, namely, IHC or WB), then this gives assurance that the test is specific in that host/tissue substrate context, but it does not address test sensitivity, or enable confident extrapolation to other host contexts. Such qualitative data can be drawn from a wide range of experimental and field studies, and relate to many different tissue/sample types (See Table C.1 in Appendix C).
Table C.1.
Summary data on performance of RT for the detection of CWD extracted from the scientific literature
| Reported status | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive samples | Negative samples | ||||||||||
| Test | Species | Tissue | IHC +ve | Presumed negativea | Total | Test result | Se % | 95 C.I. % | Sp % | 95 C.I. % | Reference |
| Bio‐Rad TeSeE WB | Wapiti, mule deer, WTD | Obex | 35 | 0 | 35 | Positive | 87.5% | 73.8–94.5% | 100% | 61–100% | Blasche et al. (2011) |
| 5 | 6 | 11 | Negative | ||||||||
| 40 | 6 | 46 | Total | ||||||||
| Bio‐Rad TeSeE ELISA | Wapiti, mule deer, WTD | Obex | 29 | 0 | 29 | Positive | 72.5% | 57.1–83.9% | 100% | 98.8–100% | Blasche et al. (2011) |
| 11 | 320 | 331 | Negative | ||||||||
| 40 | 320 | 360 | Total | ||||||||
| Bio‐Rad TeSeE s/g ELISA | Wapiti, mule deer, WTD | obex | 34 | 87 | 121 | Positive | 85% | 70.9–92.9% | 91.7% | 89.8–93.2% | Blasche et al. (2011) |
| 6 | 945 | 951 | Negative | ||||||||
| 40 | 1,032 | 1,072 | Total | ||||||||
| Bio‐Rad TeSeE WB | Wapiti, mule deer, WTD | LN | 37 | 0 | 37 | Positive | 100% | 90.5–100% | 100% | 56.5–100% | Blasche et al. (2011) |
| 0 | 5 | 5 | Negative | ||||||||
| 37 | 5 | 42 | Total | ||||||||
| Bio‐Rad TeSeE ELISA | Wapiti, mule deer, WTD | LN | 29 | 0 | 29 | Positive | 78.4% | 62.8–88.6% | 100% | 98.8–100% | Blasche et al. (2011) |
| 8 | 320 | 328 | Negative | ||||||||
| 37 | 320 | 357 | Total | ||||||||
| Bio‐Rad TeSeE s/g ELISA | Wapiti, mule deer, WTD | LN | 39 | 0 | 39 | Positive | 97.5% | 87.1–99.5% | 100% | 99.6–100% | Blasche et al. (2011) |
| 1 | 1,080 | 1,081 | Negative | ||||||||
| 40 | 1,080 | 1,120 | Total | ||||||||
| IHC +ve | IHC −ve | Total | Test result | ||||||||
| Bio‐Rad | Mule deer | RPLN | 59 | 0 | 59 | Positive | 98.3% | 91.1–99.7% | 100% | 99.6–100% | Hibler et., (2003) |
| 1 | 1,097 | 1,098 | Negative | ||||||||
| 60 | 1,097 | 1,157 | Total | ||||||||
| Wapiti | RPLN | 21 | 1 | 22 | Positive | 100% | 84.5–100% | 99.87% | 99.3–99.9% | Hibler et al. (2003) | |
| 0 | 814 | 814 | Negative | ||||||||
| 21 | 815 | 836 | Total | ||||||||
| WTD | RPLN | 3 | 0 | 3 | Positive | 100% | 43.8–100% | 100% | 97.1–100% | Hibler et al. (2003) | |
| 0 | 130 | 130 | Negative | ||||||||
| 3 | 130 | 133 | Total | ||||||||
| Bio‐Rad | Mule deer | obex | 35 | 0 | 35 | Positive | 92.1% | 79.2–97.2% | 100% | 99.6–100% | Hibler et al. (2003) |
| 3 | 958 | 961 | Negative | ||||||||
| 38 | 958 | 996 | Total | ||||||||
| Wapiti | Obex | 14 | 0 | 14 | Positive | 93.3% | 70.2–98.8% | 100% | 99.6–100% | Hibler et al. (2003) | |
| 1 | 1,028 | 1,029 | Negative | ||||||||
| 15 | 1,028 | 1,043 | Total | ||||||||
| WTD | Obex | 0 | 0 | 0 | Positive | – | – | 100% | 72.2–100% | Hibler et al. (2003) | |
| 0 | 10 | 10 | Negative | ||||||||
| 0 | 10 | 10 | Total | ||||||||
| IDEXX HerdChek CWD | Roe deer | Brain | 0 | 0 | 0 | Positive | – | – | 100% | 98.35–100% | De Bosschere et al. (2006) |
| 0 | 222 | 222 | Negative | ||||||||
| 0 | 222 | 222 | Total | ||||||||
| IDEXX HerdChek CWD | Roe deer | Spleen | 0 | 0 | 0 | Positive | – | – | 100% | 83.23–100% | De Bosschere et al. (2006) |
| 0 | 206 | 206 | Negative | ||||||||
| 0 | 206 | 206 | Total | ||||||||
Negative samples originated from Germany.
When the previous EU cervid surveillance was undertaken, it was agreed to use the tests concurrently in place for BSE and scrapie, based on their routine use in North American CWD surveillance programmes. It was also agreed to test brainstem, in order to align the laboratory testing with the concurrent procedures for cattle and small ruminants. An External Quality Assessment (EQA) exercise for rapid and confirmatory testing was put in place for participating NRL, coordinated by the EURL and the EU research network Neuroprion, together with assistance and samples from research and regulatory groups in both the US and Canada (Benestad and Gavier‐Widen, 2006).
3.4. Diagnostic methods for the detection of CWD
3.4.1. Screening tests
In the 2004 EFSA opinion (EFSA, 2004b), the data available for test performance were reviewed, and the conclusions on test specificity and sensitivity were tabulated. This opinion acknowledged the limitations on the data with regard to the differing sample panels; the lack of detailed information on any modifications that had been made to test formats for their application to cervid tissue, the lack of any detailed sample background including sample quality; whether or not the sample panels were blinded with regard to parallel test data; and the limitations resulting from sample panel size.
Data from the application of six test formats to lymphoid tissue from mule deer and WTD were also evaluated (EFSA, 2004b). The proportional split between the species, the stage of disease (clinical vs preclinical) and whether the animals had been experimentally challenged (infection status known) or wild (infection status not known) were not provided. All tests performed well (DSe: 82–100%; DSp: 99.7–100%), and on large numbers of samples (between 395 and 2,114) (see Appendices of the 2004 EFSA opinion on a surveillance programme for CWD in the EU (EFSA, 2004a)). However, in the situation where either sensitivity or specificity were less than 100%, it is not possible to assess how much of this could be attributable to the fact that some subsamples of the lymph node (LN) used for the different tests may not have had affected lymphoid follicles, since disease‐specific PrPSc accumulation does not occur uniformly across the tissue early in the disease course (i.e. sampling artefact). The principal modification of the available commercial tests for use in lymphoid tissue of cervids relates to the methods for the physical preparation of the sample, with more mechanical force being required to break down lymphoid tissue samples as compared to brain tissue. It was assumed that the tests were conducted following the manufacturers’ instructions.
Test results from brainstem samples were also provided for four of these tests, with similar sensitivity and specificity ranges (80–100%), but with much smaller numbers (between 32 and 636). Again, there was incomplete information about the animals from which the samples were obtained, with regard to how disease status was determined to provide the ‘gold standard’ for the evaluation of the sensitivity and specificity of the screening test (e.g. was the assessment calculating DSe, and was this consistent across all sample sets?). It was again assumed that the tests were conducted following the manufacturers’ tissue‐specific instructions.
Data were also assessed for wapiti samples, with three out of the four tests attaining less than 100% DSe (84.1%, 93.8% and 97.3%, respectively) but this lower performance was ascribed to the known, more limited, accumulation of PrPSc in wapiti lymphoid tissues (Spraker et al., 2006).
At the time the 2004 Opinion (EFSA, 2004a) was written, the United States Department of Agriculture had licensed four different test kits, all for use on cervid lymphoid tissue: Bio‐Rad ELISA, VMRD Dot‐blot ELISA, IDEXX EIA and the Prion Developmental Laboratories INC lateral flow colorimetric assay. The licensing was restricted by species, with WTD the only species common to all test kits, mule deer for two (Bio‐Rad and IDEXX) and with only Bio‐Rad also including wapiti.
A large field assessment of the Bio‐Rad CWD ELISA kit was published in 2003 (Hibler et al., 2003) in which over 25,000 samples from deer and wapiti were tested, with the first 4,175 being tested in parallel with IHC.
Four ELISA‐based tests (Bio‐Rad TeSeE™, Bio‐Rad TeSeE™ sheep/goat (s/g), Prionics‐Check LIA, and R‐Biopharm PrionScreen), two WB tests (Bio‐Rad TeSeE™ Western Blot and Prionics‐Check WESTERN) and one lateral flow assay (Prionics‐Check PrioSTRIP) were evaluated by Blasche et al. (2012) comparing them with IHC using samples from Rocky Mountain elk, WTD and mule deer. The number of positive samples (by IHC) used in this evaluation ranged between 53 and 93 (depending on the test and the abundance of sample material) of both obex and lymph node tissues. The results showed that the DSe and DSp of the tests varied significantly. The Bio‐Rad TeSeE™ s/g ELISA showed a high DSe for the qualitative and quantitative detection of PrPSc. However, it displayed a reduced DSp (lower ability to identify negative obex samples) and variable DSe. Similar patterns were observed in the R‐Biopharm PrionScreen. Summary of the available data on the DSe and DSp of the Bio‐Rad RT and IDEXX RT from literature and manufacturers’ data are presented in Appendix C (Tables C.2 and C.3).
Table C.2.
Summary data on performance of IDEXX HerdChek BSE Scrapie Antigen Test Kit for the detection of CWD (based on manufacturer's data)
| Reported status | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Test | Species | Tissue | IHC +ve | IHC −ve | Total | Test result | Se % | 95 C.I. % | Sp % | 95 C.I. % | Comment |
| HerdChek BSE‐Scrapie Test | Wapiti | Brain | 8 | 0 | 8 | Positive | 100% | 67.5–100% | 100% | 81.5–100% | 2013 |
| 0 | 22 | 22 | Negative | ||||||||
| 8 | 22 | 30 | Total | ||||||||
| HerdChek BSE‐Scrapie Test | Wapiti | Tonsil | 6 | 0 | 6 | Positive | 85.7% | 42–99.2% | 100% | 83.9–100% | 2013 |
| 1 | 20 | 21 | Negative | ||||||||
| 7 | 20 | 27 | Total | ||||||||
| HerdChek BSE‐Scrapie Test | Wapiti | LN | 6 | 1 | 7 | Positive | 100% | 61–100% | 95.4% | 78.2–99.2% | 2013 |
| 0 | 20 | 20 | Negative | ||||||||
| 6 | 21 | 27 | Total | ||||||||
Table C.3.
Summary data on performance of Bio‐Rad TeSeE and Bio‐Rad TeSeE SAP for the detection of CWD (based on manufacturer's data)
| Reported status | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Test | Species | IHC +ve | Presumed negative (NO IHC) | Total | Test result | Se % | 95 C.I. % | Sp % | 95 C.I. % | Comment |
| Bio‐Rad TeSeE | Mule deer | 152 | 0 | 152 | Positive | 100% | 97.5–100% | 100% | 99.3–100% | Colorado (2008) |
| 0 | 678 | 678 | Negative | |||||||
| 152 | 678 | 830 | Total | |||||||
| Bio‐Rad TeSeE SAP | Mule deer | 151 | 0 | 151 | Positive | 99.30% | 96.4–99.8% | 100% | 99.3–100% | Colorado (2008) |
| 1 | 678 | 679 | Negative | |||||||
| 152 | 678 | 830 | Total | |||||||
| Bio‐Rad TeSeE | WTD | 177 | 0 | 177 | Positive | 96.2% | 92–98.3% | 100% | 99.4–100% | Wisconsin (2008) |
| 7 | 806 | 813 | Negative | |||||||
| 184 | 806 | 990 | Total | |||||||
| Bio‐Rad TeSeE SAP | WTD | 173 | 0 | 173 | Positive | 94% | 89.2–96.8% | 100% | 99.4–100% | Wisconsin (2008) |
| 11 | 806 | 817 | Negative | |||||||
| 184 | 806 | 990 | Total | |||||||
| Bio‐Rad TeSeE | WTD | 12 | 0 | 12 | Positive | 100% | 69.8–100% | 100% | 56.1–100% | Colorado (2008) |
| 0 | 7 | 7 | Negative | |||||||
| 12 | 7 | 19 | Total | |||||||
| Bio‐Rad TeSeE SAP | WTD | 12 | 0 | 12 | Positive | 100% | 69.8–100% | 100% | 56.1–100% | Colorado (2008) |
| 0 | 7 | 7 | Negative | |||||||
| 12 | 7 | 19 | Total | |||||||
| Bio‐Rad TeSeE | Wapiti | 41 | 0 | 41 | Positive | 95.30% | 82.9–99.2% | 100% | 99.6–100% | Colorado (2008) |
| 2 | 1,222 | 1,224 | Negative | |||||||
| 43 | 1,222 | 1,265 | Total | |||||||
| Bio‐Rad TeSeE SAP | Wapiti | 42 | 0 | 42 | Positive | 97.60% | 86.2–99.9% | 100% | 99.6–100% | Colorado (2008) |
| 1 | 1,222 | 1223 | Negative | |||||||
| 43 | 1,222 | 1265 | Total | |||||||
To update the information previously gathered for the 2004 opinion, the test manufacturers known to market authorised diagnostic kits for the detection of CWD in the US were contacted, and invited to present any further specific data they have on test performance for cervids, with a specific request for information on the species, the genotype, the tissue tested, whether the disease was experimental or natural, age (and days post‐infection for experimental animals) and all comparative testing information. One manufacturer (VMRD) replied that their test was no longer being actively marketed. The other two (Bio‐Rad and IDEXX), the only companies currently marketing tests for specific use in cervids in the US, each made available a set of documents, including internal evaluations, scientific publications and certifications. A summary of the data, together with further examples of comparative test performance from the literature are given in Appendix C (Table C.1).
The sections below summarise the available information for the tests currently being actively marketed for use in cervid surveillance.
3.4.1.1. IDEXX HerdChek BSE‐Scrapie Antigen Test Kit
The precise target regions for the antibodies used in the IDEXX HerdChek BSE‐Scrapie Antigen Test Kit are proprietary, but lie within the highly conserved core region of PrP. The cervid kit instructions for use (IFU) include a sampling protocol for the subsampling of lymphoid tissue to be presented to the test (2–3 areas of the cortex of the LN). However, unlike the kit IFU for bovine or small ruminant use, no indication is given of how to preserve anatomically appropriate tissue for confirmatory testing. No instructions are given for use of the test kit on brain samples.
The IDEXX HerdChek BSE‐Scrapie Antigen Test Kit, in different formats (BSE long, BSE short and BSE ultrashort) has been evaluated against a CWD‐specific short and ultrashort profile by the Canadian BSE reference laboratory. Based on a small panel (n = 22) of experimentally infected wapiti samples (including brain, lymphoid tissues and a dilution series of one brain sample), the BSE long protocol kit was the most sensitive. Results of the evaluation of the performance of the IDEXX HerdChek BSE‐Scrapie Antigen Test Kit for the detection of CWD in cervid tissue are presented in Table C.2 of Appendix C.
This kit was also used on a panel of 206 spleen samples and 222 brain samples collected from roe deer in Belgium and compared with IHC using R524, 2G11 and 12F10 antibodies (De Bosschere et al., 2006) (see Table C.1 in Appendix C). All samples gave negative results.
3.4.1.2. Bio‐Rad TeSeE™ and TeSeE™ SAP
The precise target regions for both ELISA tests developed by Bio‐Rad Laboratories, in different formats (TeSeE™ and TeSeE™ SAP), are proprietary. According to the manufacturer, this test has been evaluated against a panel of wapiti, mule deer and WTD RPLN samples collected from Colorado (1,265 wapiti: 43 IHC positive and 1,222 negative samples; 830 mule deer: 152 IHC positive and 678 negative samples, and 19 WTD: 12 IHC positive and 7 negative samples) and from Wisconsin (990 WTD: 184 IHC positive and 806 negative samples) (see Table C.3 in Appendix C).
In the tests of samples from Colorado mule deer, Rocky Mountain wapiti, and WTD reported by Hibler et al. (2003), the relative sensitivity of the Bio‐Rad test (brELISA) ranged from 98.3% to 100% for RPLN samples and 92.1–93.3% for obex samples. The relative specificity of brELISA, depending on species, ranged from 99.9% to 100% for RPLN samples and was 100% for obex samples (Hibler et al., 2003). Overall agreement between brELISA and IHC analysis was 97.6% in RPLN samples and 95.7% in obex samples (Hibler et al., 2003). Based on these data, brELISA was determined to be an appropriate RT for screening large numbers of samples in surveys designed to detect CWD infections in deer and wapiti populations. Results of the evaluation of the performance of the Bio‐Rad TeSeE™ and Bio‐Rad TeSeE™ SAP for the detection of CWD in cervid tissue are presented in Table C.3 (Appendix C).
Further analysis testing different cut‐off points concluded that with a lower constant value added to the negative control (from 0.210 to 0.110) a kit DSe of 99.6% and a DSp of 99.7% could be estimated (Hibler et al., 2003).
3.4.2. Validation status of rapid tests for CWD
Both IDEXX and Bio‐Rad RT have been fully validated for the testing of brain from cattle and sheep, in accordance with the OIE recommendations. However, neither RT has been subjected to a full standardised and laboratory assessment for use in cervids. Moreover, the evaluations so far have been undertaken in the North American context, and predominantly on lymphoid tissue and not on brain. The large amount of field data available from the use of these tests in the US and Canada provide a good level of assurance about DSp, but there is much greater uncertainty about DSe (Appendix C). It is also very difficult to separate ASe and DSe issues when lymphoid tissues are being tested, and there is minimal information on the genetic background of the animals sampled or the stage of disease at which the samples were taken. However, there is sufficient laboratory and/or field data in North America for the tests to meet the requirements for formal ‘provisional recognition’ as defined by the OIE (See Appendix B).
