Abstract
Background:
Overexpression of c-kit, a tyrosine kinase receptor protein encoded by the protooncogene kit, has been previously reported in thymic epithelial tumors and in other neoplasms such as gastrointestinal stromal tumors, myeloproliferative disorders, melanoma, and seminoma. Mutations in the kit gene have been related to response to imatinib in gastrointestinal stromal tumor and one case report of thymic carcinoma. We studied expression of c-kit in a large retrospective series of thymic epithelial malignancies and sequenced the whole gene in a subset of patients.
Methods:
Thymic epithelial tumors from 120 patients (13 thymic carcinomas and 107 thymomas) were examined. Immunohistochemical staining with an antic-kit polyclonal antibody was performed on a tissue microarray. Mutation analyses of exons 1 to 20 were conducted by direct DNA sequencing of polymerase chain reaction products in eight thymic carcinomas, five thymomas, and one thymic carcinoma cell line.
Results:
The percentage of c-kit positive cells was significantly higher in thymic carcinoma (46%) than in thymoma (4%). Decreased disease-related survival and progression-free survival were observed in c-kit positive tumors. No mutations were detected.
Conclusion:
c-kit expression is strongly but not exclusively related to thymic carcinoma histotype, and it is of prognostic value. Mutations are very rare.
Keywords: c-kit, Thymic carcinoma, Thymoma
Thymic malignancies are rare tumors with an overall incidence of 0.15 each 100,000 persons-year; however, they are the most common anterior mediastinal neoplasm in adults, representing 50% of anterior mediastinal masses.1,2 The 2004 World Health Organization (WHO) classification recognizes five histologic subtypes of thymoma (A, AB, B1, B2, and B3) and 11 subtypes of thymic carcinomas.3 Surgery is the mainstay treatments for localized tumors. However, thymic carcinomas and less frequently thymomas display an aggressive behavior that requires combined modality treatments, including systemic therapy. Many patients with metastatic or unresectable tumors are candidates for systemic platinum-based chemotherapy, which is palliative in nature. Development of new drugs is important in thymic malignancies but is hampered by the rarity of this disease. Several molecularly targeted therapies have recently been investigated in thymic malignancies, but results of phase II studies have so far been disappointing.4
c-kit is a type III cytokine receptor expressed on the membrane of hematopoietic stem cells, mast cells, melanocytes, and interstitial cells of Cajal.5 Stem cell factor binds to c-kit inducing homodimerization of the receptor and phosphorylation of downstream intracellular molecules that regulate cell proliferation, differentiation, adhesion, and apoptosis.6 Overexpression of c-kit is observed in a spectrum of human malignancies, primarily gastrointestinal stromal tumors (GISTs), chronic myeloid leukemias, mast cell neoplasms, melanomas, and seminomas.7 c-kit mutations have also been described in several of these tumors.6,8 In GISTs, the most common mutations affect exons 11, 9, 13, and 17,6 and the site of mutation has prognostic implications in this disease. Responsiveness of GISTs to treatment with the kinase inhibitor imatinib depends largely on the exonic location of the c-kit mutation.6 Higher objective response rates to imatinib have been described in patients with mutations of exon 11 compared with patients with wild-type receptor or with mutations of exon 9.9 Mutations have been also described in patients with negative c-kit expression, evaluated by immunohistochemistry (IHC).10 Durable responses to imatinib treatment were observed in GIST patients with a very low expression of c-kit but showing exon 11 mutation.11
In thymic epithelial tumors, c-kit expression has been reported between 50% and 88% in thymic carcinoma, but it is rare in thymomas (0–5%).7,12–16 In thymic carcinoma, five mutations have been described in total in the literature.7,15–17 Recently, Girard et al.14 reported two c-kit mutations, of seven thymic carcinomas analyzed. One of these mutations (H697Y) was novel and in exon 14, a region not sequenced in previous studies.
In 2004, Strobel et al.17 reported a case of poorly differentiated epidermoid carcinoma with V560del kit mutation who responded to imatinib. In 2009, another case of thymic carcinoma with D820E kit mutation was reported that responded to sorafenib, a multitargeted tyrosine kinase inhibitor, with weak c-kit inhibitory activity.18,19 Two phase II studies have been performed with imatinib in patients with mainly thymic carcinoma.20,21 There were, however, no responses in these two studies that enrolled a total of 16 patients with thymic carcinoma. Unfortunately, only three patients were sequenced for c-kit mutations and found to have wild-type c-kit.20
The objectives of this study were to investigate kit mutation status, to extend sequencing to exons not previously evaluated in thymic epithelial malignancy (TEM), and to define the relationship between protein expression, assessed by immunohistochemistry, and prognosis.
We evaluated the c-kit expression in a series of 120 thymic epithelial tumors and confirmed a higher percentage of c-kit positive cells in thymic carcinomas (46%) than thymomas (4%). We also identified a statistically significant correlation between c-kit expression and poorer disease-related survival (DRS) and progression-free survival (PFS). We sequenced kit gene from exons 1 to 20 of eight formalin-fixed paraffin-embedded (FFPE) thymic carcinoma samples, one thymic carcinoma cell line, and five FFPE thymoma samples. No mutations were identified.
