Abstract
Fertility depends, in part, on interactions between male and female reproductive proteins inside the female reproductive tract (FRT) that mediate postmating changes in female behaviour, morphology, and physiology. Coevolution between interacting proteins within species may drive reproductive incompatibilities between species, yet the mechanisms underlying postmating–prezygotic (PMPZ) isolating barriers remain poorly resolved. Here, we used quantitative proteomics in sibling Drosophila species to investigate the molecular composition of the FRT environment and its role in mediating species-specific postmating responses. We found that (i) FRT proteomes in D. simulans and D. mauritiana virgin females express unique combinations of secreted proteins and are enriched for distinct functional categories, (ii) mating induces substantial changes to the FRT proteome in D. mauritiana but not in D. simulans, and (iii) the D. simulans FRT proteome exhibits limited postmating changes irrespective of whether females mate with conspecific or heterospecific males, suggesting an active female role in mediating reproductive interactions. Comparisons with similar data in the closely related outgroup species D. melanogaster suggest that divergence is concentrated on the D. simulans lineage. Our study suggests that divergence in the FRT extracellular environment and postmating response contribute to previously described patterns of PMPZ isolation and the maintenance of species boundaries.
Keywords: Drosophila, fertility, reproductive isolation, sexual selection, speciation
1. Introduction
In species with internal fertilization, fertility depends on complex and potentially protracted interactions between the sexes that take place within the female reproductive tract (FRT) [1–3]. These interactions are expected to be particularly dynamic in species in which females mate with multiple males, because postcopulatory sexual selection (including sexual conflict) drives the evolution of sex-specific traits that influence male competitive fertilization success and female control over paternity [4–9]. In turn, the coevolution of interacting male and female reproductive traits within populations [10] may generate postmating–prezygotic (PMPZ) incompatibilities between populations [11], potentially contributing to the formation of new species [12–15].
Interactions between male and female reproductive proteins mediate a suite of postmating changes in female behaviour, morphology, physiology, and gene expression, collectively known as ‘postmating responses' [1,2,16,17]. In particular, male-derived seminal fluid proteins (SFPs) transferred during mating trigger multiple postmating responses, including changes in female receptivity to remating, rates of ovulation and oviposition, patterns of sperm storage and usage, and female lifespan (reviewed in [1,17]). Studies in a growing number of insect taxa have characterized the transcriptomic and/or proteomic changes that occur in females after mating (e.g. fruit flies [18–22], mosquitoes [23,24], honeybees [25], butterflies and moths [26,27]). Because SFPs are among the most rapidly evolving proteins known [12,13], coevolution between functionally interacting male and female reproductive proteins is likely to result in species-specific molecular interactions that are needed to coordinate a ‘successful' female postmating response [22,26,28]. Theory predicts that divergence in these traits may represent a taxonomically widespread ‘engine of speciation' [2,14], yet it is currently unknown whether male–female molecular interactions evolve sufficiently fast to be relevant to the speciation process. Few studies have examined how divergent ejaculate–female interactions may result in different female postmating responses among closely related species or between conspecific and heterospecific crosses [22,26,28]. Consequently, our understanding of how molecular interactions between the sexes mediate reproductive outcomes relevant to the formation of PMPZ reproductive isolation remains limited.
Drosophila simulans and D. mauritiana are a model system for studying the evolution of PMPZ reproductive isolation due to their very recent evolutionary divergence and phylogenetic proximity to the genetic model D. melanogaster [29–36]. The two species diverged from a common ancestor approximately 240 000 years ago, and the common ancestor of the D. simulans clade diverged from D. melanogaster approximately 3 million years ago [37]. Drosophila simulans is a cosmopolitan, human commensal, yet contemporary populations are geographically isolated from those of D. mauritiana, which is endemic to the islands of Mauritius [29]. Previous studies have identified several physiological and behavioural mechanisms underlying PMPZ barriers in these species. First, many matings between D. simulans females and D. mauritiana males are of abnormally short duration, which interrupts sperm transfer and results in a low oviposition rate [29,34,36]. Second, although D. simulans females who do copulate for long enough with D. mauritiana males receive as many sperm as in conspecific matings [34,36], they tend to eject heterospecific ejaculates more rapidly than conspecific ejaculates [36]. Third, when D. simulans females mate with both D. simulans and D. mauritiana males, progeny are sired predominantly by the conspecific male, regardless of the order of matings, due to species-specific patterns of sperm transfer, storage, and usage [32,35,36,38]. Because our knowledge of PMPZ isolating mechanisms in this sibling species pair exceeds that of any other taxa, it is an ideal system for studying how divergence in female postmating responses may contribute to PMPZ reproductive isolation.
In this study, we take a quantitative proteomics approach to investigate the molecular mechanisms underlying male–female postmating interactions within and among species, and their potential role in mediating species-specific postmating responses. Specifically, we used highly accurate isobaric labelling to compare the protein composition of the FRT before and after mating, and after both conspecific and heterospecific inseminations in D. simulans and D. mauritiana. We then used semi-quantitative proteomics in the closely related outgroup species D. melanogaster to polarize the relative divergence of postmating responses observed on the D. simulans and D. mauritiana lineages. Our results shed light on how divergence in the FRT molecular environment may have contributed to mechanisms of conspecific sperm precedence that underlie PMPZ reproductive isolation in these closely related species [32,36].
2. Material and methods
Detailed descriptions can be found in the electronic supplementary material.
(a). Sample collection and preparation
We collected FRT samples from the following five conditions: Drosophila simulans virgins (hereafter called sim virgins), D. mauritiana virgins (mau virgins), D. simulans females mated to D. simulans males (sim × sim), D. mauritiana females mated to D. mauritiana males (mau × mau), and D. simulans females mated to D. mauritiana males (sim × mau). It was not possible to collect sufficient tissue from the reciprocal hybrid cross (D. mauritiana females mated to D. simulans males) due to the high frequency of female rejection [30]. Mated samples were collected 6 h after the end of a successful copulation. We chose 6 h as the postmating timepoint to maximize our chances of detecting proteomic differences between virgins and mated females. D. melanogaster females exhibit a peak in differential gene expression at 6 h postmating [19], and 6 h roughly corresponds to the end of the first postmating phase of FRT maturation, in which females switch to a sustained, elevated level of ovulation and fertilization [19,39].
FRTs (i.e. bursa, oviduct, seminal receptacle, spermathecae, parovaria, and associated fat body) of approximately 100 females from each condition were dissected and pooled in PBS per replicate. Two replicates were collected per condition resulting in 10 samples. Samples were solubilized in 100 µl 1 M HEPES with 2% SDS and 5% b-mercaptoethanol. Thirty micrograms of each sample was prepared and labelled with 10-plex tandem mass tags (TMT, Thermo Scientific). Samples were reduced with TCEP, alkylated with iodoacetamide, digested with trypsin, individually labelled with TMT reagents, and combined at equal amounts. The peptide mixture was fractionated using high pH reverse-phase chromatography and profiled with a linear gradient of 5–60% acetonitrile + 20 mM ammonium formate. Thirty-six fractions were initially collected and combined into 18 fractions for analysis by LC–MS/MS.