In the context of European CWD, there is currently insufficient laboratory or field data to enable the tests to meet even the requirements for formal ‘provisional recognition’, and they must therefore be used on the assumption that extrapolation from other species in a different geographical location will provide satisfactory results.
In the context of the very small number of European cases already detected in Norway, there is no indication that the DSp of the Bio‐Rad test is not robust, but no conclusion can be reached about its DSe. Currently there are no equivalent European data for the IDEXX test.
IHC and WB, now usually applied as confirmatory tests (see Section 3.5) were widely used for initial screening for TSE before rapid tests were developed (see Section 3.1), and can still be used directly as ‘screening’ tests (e.g. Miller et al., 2000; Miller and Williams, 2002), for example in situations where cold‐chain continuity for chilled or frozen samples is difficult, or where the samples should be fixed because delays in submission might compromise sample integrity.
3.4.3. Other methods with the potential to contribute to CWD detection
3.4.3.1. Protein misfolding cyclic amplification (PMCA)
Although immunoassays detect PrPSc in brain and lymphoid tissues from CWD‐affected animals with good sensitivity and high specificity (Haley et al., 2009a, 2012), they do not detect prion infectivity, and were not as sensitive as, for example, PMCA for the detection of CWD prions in subclinically infected animals or in the body fluids of infected animals (Haley et al., 2009a,b).
During PMCA, the normal form of PrP (PrPC) is converted into protease K‐resistant PrP (PrpRes) using small amounts of infectious PrPSc. Continued recruitment and conversion of PrPC by PrPSc is accomplished by sonication in a process analogous to amplification of DNA by the polymerase chain reaction (PCR). Following the development of Tg(CerPrP)1536 mice (see Section 3.6.3.2), studies were initiated to evaluate the feasibility of combining the resources of PMCA (Saborio et al., 2001) with Tg mouse models of prion disease. In seminal studies, deer PrPC expressed in the brains of Tg mice was used for amplification of CWD prions by PMCA, and using Tg(CerPrP) mice to characterise this in vitro‐generated infectivity, PMCA was shown to enable high fidelity amplification of CWD prions with apparently unaltered strain properties (Green et al., 2008b). These studies extended previous reports (Kurt et al., 2007) by showing that PMCA‐derived CWD prions induce disease and the production of PrPSc in Tg(CerPrP) mice as efficiently as prions isolated from the CNS of deer with CWD. This amplification process enhances detection sensitivity by several orders of magnitude as compared to WB and has been used to confirm the presence of CWD prions in muscle (Daus et al., 2011) and faeces (Pulford et al., 2012). Adaptation of the process whereby successive rounds of PMCA are performed using fresh brain homogenate substrate (sPMCA) – with ultimate evaluation by WB‐ added additional sensitivity to CWD detection (Angers et al., 2009; Haley et al., 2011). Furthermore, inclusion of Teflon® beads in the PMCA reaction (PMCAb) increased the sensitivity of CWD detection without compromising assay specificity. PMCAb was demonstrated to be five orders of magnitude more sensitive than bioassay in Tg mice (Johnson et al., 2012). Using this approach, CWD prions were detected in the cerebrospinal fluid of CWD‐exposed deer (Haley et al., 2013). Baculovirus‐expressed PrP has also been used as a source of substrate for PMCA conversion of CWD prions (Faburay et al., 2014).
PMCA has been used in a wide range of experimental and laboratory contexts, and has also been developed for the screening of blood samples, and to look for evidence of environmental contamination with TSE (Konold et al., 2015). However, it has been shown to generate abnormal PrP de novo under some circumstances, which could compromise specificity (Saá and Cervenakova, 2015). Additionally, there is no universal substrate for this method, with some outcomes being strain or genotype dependent to the point where substrate variation can be used as the basis for discrimination of strains (Simmons et al., 2015) and therefore, it is not a good method to use when looking for potential unknowns.
3.4.3.2. Real‐time quaking‐induced conversion (RT‐QuIC)
In the assay referred to as real‐time quaking‐induced conversion (RT‐QuIC) (Atarashi et al., 2008), prion seeds are thought to induce recombinant PrP to adopt a β‐sheet structure. Thioflavin T, added to the reaction, is incorporated into the growing amyloid causing an altered spectrofluorimetric emission pattern, which is monitored over time. As an alternative to whole brain homogenates expressing PrPC, and as part of a larger attempt to address the minimal components required for prion propagation, bacterially expressed recombinant PrP has been used as a substrate for amplification. RT‐QuIC has been extensively applied to the detection of CWD prions in tissues, body fluids and excreta, such as lymphoid tissue (Haley et al., 2014), cerebrospinal fluid (Haley et al., 2013), blood (Elder et al., 2013), saliva (Henderson et al., 2013), and urine and faeces from preclinical animals (John et al., 2013; Cheng et al., 2016). Using these methods, Henderson and colleagues estimated that urine and saliva from CWD‐infected deer both contained 1–5 LD50 (lethal dose that kills 50%) per 10 mL (Henderson et al., 2015a,b), suggesting substantial environmental contamination is likely to occur during the several‐year course of an infection.
This method is showing diagnostic promise for in vivo screening for human TSE (Orrù et al., 2017) and has also been successfully applied to the detection of disease in both WTD and elk (Haley et al., 2016a,b). Analysis of RAMALT biopsy specimens and nasal brushings collected ante‐mortem indicated that, like IHC analysis, RT‐QuIC is a relatively sensitive assay for detection of CWD prions in RAMALT biopsy specimens with potential for rapid automated testing of ante‐mortem samples for CWD. However, there is very limited data on the specificity of this method, and as a consequence, it would not be advisable to consider its use for surveillance in populations where disease has not already been confirmed.
3.5. Diagnostic methods for the confirmation of CWD
As it is the case for all other recognised TSE, confirmation of an initial ‘screening positive’ (or ‘suspect’) sample can be undertaken using immunodetection methods either in a WB format, or using IHC. The former gives some classification data (see below) based on the molecular mass and glycosylation profiles of the PrPSc, while the latter enables PrPSc accumulations to be assessed in the context of anatomical (including cellular) location. These tests should be applied to the same tissue as the screening test, and potentially to other tissues, depending on the case definition being applied (see Section 3.2). As screening tests become more sensitive, and in the absence of a true ‘gold standard’, disagreements in test outcomes should be interpreted with caution because it is difficult to distinguish between a specificity issue with the screening test (i.e. false positive results) or a sensitivity issue (false negative results) with the confirmatory test.
IHC can be used to visualise abnormalities occurring even in single cells, and its sensitivity is not adversely affected when very small depositions of PrPSc are surrounded by larger volumes of negative tissue. When applied to brain samples, it enables the neuroanatomical position of the sample to be confirmed, ensuring that the appropriate target areas have been selected for testing. In lymphoid tissue, it can also be confirmed that labelling is associated with the correct cells in the secondary follicles, offering supporting qualitative data for the specificity of the result. For this reason, IHC has historically been considered as the diagnostic ‘gold standard’ for TSE diagnosis. However, successful IHC relies on the fixation of a piece of solid tissue. In cases where the starting material is severely autolysed and has lost its normal consistency, WB may be the only confirmatory test that can be used.
Brainstem collection protocols for TSE testing in a range of species require the sample to be divided along the midline, and obex samples for testing and confirmation should be taken from the same sagittal plane (OIE, 2017b). This capitalises on the observation that even in pre‐clinical cases with limited accumulation of PrPSc, these accumulations are very precisely targeted to specific neuroanatomical areas, which are bilaterally symmetrical in distribution. The tissue homogenate and the fixed tissue used for confirmation are therefore considered to be equivalent.
In contrast, visible PrPSc accumulation in lymphoid tissue is confined to secondary follicles, and not all follicles are affected at the same time. In the earlier stages of infection, possibly only a few follicles are affected (Stack et al., 2011) and, unlike the situation in the brainstem, this is not linked to any internal anatomical features of the lymph node, so the subsampling of lymphoid tissue for either the screening test or the confirmatory test cannot be guided to improve sensitivity. It also reduces the certainty that two separate samples are equally representative of the whole tissue. Such random distribution of PrPSc within each lymph node may also account for observed differences in comparative test performance studies, making it impossible to distinguish between test sensitivity and specificity issues if the outcome is not complete agreement between tests.
In the absence of appropriate positive controls for these tests in the target European species, the suitability of mAb for use in confirmatory tests must be based on extrapolation from the North American experience, and a ‘first principles’ approach avoiding antibodies that bind at a polymorphic site (see Table 1).
3.6. Classification of isolates
The gold standard for isolate classification and strain comparison, and the ultimate measure of both TSE infectivity and susceptibility, is the bioassay in rodent models. However, some preliminary classification can be undertaken using confirmatory testing data from the original host. The assessment of whether any particular strain represents a risk to other species currently relies predominantly on experimental transmissions in either the target host species, or the appropriate transgenic (Tg) rodent models.
Experimental infections would be required in order to understand both the susceptibility and the pathobiology, including infection kinetics, transmissibility, tissue infectivity, lesion profile and biochemical characteristics of different strains of CWD prions in the various potential host species.
The susceptibility of several European species of cervids, including some widely occurring throughout Europe, such as the roe deer, is not yet established. In the absence of such data, positive controls for comparison and preliminary classification of any European isolates must be drawn from both natural and experimental cases in North America. Current data indicate that at least two strains of CWD have been detected in North American wild cervid populations as identified after passage into mice (Tamguney et al., 2006), but it cannot be assumed that these are the only ones. Strain classification and host susceptibility data ultimately inform case attribution, epidemiological patterns, outbreak control and the assessment of zoonotic potential.
There is no standardised system for ‘naming’ disease subsets within the TSE. In human TSE, nomenclature has tended to reflect the clinicians who first described the diseases (e.g. Creutzfeldt‐Jakob, Gerstmann‐Staussler‐Sheinker) or the principal presenting signs (e.g. fatal familial insomnia). In animals, presenting signs gave rise to the various historical names for TSE in sheep (scrapie, ‘la tremblante’, ‘traberkrankheit’, ‘gnubberkrankheit’, ‘prúrigo lumbar’ (OIE, 2017b)), whereas the more recently identified BSE was objectively named, based on species and pathology in the natural disease.
Subsequent advances in strain definition and classification have highlighted that ‘scrapie’ can be caused by at least three distinct field strains, whereas the BSE epidemic was driven by a single strain. The stability of this strain, and the wide range of cross‐species experimental challenge data that have been generated on BSE means that this specific strain can be identified against a range of host species backgrounds (cattle, sheep, goats, deer, humans), making it unique among the animal TSE. More recently, the picture has been further complicated by the identification of ‘atypical’ strains of both BSE and scrapie, identified as subsets of these classifications purely on the basis of species of origin.
Over the last two decades, a range of isolates from BSE and scrapie have been characterised in a variety of transgenic mice expressing bovine, ovine, caprine, porcine and human PrP, and also in bank voles. When this is added to the information on host species, genotype, and the biochemical profile of the original animal, a ‘fingerprint’ of strains of policy interest, in particular BSE, can be built up and specifically screened for. This forms the basis of Annex X to Regulation (EC) No 999/2001, as amended, for the classification of all TSE positive isolates in small ruminants, which enables any isolate that is BSE‐like on primary classification to be systematically investigated further in models that have the full range of positive controls.
CWD was the term originally given to mule deer (and later wapiti) with TSE, based on the predominant clinical signs (Williams and Young, 1980, 1992). Although certain TSE strains are associated with distinct conformers of PrPSc, not all strains that can be biologically distinguished are composed of PrPSc with recognisably different biochemical properties. The electrophoretic migration patterns of PrPSc from the brains of mice infected by either CWD1 or CWD2 were indiscernible. PrPSc associated with CWD1 and CWD2 was composed of equivalent proportions of di‐, mono‐ and a‐glycosyl forms, and had similar unfolding characteristics after treatment with guanidinium hydrochloride (Gdn.HCl). When CWD1 or CWD2 wapiti isolates were transmitted to transgenic mice expressing wapiti PrP, stabilities of the resulting PrPSc remained indistinguishable, but were distinct from PrPSc in Tg(CerPrP)1536 mice expressing deer PrP.
As described in Section 3.6.3, CWD isolates have mostly been inoculated into transgenic cervid mice (for research purposes) or humanised mice (for the assessment of zoonotic potential) (Lee et al., 2013a,b). There is therefore a less comprehensive body of data on field‐derived ‘CWD’ in laboratory terms, to enable distinction from other strains, or to identify similarities with other known TSE. Experimental inoculation of scrapie into WTD resulted in a disease with a histological lesion profile very similar to CWD, but variable WB profiles which differed from each other, and from that of the CWD control (Greenlee et al., 2011).
It is clear that there are at least two distinct CWD ‘strains’ described in North America, and possibly two distinct TSE strains within the small number of cases detected to date in Europe. How these different isolates relate to one another (and to TSE from other species) is not yet known, although rodent model isolation and classification studies are ongoing and their results will contribute to answer this question.
Experimental transmission of CWD to other (non‐cervid) species has shown mixed results. Susceptible species include several species of voles, white‐footed mice, deer mice, cats, raccoons, pigs and squirrel monkeys (Hamir et al., 2003, 2007; Race et al., 2009b, 2014; Heisey et al., 2010; Di Bari et al., 2013; Mathiason et al., 2013; Moore et al., 2017), while the CWD agent transmitted poorly to Syrian golden hamsters, ferrets, mink and squirrel monkeys (Bartz et al., 1998; Marsh et al., 2005; Sigurdson et al., 2008). Non‐Tg mice have been reported to be resistant to CWD infection (Browning et al., 2004), but limited infection of the VM/Dk inbred strain of mice with wapiti CWD prions has been reported (Lee et al., 2013a), indicating that some variable species barriers to the transmission of CWD exist.
With regard to the potential for CWD to be zoonotic, previous EFSA opinions (EFSA BIOHAZ Panel, 2011, 2015) concluded that while mouse bioassays did not provide any evidence supporting the zoonotic potential of CWD, very limited data on the experimental challenge of primates means that the human species barrier for CWD prions does not appear to be absolute. However, epidemiological investigations carried out to date make no association between the occurrence of disease in humans and exposure to CWD prions. As noted in the 2017 EFSA opinion on CWD (EFSA BIOHAZ Panel, 2017a), the tissue distribution of infectivity in CWD‐infected cervids is now known to extend beyond CNS and lymphoid tissues, so simply removing these tissues from the human food chain may not reduce potential exposure risk as greatly as believed in 2004. The removal from the food chain of the whole carcass of any infected animal would be more effective in minimising human exposure.
3.6.1. Classification using confirmatory testing data
Both WB and IHC approaches allow a comparison of labelling patterns with antibodies that recognise different epitopes of the protein, and help to elucidate specific proteinase‐K cleavage sites that are thought to be an inherent property of strains. This approach forms the basis of the discriminatory testing of small ruminant isolates, to differentiate between isolates that can be classified as scrapie, and those that are considered BSE‐like, and require further testing and possible bioassay.6 While the exclusion of BSE is not a requirement for CWD surveillance in North America, immunoblotting techniques have demonstrated the ability to discriminate BSE from CWD by looking at the lower molecular weight for the unglycosylated protein band obtained from red deer experimentally infected with BSE either oral (p.o.) or intracerebral (i.c.) inoculation, compared to that obtained for ovine scrapie, wapiti CWD or red deer CWD samples, using the Bio‐Rad TeSeE WB (Martin et al., 2009).
Some classification of isolates in both cattle and small ruminants can be achieved by comparing the molecular characteristics of the PrPres in WB, such as the mass of the unglycosylated fragment and the ratio of the mon‐ and di‐glycosylated fragments, for example between classical, atypical/Nor98 and CH1641 scrapie in sheep and goats, and BSE and scrapie (Jacobs et al., 2007; EURL, 2016).
Neuroanatomical patterns of immunolabelling and/or vacuolation (the lesion profile) have also been used to classify TSE, including the subdivision of scrapie cases with similar WB profiles (Gonzalez et al., 2003), and have played a key role in rodent bioassay interpretation (Beck et al., 2010). This approach could also be applied to cervids, although this method is somewhat limited when only the obex region of the brain is available for assessment. Preliminary data show that there are differences in the pattern of intraneuronal immunolabelling between cases in reindeer and moose in Norway.
The initial confirmatory testing data from Norway suggest that the strain identified in moose differs from that in reindeer (Pirisinu et al., 2017), but until laboratory strain‐typing studies have been completed it cannot be determined whether or not the strain identified in moose represents a new or atypical CWD strain. However, the comparison of disease immunopathology in Norwegian and North American moose suggest that there is a strain difference, with the disease in Norwegian moose being unlike anything previously reported for CWD (Pirisinu et al., 2017). Although CWD has not been reported in North American caribou, and so direct comparison is not possible, in vitro characteristics of the Norwegian reindeer isolates appear to be indistinguishable from isolates of reindeer experimentally inoculated with North American CWD derived from WTD (Mitchell et al., 2012), but a conclusion about the strain similarities or differences requires results from the ongoing bioassay experiments.
3.6.2. Classification using bioassay in potential natural host species
CWD transmission can be demonstrated after i.c. or p.o. inoculation of the natural host, but only after incubation periods of up 230 days or more (Williams and Young, 1992; Sigurdson et al., 1999; Fox et al., 2006; Wolfe et al., 2012). Oral transmission of mule deer‐derived CWD prions was also reported in captive moose in Wyoming (Kreeger et al., 2006). Other cervid species have been shown to be susceptible to CWD following experimental i.c. or p.o. challenge, including European red deer (Martin et al., 2009), muntjac deer (Nalls et al., 2013) and reindeer (Mitchell et al., 2012).
However, animal experiments always have important welfare and ethical implications. While farmed cervids adapt to experimental conditions, experiments in wild animals pose major challenges and require special considerations. These include the susceptibility of wild animals to the stress of captivity and handling, their special diet requirements, diseases related to transition from pastures (i.e. free ranging) to feeding, and the necessity for appropriate enclosures that meet their physiological and behavioural needs. In the US, such experiments have been undertaken in mule deer, WTD, wapiti, and reindeer, but in other species like moose, (despite attempts to keep moose domesticated for milk production in some countries) or in species inherently difficult to keep in captivity, like roe deer, it would be much more problematic. Roe deer and other forest living cervids normally live in or close to dense vegetation and would need part wooded and part grassland enclosures. These requirements also make it very difficult to ensure appropriate biosecurity. Thus, in Europe, animal experiments with CWD could only be conducted either under biosafety‐3 containment level or within an area already considered to be infected. In the latter situation, there would still be a risk of artificially introducing other strain(s) of CWD prions.