PATIENTS AND METHODS
Patient Samples
FFPE specimens were retrieved from the pathology department of the Istituto Clinico Humanitas (Rozzano, Milan, Italy). Consecutive cases with a diagnosis of thymoma or thymic carcinoma with available tumor tissue were selected. Tumors were reviewed and classified according to the 2004 WHO criteria.22 Tumor staging was according to the revised Masaoka system.23 Main patient characteristics are reported in Table 2. The completeness of resection was classified as R0 = complete resection, R1 = microscopic residual disease infiltrating resection margins, and R2 = macroscopic residual disease.24 This study was conducted in agreement with the Declaration of Helsinki and was approved by the institutional ethical review boards (ClinicalTrials.gov ID: NCT00965627).
TABLE 2.
Patient Characteristics and c-kit Immunohistochemistry
| Samples | c-kit + (%) | + 1 | +2 | +3 | |
|---|---|---|---|---|---|
| Age (y) | |||||
| <55 | 58 | 0 | 0 | 0 | 0 |
| ≥55 | 62 | 10 (16) | 4 | 2 | 4 |
| Sex | |||||
| M | 60 | 5 (8.3) | 1 | 2 | |
| F | 60 | 5 (8.3) | 3 | 0 | 2 |
| Tumor sample | |||||
| Primary | 98 | 10 (10) | 4 | 2 | 4 |
| Relapse | 22 | 0 | 0 | 0 | 0 |
| WHO | |||||
| A | 12 | 2 (17) | 2 | 0 | 0 |
| AB | 24 | 1 (4) | 1 | 0 | 0 |
| B1 | 24 | 0 | 0 | 0 | 0 |
| B1/B2 | 6 | 0 | 0 | 0 | 0 |
| B2 | 7 | 0 | 0 | 0 | 0 |
| B2/B3 | 10 | 0 | 0 | 0 | 0 |
| B3 | 22 | 1 (5) | 0 | 0 | 1 |
| C | 13 | 6 (46) | 1 | 2 | 3 |
| Othera | 2 | 0 | 0 | 0 | 0 |
| Subtotal | |||||
| Thymoma | 107 | 4 (4) | 3 | 0 | 1 |
| Carcinoma | 13 | 6 (46) | 1 | 2 | 3 |
| Stage | |||||
| I | 29 | 4 (14) | 2 | 1 | 1 |
| IIa | 26 | 0 | 0 | 0 | 0 |
| IIb | 17 | 1 (6) | 1 | 0 | 0 |
| IIIa | 12 | 3 (25) | 1 | 1 | 1 |
| IIIb | 3 | 0 | 0 | 0 | 0 |
| Iva | 5 | 0 | 0 | 0 | 0 |
| IVb | 10 | 2 (20) | 0 | 0 | 2 |
| Nab | 18 | 0 | 0 | 0 | 0 |
| Resection | |||||
| RO | 69 | 7 (10) | 3 | 2 | 2 |
| Rl | 22 | 2 (9) | 1 | 0 | 1 |
| R2 | 9 | 1 (11) | 0 | 0 | 1 |
| Na | 20 | 0 | 0 | 0 | 0 |
| Paraneoplasticc | |||||
| Yes | 30 | 1 (3) | 1 | 0 | 0 |
| No | 84 | 8 (10) | 3 | 2 | 3 |
| Na | 6 | 1 (17) | 0 | 0 | 1 |
One micronodular and one cystic thymoma.
Na, not assessable: data are missing at the diagnosis.
Paraneoplastic, paraneoplastic syndromes: 29 myasthenia gravis cases and one autoimmune glomerulonephritis.
WHO, World Health Organization.
Construction of Tissue Microarray
FFPE tumor specimens were assessed for quality and adequacy of fixation and storage. A tissue microarray block containing tissue from 132 thymoma or thymic carcinoma cases was generated. In brief, three punches of 0.36 mm2 (0.6 mm in diameter) were taken from different intratumoral areas in each tumor sample and arranged in the recipient tissue array block. For 21 samples, peritumoral normal tissue was also available and included in the tissue microarray. A pathologist (H.S.L.) verified the presence of tumor tissue on a hematoxylin and eosin-stained tissue microarray slide. Samples were considered adequate if tumor occupied one or more cores of three punches.
Immunohistochemistry
Expression of c-kit was analyzed by IHC. FFPE tissue microarrays were cut at 4 μm, deparaffinized with xylene, and rehydrated in graded ethanol. Antigen retrieval was performed heating the slides at 95°C for 20 minutes in Target Retrieval Solution (Dako, Carpinteria, CA). Endogenous peroxidase blocking solution (EnVision+ System HRP [DAB], Dako) was applied on the tissue for 10 minutes followed by incubation in protein-free T20 (TBS) blocking buffer (Thermo Scientific, Rockford, IL) for 1 hour. Samples were then incubated with c-kit pharmDx Polyclonal Rabbit IgG (c-kit pharmDx IHC kit, Dako) for 30 minutes in a humid chamber at room temperature. After three washes with Tris-Buffer saline Twin 20 0.5% buffer, the slides were incubated for 30 minutes with labeled polymer-HRP as a secondary antibody and then immune reactions were visualized with 3′,3′-diaminobenzidine as chromogen (EnVision+ System HRP [DAB], Dako). Slides were counterstained with hematoxylin, dehydrated, and mounted. Negative control specimens were included with Rabbit IgG Negative Control Re-agent (c-kit pharmDx IHC kit, Dako). As positive controls, specimens of FFPE P815 mouse mastocytoma cell line (c-kit pharmDx IHC kit, Dako) and mouse normal pancreas were used, according to the vendor’s instructions. Degree of immunostaining was scored as follows by one pathologist (H.S.L.) who was blinded to the patients’ information: −, no staining; 1+, staining <10% of tumor cells; 2+, staining ≥10% but <50% of tumor cells; or 3+, staining ≥50% of tumor cells.13,16 Positive were considered samples with at least 1+.