(b). LC–MS/MS analysis
LC–MS/MS was performed using a Dionex Ultimate 3000 and Lumos Orbitrap mass spectrometer (Thermo Scientific). One microlitre of each fraction was separated with a gradient of 1.6–32% acetonitrile in 0.1% formic acid. Peptides were quantified using a synchronous precursor selection MS3 method [40]. Each full MS1 scan was followed by data-dependent MS2 scans to isolate and fragment the most abundant precursor ions by collision-induced dissociation (35% normalized collision energy), and then the 10 most abundant fragment ions were selected for MS3 analysis for further fragmentation by higher-energy collisional dissociation (65% normalized collision energy).
Mass spectra were searched against the D. simulans protein database (dsim-all-translation-r2.02, FlyBase.org) using PEAKS X (Bioinformatics Solutions Inc.). Although protein divergence between D. simulans and D. mauritiana is very low (non-synonymous substitution rate, or dN = 0.007; see electronic supplementary material), we used the SPIDER algorithm [41] which allows single amino acid substitutions to account for the potential limitation of cross-species database searches. Reporter ion intensities were calculated by summing the centroided reporter ions, and protein abundances were calculated by summing the reporter ion intensities in each channel. Peptide identifications were accepted if the false discovery rate (FDR) < 1% based on the decoy-fusion approach [42], and protein identifications were accepted if the FDR < 1%.
(c). Protein database annotation and analyses
Drosophila simulans FlyBase gene (FBgn) identifiers were converted to their orthologous D. melanogaster FBgn identifiers using the FlyBase Drosophila Orthologs database (dmel_orthologs_in_drosophila_species_fb_2019_01). To compare mating-induced abundance changes of FRT proteins, we identified and removed putative male-derived proteins. Proteins were classified as male-derived if they had been previously identified as D. melanogaster sperm proteins [43,44] or D. melanogaster SFPs (N Brown, JL Sitnik, MF Wolfner 2020, personal communication), or if they showed signatures of being male-derived ejaculate proteins unique to D. simulans and/or D. mauritiana (see supplemental methods for details). Our final dataset included 3287 FRT proteins in D. simulans and D. mauritiana (available on Dryad [45]).
Raw protein abundances were log2-transformed and median-normalized using the MSnbase package in Bioconductor [46]. Protein abundances were highly correlated between replicates, with Pearson's r > 0.97 for all pairwise comparisons. Differential protein abundances were evaluated with empirical Bayes moderated t-tests using the LIMMA package in Bioconductor [47]. Proteins were classified as differentially abundant if the absolute log2-fold change was greater than 1 and FDR-adjusted p-value was less than 0.05, unless otherwise noted. To reduce the dimensionality of the data and identify the major axes of variation in FRT protein composition among conditions, we conducted a principal component analysis (PCA) on average protein abundances (log2-transformed, median-normalized) of all 3287 FRT proteins using the DEP package in Bioconductor [48].
Functional annotation was conducted using the Database for Annotation, Visualization, and Integrated Discovery v. 6.8 [49]. The full list of FRT proteins was specified as the background dataset for comparison, and enrichment was considered significant if the FDR < 5%. SignalP v. 5.0 was used to identify proteins that contained a secretory signal peptide (hereafter called secreted proteins).
(d). Comparison to semi-quantitative proteomic data from D. melanogaster
After discovering substantially different postmating responses in D. simulans and D. mauritiana, we decided to compare our TMT results to analogous, semi-quantitative proteomic results from virgin and mated D. melanogaster FRTs to polarize the observed evolutionary differences on the D. simulans and D. mauritiana lineages. Because the goal of this additional experiment was to gain insight on the possible ancestral FRT postmating response, rather than to conduct detailed quantitative comparisons between all three species, we decided that a label-free approach was appropriate.
Consistent with the simulans/mauritiana samples, mated D. melanogaster samples were collected 6 h after copulation. FRTs from approximately 150 females were dissected and pooled in PBS per replicate, and two replicates were collected per condition. Unlike the simulans/mauritiana samples, D. melanogaster females were mated to males raised on media containing isotopically labelled arginine and lysine, so mass spectrometry searches could be conducted exclusively for unlabelled female-derived proteins [50]. Samples were solubilized in 100 µl 2× Laemmli buffer with 10% TCEP. Fifteen micrograms of each sample was separated on a 1.5 mm 12% SDS–PAGE gel. Each sample was sliced into 10 bands, and then reduced with DDT, alkylated with iodoacetomide, digested with trypsin, and eluted with 0.1% formic acid for LC–MS/MS analysis.
LC–MS/MS was performed using a Dionex Ultimate 3000 and Q Exactive Orbitrap mass spectrometer (Thermo Scientific). Peptides were separated with a gradient of 1.6–32% acetonitrile in 0.1% formic acid. MS1 scans were followed by data-dependent MS2 scans to isolate and fragment the most abundant precursor ions by higher-energy collisional dissociation (25% normalized collision energy). Mass spectra were searched against the D. melanogaster protein database (dmel-all-translation-r6.30, FlyBase.org) in PEAKS X.
Because male-derived proteins were isotopically labelled, all proteins identified by PEAKS are known to be female-derived. However, to be consistent with our analyses in simulans/mauritiana, we removed proteins that had previously been identified as sperm proteins or SFPs. Our final dataset in D. melanogaster included 1909 FRT proteins (available on Dryad [45]).
Protein abundances were quantified as spectral counts corrected for protein length [51], and analysed using identical methods in Bioconductor. There was a strong correlation in protein abundances between replicates, with Pearson's r > 0.95 for all pairwise comparisons. Due to differences in quantitative accuracy between semi-quantitative and TMT proteomics, we evaluated the magnitude of interspecific postmating responses by comparing the distributions of mating-induced fold changes in FRT protein abundance for each species.
3. Results
Hierarchical clustering of average protein abundances for each condition (i.e. sim virgin, mau virgin, sim × sim, mau × mau, and sim × mau) indicate that the two species have distinct protein abundance patterns, with sim virgin clustering with sim × sim, and mau virgin clustering with mau × mau, respectively (figure 1). The heterospecific mated condition (sim female × mau male) clusters as an outgroup to sim virgin and sim × sim. The first two principal components from our PCA collectively explain nearly 90% of the variation in average protein abundances (figure 2) and are sufficient to separate the conditions by species (PC1: 63.5% of variance explained) and mating status (PC2: 26.1% of variance explained).
Figure 1.