Further difficulties of experimental infection with CWD are that the studies are prolonged and expensive and are therefore performed on limited numbers of animals resulting in an inherent variability of results related to the complexity of the biological system of the whole animal (individual variations). Notwithstanding these drawbacks, and since experimental transmission of CWD to transgenic mouse models has been successful in recapitulating the cardinal features of disease, their future use in characterising European cases of disease is likely to be of significant value.
3.6.3. Classification using bioassay in rodent models
3.6.3.1. Historical background
The biological characterisation of prion strain properties mainly relies on experimental transmissions of isolates in animal models, with ‘isolate’ referring to a primary source of prion infectivity from a naturally occurring disease. As such, isolates may contain one or more ‘strains’, which generally refers to infectivity, with distinctive phenotypic properties that have been experimentally characterised in animal models. Standard criteria for characterising and differentiating strains include the distribution and severity of PrP‐associated pathology, often revealed by labelling brain sections with anti‐PrP Ab (Bruce et al., 1989), and the time to onset of disease after inoculation, referred to as the incubation time (Bruce and Fraser, 1991; Bruce et al., 1994, 2002).
The archetypal scrapie classification work in conventional mouse models required several serial passages within a host species of consistent PrP genotype to establish the phenotypic properties of strains. Strain cloning was accomplished by subjecting a strain to serial passage at limiting dilution, with the aim of purification from minor, slower replicating strains. By varying transmission conditions, early strain typing studies of small ruminant isolates in wild‐type mice identified three classes of strains with varying stabilities (Bruce and Dickinson, 1979).
However, the use of inbred wild type mice for strain typing is associated with several drawbacks. First, the generally inefficient propagation of human and animal prion isolates in mice. Such examples of inefficient trans‐species prion transmission are referred to as species barriers (Pattison, 1965). The transmission barrier may be absolute, in which case no transmission is recorded during the life span of the mice or partial with two alternative types of expression: (a) primary transmission characterised by long incubation times and low attack rates followed by greatly reduced incubation times and high attack rates on secondary passage; (b) high attack rates on both primary and secondary passage with reduced incubation times on secondary passage. Second, since multiple prion strains may exist in a single isolate, and host genetic background can influence strain characteristics, when a strain moves from one species to another strain characteristics can alter in unpredictable ways (Bartz et al., 1998). Interspecies transmission may therefore result in selection of minor strains or strain mutation that may not represent the dominant populations of strains in the original inoculum.
These drawbacks have been largely circumvented by the use of transgenic approaches for the typing of strains from human and animal prion isolates (Telling, 2010). In this alternative approach, foreign PrP genes are expressed in mice through the use of gene replacement methods. This ensures that the PrP coding sequence is controlled by the same regulatory elements as wild type mouse PrP, in which case gene expression is expected to recapitulate authentic PrPC expression, although there are examples where this is not the case (Vickery et al., 2014; EFSA BIOHAZ Panel, 2017b). While microinjection and gene replacement models generally provide complementary results, transgenic overexpression is desirable in most cases since it results in highly reduced incubation times to fully assess the extent of a species barrier. The strain typing methods developed using either kind of transgenic mouse models are essentially similar to those used in wild‐type mice.
3.6.3.2. Transgenic rodent models
Several transgenic mouse lines expressing either wapiti/elk or deer PrP have been developed in which the species barrier to CWD has been eliminated. Prototype transgenic mice expressing deer PrP, designated Tg(CerPrP)1536+/− (Browning et al., 2004), recapitulated the cardinal neuropathological, clinical and biochemical features of CWD, an observation subsequently confirmed in comparable transgenic mouse models expressing deer or wapiti PrP (Kong et al., 2005; LaFauci et al., 2006; Tamguney et al., 2006; Meade‐White et al., 2007; Green et al., 2008a; Angers et al., 2009). Until recently, the prevalence of cervid prion strains had not been assessed. Although original studies in transgenic mice (Browning et al., 2004), and subsequent work (LaFauci et al., 2006) raised the possibility of CWD strain variation, the limited number of isolates and the lack of detailed strain analyses in those studies meant that this hypothesis remained speculative. Subsequent studies supported the feasibility of using Tg(CerPrP)1536+/– mice for characterising naturally occurring CWD strains, CWD prions generated by PMCA and novel cervid prions (Green et al., 2008b). The prevalence of CWD prion strains in a large collection of captive and wild cervids from different species and geographic locations within North America was assessed by bioassay in transgenic mice (Angers et al., 2010). The findings provided substantial evidence for two prevalent CWD prion strains, referred to as CWD1 and CWD2, with different clinical and neuropathological properties. Remarkably, primary transmissions of CWD prions from the wapiti sampled produced either CWD1 or CWD2 profiles, while transmission of sampled deer inocula favoured the production of mixed intrastudy incubation times and CWD1 and CWD2 neuropathologies. These findings indicate that wapiti may be infected with either CWD1 or CWD2, while deer brains tend to harbour CWD1/CWD2 strain mixtures. Interestingly, previous CWD transmission studies in transgenic mice suggested that cervid brain inocula might be composed of strain mixtures (Tamguney et al., 2006).
Additional previous studies also support the existence of multiple CWD strains. CWD has also been transmitted, albeit with varying efficiency, to transgenic mice expressing mouse PrP, referred to as Tga20 mice (Sigurdson et al., 2006; Tamguney et al., 2006). In the former study, a single mule deer isolate produced disease in all inoculated Tga20 mice. On successive passages, incubation times dropped to ~ 160 days. In the second study, one wapiti isolate from a total of eight deer and wapiti CWD isolates induced disease in 75% of inoculated transgenic mice overexpressing mouse PrP, referred to as Tg(MoPrP)4053 mice. It is worth noting that the distribution of lesions in both studies appeared to resemble the CWD1 pattern. Low efficiency CWD prion transmission was also recorded in hamsters and transgenic mice expressing Syrian hamster PrP (Raymond et al., 2007). In that study, during serial passage of mule deer CWD, fast and slow incubation time strains with different patterns of brain pathology and PrPSc deposition were also isolated.
In more recent studies, the transmission properties of CWD prions derived experimentally from deer of four PRNP genotypes (variations at codons 95 and 96) in Tg mice expressing wild‐type deer PrP, or PrP containing the S96 polymorphism were evaluated (Duque Velasquez et al., 2015). Disease signs, and neuropathological and PrPSc profiles in infected Tg mice expressing wild‐type PrP were similar between groups, indicating that a prion strain common to all CWD inocula was amplified. In contrast, Tg60 mice developed prion disease only when inoculated with the H95/wild‐type and H95/S96 CWD allotypes. Serial passage resulted in adaptation of a novel CWD strain referred to as H95(+). Transmission into Tg mice expressing wild‐type PrP, however, elicited two prion disease presentations consistent with a mixture of strains.
Bioassays of the first few Norwegian cases are ongoing in a range of rodent models. These include cervinised mice, which will allow direct comparison of their transmission characteristics with North American isolates, and bank voles, which are susceptible to a wide range of TSE (Espinosa et al., 2016), enabling the comparison of the characteristics of these new cervid isolates with a wide range of TSE from several species.
3.7. The occurrence of CWD in the European cervid population
The 2010 Scientific Opinion ‘on the results of the EU survey for Chronic Wasting Disease (CWD) in cervids' (EFSA BIOHAZ Panel, 2010) broadly concluded that the failure to detect CNS infection among ~ 13,000 brainstem samples from European cervids of eight species (mostly farmed and wild red deer and wild white‐tailed deer) during 2006–2010 indicated that there was not a cervid TSE epidemic in the EU, but that the occurrence of cases of TSE among cervids in the EU could not be excluded. In this current opinion these conclusions are briefly reviewed, recognising that relatively little has been done to survey for CWD in Europe since 2010. As already highlighted in a previous EFSA BIOHAZ Panel Opinion (2017a), the monitoring activities in cervids carried out throughout the EU since 2010 have been sporadic, not supported by any specific study design or targeting strategies and have examined a low number of animals (see surveillance in Europe for the period 2015–2017 in Appendix A).
The specific conclusions from 2010 have been reviewed, either individually or by logical grouping, and comments, perspective, and references added where appropriate to provide a foundation for updating those conclusions in light of reported CWD occurrence in Norway. The EFSA BIOHAZ Panel, 2010 opinion (EFSA BIOHAZ Panel, 2010) detailed available data, assumptions and analysis approaches that led to the conclusions drawn at that time. Consequently, those details will not be repeated here unless needed in the context of discussion. Similarly, the 2017 EFSA opinion on CWD (EFSA BIOHAZ Panel, 2017a) reviewed new available data in the wake of nearly simultaneous detections of CWD in reindeer and moose from separate geographic foci in Norway during 2016 and consequently those details will also not be repeated here unless needed.
The prevalence estimates reported in the 2010 opinion (EFSA BIOHAZ Panel, 2010), arrived at through a Bayesian approach, appear to have been based on the aggregated sample within respective host‐source categories. As detailed elsewhere (EFSA BIOHAZ Panel, 2017a), available biological and epidemiological data lend no support to the assumption that free‐ranging or captive cervid hosts in the EU represent a single, homogeneously mixed population. The assumption of homogeneous disease distribution was also untenable. Moreover, the prevalence estimates provided in the 2010 Opinion were obtained from national surveys that never detected a case and therefore were essentially reflecting the attained (negative) sample size and different assumed test DSe: it is remarkable that in each of the presented scenarios the most likely prevalence was zero.
Actual implementation of surveillance in 2006–2010 had limitations. The random sampling (compared with a geographically based sampling) was inappropriate for attaining a geographically representative sample from the cervid population.
It could be assumed that for logistical reasons the sampling of free‐ranging animals did not completely cover epidemiologically relevant geographic units. However, the survey data set did not provide information on geographical location of the free‐ranging animals tested, the data were limited to the country level only. Nonetheless, a few MS (the Czech Republic, Poland, the UK, Ireland and Finland) provided EFSA with additional data on the geographical distribution of samples within countries. These data confirm that there were discrepancies in the geographic representativeness of the sampling, since there were some areas with a large cervid population and a limited number of samples collected (EFSA BIOHAZ Panel, 2010). Finally, the random sampling strategy applied to wildlife resulted in the scattering of the sample collection preventing the possibility of achieving sufficient sample sizes for each local target subpopulations.
Additionally, the testing of cervids that died in nature was likely limited due to the difficulty of finding carcasses and transporting them for testing. Another potential limiting factor was the decomposition of the carcasses, because advanced autolysis may preclude anatomically accurate sampling of target areas. As a consequence, and far from the designed survey, only 8% of the total were higher‐risk animals.
3.7.1. Spatial and temporal distribution of CWD
The 2010 opinion did not explicitly define what was considered to constitute a ‘cervid TSE epidemic in the EU’. However, analysis assumptions listed in the results of that opinion (EFSA BIOHAZ Panel, 2010; Section 4.2) included ‘random sampling’ (addressed below) and ‘homogeneous distribution of the disease’. The basis for the latter assumption is unclear. None of the published accounts of CWD in North America state or imply homogeneous disease distribution over large geographic areas, and thus there is no logical basis to assume differently in Europe. On the contrary, maps (Figure 3) and publications (e.g. Miller et al., 2000; Farnsworth et al., 2006; Joly et al., 2006; Heisey et al., 2010) available in 2010 underscored the heterogeneity of CWD distribution. Moreover, guidance on CWD surveillance available at the time emphasised spatial heterogeneity as an important challenge of designing and interpreting surveys for detecting CWD (Samuel et al., 2003; Conner et al., 2007b). If homogeneous distribution of CWD was a requisite criterion for qualifying a cervid TSE ‘epidemic’, then technically neither the US nor Canadian infection events would be regarded as experiencing a cervid TSE ‘epidemic’ because the disease occurs as scattered foci of varied size in both countries. Equally, if ‘epidemic’ is defined as ‘an increase, often sudden, in the number of cases of a disease above what is normally expected in that population in that area’ (www.cdc.gov), then the term epidemic was not appropriate in the context of the 2010 EFSA opinion.
Figure 3.
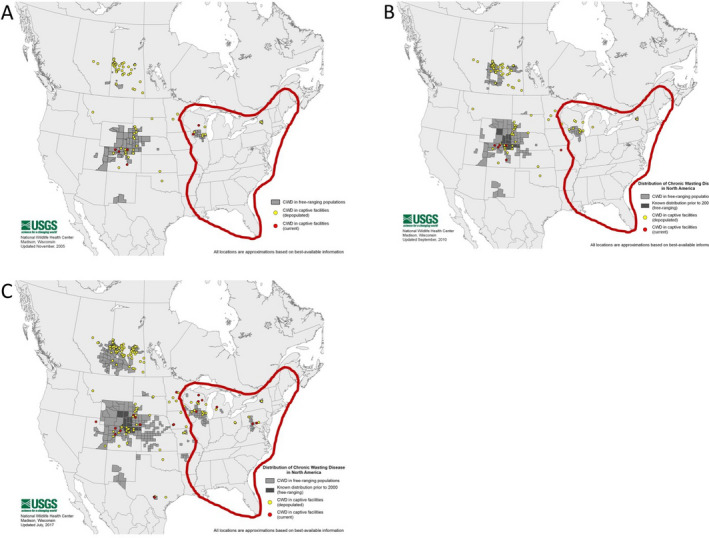
The distribution of CWD in the US in 2005 (A), 2010 (B) and 2017 (C) and area of the 28 contiguous states east of the Mississippi River spanning ~ 2.5 M km2 (within the red line) selected for comparison. Maps produced by and courtesy of the US Geological Survey's National Wildlife Health Center (https://www.nwhc.usgs.gov/disease_information/chronic_wasting_disease/)
The apparent emergence (or the evolving awareness) of CWD distribution in the Eastern US during 2005–2017 illustrates the potential spatial challenges of large‐scale, detection‐oriented surveillance involving multiple jurisdictions. The EU – with 28 MS collectively spanning > 4.4 M km2 – is similar in size and jurisdictional complexity to the eastern US (the 28 contiguous states east of the Mississippi River spanning ~ 2.5 M km2). A comparison was regarded as potentially useful in illustrating concepts related to surveillance challenges and historical inadequacies in Europe. Similarities relevant to CWD epidemiology, surveillance, and likelihood of detection between Europe and the Eastern US include landscapes with large expanses of middle latitude deciduous and mixed forest – modified and fragmented by agriculture and urban development – that provide habitat for resident, mostly non‐migratory native cervid species that vary in abundance and are hunted seasonally throughout both areas. In addition, most US and European jurisdictions allow private ownership and various commercial uses of captive cervids and have other potential sources of exposure to TSE including the import/movement of dead and living cervids and scrapie in domestic sheep and goats. These similarities were considered sufficient to make general comparisons and illustrate concepts related to CWD surveillance and its potential limitations for purposes of the current opinion. However, differences between Europe and the eastern US (e.g. topographic and climatic variation; cervid species diversity, abundance and CWD susceptibility; endemic prion strains; regulatory harmony) also could lead to different patterns of CWD emergence or detection in the former.
As in Europe during 2006–2010, CWD surveillance in the Eastern US (and throughout North America) has been organised and conducted relatively independently by each state and has relied heavily upon sampling of apparently healthy animals, particularly for free‐ranging species. Prior to 2002, CWD was not believed to occur east of the Mississippi River in the US (Belay et al., 2004; Bunk, 2004). Surveillance generally expanded in the eastern US after CWD was reported in Wisconsin in 2002, but survey designs and sampling efforts varied widely between and within jurisdictions. Over 182,000 samples from the eastern US were screened during 2002–2005, with individual state totals ranging from 0 to > 74,000. Nearly 341,000 additional samples were examined during 2005–2012. (Miller, 2017b). Much of the sampling of free‐ranging cervids was done on apparently healthy hunter‐killed deer. In ostensibly ‘CWD‐free’ states, sampling often was oriented towards perceived extra‐jurisdictional risk factors (e.g. the border of a neighbouring state where CWD already had been detected). Only one state (Wisconsin) conducted a systematic survey of the entire state, using counties as the spatial unit based on understanding of local deer biology and movements. Similarly, the scope, compliance, and rules governing surveillance of captive cervids varied widely between states. These patterns resemble those described below and previously (EFSA BIOHAZ Panel, 2010, 2017a) with respect to the 2006–2010 CWD surveillance campaign undertaken by the EU, although the number of samples (> 520,000) examined during the 2002–2012 Eastern US surveillance was 40 times greater than the 2006−2010 European effort and was concentrated on a geographic area about 57% the size of the EU.
The pattern of recognised CWD distribution in the eastern US revealed by uneven surveillance efforts suggested both local spatial spread around recognised foci and emergence of new captive or free‐ranging foci distant to previously known foci (Figure 3). Although the number of Eastern US states with detected CWD appeared to double between 2005 and 2017, at least some of the ‘new’ foci likely were present but undetected even before 2005. Similarly, although some foci appeared to expand rapidly over a few years’ time this may more likely be an artefact of spatially incomplete surveillance or spatial selection and bias of sampling based on prior distribution knowledge. In other words, sampling for the first time in areas adjacent to recently detected foci can give the superficial appearance of spatial ‘spread’ if the surveillance history is not considered. The observed pattern of CWD distribution in the eastern US that has apparently resulted from multiple foci emerging over the last two decades or longer offers a conceptual model for considering how CWD might appear to emerge in Europe given the similarities in size, complexity, and flawed surveillance approaches shared by these three geographically distinct areas where CWD has now been detected.
The distribution of CWD in North America is heterogeneous over large geographic areas. Changes in apparent geographic distribution over time are likely the result of both spatial spread (natural and anthropogenic) and detection of pre‐existing foci (Miller and Fischer, 2016). One caveat with this approach that should be borne in mind is that the cervid species, and possibly TSE strains, differ between Europe and eastern US thus the epidemiology of CWD could show differences, even though it would still be clustered.
An underappreciated aspect of CWD epidemic dynamics in North America is the timeframes over which outbreaks unfold. Although the disease has been said to ‘spread rapidly’ (Lang and Blanchong, 2012), prevalence likely remains low, and infections spatially localised, for a decade or more after the introduction into natural cervid populations (Miller et al., 2000; Wasserberg et al., 2009; Jennelle et al., 2014). One likely outcome of focusing detection on standard thresholds (e.g. 1% prevalence) in North America has been that CWD may have been present for 10–20 years before the first case was identified in a cervid population unit (Figure 4) (Miller and Fischer, 2016). Based on North American experiences, an even longer period of time likely would to be needed for a CWD outbreak to expand across an entire political jurisdiction (e.g. state, province, or country; see Figure 3). It is in part for this reason that updated recommendations on CWD surveillance in the EU include consideration of dividing cervid populations into multiple, biologically relevant spatial units within each MS in order to increase detection probabilities (EFSA BIOHAZ Panel, 2017a).
