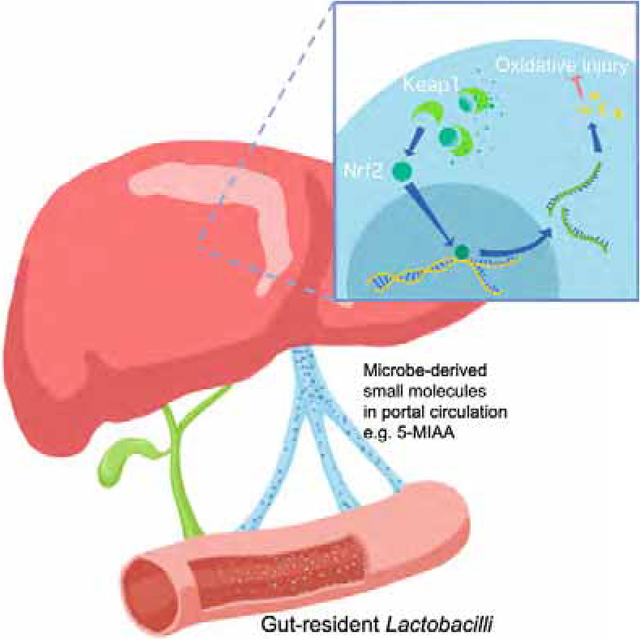
SUMMARY
Many studies have suggested a role for gut-resident microbes (the “gut microbiome”) in modulating host health, however the mechanisms by which they impact systemic physiology remain largely unknown. In this study, metabolomic and transcriptional profiling of germ-free and conventionalized mouse liver revealed an upregulation of the Nrf2 antioxidant and xenobiotic response in microbiome-replete animals. Using a Drosophila-based screening assay, we identified members of the genus Lactobacillus capable of stimulating Nrf2. Indeed, the human commensal Lactobacillus rhamnosus GG (LGG) potently activated Nrf2 in the Drosophila liver analog and the murine liver. This activation was sufficient to protect against two models of oxidative liver injury, acetaminophen overdose and acute ethanol toxicity. Characterization of the portal circulation of LGG-treated mice by tandem mass spectrometry identified a small molecule activator of Nrf2, 5-methoxyindoleacetic acid, produced by LGG. Taken together, these data demonstrate a mechanism by which intestinal microbes modulate hepatic susceptibility to oxidative injury.
Keywords: Lactobacillus, hepatotoxicity, 5-methoxyindoleacetic acid, Nrf2, microbiome
Graphical Abstract
eTOC blurb
Saeedi, Liu et al. demonstrate that the gut microbiome acts at a distance to activate host antioxidant responses in the liver. These responses can be amplified by exogenous administration of Lactobacilli and protect against oxidative liver injury. Finally, they identify a Lactobacilli-demed small molecule that mediates, in part, these effects.
INTRODUCTION
The microorganisms that reside within the human gut comprise over 100 trillion bacteria, along with myriad fungi and viruses (Human Microbiome Project Consortium, 2012). Until relatively recently, little was known about the functional consequences of this diverse and dynamic community on the host. With the advent of nucleic acid based taxonomic profiling and the development of germ-free model systems, substantial progress has been achieved in understanding the effects of the gut microbiome on host physiology and homeostasis (Integrative HMP (iHMP) Research Network Consortium, 2019). Initial investigations of host-microbiome interactions focused on the physiology and pathology of the intestine, the tissue with which these microbes are in direct contact. Multiple studies have demonstrated that intestinal disorders of diverse etiology, including inflammatory bowel disease (Sartor, 2008; Tamboli et al., 2004), irritable bowel syndrome (Carroll et al., 2011, 2012), celiac disease (Girbovan et al., 2017), and colon cancer (Couturier-Maillard et al., 2013; Sobhani et al., 2011; Zhan et al., 2013), are profoundly influenced by the microbiome. Recently, however, investigators have begun to appreciate that the effects of gut-resident bacteria are not limited to intestinal tissue, but can have far-reaching effects on distal organs, including the liver, pancreas, heart, and even the brain (Ho et al., 2015).
Of these systemic organs, the liver is situated in a unique position to receive signals from the intestinal microbiome. Ingested small molecules and peptides derived from non-self foreign substances in the gut (xenobiotics) are both passively and actively absorbed by the intestinal epithelium and transported into the portal circulation. This component of the circulatory system collects blood from the majority of the intestine and delivers it directly to the liver, the principal metabolic and detoxification hub of the mammalian organism. The liver, therefore, serves a critical gatekeeper function at the interface between a flood of foreign -xenobiotic- substances and a finely tuned systemic milieu.
It has long been appreciated that the xenobiotic detoxification functions of the liver evolved to deal with this exposure to diet-derived foreign material arriving from the intestine. Several transcriptional regulators, including pregnane X receptor (PXR), constitutive active/androstane receptor (CAR), aryl hydrocarbon receptor (AHR), among others, can acutely sense the presence of xenobiotic ligands and mediate the subsequent cellular response (Larigot et al., 2018; Timsit and Negishi, 2007). Additionally, recent studies have revealed that these pathways are not only sensitive to changes in the diet but may be altered by peptides and small molecules derived from the gut-resident microbiota (Macpherson et al., 2016). This is due to the diverse biochemical activities encoded in microbial genomes that play key roles in the biotransformation and detoxification of ingested nutrients and pharmacological agents (Klaassen and Cui, 2015). Multiple signaling pathways, including those involved in bile acid biosynthesis (Dawson and Karpen, 2015; Foley et al., 2019; Jiang et al., 2015), the urea cycle (Stewart and Smith, 2005), and choline metabolism (Romano et al., 2015; Schugar et al., 2018; Wang et al., 2011b), have been shown to be sensitive to changes to the microbial communities of the gut. Consistently, the microbiome appears to modify susceptibility to several hepatic diseases, including non-alcoholic fatty liver disease (NAFLD) (de Faria Ghetti et al., 2018; Perumpail et al., 2019), alcoholic liver disease (ALD) (Cassard and Ciocan, 2018), hepatocellular carcinoma (HCC) (Roderburg and Luedde, 2014; Wan and El-Nezami, 2018), among others. Thus, the microbiome-gut-liver relationship is emerging as an important factor in multiple physiological and pathological states.
In this study, we took a global approach to identify the signaling pathways in the liver that are modified by the microbiome. We performed metabolomic analysis on hepatic tissue from germ-free or conventionalized (microbiome-replete) mice. In conventionalized animals, we observed a significant increase in metabolites associated with antioxidant and xenobiotic response pathways. A highly conserved master regulator of these responses is the basic leucine zipper transcription factor Nuclear factor erythroid-derived 2-like 2 (Nrf2) (Moi et al., 1994). Indeed, subsequent transcriptomic analysis of liver from the same animals revealed significant inductions of a number of canonical Nrf2 target transcripts, but not of other classical regulators of the detoxification response such as PXR, CAR, or AHR.
To determine the microbial populations responsible for the observed activation of hepatic Nrf2, we utilized a transgenic Nrf2 reporter Drosophila melanogaster screening assay (Sykiotis and Bohmann, 2008). Our results reveal that members of the genus Lactobacillus were capable of activating systemic Nrf2 signaling. Furthermore, oral gavage of the representative Lactobacillus, Lactobacillus rhamnosus GG (LGG), induced Nrf2 in the liver of conventional mice and this activation was sufficient to protect against two different models of acute oxidative liver injury. Finally, we performed ultra-high resolution LC MS/MS to characterize the small molecule complement present in the portal circulation of mice treated with LGG and identified 5-methoxyindoleacetic acid (5-MIAA) as a novel LGG-derived small molecule capable of activating hepatic Nrf2. These data address key gaps in our understanding of the commensal-liver axis and provide further insight into the role of the intestinal microbiome in hepatic health and homeostasis.
RESULTS
Conventionalization of germ-free mice induces the hepatic xenobiotic and antioxidant response
Given the taxonomic complexity of the microbiome and the diverse functional consequences of dysbiosis on host hepatic physiology, we utilized a non-biased metabolomics-based approach to outline the broad impact of commensal-liver interactions. Germ-free (GF) mouse littermates were separated at weaning (3 weeks of age) into two groups with one group maintained in sterile isolators, while the other group was transferred to cages with bedding obtained from conventionally raised specific pathogen free (SPF) mice (Figure 1A). These mice were then allowed three weeks to acquire and establish a microbiome (“conventionalization”) (CV) before sacrifice and metabolomic analysis (Go et al., 2014; Liu et al., 2016) of mitochondrial fractions obtained from liver tissue from both groups (Figure 1B–C). Pathway analysis of altered features using Mummichog analysis (Li et al., 2013) revealed that conventionalization elicited distinct shifts in metabolic pathways previously shown to be modulated by the microbiota (Figure 1D). More specifically, pathways for hepatic amino acid metabolism (Lin et al., 2017), bile acid biosynthesis (Dawson and Karpen, 2015; Foley et al., 2019), and the urea cycle (Ramezani et al., 2016; Stewart and Smith, 2005) were altered by the microbiome. These analyses also revealed an induction of regulatory networks involved in drug metabolism and the antioxidant response, including alterations in cytochrome P450 metabolites and glutathione synthesis (Figure 1D).
Figure 1. Conventionalization of germ-free mice induces the hepatic antioxidant and xenobiotic response.
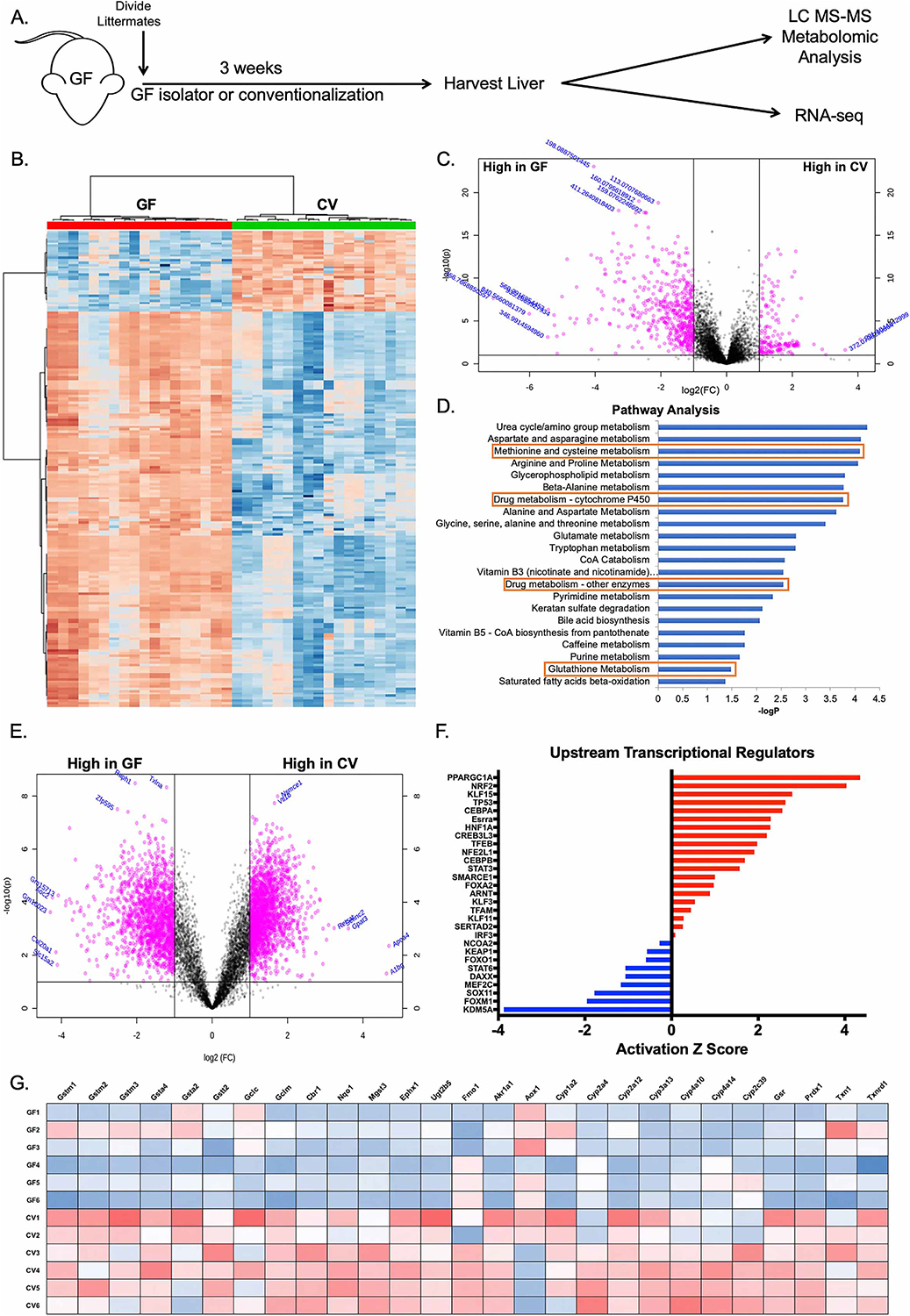
(A) Schematic of experimental design. Germ-free (GF) littermates were separated at weaning and either maintained GF or conventionalized (CV) with bedding from conventionally raised Specific pathogen free (SPF) mice for three weeks prior to sacrifice and harvest of liver tissue. (B) Hierarchical clustering of the top 200 altered features identified from ultra-high resolution LC-MS analysis of liver tissue from experiment described in (A). (C) Volcano plot of features identified in LC-MS analysis of liver tissue from experiment described in (A), where pink indicates p<0.05 as calculated by t-test. (D) Mummichog pathway analysis for the identification of pathways induced in CV relative to GF. Red boxes highlight pathways involved in antioxidant and xenobiotic metabolism. (E) Volcano plot of transcript enrichment detected by RNAseq analysis of liver tissue of GF and CV mice. Pink data points denote transcripts that were significantly enriched in GF or conventional mice to a confidence level of p<0.05 as determined by t-test. (F) Ingenuity Pathway Analysis of RNAseq identifies activated (red) and repressed (blue) transcriptional regulators in CV relative to GF mice. (G) Heatmap of genes under the transcriptional control of Nrf2 identified in RNAseq analysis. n=6 mice per condition (GF or CV), 3 male and 3 female.
