SUMMARY
We compare immunogenicity and protective efficacy of an HIV vaccine comprised of env and gag DNA and Env (Envelope) proteins by co-administration of the vaccine components in the same muscles or by separate administration of DNA + protein in contralateral sites in female rhesus macaques. The 6-valent vaccine includes gp145 Env DNAs, representing six sequentially isolated Envs from the HIV-infected individual CH505, and matching GLA-SE-adjuvanted gp120 Env proteins. Interestingly, only macaques in the co-administration vaccine group are protected against SHIV CH505 acquisition after repeated low-dose intravaginal challenge and show 67% risk reduction per exposure. Macaques in the co-administration group develop higher Env-specific humoral and cellular immune responses. Non-neutralizing Env antibodies, ADCC, and antibodies binding to FcγRIIIa are associated with decreased transmission risk. These data suggest that simultaneous recognition, processing, and presentation of DNA + Env protein in the same draining lymph nodes play a critical role in the development of protective immunity.
Graphical Abstract
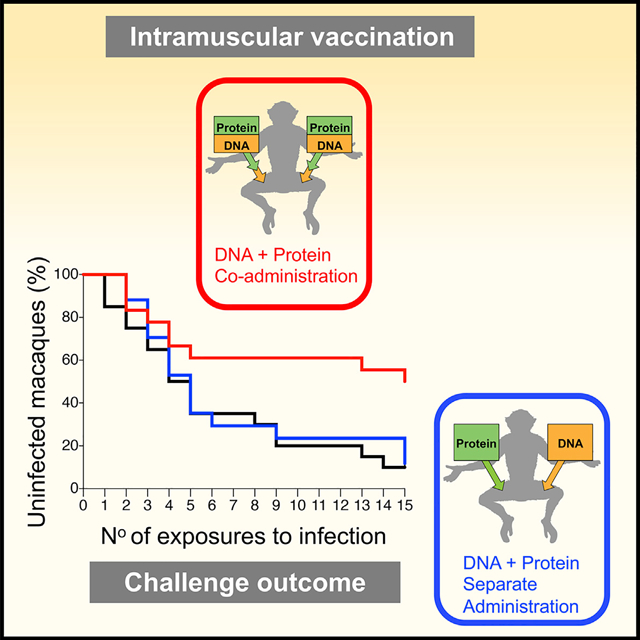
In Brief
Felber et al. show that immune responses induced by an HIV DNA and protein combination vaccine co-administered simultaneously in the same anatomical site reduce infection risk upon pathogenic SHIV challenge, compared to separate delivery of the same DNA and protein components in contralateral sites.
INTRODUCTION
Development of an HIV vaccine is a public health priority with 1.7 million new infections worldwide in 2018 (https://www.unaids.org/en/resources/fact-sheet). The only HIV vaccine trial to show a protective effect in humans was RV144, which demonstrated modest (31.2%) estimated vaccine efficacy (Rerks-Ngarm et al., 2009), using a canarypox vector (ALVAC) expressing HIV genes as a prime, followed by booster immunizations with ALVAC plus recombinant HIV gp120 Env glycoproteins (AIDSVAX B/E) (Rerks-Ngarm et al., 2009). Immune correlates analysis identified non-neutralizing antibodies (Abs) against the Env-variable V1V2 region and Abs-mediating cellular cytotoxicity (ADCC) as correlates of reduced risk of infection (Corey et al., 2015; Haynes et al., 2012; Karasawas et al., 2012; Rolland et al., 2012; Zolla-Pazner et al., 2013). Several vaccine regimens are presently being tested in phase-I and phase-II/III human trials as well as in rhesus macaque (RM) models with the aim to improve vaccine efficacy, durability, and breadth (Ackerman et al., 2018; Bradley et al., 2017; Tomaras and Plotkin, 2017). Among the different vaccine strategies being explored, DNA is a promising platform due to its simplicity, scalability, and possibility for repeated applications without eliciting immunity against the vector (Felber et al., 2014; Flingai et al., 2013; Villarreal et al., 2013). Use of RNA/codon-optimized HIV/SIV DNA vaccines in the RM model can induce robust and durable T cell responses, which efficiently disseminate into mucosal sites (Hirao et al., 2008; Muthumani et al., 2003, 2013; Patel et al., 2013; Rosati et al., 2009, 2005; Valentin et al., 2014; Vargas-Inchaustegui et al., 2014). Whereas SIV/HIV DNA vaccines elicit robust cellular responses, the levels of humoral immune responses after intramuscular (IM) administration are modest. This limitation could be alleviated by boosting with Env protein, which increases the magnitude of the humoral responses, although the durability of these responses was limited (Jalah et al., 2014; Patel et al., 2013), a common problem in HIV/SIV vaccines. A combined vaccine regimen of simultaneous co-administration of DNA + protein in the same anatomical sites (Jalah et al., 2014; Li et al., 2013; Patel et al., 2013) induced cellular and humoral immune responses with superior quality, magnitude, and longevity, which efficiently disseminated to mucosal sites and provided durable immunity. RMs vaccinated with the SIV DNA + protein vaccine showed delay in SIV acquisition and efficient control of viremia, preventing progression toward AIDS (Patel et al., 2013; Singh et al., 2018). The robust humoral immunity induced by the DNA + protein combination vaccine, detectable after the first vaccination, was reported in mice and RMs (Jalah et al., 2014; Li et al., 2013; Patel et al., 2013; Singh et al., 2018; Valentin et al., 2014; Vargas-Inchaustegui et al., 2014) and has been corroborated in additional studies in rabbits and RMs (Jaworski et al., 2012; Krebs et al., 2014; Pissani et al., 2014; Zolla-Pazner et al., 2016).
In the present study, we explored whether vaccination with DNA + protein combination vaccines using a series of well-characterized sequentially isolated CH505-Env sequences would be able to stimulate B cell lineages, resulting in development of broadly neutralizing Abs (bNAbs) and/or non-neutralizing Abs able to protect female RMs from intravaginal infection using a pathogenic SHIV challenge. The Env immunogens were obtained from patient CH505 infected with clade C HIV-1, who developed bNAbs recognizing the CD4-binding site (CD4bs), associated with the co-evolution of two B cell lineages (CH103 and CH235) (Bonsignori et al., 2016, 2017; Fera et al., 2014; Gao et al., 2014; LaBranche et al., 2019; Liao et al., 2013; Saunders et al., 2017b), and from whom longitudinal virus isolates were available for over six years, beginning with acute infection and extending through chronic infection. Selected sequentially isolated Env were employed as DNA + protein vaccine in this study to test the hypothesis that such a vaccine could reproduce the development of bNAbs observed in patient CH505 (Saunders et al., 2017b; Williams et al., 2017).