Figure 4.
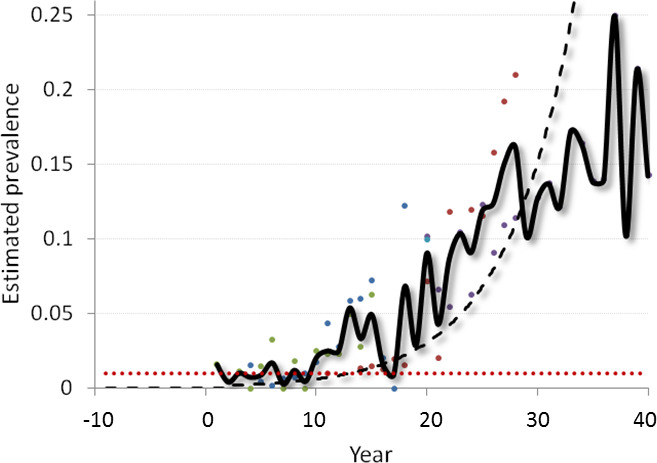
Observed and predicted CWD prevalence in mule deer herds of Colorado (US)
- Individual points are surveillance data from five Colorado mule deer herds, collected over 15–21 years. Each herd is a different colour. Because outbreaks were asynchronous, prevalence of individual herd data segments were aligned by time points rather than by calendar year. Values were then averaged across each generic ‘year’ to generate a composite epidemic curve (solid black line). The standard survey design (detecting 1% prevalence with 95% confidence; dotted red line).
Field and modelling data from North America suggest that CWD epidemics develop relatively slowly as compared to other infectious diseases in wildlife. Data from Colorado, US, as displayed in Figure 4, illustrate this concept. The individual points are surveillance data from five Colorado mule deer herds, collected over 15–21 years. Each herd is a different colour. Because outbreaks were asynchronous, prevalence of individual herd data segments were aligned by time points rather than by calendar year. Values were then averaged across each generic ‘year’ to generate a composite epidemic curve (solid black line). Comparing observed and modelled (dashed black line; from Miller et al., 2000) epidemic dynamics illustrates the potential lag of a decade or more between a local outbreak's outset and its detection using the standard survey design (detecting 1% prevalence with 95% confidence; dotted red line).
The temporal dynamics of CWD in European cervid species are unknown given that foci were first detected only in the year before the current opinion was written. However, the pattern of PrPSc distribution seen thus far among infected reindeer (n = 8), with lymphoid tissue involvement in all infected individuals but CNS involvement in only 5 out of 8 (62.5%), supports the assumption that pathogenesis resembles that described for CWD in North American deer and wapiti, as well as for scrapie in sheep. It follows that natural (e.g. horizontal) CWD transmission and commensurate epidemic dynamics similar to that described for North American cervids may be reasonably assumed. Comparing the point estimate of CWD prevalence among ‘adult’ (> 1 year old) wild reindeer harvested (including hunted and found dead/injured/diseased) in Nordfjella 1 in 2016 (3/310: 0.97%. 95% C.I.: 0.2–2.8%) and during 2017 (up to 27 November 2017: 5/738: 0.68%.95% C.I.: 0.22–1.6%) (Hopp, 2017) the epidemic curve for mule deer (Figure 4), it appears plausible that CWD became established in Norway more than a decade ago.
3.7.2. Strategies to enhance detection of CWD
As highlighted in the recent EFSA Opinion on CWD (EFSA BIOHAZ Panel, 2017a) and in Section 3.7.1, the experience gained from the occurrence of CWD in North America and Norway may help in identifying the most efficient strategies to enhance the detection of CWD, if present.
It has been reported that after introduction in a new area or subpopulation, CWD tends to show a scattered and heterogeneous geographical distribution (clustered foci consistent with a hot spots and sparks model) and an initial and long‐lasting low prevalence. Like BSE and scrapie, within the tested population, ‘high risk’ animal categories (i.e. associated with higher levels of prevalence and therefore higher probability of being detected) may be identified and specifically targeted. Therefore, a two‐tier strategy may contribute in better addressing the mentioned spatio‐temporal dynamics of a CWD new incursion.
The effort to obtain random samples at country level may prove to be inappropriate at local level since they might be sparse and numerically insufficient. In other words, if a nationally representative sample may be sufficiently large to detect a relatively low (but geographically homogeneous) design prevalence, it may be inefficient for detecting a disease with a geographically heterogeneous presentation. In localised subpopulations, the scarce number of available animals to be shot may be insufficient to detect the disease. As explained in the 2017 EFSA opinion (EFSA BIOHAZ Panel, 2017a), using a representative sample of primary sampling units (multiple, biologically relevant spatial units within each country/region) and the convenience sampling of targeted high‐risk animals within them maximises the sensitivity of the surveillance both at national and local levels.
As described in the 2010 opinion (EFSA BIOHAZ Panel, 2010), the sampling conducted to support the 2006–2010 EU CWD survey was not totally random. However, practical limitations on sampling were compounded by additional assumptions that served to focus efforts mainly on two species, red deer and WTD, because of perceived risk, namely, known susceptibility, import from North America, or widespread supplemental feeding (EFSA, 2004a). However, the result was that relatively few reindeer (n = 76) and moose (n = 266) were examined.
On the other hand, to prevent a new incursion of disease passing undetected due to the temporal dynamics of CWD, a long‐term monitoring effort must be employed. The results of the 2006–2010 EU survey may have had a negative effect in justifying its discontinuation and in decreasing the attention to the potential for the CWD introduction and spread. The EU survey was based on a set of well‐defined criteria that, even if not totally adequate, entitled MS to carry out a systematic monitoring activity. After the discontinuation, apart from passive surveillance, only voluntary monitoring limited and geographically restricted to few MS has been carried out as described in the current opinion (see Table A.2 in Appendix A). In particular, between 2011 and 2014 the testing activity was low; in 2015 only two MS reported test results on cervids and in 2016, despite the news from Norway (see Appendix A), only seven MS carried out some monitoring with circa 2,700 animals tested EU‐wide (mostly in Romania).
Table A.2.
Results of the TSE surveillance in cervids in the EU and EFTA countries in 2015 and 2016 (source: 2015 and 2016 TSE EU Summary Reports) ex cluding Norway
| Wild/semi‐domesticated | Captive/farmed | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Country | Wild Eurasian tundra reindeer | Semi‐domesticated Eurasian tundra reindeer | Finnish (Eurasian) forest reindeer | Moose | Roe deer | White‐tailed deer | Red deer | Fallow deer | Eurasian tundra reindeer | Finnish (Eurasian forest reindeer) | Moose | Roe deer | White‐tailed deer | Red deer | Fallow deer | Other | Unknown |
| AT | 1 | 1 | |||||||||||||||
| EE | 1 | ||||||||||||||||
| FI | 5/3a | 26/6a | 7 | 12/4a | 0/1a | 6 | |||||||||||
| HU | 6 | 1 | 1 | 1 | |||||||||||||
| IT | 49 | 24 | 1 | ||||||||||||||
| RO | 459 | 7 | 2,034 | ||||||||||||||
| SE | 60 | 7 | 3 | ||||||||||||||
| IS | 14 | ||||||||||||||||
2016 data/2015 data. If one single figure, it refers to 2016. Empty cell means no testing.
With regard to the advantages provided by a targeted surveillance strategy, experiences with CWD in North America (Miller et al., 2000; Samuel et al., 2003; Walsh and Miller, 2010), and ruminant TSE in Europe (Doherr et al., 1999; Hagenaars et al., 2010; Arnold and Ortiz‐Pelaez, 2014; EFSA BIOHAZ Panel, 2014) underscore the value of focusing detection‐motivated surveillance on potentially symptomatic animals or, somewhat more generically, on ‘fallen stock’ (that show the highest prevalence among the tested animals). This approach was reviewed in detail recently (EFSA BIOHAZ Panel, 2017a). The first reindeer, the first moose and the only red deer detected in Norway came from these ‘high risk’ sample sources, underscoring the merits of focusing future surveys on this approach for enhancing detection probability in areas where CWD may occur.
When considering the most efficient selection of tissues to be collected, variations in patterns of tissue PrPSc deposition across CWD cases encountered in Europe and elsewhere illustrate why both lymphoid and brainstem tissues should be collected and screened wherever feasible. For North American‐type CWD, single‐tissue screening results in relatively modest compromises to DSe among apparently healthy animals and which are minimised when surveillance focuses on fallen or suspect animals. But limited experiences in Europe thus far illustrate the merits of screening both lymphoid and brainstem tissues. Sampling only brainstem, as done for the 2006–2010 European CWD survey, could reduce sensitivity. This would be especially problematic in samples from young, apparently healthy animals. Alternatively, sampling only lymphoid tissue also could reduce sensitivity or, in cases like the Norwegian moose and red deer, could preclude diagnosis altogether. Based on available data, the 2006–2010 focus on brainstem samples may have offered sensitivity of 57% for reindeer‐type cases in a sample of apparently healthy animals (4/7), but would not have missed cases in symptomatic Norwegian reindeer‐type cases (1/1) and symptomatic Norwegian moose‐type cases (1/1).
An unresolved source of uncertainty is linked to the potential role of the still unknown strains that may have a different anatomical distribution of the PrPSc accumulation, or altered test performance, leading to unpredictable consequences for the DSe similar to the issues encountered with atypical/Nor98 scrapie (see Section 3.1).
4. Answers to the ToRs
4.1. Answer to ToR 4
Are the conclusions and recommendations in the EFSA opinion of June 2004 on diagnostic methods for CWD still valid? If not, an update should be provided.
The conclusions of the 2004 EFSA scientific opinion were as follows:
All rapid tests listed in this report would appear able to detect a case of TSE in European cervids in a properly defined surveillance program. However, since these tests have not been validated for European cervids it is not possible to recommend specific test(s).
Evaluation of data submitted for six tests suggest that these tests could also be used for screening for CWD in European cervid species. IHC and Western blotting tests should be used to confirm a diagnosis of CWD. Precise evaluation of the specificity and sensitivity of these tests for CWD is impossible in the context of the European cervid populations because of the paucity of positive samples.
At present, biological strain typing by transmission to laboratory rodents is the only definite method which could differentiate between CWD and BSE/scrapie. Current molecular and IHC methods show potential to differentiate these diseases but require further evaluation; ideally both biological and molecular methods require prior validation using tissues from TSE‐infected cervids of European species.
Experimental studies are essential to understand the pathogenesis and to ascertain tissue infectivity of TSEs in European cervids before large scale surveillance could be expected to give reliable results. Such experimental studies should start in parallel with any planned surveillance.
The recommendations of the 2004 EFSA scientific opinion were as follows:
-
To initiate as soon as possible an EU‐wide experimental screening, targeting at‐risk groups of animals, using a rapid test and confirmatory methods. Initially, such a survey should focus on farmed deer and fallen stock cervid species in Europe older than 18 months, in particular targeting:
Red deer (Cervus elaphus elaphus); due to their close genetic relationship to Rocky Mountain elk (Cervus elaphus nelsoni) it is most likely that their PrPres distribution may be found to be identical.
White tailed deer (Odocoileus virginianus) population e.g. in Finland and Sweden.
Animals likely to have been exposed to BSE and/or scrapie in regions or countries (e.g. Britain) where BSE or scrapie appeared at high level and where farmed deer are known to have been given compound feed.
Farmed and free ranging cervids with observed neurological symptoms, sick or in poor condition.
-
In addition, an EU wide survey:
Should include all forms of TSE and not only focus on CWD.
Needs statistical planning, e.g. as expressed in the SSC opinion on “Requirements for statistically authoritative BSE/TSE surveys” (SSC, 2001 ).
Should seek to match a cut‐off value of prevalence of at least 0.5% (for risk populations) or at least 1% for other populations. Such detection limits were sufficient in North America (e.g. Wisconsin) to detect CWD, even though the disease is endemic only in a certain region.
Implies different sampling techniques are needed for selected locations and countries and that the accessibility to the potential animals to be tested has to be considered.
Needs in parallel, research‐based data to be collected on the pathogenesis of TSEs in European cervids and the tissue distribution of PrP. Therefore, it would be advisable to initiate experimental inoculation studies (in vitro and in vivo) and “natural route” transmission studies including introduction of sentinel animals to known ‘infected’ cervid herds. These studies could also include genetic analysis of prion protein genes from European cervids.
Testing for PrP res in cervids is an essential step in detecting and potentially in controlling the disease. Sampling is a key event and because there might be differences in sensitivity and specificity between the materials sampled and in the tissue distribution of PrP, it is recommended that both retropharyngeal lymph nodes (cortex area) and brain stem samples (including dorsal vagus nucleus) are included in the testing.
To ensure proper sampling it is advisable that the entire head of animals is sent to veterinary laboratories with a pathology unit.
Although biological strain typing is currently the only method which could possibly differentiate between CWD and BSE/scrapie, it is recommended to further support and/or initiate research on molecular methods for discrimination of the different TSEs. Ideally both biological and molecular methods would require prior validation using tissues from cervids of European species infected experimentally with TSEs.
Existence of genetic resistance, as is the case in scrapie in sheep, should be further explored.
lthough some tests have been validated in North America for use in CWD, this information may only be regarded as one basis for a survey for TSE in cervids of Europe. The cervid species to be tested in Europe are not all closely related to the North American cervid species.
Even though human TSE‐exposure risk through consumption of game from European cervids can be assumed to be minor, if at all existing, no final conclusion can be drawn due to the overall lack of scientific data. In particular the US data do not clearly exclude the possibility of human (sporadic or familial) TSE development due to consumption of venison. The Working Group thus recognizes a potential risk to consumers if a TSE would be present in European cervids. It might be prudent considering appropriate measures to reduce such a risk, e.g. excluding tissues such as CNS and lymphoid tissues from the human food chain, which would greatly reduce any potential risk for consumers. However, it is stressed that currently, no data regarding a risk of TSE infections from cervid products are available.
The conclusions and recommendations of the 2004 EFSA opinion are updated as follows:
The conclusions (1, 2, 3, 4) and recommendations (3, 5, 6, 7) of the 2004 EFSA opinion on diagnostic methods for CWD remain valid.
The available data on the performance of authorised RT for the detection of CWD in cervids in NA are not comprehensive and are much more limited than those supporting their validated use for the detection of BSE in cattle and scrapie in sheep.
For the Bio‐Rad RT and the IDEXX RT, the evaluation undertaken in North America relates primarily to the testing of lymphoid tissue, and data on the source of samples used for these evaluations (with regard to host species, genotype, stage of pathogenesis and strain) are incomplete. Both of these tests were fully evaluated prior to use in the EU statutory surveillance programmes for cattle and small ruminants. Equivalent data do not exist for cervids. However, the data available in North America for both the Bio‐Rad RT and the IDEXX RT would fulfil the criteria for OIE provisional recognition.
While validation requirements for sample size to achieve provisional recognition are lower for tests being applied to wildlife populations, it is still not possible to demonstrate that any test has met the requirements for provisional recognition in Europe.
The current lack of sufficient information on the strain(s) that might be circulating in Europe and the absence of a panel of representative samples make it unfeasible to estimate the DSe of any test for European cervid samples.
No direct comparison of test performance (i.e. parallel testing on the same panel of samples) can be made from the data available so there is no possibility to identify any differences between the two RT available on the market.
To obtain the greatest surveillance sensitivity possible, samples should be collected by trained personnel, and both brainstem and lymphoid tissue should be tested from each animal.
The experience in Norway so far shows that the Bio‐Rad RT (TeSeE™ SAP) has detected cases of CWD in reindeer, moose and red deer. It has also been shown that the antibodies raised against the core or C‐terminal parts of the prion protein used for IHC and WB were able to detect and confirm these cases.
Established immunoblotting techniques can discriminate between experimental cervid BSE and CWD. However, there is only limited information on the biological and molecular characteristics that define different strains in the North American cervid population against which the European isolates could be classified.
Experimental inoculations in a range of relevant cervid species would inform directly on disease pathogenesis, strain characterisation and host susceptibility. However, they are not easy to perform in non‐domesticated species. They would require the maintenance of experimentally infected individuals in high biosafety facilities for a long period of time, and would raise practical and welfare issues for such animals.
The surveillance programme proposed in the 2017 EFSA opinion supersedes the specifications of the EU‐wide survey proposed in the 2004 EFSA opinion. However, the finding of the first case of CWD in red deer in Norway means that the surveillance scheme as in Reg. 999/2001,7 as amended, no longer covers all the MS in which cervid species found to be susceptible to a strain of CWD present in Europe are located.
Since 2004, very little additional data have been gathered on cervid PRNP gene polymorphisms in Europe. It is essential to acquire such information to better understand species and strain susceptibility, to underpin robust targeted surveillance and to inform future control strategies for CWD in farmed cervids.
The tissue distribution of infectivity in CWD‐infected cervids is now known to extend beyond CNS and lymphoid tissues. While the removal of these specific tissues from the food chain would reduce human dietary exposure to infectivity, exclusion from the food chain of the whole carcass of any infected animal would be required to eliminate human dietary exposure.
4.2. Answer to ToR 5
EFSA is asked to update the conclusions of the 2010 EFSA opinion on the results of the EU survey on CWD in cervids, as regards the occurrence of CWD in the cervid population in the EU.
The conclusions of the 2010 EFSA scientific opinion were as follows:
The EU CWD/TSEs survey carried out in 2006–2010 was designed taking into account recommendations from an earlier 2004 EFSA opinion.
Approximately 13,000 brain stem samples were collected from cervids of different species in 21 EU Member States and Norway. No TSE positive results were found.
The lack of one positive TSE test in the farmed and wild red deer and wild white‐tailed deer which were sampled indicates that there is not a cervid TSE epidemic in the EU.
Since the assumption of a random sampling is not fulfilled, a quantitative estimate of the true prevalence with confidence intervals has limitations and needs to be interpreted with care. This is because, with regards to TSEs in cervids, the true prevalence may be different in presently unsampled areas.