Parallel transcriptomic analysis using RNAseq on the same liver tissue demonstrated a significantly altered profile in the hepatic transcriptome of CV mice relative to GF (Figure 1E). Analysis of upstream transcriptional regulators of the altered transcripts by Ingenuity Pathway Analysis showed an induction of genes in CV mice known to be regulated by Nrf2, a master regulator of the cellular antioxidant response (Figure 1F). Further analysis of the transcriptional profiles of CV and GF livers revealed an induction of many Nrf2 target transcripts in CV mice involved in the hepatic antioxidant and xenobiotic response (Figure 1G). Collectively, these data demonstrate that colonization with gut-resident microbes has significant influences on hepatic oxidative metabolic processes, and by inference, gut-liver signaling.
Symbiotic Lactobacilli sp. activate the Nrf2 pathway in the Drosophila melanogaster fat body
Next, we sought to determine which bacterial members of the microbiome were responsible for the observed induction of Nrf2 signaling. Because of the multi-dimensional nature of the gut-liver axis, we utilized the model organism Drosophila melanogaster to screen for Nrf2 inducing bacteria. Drosophila have an intestinal system similar to that of vertebrates, with a single layer of epithelium lining the gut lumen (Douglas, 2018) and also possess an organ analogous to the vertebrate liver known as the fat body (Søndergaard, 1993).
Using a transgenic Nrf2 reporter fly (gstD1-GFP) (Sykiotis and Bohmann, 2008), in which GFP is expressed in response to Nrf2 activation and binding to a consensus ARE (Figure 2A), we assessed the capacity of candidate Gram-positive and Gram-negative commensal bacteria to activate Nrf2 signaling in the Drosophila fat body (Figure 2B). Early third-instar germ-free larvae were allowed to feed on either sterile food or food inoculated with 2×108 CFU/gram of bacteria. 16 hours later, the fat body was dissected and analyzed using fluorescence microscopy. Of the bacteria assayed, Lactobacilli were the most robust inducers of Nrf2 in the fat body (Figure 2C–D). These included the Drosophila commensal Lactobacillus plantarum and the human commensal and probiotic Lactobacillus rhamnosus GG (LGG). These data are in agreement with previous reports from our research group showing that oral administration of L. plantarum or LGG can activate Nrf2 in the intestinal epithelium of both flies and mice (Jones et al., 2015). Moreover, these Drosophila data demonstrate that a specific subset of bacteria can exert effects on Nrf2 beyond the confines of the gut in tissues that do not experience direct cell-microbe contact.
Figure 2. Symbiotic Lactobacilli sp. activate the Nrf2 pathway in the Drosophila melanogaster fat body.
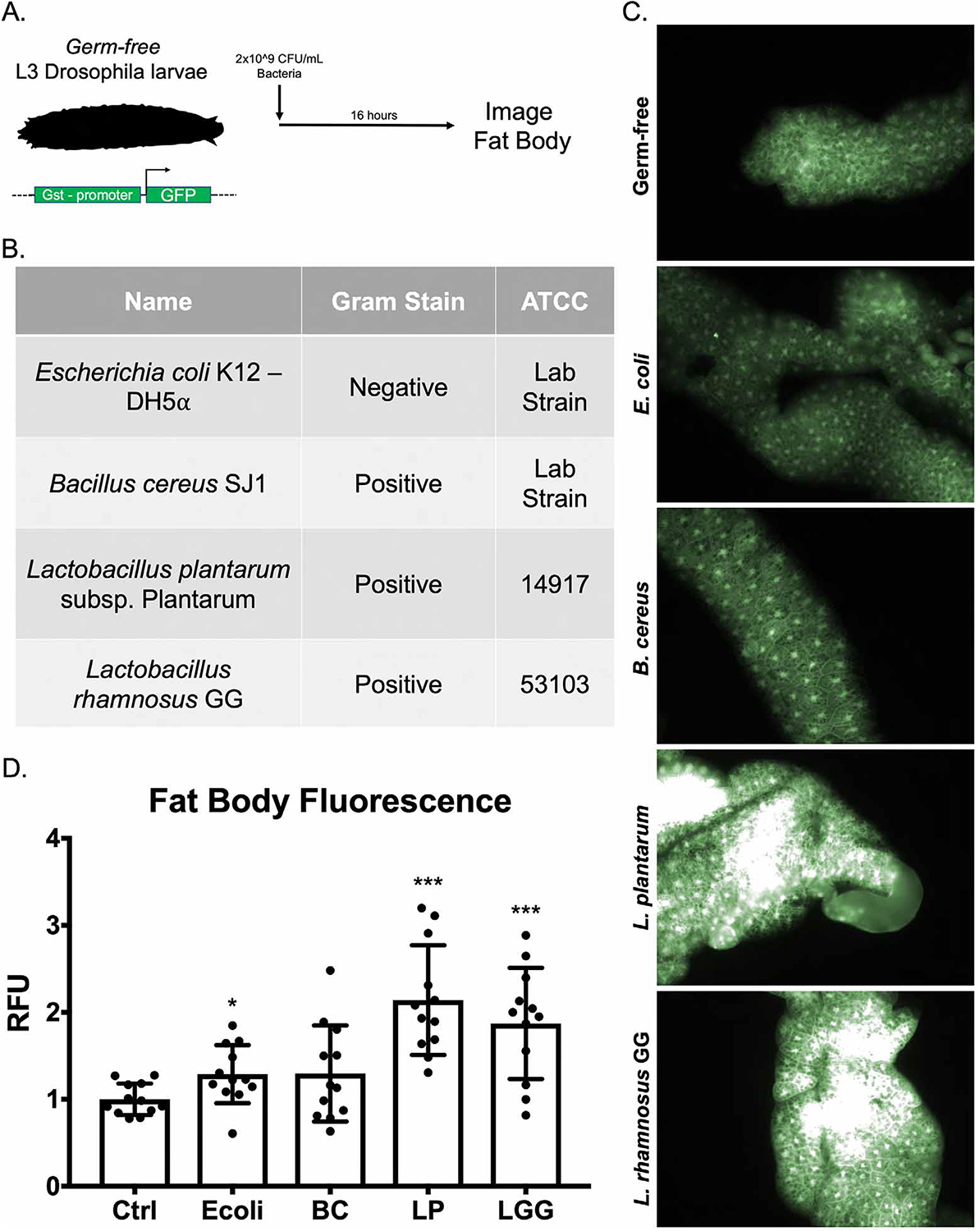
(A) Schematic of experimental system. (B) Identities and characteristics of assayed bacteria. (C) Fluorescent images of Drosophila fat body from transgenic flies described in (A) after 16-hour treatment with the indicated bacteria. (D) Quantification of fat body fluorescence detected in (C) by Image J software analysis. *p<0.05, **p<0.01, ***p<0.002 as determined by one-way ANOVA and post-hoc t-test. n=12 flies per condition.
Oral administration of Lactobacillus rhamnosus GG activates Nrf2 signaling in the murine liver
To corroborate these findings in vertebrates, we performed a two week regimen of daily oral gavage of conventional (microbiome-replete) C57BL/6 mice with vehicle (HBSS), LGG, or Bacillus cereus (BC), a Gram-positive commensal that does not induce Nrf2 in the intestine (Jones et al., 2013). Immunofluorescence for Nrf2 in hepatic sections from these animals reveals a faint signal in both the HBSS and BC treated animals (Figure 3A). The LGG treatment group, however, demonstrated a significant increase in Nrf2 immunoreactivity as well as a prominent punctate staining pattern that colocalized with the nuclear stain DAPI (Figure 3A–B). These data indicate an overall stabilization and subsequent nuclear translocation of Nrf2 in hepatocytes in response to daily gavage of LGG. Furthermore, this response appears to be specific to LGG, as BC had no effect on levels of Nrf2 immunofluorescence or DAPI colocalization (Figure 3A–B). To investigate the zonal distribution of this Nrf2 activation, we costained for Nrf2 and the glutamine synthetase, a marker of pericentral hepatocytes (zone 3). These data demonstrate that there appears to be a gradient of Nrf2 activation in LGG treated liver that is higher in zones 1 and 2, and slightly diminished in zone 3 (Figure S1). This is consistent with the pattern expected from a microbe-derived signal arising from the portal circulation. To determine if the observed Nrf2 stabilization results in increased Nrf2 activity, we performed RT-PCR for known Nrf2 target transcripts in livers from HBSS, LGG, or BC treated mice. In agreement with the immunofluorescence, HBSS and BC had similar steady state mRNA levels of the Nrf2 target transcripts Nqo1, Trx1, Gsta1, and Gclc. LGG treatment, however, led to a significant increase in the levels of all of the analyzed targets (Figure 3C). These data support our findings in Drosophila that implicate Lactobacilli in systemic activation of Nrf2.
Figure 3. Oral administration of Lactobacillus rhamnosus GG activates Nrf2 signaling in murine liver.