We tested two vaccination regimens differing only in the modes of administration of the vaccine components, to determine whether Ab development was affected when the Env DNA + protein vaccine components were co-administered at the same anatomical sites, so that both immunogens targeted the same draining lymph nodes, in comparison with RMs vaccinated with the same components injected separately at contralateral sites, such that the DNA and protein vaccine components targeted different draining lymph nodes (LNs). Our results showed that the co-administration of the vaccine components in the same anatomical sites achieved higher Env-specific immune responses and greater protection from pathogenic tier-2 SHIV intravaginal challenges. In the absence of NAbs to the tier-2 challenge virus, our study demonstrated that non-neutralizing Abs, ADCC activity and HIV Env Ab binding to FcγRIIIa contributed to the reduced infection risk.
RESULTS
DNA + Protein Co-administration in the Same Anatomical Sites Protects from Infection
We compared immunogenicity and protective efficacy in Indian RMs of an HIV vaccine comprising DNA and protein co-administering the vaccine components in the same anatomical sites (termed “co-administration”) or administering the vaccine components in contralateral sites (termed “separate administration”) (Figure 1A). RMs in the co-administration protocol received DNA in the left and right thighs, followed immediately by injection of the protein vaccine into the same muscles. RMs in the separate administration protocol received DNA in the left thigh and protein in the right thigh. The two vaccine groups (20 female RMs each; Table S1) received the same series of immunogens and the same vaccine dose, maintaining the anatomical sites for DNA and protein immunization constant throughout the study. The 6-valent Env vaccine (Figure S1) comprised DNA plasmids expressing membrane-anchored gp145-Env and the matching gp120 proteins adjuvanted in GLA-SE (Toll-like receptor 4 [TLR-4] agonist) (Kramer et al., 2018). The six vaccinations (Figure 1B) comprised a mixture of CH505.M5 and CH505.M11 (vaccination 1) aiming to initiate the two distinct B cell lineages, CH505.w20.14 (vaccination 2), CH505.w30.20 (vaccination 3), CH505.w30.12 (vaccination 4), CH505.w136.B18 (vaccination 5), and a mixture of the four Envs used in vaccinations 2–5 (vaccination 6). The vaccine also included SIV gag DNA and macaque IL-12 DNA as molecular vaccine adjuvant. The DNA vaccine was administered via the IM route followed by in vivo electroporation (EP) (Cellectra 5P, Inovio Pharmaceuticals). The vaccinations were separated by 2–5 months to allow contraction of immune responses and promote Ab affinity maturation before re-boosting (Figure 1B). Five weeks after the last vaccination, RMs were exposed weekly to repeated low-dose intravaginal challenges using a titrated stock of SHIV.CH505 (Li et al., 2016). Nine of the 18 RMs (50%) in the co-administration group remained uninfected after 15 weekly exposures (Figure 1C), compared with two of 17 RMs (12%) in the separate administration group and two of 20 (10%) in the control group (Table S1). Statistical comparison demonstrated a significant delay in SHIV.CH505 acquisition in RMs from the co-administration vaccine group (p = 0.036, exact log rank test) compared to the separate administration group, with a 67% reduction in per exposure acquisition risk in the co-administration group relative to controls.
Figure 1. Co-administration Group Shows Significant Protection from SHIV.CH505 Infection.
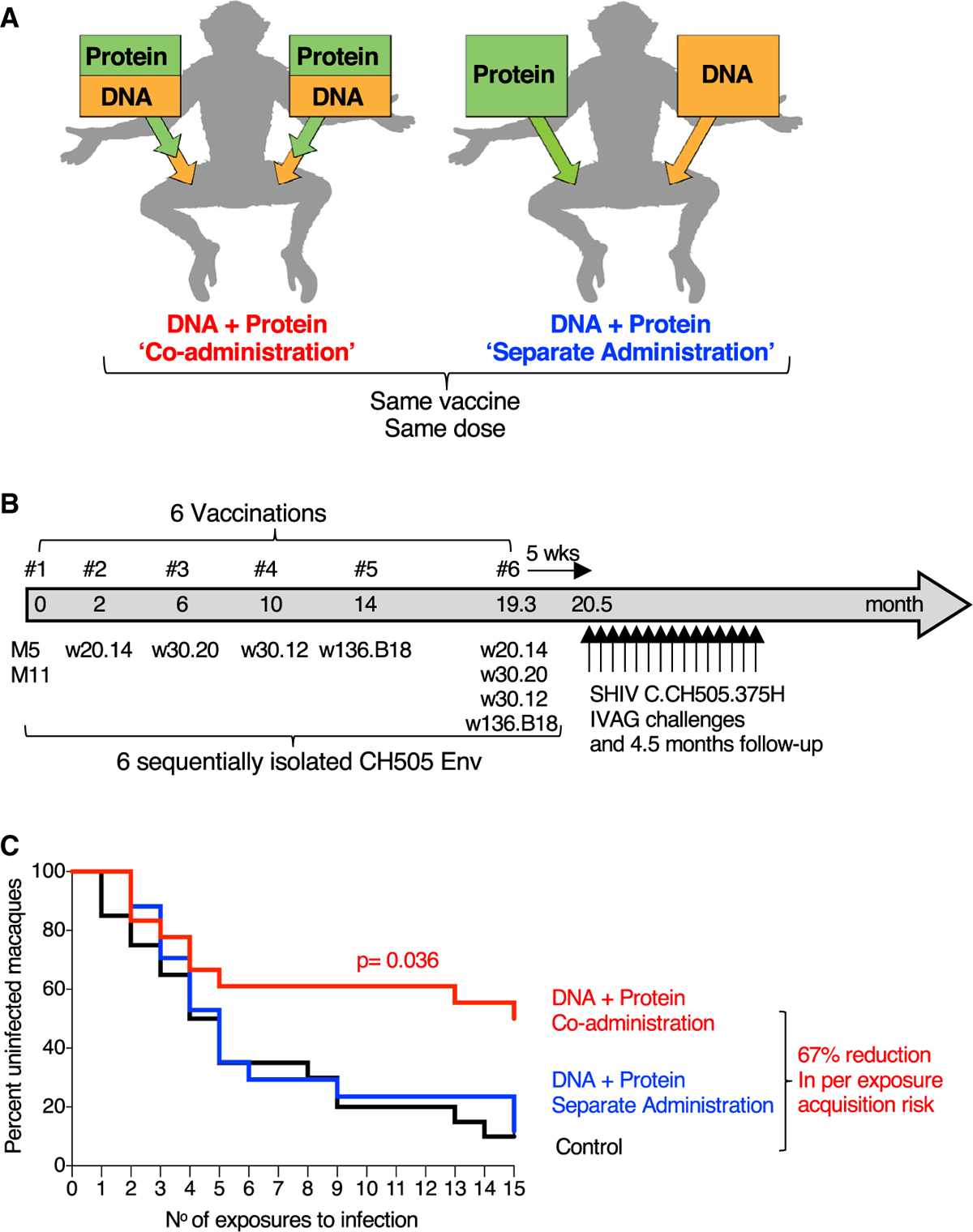
(A) Schematic representation of vaccine delivery of the two components (DNA and protein) in the two vaccination regimens, “Co-administration” in the same anatomical sites and “separate administration” in contralateral sites. The co-administration group received DNA delivered via IM/EP followed immediately by IM injection of the adjuvanted protein. The separate administration group received the vaccine components in different anatomical sites with DNA delivered by IM/EP in the left site and protein IM in the right site. The same vaccine components and the same total vaccine dose were used for both regimens.
