Summary
A novel SARS-like coronavirus (SARS-CoV-2) recently emerged and is rapidly spreading in humans 1,2. A key to tackling this epidemic is to understand the virus’s receptor recognition mechanism, which regulates its infectivity, pathogenesis and host range. SARS-CoV-2 and SARS-CoV recognize the same receptor - human ACE2 (hACE2) 3,4. Here we determined the crystal structure of SARS-CoV-2 receptor-binding domain (RBD) (engineered to facilitate crystallization) in complex of hACE2. Compared with SARS-CoV RBD, a hACE2-binding ridge in SARS-CoV-2 RBD takes a more compact conformation; moreover, several residue changes in SARS-CoV-2 RBD stabilize two virus-binding hotspots at the RBD/hACE2 interface. These structural features of SARS-CoV-2 RBD enhance its hACE2-binding affinity. Additionally, we showed that RaTG13, a bat coronavirus closely related to SARS-CoV-2, also uses hACE2 as its receptor. The differences among SARS-CoV-2, SARS-CoV and RaTG13 in hACE2 recognition shed light on potential animal-to-human transmission of SARS-CoV-2. This study provides guidance for intervention strategies targeting receptor recognition by SARS-CoV-2.
Keywords: SARS-CoV-2, SARS-CoV, receptor-binding domain, receptor-binding motif, angiotensin-converting enzyme 2, X-ray crystallography
Introduction
The sudden emergence and rapid spread of SARS-CoV-2 is endangering global health and economy 1,2. SARS-CoV-2 has caused more infections, deaths, and economic disruptions than did the 2002-2003 SARS-CoV 5,6. The origin of SARS-CoV-2 remains a mystery. Bats are considered the original source of SARS-CoV-2 because a closely related coronavirus, RaTG13, has been isolated from bats 7. However, the molecular events that led to the possible bat-to-human transmission of SARS-CoV-2 are unknown. Also lacking are clinically approved vaccines or drugs that specifically target SARS-CoV-2. Receptor recognition by coronaviruses is an important determinant of viral infectivity, pathogenesis, and host range 8,9. It presents a major target for vaccination and antiviral strategies 10. Here we elucidate the structural and biochemical mechanisms of receptor recognition by SARS-CoV-2.
Receptor recognition by SARS-CoV has been extensively studied. A virus-surface spike protein mediates coronavirus entry into host cells. SARS-CoV spike protein contains a receptor-binding domain (RBD) that specifically recognizes angiotensin-converting enzyme 2 (ACE2) as its receptor 3,4. We previously determined a series of crystal structures of SARS-CoV RBD from different strains complexed with ACE2 from different hosts 3,11,12. These structures showed that SARS-CoV RBD contains a core and a receptor-binding motif (RBM); the RBM mediates contacts with ACE2. The surface of human ACE2 (hACE2) contains two virus-binding hotspots that are critical for SARS-CoV binding. Several naturally selected mutations in SARS-CoV RBM surround these hotspots and regulate the infectivity, pathogenesis, and cross-species and human-to-human transmissions of SARS-CoV 3,11,12.
Because of the sequence similarity between SARS-CoV and SARS-CoV-2 spikes, we recently predicted that SARS-CoV-2 also uses hACE2 as its receptor 13, which has been validated by other studies 7,14-16. Here we determined the structural basis for receptor recognition by SARS-CoV-2 and compared the hACE2-binding affinity among SARS-CoV-2, SARS-CoV and RaTG13. Our findings identify the molecular and structural features of SARS-CoV-2 RBM that result in tight hACE2 binding. They provide insights into the animal origin of SARS-CoV-2, and can guide intervention strategies targeting SARS-CoV-2/hACE2 interactions.
Results
To understand the structural basis for hACE2 recognition by SARS-CoV-2, we aimed to crystallize the SARS-CoV-2 RBD/hACE2 complex. Our strategy was informed by previous crystallization of the SARS-CoV RBD/hACE2 complex 3. In this crystal form, the core of SARS-CoV RBD (along with hACE2 surface) was mainly involved in crystal lattice contact; the critical hACE2-binding residues in SARS-CoV RBM were buried at the RBD/hACE2 interface and did not affect crystallization. To facilitate crystallization, we designed a chimeric RBD that uses the core from SARS-CoV RBD as the crystallization scaffold and the RBM from SARS-CoV-2 as the functionally relevant unit (Fig.1a; Extended Data Fig.1). To further enhance crystallization, we improved the hACE2-binding affinity of the chimeric RBD by keeping a short loop from SARS-CoV RBM, which maintains a strong salt bridge between Arg426 from the RBD and Glu329 from hACE2 (Extended Data Fig.2a). This loop sits on the side of the binding interface, away from the main binding interface. We expressed and purified the chimeric RBD and hACE2, and crystallized their complex under the same condition and in the same crystal form as those for the SARS-CoV RBD/hACE2 complex. Based on X-ray diffraction data, we determined the structure of the chimeric RBD/hACE2 complex by molecular replacement using the structure of the SARS-CoV RBD/hACE2 complex as the search template. We refined the structure to 2.68 Å (Extended Data Table.1; Extended Data Fig.3). The structure of this chimeric RBD/hACE2 complex, particularly in the RBM region, is highly similar to another recently determined structure of the SARS-CoV-2 wild-type RBD/hACE2 complex 17, confirming that the chimeric RBD is a successful design.
Figure 1: Structure of SARS-CoV-2 chimeric RBD complexed with human ACE2.
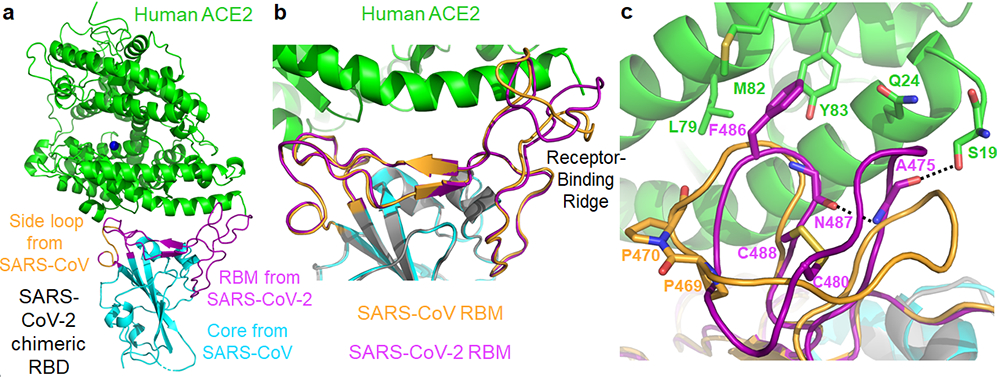
(a) Crystal structure of SARS-CoV-2 chimeric RBD complexed with ACE2. ACE2 is in green. RBD core is in cyan. RBM is in magenta. A side loop in RBM is in orange. A zinc ion in ACE2 is in blue. (b) Comparison of the conformations of the ridge in SARS-CoV-2 RBM (purple) and SARS-CoV RBM (orange). (c) Comparison of the conformations of the ridge from another angle of view. In SARS-CoV RBM, a proline-proline-alanine motif is shown. In SARS-CoV-2 RBM, a newly formed hydrogen bond, Phe486, and some of the ridge’s interactions with the N-terminal helix of ACE2 are shown.