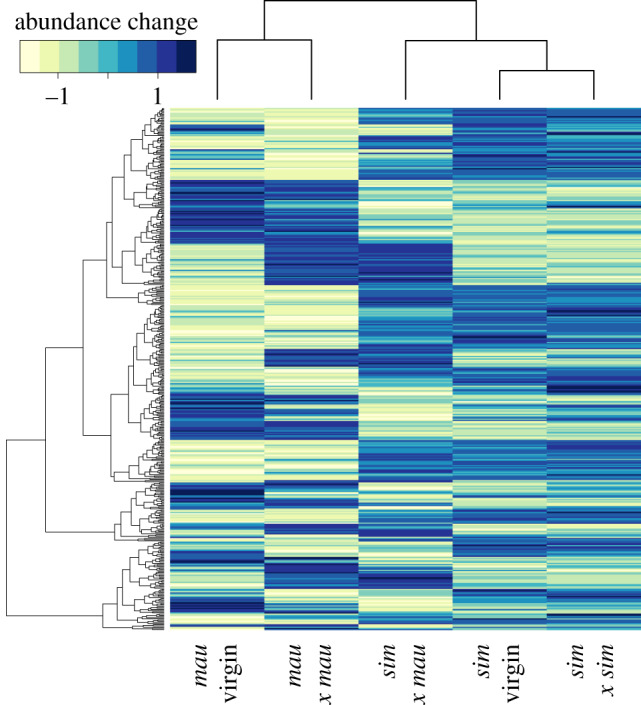
Clustering of FRT protein composition. For illustrative purposes, the heatmap shows average protein abundances for the 442 FRT proteins identified as significantly different between virgins, mated females, and/or after mating in D. simulans and D. mauritiana. Differentially abundant proteins have absolute log2-fold change greater than 1 and FDR-adjusted p-value < 0.05 in at least one pairwise comparison. The hierarchical clustering of conditions (columns) and proteins (rows) is based on Euclidean distance. Heat map colours indicate relative change in protein abundance normalized for each protein (yellow, decrease; blue, increase). (Online version in colour.)
Figure 2.
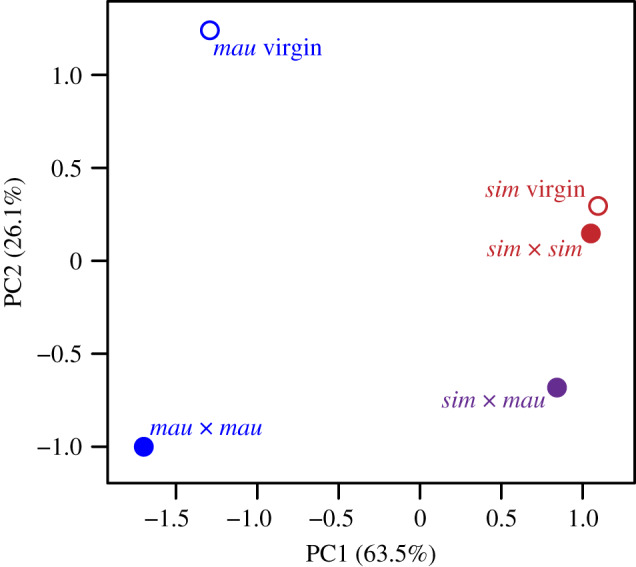
Principal component analysis of FRT protein composition. The first two principal components explain nearly 90% of the variation in average protein abundances and are sufficient to separate conditions based on species (PC1) and mating status (PC2). (Online version in colour.)
(a). Protein compositions of virgin and mated female reproductive tracts differ between sibling species
We identified 244 proteins as differentially abundant between D. simulans and D. mauritiana virgin FRTs. For simplicity, we refer to the proteins that are significantly more abundant in either sim virgin or mau virgin, respectively, as species-biased in virgins. Among these, there is a significant enrichment of secreted proteins (figure 3; FDR < 0.001% for both sim virgin-biased and mau virgin-biased). Almost half (42%) of the proteins that are species-biased are secreted proteins, while only 14% of the remaining proteins are secreted proteins (χ2 = 128.12, p < 0.0001). These results indicate that the virgin FRT of D. simulans and D. mauritiana have distinct compositions of secreted proteins, which may differentiate the FRT environments in each species. The proteins that are species-biased in virgins are also enriched for distinct biological categories. Specifically, the proteins that are significantly more abundant in sim virgins are enriched for cytochrome p450 enzymes (FDR < 0.01%) and serine proteases (FDR = 1.4%), whereas the proteins that are significantly more abundant in mau virgins are enriched for proteins involved in innate immunity (FDR = 0.73%).
Figure 3.
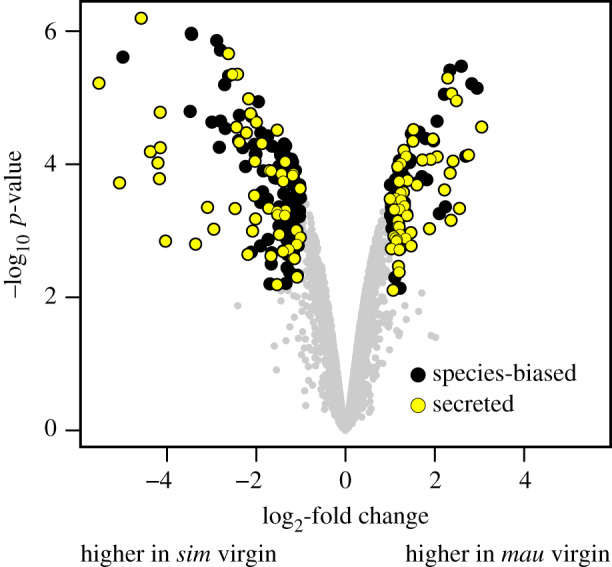
Enrichment of secreted species-biased proteins in D. simulans and D. mauritiana virgin FRTs. Proteins that are species-biased (i.e. significantly more abundant in either sim virgin or mau virgin) are highlighted in black. Species-biased proteins that are secreted are highlighted in yellow. (Online version in colour.)
We identified 320 proteins as differentially abundant between D. simulans and D. mauritiana mated FRTs. Half of these (49%) overlap with those that are species-biased in virgins. Consistent with the virgin comparisons, there is a significant enrichment of secreted proteins among the proteins that are species-biased in mated FRTs (FDR < 0.001% for sim × sim biased; FDR = 0.02% for mau × mau biased). There is a strong correlation in the level of species bias between the virgin and mated conditions (r = 0.64, p < 0.001), meaning that proteins that are significantly more abundant in sim virgin compared to mau virgin are also more abundant in sim × sim compared to mau × mau, and vice versa. These results indicate that there are differences between species in the FRT environment that are independent of mating status (see also figure 2).
(b). Mating induces substantial changes to female reproductive tract protein abundances in D. mauritiana but not D. simulans
There is a dramatic difference in the postmating response between the two species (figure 4). In D. mauritiana, 105 proteins (3.2% of total) are identified as significantly responsive to mating, whereas in D. simulans, only four proteins (0.1% of total) are identified as significantly responsive to mating. There is also significantly less variance of mating-induced fold changes in D. simulans compared to D. mauritiana (figure 5; Levene's test: F = 532.1, p < 0.001). Of the 105 proteins that are responsive to mating in D. mauritiana, 64 change in the same direction in D. simulans, which is a greater number than expected by chance (binomial test: p = 0.03). These results indicate that the FRT proteome in D. simulans responds to mating in a similar fashion as in D. mauritiana, just to a substantially lesser degree (see also figure 2). Thus, mating induces substantial changes to FRT protein abundances in D. mauritiana, but limited change in D. simulans.
Figure 4.