Cell Line
Thymic carcinoma cell line T1889 was kindly provided by Marco Breinig.25 The cells were cultured in RPMI 1640 containing 25 mM Hepes, 200 mM l-glutamine (Gibco, Invitrogen, Grand Island, NY), 50 U/ml penicillin, 50 U/ml streptomycin (Invitrogen), and 10% heat-inactivated calf serum (Invitrogen), and grown in a 37°C incubator with humidified 5% CO2 atm.
Genomic DNA Sequencing
A hematoxylin and eosin-stained slide was obtained for each sample. A pathologist verified the presence of tumor tissue, marking the areas with more than 80% cancer cells on the paraffin blocks. Five cores of tissue were punched from the marked area on FFPE blocks using a 0.6 mm tissue microarray needle by a depth of approximately 1 mm. DNA was extracted using DNeasy kit (Qiagen Inc., Valencia, CA). Samples were sequenced as described previously.26 Briefly, coding sequences were amplified by polymerase chain reaction (PCR) using AmpliTaq Gold PCR Master Mix (Applied Biosystems, Foster City, CA). Primers tagged with M13 sequence were used to sequence exons 1 to 20. The sequences of the primers are listed in Table 1. A total of 40 cycles were performed using Veriti 96-Well Thermal Cycler (Applied Biosystems) at 94°C for 30 seconds, 60°C for 45 seconds, and 72°C for 45 seconds. Polymerase chain reaction products underwent ExoSAP-IT (USB, Cleveland, OH) purification. The purified product was directly sequenced using a BigDye terminator v 3.1 cycle sequencing kit (Applied Biosystems) and 3730xl DNA Analyzer (Applied Biosystems). Data were analyzed using Mutation Surveyor v 3.23 (SoftGenetics LLC, State College, PA).
TABLE 1.
Primers Used for Polymerase Chain Reactions
| Exon | Forward Sequence | Reverse Sequence |
|---|---|---|
| 1 | TTAACACGTCGAAAGAGCAGG | AGTCCTCTCTCCGGATGCAC |
| 2 | AAATAGCAGGGCAGCTTTGTC | GCTCAGTCATCCATATGTCATCC |
| 3 | GCCACTAGTCATGAAAGGCAAC | CACTAAGGTGGATCAACGAGAAG |
| 4 | GGATGCTTGATTTAATTGCTGG | TGACAGACGCACTAGTCGAGG |
| 5 | TGACAGACTTGTCATGATGCTTT | AACAGCTTTCCACTTTCCTCC |
| 6 | GCCAATTAATACTGGAAATCAACC | GGATGAGGACATAGGAGGCAG |
| 7 | TCCCAGATGGAATATGTGTGTG | CACAAGTTGAGTCCTTGCAGC |
| 8 | CAGCCTCAGGAAGGTTGTAGG | GCTAGAAATTGCATGATAAATCCAGA |
| 9 | CACTAGGTCACCAAAGTGCTTATTC | GAAATGACATGGTCAATGTTGG |
| 10 | TCACATAGCTTTGCATCCTGC | TCATACATGGGTTTCTGTGGG |
| 11 | TGCTGATTGGTTTCGTAATCG | AAACAAAGGAAGCCACTGGAG |
| 12 | GAAACTGCACAAATGGTCCTTC | AGTTCAGACATGAGGGCTTCC |
| 13 | AAGATGCTCAAGCGTAAGTTCC | ACTAGGGTATGTCCTGGGCTG |
| 14 | TATTAATGGCCATGACCACCC | CAGCCTTGATTGCAAACCC |
| 15 | ACACCTAGTTTCTGGGCATGG | AGAACTGGTCTGCATCATTGC |
| 16 | AGCCTTTGGTATGTCATTGCC | TTCCAAAGAGACAGCAGTTGG |
| 17 | TCCCTATGAATGAAAGCAGTCC | TGTGTGATATCCCTAGACAGGATTTAC |
| 18 | ACTCCACATTTCAGCAACAGC | TCTTACATTTCAGCAGGTGCG |
| 19 | ACGTTTGAAAGTGACGTCTGG | AACCCTCAACATCTGGGTTTC |
| 20 | GCTGGATGCCCATACATTTG | AAGCCCAATTTGCAACCTAAG |
Statistical Analysis
Clinical and biologic features were compared using the Fisher’s exact test or χ2 tests, when appropriate. Survival curves were generated using the Kaplan-Meier method, and differences between curves were analyzed using the Log-Rank and Breslow test. DRS was calculated from the date of surgery to the death date. Patients alive or dead for causes other than thymoma were censored. PFS was calculated from the date of surgery to date of progression assessed according to RECIST criteria. Patients without evidence of progression were censored. All tests were performed using SPSS version 17 (SPSS Inc., Chicago, IL).