The overall structure of the chimeric RBD/hACE2 complex is similar to that of the SARS-CoV RBD/hACE2 complex (Fig.1a). Like SARS-CoV RBM, SARS-CoV-2 RBM forms a gently concave surface with a ridge on one side; it binds to the exposed outer surface of the claw-like structure of hACE2 (Fig.1a). Surprisingly, the strong salt bridge between SARS-CoV RBD and hACE2 became a weaker (as judged by the longer distance of the interaction), but still energetically favorable, N-O bridge between Arg439 from the chimeric RBD and Glu329 from hACE2 18 (Extended Data Fig.2b). Compared with SARS-CoV RBM, SARS-CoV-2 RBM forms a larger binding interface and more contacts with hACE2 (Extended Data Fig.4a; Extended Data Fig.4b). Our structural model also contained glycans attached to four hACE2 sites and one RBD site (Extended Data Fig.5a). The glycan attached to Asn90 from hACE2 forms a hydrogen bond with Arg408 from the RBD core (Extended Data Fig.5b); this glycan-interacting arginine is conserved between SARS-CoV-2 and SARS-CoV (Extended Data Fig.1). The overall structural similarity in hACE2 binding by SARS-CoV-2 and SARS-CoV supports a close evolutionary relationship between the two viruses.
We measured the binding affinities between each of the three RBDs (SARS-CoV-2, chimeric, and SARS-CoV) and hACE2 using surface plasmon resonance (SPR) (Extended Data Fig.4c; Extended Data Fig.6). The results showed that the chimeric RBD has higher hACE2-binding affinity than SARS-CoV-2 RBD, consistent with the introduced N-O bridge between the chimeric RBD and hACE2. Both the chimeric and SARS-CoV-2 RBDs have significantly higher hACE2-binding affinity than SARS-CoV RBD. These Kd values are consistent with other SPR studies 12,19, although the exact Kd values vary depending on the specific approaches of each SPR experiment (Extended Data Table.2). Here we investigate the structural differences between SARS-CoV-2 and SARS-CoV RBMs that account for their different hACE2-binding affinities.
A significant structural difference between SARS-CoV-2 and SARS-CoV RBMs is the conformations of the loops in the hACE2-binding ridge (Fig.1b, 1c). In both RBMs, one of the ridge loops contains a critical disulfide bond and the region between the disulfide-bond-forming cysteines is variable (Fig.1c; Extended Data Fig.1). Specifically, human and civet SARS-CoV strains and bat coronavirus Rs3367 all contain a three-residue motif proline-proline-alanine in this loop; the tandem prolines allow the loop to take a sharp turn. In contrast, SARS-CoV-2 and bat coronavirus RaTG13 both contain a four-residue motif glycine-valine/glutamine-glutamate/threonine-glycine; two relatively bulky residues and two flexible glycines allow the loop to take a different conformation (Fig.1c; Extended Data Fig.1). Because of these structural differences, an additional main-chain hydrogen bond forms between Asn487 and Ala475 in SARS-CoV-2 RBM, causing the ridge to take a more compact conformation and the loop containing Ala475 to move closer to hACE2 (Fig.1c). Consequently, the ridge in SARS-CoV-2 RBM forms more contact with the N-terminal helix of hACE2 (Extended Data Fig.4b). For example, N-terminal residue Ser19 from hACE2 forms a new hydrogen bond with the main chain of Ala475 from SARS-CoV-2 RBM, and Gln24 from the N-terminal helix of hACE2 also forms new contact with SARS-CoV-2 RBM (Fig.1c; Extended Data Fig.4b). Moreover, compared with the corresponding Leu472 from SARS-CoV RBM, Phe486 from SARS-CoV-2 RBM points to a different direction and inserts into a hydrophobic pocket involving Met82, Leu79, and Tyr83 from hACE2 (Fig.1c, 2a, 2b). Compared with SARS-CoV RBM, these structural changes in SARS-CoV-2 RBM are more favorable for hACE2 binding.
Figure 2: Structural details at the interface between SARS-CoV-2 RBM and human ACE2.
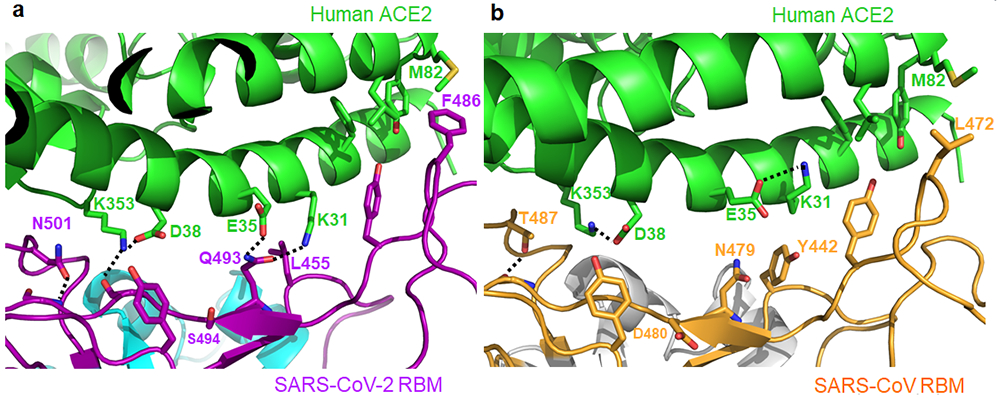
(A) Interface between SARS-CoV-2 RBM and human ACE2. (B) Interface between SARS-CoV RBM and human ACE2.