(B) Vaccination schedule indicating the sequentially isolated CH505 immunogens used. Five weeks after the last vaccination, the animals were exposed weekly to repeated low-dose vaginal challenges using SHIV.CH505.
(C) Kaplan-Meier curves show the viral acquisition rate after repeated low-dose SHIV.CH505 challenges of the two vaccine groups (n = 18 and 17, respectively) and the control group (n = 20). The RMs were exposed to 15 weekly intravaginal challenges. Infection was defined by two consecutive positive plasma VL measurements. No RMs were censored. p value, exact log-rank test.
The Co-administration Regimen Induced Higher Env-Specific Humoral and Cellular Immune Responses
An analysis of humoral immune responses (Tables S2 and S3) was performed to evaluate differences among the vaccine groups and to explore their potential contribution to reducing the risk of infection. Measurements of CH505 Env Abs in plasma showed rapid onset of responses, detectable after the first vaccination in both vaccine groups, reaching peak levels by the fourth vaccination, with comparable kinetics and similar boosting upon the fifth and sixth vaccinations (Figure 2A). These data are in agreement with our previous findings showing rapid and robust development of SIV Env Ab responses in RMs when protein and DNA were co-administered in the same muscle in the priming vaccination (Li et al., 2013; Patel et al., 2013; Singh et al., 2018). Throughout the study, the co-administration group showed significantly higher Env Ab responses against a panel of CH505 Env variants compared to the separate administration group (Figures 2A–2C).
Figure 2. Co-administration Regimen Induced Higher Env Immune Responses.
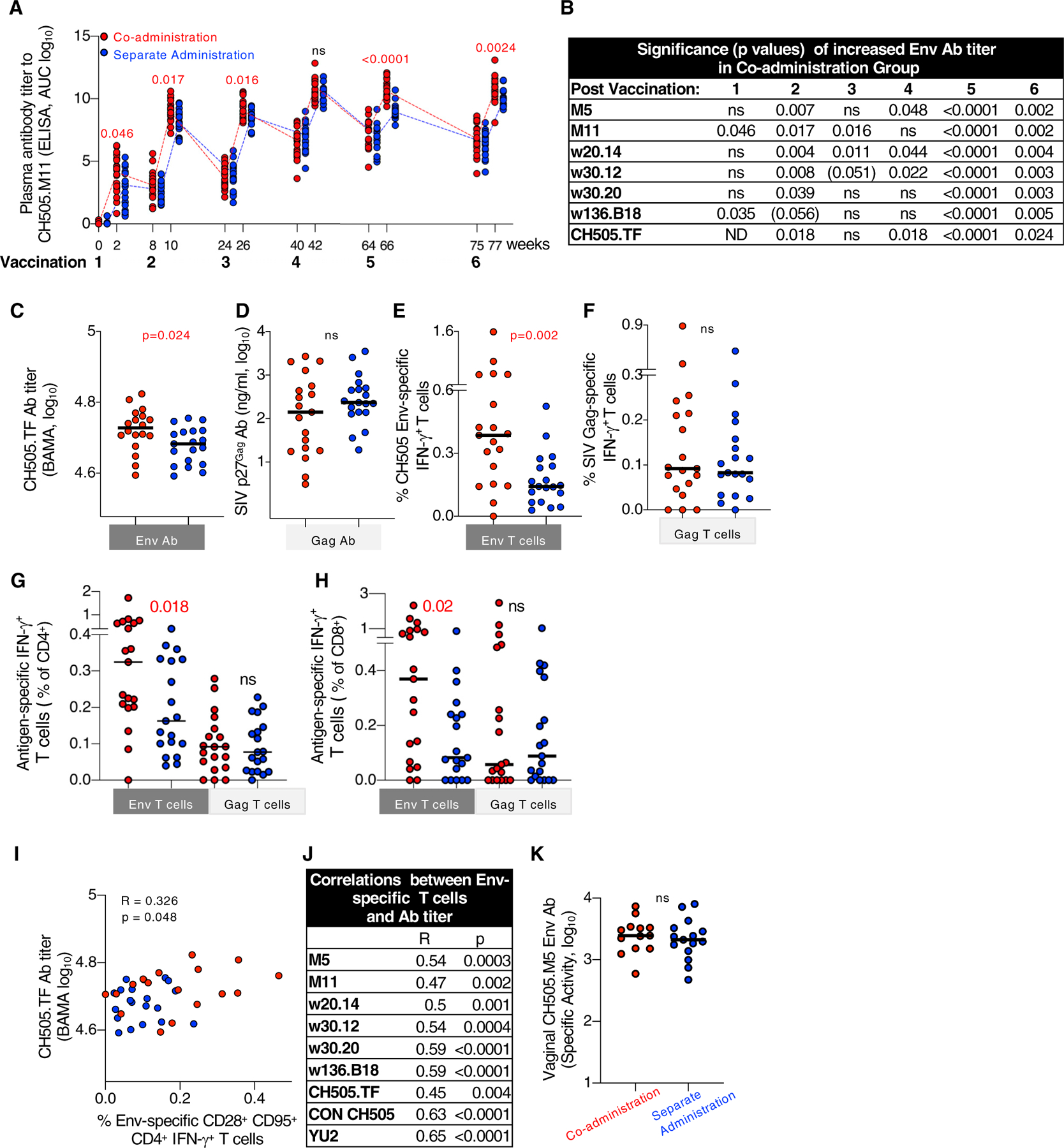
(A) Kinetics of vaccine-induced CH505.M11 Env antibodies (Abs) were measured by ELISA and Env Ab titers of individual RMs are shown as area under the curve (AUC) at the time of vaccination and 2 weeks later. Red symbols, co-administration group; blue symbols, separate administration vaccine group. Red numbers indicate p values showing significant higher values measured in the co-administration group.