RESULTS
c-kit IHC
Patient characteristics of results of c-kit expression and grading are reported in Table 2. Examples of positive and negative samples are shown in Figures 1A, B. There was no c-kit expression by IHC in the normal residual thymus of nine evaluable patients. Ten (8.3%) of 120 evaluable samples were positive for c-kit expression. This included 6 of 13 thymic carcinomas tested (46%) and 4 of 107 thymomas (4%). c-kit expression was significantly more frequent in thymic carcinoma than in thymoma (Fisher’s exact test, p < 0.0001). There was no difference in c-kit expression between early versus advanced stages, level of completeness of resection, and primary versus relapsed tumors. However, c-kit-positive samples were all found in primary tumors (98 cases) and none in relapsed tumors (22 cases). All patients with tumors showing c-kit expression were older than the median age of 55 years (Fisher’s exact test, p = 0.001).
FIGURE 1.
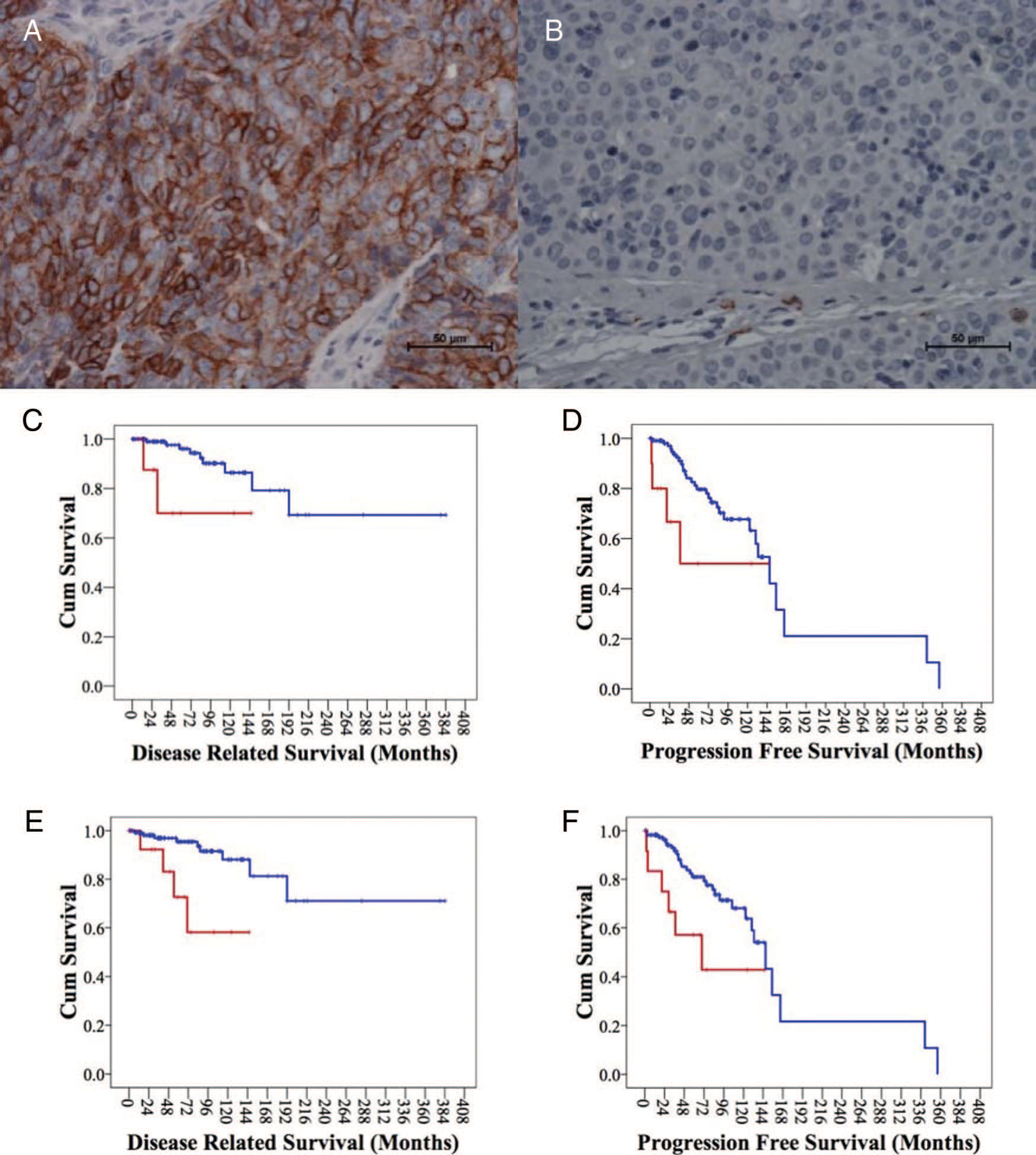
A, Membrane c-kit positive staining, ×40 original magnification. B, c-kit negative staining, ×40 original magnification. C, Kaplan-Meier plot for disease-related survival of patients positive or negative for c-kit expression. D, Progression-free survival related to c-kit expression. E, Disease-related survival of thymoma and thymic carcinoma patients. F, Progression-free survival of thymoma and thymic carcinoma patients.
c-kit expression was associated with a worse DRS (Log-Rank, p = 0.028 and Breslow, p = 0.002). The 10-year DRS was 90% for c-kit-negative patients and 71% for c-kit-positive patients (Figure 1C). A statistically significant worse PFS was observed using the Breslow test (p = 0.001) but not using the Log-Rank test (p = 0.061). The 10-year PFS was 70% for c-kit-negative patients and 50% for c-kit-positive patients (Figure 1D).