Considering the practical issues inherent to collection of samples in EU wild cervids, achieving a survey that would allow a quantitative estimate of the true prevalence of TSEs in these species in the EU would remain extremely difficult
Considering the spreading of CWD within and from clusters in North America, the limitations of the sampling performed in the EU CWD/TSEs survey and the known susceptibility of certain cervid species to CWD, occurrence of cases of TSEs, especially in remote and presently unsampled geographic areas, may not be excluded in cervids in the EU.
The recommendations of the 2010 EFSA scientific opinion were as follows:
Further experimental studies should be considered to assess the susceptibility of the various European cervid species to CWD and other TSE agents.
PrP genetic diversity of EU cervid species should be investigated and compared to data described in North American cervid population.
Testing for TSEs should be incorporated as part of wildlife disease monitoring programmes for cervids carried out in different EU Member States in order to ensure a continuous vigilance on the possible emergence of TSEs in EU cervids.
The design of monitoring programmes for TSEs in cervids should take into account both the objectives to be achieved and the new scientific knowledge available (risk categories, test sensitivity, target tissue, age of the animals, susceptibility of North American moose etc.).
The conclusions and recommendations of the 2010 EFSA scientific opinion are updated as follows:
The conclusions (1, 2, 4, 5, 6) and all recommendations (1, 2, 3, 4) of the 2010 EFSA opinion remain valid.
Cases of CWD have now been detected in wild Norwegian reindeer, moose and red deer, confirming the long‐held suspicion that at least some European cervid species are naturally susceptible.
The 2010 EFSA opinion concluded that available monitoring data did not support the presence of an ‘epidemic’, without providing a clear definition of the term. Current available data do not preclude the possibility that CWD was present in Norway and perhaps elsewhere in Europe before the 2006–2010 EU CWD survey was conducted, whether in epidemic form or not.
Shortcomings in both the design and subsequent implementation of the 2006–2010 EU CWD survey limited the reliability of inferences that could be made about the potential occurrence of CWD in Europe, including the lack of positive tests in red deer and WTD being extrapolated in the conclusion to all cervids, which has now been challenged by the finding of CWD in two other species, reindeer and moose, as well as in red deer.
Following the completion of the 2006–2010 EU survey, and despite the very low testing activity in Europe, CWD was detected for the first time in a diseased deer in Norway, highlighting the importance of risk‐based surveillance. This finding has triggered a substantial increase in the testing effort in this country in 2016 and 2017.
Comparing the point estimate of CWD prevalence among ‘adult’ (> 1 year old) reindeer harvested in Nordfjella 1 in 2016 and during 2017 to an epidemic curve for mule deer in the US, it appears plausible that CWD could have been established in Norway more than a decade ago.
The 2017 EFSA opinion surveillance recommendations were intended to improve the probability of detecting additional foci of CWD, should they be present in Norway or in a number of predefined European countries. Given compliance, the recommendations (EFSA BIOHAZ Panel, 2017a), focusing on the sampling of high‐risk individuals, and developing spatial sampling frameworks relevant to the populations being monitored, will improve the reliability and value of data arising from the CWD surveillance.
Very few studies have addressed the genetic diversity of the PRNP gene in cervid populations of Europe. It appears that the genetic diversity is very much linked to the deer species, with reindeer and red deer more diverse than others. However, the available data, drawn from a limited number of animals and geographical sources precludes any conclusion on the presence and frequency of polymorphic alleles in the PRNP gene of European cervids.
There are at least two distinct CWD ‘strains’ described in North America, and possibly two distinct TSE strains within the small number of cases detected to date in Europe, one of them, in moose and red deer, seemingly distinguishable from the strains described in North America. The results of the ongoing rodent model isolation and characterisation studies will enable these isolates to be classified in the context of other naturally occurring TSE.
5. Recommendations
To incorporate sampling and testing for CWD into any wildlife surveillance programmes. Such programmes would need to take into account the knowledge gained in the CWD field and apply it to the surveillance strategies as suggested by OIE. In particular, surveillance should focus on clinical suspects and other high‐risk animals.
To increase awareness and to disseminate information about CWD in appropriate forums in the EU in order to improve the reporting of suspect cases.
To use only trained personnel for sample collection, to ensure accurate, optimised sampling of the target tissues, while minimising sampling‐related human exposure and potential environmental contamination.
To avoid any test or detection method that uses antibodies for which the epitope is known to be polymorphic in cervids, unless successful binding in positive animals can be demonstrated.
To retain residual samples, including relevant metadata, from all positive animals and from as many tissues as possible, for isolate classification and future test evaluation, epidemiology, or research purposes.
To design and conduct ad hoc studies on the European CWD outbreaks to identify any relevant differences influencing the epidemiology of the disease, including transmissibility, species susceptibility and strain variability, compared to that of North American CWD.
To conduct surveys to investigate the presence and frequency of polymorphisms potentially associated with resistance in the European cervid population.
To keep the performance of all tests under review, and revise testing protocols as required, in the context of emerging data on European‐specific host/strain combinations, and any related information on species susceptibility and tissue distribution of disease‐related PrP.
Abbreviations
- Ab
antibody
- ASe
analytical sensitivity
- ASp
analytical specificity
- BIOHAZ
EFSA Panel on Biological Hazards
- brELISA
Bio‐Rad ELISA
- BSE
bovine spongiform encephalopathy
- CC
conjugate concentrate
- CFIA
Canadian Food Inspection Agency
- CNS
central nervous system
- CRL
Community reference laboratory
- CWD
chronic wasting disease
- DSe
diagnostic Sensitivity
- DSp
diagnostic Specificity
- EEA
European Economic Area
- EIA
Enzyme‐linked immunoassay
- ELISA
enzyme‐linked immunosorbent assay
- EQA
External Quality Assurance
- EURL
European Reference Laboratory
- Gdn.HCl
guanidine hydrochloride
- i.c.
intracerebral inoculation
- IFU
Instruction for use
- LD50
lethal dose 50%
- LN
lymph node
- LRS
lymphoreticular system
- mAb
monoclonal antibodies
- MS
Member state
- NA
North America
- NRL
National reference laboratory
- OIE
International Organization of Animal Health
- p.o.
Per os (oral inoculation)
- PCR
polymerase chain reaction
- PMCA
protein misfolding cyclic amplification
- PRNP
gene encoding for the major prion protein PrP
- PrP
host‐encoded prion protein
- PrPc
cellular PrP
- PrPCWD
CWD‐associated prion
- PrPres
proteinase‐K resistant prion protein
- PrPSc
abnormal disease‐associated isoform of the protein PrP
- RAMALT
rectoanal mucosa‐associated lymphoid tissue
- RPLN
retropharyngeal lymph node
- RT
rapid test
- RT‐QuIC
real‐time quaking‐induced conversion
- sPMCA
serial PMCA
- SSC
Scientific Steering Committee of the EC
- ToR
Term of reference
- TSE
transmissible spongiform encephalopathy/encephalopathies
- WB
western Blot
- WG
Working group
- WTD
white‐tailed deer
Appendix A – TSE surveillance data in cervids in Europe (2015–2017)
Appendix B – Validation of diagnostic tests
Stages of validation
Stage 1 of the validation is the assessment of the analytical performance of the assay (benchmark parameters) and includes studies on repeatability, ASe (limit of detection of the analyte) and ASp.
Stage 2 estimates the diagnostic performance of the assay. The diagnostic sensitivity (DSe) is the proportion of samples from known infected reference animals that test positive in an assay and the diagnostic specificity (DSp) is the proportion of samples from known uninfected reference animals that test negative in an assay. These estimates result from testing a panel of samples from reference animals with known history and infection status for the particular disease and from the region in which the test will be applied. The number of known positive and known negative samples to be tested in Stage 2 will depend on the likely values of DSe and DSp of the candidate test under validation and the desired confidence level for the estimates.
Provisional recognition can be given to an assay that has been evaluated through Stage 1 for critical assay benchmark parameters (ASe, ASp and repeatability) and also for which preliminary estimate of DSp and DSe has been obtained based on a small panel of reference samples, representing partial completion of Stage 2. Provisional recognition of an assay means that the test has not been fully evaluated for diagnostic performance characteristics.
Stage 3 evaluates the reproducibility (the ability of the test to provide consistent results when applied in different laboratories) and augmented repeatability (the level of agreement between results of replicates of a sample within and between runs in a given laboratory) estimates. Measurements of precision can be estimated for the reproducibility and the repeatability data.
Once Stage 3 of the validation pathway is completed, and if the earlier stages have been entirely and satisfactorily completed, the assay may be designated as ‘validated for the original intended purpose’. Continuous monitoring of the test performance is necessary in order to retain its designation as validated.
Stage 4 consists of programme implementation of the assay and interpretation of tests results. The successful utilisation of the test confers additional and important evidence for its performance in accordance to the expectations. Interpretation of test results (predictive values) is dependent on knowledge of prevalence, i.e. the true prevalence of the true diagnostic status in the targeted population.
International recognition of an assay: assays are recognised internationally by the OIE when they are designated as prescribed or alternate tests for trade purposes, which is frequently based on evidence of their value on a national, regional or international basis. The OIE Register8 lists commercial diagnostic kits that have gone through the OIE procedure for validation and certification of diagnostic assays. Tests listed in the OIE Register are certified as fit for a specific purpose if they have completed Validation Stages 1, 2 and 3. The OIE Register includes 2 listed TSE tests: the Prionics AG ‐ Check Western and the TeSeE™ Western Blot (Bio‐Rad).
In the 2002 Opinion of the Scientific Steering Committee (SSC) on design of a field trial for the evaluation of new rapid BSE post mortem tests (SSC, 2002a), it was recommended for the estimation of sensitivity relative to approved tests that 200 true positive samples should be tested by a new RT compared to approved tests, which would ensure with a 95% probability that the sensitivity of the new RT is not below 98.5% compared with the approved test. A total of 10,000 consecutive samples from healthy slaughtered animals that are tested negative using an approved test should be used for the estimation of specificity in comparison with approved tests.
In an EC Scientific Steering Committee report on a programme for the evaluation of rapid post‐mortem tests to detect TSE in small ruminants (SSC, 2002b), the recommendation of the OIE on the sample size for the qualitative evaluation of diagnostic tests: 300 positives and 1,000 negatives is acknowledged. This allows a sensitivity of > 99.2% assuming the test screens a population with at least 1000 positive animals. A 100% success in testing 1,000 samples leads to a specificity of 99.8% within a 95% confidence range.
Validation of tests for use in wild populations
Diagnostic testing of wildlife can be more challenging than in domestic animals for various reasons, for example, little or no existing information (i.e. no previous screenings), difficulties in acquiring samples, poor sample representativeness (i.e. season, sex, age), poor sample quality (i.e. autolysis), or poor knowledge of epidemiology and pathogenesis of infectious diseases in a particular wildlife species. Recognising that completion of the validation process may not be possible, guidance is given for following the validation pathway to a point where the test can be provisionally accepted to provide confidence in results and for use in specific applications in a regional or national context.
A description of test validation specific for wildlife species is presented in the OIE Terrestrial Manual Chapter 3.6.7, ‘Principles and methods for the validation of diagnostic tests for infectious diseases applicable to wildlife’ (OIE, 2017a). The direct pathogen identification techniques should in theory not be affected by the species of the host. However, there are some host‐species variation in the pathobiology of infections affecting for example the amount and distribution of pathogens in the tissues/organs of the hosts. Many diagnostic tests that have been developed and validated for domestic animals have not been validated for wildlife species and it remains uncertain if there are any important differences in their DSe and DSp when applied to samples from wildlife species. Thus, in cases in which full validation is not feasible, the best possible alternative may be to determine the fitness of the assay in a reduced number of reference samples of wildlife species. These initial estimates of a test performance may give sufficient information for government or regional authorities to accept that the test can be provisionally recognised for application in wild animals, for example for pathogen surveillance within a country or region.
More specifically, according to the OIE standard for validation of tests applicable to wildlife, either a new test needs to be developed for the wildlife species or a pre‐existing test that has been validated for one species may be adapted to and evaluated in other related species with minor or no modifications. In all cases, the intended purpose and application of the test needs to be established and defined before it is developed and validated since this may affect the selection of suitable reference samples and the validation results.
The two scenarios (Table B.1 and Figure B.1) described in the Standard are: the lack of availability (Pathway 1) or the availability of a validated test in another related species (Pathway 2) for the same disease agent. Taxonomic relatedness of species is of primary importance when selecting pathway 1 or 2. The OIE guidelines do not specify the level of taxonomic relatedness that is needed in order to select Pathway 2. Thus, it is not stated if the relatedness should be at the family level (for example, for CWD tests family Cervidae), at the subfamily level (for CWD tests: Cervinae and Capreolinae (or Odocoileinae), or at the species level. However, it does point out that the taxonomic relatedness is particularly important when indirect methods (detection of antibody or cell immunity response) are used. Furthermore, other criteria, including behaviour of the species, variation in strains of pathogens or ecology of the diseases, should also be taken into account. Most often for tests to be applied to wildlife, Pathway 1 is appropriate because of the lack of validated tests in closely related species. If Pathway 2 is selected, justification needs to be provided by documenting the existence of a validated test. For the steps of Stage 1 of validation (estimation of analytical characteristics), all the characteristics of the Stage 1 should be evaluated in the Pathway 1 while in Pathway 2 repeatability and preliminary reproducibility will not need to be reassessed until Stage 3. Stage 2 (estimation of diagnostic characteristics) is divided into Stage 2a and Stage 2b. For Stage 2a in Pathway 1, a minimum of 30 positive reference samples and 30 negative reference samples need to be tested, while for Pathway 2a minimum of 10 positive reference samples and 10 negative reference samples are required. Fulfilment of Stage 2a of the validation pathway can provide enough information for conferring the test provisional recognition, so that the test can start to be applied to the wildlife population. As more data are collected from the testing, the confidence of the results increases.
Table B.1.
Validation pathway: steps required to meet validation criteria described in the OIE Validation Standard and to estimate test characteristics. Requirements in the different stages need to be fulfilled with an acceptable outcome
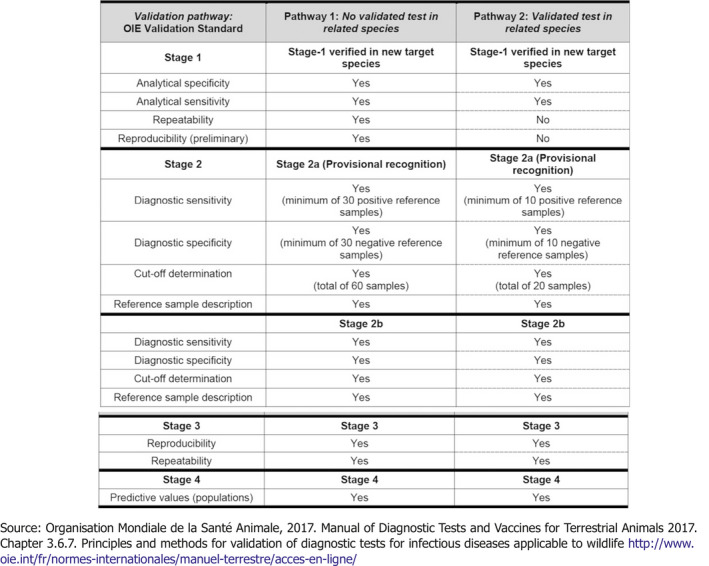
Figure B.1.
Flow chart of pathways and stages of test validation in wildlife when a previously validated test exists or does not exist
- Source: Organisation Mondiale de la Santé Animale, 2017. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2017. Chapter 3.6.7. Principles and methods for validation of diagnostic tests for infectious diseases applicable to wildlife http://www.oie.int/fr/normes-internationales/manuel-terrestre/acces-en-ligne/
As for domestic animals, to complete Stage 2 (Stage 2b) the sample size needs to be selected based on expected values for DSe and DSp, the desired confidence level and error margin.
For the application of diagnostic tests for CWD in Europe, it should be recognised that it is not currently possible to validate the tests fulfilling Stage 2 of the validation pathway because appropriate positive reference samples from the target population are scarce (fewer than 10 cases of CWD have so far been diagnosed in Europe in a single species) or not available for all the species to be tested (for example, positive samples from roe deer since CWD has never been detected in roe deer).
Appendix C – Summary data on the performance of rapid tests for the detection of CWD
Suggested citation: EFSA Panel on Biological Hazards (BIOHAZ) , Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Fernández Escámez PS, Gironés R, Herman L, Koutsoumanis K, Lindqvist R, Nørrung B, Robertson L, Ru G, Sanaa M, Skandamis P, Snary E, Speybroeck N, Kuile BT, Threlfall J, Wahlström H, Benestad S, Gavier‐Widen D, Miller MW, Telling GC, Tryland M, Latronico F, Ortiz‐Pelaez A, Stella P and Simmons M, 2018. Scientific opinion on chronic wasting disease (II). EFSA Journal 2018;16(1):5132, 59 pp. 10.2903/j.efsa.2018.5132
Requestor: European Commission
Question number: EFSA‐Q‐2016‐00411
Panel members: Antonia Ricci, Ana Allende, Declan Bolton, Marianne Chemaly, Robert Davies, Pablo Salvador Fernández Escámez, Rosina Gironés, Lieve Herman, Kostas Koutsoumanis, Roland Lindqvist, Birgit Nørrung, Lucy Robertson, Moez Sanaa, Panagiotis Skandamis, Emma Snary, Niko Speybroeck, Benno Ter Kuile, Giuseppe Ru, Marion Simmons, John Threlfall and Helene Wahlström.
Adopted: 6 December 2017
Reproduction of the images listed below is prohibited and permission must be sought directly from the copyright holder:
Figure 3: © USGS; Table B.1; Figure B.1: © OIE
Notes
Commission Decision of 19 March 2007 on a survey for chronic wasting disease in cervids. OJ L 84/37 24.3.2007.
Regulation (EC) No 999/2001 of the European Parliament and of the Council of 22 May 2001 laying down rules for the prevention, control and eradication of certain transmissible spongiform encephalopathies. OJ L 147, 31.5.2001, p. 1–40.