(B) Summary of the differences in Env Ab titers measured after each vaccination against a panel of CH505 gp120 proteins. p values denote statistically significant higher values in the co-administration group; p values (p = 0.051- < 0.06) indicate trend; ns, not significant. ND, not determined.
(C–F) Comparison of HIV Env and SIV Gag Abs and T cell responses between the vaccine groups after the last vaccination.
(C and D) Ab responses measured to (C) HIV CH505.TF gp120 Env and (D) SIV Gag.
(E and F) T cell responses (% antigen-specific IFN-γ+) measured with peptides covering (E) all CH505 gp145 Env proteins used in the vaccine (F) SIV p57Gag. Similar data were obtained from samples analyzed after fifth vaccination.
(G and H) Antigen-specific IFN-γ+ T cells are presented as a percentange of CD4+ (G) and CD8+ (H) T cell subsets.
(I and J) Direct correlation between the CH505 Env-specific CM-like CD4+ IFN-γ+ T cells (presented as a percentage of the CD3+ cells) and (I) CH505.TF Ab levels after sixth vaccination and (J) summary of correlations with different Env Abs after fifth vaccination. R and p values (Spearman) are given.
(K) Abs to CH505.M5 Env were measured in vaginal secretion after vaccination 6. Values are presented as specific binding activity (SA) and were calculated as mean fluorescence intensity (MFI) 3 dilution/total IgG concentration (micrograms per milliliter). p values (Mann-Whitney test) are given for all comparisons.
In contrast to the differences in Env Ab titers, no differences in Gag Ab levels were found between the two groups (Figure 2D). Of note, Gag was not included as protein, but was only produced from the DNA. Measurements of cellular immunity at the same time point in PBMCs (peripheral mononuclear cells) showed significantly higher levels of Env-specific T cell responses in the co-administration group (Figure 2E). In contrast, the magnitude of Gag-specific T cell responses was similar in both groups (Figure 2F). Analysis of T cell subsets showed the presence of both CD4+ and CD8+ Env and Gag-specific IFN-γ+ T cells, respectively (Figures 2G and 2H), with higher Env-specific CD4+ and CD8+ T cells in the co-administration group. Interestingly, we found a direct correlation between the frequency of Env-specific CD95+CD28+ (central memory [CM]-like) CD4+ T cells and the Env-Ab titers (Figures 2I and 2J), suggesting a role for CD4+ T cell help in the development of Env-specific Ab.
We next explored potential differences in the mucosal distribution and binding specificities of the Env Abs induced by the two vaccine regimens. Env-specific immunoglobulin G (IgG) induced by both regimens disseminated to mucosal surfaces and was found in vaginal secretions (Figure 2K). Similarly low levels of CH505-specific IgA were found in serum in both groups (Figures S2A and S2B). Vaccine-specific IgA was not measured in mucosal samples due to scarcity of available material and also due to the results of previous studies with similar vaccine formulations, which demonstrated that vaginal IgA was sporadically detectable or low in RMs that received the DNA + protein vaccinees (Vargas-Inchaustegui et al., 2014; Singh et al., 2018).
The avidity of Env-specific serum Abs to CH505 gp120 Env and to other Envs (clade C and clade B) was similar between the groups (Figures S2C and S2D). Linear peptide mapping showed an overall comparable magnitude and binding specificities to CH505 peptides (Figures S3A and S3B), with dominant responses to V3 peptides, in accord with our previous report (Shen et al., 2015), and readily detectable responses recognizing V2 peptides. We found overall similar cross-clade linear peptide responses with a strong recognition of V2 and C3 regions. We further compared the ability of the Abs to recognize V1V2, a correlate of reduced risk of infection in RV144 (Haynes et al., 2012; Rolland et al., 2012), and found binding to cyclic V2 from different clades as early as after the second vaccination (Figure S3C). The Env-specific Abs showed robust cross-clade recognition of scaffolded gp70-V1V2 protein in plasma (Figure S3D) and vaginal secretions (Figure S3E). A magnitude-breadth (MB) score, calculated based on binding magnitude for every V1V2 scaffold protein (Figure S3F), showed no difference between the groups and there was no correlation between the MB score and the number of exposures to infection.
Blocking experiments with NAbs (CH235, CH106) targeting the CD4bs or sCD4 resulted in a reduction of the plasma Ab binding to CH505.TF gp120, demonstrating that a portion of the Ab responses was able to recognize the CD4bs (Figure S2E). The presence of CD4bs activity was found as early as after the second vaccination in both groups, and this specificity was boosted to similar levels after each vaccination, but did not correlate with protection.
Thus, RMs enrolled in the co-administration vaccine regimen developed higher plasma Abs and cellular anti-Env responses compared to RMs in the separate administration regimen, although the mucosal dissemination and Ab binding specificities were similar in both groups.