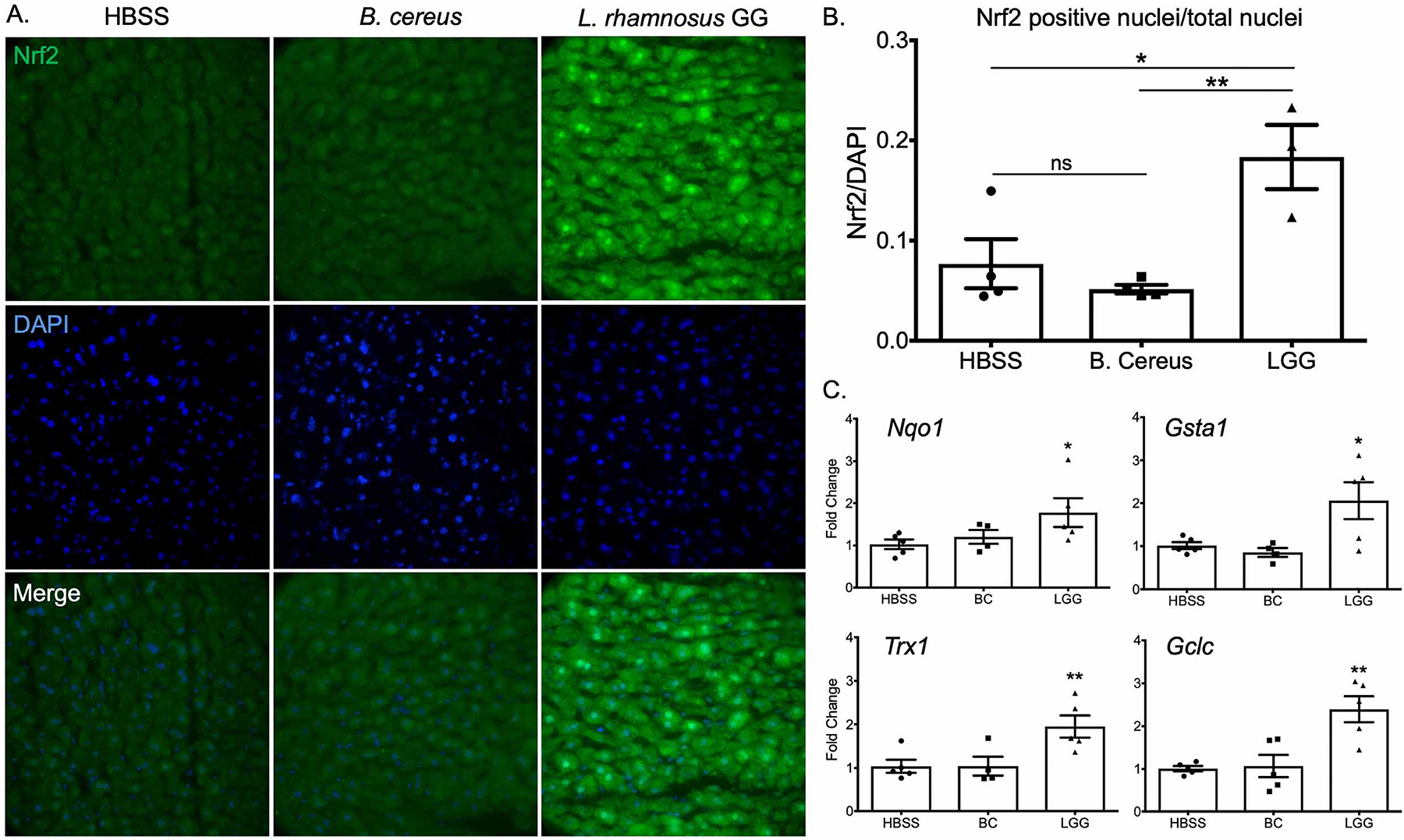
(A) Immunofluorescence analysis using an antibody against Nrf2 on liver histological sections prepared from animals orally administered HBSS (vehicle), or 2×108 CFU BC or LGG daily for two weeks. Images were acquired at 60x magnification. (Green=Nrf2) (Blue=DAPI). (B) Quantification of nuclear co-localization from images in A. n=3 animals per treatment. (C) RT-PCR of liver tissue for the steady state transcript levels of Nrf2 targets involved in the cellular antioxidant and xenobiotic response. Each experiment included a minimum of n=3 per treatment. *p<0.05, **p<0.01, ***p<0.002 as determined by one-way ANOVA and post-hoc t-test. n=3 mice per condition.
Lactobacillus rhamnosus GG protects the Drosophila fat body against toxicity in a Nrf2-dependent manner
Given the effect of LGG administration on Nrf2 activity and expression of key antioxidant enzymes, we hypothesized that LGG would be protective against acute oxidative liver injury. First, we utilized a Drosophila model of oxidative injury. Five-day old adult Drosophila were fed acetaminophen (APAP), an acute oxidative stressor in mammals (Bunchorntavakul and Reddy, 2018), or the herbicide paraquat (PQ) which is known to induce oxidative stress in the fly (Hosamani and Muralidhara, 2013). We demonstrate that feeding of either PQ or APAP resulted in dose-dependent mortality in adult Drosophila (Figure 4A). To determine the effect of these compounds on fat body signaling, we again utilized the gstD1-GFP Nrf2 reporter fly. Third-instar larvae were fed either 15 mM PQ or 100 mM APAP overnight, followed by dissection and fluorescent evaluation of the fat body. Indeed, both PQ and APAP robustly induced dNrf2 signaling in the fat body at this time point, relative to vehicle controls (Figure 4B), consistent with a production of ROS and subsequent activation of dNrf2.
Figure 4. Lactobacillus rhamnosus GG protects the Drosophila fat body against toxicity in a Nrf2 dependent manner.
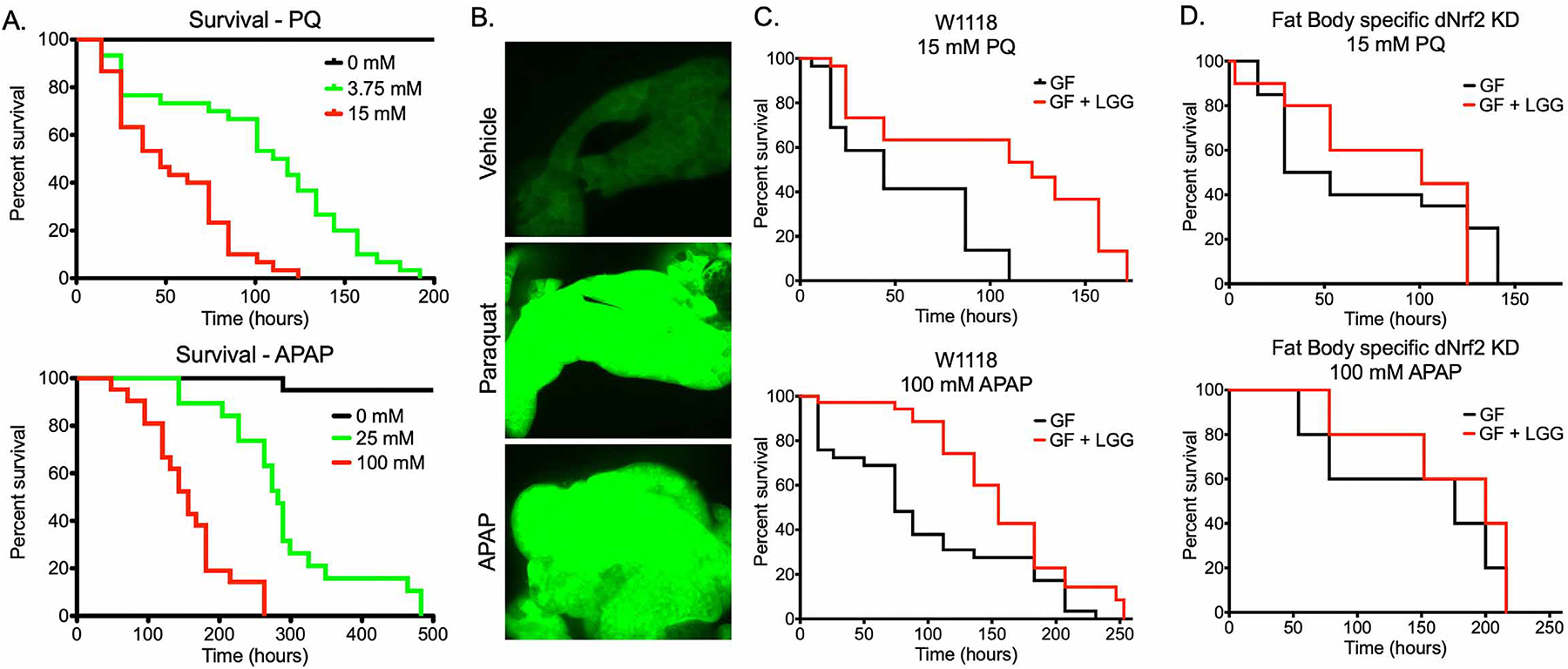
(A) Survival of adult Drosophila exposed to the indicated doses of either paraquat (PQ) or acetaminophen (APAP). Log-Rank test p<0.0001, n=20. (B) Fluorescent imaging of fat bodies of gstD1-gfp larvae following 16 hours exposure to 15 mM PQ or 100 mM APAP. (C) Survival of either germ-free (GF) or Lactobacillus rhamnosus GG (LGG) mono-associated adult Drosophila exposed to 15 mM PQ or 100 mM APAP. PQ: Log-Rank p<0.0001, n=30. APAP: Log-Rank p<0.01, n=30. (D) Survival of GF or LGG mono-associated adult female Drosophila of the genotype yolk-GAL4 UAS-cncCIR (Fat body specific Drosophila (d)Nrf2 knockdown) exposed to 15 mM PQ or 100 mM APAP. PQ: Log-Rank p=0.7799, n=20. APAP: Log-Rank p=0.4765, n=20.
We next sought to determine the effect of LGG colonization on PQ or APAP toxicity. Five-day old germ-free flies were collected and fed for 3 days on germ-free food or LGG colonized food, followed by challenge with PQ or APAP. LGG treatment significantly attenuated PQ and APAP toxicity (Fig 4C). To determine the involvement of dNrf2 signaling in LGG-mediated protection, we specifically depleted dNrf2 in the fat body and maintained GF or monocolonized with LGG. In this context, there was no significant difference between the mortality rates of GF and LGG treated flies in response to APAP or PQ (Figure 4D). These data demonstrate that LGG mediated dNrf2 activation is the mechanism by which LGG protects against oxidative fat body injury in Drosophila.
Lactobacillus rhamnosus GG administration attenuates acetaminophen hepatotoxicity in conventional mice
We next sought to corroborate these findings in a mammalian model of acetaminophen hepatotoxicity. To this end, conventionally raised C57BL/6 mice were orally administered either vehicle (HBSS) or LGG for 2 weeks. These treatments had no discernable effect on overall liver histology as assessed by H&E (Figure 5A). To determine the effect of LGG treatment on histologic hallmarks of APAP toxicity, HBSS and LGG pretreated mice were given a sub-lethal oral dose of 300 mg/kg APAP. HBSS-treated animals 24 hours after APAP had significant centrilobular necrosis, whereas liver injury in the LGG-treated groups was significantly attenuated (Figure 5A–B). In agreement with these results, serum ALT (a marker of hepatocyte injury) was elevated in the HBSS group after acetaminophen overdose but was markedly diminished in LGG treated animals (Figure 5C). To evaluate hepatic oxidative stress, we determined the ratio of total (GSH) to oxidized (GSSG) glutathione in the liver 24 hours after APAP overdose. Liver tissues from LGG pretreated animals had a significantly higher ratio of GSH to GSSG than those from HBSS pretreated animals (Figure 5D), indicating diminished hepatocellular oxidative stress. These data demonstrate the protective effect of LGG administration against the oxidative liver injury induced by acetaminophen. This protection does not appear to be due to altered APAP metabolism, as the mRNA levels of the cytochrome P450 enzymes responsible for its bioconversion were unchanged by LGG administration (Figure S2). In addition, immunohistochemical analysis for the proliferative marker PCNA 24 hours after APAP revealed no significant difference between HBSS and LGG treated animals (Figure S3A–B). These data suggest that an increase in hepatic repair is not the primary mechanism for the attenuated hepatic injury in LGG treated animals. Furthermore, the protective effect of LGG can be overwhelmed by higher doses of acetaminophen, as an overdose of 600 mg/kg APAP resulted in a similar level of injury between HBSS and LGG treated animals (Figure S4A–C).