Commission Regulation (EC) No 36/2005 of 12 January 2005 amending Annexes III and X to Regulation (EC) No 999/2001 of the European Parliament and of the Council as regards epidemio‐surveillance for transmissible spongiform encephalopathies in bovine, ovine and caprine animals. OJ L 10, 13.1.2005, p. 9–17. TSE strain characterisation in small ruminants a technical handbook for national reference laboratories in the EU. https://science.vla.gov.uk/tse-lab-net/documents/tse-oie-rl-handbook.pdf
Commission Regulation (EU) 2017/1972 of 30 October 2017 amending Annexes I and III to Regulation (EC) No 999/2001 of the European Parliament and of the Council as regards a surveillance programme for chronic wasting disease in cervids in Estonia, Finland, Latvia, Lithuania, Poland and Sweden and repealing Commission Decision 2007/182/EC.
References
- Angers RC, Seward TS, Napier D, Green M, Hoover E, Spraker T, O'Rourke K, Balachandran A and Telling GC, 2009. Chronic wasting disease prions in elk antler velvet. Emerging Infectious Diseases, 15, 696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers RC, Kang HE, Napier D, Browning S, Seward T, Mathiason C, Balachandran A, McKenzie D, Castilla J, Soto C, Jewell J, Graham C, Hoover EA and Telling GC, 2010. Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science, 328, 1154–1158. 10.1126/science.1187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M and Ortiz‐Pelaez A, 2014. The evolution of the prevalence of classical scrapie in sheep in Great Britain using surveillance data between 2005 and 2012. Preventive Veterinary Medicine, 117, 242–250. [DOI] [PubMed] [Google Scholar]
- Arnold ME, Ryan JBM, Konold T, Simmons MM, Spencer YI, Wear A, Chaplin M, Stack M, Czub S, Mueller R, Webb PR, Davis A, Spiropoulos J, Holdaway J, Hawkins SAC, Austin AR and Wells GAH, 2007. Estimating the temporal relationship between PrPSc detection and incubation period in experimental bovine spongiform encephalopathy of cattle. Journal of General Virology, 88, 3198–3208. [DOI] [PubMed] [Google Scholar]
- Atarashi R, Wilham JM, Christensen L, Hughson AG, Moore RA, Johnson LM, Onwubiko HA, Priola SA and Caughey B, 2008. Simplified ultrasensitive prion detection by recombinant PrP conversion with shaking. Nature Methods, 5, 211–212. 10.1038/nmeth0308-211 [DOI] [PubMed] [Google Scholar]
- Baeten LA, Powers BE, Jewell JE, Spraker TR and Miller MW, 2007. A natural case of chronic wasting disease in a free‐ranging moose (Alces alces shirasi). Journal of Wildlife Diseases, 43, 309–314. [DOI] [PubMed] [Google Scholar]
- Balachandran A, Harrington NP, Algire J, Soutyrine A, Spraker TR, Jeffrey M, González L and O'Rourke KI, 2010. Experimental oral transmission of chronic wasting disease to red deer (Cervus elaphus elaphus): early detection and late stage distribution of protease‐resistant prion protein. The Canadian Veterinary Journal 51, 169. [PMC free article] [PubMed] [Google Scholar]
- Baron T, Biacabe AG, Arsac JN, Benestad S and Groschup MH, 2007. Atypical transmissible spongiform encephalopathies (TSEs) in ruminants. Vaccine, 25, 5625–5630. [DOI] [PubMed] [Google Scholar]
- Bartz JC, Marsh RF, McKenzie DI and Aiken JM, 1998. The host range of chronic wasting disease is altered on passage in ferrets. Virology, 251, 297–301. 10.1006/viro.1998.9427 [DOI] [PubMed] [Google Scholar]
- Baszler TV, Kiupel M, Williams ES, Thomsen BV, Gidlewski T, Montgomery DL, O'Rourke KI and Hall SM, 2006. Comparison of two automated immunohistochemical procedures for the diagnosis of scrapie in domestic sheep and chronic wasting disease in North American white‐tailed deer (Odocoileus virginianus) and mule deer (Odocoileus hemionus). Journal of Veterinary Diagnostic Investigation 18, 147–155. [DOI] [PubMed] [Google Scholar]
- Beck KE, Sallis RE, Lockey R, Simmons MM and Spiropoulos J, 2010. Ovine PrP genotype is linked with lesion profile and immunohistochemistry patterns after primary transmission of classical scrapie to wild‐type mice. Journal of Neuropathology and Experimental Neurology, 69, 483–497. [DOI] [PubMed] [Google Scholar]
- Belay ED, Maddox RA, Williams ES, Miller MW, Gambetti P and Schonberger LB, 2004. Chronic wasting disease and potential transmission to humans. Emerging Infectious Diseases 10, 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benestad S, 2017a. Re: lymphoid tissues. Message to Angel Ortiz Pelaez. 30 October 2017.
- Benestad S, 2017b. Re: Antibodies.pptx. Message to Angel Ortiz Pelaez. 30 October 2017.
- Benestad S, 2017c. Re: new table in version 5.1. Message to Angel Ortiz Pelaez. 24 November 2017.
- Benestad S and Gavier‐Widen D, 2006. Results of the European proficiency testing for the diagnosis of CWD. Chronic wasting disease (CWD): current knowledge and European perspective‐2006. Torino, 3 October 2006. Available online: http://www.neuroprion.org/resources/pdf_docs/conferences/prion2006/abstract_book.pdf (Accessed: November 2017)
- Benestad SL, Sarradin P, Thu B, Schönheit J, Tranulis MA and Bratberg B, 2003. Cases of scrapie with unusual features in Norway and designation of a new type, Nor98. The Veterinary Record, 153, 202–208. [DOI] [PubMed] [Google Scholar]
- Benestad SL, Mitchell G, Simmons M, Ytrehus B and Vikøren T, 2016. First case of chronic wasting disease in Europe in a Norwegian free‐ranging reindeer. Veterinary Research, 47, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasche T, Schenck EV, Balachandran A, Miller MW, Langenberg J, Frolich K and Steinbach F, 2012. Rapid detection of CWD PrP: comparison of tests designed for the detection of BSE or scrapie. Transboundary and Emerging Diseases, 59, 405–415. [DOI] [PubMed] [Google Scholar]
- Browning SR, Mason GL, Seward T, Green M, Eliason GA, Mathiason C, Miller MW, Williams ES, Hoover E and Telling GC, 2004. Transmission of prions from mule deer and elk with chronic wasting disease to transgenic mice expressing cervid PrP. Journal of Virology, 78, 13345–13350. 10.1128/JVI.78.23.13345-13350.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce ME and Dickinson AG, 1979. Biological stability of different classes of scrapie agent. In: Prusiner SB and Hadlow WJ (eds.). Slow Transmissible Diseases of the Nervous System, Vol. 2, Academic Press, New York. pp. 71–86. [Google Scholar]
- Bruce ME and Fraser H, 1991. Scrapie strain variation and its implications. Scrapie strain variation and its implications. Current Topics in Microbiology and Immunology, 172, 125–137. [DOI] [PubMed] [Google Scholar]
- Bruce ME, McBride PA and Farquhar CF, 1989. Precise targeting of the pathology of the sialoglycoprotein, PrP, and vacuolar degeneration in mouse scrapie. Neuroscience Letters, 102, 1–6. [DOI] [PubMed] [Google Scholar]
- Bruce M, Chree A, McConnell I, Foster JP, Pearson G and Fraser H, 1994. Transmission of bovine spongiform encephalopathy and scrapie to mice: strain variation and the species barrier. Philosophical Transactions of the Royal Society of London B: Biological Sciences, 343, 405–411. [DOI] [PubMed] [Google Scholar]
- Bruce ME, Boyle A, Cousens S, McConnell I, Foster J, Goldmann W and Fraser H, 2002. Strain characterization of natural sheep scrapie and comparison with BSE. Journal of General Virology, 83, 695–704. [DOI] [PubMed] [Google Scholar]
- Brun A, Castilla J, Ramírez MA, Prager K, Parra B, Salguero FJ, Shiveral D, Sánchez C, Sánchez‐Vizcaíno JM, Douglas A and Torres JM, 2004. Proteinase K enhanced immunoreactivity of the prion protein‐specific monoclonal antibody 2A11. Neuroscience Research, 48, 75–83. [DOI] [PubMed] [Google Scholar]
- Bunk S, 2004. Chronic wasting disease—prion disease in the wild. PLoS Biology, 2, e121. 10.1371/journal.pbio.0020121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Miller MW, Bulgin MS, Sorenson‐Melson S, Balachandran A, Chiu A and Rubenstein R, 2008. PrP antibody binding‐induced epitope modulation evokes immunocooperativity. Journal of Neuroimmunology, 205, 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YC, Hannaoui S, John TR, Dudas S, Czub S and Gilch S, 2016. Early and non‐invasive detection of chronic wasting disease prions in elk feces by real‐time quaking induced conversion. PLoS ONE, 11, e0166187. 10.1371/journal.pone.0166187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner MM, Miller MW, Ebinger MR and Burnham KP, 2007a. A meta‐baci approach for evaluating management intervention on chronic wasting disease in mule deer. Ecological Applications, 17, 140–153. [DOI] [PubMed] [Google Scholar]
- Conner MM, Gross JE, Cross PC, Gillies R, Samuel MD and Miller MW, 2007b. Scale‐dependent approaches to modeling spatial epidemiology of chronic wasting disease. Utah Division of Wildlife Resources [Google Scholar]
- Dagleish MP, Martin S, Steele P, Finlayson J, Sisó S, Hamilton S, Chianini F, Reid HW, González L and Jeffrey M, 2008. Experimental transmission of bovine spongiform encephalopathy to European red deer (Cervus elaphus elaphus). BMC Veterinary Research, 4, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagleish MP, Martin S, Steele P, Finlayson J, Eaton SL, Sisó S, Stewart P, Fernandez‐Borges N, Hamilton S, Pang Y, Chianini F, Reid HW, Goldmann W, Gonzalez L, Castilla J and Jeffrey M, 2015. Susceptibility of European red deer (Cervus elaphus elaphus) to alimentary challenge with bovine spongiform encephalopathy. PLoS ONE, 10, e0116094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake RP, Truscott TC, Özyiğit MÖ, Zhuang D, Schneider DA and O'Rourke KI, 2013. Accumulation profiles of PrPSc in hemal nodes of naturally and experimentally scrapie‐infected sheep. BMC Veterinary Research, 9, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daus ML, Breyer J, Wagenfuehr K, Wemheuer WM, Thomzig A, Schulz‐Schaeffer WJ and Beekes M, 2011. Presence and seeding activity of pathological prion protein (PrP(TSE)) in skeletal muscles of white‐tailed deer infected with chronic wasting disease. PLoS ONE, 6, e18345. 10.1371/journal.pone.0018345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidowitz E, Eljuga L, Dover K, Tian J and Grossman A, 2005. Concentration of prion protein from biological samples to increase the limits of detection by immunoassay. Biotechnology and Applied Biochemistry, 41, 247–253. [DOI] [PubMed] [Google Scholar]
- De Bosschere H, Saegerman C, Neukermans A, Berkvens D, Casaer J, Vanopdenbosch E and Roels S, 2006. First chronic wasting disease (CWD) surveillance of roe deer (Capreolus capreolus) in the northern part of Belgium. Veterinary Quarterly, 28, 54–60. [PubMed] [Google Scholar]
- Di Bari MA, Nonno R, Castilla J, D'Agostino C, Pirisinu L, Riccardi G, Conte M, Richt J, Kunkle R, Langeveld J, Vaccari G and Agrimi U, 2013. Chronic wasting disease in bank voles: characterisation of the shortest incubation time model for prion diseases. PLoS Pathogens, 9, e1003219. 10.1371/journal.ppat.1003219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherr MG, Oesch B, Moser M, Vandevelde M and Heim D, 1999. Targeted surveillance for bovine spongiform encephalopathy. The Veterinary Record, 145, 672. [PubMed] [Google Scholar]
- Duque Velasquez C, Kim C, Herbst A, Daude N, Garza MC, Wille H, Aiken J and McKenzie D, 2015. Deer prion proteins modulate the emergence and adaptation of chronic wasting disease strains. Journal of Virology, 89, 12362–12373. 10.1128/JVI.02010-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority), 2004a. Opinion on a surveillance programme for Chronic Wasting Disease in the European Union. EFSA Journal 2004;2(7):70, 7 pp. 10.2903/j.efsa.2004.70 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2004b. EFSA Scientific Report (2004) 18, 1‐13 on the Evaluation of Seven New Rapid post mortem BSE Tests. EFSA Journal 2004;2(11):18r, 13 pp. 10.2903/j.efsa.2004.18r [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2005a. EFSA Scientific Report (2005) 48, 1‐10 on the Evaluation of Two Rapid post mortem BSE Tests. EFSA Journal 2005;3(9):48r, 10 pp. http://onlinelibrary.wiley.com/doi/10.2903/j.efsa.2005.48r/full [Google Scholar]
- EFSA (European Food Safety Authority), 2005b. EFSA Scientific Report (2005) 31, 1‐17 on the Evaluation of Rapid post mortem TSE Tests intended for Small Ruminants. EFSA Journal 2005;3(6):31r, 17 pp. 10.2903/j.efsa.2005.31r [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2005c. EFSA Scientific Report (2005) 49, 1‐16 on the Evaluation of Rapid post mortem TSE Tests intended for Small Ruminants. EFSA Journal 2005;3(10):49r, 16 pp. http://onlinelibrary.wiley.com/doi/10.2903/j.efsa.2005.49r/full [Google Scholar]
- EFSA (European Food Safety Authority), 2006. Scientific Report of the Scientific Expert Working Group of the European Food Safety Authority on Transmissible Spongiform Encephalopathy (TSE). Adopted on 27 November 2006. Question No EFSA‐Q‐2003‐084. EFSA Journal 2006;4(12):95, 14 pp. 10.2903/j.efsa.2006.95r [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2007a. Scientific Opinion of the Panel on Biological Hazards on a request from the European Commission on a protocol for the evaluation of rapid post mortem tests to detect TSE in small ruminants. EFSA Journal 2007;5(6):509, 31 pp. 10.2903/j.efsa.2007.509 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2007b. Scientific Opinion of the Panel on Biological Hazards on a request from the European Commission on a protocol for the evaluation of new rapid BSE post mortem tests. EFSA Journal 2007;5(6):508, 20 pp. 10.2903/j.efsa.2007.508 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2009. Scientific Opinion on ASe of approved TSE rapid tests. EFSA Journal 2009;7(12):1436, 32 pp. 10.2903/j.efsa.2009.1436 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2010. Scientific Opinion on the results of the EU survey for Chronic Wasting Disease (CWD) in cervids. EFSA Journal 2010;8(10):1861, 29 pp. 10.2903/j.efsa.2010.1861 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2011. Joint Scientific Opinion on any possible epidemiological or molecular association between TSEs in animals and humans. EFSA Journal 2011;9(1):1945, 111 pp. 10.2903/j.efsa.2011.1945 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2012. Scientific Opinion on the evaluation of new TSE rapid tests submitted in the framework of the Commission Call for expression of interest 2007/S204‐247339. EFSA Journal 2012;10(5):2660, 32 pp. 10.2903/j.efsa.2012.2660 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2014. Scientific Opinion on the scrapie situation in the EU after 10 years of monitoring and control in sheep and goats. EFSA Journal 2014;12(7):3781, 155 pp. 10.2903/j.efsa.2014.378 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2015. Scientific Opinion on a request for a review of a scientific publication concerning the zoonotic potential of ovine scrapie prions. EFSA Journal 2015;13(8):4197, 58 pp. 10.2903/j.efsa.2015.4197 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Fernandez Escamez PS, Girones R, Herman L, Koutsoumanis K, Lindqvist R, Nørrung B, Robertson L, Sanaa M, Skandamis P, Snary E, Speybroeck N, Kuile BT, Threlfall J, Wahlström H, Benestad S, Gavier‐Widen D, Miller MW, Ru G, Telling GC, Tryland M, Ortiz‐Pelaez A and Simmons M, 2017a. Scientific opinion on chronic wasting disease (CWD) in cervids. EFSA Journal 2017;15(1):4667, 62 pp. 10.2903/j.efsa.2017.4667 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Fernández Escámez PS, Gironés R, Herman L, Koutsoumanis K, Lindqvist R, Nørrung B, Robertson L, Ru G, Sanaa M, Skandamis P, Speybroeck N, Simmons M, Kuile BT, Threlfall J, Wahlström H, Acutis P‐L, Andreoletti O, Goldmann W, Langeveld J, Windig JJ, Ortiz‐Pelaez A and Snary E, 2017b. Scientific Opinion on genetic resistance to transmissible spongiform encephalopathies(TSE) in goats. EFSA Journal 2017;15(8):4962, 79 pp. 10.2903/j.efsa.2017.4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder AM, Henderson DM, Nalls AV, Wilham JM, Caughey BW, Hoover EA, Kincaid AE, Bartz JC and Mathiason CK, 2013. In vitro detection of prionemia in TSE‐infected cervids and hamsters. PLoS ONE, 8, e80203. 10.1371/journal.pone.0080203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JC, Nonno R, Di Bari M, Aguilar‐Calvo P, Pirisinu L, Fernández‐Borges N, Vanni I, Vaccari G, Marín‐Moreno A, Frassanito P, Lorenzo P, Agrimi U and Torres JM, 2016. PrPC governs susceptibility to prion strains in bank vole, while other host factors modulate strain features. Journal of Virology, 90, 10660–10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EURL (European Union Reference Laboratory (EURL) for TSE), 2016. TSE strain characterisation in small ruminants. A technical handbook for national reference laboratories in the EU. Available online: https://science.vla.gov.uk/tse-lab-net/documents/tse-oie-rl-handbook.pdf [Accessed: November 2017]
- European Commission , 1999. DG XXIV Consumer Policy and Consumer Health Protection Report on “The evaluation of tests for the diagnosis of Transmissible Spongiform Encephalopathy in bovines” on 8 July 1999.