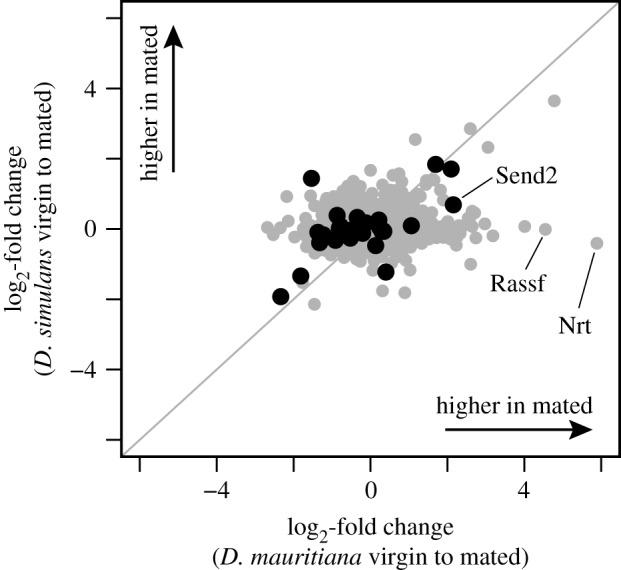
Relationship between the mating-induced changes in protein abundance for FRT proteins in D. mauritiana versus D. simulans. Proteins with serine-type endopeptidase activity are highlighted in black. There is greater spread along the x-axis than y-axis, indicating that there are more proteins with substantial mating-induced abundance changes in D. mauritiana compared to D. simulans. The grey line represents a 1 : 1 relationship, or perfect correlation between species in the female postmating response.
Figure 5.
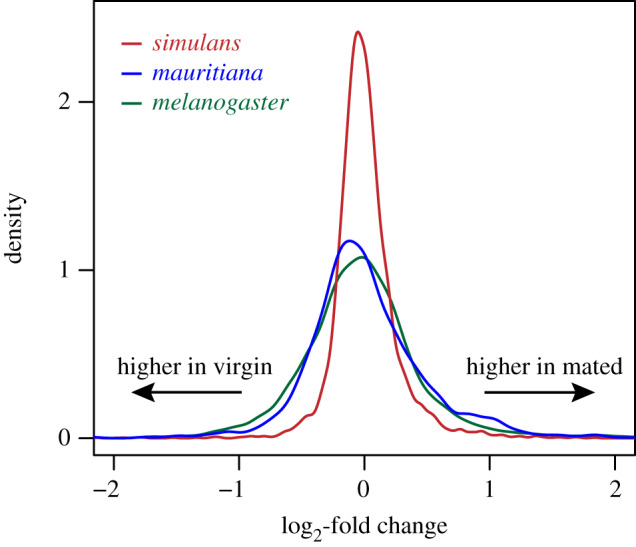
Distributions of mating-induced fold changes in FRT protein abundances in D. melanogaster (green), D. mauritiana (blue), and D. simulans (red). There is significantly less variance in the log2-fold changes between virgin and mated FRT protein abundances in D. simulans compared to D. melanogaster (Levene's test: F = 438.9, p < 0.001) and D. mauritiana (F = 532.1, p < 0.001), but no difference in the variance between D. melanogaster and D. mauritiana (F = 1.77, p = 0.2). (Online version in colour.)
Among the proteins that decrease in abundance after mating in D. mauritiana, there is a significant enrichment of secreted proteins (FDR = 0.10%) and proteins with serine-type endopeptidase activity (FDR = 0.06%). Decreased serine endopeptidase abundance in mated females is consistent with the hypothesis that these proteases are negatively regulated by the male ejaculate, potentially to reduce enzymatic activity in the FRT that could degrade SFPs [20,52]. Interestingly, Spermathecal endopeptidase 2 (Send2)—a well-studied serine protease that is strongly upregulated after mating in D. melanogaster [53,54]—is a notable exception to this pattern and exhibits dramatically different abundance patterns in D. simulans and D. mauritiana. Send2 is at low abundance in the D. mauritiana virgin FRT but is strongly induced after mating (log2-fold change = 2.2; p = 0.03), whereas Send2 is at high abundance in the D. simulans virgin FRT and changes very little after mating (log2-fold change = 0.7; p = 0.68).
We also note that the two proteins that are the most uniquely responsive to mating in D. mauritiana (Nrt and Rassf) are involved in cell–cell adhesion, suggesting that this molecular capability makes them important players in mediating species-specific male–female cell–cell interactions [55]. There is no evidence for functional enrichment among the proteins that increase in abundance after mating in D. mauritiana, and we were unable to conduct enrichment analyses for D. simulans due to the very small number of differentially abundant proteins.
(c). Postmating proteomic responses are not perturbed after heterospecific inseminations
To evaluate whether heterospecific inseminations elicit a unique postmating response, we compared the mating-induced fold changes in FRT protein abundances following conspecific (sim virgin versus sim × sim) and heterospecific (sim virgin versus sim × mau) inseminations. There is a significant correlation between the postmating fold changes (r = 0.38, p < 0.001; see electronic supplementary material, figure S1), indicating that an ejaculate from a D. mauritiana male does not elicit a substantively different postmating response in D. simulans females. (We note that the relatively weak correlation, albeit significant, reflects the fact that most proteins exhibit very little abundance change after mating.) Similarly, there are no proteins that are differentially abundant between mated females after conspecific (sim × sim) versus heterospecific (sim × mau) matings. These results suggest that regardless of whether a D. simulans female mates with a conspecific or heterospecific male, the FRT proteome remains largely unchanged.
(d). D. melanogaster postmating response suggests a derived evolutionarily condition in D. simulans
Because D. simulans, D. mauritiana, and D. melanogaster shared a most recent common ancestor approximately 3 Ma [37], D. melanogaster is informative with regard to inferring the possible ancestral FRT postmating response. There is a comparably high number of proteins that are responsive to mating (absolute log2-fold change greater than 1) in D. melanogaster (5.9% of total) and D. mauritiana (5.6%), but far fewer in D. simulans (1.2%). There is also no difference between D. melanogaster and D. mauritiana in the variance of mating-induced fold changes (figure 5; F = 1.77, p = 0.2), but significantly more variance in D. melanogaster compared to D. simulans (F = 438.9, p < 0.001). While the breadth and magnitude of the postmating response appears to be conserved between D. melanogaster and D. mauritiana, the specific proteins that are responsive to mating are largely different. Only 11 proteins are responsive to mating in both species (out of 112 and 183 mating-responsive proteins in D. melanogaster and D. mauritiana, respectively), but all change in the same direction. Five of these are identified as having serine-type endopeptidase activity (FDR < 0.001%). Given the similar magnitude of postmating changes in D. melanogaster and D. mauritiana, our results suggest that prominent changes to the FRT proteome is likely to be the ancestral postmating response, and the limited changes in D. simulans is a derived condition.
4. Discussion
Resolving the molecular basis of interactions between the sexes is important for understanding fundamental reproductive processes, as these interactions impact fertility and competitive fertilization success within populations [56–59], as well as reproductive isolation between divergent populations and species [13–15]. Here, we used quantitative proteomics in a well-studied speciation model system [29–31] to compare the molecular composition of the FRT environment in virgin and mated females and its role in mediating species-specific postmating responses. Our findings reveal substantial divergence in the virgin FRT proteomes between sibling Drosophila species, as well as dramatic differences among species in the molecular postmating female response. Specifically, we found that the FRT proteomes in D. simulans and D. mauritiana virgin females express unique combinations of secreted proteins and are enriched for distinct functional categories. Furthermore, at 6 h after mating, there were substantial differences in FRT protein abundances between virgin and mated females in D. mauritiana and D. melanogaster, but very few changes in D. simulans. These differences are particularly striking given that D. simulans and D. mauritiana diverged from a common ancestor only 240 000 years ago [37], but are consistent with the hypothesis that female postmating responses diverge rapidly among lineages and contribute to PMPZ isolation [2,52].