Patients with thymic carcinoma showed a statistically significant worse DRS than thymomas (Log-Rank, p = 0.002 and Breslow, p = 0.003). Median survival was not reached for both groups. The 10-year DRS was 60% for thymic carcinomas and 91% for thymomas (Figure 1E). Also, the PFS was significantly worse for thymic carcinomas (Log-Rank, p = 0.021 and Breslow, p = 0.002). The estimated median PFS was 69.4 months for thymic carcinoma (confidence interval: 0–142.7) and 147 months for thymomas (confidence interval: 121.7–172.4). The 10-year PFS was 43% for thymic carcinoma and 72% for thymomas (Figure 1F). By multivariate analysis, c-kit expression and the difference between thymic carcinoma and thymoma were neither independent prognostic factors for DRS (p = 0.67 and 0.22, respectively) nor for PFS (p = 0.79 and p = 0.23, respectively).
kit Sequencing
We sequenced kit in eight thymic carcinomas and five thymomas from FFPE samples with a success rate of 77.5%. Histologic characteristics of the tumors that underwent sequencing are reported in Table 3. We did not detect mutations in any of the tumor samples and in the T1889 thymic carcinoma cell line, previously reported to be negative for c-kit expression.25 We observed the previously reported single-nucleotide polymorphism corresponding to M541L in one type A thymoma and in one thymic carcinoma.
TABLE 3.
Thymic Carcinoma and Thymoma Histology, Mutation, and c-kit Immunohistochemistry
| Sample | Histotype | c-kit IHC | kit Mutation |
|---|---|---|---|
| I-Thy 1 | Squamous cell carcinoma | + 3 | No |
| I-Thy 2 | Mucinous adenocarcinoma | 0 | No |
| I-Thy 3 | Squamous cell carcinoma | + 2 | – |
| I-Thy 4 | Mucoepidermoid carcinoma | 0 | No |
| I-Thy 5 | Squamous cell carcinoma | 0 | No |
| I-Thy 6 | Neuroendocrine carcinoma | 0 | – |
| I-Thy 7 | Squamous cell carcinoma | + 1 | – |
| I-Thy 8 | Squamous cell carcinoma | + 3 | No |
| I-Thy 9 | Squamous cell carcinoma | + 2 | No |
| I-Thy 10 | Neuroendocrine carcinoma | 0 | No |
| I-Thy 11 | Mucinous adenocarcinoma | + 3 | No |
| I-Thy 12 | Undifferentiated carcinoma | 0 | – |
| I-Thy 13 | Undifferentiated carcinoma | 0 | – |
| I-Thy 14 | Thymoma type A | + 1 | No |
| I-Thy 15 | Thymoma type B3 | 0 | No |
| I-Thy 16 | Thymoma type AB | + 1 | No |
| I-Thy 17 | Thymoma type A | + l | No |
| I-Thy 18 | Thymoma type B3 | 0 | No |
| T-1889 | Undifferentiated carcinoma cell line | 025 | No |
IHC, immunohistochemistry.
DISCUSSION
In our study of 120 patients with thymic malignancies, we confirmed a significantly higher incidence of c-kit-positive cells in thymic carcinoma compared with thymoma. This is one of the largest series reporting on expression of c-kit in TEM. We observed c-kit expression in 46% of thymic carcinomas and in 4% of thymomas. The low expression in thymomas is in line with previous reports where expression ranged between 0% and 5%.7,12,13,15,16 Overall, considering all cases reported in the literature, only 7 of 366 thymomas analyzed resulted positive for c-kit expression (2%) (Table 4). c-kit expression in thymic carcinomas has been reported to range between 50% and 88%.7,12,13,15,16 Overall, 95 of 127 (75%) thymic carcinomas resulted positive for c-kit expression.7,12–18,20,21,27 Thymic epithelial cells are considered the cells of origin for both thymoma and thymic carcinoma, and normal thymic epithelium does not express c-kit.28 Our study confirmed the lack of c-kit expression in residual normal thymus of nine thymoma patients.
TABLE 4.