Compared with the SARS-CoV RBM/hACE2 interface, subtle yet functionally important structural changes take place near the two virus-binding hotspots at the SARS-CoV-2 RBM/hACE2 interface (Fig.2a, 2b). At the SARS-CoV/hACE2 interface, we previously identified two virus-binding hotspots 11,12: hotspot Lys31 (i.e., hotspot-31) consists of a salt bridge between Lys31 and Glu35, and hotspot Lys353 (i.e., hotspot-353) consists of a salt bridge between Lys353 and Asp38. Both salt bridges are weak, as judged by the relatively long distance of these interactions. Burial of these weak salt bridges in hydrophobic environments upon virus binding would enhance their energy due to reduction of the dielectric constant. This process is facilitated through the hotspots’ interactions with nearby RBD residues. First, at the SARS-CoV RBM/hACEC2 interface, hotspot-31 requires support from Tyr442 in SARS-CoV RBM (Fig.2b). In comparison, at the SARS-CoV-2 RBM/hACE2 interface, Leu455 from SARS-CoV-2 RBM (corresponding to Tyr442 from SARS-CoV RBM) has a less bulky side chain, providing less support for Lys31 from hACE2. As a result, the hotspot-31 structure has rearranged: the salt bridge between Lys31 and Glu35 breaks apart, and each of the residues forms a hydrogen bond with Gln493 from SARS-CoV-2 RBM (Fig.2a). Second, at the SARS-CoV RBM/hACEC2 interface, hotspot-353 requires support from the side-chain methyl group of Thr487 in SARS-CoV RBM, whereas the side-chain hydroxyl group of Thr487 forms a hydrogen bond with the RBM main chain (which fixes the conformation of the Thr487 side chain) (Fig.2b). In comparison, at the SARS-CoV-2 RBM/hACE2 interface, Asn501 from SARS-CoV-2 RBM also has its conformation fixed through a hydrogen bond between its side chain and the RBM main chain; correspondingly, its side chain provides less support to hotspot-353 than the corresponding Thr487 from SARS-CoV RBM does (Fig.2a). Consequently, lys353 from hACE2 takes a slightly different conformation, forming a hydrogen bond with the main chain of the SARS-CoV-2 RBM while maintaining the salt bridge with Asp38 from hACE2 (Fig.2a). Thus, both hotspots have adjusted to the reduced support from nearby RBD residues, yet still become well stabilized at the SARS-CoV-2 RBM/hACE2 interface.
To corroborate the structural observations, we characterized hACE2-binding affinities of SARS-CoV-2 spike bearing critical ACE2-interacting mutations. To this end, protein pull-down assay was performed, with purified recombinant hACE2 as the bait and cell-associated SARS-CoV-2 spike as the target (Fig.3a). For cross validation, we used hACE2 with two different tags, His6 and Fc. The SARS-CoV-2 spike contained one of the following RBM changes: 481-487 (481-NGVEGFN-487 in SARS-CoV-2 were mutated to TPPALN as in SARS-CoV), Q493N (Gln493 in SARS-CoV-2 was mutated to an asparagine as in human SARS-CoV), Q493Y (Gln493 in SARS-CoV-2 was mutated to a tyrosine as in bat RaTG13), N501T (Asn501 in SARS-CoV-2 was mutated to a threonine as in human SARS-CoV), and N501D (Asn501 in SARS-CoV-2 was mutated to an aspartate as in bat RaTG13). The results showed that all of these introduced mutations reduced the hACE2-binding affinity of SARS-CoV-2 spike. They confirm that the structural features of SARS-CoV-2 RBM, including the ACE2-binding ridge and the hotspots-stabilizing residues, all contribute critically to the high hACE2-binding affinity of SARS-CoV-2.
Figure 3: Biochemical data showing the interactions between SARS-CoV-2 or bat RaTG13 spike and human ACE2.
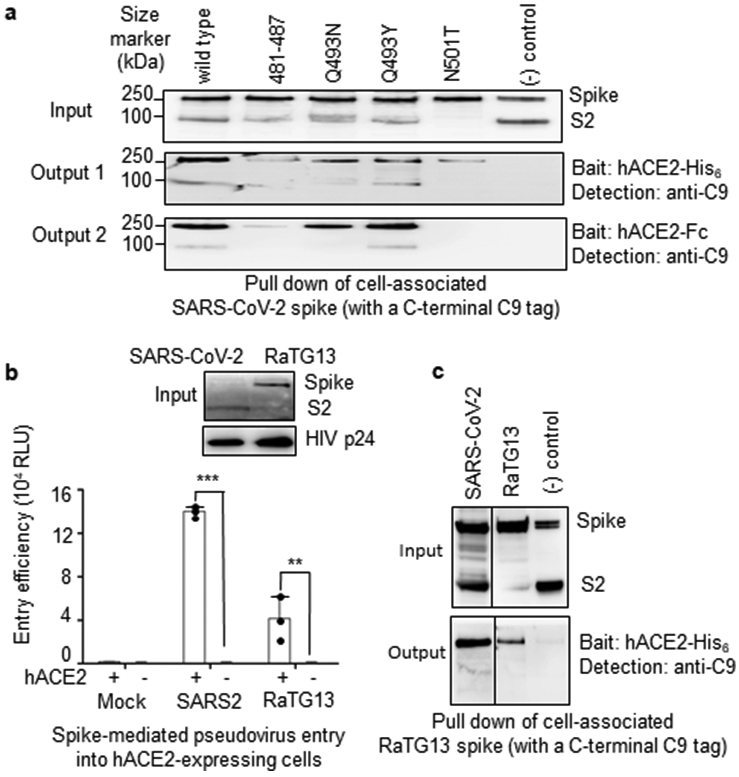
(A) Protein pull-down assay using hACE2 as the bait and cell-associated SARS-CoV-2 spike molecules (wild type and mutants) as the targets. Top panel: cell-expressed SARS-CoV-2 spike. Middle panel: pull-down result using His6-tagged hACE2. Bottom panel: pull-down result using Fc-tagged hACE2. MERS-CoV spike was used as a negative control. (B) Entry of SARS-CoV-2 and bat RaTG13 pseudoviruses into hACE2-expressing cells. Top: packaged SARS-CoV-2 and bat RaTG13 pseudoviruses. HIV p24 was detected as an internal control. Bottom: pseudovirus entry efficiency. Mock: no pseudoviruses. Error bars indicate +1 S.D. Two-tailed t-test comparing SARS-CoV-2 (with hACE2; n=3 independent samples) with SARS-CoV-2 (no hACE2; n=4 independent samples) showed a significant difference, p<1.16×10−8. Two-tailed t-test comparing RaTG13 (with hACE2; n=3 independent samples) with RaTG13 (no hACE2; n=4 independent samples) showed a significant difference, p=.0097. Individual data points are in black dots. *** p<.001. ** p<.01.(C) Protein pull-down assay using hACE2 as the bait and cell-associated RaTG13 spike as the target. All experiments were repeated independently three times with similar results.