Figure 5. Lactobacillus rhamnosus GG administration attenuates acetaminophen hepatotoxicity in conventional mice.
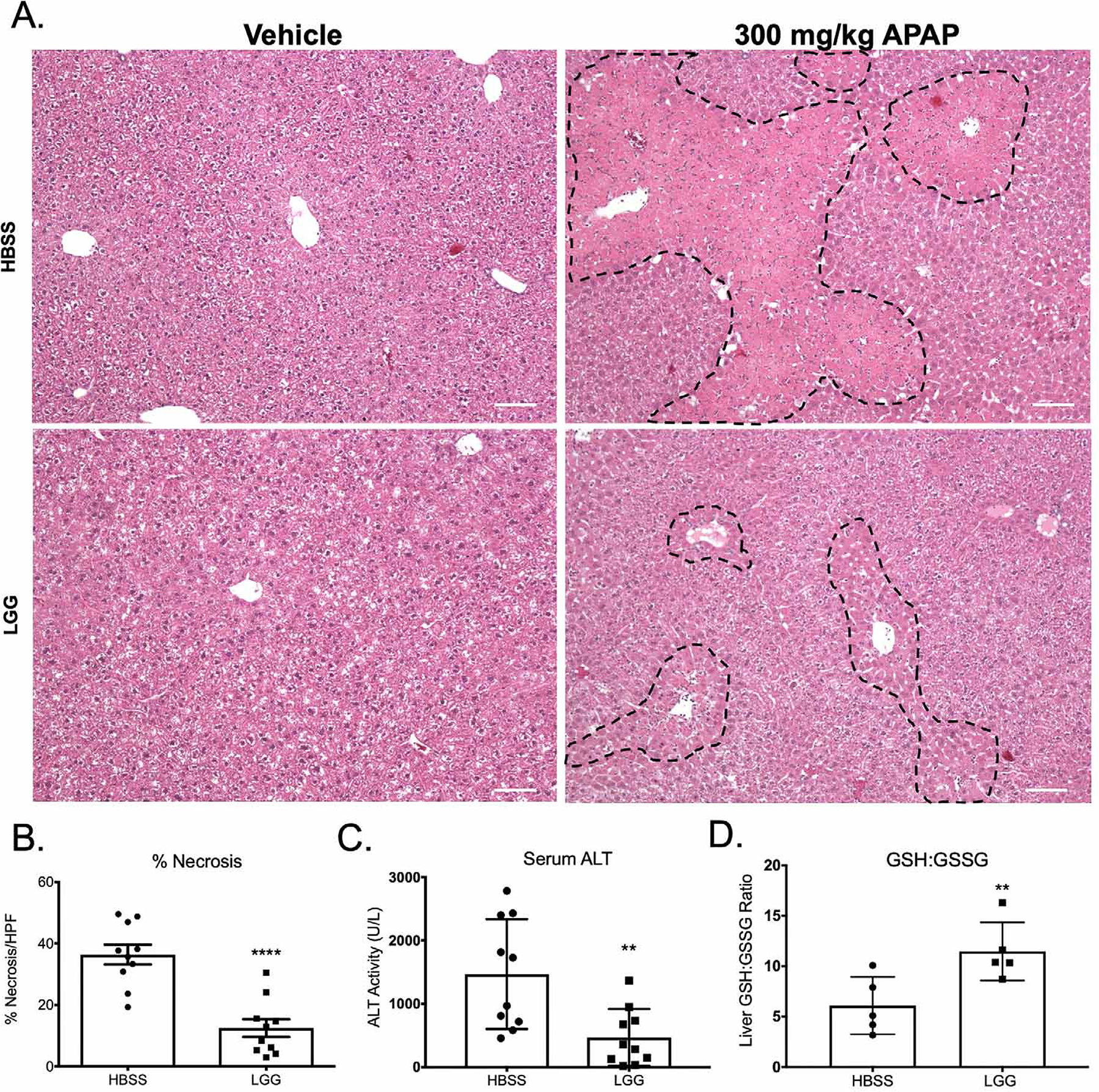
(A) Representative H&E images of mouse livers 24 hours after oral administration of 300 mg/kg APAP or vehicle control. Where indicated, mice received a daily oral gavage of either HBSS (vehicle) or 2×108 CFU Lactobacillus rhamnosus GG (LGG) for two weeks prior to APAP administration. Characteristic centrilobular necrosis is outlined by dashed lines. (B) Quantification of percent necrosis per high power field of histology from (A). n=5 mice per group. (C) Serum ALT levels at 24 hours after oral administration of 300 mg/kg APAP or vehicle control of mice described in (A). (D) Ratio of hepatic GSH to GSSG of livers 24 hours post APAP overdose as described in (A). **p<0.01 ****p<0.0001 as determined by t-test. n=10 mice per group (A-C). Scale bar (white) = 100 μm.
Hepatic Nrf2 mediates Lactobacillus rhamnosus GG protection against acetaminophen hepatotoxicity
Our findings agree with an existing body of literature that describes the protective effects of LGG against oxidative liver injury (Chen et al., 2016; de Faria Ghetti et al., 2018; Forsyth et al., 2009; Kim et al., 2016; Ritze et al., 2014; Wang et al., 2011a). The mechanisms proposed for this protection vary from an improvement in barrier function (Chen et al., 2016; Forsyth et al., 2009; Wang et al., 2011a) to alterations in bile acid signaling (Kim et al., 2016). Indeed, in our system, LGG led to a significant increase in intestinal Zo1 transcript (Figure S5A) and an improvement in intestinal epithelial barrier function (Figure S5B). To specifically investigate the role of LGG-mediated Nrf2 activation in facilitating the observed hepatoprotection, we generated liver-specific Nrf2 knockout mice using Albumin-Cre+/− × Nrf2fl/fl mice. WT (Nrf2fl/fl) and KO (Albumin-Cre × Nrf2fl/fl) mice were gavaged with either HBSS or LGG as described above for 2 weeks, followed by oral administration of 300 mg/kg APAP. Histologic analysis of Nrf2fl/fl animals 24 hours after APAP administration revealed significant centrilobular necrosis in HBSS treated animals that was attenuated in LGG treated animals (Figure 6A) in agreement with our previous results (Figure 5B). However, in Albumin-Cre+/− × Nrf2fl/fl mice, both HBSS and LGG treated animals exhibited similar tissue necrosis (Figure 6A, quantified in Figure 6B). Consistently, serum ALT levels and hepatic GSH:GSSG ratio from Albumin-Cre × Nrf2fl/fl animals were not different between HBSS and LGG treated mice. (Figure 6C–D). Note that elevation of Nrf2 target genes was maintained in LGG treated animals relative to HBSS in WT animals post-APAP(Figure S6A). This elevation was lost in Nrf2 KO animals (Figure S6B).
Figure 6. Hepatic Nrf2 mediates Lactobacillus rhamnosus GG protection against acetaminophen hepatotoxicity.
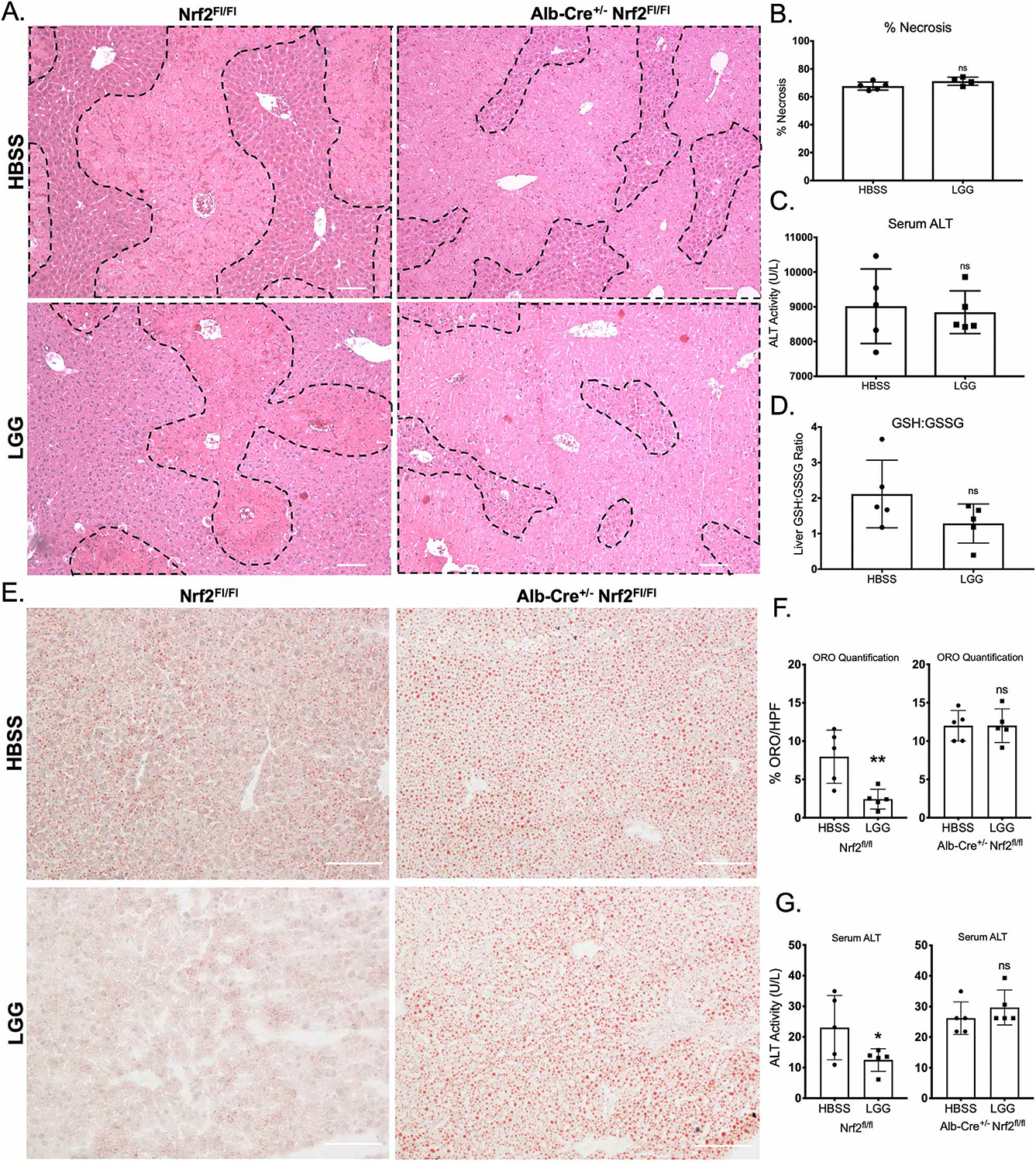
(A) Representative H&E of mouse livers 24 hours after oral administration of 300 mg/kg APAP or vehicle control from WT or littermate albumin-Cre × Nrf2fl/fl liver-specific Nrf2 knockout mice. Where indicated, mice received a daily oral gavage of either HBSS (vehicle) or 2×108 CFU Lactobacillus rhamnosus GG (LGG) for two weeks prior to APAP administration. (B) Quantification of percent necrosis/HPF from histology in (A). (C) Serum ALT activity at 24 hours post overdose of mice described in (A). (D) Ratio of hepatic GSH to GSSG 24 hours post overdose as described in (A). (E) Representative Oil red O staining for lipid content of mouse livers pretreated with HBSS or LGG by daily oral gavage for 2 weeks, 6 hours after a single oral dose of 6 g/kg ethanol, in WT or littermate albumin-Cre × Nrf2fl/fl liver-specific Nrf2 knockout mice. (F) Quantification or Oil red O staining from histologic images from (E). (G) Serum ALT activity after ethanol overdose from mice in (E). *p<0.05 **p<0.01 ns=not significant as determined by t-test. n=5 mice per group. Scale bar (white) = 100 μm.