- Faburay B, Tark D, Kanthasamy AG and Richt JA, 2014. In vitro amplification of scrapie and chronic wasting disease PrP(res) using baculovirus‐expressed recombinant PrP as substrate. Prion, 8, 393–403. 10.4161/19336896.2014.983753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth ML, Hoeting JA, Hobbs NT and Miller MW, 2006. Linking chronic wasting disease to mule deer movement scales: a hierarchical Bayesian approach. Ecological Applications 16, 1026–1036. [DOI] [PubMed] [Google Scholar]
- Fox KA, Jewell JE, Williams ES and Miller MW, 2006. Patterns of PrPCWD accumulation during the course of chronic wasting disease infection in orally inoculated mule deer (Odocoileus hemionus). Journal of General Virology, 87, 3451–3461. [DOI] [PubMed] [Google Scholar]
- Gonzalez L, Martin S and Jeffrey M, 2003. Distinct profiles of PrPd immunoreactivity in the brain of scrapie‐and BSE‐infected sheep: implications for differential cell targeting and PrP processing. Journal of General Virology 84, 1339–1350. [DOI] [PubMed] [Google Scholar]
- Gonzalez L, Jeffrey M and Siso S, 2005. Diagnosis of preclinical scrapie in samples of rectal mucosa. The Veterinary Record, 156, 846–847. [DOI] [PubMed] [Google Scholar]
- González L, Dagleish MP, Martin S, Dexter G, Steele P, Finlayson J and Jeffrey M, 2008. Diagnosis of preclinical scrapie in live sheep by the immunohistochemical examination of rectal biopsies. The Veterinary Record 162, 397–403. [DOI] [PubMed] [Google Scholar]
- Green KM, Browning SR, Seward TS, Jewell JE, Ross DL, Green MA, Williams ES, Hoover EA and Telling GC, 2008a. The elk PRNP codon 132 polymorphism controls cervid and scrapie prion propagation. Journal of General Virology, 89, 598–608. 10.1099/vir.0.83168-0 [DOI] [PubMed] [Google Scholar]
- Green KM, Castilla J, Seward TS, Napier DL, Jewell JE, Soto C and Telling GC, 2008b. Accelerated high fidelity prion amplification within and across prion species barriers. PLoS Pathogens, 4, e1000139. 10.1371/journal.ppat.1000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee JJ, Smith JD and Kunkle RA, 2011. White‐tailed deer are susceptible to the agent of sheep scrapie by intracerebral inoculation. Veterinary Research 42, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenaars TJ, Melchior MB, Bossers A, Davidse A, Engel B and van Zijderveld F, 2010. Scrapie prevalence in sheep of susceptible genotype is declining in a population subject to breeding for resistance. BMC Veterinary Research, 6, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley NJ, Mathiason CK, Zabel MD, Telling GC and Hoover EA, 2009a. Detection of sub‐clinical CWD infection in conventional test‐negative deer long after oral exposure to urine and feces from CWD+ deer. PLoS ONE, 4, e7990. 10.1371/journal.pone.0007990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley NJ, Seelig DM, Zabel MD, Telling GC and Hoover EA, 2009b. Detection of CWD prions in urine and saliva of deer by transgenic mouse bioassay. PLoS ONE, 4, e4848. 10.1371/journal.pone.0004848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley NJ, Mathiason CK, Carver S, Zabel M, Telling GC and Hoover EA, 2011. Detection of CWD prions in salivary, urinary, and intestinal tissues of deer: potential mechanisms of prion shedding and transmission. Journal of Virology, 85, 6309–6318. 10.1128/JVI.00425-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley NJ, Mathiason CK, Carver S, Telling GC, Zabel MD and Hoover EA, 2012. Sensitivity of protein misfolding cyclic amplification versus immunohistochemistry in ante‐mortem detection of chronic wasting disease. Journal of General Virology, 93, 1141–1150. 10.1099/vir.0.039073-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley NJ, Van de Motter A, Carver S, Henderson D, Davenport K, Seelig DM, Mathiason C and Hoover E, 2013. Prion‐seeding activity in cerebrospinal fluid of deer with chronic wasting disease. PLoS ONE, 8, e81488. 10.1371/journal.pone.0081488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley NJ, Carver S, Hoon‐Hanks LL, Henderson DM, Davenport KA, Bunting E, Gray S, Trindle B, Galeota J, LeVan I, Dubovos T, Shelton P and Hoover EA, 2014. Detection of chronic wasting disease in the lymph nodes of free‐ranging cervids by real‐time quaking‐induced conversion. Journal of Clinical Microbiology, 52, 3237–3243. 10.1128/JCM.01258-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley NJ, Siepker C, Walter WD, Thomsen BV, Greenlee JJ, Lehmkuhl AD and Richt JA, 2016a. Antemortem detection of chronic wasting disease prions in nasal brush collections and rectal biopsy specimens from white‐tailed deer by real‐time quaking‐induced conversion. Journal of Clinical Microbiology, 54, 1108–1116. 10.1128/jcm.02699-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley NJ, Siepker C, Hoon‐Hanks LL, Mitchell G, Walter WD, Manca M, Monello RJ, Powers JG, Wild MA, Hoover EA, Caughey B and Richt JA, 2016b. Seeded amplification of chronic wasting disease prions in nasal brushings and recto‐anal mucosa‐associated lymphoid tissues from Elk by real‐time quaking‐induced conversion. Journal of Clinical Microbiology, 54, 1117–1126. 10.1128/JCM.02700-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamir AN, Miller JM, Cutlip RC, Stack MJ, Chaplin MJ, Jenny AL and Williams ES, 2003. Experimental inoculation of scrapie and chronic wasting disease agents in raccoons (Procyon lotor). The Veterinary Record, 153, 121–123. [DOI] [PubMed] [Google Scholar]
- Hamir AN, Miller JM, Cutlip RC, Kunkle RA, Jenny AL, Stack MJ and Chaplin MJ, 2004. Transmission of sheep scrapie to elk (Cervus Elaphus Nelsoni) by intracerebral inoculation: final outcome of the experiment. Journal of Veterinary Diagnostic Investigation, 16, 316–321. [DOI] [PubMed] [Google Scholar]
- Hamir AN, Gidlewski T, Spraker TR, Miller JM, Creekmore L, Crocheck M, Cline T and O'Rourke KI, 2006. Preliminary observations of genetic susceptibility of elk (Cervus elaphus nelsoni) to chronic wasting disease by experimental oral inoculation. Journal of Veterinary Diagnostic Investigation, 18, 110–114. [DOI] [PubMed] [Google Scholar]
- Hamir AN, Miller JM, Kunkle RA, Hall SM and Richt JA, 2007. Susceptibility of cattle to first‐passage intracerebral inoculation with chronic wasting disease agent from white‐tailed deer. Veterinary Pathology, 44, 487–493. 10.1354/vp.44-4-487 [DOI] [PubMed] [Google Scholar]
- Heisey DM, Mickelsen NA, Schneider JR, Johnson CJ, Johnson CJ, Langenberg JA, Bochsler PN, Keane DP and Barr DJ, 2010. Chronic wasting disease (CWD) susceptibility of several North American rodents that are sympatric with cervid CWD epidemics. Journal of Virology, 84, 210–215. 10.1128/JVI.00560-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson DM, Manca M, Haley NJ, Denkers ND, Nalls AV, Mathiason CK, Caughey B and Hoover EA, 2013. Rapid antemortem detection of CWD prions in deer saliva. PLoS ONE, 8, e74377. 10.1371/journal.pone.0074377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson DM, Davenport KA, Haley NJ, Denkers ND, Mathiason CK and Hoover EA, 2015a. Quantitative assessment of prion infectivity in tissues and body fluids by real‐time quaking‐induced conversion. Journal of General Virology, 96, 210–219. 10.1099/vir.0.069906-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson DM, Denkers ND, Hoover CE, Garbino N, Mathiason CK and Hoover EA, 2015b. Longitudinal detection of prion shedding in saliva and urine by chronic wasting disease‐infected deer by real‐time quaking‐induced conversion. Journal of Virology, 89, 9338–9347. 10.1128/JVI.01118-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibler CP, Wilson KL, Spraker TR, Miller MW, Zink RR, DeBuse LL, Andersen E, Schweitzer D, Kennedy JA, Baeten LA, Smeltzer JF, Salman MD and Powers BE, 2003. Field validation and assessment of an enzyme‐linked immunosorbent assay for detecting chronic wasting disease in mule deer (Odocoileus hemionus), white‐tailed deer (Odocoileus virginianus), and Rocky Mountain elk (Cervus elaphus nelsoni). Journal of Veterinary Diagnostic Investigation, 15, 311–319. [DOI] [PubMed] [Google Scholar]
- Hopp P, 2017. FW: SV: antall reinsdyr fra Nordfjella?. Message to Angel Ortiz Pelaez. 29 November 2017.
- Huang H, Rendulich J, Stevenson D, O'Rourke K and Balachandran A, 2005. Evaluation of Western blotting methods using samples with or without sodium phosphotungstic acid precipitation for diagnosis of scrapie and chronic wasting disease. Canadian Journal of Veterinary Research 69, 193. [PMC free article] [PubMed] [Google Scholar]
- Jacobs JG, Langeveld JP, Biacabe AG, Acutis PL, Polak MP, Gavier‐Widen D, Buschmann A, Caramelli M, Casalone C, Mazza M and Groschup M, 2007. Molecular discrimination of atypical bovine spongiform encephalopathy strains from a geographical region spanning a wide area in Europe. Journal of Clinical Microbiology 45, 1821–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennelle CS, Henaux V, Wasserberg G, Thiagarajan B, Rolley RE and Samuel MD, 2014. Transmission of chronic wasting disease in Wisconsin white‐tailed deer: implications for disease spread and management. PLoS ONE 9, e91043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell JE, Conner MM, Wolfe LL, Miller MW and Williams ES, 2005. Low frequency of PrP genotype 225SF among free‐ranging mule deer (Odocoileus hemionus) with chronic wasting disease. The Journal of General Virology, 86(Pt 8), 2127–2134. [DOI] [PubMed] [Google Scholar]
- John TR, Schatzl HM and Gilch S, 2013. Early detection of chronic wasting disease prions in urine of pre‐symptomatic deer by real‐time quaking‐induced conversion assay. Prion, 7, 253–258. 10.4161/pri.24430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CJ, Aiken JM, McKenzie D, Samuel MD and Pedersen JA, 2012. Highly efficient amplification of chronic wasting disease agent by protein misfolding cyclic amplification with beads (PMCAb). PLoS ONE, 7, e35383. 10.1371/journal.pone.0035383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly DO, Samuel MD, Langenberg JA, Blanchong JA, Batha CA, Rolley RE, Keane RD and Ribic CA, 2006. Spatial epidemiology of chronic wasting disease in Wisconsin white‐tailed deer. Journal of Wildlife Diseases, 42, 578–588. [DOI] [PubMed] [Google Scholar]
- Keane DP, Barr DJ, Keller JE, Hall SM, Langenberg JA and Bochsler PN, 2008a. Comparison of retropharyngeal lymph node and obex region of the brainstem in detection of chronic wasting disease in white‐tailed deer (Odocoileus virginianus). Journal of Veterinary Diagnostic Investigation, 20, 58‐60A. [DOI] [PubMed] [Google Scholar]
- Keane DP, Barr DJ, Bochsler PN, Hall SM, Gidlewski T, O'rourke KI, Spraker TR and Samuel MD, 2008b. Chronic wasting disease in a Wisconsin white‐tailed deer farm. Journal of Veterinary Diagnostic Investigation, 20, 698–703. [DOI] [PubMed] [Google Scholar]
- Keane D, Barr D, Osborn R, Langenberg J, O'rourke K, Schneider D andBochsler P, 2009. Validation of use of rectoanal mucosa‐associated lymphoid tissue for immunohistochemical diagnosis of chronic wasting disease in white‐tailed deer (Odocoileus virginianus). Journal of Clinical Microbiology, 47, 1412–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Huang S, Zou W, Vanegas D, Wang M, Wu D, Yuan J, Zheng M, Bai H, Deng H, Chen K, Jenny AL, O'Rourke K, Belay ED, Schonberger LB, Petersen RB, Sy MS, Chen SG and Gambetti P, 2005. Chronic wasting disease of elk: transmissibility to humans examined by transgenic mouse models. Journal of Neuroscience, 25, 7944–7949. 10.1523/JNEUROSCI.2467-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konold T, Hawkins SA, Thurston LC, Maddison BC, Gough KC, Duarte A and Simmons HA, 2015. Objects in contact with classical scrapie sheep act as a reservoir for scrapie transmission. Frontiers in Veterinary Science 2, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreeger TJ, Montgomery DL, Jewell JE, Schultz W and Williams ES, 2006. Oral transmission of chronic wasting disease in captive Shira's moose. Journal of Wildlife Diseases, 42, 640–645. 10.7589/0090-3558-42.3.640 [DOI] [PubMed] [Google Scholar]
- Kuiken T, Ryser‐Degiorgis M‐P, Gavier‐Widén D and Gortázar C, 2011. Establishing a European network for wildlife health surveillance. Revue Scientifique et Technique of the Office International des Epizooties, 30, 755–761. [DOI] [PubMed] [Google Scholar]
- Kurt TD, Perrott MR, Wilusz CJ, Wilusz J, Supattapone S, Telling GC, Zabel MD and Hoover EA, 2007. Efficient in vitro amplification of chronic wasting disease PrPRes . Journal of Virology, 81, 9605–9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFauci G, Carp RI, Meeker HC, Ye X, Kim JI, Natelli M, Cedeno M, Petersen RB, Kascsak R and Rubenstein R, 2006. Passage of chronic wasting disease prion into transgenic mice expressing Rocky Mountain elk (Cervus elaphus nelsoni) PrPC . Journal of General Virology, 87, 3773–3780. 10.1099/vir.0.82137-0 [DOI] [PubMed] [Google Scholar]
- Lang KR and Blanchong JA, 2012. Population genetic structure of white‐tailed deer: understanding risk of chronic wasting disease spread. The Journal of Wildlife Management, 76, 832–840. [Google Scholar]
- Lee YH, Sohn HJ, Kim MJ, Kim HJ, Park KJ, Lee WY, Yun EI, Tark DS, Choi YP, Cho IS and Balachandran A, 2013a. Experimental chronic wasting disease in wild type VM mice. Journal of Veterinary Medical Science, 75, 1107–1110. [DOI] [PubMed] [Google Scholar]
- Lee YH, Sohn HJ, Kim MJ, Kim HJ, Lee WY, Yun EI, Tark DS, Cho IS and Balachandran A, 2013b. Strain characterization of the Korean CWD cases in 2001 and 2004. Journal of Veterinary Medical Science, 75, 95–98. [DOI] [PubMed] [Google Scholar]
- Madsen‐Bouterse SA, Schneider DA, Dassanayake RP, Truscott TC, Zhuang D, Kumpula‐McWhirter N and O'Rourke KI, 2015. PRNP variants in goats reduce sensitivity of detection of PrPSc by immunoassay. Journal of Veterinary Diagnostic Investigation, 27, 332–343. [DOI] [PubMed] [Google Scholar]
- Marsh RF, Kincaid AE, Bessen RA and Bartz JC, 2005. Interspecies transmission of chronic wasting disease prions to squirrel monkeys (Saimiri sciureus). Journal of Virology, 79, 13794–13796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Jeffrey M, Gonzalez L, Siso S, Reid HW, Steele P, Dagleish MP, Stack MJ, Chaplin MJ and Balachandran A, 2009. Immunohistochemical and biochemical characteristics of BSE and CWD in experimentally infected European red deer (Cervus elaphus elaphus). BMC Veterinary Research, 5, 26. 10.1186/1746-6148-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masujin K, Shimada K, Kimura KM, Imamura M, Yoshida A, Iwamaru Y, Mohri S and Yokoyama T, 2007. Applicability of current bovine spongiform encephalopathy (BSE) diagnostic procedures for chronic wasting disease (CWD). Microbiology and Immunology, 51, 1039–1043. [DOI] [PubMed] [Google Scholar]
- Mathiason CK, Hays SA, Powers J, Hayes‐Klug J, Langenberg J, Dahmes SJ, Osborne DA, Miller KV, Warren RJ, Mason GL and Hoover EA, 2009. Infectious prions in pre‐clinical deer and transmission of chronic wasting disease solely by environmental exposure. PLoS ONE, 4, e5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiason CK, Nalls AV, Seelig DM, Kraft SL, Carnes K, Anderson KR, Hayes‐Klug J and Hoover EA, 2013. Susceptibility of domestic cats to chronic wasting disease. Journal of Virology, 87, 1947–1956. 10.1128/JVI.02592-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade‐White K, Race B, Trifilo M, Bossers A, Favara C, Lacasse R, Miller M, Williams E, Oldstone M, Race R and Chesebro B, 2007. Resistance to chronic wasting disease in transgenic mice expressing a naturally occurring allelic variant of deer prion protein. Journal of Virology, 81, 4533–4539. 10.1128/JVI.02762-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MW, 2017a. FW: IDEXX test in mule deer or moose. Message to Angel Ortiz Pelaez. 29 November 2017.
- Miller MW, 2017b. Re: Eastern US CWD surveillance data. Message to Angel Ortiz Pelaez. 17 November 2017.