Having compared hACE2 recognition by SARS-CoV-2 and SARS-CoV, we further investigated hACE2 binding by bat RaTG13. To this end, we performed a pseudovirus entry assay in which retroviruses pseudotyped with RaTG13 spike (i.e., RaTG13 pseudoviruses) were used to enter hACE2-expressing human cells (Fig.3b). The results showed that RaTG13 pseudovirus entry into cells depends on hACE2. Additionally, RaTG13 spike was not cleaved on pseudovirus surface. SARS-CoV-2 pseudovirus entry also depends on hACE2, but its spike was cleaved to S2 on pseudovirus surface (likely because of a furin site insertion 16) (Fig.3b). Moreover, we performed a protein pull-down assay using hACE2 as the bait and cell-associated RaTG13 spike as the target (Fig.3c). The result showed that RaTG13 spike was pulled down by hACE2. Therefore, like SARS-CoV-2, bat RaTG13 binds hACE2 and can use hACE2 as its entry receptor.
Discussion
The current SARS-CoV-2 outbreak has become a global pandemic. Our previous structural studies on SARS-CoV have established receptor recognition as an important determinant of SARS-CoV infectivity, pathogenesis, and host range 9. Based on the newly discovered structural information presented here, along with biochemical data, we discuss the receptor recognition and evolution of SARS-CoV-2.
How well does SARS-CoV-2 recognize hACE2 as compared with SARS-CoV? First, our study finds that compared with SARS-CoV, SARS-CoV-2 RBM contains structural changes in the hACE2-binding ridge, largely caused by a four-residue motif (residues 482-485: Gly-Val-Glu-Gly). This structural change allows the ridge to become more compact and form better contact with the N-terminal helix of hACE2 (Fig.1b, 1c). Second, Phe486 from SARS-CoV-2 RBM inserts into a hydrophobic pocket (Fig.1c). The corresponding residue in SARS-CoV RBM is a leucine, which likely forms weaker contact with hACE2 due to its smaller side chain. Third, both virus-binding hotspots have become more stabilized at the RBM/hACE2 interface through interactions with SARS-CoV-2 RBM. As our previous studies showed 11,12, these hotspots on hACE2 are critical for coronavirus binding because they involve two lysine residues that need to be accommodated properly in hydrophobic environments. Neutralizing the charges of the lysines is key to the binding of coronavirus RBDs to hACE2. SARS-CoV-2 RBM has evolved strategies to stabilize the two hotspots: Gln493 and Leu455 stabilize hotspot-31, whereas Asn501 stabilizes hotspot-353. Our biochemical data confirm that SARS-CoV-2 RBD has significantly higher hACE2-binding affinity than SARS-CoV RBD and that the above structural features of SARS-CoV-2 RBM contribute to SARS-CoV-2’s high hACE2-binding affinity. Thus, both structural and biochemical data reveal that SARS-CoV-2 RBD recognizes hACE2 better than SARS-CoV RBD does.
How did SARS-CoV-2 transmit from bats to humans? First, we found that bat RaTG13 uses hACE2 as its receptor, suggesting that RaTG13 may infect humans (Fig.3c). Second, like SARS-CoV-2, bat RaTG13 RBM contains a similar four-residue motif in the ACE2-binding ridge (Extended Data Table.3), supporting that SARS-CoV-2 may have evolved from RaTG13 or a RaTG13-related bat coronavirus. (Extended Data Fig.7). Third, the L486F, Y493Q and D501N residue changes from RaTG13 to SARS-CoV-2 enhance hACE2 recognition and may have facilitated the bat-to-human transmission of SARS-CoV-2 (Extended Data Table.3; Extended Data Fig.7). Note that a lysine-to-asparagine mutation at the 479 position in SARS-CoV RBD (corresponding to the 493 position in SARS-CoV-2 RBD) allowed SARS-CoV to get into human populations 3. Fourth, Leu455 contributes favorably to hACE2 recognition, and it is conserved between RaTG13 and SARS-CoV-2; its presences in SARS-CoV-2 RBM may be important for the bat-to-human transmission of SARS-CoV-2 (Extended Data Table.3; Extended Data Fig.7). Note that host and viral factors other than receptor recognition also play important roles in the cross-species transmission of coronaviruses 20,21. Nevertheless, the identified receptor-binding features of SARS-CoV-2 RBM may have facilitated SARS-CoV-2 to transmit from bats to humans (Extended Data Fig.7).
Are intermediate hosts involved in the potential bat-to-human transmission of SARS-CoV-2? Because bat coronavirus RaTG13 binds hACE2, one possibility is that there is not an intermediate host. Alternatively, pangolins have been proposed to be an intermediate host 22. The structural information provided in this study allows us to inspect and understand the critical RBM residues in coronaviruses isolated from pangolins. Two coronaviruses, CoV-pangolin/GD and CoV-pangolin/GX, have been isolated from pangolins at two different locations in China: Guangdong (GD) and Guangxi (GX). The RBM of CoV-pangolin/GD contains Leu455, the 482-485 loop, Phe486, Gln493 and Asn501 (Extended Data Table.3), all of which are favorable for hACE2 recognition. The RBM of CoV-pangolin/GX contains Leu455 and the 482-485 loop, both of which are favorable for hACE2 recognition; it also contains Leu486, Glu493 and Thr501 (Extended Data Table.3), all of which are less favorable for hACE2 recognition. Therefore, CoV-pangolin/GD potentially recognizes hACE2 well, while CoV-pangolin/GX does not. Hence, GD pangolins, but not GX pangolins, could potentially pass coronaviruses to humans. But, again, many other factors determine the cross-species transmission of coronaviruses 20,21, and the above analysis will need to be verified experimentally.
What does this study inform on intervention strategies? First, neutralizing monoclonal antibodies targeting SARS-CoV-2 RBM can prevent the virus from binding to hACE2, and hence are promising antiviral drugs. Our structure has laid out all of the functionally important epitopes in SARS-CoV-2 RBM that can potentially be targeted by neutralizing antibody drugs. Hence this study can guide the development and optimization of these antibody drugs. Second, the RBD itself can function as a subunit vaccine 10,23. The functionally important epitopes in SARS-CoV-2 RBM, identified in this study, can guide structure-based design of highly efficacious RBD vaccines. We previously developed such a structure-based strategy for subunit vaccine design 24. This strategy may be helpful in designing SARS-CoV-2 RBD vaccines. Overall, this study can guide structure-based intervention strategies that target receptor recognition by SARS-CoV-2.