Several studies have implicated the microbiome in susceptibility to acute ethanol toxicity and demonstrated efficacy of LGG administration in this model of acute hepatic oxidative stress (Chen et al., 2015; Wang et al., 2012). As Nrf2 has also been implicated in the pathogenesis of alcohol induced liver injury, we sought to determine the role of Nrf2 in mediating the protective effects of LGG in this system. Nrf2fl/fl and Albumin-Cre+/− × Nrf2fl/fl were treated with oral HBSS or LGG for two weeks as described above, followed by a single oral dose of 6g/kg ethanol. Livers harvested from HBSS pretreated mice 6 hours post ethanol administration exhibited significant steatosis, a histologic hallmark of acute ethanol hepatotoxicity, as visualized by Oil red O staining (Figure 6E). LGG pretreatment in Nrf2fl/fl animals significantly reduced the amount of hepatic steatosis and serum ALT (Figure 6E–G). However, no difference was observed in either steatosis or serum ALT between HBSS and LGG treated groups in the Albumin-Cre+/− × Nrf2fl/fl mice (Figure 6E–G). These data demonstrate that Nrf2 is required for the hepatoprotective effects of LGG in the setting of acute oxidative stress in two distinct modes of injury, APAP overdose and ethanol toxicity.
Lactobacillus rhamnosus GG mediated generation of 5-methoxyindoleacetic acid activates hepatic Nrf2
To determine the mechanism by which gut-resident LGG activates Nrf2 at a distance, we characterized the small molecule milieu present in the portal circulation of LGG-treated animals relative to HBSS treated controls. Portal blood was collected 12 hours after oral administration and mass spectrometry performed (Figure 7A). Subsequent tandem mass spectrometry identified candidate small molecules that were significantly enriched in the portal circulation of LGG-treated animals, of which 6 were positively identified (Figure 7B). To determine the effect of these small molecules on Nrf2 activation, we utilized a stably-transduced ARE-Luciferase HepG2 cell line as a Nrf2 reporter system. Of the small molecules enriched in the portal serum of LGG treated animals, only 5-Methoxyindoleacetic acid (5-MIAA) resulted in an increase in reporter activity (Figure S7A, Figure 7C). To corroborate these findings, we determined the steady state mRNA levels of the known Nrf2 target transcripts NQO1, HMOX1, and GCLC after a 12-hour stimulus with 5-MIAA. We observed a significant induction of all of these targets in response to 5-MIAA relative to methanol (vehicle control) (Figure 7D). To confirm that this response was due to an increase in Nrf2 stabilization and nuclear translocation, we treated HepG2 cells with 5 mM 5-MIAA for 6 hours and western-blotted for Nrf2 in the nuclear fraction (Figure 7E). Indeed, 5-MIAA treatment significantly increased the levels of nuclear Nrf2 relative to methanol control (Figure 7F). Using mass spectrometry-based quantification of the portal circulation of LGG-treated animals, 5-MIAA was found at a concentration of 2 uM (Figure S7B–C). A similar concentration of 5 uM resulted in an increase in ARE-luciferase in-vitro of 1.6–2 fold (Figure 7C), which is similar to the increase in Nrf2 target genes observed in-vivo after LGG gavage (Figure 3C).
Figure 7. Lactobacillus rhamnosus GG mediated generation of 5-methoxyindoleacetic acid activates hepatic Nrf2.
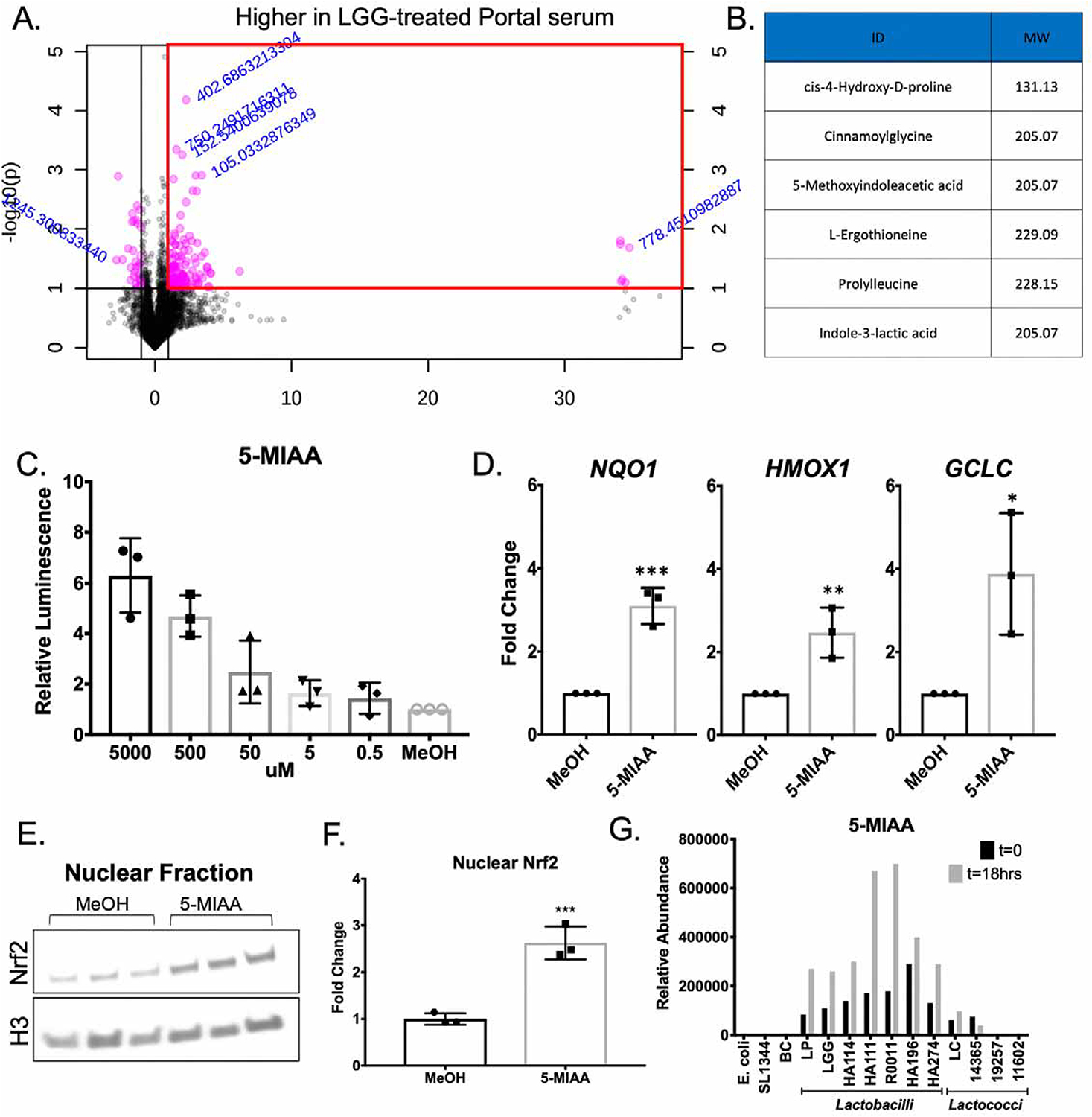
(A) Features identified by ultra-high resolution mass spectrometry of serum collected from the hepatic portal vein of mice at 12 hours after oral administration of Lactobacillus rhamnosus GG (LGG) or vehicle control. Red box outlines features increased by >2 fold in portal serum from LGG treated mice relative to vehicle treated. (B) List of matched metabolites from MS2 spectra increased in the portal serum of mice administered LGG as described in (A). (C) Luciferase activity in extracts of HepG2 cells which harbors a Nrf2 responsive promoter transcriptionally fused to luc upon exposure to 5-Methoxyindoleacetic acid (5-MIAA) for 12 hours. (D) RT-PCR for steady state mRNA levels of genes under the transcriptional control of Nrf2 after a 12-hour exposure of HepG2 cells to 5-MIAA or methanol (vehicle control). (E) Immunoblot analysis using antibodies against Nrf2 or Histone H3 (loading control) in nuclear fractions purified from HepG2 cells +/− 5-MIAA for 6 hours. (F) Densitometry analysis of data in (E). *p<0.05, **p<0.01, ***p<0.002 as determined by t-test. n=3 independent replicates. (G) Mass spectrometry for 5-MIAA from the culture supernatant of listed bacteria at 0 and 18 hours after inoculation.
Finally, to determine the microbial origin of 5-MIAA we cultured an array of representative gut microbes for 18 hours under appropriate growth conditions. Subsequent targeted mass spectrometry for 5-MIAA showed an accumulation of 5-MIAA in LGG, LP, and all of the other Lactobacillus culture supernatants tested over the course of 18 hours, while the Gram negative commensal E. coli, the Gram negative pathogen S. Typhimurium, and several strains of the genus Lactococcus did not produce 5-MIAA in this time frame (Figure 7G). Together, these data demonstrate that the Lactobacillus-derived small molecule 5-MIAA can activate hepatic Nrf2 and promote transcription of genes involved in the cellular antioxidant response.
DISCUSSION
In this study, we demonstrate a previously unappreciated role for the microbiome in altering hepatic xenobiotic and antioxidant responses via activation of the transcription factor Nrf2. Furthermore, we identify Lactobacilli sp. as mediators of this effect in part due to their production of 5-methoxyindole acetic acid, which we detected in both culture supernatant from in-vitro cultures of Lactobacilli sp. and in serum extracted from the portal circulation of LGG gavaged animals. Activation of Nrf2 by LGG in-vivo was functionally protective against oxidative liver injury, as administration of exogenous LGG protected against both acetaminophen-induced hepatotoxicity and acute ethanol toxicity in a Nrf2 dependent manner.
Our data demonstrate commensal-mediated activation of a highly conserved xenobiotic response pathway in two model organisms, Drosophila melanogaster and Mus musculus. Insects and mammals are highly evolutionarily divergent, yet this signaling is mechanistically conserved, providing further evidence for the importance of gut-resident microbes in modulating systemic health and susceptibility to disease. Our group has demonstrated that ingested microbes can induce activation of Nrf2 and confer cytoprotection of the gut epithelium of both flies and mice when directly contacted by bacteria (Jones et al., 2015), suggesting the Nrf2 pathway may have evolved a secondary role in perception and response to the microbiota. The present work shows microbes can stimulate cytoprotective effects systemically, and affect a central nexus of biochemical processing with profound effects on host metabolism of ingested xenobiotics.
Recent research has established a role for the gut microbiome in influencing human health and homeostasis, even in organs anatomically distant from the intestine. Indeed, many systemic human diseases affecting hepatic, cardiovascular, and even neuronal tissues can be modulated by the collective microbial structure within the gut (and likely other tissues) (Ho et al., 2015). This realization has prompted attempts to modulate the microbiome therapeutically through the use of probiotics, antibiotics, and fecal microbiota transplant (FMT). Of these, probiotics hold significant promise, with fewer negative sequelae than antibiotics and without the cost and invasiveness of FMT. However, the likely diverse mechanisms by which specific bacteria and their products elicit their distinct beneficial effects remain unknown. Additionally, probiotics have only been shown to be efficacious in very specific clinical scenarios. Therefore, identification of discrete molecular pathways affected by probiotic administration would allow for the more targeted use of probiotics or exploitation of microbial derived small molecules.