Summary of Published Reports of c-kit Expression and Mutation in TEMs
| Reports | Samples | IHC − | IHC + | Mutation − | Mutation + | Described Mutations |
|---|---|---|---|---|---|---|
| Retrospective studies | ||||||
| Henley et al.12 | Thymoma | 19 | 1 (5%) | |||
| T. carcinoma | 4 | 11 (73%) | ||||
| Pan et al.36 | Thymoma | 110 | 0 | |||
| T. carcinoma | 3 | 19 (86%) | 21 | |||
| Nakagawa et al.13 | Thymoma | 48 | 2 (4%) | |||
| T. carcinoma | 4 | 16 (80%) | ||||
| Tsuchida et al.16 | Thymoma | 20 | 0 | |||
| T. carcinoma | 6 | 11 (65%) | 9 | |||
| Yoh et al.15 | Thymoma | 24 | 0 | 22 | ||
| T. carcinoma | 15 (88%) | 10 | 1 | L576P | ||
| Girard et al.14 | Thymoma | 33 | 0 | 38 | ||
| T. carcinoma | 3 | 3 (50%) | 5 | 2 | V560del | |
| Phase II studies | ||||||
| Salter et al.27 | T. carcinoma | 2 | 9 | |||
| Giaceone et al.20 | Thymoma | 2 | 0 | 2 | ||
| T. carcinoma | 1 | 1 | 1 | |||
| Case reports | ||||||
| Strobel et al.17 | T carcinoma | 1 | 1 | V560del | ||
| Bisagni et al.18 | T. carcinoma | 1 | 1 | D820E | ||
| Li et al37 | T. carcinoma | 1 | ||||
| Vasamiliette et al.28 | T. carcinoma | 1 | ||||
| Our series | Thymoma | 103 | 4 (4%) | 5 | ||
| T. carcinoma | 7 | 6 (46%) | 8 | |||
| Total | Thymoma | 359 | 7 (2%) | 67 | 0 | |
| Thymic carcinoma | 32 | 95 (75%) | 54 | 5 | ||
IHC, immunohistochemistry; TEM, thymic epithelial malignancy.
The variability of c-kit expression in thymic carcinomas may be related to the small number of cases analyzed in single studies, ranging between 6 and 22. Although most of the studies used the same antibody that we also used, not all studies considered only membranous staining as positive, which may have resulted in the higher percentage of positivity. Moreover, these results also underline the histologic heterogeneity of the thymic carcinoma group for which the 2004 WHO classification describes 11 subtypes.22 c-kit expression seems to be frequent in squamous cell thymic carcinoma in all published reports. However, expression has been described in other subtypes as well, including lymphoepithelioma-like,12 undifferentiated carcinomas,12 and basaloid carcinoma.13 In our study, one of two mucinous adenocarcinomas stained strongly positive.
For the first time, we report a worse DRS and PFS for patients with c-kit expressing tumors. These data suggest a negative prognostic role for c-kit expression especially within the first 3 years (Figure 1D). c-kit expression has also been reported as a poor prognostic factor in small cell lung cancer.29,30 We used DRS and PFS as prognostic endpoint rather than overall survival because of the long expected survival after radical surgery. The median DRS was not reached for patients with either c-kit-positive or negative tumors. However, by multivariate analysis, c-kit expression was not an independent prognostic factor for TEM histotype in our study.
The 10-year DRS (60%) that we observed in thymic carcinoma differs from previously reported overall survival (around 40%) in a similar surgical series.32 However, in our study, we assessed DRS and not overall survival, which in a relatively indolent disease may not be accurate. Moreover, in the study by Chen et al.31 thymic carcinomas were diagnosed in higher stages of the disease compared with our series (83% versus 69% stages III and IV, respectively).
We did not observe any c-kit positive tumors in patients who underwent surgery for relapse. Although c-kit expression is strongly related to thymic carcinoma histology, even if the relapse rate is high in thymic carcinoma, these tumors are mostly not resectable due to their more aggressive and infiltrative behavior. If fact, no thymic carcinoma was resected on relapse, in our series.
Only a few c-kit mutations have been reported in the literature in TEM and exclusively in thymic carcinomas. Table 4 summarizes the studies that have investigated c-kit mutations in TEMs. No kit mutations were reported in thymomas among 88 cases sequenced. In thymic carcinoma, only five mutations have been described in 59 cases studied (9%). However, this includes also case reports of two patients who responded to c-kit tyrosine kinase inhibitors.17,18
In 2004, Strobel et al.17 reported a V560del mutation of kit in a case of metastatic poorly differentiated epidermoid carcinoma. This patient experienced a response to treatment with imatinib that lasted 6 months. In 2009, Bisagni et al. reported a case of an undifferentiated thymic carcinoma carrying mutation D820E encoded by kit exon 17. The patient was treated with sorafenib, and the authors reported a partial response that lasted over 15 months.18 The mutation D820E of c-kit activation loop has been previously reported as an acquired resistant mutation to imatinib and sunitinib treatment in GISTs.32,33
Girard et al.14 recently reported two mutations of seven thymic carcinomas. The authors sequenced exons 10 and 14 in addition to 9, 11, 13, and 17 evaluated in previous studies. Interestingly, one of the mutations, H697Y, was in exon 14. H697Y showed higher sensitivity to sunitinib than imatinib in vitro when transfected in Ba/F3 cells. These results underline the importance to extend the analysis to regions of kit in thymic carcinomas beyond the most frequent sites of mutations in GISTs: exon 9, 11, 13, and 17.6 We sequenced kit from exons 1 to 20 in five thymic carcinomas and three thymomas expressing the receptor, and in five that did not express c-kit, and failed to identify any mutations. Pan et al.7 in 2004 and Tsuchida et al.16 in 2008 sequenced these exons in 21 and nine thymic carcinomas, respectively, reporting no mutations. Yoh et al.15 in 2008 identified the L576P kit mutation in exon 17 of a thymic carcinoma. This mutation was previously described in GIST to be activating and resistant to imatinib.35
In GISTs, kit mutations have been described both in c-kit positive tumors (the majority) and in c-kit negative tumors.10 Durable responses to imatinib were also observed in patients with exon 11 mutation and a very low expression of c-kit.11
Given the frequent expression of c-kit reported in thymic carcinomas, and the case reports of response to imatinib, two small phase II studies were performed with this drug. A phase II study including five thymic carcinomas and two B3 thymomas did not show any activity of this agent.20 Salter et al.21 reported a series of 11 thymic carcinomas treated with imatinib where only three obtained a stable disease as best response. Unfortunately, only one patient with thymic carcinoma was sequenced in these studies.