Non-neutralizing Abs Contribute to Reduction of Risk of Infection
A role for Env Abs in decreased transmission risk was supported by univariate analyses showing direct correlation (or trend) of early CH505 bAb (binding Ab) responses (after third and fourth vaccinations) and delayed virus acquisition including all vaccinees (Figures 3A and 3B). To identify potential immune mechanisms mediating decreased transmission risk, we analyzed the functional properties and characteristics of Env Ab responses (Tables S2 and S3), including Ab effector functions, previously associated with protection (Ackerman et al., 2018; Barouch et al., 2015; Bradley et al., 2017; Florese et al., 2009; Haynes et al., 2012; Hidajat et al., 2009; Neidich et al., 2019; Pittala et al., 2019; Xiao et al., 2010). We performed an extensive functional analysis of the Env Abs, including NAbs (Montefiori, 2009), recognition of Env on infected cells (Ferrari et al., 2011) and a series of Ab-dependent innate immune functions (Barouch et al., 2015; Lai et al., 2012; Pollara et al., 2011; Vaccari et al., 2016). We also determined the CH505-specific Ab glycan structures (Barouch et al., 2015; Vaccari et al., 2016) and used multiplexed Fc Array assay to simultaneously probe Fc and Fab characteristics of virus-specific Abs (Brown et al., 2017, 2015). We focused on measurements of effector functions, because they have been previously associated with protection in some challenge studies (Ackerman et al., 2018; Barouch et al., 2015; Bradley et al., 2017; Florese et al., 2009; Haynes et al., 2012; Hidajat et al., 2009; Neidich et al., 2019; Xiao et al., 2010).
Figure 3. Non-neutralizing Abs Contribute to Reduced Risk of Infection.
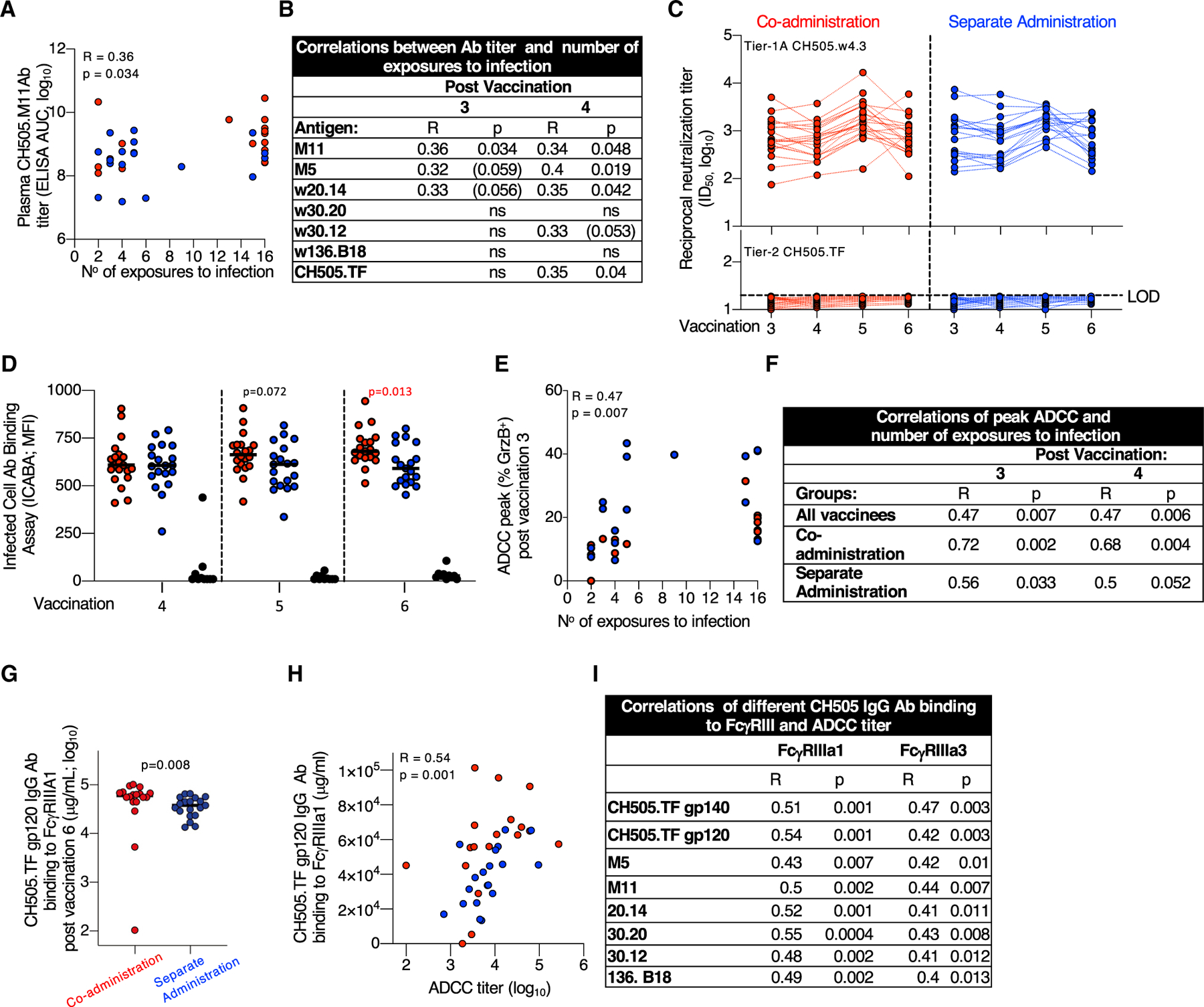
(A and B) Direct correlation of CH505 bAb titers (AUC, log10) and the number of SHIV.CH505 exposures to infection plotted using plasma after (A) third vaccination and (B) summarized for all Env antigens after third and fourth vaccinations. No correlations were found after the fifth and sixth vaccinations. p values (Spearman) are listed, p values in parentheses indicate trend.
(C) NAbs to tier-1A CH505.w4.3 (upper panel) and the tier-2 CH505.TF (lower panel) measured in serum 2 weeks after the third to sixth vaccinations. Black dotted line indicates the limit of detection (LOD) of neutralization in the TZM-bl assay. CH505.w4.3 differs from the tier-2 CH505.TF by a single point mutation (W663G), located in the MPER (membrane-proximal extracellular region).
(D) Binding of Env-Abs exposed on the surface of HIV.CH505 infected cells measured by the infected cell Ab binding assay (ICABA) showing MFI of the two vaccine groups. p values are from Wilcoxon test (statistical analysis software).