Previous studies in mice have shown that the commercially available probiotic Lactobacillus rhamnosus GG can improve outcomes in several clinically relevant models of liver injury, including NAFLD (Kim et al., 2016; Perumpail et al., 2019; Ritze et al., 2014) and ALD (Chen et al., 2016; Forsyth et al., 2009; Wang et al., 2011a). Mechanistically, these effects were attributed to the ability of LGG to alter bile acid signaling and promote barrier function. Indeed, we and others have shown Lactobacilli are quite effective at enhancing intestinal epithelial barrier function (Swanson et al., 2011), suppressing innate inflammatory signaling pathways (Collier-Hyams et al., 2005), and stimulating proliferation (Jones et al., 2013), which clearly could contribute to the widely known beneficial effects of treating mice with this microbe. The data presented here add to this body of literature by establishing a novel mechanism by which LGG can alter hepatic networks and consequently modify susceptibility to disease. We identify 5-MIAA as a LGG-derived small molecule inducer of Nrf2, though it is likely that many other small molecules are present within the portal circulation in response to LGG that we could not identify using our specific metabolomic platform. Future studies investigating the proteome, lipidome, and transcriptome of the portal circulation of mice exposed to a range of bacteria will be necessary for us to gain a full understanding of the microbiome-gut-liver axis.
We utilized acetaminophen toxicity, an acute model of oxidative liver injury, to demonstrate the functional importance of LGG-mediated hepatic Nrf2 activation. These data demonstrate that Nrf2 mediates the hepatoprotection observed in LGG pretreated animals after APAP overdose. Importantly, LGG administration does not alter the expression of the cytochrome P450 enzymes necessary for the conversion of APAP to its toxic metabolic byproduct, NAPQI. Additionally, while no significant alteration in hepatocyte proliferation (as measured by PCNA immunohistochemistry) was observed between vehicle and LGG treated mice at 24 hours post-APAP, there was a trend toward more proliferation in LGG-treated animals. These data raise the possibility that hepatic repair mechanisms may be altered and may play a role in models of chronic liver injury. While acetaminophen hepatotoxicity is a clinically important problem (Bunchorntavakul and Reddy, 2018; Larson et al., 2005), the necessity for pretreatment with LGG largely diminishes its therapeutic potential. Nrf2 is protective against many liver insults, and is implicated in the pathogenesis of more chronic conditions such as non-alcoholic fatty liver disease (Chambel et al., 2015), ALD (Lamlé et al., 2008), and HCC (Yates and Kensler, 2007). As these injuries occur over a much longer time frame than acetaminophen overdose, it is possible that probiotics can be used to therapeutically boost Nrf2 activity and the subsequent cytoprotective responses after diagnosis. Several studies have begun to elucidate the utility of LGG in protection from these chronic liver injuries, however the mechanistic importance of Nrf2 has not been investigated. More generally, our observation that specific bacteria can induce hepatic detoxification pathways has additional implications for clinical medicine. It is well known that specific bacterial members of the microbiota can directly metabolize specific ingested pharmacological agents and xenobiotics, resulting in inactivation (or transformation to a toxic form) (Klaassen and Cui, 2015). Our results suggest that composition of the microbiota, or supplemented bacteria can alter host bioprocessing of xenobiotics with potentially greater global effects individual pharmacological agents given the wide spectrum of activities induced by the Nrf2 pathway.
In summary, these findings add to our understanding of the functional consequences of the microbiome-gut-liver axis. Furthermore, we establish Lactobacilli as potent drivers of hepatic Nrf2 and add to the significant body of literature supporting its potential as a hepatoprotective agent. These data also indicate that the microbiota, aside from specific intrinsic encode biocatalytic activities, are capable of inducing host bioprocessing pathways, with potential consequences on ingested xenobiotics and pharmacological agents.
Limitations of Study
While we demonstrate in this study that Lactobacilli are capable of activating Nrf2 at a distance via the production of the small molecule 5-MIAA, there are several key limitations that should be noted. First, we only tested a few representative microbes for systemic Nrf2 activation in our Drosophila model. These microbes do not cover the gut microbiome as a whole, and it would be interesting to determine what other bacterial taxa have this capability. Drosophila have a much more limited microbiome than mammals, with an abundance of aerobes and facultative anaerobes present in the gut and a complete absence of anaerobes (Douglas, 2018). This discrepancy prevents a full screen of all the critical bacterial species present in the murine or human intestine. Another limitation of this study is that analysis of portal serum from LGG treated mice revealed 133 differentially abundant features, of which only 6 were identified. It will be interesting to identify those other small molecules and characterize their Nrf2 activating capacity. Additionally, we only assessed small molecules in this study. Future studies should include analysis of other components of the portal circulation via proteomics and lipidomics.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andrew S. Neish (aneish@emory.edu). This study did not result in the generation of novel reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Germ-free C57BL/6 animals were obtained from Taconic and maintained in germ-free isolators under the care of the Emory Gnotobiotic core. Conventional animals were obtained from Jackson Laboratories. Nrf2fl/fl animals were obtained from Taconic (Model 13107) and crossed with Alb-Cre hemizygous mice from Jackson Laboratories (Stock #003574). Nrf2 KO mice were Alb-Cre+/−, Nrf2fl/fl and WT controls were littermate Alb-Cre−/−, Nrf2fl/fl. Gavage of commensals and probiotics was performed as described previously(Jones et al., 2015). Briefly, 2×10^8 CFU of LGG or BC was prepared as described below and resuspended in 100 uL HBSS and administered via oral gavage to mice. This was performed daily for 14 days prior to sacrifice or administration of APAP or EtOH. All conventional mice utilized for this study were male, 8–10 weeks of age. Mice were maintained as per the approved protocol by the Emory University Animal Care and Use Committee. Mice were maintained at an average temperature of 22° C and a standard 12 hour light/dark cycle (7 am – 7 pm). Mice were fed a standard chow diet (Purina Lab Diets Cat# 5001). Health Status checks were performed daily by veterinary technicians.
Conventionalization
GF Littermates were divided at weaning (3 weeks of age) and half (3 males, 3 females) were maintained in the germ-free isolators for three weeks. The other half of the litter (3 males, 3 females) was transferred out of the isolators and placed in cages containing dirty bedding from the specific pathogen free (SPF) colony within the same mouse facility. These mice were given three weeks to acquire a microbiome prior to sacrifice and analysis of liver tissue. As all follow up studies in conventional mice were performed in males, no analysis of the sex-specific effects of conventionalization was performed for this study.
Drosophila
Germ-free Drosophila were generated by collecting freshly laid eggs, then incubating in 50% bleach for 5 minutes followed by three 5 minute washes in sterile water. 5 days post-eclosure, GF Drosophila were allowed to feed on either GF food or food containing 2×10^8 CFU LGG/mL for 3 days. Monocolonized Drosophila were then used for experiments. dNrf2 knockdown Drosophila were generated using the Gal4 UAS system described previously(Duffy, 2002). Briefly, virgin Yolk-Gal4/cyo Drosophila were crossed with male W1118 (control), or UAS-CNCC IR (dNrf2 knockdown). Straight-winged female offspring were collected 5 days post-eclosure and used for experiments. gstD1-GFP Drosophila were made germ-free, and early third instar larvae were allowed to feed on GF, E. coli, B. cereus, LGG, or L. plantarum colonized food for 16 hours. All studies in adult Drosophila were initiated at 5 days post-eclosure. Drosophila were maintained on a 12 hr light/dark cycle (7 am – 7 pm) at 25° C.
Cell lines and Culture
HepG2 cells stably transduced with the ARE-Luciferase construct were purchased from BPS Bioscience. These cells are male in origin. Cells were cultured in Growth medium 1K (MEM medium supplemented with 10% FBS, 1% non-essential amino acids, 1 mM Na-pyruvate, 1% Penicillin/Streptomycin) at 37° C and 5% CO2.
METHOD DETAILS
Mass spectrometry
Liver samples were collected from GF and CV animals (n = 6 per group), weighed, and metabolites extracted by protein precipitation using acetonitrile (10x volume:tissue). Untargeted metabolomics profiling was performed using liquid chromatography (HILIC or C18 analytical separation) coupled to a Thermo Scientific High-Field Qexactive mass spectrometer (HILIC/ESI+, C18/ESI-, 85–1,275 m/z, 120k resolution). Spectral features (m/z, retention time) corresponding to identified and uncharacterized metabolites were integrated and aligned using apLCMS/xMSanalyzer software. Statistical analysis was performed using Metaboanalyst(Xia and Wishart, 2016) and pathway enrichment analysis using Mummichog software(Li et al., 2013) and used in combination with a library of > 500 chemicals verified by m/z, retention time and MS/MS by authenticated standards. Candidate molecule were identified following ion dissociation experiments on a Thermo Scientific Fusion mass spectrometer, and spectral library matching to the mzCloud library using Compound Discoverer 3.0.
RNAsequencing
RNA-Seq analyses were conducted at the Yerkes NHP Genomics Core on GF (n = 6) and CV liver (n = 6). RNA was isolated from snap frozen liver tissues using the Qiagen RNeasy mini kit (Qiagen) and assessed for integrity and quantity using an Agilent Bioanalyzer (Agilent Technologies) and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). Libraries were prepared using the Illumina TruSeq mRNA stranded kit as per the manufacturer’s instructions. Briefly, 500–1,000 ng of Globin-depleted RNA was used for library preparation. ERCC synthetic spike-in controls 1 or 2 (Ambion) were added to each total RNA sample and processed in parallel. Amplified libraries were validated using the Agilent 4200 TapeStation and quantified using a Qubit fluorometer. Libraries were normalized, pooled, followed by clustering on a HiSeq 3000/4000 flowcell using the Illumina cBot. The clustered flowcell was then sequenced on the Illumina HiSeq 3000 system employing a single-end 101-cycle run, with multiplexing to achieve approximately 20 million reads per sample. Transcript abundance was estimated using htseq-count v0.6.1p1 and differential expression analyses were performed using DESeq2. The upstream transcriptional regulator module in the Ingenuity Pathway Analysis software was used to determine potential transcription factors involved in the observed transcriptional profiles from GF and CV mouse livers.
Bacterial cultures and growth conditions
Lactobacillus sp. and Lactococcus sp. were grown in de Man, Rogosa, and Sharpe (MRS) broth at 37 degrees Celsius without shaking. Bacillus cereus was grown in brain heart infusion (BHI) media with shaking at 37 degrees Celsius. Escherichia coli K12 and Salmonella typhimurium were grown in LB media with shaking at 37 degrees Celsius for 16 hours prior to administration. For oral gavage, all bacteria were cultured for 16 hours, centrifuged at 3000xg for 5 minutes, the supernatant aspirated, and the pellet washed with one volume HBSS. This was repeated for a total of 3 washes. The bacteria were then resuspended to a final concentration of 2×10^9 CFU/mL and 100 uL of this was gavaged to animals as described above. For Drosophila monocolonization, bacteria were resuspended in food at a final concentration of 2×10^8 CFU/mL.
Drosophila fat body challenge
For evaluation of dNrf2 activation in the fat body in response to paraquat (PQ) and acetaminophen (APAP), early third instar GST-GFP larvae were fed food containing either H2O (vehicle), 15 mM PQ, or 100 mM APAP. 16 hours later, fat bodies were dissected, fixed, and imaged as described above. For survival assays, 5 day old GF, CV, or LGG monocolonized Drosophila were transferred to fresh vials containing either H2O (vehicle), 15 mM PQ, or 100 mM APAP dissolved in 5% sucrose and 1% agar. Flies were maintained at 25 degrees Celsius and mortality recorded.