Strengths of this study are represented by the large samples size and by the prognostic implication of c-kit expression, reported for the first time in TEM. On the other hand, the relative small number of c-kit-positive thymic carcinomas sequenced and the great difference in the number of samples that were c-kit positive versus negative have made the difference in survival less impressive.
TEMs are rare tumors, and thymic carcinoma represents approximately only 10 to 20% of them.22 Because of the rarity of thymic carcinomas and that responses have only been described in case reports, and the frequency of reported mutations is probably less than 10% in thymic carcinomas, this represents only approximately five cases potentially diagnosed in the United States every year. This would make a phase II study in thymic carcinomas with c-kit mutations very difficult to accrue.
ACKNOWLEDGMENTS
Supported by the NIH/NCI intramural program.
The authors thank Dr. Marco Brenig for providing the T1889 thymic carcinoma cell line and and Dr. Maria Merino for optimization of immunohistochemistry.
Footnotes
Disclosure: The authors have no financial conflicts of interest.
REFERENCES
- 1.Engels EA, Pfeiffer RM. Malignant thymoma in the United States: demographic patterns in incidence and associations with subsequent malignancies. Int J Cancer 2003;105:546–551. [DOI] [PubMed] [Google Scholar]
- 2.Takeda S, Miyoshi S, Akashi A, et al. Clinical spectrum of primary mediastinal tumors: a comparison of adult and pediatric populations at a single Japanese institution. J Surg Oncol 2003;83:24–30. [DOI] [PubMed] [Google Scholar]
- 3.Okumura M, Ohta M, Tateyama H, et al. The World Health Organization histologic classification system reflects the oncologic behavior of thymoma: a clinical study of 273 patients. Cancer 2002;94:624–632. [DOI] [PubMed] [Google Scholar]
- 4.Rajan A, Giaccone G. Treatment of advanced thymoma and thymic carcinoma. Curr Treat Options Oncol 2008;9:277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edling CE, Hallberg B. c-Kit—a hematopoietic cell essential receptor tyrosine kinase. Int J Biochem Cell Biol 2007;39:1995–1998. [DOI] [PubMed] [Google Scholar]
- 6.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol 2004;22:3813–3825. [DOI] [PubMed] [Google Scholar]
- 7.Pan CC, Chen PC, Chiang H. Overexpression of KIT (CD117) in chromophobe renal cell carcinoma and renal oncocytoma. Am J Clin Pathol 2004;121:878–883. [DOI] [PubMed] [Google Scholar]
- 8.Inokuchi K, Yamaguchi H, Tarusawa M, et al. Abnormality of c-kit oncoprotein in certain patients with chronic myelogenous leukemia—potential clinical significance. Leukemia 2002;16:170–177. [DOI] [PubMed] [Google Scholar]
- 9.Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol 2003;21:4342–4349. [DOI] [PubMed] [Google Scholar]
- 10.Medeiros F, Corless CL, Duensing A, et al. KIT-negative gastrointestinal stromal tumors: proof of concept and therapeutic implications. Am J Surg Pathol 2004;28:889–894. [DOI] [PubMed] [Google Scholar]
- 11.Bauer S, Corless CL, Heinrich MC, et al. Response to imatinib mesylate of a gastrointestinal stromal tumor with very low expression of KIT. Cancer Chemother Pharmacol 2003;51:261–265. [DOI] [PubMed] [Google Scholar]
- 12.Henley JD, Cummings OW, Loehrer PJ Sr. Tyrosine kinase receptor expression in thymomas. J Cancer Res Clin Oncol 2004;130:222–224. [DOI] [PubMed] [Google Scholar]
- 13.Nakagawa K, Matsuno Y, Kunitoh H, et al. Immunohistochemical KIT (CD117) expression in thymic epithelial tumors. Chest 2005;128:140–144. [DOI] [PubMed] [Google Scholar]
- 14.Girard N, Shen R, Guo T, et al. Comprehensive genomic analysis reveals clinically relevant molecular distinctions between thymic carcinomas and thymomas. Clin Cancer Res 2009;15:6790–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoh K, Nishiwaki Y, Ishii G, et al. Mutational status of EGFR and KIT in thymoma and thymic carcinoma. Lung Cancer 2008;62:316–320. [DOI] [PubMed] [Google Scholar]
- 16.Tsuchida M, Umezu H, Hashimoto T, et al. Absence of gene mutations in KIT-positive thymic epithelial tumors. Lung Cancer 2008;62:321–325. [DOI] [PubMed] [Google Scholar]