- Miller MW and Conner MM, 2005. Epidemiology of chronic wasting disease in free‐ranging mule deer: spatial, temporal, and demographic influences on observed prevalence patterns. Journal of Wildlife Diseases, 41, 275–90. [DOI] [PubMed] [Google Scholar]
- Miller MW and Fischer JR, 2016. The first five (or more) decades of chronic wasting disease: lessons for the five decades to come. Transactions of the North American Wildlife and Natural Resources Conference 81, in press. Available online: http://cpw.state.co.us/Documents/Research/CWD/Miller-Fischer_CWDlessons.pdf [Accessed: November 2017]
- Miller MW and Williams ES, 2002. Detection of PrP(CWD) in mule deer by immunohistochemistry of lymphoid tissues. The Veterinary Record, 151, 610–612. [DOI] [PubMed] [Google Scholar]
- Miller MW, Williams ES, McCarty CW, Spraker TR, Kreeger TJ, Larsen CT and Thorne ET, 2000. Epizootiology of chronic wasting disease in free‐ranging cervids in Colorado and Wyoming. Journal of Wildlife Diseases, 36, 676–690. [DOI] [PubMed] [Google Scholar]
- Mitchell GB, Sigurdson CJ, O'Rourke KI, Algire J, Harrington NP, Walther I, Spraker TR and Balachandran A, 2012. Experimental oral transmission of chronic wasting disease to reindeer (Rangifer tarandus tarandus). PLoS ONE, 7, e39055. 10.1371/journal.pone.0039055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monello RJ, Powers JG, Hobbs NT, Spraker TR, O'Rourke KI and Wild MA, 2013. Efficacy of antemortem rectal biopsies to diagnose and estimate prevalence of chronic wasting disease in free‐ranging cow elk (Cervus elaphus nelsoni). Journal of Wildlife Diseases, 49, 270–278. [DOI] [PubMed] [Google Scholar]
- Moore SJ, West Greenlee MH, Kondru N, Manne S, Smith JD, Kunkle RA, Kanthasamy A and Greenlee JJ, 2017. Experimental transmission of the chronic wasting disease agent to swine after oral or intracranial inoculation. Journal of Virology, 91, e00926‐17. 10.1128/JVI.00926-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls AV, McNulty E, Powers J, Seelig DM, Hoover C, Haley NJ, Hayes‐Klug J, Anderson K, Stewart P, Goldmann W, Hoover EA and Mathiason CK, 2013. Mother to offspring transmission of chronic wasting disease in reeves’ muntjac deer. PLoS ONE, 8, e71844. 10.1371/journal.pone.0071844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols TA, Spraker TR, Gidlewski T, Powers JG, Telling GC, VerCauteren KC and Zabel MD, 2012. Detection of prion protein in the cerebrospinal fluid of elk (Cervus canadensis nelsoni) with chronic wasting disease using protein misfolding cyclic amplification. Journal of Veterinary Diagnostic Investigation 24, 746–749. [DOI] [PubMed] [Google Scholar]
- OIE (World Organisation for Animal Health), 2009. Terrestrial Animal Health Code, 18th Ed. Glossary. OIE, Paris. Available online: http://www.oie.int/index.php?id=169&L=0&htmfile=glossaire.htm [Accessed: November 2017]
- OIE (World Organisation for Animal Health), 2010. Training manual on wildlife diseases and surveillance: workshop for OIE National Focal Points for Wildlife. OIE, Paris. Available online: http://www.oie.int/fileadmin/Home/eng/Internationa_Standard_Setting/docs/pdf/WGWildlife/A_Training_Manual_Wildlife.pdf [Accessed: August 217]
- OIE (World Organisation for Animal Health), 2017a. Improving wildlife surveillance for its protection while protecting us from the diseases it transmits. OIE, Paris. Available online: http://www.oie.int/en/for-the-media/editorials/detail/article/improving-wildlife-surveillance-for-its-protection-while-protecting-us-from-the-diseases-it-transmit [Accessed: August 2017]
- OIE (World Organisation for Animal Health), 2017b. Manual of diagnostic tests and vaccines for terrestrial animals 2017. Available online: http://www.oie.int/en/international-standard-setting/terrestrial-manual/access-online [Accessed: November 2017]
- Organisation Mondiale de la Santé Animale , 2017. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2017. Available online: http://www.oie.int/fr/normes-internationales/manuel-terrestre/acces-en-ligne/ [Accessed: November 2017]
- O'rourke KI, Spraker TR, Zhuang D, Greenlee JJ, Gidlewski TE and Hamir AN, 2007. Elk with a long incubation prion disease phenotype have a unique PrPd profile. NeuroReport, 18, 1935–1938. [DOI] [PubMed] [Google Scholar]
- Orrù CD, Groveman BR, Hughson AG, Manca M, Raymond LD, Raymond GJ, Campbell KJ, Anson KJ, Kraus A and Caughey B, 2017. RT‐QuIC assays for prion disease detection and diagnostics. Methods in Molecular Biology, 1658, 185–203. [DOI] [PubMed] [Google Scholar]
- Pattison IH, 1965. Experiments with scrapie with special reference to the nature of the agent and the pathology of the disease. In: Gajdusek DC, Gibbs JR CJ and Alpers MP (eds.). Slow, Latent and Temperate Virus Infections, NINDB Monograph 2. U.S. Government Printing, Washington, DC, pp. 249–257. [Google Scholar]
- Peletto S, Perucchini M, Acín C, Dalgleish MP, Reid HW, Rasero R, Sacchi P, Stewart P, Caramelli M, Ferroglio E and Bozzetta E, 2009. Genetic variability of the prion protein gene (PRNP) in wild ruminants from Italy and Scotland. Journal of Veterinary Science, 10, 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, Miller JM, Jenny AL, Peterson TL and Carmichael KP, 2000. Immunohistochemical diagnosis of chronic wasting disease in preclinically affected elk from a captive herd. Journal of Veterinary Diagnostic Investigation, 12, 579–582. [DOI] [PubMed] [Google Scholar]
- Pirisinu L, Tran L, Mitchell G, Balachandran A, Baron T, Casalone C, Bari M, Agrimi U, Nonno R and Benestad S, 2017. Chronic Wasting Disease in European moose is associated with PrPSc features different from North American CWD. Proceedings of the 2017 Prion Conference. 23–26 May 2017. Edinburgh, UK.
- Pulford B, Spraker TR, Wyckoff AC, Meyerett C, Bender H, Ferguson A, Wyatt B, Lockwood K, Powers J, Telling GC, Wild MA and Zabel MD, 2012. Detection of PrPCWD in feces from naturally exposed Rocky Mountain elk (Cervus elaphus nelsoni) using protein misfolding cyclic amplification. Journal of Wildlife Diseases, 48, 425–434. [DOI] [PubMed] [Google Scholar]
- Race BL, Meade‐White KD, Ward A, Jewell J, Miller MW, Williams ES, Chesebro B and Race RE, 2007. Levels of abnormal prion protein in deer and elk with chronic wasting disease. Emerging Infectious Diseases, 13, 824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race B, Meade‐White K, Race R and Chesebro B, 2009a. Prion infectivity in fat of deer with chronic wasting disease. Journal of Virology, 83, 9608–9610. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2738259/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race B, Meade‐White KD, Miller MW, Barbian KD, Rubenstein R, LaFauci G, Cervenakova L, Favara C, Gardner D, Long D, Parnell M, Striebel J, Priola SA, Ward A, Williams ES, Race R and Chesebro B, 2009b. Susceptibilities of nonhuman primates to chronic wasting disease. Emerging Infectious Diseases, 15, 1366–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race B, Meade‐White KD, Phillips K, Striebel J, Race R and Chesebro B, 2014. Chronic wasting disease agents in nonhuman primates. Emerging Infectious Diseases, 20, 833–837. 10.3201/eid2005.130778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond GJ, Raymond LD, Meade‐White KD, Hughson AG, Favara C, Gardner D, Williams ES, Miller MW, Race RE and Caughey B, 2007. Transmission and adaptation of chronic wasting disease to hamsters and transgenic mice: evidence for strains. Journal of Virology, 81, 4305–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein R, Chang B, Gray P, Piltch M, Bulgin MS, Sorensen‐Melson S and Miller MW, 2010. A novel method for preclinical detection of PrPSc in blood Journal of General Virology, 91, 1883–1892. [DOI] [PubMed] [Google Scholar]
- Rubenstein R, Chang B, Gray P, Piltch M, Bulgin MS, Sorensen‐Melson S and Miller MW, 2011. Prion disease detection, PMCA kinetics, and IgG in urine from sheep naturally/experimentally infected with scrapie and deer with preclinical/clinical chronic wasting disease. Journal of Virology, 85, 9031–9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saá P and Cervenakova L, 2015. Protein misfolding cyclic amplification (PMCA): current status and future directions. Virus Research 207, 47–61. [DOI] [PubMed] [Google Scholar]
- Saborio GP, Permanne B and Soto C, 2001. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature, 411, 810–813. [DOI] [PubMed] [Google Scholar]
- Samuel MD, Joly DO, Wild MA, Wright SD, Otis DL, Werge RW and Miller MW, 2003. Surveillance strategies for detecting chronic wasting disease in free‐ranging deer and Elk. Results of a CWD Surveillance Workshop Madison, Wisconsin December 10–12, 2002. Available online: http://www.nwhc.usgs.gov/publications/fact_sheets/pdfs/cwd/CWD_Surveillance_Strategies.pdf [Accessed: November 2017]
- Selariu A, Powers JG, Nalls A, Brandhuber M, Mayfield A, Fullaway S, Wyckoff CA, Goldmann W, Zabel MM, Wild MA, Hoover EA and Mathiason CK, 2015. In utero transmission and tissue distribution of chronic wasting disease‐associated prions in free‐ranging Rocky Mountain elk. Journal General Virology, 96, 3444–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson CJ, Williams ES, Miller MW, Spraker TR, O'Rourke KI and Hoover EA, 1999. Oral transmission and early lymphoid tropism of chronic wasting disease PrPres in mule deer fawns (Odocoileus hemionus). The Journal of General Virology, 80(Pt. 10), 2757–2764. [DOI] [PubMed] [Google Scholar]
- Sigurdson CJ, Barillas‐Mury C, Miller MW, Oesch B, van Keulen LJM, Langeveld JPM and Hoover EA, 2002. PrPCWD lymphoid cell targets in early and advanced chronic wasting disease of mule deer. Journal of General Virology, 83, 2617–2628. [DOI] [PubMed] [Google Scholar]
- Sigurdson CJ, Manco G, Schwarz P, Liberski P, Hoover EA, Hornemann S, Polymenidou M, Miller MW, Glatzel M and Aguzzi A, 2006. Strain fidelity of chronic wasting disease upon murine adaptation. Journal of Virology, 80, 12303–12311. 10.1128/JVI.01120-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson CJ, Mathiason CK, Perrott MR, Eliason GA, Spraker TR, Glatzel M, Manco G, Bartz JC, Miller MW and Hoover EA, 2008. Experimental chronic wasting disease (CWD) in the ferret. Journal of Comparative Pathology, 138, 189–196. 10.1016/j.jcpa.2008.01.004 [DOI] [PubMed] [Google Scholar]
- Simmons MM, Chaplin MJ, Vickery CM, Simon S, Davis L, Denyer M, Lockey R, Stack MJ, O'Connor MJ, Bishop K, Gough KC, Maddison BC, Thorne L and Spiropoulos J, 2015. Does the presence of scrapie affect the ability of current statutory discriminatory tests to detect the presence of bovine spongiform encephalopathy? Journal of Clinical Microbiology 53, 2593–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisó S, González L and Jeffrey M, 2010. Neuroinvasion in prion diseases: the roles of ascending neural infection and blood dissemination. Interdisciplinary Perspectives on Infectious Diseases, 2010, Article ID 747892, 16 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spraker TR, Miller MW, Williams ES, Getzy DM, Adrian WJ, Schoonveld GG, Spowart RA, O'Rourke KI, Miller JM and Merz PA, 1997. Spongiform encephalopathy in free‐ranging mule deer (Odocoileus hemionus), white‐tailed deer (Odocoileus virginianus) and Rocky Mountain elk (Cervus elaphus nelsoni) in northcentral Colorado. Journal of Wildlife Diseases, 33, 1–6. [DOI] [PubMed] [Google Scholar]
- Spraker TR, Zink RR, Cummings BA, Sigurdson CJ, Miller MW and O'Rourke KI, 2002. Distribution of protease‐resistant prion protein and spongiform encephalopathy in free‐ranging mule deer (Odocoileus hemionus) with chronic wasting disease. Veterinary Pathology, 39, 546–556. [DOI] [PubMed] [Google Scholar]
- Spraker TR, Balachandran A, Zhuang D and O'Rourke KI, 2004. Variable patterns of PrP‐CWD distribution in obex and cranial lymphoid tissues of Rocky Mountain elk with nonclinical chronic wasting disease. The Veterinary Record, 155, 295–302. [DOI] [PubMed] [Google Scholar]
- Spraker TR, Gidlewski TL, Balachandran A, VerCauteren KC, Creekmore L and Munger RD, 2006. Detection of PrPCWD in postmortem rectal lymphoid tissues in Rocky Mountain elk (Cervus elaphus nelsoni) infected with chronic wasting disease. Journal of Veterinary Diagnostic Investigation, 18, 553–557. [DOI] [PubMed] [Google Scholar]
- Spraker TR, VerCauteren KC, Gidlewski T, Schneider DA, Munger R, Balachandran A and O'Rourke KI, 2009. Antemortem detection of PrPCWD in preclinical, ranch‐raised rocky mountain Elk (Cevvus Elaphus Nelsoni) by biopsy of the rectal mucosa. Journal of Veterinary Diagnostic Investigation, 21, 15–24. [DOI] [PubMed] [Google Scholar]
- SSC (Scientific Steering Committee), 2001. Requirements for statistically authoritative BSE/TSE surveys. Adopted by the Scientific Steering Committee at its meeting of 29–30 November 2001.
- SSC (Scientific Steering Committee), 2002a. Opinion of the SSC on design of a field trial for the evaluation of new rapid BSE post mortem tests. Adopted on 22 February 2002.
- SSC (Scientific Steering Committee), 2002b. Opinion on a programme for the evaluation of rapid post mortem tests to detect TSE in small ruminants adopted by the scientific steering committee at its meeting of 7–8 November 2002.
- SSC (Scientific Steering Committee), 2003. Opinion of the scientific steering committee on the field trial evaluation of two new rapid BSE post mortem tests results achieved using the LIA Test (Prionics) and the aCDI Test (InPro) in the field trial. Adopted on 6 March 2003.
- Stack MJ, Moore SJ, Vidal‐Diez A, Arnold ME, Jones EM, Spencer YI, Webb P, Spiropoulos J, Powell L, Bellerby P, Thurston L, Cooper J, Chaplin MJ, Davis LA, Everitt S, Focosi‐Snyman R, Hawkins SAC, Simmons MM and Wells GAH, 2011. Experimental bovine spongiform encephalopathy: detection of PrP Sc in the small intestine relative to exposure dose and age. Journal of Comparative Pathology, 145, 289–301. [DOI] [PubMed] [Google Scholar]
- Tamguney G, Giles K, Bouzamondo‐Bernstein E, Bosque PJ, Miller MW, Safar J, DeArmond SJ and Prusiner SB, 2006. Transmission of elk and deer prions to transgenic mice. Journal of Virology, 80, 9104–9114. 10.1128/JVI.00098-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telling GC, 2010. Nucleic acid‐free mutation of prion strains. Prion, 4, 252–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LA, Marsh S, Ryder SJ, Hawkins SA, Wells GA and Spencer YI, 2003. Detection of disease‐specific PrP in the distal ileum of cattle exposed orally to the agent of bovine spongiform encephalopathy. The Veterinary Record, 152, 387–392. [DOI] [PubMed] [Google Scholar]
- Thomsen BV, Schneider DA, O'Rourke KI, Gidlewski T, McLane J, Allen RW, McIsaac AA, Mitchell GB, Keane DP, Spraker TR and Balachandran A, 2012. Diagnostic accuracy of rectal mucosa biopsy testing for chronic wasting disease within white‐tailed deer (Odocoileus virginianus) herds in North America: effects of age, sex, polymorphism at PRNP codon 96, and disease progression. Journal of Veterinary Diagnostic Investigation, 24, 878–887. [DOI] [PubMed] [Google Scholar]
- Vickery CM, Lockey R, Holder TM, Thorne L, Beck KE, Wilson C, Denyer M, Sheehan J, Marsha S, Webb PR, Dexter I, Norman A, Popescu E, Schneider A, Holden P, Griffiths PC, Plater JM, Dagleish MP, Martin S, Telling GC, Simmons MM and John Spiropoulos J, 2014. Assessing the susceptibility of transgenic mice overexpressing deer prion protein to bovine spongiform encephalopathy. Journal of Virology, 88, 1830–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DP and Miller MW, 2010. A weighted surveillance approach for detecting chronic wasting disease foci. Journal of Wildlife Diseases, 46, 118–135. [DOI] [PubMed] [Google Scholar]
- Wasserberg G, Osnas EE, Rolley RE and Samuel MD, 2009. Host culling as an adaptive management tool for chronic wasting disease in white‐tailed deer: a modelling study. Journal of Applied Ecology, 46, 457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wik L, Mikko S, Klingeborn M, Stéen M, Simonsson M and Linné T, 2012. Polymorphisms and variants in the prion protein sequence of European Moose (Alces alces), Reindeer (Rangifer tarandus), Roe Deer (Capreolus capreolus) and Fallow Deer (Dama dama) in Scandinavia. Prion, 6, 256–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild MA, Spraker TR, Sigurdson CJ, O'Rourke KI and Miller MW, 2002. Preclinical diagnosis of chronic wasting disease in captive mule deer (Odocoileus hemionus) and white‐tailed deer (Odocoileus virginianus) using tonsillar biopsy. Journal of General Virology, 83, 2629–2634. [DOI] [PubMed] [Google Scholar]
- Williams ES and Young S, 1980. Chronic wasting disease of captive mule deer: a spongiform encephalopathy. Journal of Wildlife Diseases, 16, 89–98. [DOI] [PubMed] [Google Scholar]
- Williams ES and Young S, 1982. Spongiform encephalopathy of Rocky Mountain Elk. Journal of Wildlife Diseases, 18, 465–471. [DOI] [PubMed] [Google Scholar]
- Williams ES and Young S, 1992. Spongiform encephalopathies in Cervidae. Revue Scientifique Et Technique‐Office International Des Epizooties, 11, 551–567. [DOI] [PubMed] [Google Scholar]
- Williams ES and Young S, 1993. Neuropathology of chronic wasting disease of mule deer (Odocoileus hemionus) and Elk (Cervus elaphus nelsoni). Veterinary Pathology, 30, 36–45. [DOI] [PubMed] [Google Scholar]
- Wolfe LL, Spraker TR, Gonzalez L, Dagleish MP, Sirochman TM, Brown JC, Jeffrey M and Miller MW, 2007. PrPCWD in rectal lymphoid tissue of deer (Odocoileus spp.). Journal of General Virology, 88, 2078–2082. [DOI] [PubMed] [Google Scholar]
- Wolfe LL, Kocisko DA, Caughey B and Miller MW, 2012. Assessment of prospective preventive therapies for chronic wasting disease in mule deer. Journal of Wildlife Diseases, 48, 530–533. 10.7589/0090-3558-48.2.530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe LL, Fox KA and Miller MW, 2014. “Atypical” chronic wasting disease in PRNP genotype 225FF mule deer. Journal of Wildlife Diseases, 50, 660–665. [DOI] [PubMed] [Google Scholar]
- Wyckoff AC, Galloway N, Meyerett‐Reid C, Powers J, Spraker T, Monello RJ, Pulford B, Wild M, Antolin M, VerCauteren K and Zabel M, 2015. Prion amplification and hierarchical bayesian modeling refine detection of prion infection. Scientific Reports, 5, 8358. 10.1038/srep08358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Dudas S, Flitton R, Pickles T, Colwell K, Fang Y, Graham C and Czub S, 2011. Bio. 175: early detection of PrPcwd in rapid test‐negative Elk using a modified western blot. Prion, 5, 94–95. [Google Scholar]