Below, we discuss how the molecular composition of the FRT environment and postmating female responses differ between D. simulans and D. mauritiana, and we postulate how such differences may contribute to previously described mechanisms of PMPZ reproductive isolation between these species [32,36]. Before doing so, we note, first, that the observed differences in protein abundances reflect the combined effects of changes in gene expression, post-transcriptional regulation, and protein degradation. Due to enriched proteolytic activity in the FRT [52,60] and among SFPs [1,17], we predict that protein cleavage, and ultimately degradation, underlies at least some of the mating-induced decreases in protein abundance. For mating-induced increases in protein abundance, we predict that both gene expression and post-transcriptional regulation are important. Previous studies in D. melanogaster have shown that many genes are significantly upregulated after mating, but also that gene expression levels may not be strongly correlated with protein abundance [19,20,61].
Second, we note that interspecific variation in postmating proteomic responses may reflect differences in both the number of mating-responsive proteins and the timing of the transition from an ‘unmated' to ‘mated' state. That is, the limited changes observed in the D. simulans FRT proteome are consistent with a scenario in which the temporal progression of postmating events is significantly faster (or slower) in D. simulans females compared to D. mauritiana or D. melanogaster females. We consider differences in the postmating timeline to be an exciting (non-mutually exclusive) alternative for how postmating responses may differ among species. The fact that these species differ in how long it takes females to become receptive to remating ([35], see also below) suggests that differences in the timing of postmating transitions are indeed an important axis of evolutionary change. The current study greatly expands our understanding of how molecular postmating female responses differ among species, but future studies that compare additional postmating timepoints are needed to fully resolve the postmating response timeline and contextualize the 6 h timepoint in each species.
Lastly, we note that it is not possible to determine the extent to which divergence in postmating responses is attributable to sexual selection, natural selection, and/or genetic drift. We find that interspecific differences in the FRT environment (both in virgin and mated females) are significantly enriched for functional categories that are known to play important roles in reproduction and sexual selection (e.g. serine proteases, immunity proteins) [52,62,63], which seems inconsistent with a model of drift. However, although our results suggest a role for sexual selection, the specific agent(s) of selection underlying this divergence is unknown.
Species-specific combinations of secreted proteins may be critical to the coordination of postmating processes [39]. For example, the specialized secretory glands of the Drosophila FRT produce proteins necessary for recruiting sperm to the sperm-storage organs (i.e. seminal receptacle and spermathecae), maintaining sperm mobility during storage and moving eggs through the reproductive tract [54,64,65]. We find that secreted proteins are over-represented among the species-biased proteins in both the virgin and mated conditions and are also over-represented among the proteins that are responsive to mating. These findings indicate that the extracellular environment of the FRT has diverged between D. simulans and D. mauritiana and suggest that secreted proteins contribute to the successful coordination of postmating responses in a species-specific manner.
Previous studies have indeed shown that D. simulans and D. mauritiana females store and use sperm differently when females mate with multiple males: D. simulans females exhibit opposing patterns of fertilization bias between the seminal receptacle (first-male bias) and spermathecae (second-male bias), whereas D. mauritiana females exhibit no fertilization bias in either storage organ [35,38]. Consequently, following both conspecific and heterospecific matings, D. simulans females are able to strategically alter their use of sperm from different sperm-storage organs to bias paternity in favour of conspecific sperm [36,38]. To the extent that different combinations of secreted proteins result in different patterns of sperm storage and usage, the species-specific compositions of secreted proteins observed here in D. simulans and D. mauritiana may represent the molecular mechanism underlying the female mediation of conspecific sperm precedence [36,38]. The fact that male-derived proteins directly interact with these female secretions also sets the stage for a potential coevolutionary arms race. For example, sperm competition is expected to favour the evolution of male proteins that manipulate the female's use of sperm to enhance the male's paternity, and sexual conflict may drive the counter-adaptation of female proteins in order for females to maintain their control of fertilization outcomes [5,13,66]. Sexually antagonistic coevolution of interacting male and female reproductive proteins may, therefore, ensue and result in the evolution of ejaculate–female incompatibilities that contribute to PMPZ reproductive isolation [8,14,15].
Differences in females' ability to process SFPs also may have contributed to the divergent postmating responses observed between D. simulans and D. mauritiana. If D. simulans females are more efficient at processing SFPs than D. mauritiana females, then D. simulans SFPs may have a more restricted time to act and thus limited scope to elicit postmating responses [1]. We find that the proteins that are more abundant in virgin D. simulans FRTs compared to D. mauritiana FRTs are over-represented by serine-type endopeptidases and cytochrome p450 enzymes—two classes of enzymes that may be involved in the degradation of male proteins. Serine-type endopeptidases regulate the cleavage of SFPs to their active forms [67,68] and are important in SFP degradation [52,69]. Cytochrome p450 enzymes are associated with oxidative degradation of endogenous and exogenous toxins [70], so their enrichment may aid in the oxidative cleavage of male-derived compounds introduced during mating [18,71–73]. We hypothesize that greater endopeptidase and cytochrome p450 abundance, and hence activity, in virgin D. simulans FRTs underlies its unique, relatively limited postmating response.
In further support of this ‘differential SFP-processing' hypothesis, we find that (i) the proteins that decrease after mating in D. mauritiana are over-represented by serine-type endopeptidases, and (ii) the abundance of an important spermathecal endopeptidase (Send2) is higher in virgin D. simulans females than in mated D. mauritiana females. SFPs are known to reduce a female's receptivity to remating in D. melanogaster [1,17,18], so it is noteworthy that the remating interval for D. simulans females (mean ± s.e.: 2.7 ± 0.04 days) is shorter than for either D. mauritiana females (3.5 ± 0.07 days) or D. melanogaster females (3.5 ± 0.03 days) [35]. Although targeted enzymatic experiments should confirm whether species differ in their rates of SFP degradation, future studies should also focus more broadly on unravelling the complexity of female × male molecular interactions by resolving protein interaction networks, including those between proteolytic enzymes and SFPs.
SFPs are known to be important agents for eliciting postmating changes in female behaviour, morphology, and physiology [1,17], so there was an a priori expectation that rapid evolution of seminal fluid composition could contribute to differences in postmating responses between D. simulans and D. mauritiana. However, there are several reasons why the divergence of male-transferred proteins cannot fully explain the observed divergence in female postmating responses. First, divergence in male-transferred proteins cannot account for the substantive interspecific differences observed in the FRT environment of virgin females. Second, the postmating response of D. simulans FRTs was largely the same irrespective of whether females mated to conspecific or heterospecific males. Third, there has been very little amino acid sequence divergence of SFPs between D. simulans and D. mauritiana (dN = 0.01; [45]). Nonetheless, we cannot rule out the possibility that even limited SFP divergence contributes to the interspecific differences in postmating responses. Previous studies in D. melanogaster have shown that intraspecific SFP allelic variation can result in different postmating outcomes, including patterns of sperm displacement, male mating rate, female postmating refractorines, and female fecundity [74–76].