Methods
Plasmids
SARS-CoV-2 spike (GenBank accession number QHD43416.1), SARS-CoV Spike (GenBank accession number AFR58740.1), RaTG13 Spike (GenBank accession number QHR63300.2), and human ACE2 (GenBank accession number NM_021804) were all synthesized (GenScript Biotech). SARS-CoV-2, SARS-CoV, chimeric RBDs (see Extended Data Fig.1 for residue ranges of RBDs), and human ACE2 ectodomain (residues 1-615) were subcloned into pFastBac vector (Life Technologies) with a N-terminal honeybee melittin signal peptide and a C-terminal His6 tag. Human ACE2 ectodomain (residues 1-615) with a C-terminal Fc tag was also constructed.
Protein expression and purification
All of the proteins were prepared from sf9 insect cells using the Bac-to-Bac system (Life Technologies) as previously described 3. Briefly, the His6-tagged proteins were harvested from cell culture medium, purified on Ni-NTA column, purified further on Superdex200 gel filtration column (GE Healthcare), and stored in a buffer containing 20 mM Tris pH7.2 and 200 mM NaCl. The Fc-tagged protein was purified in the same way as the His6-tagged proteins, except that protein A column replaced Ni-NTA column in the procedure.
Crystallization and structure determination
To purify the RBD/ACE2 complex, human ACE2 and RBD were incubated together, and then the complex was purified on Superdex200 gel filtration chromatography. RBD/ACE2 crystals were grown in sitting drops at room temperature over wells containing 100 mM Tris (pH 8.5), 18-20% PEG 6000, and 100 mM NaCl. Crystals were soaked briefly in 100 mM Tris (pH 8.5), 30% PEG 6000, 100 mM NaCl, and 30% ethylene glycol before being flash-frozen in liquid nitrogen. X-ray diffraction data were collected at the Advanced Photon Source beamline 24-ID-E. The structure was determined by molecular replacement using the structure of human ACE2 complexed with SARS-CoV RBD as the search template (Protein Data Bank accession code 2AJF). Structure data and refinement statistics are shown in Extended Data Table.1.
Protein-protein binding assay
Surface plasmon resonance assay using a Biacore 2000 system (GE Healthcare) were carried out as described previously 12. Briefly, different RBDs were covalently immobilized to a CM5 sensor chip via their amine groups (GE Healthcare). The running buffer contained 10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA and 0.05% Tween 20. Serial dilutions of purified recombinant human ACE2 were injected ranging in concentration from 5 to 80 nM for SARS-CoV-2 RBD and chimeric RBD, and 20-320 nM for SARS-CoV RBD. The resulting data were fit to a 1:1 binding model using Biacore Evaluation Software (GE Healthcare).
Protein pull-down assay was performed using a Dynabeads™ His-Tag Isolation and Pull-down kit (Invitrogen) and a Dynabeads™ Protein A for Immunoprecipitation kit (Invitrogen) according to the manufacturers’ manual. Briefly, 150 μl indicated Dynabeads were washed with PBS buffer (phosphate-buffered saline) and incubated with either 5 μg human ACE2-His6 (human ACE2 with a C-terminal His6 tag) or 5 μg human ACE2-Fc (human ACE2 with a C-terminal Fc tag) on a roller at room temperature for 30 min. After incubation, human ACE2-bound beads were washed 3 times with 1 mL PBST buffer (PBS buffer plus 0.05% Tween-20) on a roller for 10 min and then were aliquoted into different tubes for use. To prepare cell-associated coronavirus spike protein, HEK293T cells were transfected with pcDNA3.1(+) plasmid encoding coronavirus spike (containing a C-terminal C9 tag); 48 hours after transfection, the spike-expressing cells were lysed using a sonicator in immunoprecipitation assay buffer (20 mM Tris-HCl, pH7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton-X-100, supplemented with protease inhibitors) and centrifuged at 12,000 × g for 2 min. The supernatants (containing solubilized SARS-CoV-2 spike) were transferred to mix with the human ACE2-bound beads in 2 mL tubes separately (spike was in excess of human ACE2). After one-hour incubation on a roller at room temperature, beads were washed 3 times with PBST buffer and the bound proteins were eluted using elution buffer (300 mM imidazole, 50 mM sodium phosphate pH 8.0, 300 mM NaCl, 0.01% Tween-20 for human ACE2-His6-bound beads; 0.1 M citric acid pH 2.7 for human ACE2-Fc-bound beads). The samples were then subjected to SDS-PAGE and analyzed through Western blotting using a anti-C9 tag antibody.
Coronavirus-spike-mediated pseudovirus entry assay
Pseudovirus entry assay was performed as described previously 21. Briefly, HEK293T cells were co-transfected with a luciferase-expressing HIV-1 genome plasmid (pNL4-3.luc.RE) and a plasmid encoding SARS-CoV-2 spike or RaTG13 spike. Pseudoviruses were harvested 72 hours post transfection, and were used to enter recipient cells (HEK293T cells exogenously expressing human ACE2). After incubation of pseudoviruses with recipient cells at 37°C for 6 hours, medium was changed and cells were incubated for an additional 60 hours. Cells were then washed with PBS buffer and lysed. Aliquots of cell lysates were transferred to Optiplate-96 (PerkinElmer), followed by addition of luciferase substrate. Relative light units (RLUs) were measured using EnSpire plate reader (PerkinElmer). All measurements were carried out on at least three independent biological samples.
Analyses of protein contact residues and protein buried surface areas
Protein contact residues were analyzed using the LigPlot+ program (Version v.1.4.5) (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/). Protein buried surface areas were analyzed using PDBePISA tool (http://pdbe.org/pisa/).
Extended Data
Extended Data Figure 1:

Sequence alignment of the RBDs from SARS-CoV and SARS-like viruses. RBM is in purple. Previously identified critical ACE2-binding residues are in blue. The seven RBM residues that differ between SARS-CoV-2 wild-type RBD and SARS-CoV-2 chimeric RBD are shaded. A critical arginine on the side loop of SARS-CoV RBM that forms a strong salt bridge with human ACE2 is in green. Another arginine in the core structure that interacts with glycan is in cyan. The residues on the variable loop between two disulfide-bond-forming cysteines in the ACE2-binding ridge are in red. The significant motif changes in the ACE2-binding ridge are underlined. GenBank accession numbers are: QHD43416.1 for SARS-CoV-2 spike; AFR58742 for SARS-CoV spike; AY304486.1 for civet SARS-CoV spike; MG916901.1 for bat Rs3367 spike; QHR63300.2 for bat RaTG13 spike. Two coronaviruses, CoV-pangolin/GD and CoV-pangolin/GX, were isolated from pangolins at two different locations in China, Guangdong (GD) and Guangxi (GX); their RBD sequences were from reference 22.