(E and F) Correlation between ADCC peak (maximum % GrzB activity) and number of SHIV CH505 exposures to infection (E) after third vaccination and (F) summary of correlations measured after third and fourth vaccinations. No correlations were found with the peak ADCC after fifth and sixth vaccinations. p values (Spearman) are given.
(G) Binding of CH505-Env-specific Abs to FcγRIIIa1.
(H and I) Direct correlations between ADCC titers and CH505.TF-specific Ab binding to FcγRIIIa1 and FcγRIIIa3 showing correlation between (H) CH505.TF gp120 Abs and FcγRIIIa1 and (I) different CH505 Abs binding to FcγRIIIa3 after sixth vaccination. Spearman R and p values are given.
We found similar robust levels of NAbs against the tier-1A CH505.w4.3 (Figure 3C, upper panel) after each vaccination in both groups. NAbs to CH505.w4.3 did not correlate with decreased infection risk. No NAbs against the tier-2 CH505.TF (bottom panel) was detected. Nab assays using CH505 lineage viruses, SHIV (CH505.375H, SF162P3), mutant viruses sensitive to CD4bs or V2 bNAbs (LaBranche et al., 2019; LaBranche et al., 2018), were also negative except for sporadic RMs with weak positive titers (Table S4): four showed weak CD4bs NAb activity; one showed weak V2 NAb activity. These data demonstrate that NAbs did not contribute to vaccine protection, confirming contribution(s) of non-neutralizing Abs in preventing infection.
The ability of Abs to recognize CH505 Env on the surface of HIVCH505-TF-infected cells (infected cell Ab binding assay [ICABA]; Figure S4A) was tested in plasma samples collected after fourth to sixth vaccinations using a flow cytometry-based assay (ICABA values as mean fluorescence intensity [MFI] [Figure 3D] and percentage [%] binding [Figure S4B]). We found robust binding of the Abs to surface-anchored Env, with RMs from the co-administration group showing a trend for better recognition of the CH505-infected cells after the fifth vaccination and significantly stronger binding after the sixth vaccination (p = 0.013, Mann-Whitney t test). Although the induced Abs were not able to neutralize the challenge virus in the TZM-bl assay, these Abs have the ability to recognize CH505 Envs exposed on infected cells.
Next, we asked whether the Abs were able to mediate ADCC, which was measured by the flow cytometry using CH505-gp120-coated cells (third to sixth vaccinations). Univariate analyses showed correlations between peak ADCC activity (measured as % GrzB+ cells) and resistance to infection. This correlation was strongest within the co-administration group after the third and fourth vaccinations (Figures 3E and 3F), but was not durable with continued fifth and sixth vaccinations.
Because ADCC function depends on Abs binding to FcγRIII, the main Fc receptor expressed by natural killer (NK) cells, we used a multiplexed Fc Array assay to probe Fc characteristics of the HIV-specific Abs (Figure S5). Interrogating the role of the Fc portion from the Env Abs, previously identified as contributing to control of infection in human and RM vaccine studies (Ackerman et al., 2018; Bradley et al., 2017; Haynes et al., 2012; Neidich et al., 2019), we found significantly higher binding of multiple CH505 Abs to FcγRIIIa1 in RMs from the co-administration group (Figure 3G), measured after the last vaccination. Univariate analyses further showed direct correlations between the CH505-specific Abs binding to both FcγRIIIa1 and FcγRIIIa3 and the ADCC titers measured using CH505 Env-coated cells (Figures 3H and 3I).
CH505-specific Ab-mediated NK activation (ADNKA), neutrophil and monocyte phagocytic activity (ADNP and ADCP, respectively), and Ab-dependent complement deposition (ADCD), measured before the challenge, showed no difference in the functional scores between the vaccine groups or the protected and infected RMs in the co-administration group and no correlation with delayed virus acquisition.
Together, these data showed that the vaccine-induced Abs recognize CH505 Envs on the surface of infected cells (Figure 3D; Figure S4) and disseminate to mucosal surfaces (Figure 2K; Figure S3E)—a critical feature to inactivate the incoming virus. Plasma Ab titers (Figures 1A–1C) and peak ADCC activity (Figures 3E and 3F) correlate with delayed virus acquisition, but the magnitude of Env-specific Abs failed to predict the peak granzyme activity measured in the ADCC assay.
No Differences Found in Systemic Effects of Vaccination
We monitored activation/inflammation induced in both groups by measuring plasma levels of several cytokines before vaccination (pre) and after the first vaccination. We did not observe any changes in the levels of the pro-inflammatory cytokines inter-leukin-1 (IL-1), IL-12, TNF-α, GM-CSF, and IFN-γ. We found increased levels of several chemokines associated with immune activation, including Eotaxin/CCL11, Eotaxin-2/CCL24, FLT3L, I-TAC/CXCL11, IP-10/CXCL10, IL-1Ra, MIP-1α/CCL3, MIP-1β/CCL4, and MDC/CCL24 (Figure S6). The levels of these chemokines were similar between the vaccine groups (Figure S6B). We also observed similarly low levels of proliferating (Ki67+) total T cells in blood in both vaccine groups after the last vaccination (Figure S6C), showing a lack of systemic activation between the vaccination regimens.
Multivariate Analysis to Discriminate between Vaccine Groups
Because several univariate measurements (Env-specific humoral and cellular immunity) showed statistically significant differences between the vaccine groups over time (Figures 2 and 3), we sought to define how accurately immunogenicity data could distinguish between the co-administration and separate administration groups using a multivariate approach (Pittala et al., 2019) including all the immunological parameters outlined in Tables S2 and S3, using data collected after each vaccination, when available. Regularized binomial logistic regression (Cox, 1958) models were used to discriminate between animals in the two vaccine groups at each of the different time points. As early as after the first vaccination, but not at baseline, and through the fifth vaccination, but not after the sixth, immunogenicity data could be used to robustly classify macaques according to vaccination regimen (Figure 4). Peak accuracy (98%) was observed after the third vaccination with a sparse, simplified feature set. We found good consistency in the ability of simplified feature sets to define signatures of group-specific immunogenicity that were robust across longitudinal time points during the immunization series (Table S5). In sum, multivariate comparison of the co-administration vaccine protocol versus the separate administration protocol showed Env Abs as optimal discriminatory features between the two vaccine groups, with good accuracy.