Our finding that the D. simulans FRT proteome exhibited limited changes after mating, regardless of whether the female received a conspecific or heterospecific ejaculate, suggests that the D. mauritiana ejaculate alone is not responsible for the observed divergence in postmating responses. This study, therefore, advances our general understanding about the role that females play in mediating reproductive interactions [77]. Specifically, our results suggest females control their developmental switch from an ‘unmated' to ‘mated' state once it has been induced by mating. Our findings contrast with those from a transcriptomics study on the sibling species D. mojavensis and D. arizonae that found very few genes are differentially expressed in the D. mojavensis FRT after conspecific matings, but many genes are differentially expressed after heterospecific matings [22]. We do not know if the discrepancy reflects different methodologies (i.e. expression of transcripts versus proteins) and/or different mating systems between the species pairs. Future studies are needed to determine how species differ in (i) the intensity of postcopulatory sexual selection in natural populations, (ii) the degree to which females determine reproductive outcomes, (iii) the potency of male-derived proteins in eliciting female postmating responses, and (iv) how male–female molecular interactions are disrupted following heterospecific crosses.
By applying a comprehensive proteomic approach to compare female postmating responses among species, our study represents a step forward in unravelling the molecular mechanisms underlying postmating interactions between the sexes and their potential role in mediating species-specific postmating responses. Our research highlights the value of this approach to resolving complex ejaculate–female interactions, their divergence among populations and species, and their contribution to PMPZ reproductive isolation.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Mike Deery and Renata Feret at the Cambridge Centre for Proteomics, Kirill Borziak and Emma Whittington for help with analyses, Sharleen Buel for technical support, and Yasir Ahmed-Braimah, Martin Garlovsky, Jane Pascar, and Zeeshan Syed for feedback on earlier drafts of the manuscript.
Data accessibility
Mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD17708 (simulans/mauritiana dataset) and PXD17730 (melanogaster dataset). Additional data and R code are available at the Dryad Digital Repository: https://doi.org/10.5061/dryad.8cz8w9gm8 [45].
Authors' contributions
All authors designed the study; E.L.M. and C.E.M. collected the samples; E.L.M. analysed the data; E.L.M. wrote the initial manuscript; all authors revised the manuscript and approved the final version.
Competing interests
The authors declare no competing interests.
Funding
This research was funded by the National Science Foundation (DEB 1655840 to S.D. and S.P.) and the National Institutes of Health (NICHD R21HD088910 to S.D. and S.P.).
References
- 1.Ravi RK, Wolfner MF. 2007. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr. Comp. Biol. 47, 427–445. ( 10.1093/icb/icm046) [DOI] [PubMed] [Google Scholar]
- 2.Pitnick S, Wolfner MF, Suarez SS. 2009. Ejaculate–female and sperm–female interactions. In Sperm biology (eds Birkhead TR, Hosken DJ, Pitnick S), pp. 247–304. London, UK: Academic Press. [Google Scholar]
- 3.Pitnick S, Wolfner MF, Dorus S. 2020. Post-ejaculatory modifications to sperm (PEMS). Biol. Rev. 95, 365–392. ( 10.1111/brv.12569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker GA. 1970. Sperm competition and its evolutionary consequences in the insects. Biol. Rev. 45, 525–567. ( 10.1111/j.1469-185X.1970.tb01176.x) [DOI] [Google Scholar]
- 5.Parker GA. 1979. Sexual selection and sexual conflict. In Sexual selection and reproductive competition in insects, pp. 123–166. New York, NY: Academic Press. [Google Scholar]
- 6.Eberhard WG. 1996. Female control: sexual selection by cryptic female choice. Princeton, NJ: Princeton University Press. [Google Scholar]
- 7.Simmons LW. 2001. Sperm competition and its evolutionary consequences in the insects. Princeton, NJ: Princeton University Press. [Google Scholar]
- 8.Arnqvist G, Rowe L. 2005. Sexual conflict. Princeton, NJ: Princeton University Press. [Google Scholar]
- 9.Firman RC, Gasparini C, Manier MK, Pizzari T. 2017. Postmating female control: 20 years of cryptic female choice. Trends Ecol. Evol. 32, 368–382. ( 10.1016/j.tree.2017.02.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitnick S, Miller GT, Schneider K, Markow TA. 2003. Ejaculate-female coevolution in Drosophila mojavensis. Proc. R. Soc. Lond. B 270, 1507–1512. ( 10.1098/rspb.2003.2382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knowles LL, Markow TA. 2001. Sexually antagonistic coevolution of a postmating-prezygotic reproductive character in desert Drosophila. Proc. Natl Acad. Sci. USA 98, 8692–8696. ( 10.1073/pnas.151123998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swanson WJ, Vacquier VD. 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137–144. ( 10.1038/nrg733) [DOI] [PubMed] [Google Scholar]
- 13.Clark NL, Aagaard JE, Swanson WJ. 2006. Evolution of reproductive proteins from animals and plants. Reproduction 131, 11–22. ( 10.1530/rep.1.00357) [DOI] [PubMed] [Google Scholar]
- 14.Howard DJ, Palumbi SR, Birge LM, Manier MK. 2009. Sperm and speciation. In Sperm biology (eds Birkhead TR, Hosken DJ, Pitnick S), pp. 367–403. London, UK: Academic Press. [Google Scholar]
- 15.McDonough CE, Whittington E, Pitnick S, Dorus S. 2016. Proteomics of reproductive systems: towards a molecular understanding of postmating, prezygotic reproductive barriers. J. Proteom. 135, 26–37. ( 10.1016/j.jprot.2015.10.015) [DOI] [PubMed] [Google Scholar]
- 16.Sirot LK, LaFlamme BA, Sitnik JL, Rubinstein CD, Avila FW, Chow CY, Wolfner MF. 2009. Molecular social interactions: Drosophila melanogaster seminal fluid proteins as a case study. Adv. Genet. 68, 23–56. ( 10.1016/S0065-2660(09)68002-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF. 2011. Insect seminal fluid proteins: identification and function. Annu. Rev. Entomol. 56, 21–40. ( 10.1146/annurev-ento-120709-144823) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGraw LA, Gibson G, Clark AG, Wolfner MF. 2004. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr. Biol. 14, 1509–1514. ( 10.1016/j.cub.2004.08.028) [DOI] [PubMed] [Google Scholar]
- 19.Mack PD, Kapelnikov A, Heifetz Y, Bender M. 2006. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proc. Natl Acad. Sci. USA 103, 10 358–10 363. ( 10.1073/pnas.0604046103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapelnikov A, Zelinger E, Gottlieb Y, Rhrissorrakrai K, Gunsalus KC, Heifetz Y. 2008. Mating induces an immune response and developmental switch in the Drosophila oviduct. Proc. Natl Acad. Sci. USA 105, 13 912–13 917. ( 10.1073/pnas.0710997105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prokupek AM, Kachman SD, Ladunga I, Harshman LG. 2009. Transcriptional profiling of the sperm storage organs of Drosophila melanogaster. Insect. Mol. Biol. 18, 465–475. ( 10.1111/j.1365-2583.2009.00887.x) [DOI] [PubMed] [Google Scholar]