Extended Data Figure 2:
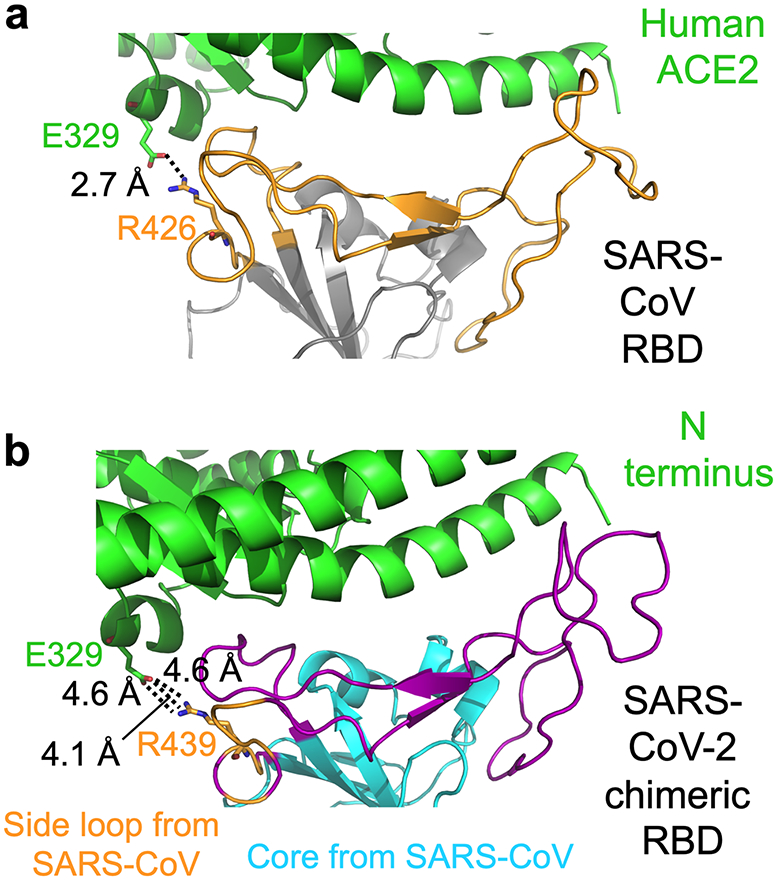
Interface between SARS-CoV-2 or SARS-CoV RBM and human ACE2. (a) Interface between SARS-CoV RBD and human ACE2, showing a strong salt bridge between Arg426 on the side loop in the RBM and Glu329 from human ACE2. Core structure is in grey. RBM is in orange. (b) Interface between SARS-CoV-2 chimeric RBD and human ACE2, showing a weaker, but still energetically favorable, N-O bridge between Arg439 on the side loop in the RBM and Glu329 from human ACE2. The interaction between Arg439 on the side loop in the RBM and Glu329 from human ACE2 is non-natural in SARS-CoV-2 (i.e., resulting from the design of the SARS-CoV-based chimera).
Extended Data Figure 3:

Comparison of human ACE2 binding by SARS-CoV RBD, SARS-CoV-2 wild-type RBD, and SARS-CoV-2 chimeric RBD. (a) Buried surface areas at SARS-CoV RBM/human ACE2 and SARS-CoV-2 RBM/human ACE2 interfaces. In the crystals for both SARS-CoV RBD/ACE2 complex and chimeric RBD/ACE2 complex, two copes of each complex were present in one asymmetric unit. Numbers for both copies of the complexes are shown. The interaction between Arg439 on the side loop in the RBM and Glu329 from human ACE2 was excluded in the calculation of buried surface area for SARS-CoV-2. (b) List of contact residues from RBM and ACE2 that are directly involved in RBM/ACE2 binding. The engineered Arg439 in the chimeric RBD is in orange. Contact residues from SARS-CoV RBM/ACE2 are taken from PDB 2AJF. (c) Binding affinities between the RBDs and human ACE2 as measured using surface plasmon resonance.
Extended Data Figure 4:

Composite omit map of the interface between SARS-CoV-2 RBM and human ACE2. Contour level is 1 sigma.
Extended Data Figure 5:
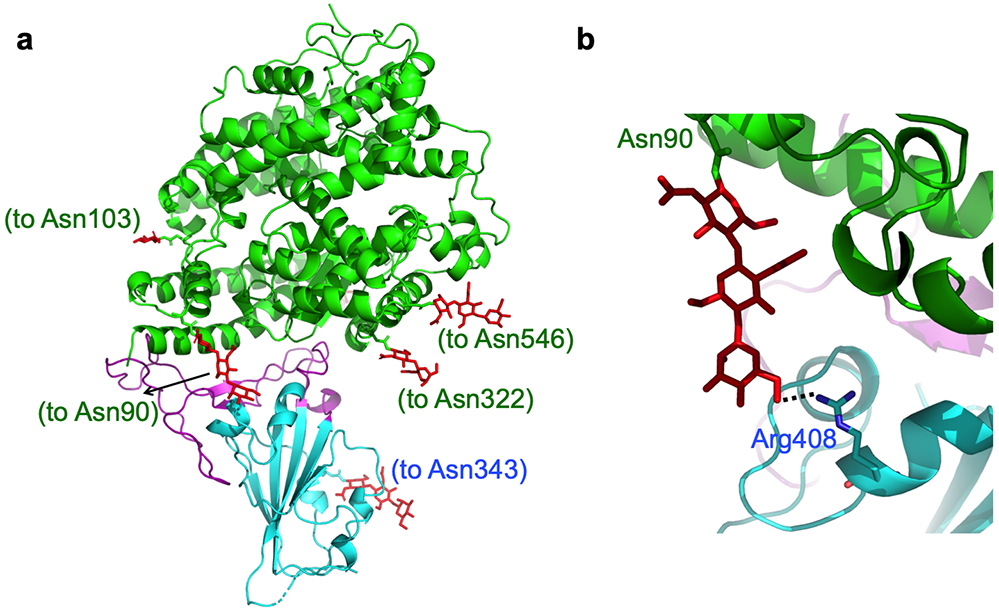
Glycans built into the SARS-CoV-2 chimeric RBD/hACE2 structure. (A) Distribution of glycans in the structure. Glycans are in red. The residues that the glycans attach to are in parentheses. (B) Interaction between a glycan attached the ACE2 residue 90 and Arg408 from the RBD.
Extended Data Figure 6:

Measurement of binding affinities between RBDs and human ACE2 by surface plasmon resonance assay using Biacore. Purified recombinant RBDs were covalently immobilized to the sensor chip via their amine groups, and purified recombinant hACE2 flowed by. Here hACE2 was diluted to different concentrations (from 5 to 80 nM for SARS-CoV-2 RBD and chimeric RBD, and 20-320 nM for SARS-CoV RBD) before being injected. The resulting data were fit to a 1:1 binding model. Each experiment was repeated independently twice with similar results. Each time, five different protein concentrations were used to calculate the Kd values.