Figure 4. Immunogenicity Profiles Robustly Distinguishes Animals Vaccinated with the Co-administration and Separate Administration Vaccine Regimens.
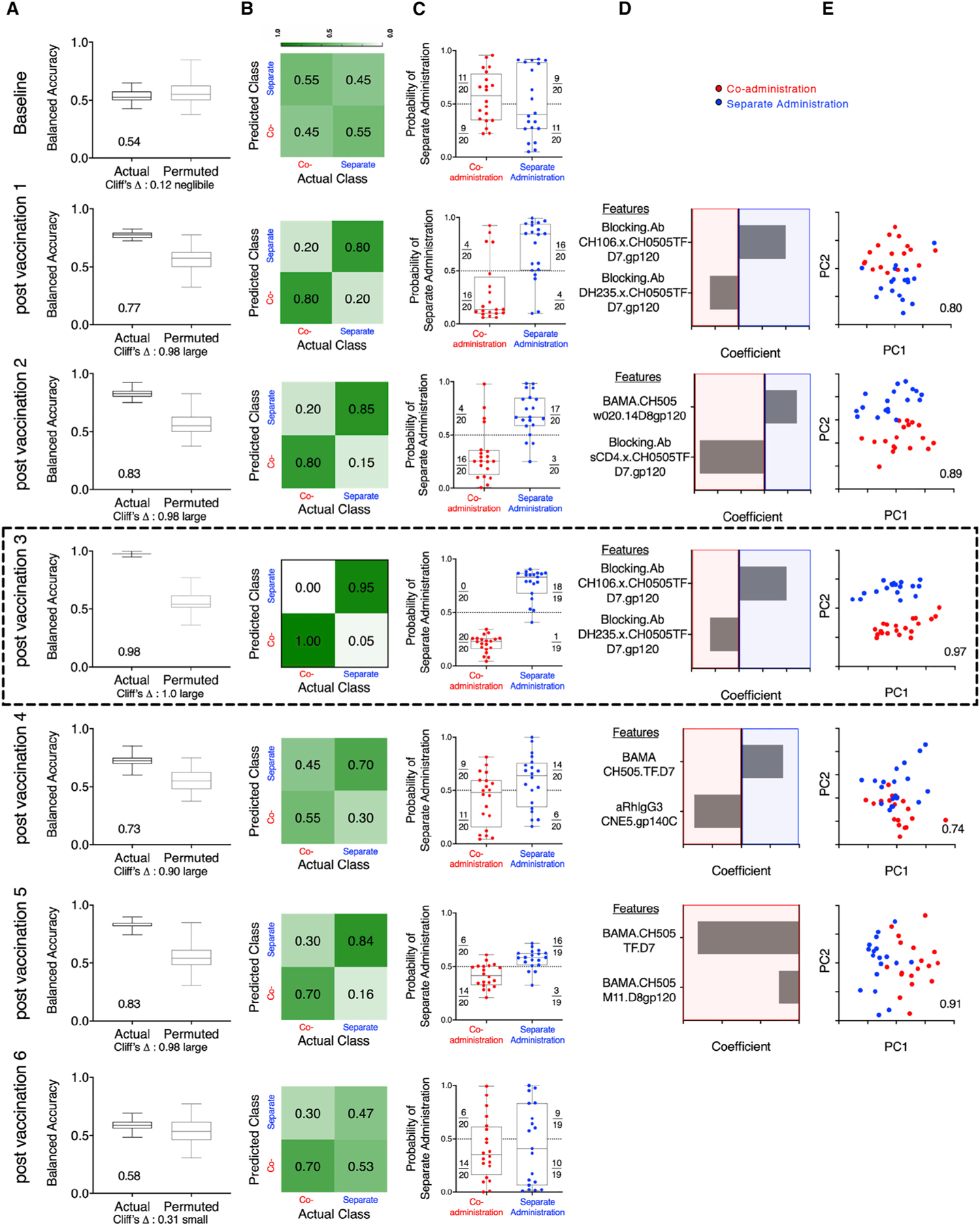
(A) Balanced accuracy of models learned on training data when applied to test data across 100 replicates of 10-fold cross validation. Models learned from actual data (left) perform substantially better (Cliff’s delta, 1.0) than those learned from permuted data (right). Average accuracy across cross-validation runs is reported in inset.
(B) Confusion matrix depicting the proportion of animals in each study arm predicted correctly/incorrectly.
(C) Model confidence, defined by probability of belonging to the separate side class. Dotted line indicates the decision boundary.
(D) Feature contributions to the simplified final model.
(E) Principal-component (PC) biplot of features contributing to the simplified final model. Simplified model accuracy is reported in inset. Animals are represented as dots, with color indicating vaccine arm. Classification performance over time could not be strictly compared, as different immunogenicity tests were performed at different time points, and in some cases, samples from all macaques were not available for all time points for all tests (Table S2). Simplified forms of the final models learned on one time point were applied to data from other time points and demonstrated good consistency in defining signatures of group-specific immunogenicity that were robust across longitudinal time points during the immunizations (Table S5). The data from the analysis performed after third vaccination showing peak accuracy is shown boxed.
Multivariate Modeling of Correlates of Infection Risk
To define potential correlates of risk, RMs were classified as “susceptible” or “resistant” depending on whether or not they were infected after 10 exposures (Figure 5A). First, we focused our analysis on RMs that received the co-administration regimen (Figures 5B and 5F) and found that peak ADCC (maximum % GrzB [granzyme B] activity) contributed predominantly to prediction of protection observed in the RMs from this group (Figures 5E and 5F).
Figure 5. Immune Response Profiles Robustly Distinguish Resistant Animals in the Co-administration Group.