- 17.Strobel P, Hartmann M, Jakob A, et al. Thymic carcinoma with overexpression of mutated KIT and the response to imatinib. N Engl J Med 2004;350:2625–2626. [DOI] [PubMed] [Google Scholar]
- 18.Bisagni G, Rossi G, Cavazza A, et al. Long lasting response to the multikinase inhibitor bay 43–9006 (Sorafenib) in a heavily pretreated metastatic thymic carcinoma. J Thorac Oncol 2009;4:773–775. [DOI] [PubMed] [Google Scholar]
- 19.Wilhelm SM, Carter C, Tang L, et al. BAY 43–9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res 2004;64:7099–7109. [DOI] [PubMed] [Google Scholar]
- 20.Giaccone G, Rajan A, Ruijter R, et al. Imatinib mesylate in patients with WHO B3 thymomas and thymic carcinomas. J Thoracic Oncol 2009;4: 1270–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salter JT, Lewis D, Yiannoutsos C, et al. Imatinib for the treatment of thymic carcinoma. J Clin Oncol 2008;26:8116. [Google Scholar]
- 22.Muller-Hermelink H, Curtis C. Tumors of the Lung, Pleura, Thymus and Heart, World Health Organization Classification of Tumours. Lyon, IARC Press, 2004. Pp 146–197. [Google Scholar]
- 23.Masaoka A, Yamakawa Y, Niwa H, et al. Thymectomy and malignancy. Eur J Cardiothorac Surg 1994;8:251–253. [DOI] [PubMed] [Google Scholar]
- 24.Strobel P, Bauer A, Puppe B, et al. Tumor recurrence and survival in patients treated for thymomas and thymic squamous cell carcinomas: a retrospective analysis. J Clin Oncol 2004;22:1501–1509. [DOI] [PubMed] [Google Scholar]
- 25.Ehemann V, Kern MA, Breinig M, et al. Establishment, characterization and drug sensitivity testing in primary cultures of human thymoma and thymic carcinoma. Int J Cancer 2008;122:2719–2725. [DOI] [PubMed] [Google Scholar]
- 26.Allander SV, Nupponen NN, Ringner M, et al. Gastrointestinal stromal tumors with KIT mutations exhibit a remarkably homogeneous gene expression profile. Cancer Res 2001;61:8624–8628. [PubMed] [Google Scholar]
- 27.Vasamiliette J, Hohenberger P, Schoenberg S, et al. Treatment monitoring with (18)F-FDG PET in metastatic thymoma after (90)Y-Dotatoc and selective internal radiation treatment (SIRT). Hell J Nucl Med 2009;12:271–273. [PubMed] [Google Scholar]
- 28.Natali PG, Nicotra MR, Sures I, et al. Expression of c-kit receptor in normal and transformed human nonlymphoid tissues. Cancer Res 1992; 52:6139–6143. [PubMed] [Google Scholar]
- 29.Rohr UP, Rehfeld N, Pflugfelder L, et al. Expression of the tyrosine kinase c-kit is an independent prognostic factor in patients with small cell lung cancer. Int J Cancer 2004;111:259–263. [DOI] [PubMed] [Google Scholar]
- 30.Blackhall FH, Pintilie M, Michael M, et al. Expression and prognostic significance of kit, protein kinase B, and mitogen-activated protein kinase in patients with small cell lung cancer. Clin Cancer Res 2003;9: 2241–2247. [PubMed] [Google Scholar]
- 31.Chen G, Marx A, Wen-Hu C, et al. New WHO histologic classification predicts prognosis of thymic epithelial tumors: a clinicopathologic study of 200 thymoma cases from China. Cancer 2002;95:420–429. [DOI] [PubMed] [Google Scholar]
- 32.Guo T, Hajdu M, Agaram NP, et al. Mechanisms of sunitinib resistance in gastrointestinal stromal tumors harboring KITAY502–3ins mutation: an in vitro mutagenesis screen for drug resistance. Clin Cancer Res 2009;15:6862–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinomura Y, Kinoshita K, Tsutsui S, et al. Pathophysiology, diagnosis, and treatment of gastrointestinal stromal tumors. J Gastroenterol 2005; 40:775–780. [DOI] [PubMed] [Google Scholar]
- 34.Conca E, Negri T, Gronchi A, et al. Activate and resist: L576P-KIT in GIST. Mol Cancer Ther 2009;8:2491–2495. [DOI] [PubMed] [Google Scholar]
- 35.Pan CC, Chen PC, Chiang H. KIT (CD117) is frequently overexpressed in thymic carcinomas but is absent in thymomas. J Pathol 2004;202: 375–381. [DOI] [PubMed] [Google Scholar]
- 36.Li XF, Chen Q, Huang WX, et al. Response to sorafenib in cisplatin-resistant thymic carcinoma: a case report. Med Oncol 2009;26:157–160. [DOI] [PubMed] [Google Scholar]