Extended Data Figure 7:
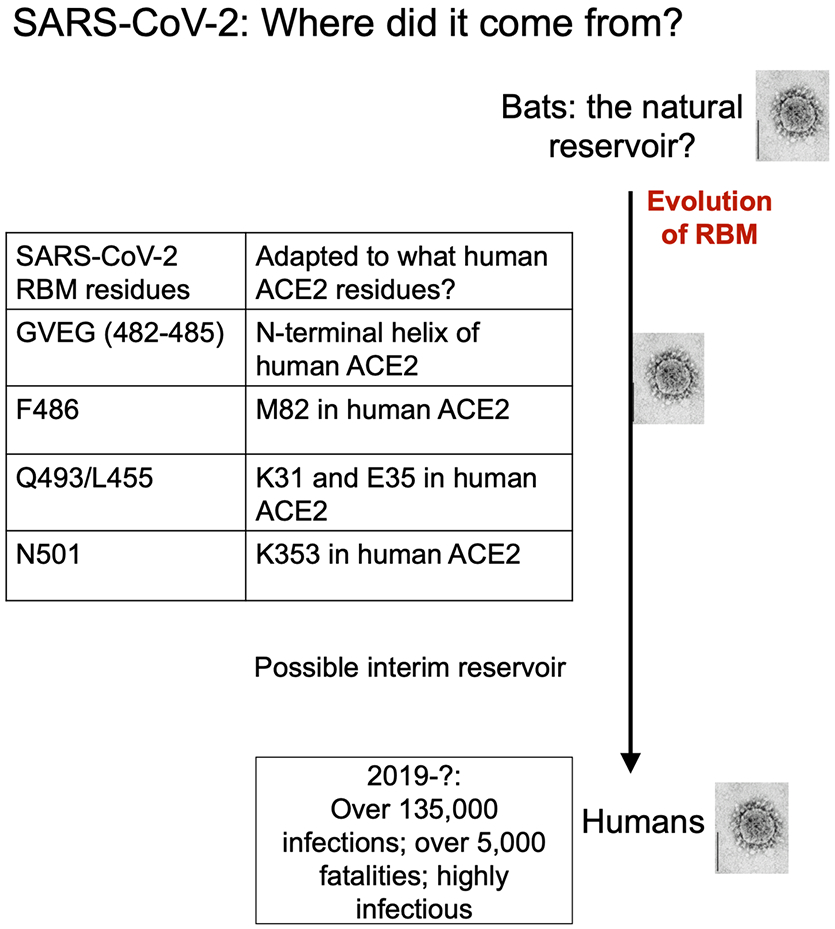
Summary of human ACE2 adaptation and evolution of SARS-CoV-2.
Extended Data Table 1:
Crystallization data collection and refinement statistics.
| Data collection | |
|---|---|
| Space group | P1211 |
| Unit cell dimensions | |
| a, b, c (Å) | 80.435, |
| 118.034, | |
| 112.075 | |
| α, β, γ (°) | 90, 93.12, 90 |
| Resolution (Å) | 59–2.68 |
| (2.78–2.68)* | |
| Rsym or Rmerge | 0.0807 (1.47) |
| I/σI | 12.08 (1.12) |
| Completeness (%) | 98.96 (98.97) |
| Redundancy | 3.9 (4.0) |
| Refinement | |
| Resolution (Å) | 59–2.68 |
| (2.78–2.68)* | |
| No. reflections | 58219 (5774) |
| Rwork / Rfree | 0.197/0.228 |
| No. atoms | 13180 |
| Protein | 12782 |
| Ligand/ion | 372 |
| Water | 26 |
| B-factors | 108.85 |
| Protein | 108.05 |
| Ligand/ion | 138.01 |
| Water | 82.32 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.60 |
Values in parentheses are for highest-resolution shell.
Data processing was done using HKL2000 25. Molecular replacement and model refinement were done using PHENIX and CCP4 26,27. Model building was done using COOT 28. Structural figures were made using PYMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.).
26 crystals were used for X-ray data collection. Each crystal resulted in one set of X-ray data. The best dataset (as judged by data statistics) was used for structure determination and refinement.
Extended Data Table 2:
Summary of binding affinities measured by different studies.
| Protein coated | Kd (M) | Coating method | Detection method |
References |
|---|---|---|---|---|
| SARS-CoV-S1-Fc tag | 1.7×10−9 | Covalently immobilized (via amine group) to sensor chip | SPR | 29 |
| SARS-CoV-RBD-Fc tag | 1.62×10−8 | Non-covalently immobilized (via Fc tag) to sensor chip | SPR | 30 |
| SARS-CoV-RBD-His tag | 1.52×10−7 | Covalently immobilized (via amine group) to sensor chip | SPR | 12 |
| Human ACE2-His tag | 2.09 ×10−8 | Covalently immobilized (via amine group) to sensor chip | SPR | |
| SARS-CoV-RBD-His tag | 3.258×10−7 | Non-covalently immobilized (via His tag) to sensor chip | SPR | 19 |
| SARS-CoV-2-spike-His tag | 1.47×10−8 | Non-covalently immobilized (via His tag) to sensor chip | SPR | |
| SARS-CoV-spike-His tag | 7.7 ×10−9 | Non-covalently immobilized (via His tag) to sensor chip | Blitz | 16 |
| SARS-CoV-2-spike-His tag | 2.9 ×10−9 | Non-covalently immobilized (via His tag) to sensor chip | Blitz | |
| SARS-CoV-S1 | Similar binding affinity | Serial dilution coated on plates | ELISA | 31 |
| SARS-CoV-2-S1 | Serial dilution coated on plates | ELISA | ||
| SARS-CoV-RBD-His tag | 1.85×10−7 | Covalently immobilized (via amine group) to sensor chip | SPR | Current study |
| SARS-CoV-2-RBD-His tag | 4.42×10−8 | Covalently immobilized (via amine group) to sensor chip | SPR |
Protein-protein binding affinities are more accurately measured using SPR than using ELISA 12,16,19,29-31, as ELISA often causes artifacts in protein binding 32. Kd values measured using SPR depend on how the proteins are coated. Non-covalently immobilized proteins via Fc tag or His tag (on the opposite side to ligand-binding sites) have the advantage over covalently immobilized proteins via amine groups because the former have the ligand-binding sites fully exposed. However, non-covalently immobilized proteins risk dissociating from sensor chips, leading to under-evaluated Kd. Covalently immobilized proteins via amine groups do not dissociate from sensor chips, but they are attached to sensor chips in many orientations; for some of these orientations, the ligand-binding sites are not approachable, leading to under-evaluated Kd. Compared with large proteins, the ligand-binding sites on covalently immobilized small proteins are more likely to be buried, leading to under-evaluated Kd. Compared with RBD/hACE2 binding, the spike protein/hACE2 binding is more complex: the RBD in the spike switches between standing up (to expose RBM for hACE2 binding) and lying down (to hide RBM) conformations 16,19, complicating interpretation of measured Kd values. Therefore, Kd values measured from different SPR studies vary, depending on which protein is coated as well as the size and shape of proteins. In a 2012 study 12, the Kd value was higher when the RBD was coated than when the ACE2 was coated. For the current study, we could not coat ACE2 because ACE2 dissociated from sensor chips in regeneration buffer (reason unknown). So we coated the RBD, and the measure Kd value was comparable to that from the 2012 study.