- 22.Bono JM, Matzkin LM, Kelleher ES, Markow TA. 2011. Postmating transcriptional changes in reproductive tracts of con- and heterospecifically mated Drosophila mojavensis females. Proc. Natl Acad. Sci. USA 108, 7878–7883. ( 10.1073/pnas.1100388108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers DW, Whitten MMA, Thailayil J, Soichot J, Levashina EA, Catteruccia F. 2008. Molecular and cellular components of the mating machinery in Anopheles gambiae females. Proc. Natl Acad. Sci. USA 105, 19 390–19 395. ( 10.1073/pnas.0809723105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alfonso-Parra C, Ahmed-Braimah YH, Degner EC, Avila FW, Villarreal SM, Pleiss JA, Wolfner MF, Harrington LC. 2016. Mating-induced transcriptome changes in the reproductive tract of female Aedes aegypti. PLoS Negl. Trop. Dis. 10, e0004451 ( 10.1371/journal.pntd.0004451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kocher SD, Richard F-J, Tarpy DR, Grozinger CM. 2008. Genomic analysis of post-mating changes in the honey bee queen (Apis mellifera). BMC Genom. 9, 232 ( 10.1186/1471-2164-9-232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Wathiqui N, Lewis SM, Dopman EB. 2014. Using RNA sequencing to characterize female reproductive genes between Z and E strains of European Corn Borer moth (Ostrinia nubilalis). BMC Genom. 15, 189 ( 10.1186/1471-2164-15-189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meslin C, Plakke MS, Deutsch AB, Small BS, Morehouse NI, Clark NL. 2015. Digestive organ in the female reproductive tract borrows genes from multiple organ systems to adopt critical functions. Mol. Biol. Evol. 32, 1567–1580. ( 10.1093/molbev/msv048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thailayil J, Gabrieli P, Caputo B, Bascuñán P, South A, Diabate A, Dabire R, della Torre A, Catteruccia F. 2018. Analysis of natural female post-mating responses of Anopheles gambiae and Anopheles coluzzii unravels similarities and differences in their reproductive ecology. Sci. Rep. 8, 6594 ( 10.1038/s41598-018-24923-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coyne JA. 1993. The genetics of an isolating mechanism between two sibling species of Drosophila. Evolution 47, 778–788. ( 10.1111/j.1558-5646.1993.tb01233.x) [DOI] [PubMed] [Google Scholar]
- 30.Coyne JA. 1989. Genetics of sexual isolation between two sibling species, Drosophila simulans and Drosophila mauritiana. Proc. Natl Acad. Sci. USA 86, 5464–5468. ( 10.1073/pnas.86.14.5464) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coyne JA, Orr HA. 1998. The evolutionary genetics of speciation. Phil. Trans. R. Soc. Lond. B 353, 287–305. ( 10.1098/rstb.1998.0210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price CSC. 1997. Conspecific sperm precedence in Drosophila. Nature 388, 663–666. ( 10.1038/41753) [DOI] [PubMed] [Google Scholar]
- 33.Price CSC, Kim CH, Posluszny J, Coyne JA. 2000. Mechanisms of conspecific sperm precedence in Drosophila. Evolution 54, 2028–2037. ( 10.1111/j.0014-3820.2000.tb01246.x) [DOI] [PubMed] [Google Scholar]
- 34.Price CSC, Kim CH, Gronlund CJ, Coyne JA. 2001. Cryptic reproductive isolation in the Drosophila simulans species complex. Evolution 55, 81–92. ( 10.1111/j.0014-3820.2001.tb01274.x) [DOI] [PubMed] [Google Scholar]
- 35.Manier MK, Belote JM, Berben KS, Lüpold S, Ala-Honkola O, Collins WF, Pitnick S. 2013. Rapid diversification of sperm precedence traits and processes among three sibling Drosophila species. Evolution 67, 2348–2362. ( 10.1111/evo.12117) [DOI] [PubMed] [Google Scholar]
- 36.Manier MK, Lüpold S, Belote JM, Starmer WT, Berben KS, Ala-Honkola O, Collins WF, Pitnick S. 2013. Postcopulatory sexual selection generates speciation phenotypes in Drosophila. Curr. Biol. 23, 1853–1862. ( 10.1016/j.cub.2013.07.086) [DOI] [PubMed] [Google Scholar]
- 37.Garrigan D, Kingan SB, Geneva AJ, Andolfatto P, Clark AG, Thornton KR, Presgraves DC. 2012. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22, 1499–1511. ( 10.1101/gr.130922.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manier MK, Lüpold S, Pitnick S, Starmer WT. 2013. An analytical framework for estimating fertilization bias and the fertilization set from multiple sperm-storage organs. Am. Nat. 182, 552–561. ( 10.1086/671782) [DOI] [PubMed] [Google Scholar]
- 39.Carmel I, Tram U, Heifetz Y. 2016. Mating induces developmental changes in the insect female reproductive tract. Curr. Opin. Insect. Sci. 13, 106–113. ( 10.1016/j.cois.2016.03.002) [DOI] [PubMed] [Google Scholar]
- 40.McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. 2014. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158. ( 10.1021/ac502040v) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han Y, Ma B, Zhang K. 2005. SPIDER: software for protein identification from sequence tags with de novo sequencing error. J. Bioinform. Comput. Biol. 3, 697–716. ( 10.1109/CSB.2004.1332434) [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, et al. 2012. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics 11, M111.010587 ( 10.1074/mcp.M111.010587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorus S, Busby SA, Gerike U, Shabanowitz J, Hunt DF, Karr TL. 2006. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat. Genet. 38, 1440–1445. ( 10.1038/ng1915) [DOI] [PubMed] [Google Scholar]
- 44.Wasbrough ER, Dorus S, Hester S, Howard-Murkin J, Lilley K, Wilkin E, Polpitiya A, Petritis K, Karr TL. 2010. The Drosophila melanogaster sperm proteome-II (DmSP-II). J. Proteom. 73, 2171–2185. ( 10.1016/j.jprot.2010.09.002) [DOI] [PubMed] [Google Scholar]
- 45.McCullough EL, McDonough CE, Pitnick S, Dorus S. 2020. Data from: Quantitative proteomics reveals rapid divergence in the postmating response of female reproductive tracts among sibling spcies Dryad Digital Repository. ( 10.5061/dryad.8cz8w9gm8) [DOI] [PMC free article] [PubMed]
- 46.Gatto L, Lilley KS. 2011. MSnbase: an R/Bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics 28, 288–289. ( 10.1093/bioinformatics/btr645) [DOI] [PubMed] [Google Scholar]
- 47.Smyth GK. 2005. limma: linear models for microarray data. In Bioinformatics and computational biology solutions using R and bioconductor (eds Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S), pp. 397–420. New York, NY: Springer. [Google Scholar]
- 48.Zhang X, Smits AH, van Tilburg GB, Ovaa H, Huber W, Vermeulen M.. 2018. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 13, 530–550. ( 10.1038/nprot.2017.147) [DOI] [PubMed] [Google Scholar]
- 49.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. ( 10.1038/nprot.2008.211) [DOI] [PubMed] [Google Scholar]