Immunofluorescence
Liver samples for immunofluorescence were embedded in OCT (Sakura Rinetek USA) and snap frozen. 5 uM sections were fixed in 4% paraformaldehyde. Samples were permeabilized in 0.5% triton-X 100 in PBS and blocked with 5% bovine serum albumin (Sigma Aldrich). The Nrf2 polyclonal rabbit antibody (Cell-signaling) and the glutamine synthetase mouse monoclonal antibody (Invitrogen) were used at 1:100 in PBS with 0.1% Triton-X and 5% BSA. Samples were incubated in primary antibody overnight at 4 degrees Celsius. Samples were then washed three times and incubated for 1 hour in Alexa-fluor 488 goat anti-rabbit or Alexa-fluor 555 goat anti-mouse secondary antibodies (Invitrogen). Samples were again washed and incubated with DAPI at a concentration of 1:10000 in PBS for 5 minutes. Samples were mounted using Prolong diamond antifade and imaged on Nikon eclipse 80i microscope fitted with a R1 Retiga Q Imaging camera. The images in Figure 3 were obtained at 60x magnification at an excitation/emission of 493/519 (Alexa-fluor 488 for Nrf2) and 358/461 (DAPI for nuclei). In Figure S1, the images were obtained at 20x magnification at an excitation/emission of 493/519 (Alexa-fluor 488 for Nrf2) and 556/573 (Alexa-fluor 555 for GS). Colocalization analysis was performed using ImageJ (NIH). For imaging of Drosophila fat body, fat bodies were dissected, fixed in 4% PFA and 0.1% NP40 in PBS for 5 minutes, followed by 3 PBS washes. Fat bodies were whole-mounted on glass microscope slides and imaged using Nikon eclipse 80i microscope fitted with a R1 Retiga Q Imaging camera. Quantification of fluorescence intensity was performed using ImageJ software (NIH).
Transcriptional analysis
For transcriptional analysis, whole liver was dissected and snap frozen in liquid nitrogen. Liver was then homogenized using a liquid nitrogen cooled mortar and pestle. 50 mg of liver tissue was mechanically disrupted in Trizol (Invitrogen) using a MagnaLyser with MagnaLyser beads (Roche). For in-vitro samples from HepG2 cells, cells were lysed directly into Trizol. RNA was prepared according to Trizol manufacturer’s instructions. RT-PCR was performed using SybrGreen supermix (Bio-Rad) using the primers listed in Table S1. Fold change determined using 2^ddCt method (Schmittgen and Livak, 2008).
FITC-Dextran Permeability Assay
Intestinal permeability in response to LGG was measured using the FITC-Dextran permeability assay as described previously (Karhausen et al., 2004). Mice were administered 0.6 mg/g body weight FITC-Dextran (MW 4 kDa, Sigma-Aldrich) by oral gavage. Mice were sacrificed 4 hours later and FITC concentration in the systemic serum was measured.
Acetaminophen challenge
Mice were fasted overnight (16 hours) with free access to water. Mice were then gavaged with 300 mg/kg or 600 mg/kg acetaminophen dissolved in HBSS with 50% polyethylene glycol. Mice were monitored for distress throughout the course of the assay. At 24 hours post treatment, mice were sacrificed, serum collected for biochemical assays (see below), and liver harvested for histology (FFPE). Histologic analysis of H&E stained sections was performed in a blinded fashion by a board-certified pathologist (BSR). ALT activity was assayed using the ALT Infinity assay (Thermo Scientific) according to manufacturer’s instructions. GSH and GSSG were measured from liver tissue using a glutathione assay kit (Cayman Chemical) according to manufacturer’s instructions. Formalin-fixed paraffin embedded samples were stained with haemotoxylin and eosin and imaged at 10x. 5 images were obtained from each section and percent necrosis calculated by a board-certified pathologist (Brian Robinson, MD/PhD). For PCNA immunohistochemistry, FFPE livers were sectioned at 5 um and stained with a mouse monoclonal PCNA antibody (Cell Signaling) as described previously (Wolfe et al., 2011).
Ethanol challenge
Ethanol challenge was carried out as described previously (Chen et al., 2015; Wang et al., 2012). Briefly, mice were fasted overnight (16 hours) with free access to water. Mice were then given an oral gavage of 6 mg ethanol per kg body weight. 6 hours later animals were sacrificed, serum collected for ALT activity assay as described above, and liver harvested. For histologic analysis of steatosis, 5 um thick liver cryosections were formalin fixed and stained with Oil red O. Images were obtained at 20X.
ARE-Luciferase Assay
HepG2 cells stably transduced with the ARE-Luciferase construct were purchased from BPS Bioscience. Cells were exposed to serial dilutions of the indicated compounds for 12 hours and luciferase detected using the One-Step Luciferase assay system (BPS Bioscience) on a HTX Synergy plate reader (BioTek). Data are represented relative to vehicle control.
Western blot analysis
For analysis of Nrf2 nuclear translocation, HepG2 cells were exposed to 5 mM 5-MIAA for 6 hours. Nuclei were prepared using the NE-PER kit (Thermo-Fisher) according to manufacturer’s instructions. Equal protein (as determined by BCA assay (Thermo Scientific)) was run on 4–20% gradient gels and transferred to nitrocellulose membranes. Primary antibodies were incubated in 5% non-fat dry milk in TBS + 0.1% Tween overnight at 4 degrees Celsius at a dilution of 1:1000. The Nrf2 antibody used was rabbit anti-Nrf2 (Abcam). HRP-conjugated anti-rabbit secondary antibodies (GE Healthcare) were used along with Pico substrate (Thermo Scientific). Blots were imaged using KwikQuant digital imager. Densitometry analysis was performed using Image-J software (NIH). Data were normalized to H3 signal.
QUANTIFICATION AND STATISTICAL ANALYSIS
Values are presented as mean +/− SEM. Statistical analyses were performed using GraphPad Prism software. For comparisons of two groups, t-test was used. For Drosophila survival studies, the Log-Rank test was used to determine statistical significance. For comparisons of groups of 3 or more, one-way ANOVA was used, followed by Dunnett’s multiple comparison test. For unpaired t-tests, F tests were performed to determine equal variances between groups. For one-way ANOVA, the Brown-Forsythe test was performed to determine equal variances between groups. Statistical parameters are stated within the figure legends.
DATA AND CODE AVAILABILITY
The RNA sequencing data generated in this study are available in the Gene Expression Omnibus repository under the accession number GSE145012.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Nrf2 | Cell Signaling Technology | Cat#12721 |
| Anti-glutamine synthetase | Abcam | Cat#ab16802 |
| Alexa Fluor goat anti-rabbit 488 | Thermo Fisher | Cat#A-11034 |
| Alexa Fluor goat anti-mouse 555 | Thermo Fisher | Cat#A32727 |
| Anti-PCNA | Cell Signaling | Cat#2586 |
| Bacterial and Virus Strains | ||
| Lactobacillus rhamnosus GG | ATCC | 14917 |
| Lactobacillus plantarum subsp. Plantarum | ATCC | 53103 |
| Escherechia coli K12 – DH5a | Neish Lab, Emory University | N/A |
| Bacillus cereus SJ1 | Neish Lab, Emory University | N/A |
| Salmonella typhimurium 1344 | Neish Lab, Emory University | NA |
| Lactobacillus rhamnosus HA114 | R. Jones Lab, Emory University | N/A |
| Lactobacillus rhamnosus HA111 | R. Jones Lab, Emory University | N/A |
| Lactobacillus rhamnosus R0011 | R. Jones Lab, Emory University | N/A |
| Lactobacillus paracasei HA196 | R. Jones Lab, Emory University | N/A |
| Lactobacillus paracasei HA274 | R. Jones Lab, Emory University | N/A |
| Lactococcus lactis subs. cremoris | R. Jones Lab, Emory University | N/A |
| Lactococcus lactis subs. cremoris | ATCC | 19257 |
| Lactococcus lactis subs. cremoris | ATCC | 14365 |
| Lactococcus lactis subs. cremoris | ATCC | 11602 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tissue-Tek OCT compound | Sakura Finetek USA | Cat#4583 |
| 2-[4-(Aminoiminomethyl)phenyl]-1H-Indole-6-carboximidamide hydrochloride (DAPI) | Tocris | Cat#5748 |
| Bovine Serum Albumin | Sigma Aldrich | Cat#A2153 |
| Methyl viologen dichloride hydrate (paraquat) | Sigma Aldrich | Cat#856177 |
| Acetaminophen | Sigma Aldrich | Cat#A7085 |
| Oil Red O | Sigma Aldrich | Cat#3125–12 |
| Cis-4-Hydroxy-D-Proline | Sigma Aldrich | Cat#H5877 |
| N-cinnamoylglycine | Sigma Aldrich | Cat#98956 |
| L-ergotheionine | Sigma Aldrich | Cat#E7521 |
| Pro-Leu | Sigma Aldrich | Cat#P1130 |
| Indol-3-Lactic Acid | Sigma Aldrich | Cat#I5508 |
| 5-Methoxy-3-indole acetic Acid | Sigma Aldrich | Cat#M14935 |
| Fluoroscein isothiocyanate (FITC)-dextran avg. MW 3–5 kDa | Sigma Aldrich | Cat#FD4 |
| Critical Commercial Assays | ||
| iQ Sybr Green Supermix | Bio-rad | Cat#1708880 |
| Infinity ALT (GPT) Liquid Stable Reagent | Thermo Scientific | Cat#TR71121 |
| Glutathione Assay Kit | Cayman Chemical | Cat#703002 |
| ONE-Step Luciferase Assay System | BPS Bioscience | Cat#60690–1 |
| NE-PER Nuclear and Cytoplasmic Extraction Reagents | Thermo Fisher | Cat#78833 |
| Mouse specific HRP/DAB (ABC) Detection IHC | Abcam | Cat#ab64259 |
| Deposited Data | ||
| RNAseq Data | Gene Expression Omnibus | GSE145012 |
| Experimental Models: Cell Lines | ||
| Human: HepG2 ARE Reporter Cell Line | BPS Bioscience | Cat#60513 |
| Human: HepG2 | ATCC | HB-8065 |
| Experimental Models: Organisms/Strains | ||
| GF mouse: C57Bl/6 | R. Jones lab, Emory University | N/A |
| Drosophila melanogaster W1118 | R. Jones lab, Emory University | N/A |
| Mouse: C57/Bl6J | Jackson Laboratories | 000664 |
| Drosophila melanogaster Gstd-GFP | R. Jones lab, Emory University | N/A |
| Drosophila melanogaster Yolk-Gal4 | R. Jones lab, Emory University | N/A |
| Drosophila melanogaster cncCIR | R. Jones lab, Emory University | N/A |
| Mouse: B6.Cg-Speer6-ps1Tg(Alb-cre)21Mgn/J | Jackson Laboratories | JAX: 003574 |
| Mouse: C57BL/6-Nfe2l2tm1.1 Mrl | Taconic | TAC: 13107 |
| Oligonucleotides | ||
| Primers for RT-PCR analysis, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| Software and Algorithms | ||
| MetaboAnalyist | Xia Lab, McGill University | https://www.metaboanalyst.ca |
| apLCMS/xMSanalyzer | Jones Lab, Emory University | http://web1.sph.emory.edu/apLCMS/ |
| Mummichog | Jones Lab, Emory University | https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003123 |
| DESeq2 | Anders Lab, Harvard School of Public Health | http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Ingenuity Pathway Analysis | QIAGEN Bioinformatics | https://digitalinsights.qiagen.com/products-overview/analysis-and-visualization/qiagen-ipa/ |
| ImageJ | Open source | https://imagej.nih.gov/ij/download.html |
| Prism v. 7 and v. 8 | Graphpad | https://www.graphpad.com |
| Other | ||
Highlights.
The gut microbiome induces the Nrf2 antioxidant response pathway in the liver
Gut-resident Lactobacilli induce hepatic Nrf2 in both Drosophila and mice
Oral delivery of Lactobacillus rhamnosus GG protects against oxidative liver injury
Lactobacilli-demed 5-methoxyindoleacetic acid activates Nrf2
Context and Significance.