Extended Data Table 3:
Critical human ACE2-binding residues in SARS-CoV-2 and SARS-CoV RBDs.
| Viral RBD | Year | 442 | 468-471 | 472 | 479 | 480 | 487 |
|---|---|---|---|---|---|---|---|
| SARS-human | 2002 | Y | P-PA | L | N | D | T |
| SARS-civet | 2002 | Y | P-PA | L | K | D | S |
| CoV-pangolin/GD | 2020 | L (455) | GVEG (482-485) | F(486) | Q (493) | S (494) | N (501) |
| CoV-pangolin/GX | 2020 | L (455) | GQVG (482-485) | L (486) | E (493) | R (494) | T (501) |
| Rs3367-bat | 2013 | S (443) | P-PA (469-472) | F (473) | N (480) | D (481) | N (488) |
| RaTG13-bat | 2020 | L (455) | GQTG (482-485) | L (486) | Y (493) | R (494) | D (501) |
| SARS-CoV-2 | 2019 | L (455) | GVEG (482-485) | F (486) | Q (493) | S (494) | N (501) |
Acknowledgements
This work was supported by NIH grants R01AI089728 and R01AI110700 (to F.L.) and R35GM118047 (to H.A.). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are supported by an NIH grant P30GM124165. We thank staff at Advanced Photon Source beamline 24-ID-E for assistance in data collection and Professor Yuhong Jiang for statistical consultation and edits to the manuscript.
Footnotes
Data availability statement
Coordinates and structure factors have been deposited to the Protein Data Bank with accession number 6VW1.
Competing interests statement
We declare that none of the authors have financial or non-financial competing interests.
References
- 1.Li Q et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med, doi: 10.1056/NEJMoa2001316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet, doi: 10.1016/s0140-6736(20)30183-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F, Li WH, Farzan M & Harrison SC Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309, 1864–1868, doi: 10.1126/science.1116480 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Li WH et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454, doi: 10.1038/nature02145 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee N et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. New England Journal of Medicine 348, 1986–1994 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Peiris JSM et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361, 1319–1325 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, doi: 10.1038/s41586-020-2012-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perlman S & Netland J Coronaviruses post-SARS: update on replication and pathogenesis. Nature Reviews Microbiology 7, 439–450, doi: 10.1038/nrmicro2147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F Structure, Function, and Evolution of Coronavirus Spike Proteins. Annual review of virology 3, 237–261, doi: 10.1146/annurev-virology-110615-042301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du LY et al. The spike protein of SARS-CoV - a target for vaccine and therapeutic development. Nature Reviews Microbiology 7, 226–236, doi: 10.1038/nrmicro2090 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. Journal of Virology 82, 6984–6991, doi: 10.1128/jvi.00442-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu KL, Peng GQ, Wilken M, Geraghty RJ & Li F Mechanisms of Host Receptor Adaptation by Severe Acute Respiratory Syndrome Coronavirus. Journal of Biological Chemistry 287, 8904–8911, doi: 10.1074/jbc.M111.325803 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan Y, Shang J, Graham R, Baric RS & Li F Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. J Virol, doi: 10.1128/jvi.00127-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letko M, Marzi A & Munster V Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature microbiology, doi: 10.1038/s41564-020-0688-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell, doi: 10.1016/j.cell.2020.02.052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walls AC et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell, doi: 10.1016/j.cell.2020.02.058 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan J et al. Crystal structure of the 2019-nCoV spike receptor-binding domain bound with the ACE2 receptor. bioRxiv, 2020.2002.2019.956235, doi: 10.1101/2020.02.19.956235 (2020). [DOI] [Google Scholar]
- 18.Pylaeva S, Brehm M & Sebastiani D Salt Bridge in Aqueous Solution: Strong Structural Motifs but Weak Enthalpic Effect. Scientific reports 8, 13626, doi: 10.1038/s41598-018-31935-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrapp D et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, doi: 10.1126/science.abb2507 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui J, Li F & Shi ZL Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17, 181–192, doi: 10.1038/s41579-018-0118-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y et al. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci U S A 111, 12516–12521, doi: 10.1073/pnas.1405889111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao K et al. Isolation and Characterization of 2019-nCoV-like Coronavirus from Malayan Pangolins. bioRxiv, 2020.2002.2017.951335, doi: 10.1101/2020.02.17.951335 (2020). [DOI] [Google Scholar]
- 23.Du L et al. MERS-CoV spike protein: a key target for antivirals. Expert opinion on therapeutic targets 21, 131–143, doi: 10.1080/14728222.2017.1271415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du L et al. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nature communications 7, 13473, doi: 10.1038/ncomms13473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Extended Data:
- 25.Otwinowski Z & Minor W in Macromolecular Crystallography, Pt A Vol. 276 Methods in Enzymology 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Liebschner D et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta crystallographica. Section D, Structural biology 75, 861–877, doi: 10.1107/s2059798319011471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winn MD et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography 67, 235–242, doi: 10.1107/s0907444910045749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography 60, 2126–2132, doi: 10.1107/s0907444904019158 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Sui JH et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proceedings of the National Academy of Sciences of the United States of America 101, 2536–2541 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li WH et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. Embo Journal 24, 1634–1643, doi: 10.1038/sj.emboj.7600640 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun C et al. SARS-CoV-2 and SARS-CoV Spike-RBD Structure and Receptor Binding Comparison and Potential Implications on Neutralizing Antibody and Vaccine Development. bioRxiv, 2020.2002.2016.951723, doi: 10.1101/2020.02.16.951723 (2020). [DOI] [Google Scholar]
- 32.Pesce AJ & Michael JG Artifacts and limitations of enzyme immunoassay. Journal of immunological methods 150, 111–119, doi: 10.1016/0022-1759(92)90070-a (1992). [DOI] [PubMed] [Google Scholar]