- 50.Cuomo A, Sanfilippo R, Vaccari T, Bonaldi T. 2014. Proteomics meets genetics: SILAC labeling of Drosophila melanogaster larvae and cells for in vivo functional studies. In Stable isotope labeling by amino acids in cell culture (SILAC): methods and protocols (ed. Warscheid B.), pp. 293–311. New York, NY: Springer. [DOI] [PubMed] [Google Scholar]
- 51.Bubis JA, Levitsky LI, Ivanov MV, Tarasova IA, Gorshkov MV. 2017. Comparative evaluation of label-free quantification methods for shotgun proteomics. Rapid Commun. Mass Spectrom. 31, 606–612. ( 10.1002/rcm.7829) [DOI] [PubMed] [Google Scholar]
- 52.Kelleher ES, Pennington JE. 2009. Protease gene duplication and proteolytic activity in Drosophila female reproductive tracts. Mol. Biol. Evol. 26, 2125–2134. ( 10.1093/molbev/msp121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawniczak MKN, Begun DJ. 2007. Molecular population genetics of female-expressed mating-induced serine proteases in Drosophila melanogaster. Mol. Biol. Evol. 24, 1944–1951. ( 10.1093/molbev/msm122) [DOI] [PubMed] [Google Scholar]
- 54.Schnakenberg SL, Matias WR, Siegal ML. 2011. Sperm-storage defects and live birth in Drosophila females lacking spermathecal secretory cells. PLoS Biol. 9, e1001192 ( 10.1371/journal.pbio.1001192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dorus S, Skerget S, Karr TL. 2012. Proteomic discovery of diverse immunity molecules in mammalian spermatozoa. Syst. Biol. Reprod. Med. 58, 218–228. ( 10.3109/19396368.2012.700442) [DOI] [PubMed] [Google Scholar]
- 56.Clark AG, Begun DJ, Prout T. 1999. Female×male interactions in Drosophila sperm competition. Science 283, 217–220. ( 10.1126/science.283.5399.217) [DOI] [PubMed] [Google Scholar]
- 57.Chow CY, Wolfner MF, Clark AG. 2010. The genetic basis for male×female interactions underlying variation in reproductive phenotypes of Drosophila. Genetics 186, 1355–1365. ( 10.1534/genetics.110.123174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reinhart M, Carney T, Clark AG, Fiumera AC. 2015. Characterizing male–female interactions using natural genetic variation in Drosophila melanogaster. J. Hered. 106, 67–79. ( 10.1093/jhered/esu076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delbare SYN, Chow CY, Wolfner MF, Clark AG. 2017. Roles of female and male genotype in post-mating responses in Drosophila melanogaster. J. Hered. 108, 740–753. ( 10.1093/jhered/esx081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelleher ES, Swanson WJ, Markow TA. 2007. Gene duplication and adaptive evolution of digestive proteases in Drosophila arizonae female reproductive tracts. PLoS Genet. 3, e148 ( 10.1371/journal.pgen.0030148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vicens A, Borziak K, Karr TL, Roldan ERS, Dorus S. 2017. Comparative sperm proteomics in mouse species with divergent mating systems. Mol. Biol. Evol. 34, 1403–1416. ( 10.1093/molbev/msx084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong A, Turchin MC, Wolfner MF, Aquadro CF. 2007. Evidence for positive selection on Drosophila melanogaster seminal fluid protease homologs. Mol. Biol. Evol. 25, 497–506. ( 10.1093/molbev/msm270) [DOI] [PubMed] [Google Scholar]
- 63.Lawniczak MKN, Barnes AI, Linklater JR, Boone JM, Wigby S, Chapman T. 2007. Mating and immunity in invertebrates. Trends Ecol. Evol. 22, 48–55. ( 10.1016/j.tree.2006.09.012) [DOI] [PubMed] [Google Scholar]
- 64.Allen AK, Spradling AC. 2008. The Sf1-related nuclear hormone receptor Hr39 regulates Drosophila female reproductive tract development and function. Development 135, 311–321. ( 10.1242/dev.015156) [DOI] [PubMed] [Google Scholar]
- 65.Sun J, Spradling AC. 2013. Ovulation in Drosophila is controlled by secretory cells of the female reproductive tract. Elife 2, e00415 ( 10.7554/eLife.00415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chapman T, Arnqvist G, Bangham J, Rowe L. 2003. Sexual conflict. Trends Ecol. Evol. 18, 41–47. ( 10.1016/S0169-5347(02)00004-6) [DOI] [Google Scholar]
- 67.Ram K Ravi, Sirot LK, Wolfner MF. 2006. Predicted seminal astacin-like protease is required for processing of reproductive proteins in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 103, 18 674–18 679. ( 10.1073/pnas.0606228103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.LaFlamme BA, Ravi Ram K, Wolfner MF. 2012. The Drosophila melanogaster seminal fluid protease ‘seminase’ regulates proteolytic and post-mating reproductive processes. PLoS Genet. 8, e1002435 ( 10.1371/journal.pgen.1002435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pilpel N, Nezer I, Applebaum SW, Heifetz Y. 2008. Mating-increases trypsin in female Drosophila hemolymph. Insect. Biochem. Mol. Biol. 38, 320–330. ( 10.1016/j.ibmb.2007.11.010) [DOI] [PubMed] [Google Scholar]
- 70.Feyereisen R. 2012. Insect CYP genes and P450 enzymes. In Insect molecular biology and biochemistry, pp. 236–316. San Diego, CA: Academic Press. [Google Scholar]
- 71.Lung O, Tram U, Finnerty CM, Eipper-Mains MA, Kalb JM, Wolfner MF. 2002. The Drosophila melanogaster seminal fluid protein Acp62F is a protease inhibitor that is toxic upon ectopic expression. Genetics 160, 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Panhuis TM, Swanson WJ. 2006. Molecular evolution and population genetic analysis of candidate female reproductive genes in Drosophila. Genetics 173, 2039–2047. ( 10.1534/genetics.105.053611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mueller JL, Page JL, Wolfner MF. 2007. An ectopic expression screen reveals the protective and toxic effects of Drosophila seminal fluid proteins. Genetics 175, 777–783. ( 10.1534/genetics.106.065318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clark AG, Aguadé M, Prout T, Harshman LG, Langley CH. 1995. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics 139, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fiumera AC, Dumont BL, Clark AG. 2005. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics 169, 243–257. ( 10.1534/genetics.104.032870) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang R, Clark AG, Fiumera AC. 2013. Natural genetic variation in male reproductive genes contributes to nontransitivity of sperm competitive ability in Drosophila melanogaster. Mol. Ecol. 22, 1400–1415. ( 10.1111/mec.12113) [DOI] [PubMed] [Google Scholar]
- 77.Lüpold S, Pitnick S, Berben KS, Blengini CS, Belote JM, Manier MK. 2013. Female mediation of competitive fertilization success in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 110, 10 693–10 698. ( 10.1073/pnas.1300954110) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- McCullough EL, McDonough CE, Pitnick S, Dorus S. 2020. Data from: Quantitative proteomics reveals rapid divergence in the postmating response of female reproductive tracts among sibling spcies Dryad Digital Repository. ( 10.5061/dryad.8cz8w9gm8) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD17708 (simulans/mauritiana dataset) and PXD17730 (melanogaster dataset). Additional data and R code are available at the Dryad Digital Repository: https://doi.org/10.5061/dryad.8cz8w9gm8 [45].