Recent studies have demonstrated that the gut microbiome can impact the function of tissues beyond the intestine. How microbes elicit these effects and whether they can be manipulated to treat disease, however, remains largely unknown. Researchers at Emory University demonstrate that the microbiome acts to increase the antioxidant tone of the liver via activation of the transcription factor Nrf2. This can be augmented by the administration of Lactobacillus rhamnosus GG, which produces a small molecule (5-methoxyindoleacetic acid) that travels from the gut to the liver to activate Nrf2. This signaling is sufficient to protect the liver from acute acetaminophen and ethanol toxicity. These findings demonstrate a discrete mechanism by which gut-resident microbiota impact the health of organs beyond the intestine.
ACKNOWLEDGEMENTS
Funding: This work was supported by the National Institute for Diabetes and Digestive and Kidney Diseases (R01 award AI64462, F30 award DK117570).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors have no interests to disclose
REFERENCES
- Bunchorntavakul C, and Reddy KR (2018). Acetaminophen (APAP or N-Acetyl-p-Aminophenol) and Acute Liver Failure. Clin Liver Dis 22, 325–346. [DOI] [PubMed] [Google Scholar]
- Carroll IM, Ringel-Kulka T, Keku TO, Chang Y-H, Packey CD, Sartor RB, and Ringel Y (2011). Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am. J. Physiol. Gastrointest. Liver Physiol 301, G799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll IM, Ringel-Kulka T, Siddle JP, and Ringel Y (2012). Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil 24, 521–530, e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassard A-M, and Ciocan D (2018). Microbiota, a key player in alcoholic liver disease. Clin Mol Hepatol 24, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambel SS, Santos-Gonçalves A, and Duarte TL (2015). The Dual Role of Nrf2 in Nonalcoholic Fatty Liver Disease: Regulation of Antioxidant Defenses and Hepatic Lipid Metabolism. Biomed Res Int 2015, 597134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Miyamoto Y, Mazagova M, Lee K-C, Eckmann L, and Schnabl B (2015). Microbiota Protects Mice Against Acute Alcohol-Induced Liver Injury. Alcohol. Clin. Exp. Res 39, 2313–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R-C, Xu L-M, Du S-J, Huang S-S, Wu H, Dong J-J, Huang J-R, Wang X-D, Feng W-K, and Chen Y-P (2016). Lactobacillus rhamnosus GG supernatant promotes intestinal barrier function, balances Treg and TH17 cells and ameliorates hepatic injury in a mouse model of chronic-binge alcohol feeding. Toxicol. Lett 241, 103–110. [DOI] [PubMed] [Google Scholar]
- Collier-Hyams LS, Sloane V, Batten BC, and Neish AS (2005). Cutting edge: bacterial modulation of epithelial signaling via changes in neddylation of cullin-1. J. Immunol 175, 4194–4198. [DOI] [PubMed] [Google Scholar]
- Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, Huot L, Grandjean T, Bressenot A, Delanoye-Crespin A, et al. (2013). NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest 123, 700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson PA, and Karpen SJ (2015). Intestinal transport and metabolism of bile acids. J. Lipid Res 56, 1085–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE (2018). The Drosophila model for microbiome research. Lab Anim (NY) 47, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB (2002). GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis 34, 1–15. [DOI] [PubMed] [Google Scholar]
- de Faria Ghetti F, Oliveira DG, de Oliveira JM, de Castro Ferreira LEVV, Cesar DE, and Moreira APB (2018). Influence of gut microbiota on the development and progression of nonalcoholic steatohepatitis. Eur J Nutr 57, 861–876. [DOI] [PubMed] [Google Scholar]
- Foley MH, O’Flaherty S, Barrangou R, and Theriot CM (2019). Bile salt hydrolases: Gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 15, e1007581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth CB, Farhadi A, Jakate SM, Tang Y, Shaikh M, and Keshavarzian A (2009). Lactobacillus GG treatment ameliorates alcohol-induced intestinal oxidative stress, gut leakiness, and liver injury in a rat model of alcoholic steatohepatitis. Alcohol 43, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girbovan A, Sur G, Samasca G, and Lupan I (2017). Dysbiosis a risk factor for celiac disease. Med. Microbiol. Immunol 206, 83–91. [DOI] [PubMed] [Google Scholar]
- Go Y-M, Uppal K, Walker DI, Tran V, Dury L, Strobel FH, Baubichon-Cortay H, Pennell KD, Roede JR, and Jones DP (2014). Mitochondrial metabolomics using high-resolution Fourier-transform mass spectrometry. Methods Mol. Biol 1198, 43–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JTK, Chan GCF, and Li JCB (2015). Systemic effects of gut microbiota and its relationship with disease and modulation. BMC Immunol. 16, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosamani R, and Muralidhara (2013). Acute exposure of Drosophila melanogaster to paraquat causes oxidative stress and mitochondrial dysfunction. Arch. Insect Biochem. Physiol 83, 25–40. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Integrative HMP (iHMP) Research Network Consortium (2019). The Integrative Human Microbiome Project. Nature 569, 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Xie C, Li F, Zhang L, Nichols RG, Krausz KW, Cai J, Qi Y, Fang Z-Z, Takahashi S, et al. (2015). Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Invest 125, 386–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA, et al. (2013). Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 32, 3017–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Desai C, Darby TM, Luo L, Wolfarth AA, Scharer CD, Ardita CS, Reedy AR, Keebaugh ES, and Neish AS (2015). Lactobacilli Modulate Epithelial Cytoprotection through the Nrf2 Pathway. Cell Rep 12, 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhausen J, Furuta GT, Tomaszewski JE, Johnson RS, Colgan SP, and Haase VH (2004). Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J. Clin. Invest 114, 1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Park K-Y, Ji Y, Park S, Holzapfel W, and Hyun C-K (2016). Protective effects of Lactobacillus rhamnosus GG against dyslipidemia in high-fat diet-induced obese mice. Biochem. Biophys. Res. Commun 473, 530–536. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, and Cui JY (2015). Review: Mechanisms of How the Intestinal Microbiota Alters the Effects of Drugs and Bile Acids. Drug Metab. Dispos 43, 1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamlé J, Marhenke S, Borlak J, von Wasielewski R, Eriksson CJP, Geffers R, Manns MP, Yamamoto M, and Vogel A (2008). Nuclear factor-eythroid 2-related factor 2 prevents alcohol-induced fulminant liver injury. Gastroenterology 134, 1159–1168. [DOI] [PubMed] [Google Scholar]
- Larigot L, Juricek L, Dairou J, and Coumoul X (2018). AhR signaling pathways and regulatory functions. Biochim Open 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AM, Polson J, Fontana RJ, Davern TJ, Lalani E, Hynan LS, Reisch JS, Schiødt FV, Ostapowicz G, Shakil AO, et al. (2005). Acetaminophen-induced acute liver failure: results of a United States multicenter, prospective study. Hepatology 42, 1364–1372. [DOI] [PubMed] [Google Scholar]
- Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Jones DP, and Pulendran B (2013). Predicting network activity from high throughput metabolomics. PLoS Comput. Biol 9, e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Liu W, Piao M, and Zhu H (2017). A review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids 49, 2083–2090. [DOI] [PubMed] [Google Scholar]
- Liu KH, Walker DI, Uppal K, Tran V, Rohrbeck P, Mallon TM, and Jones DP (2016). High-Resolution Metabolomics Assessment of Military Personnel: Evaluating Analytical Strategies for Chemical Detection. J. Occup. Environ. Med 58, S53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Heikenwalder M, and Ganal-Vonarburg SC (2016). The Liver at the Nexus of Host-Microbial Interactions. Cell Host Microbe 20, 561–571. [DOI] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, and Kan YW (1994). Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NFE2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. U.S.A 91, 9926–9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumpail BJ, Li AA, John N, Sallam S, Shah ND, Kwong W, Cholankeril G, Kim D, and Ahmed A (2019). The Therapeutic Implications of the Gut Microbiome and Probiotics in Patients with NAFLD. Diseases 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani A, Massy ZA, Meijers B, Evenepoel P, Vanholder R, and Raj DS (2016). Role of the Gut Microbiome in Uremia: A Potential Therapeutic Target. Am. J. Kidney Dis 67, 483–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritze Y, Bárdos G, Claus A, Ehrmann V, Bergheim I, Schwiertz A, and Bischoff SC (2014). Lactobacillus rhamnosus GG protects against non-alcoholic fatty liver disease in mice. PLoS ONE 9, e80169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderburg C, and Luedde T (2014). The role of the gut microbiome in the development and progression of liver cirrhosis and hepatocellular carcinoma. Gut Microbes 5, 441–445. [DOI] [PubMed] [Google Scholar]
- Romano KA, Vivas EI, Amador-Noguez D, and Rey FE (2015). Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio 6, e02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor RB (2008). Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, and Livak KJ (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schugar RC, Willard B, Wang Z, and Brown JM (2018). Postprandial gut microbiota-driven choline metabolism links dietary cues to adipose tissue dysfunction. Adipocyte 7, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran Van Nhieu J, and Furet JP (2011). Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE 6, e16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søndergaard L (1993). Homology between the mammalian liver and the Drosophila fat body. Trends Genet. 9, 193. [DOI] [PubMed] [Google Scholar]
- Stewart GS, and Smith CP (2005). Urea nitrogen salvage mechanisms and their relevance to ruminants, non-ruminants and man. Nutr Res Rev 18, 49–62. [DOI] [PubMed] [Google Scholar]
- Swanson PA, Kumar A, Samarin S, Vijay-Kumar M, Kundu K, Murthy N, Hansen J, Nusrat A, and Neish AS (2011). Enteric commensal bacteria potentiate epithelial restitution via reactive oxygen species-mediated inactivation of focal adhesion kinase phosphatases. Proc. Natl. Acad. Sci. U.S.A 108, 8803–8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis GP, and Bohmann D (2008). Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell 14, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamboli CP, Neut C, Desreumaux P, and Colombel JF (2004). Dysbiosis as a prerequisite for IBD. Gut 53, 1057. [PMC free article] [PubMed] [Google Scholar]
- Timsit YE, and Negishi M (2007). CAR and PXR: the xenobiotic-sensing receptors. Steroids 72, 231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan MLY, and El-Nezami H (2018). Targeting gut microbiota in hepatocellular carcinoma: probiotics as a novel therapy. Hepatobiliary Surg Nutr 7, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kirpich I, Liu Y, Ma Z, Barve S, McClain CJ, and Feng W (2011a). Lactobacillus rhamnosus GG treatment potentiates intestinal hypoxia-inducible factor, promotes intestinal integrity and ameliorates alcohol-induced liver injury. Am. J. Pathol 179, 2866–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu Y, Sidhu A, Ma Z, McClain C, and Feng W (2012). Lactobacillus rhamnosus GG culture supernatant ameliorates acute alcohol-induced intestinal permeability and liver injury. Am. J. Physiol. Gastrointest. Liver Physiol 303, G32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, et al. (2011b). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A, Thomas A, Edwards G, Jaseja R, Guo GL, and Apte U (2011). Increased activation of the Wnt/β-catenin pathway in spontaneous hepatocellular carcinoma observed in farnesoid X receptor knockout mice. J. Pharmacol. Exp. Ther 338, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, and Wishart DS (2016). Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr Protoc Bioinformatics 55, 14.10.1–14.10.91. [DOI] [PubMed] [Google Scholar]
- Yates MS, and Kensler TW (2007). Keap1 eye on the target: chemoprevention of liver cancer. Acta Pharmacol. Sin 28, 1331–1342. [DOI] [PubMed] [Google Scholar]
- Zhan Y, Chen P-J, Sadler WD, Wang F, Poe S, Núñez G, Eaton KA, and Chen GY (2013). Gut microbiota protects against gastrointestinal tumorigenesis caused by epithelial injury. Cancer Res. 73, 7199–7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data generated in this study are available in the Gene Expression Omnibus repository under the accession number GSE145012.