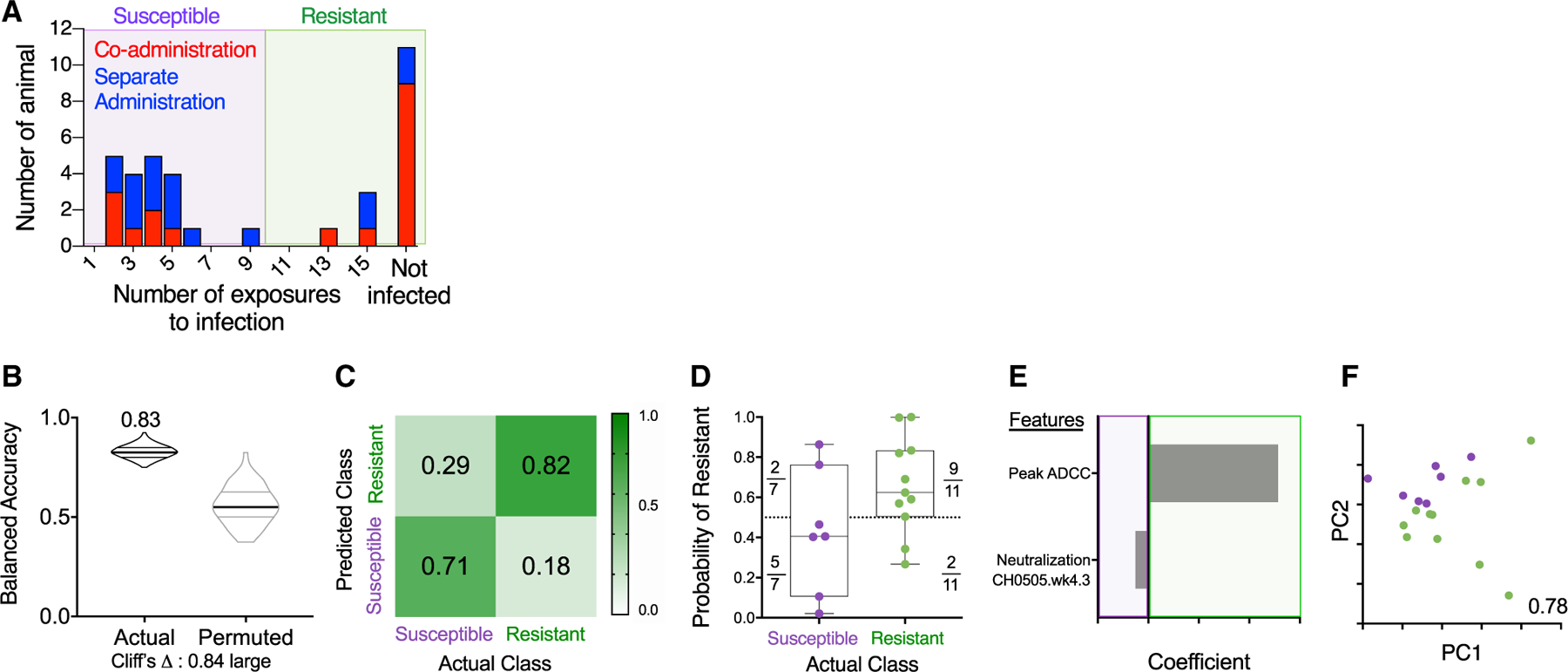
(A) Histogram of challenge outcomes for all RMs with sensitive and resistant animals defined as those infected (purple box) or not infected (green box) prior to the 10th challenge.
(B–F) Models of infection resistance learned from immunogenicity data post-third immunization for the co-administration group. A classifier was trained to use immunogenicity data to distinguish between animals based on challenge resistance.
(B) Balanced accuracy of models learned on training data when applied to test data. Models learned from actual data (left) perform substantially better (Cliff’s delta, 0.84) than those learned from permuted data (right). Robust performance and good overall average accuracy (83%) in the setting of repeated cross validation was achieved.
(C) Confusion matrix depicting the proportion of animals in each study arm predicted correctly/incorrectly.
(D) Model confidence defined by probability of belonging to the assigned class.
(E and F) A final model was trained on the complete data and simplified to a sparse, two-feature model.
(E) Coefficients of features in the simplified final model.
(F) PC biplot of features contributing to the simplified final model with accuracy reported in inset. Resistant RMs are indicated in green; susceptible RMs are indicated in purple.
Because Ab effector functions have previously been associated with vaccine-mediated protection (Ackerman et al., 2018; Barouch et al., 2015; Bradley et al., 2017; Florese et al., 2009; Haynes et al., 2012; Hidajat et al., 2009; Neidich et al., 2019; Xiao et al., 2010), we next considered whether the relationship between peak ADCC and time to infection already observed using univariate analysis after the third and fourth vaccinations (Figures 3E and 3F) was a correlate of decreased infection risk among RMs from both vaccine regimens using a multivariate approach. We found that, across all time points for which data were available, peak ADCC activity correctly classified an average 2/3 of RMs (Table S5), confirming the robustness of the relationship between peak ADCC over time and resistance to infection in the co-administration group. These results suggest that ADCC serves as a biomarker of vaccine efficacy over time, although, as was observed in immunization group models, predictive power was lost when applied to post-sixth immunization profiles. A number of other Ab functions (Table S2), including complement deposition and phagocytosis, were evaluated only at this post-sixth immunization time point; none were observed to correlate with resistance to infection.
We explored additional potential correlates of protection as previously reported (Ackerman et al., 2018; Bradley et al., 2017; Haynes et al., 2012; Neidich et al., 2019), such as Fc Array measures in a multivariate analysis including FcγRIIIa and FcγRII. Of note, we found higher binding of CH505-specific Abs to FcγRIIIa in the co-administration group and a significant correlation with ADCC using univariate analysis (Figures 3G–3I). While longitudinal profiles differed, elevated FcγRII-and FcγRIII-binding Abs were observed among resistant animals for multiple distinct antigen-specificities across multiple time points (Figure S7).
We then explored whether, after the sixth immunization, models learned exclusively from FcγRIII-binding Ab responses, which were assessed across a wide range of Env-sequence variants, could predict challenge resistance across all immunized RMs (Figure 6). A sparse, three-feature final model using FcγRIIIa-binding to Abs specific to CH505.TF gp140, CH505.w20.14 gp120, and CH505.w30.12 could classify the animals according to resistance to infection with greater than 80% accuracy (Figures 6D and 6E). Thus, our data showed a contribution of ADCC (Figures 3 and 5) and the ability of CH505 Env-specific Abs to interact with FcγRIIIa (Figures 3 and 6) to be associated with, and predictive of, reduced risk of infection. While targeted experiments are necessary to evaluate the mechanistic relevance of these features to challenge resistance, this model is consistent with other reports (Ackerman et al., 2018; Neidich et al., 2019). In conclusion, across immunization groups and over time, Env Abs, ADCC, and FcγRIIIa-binding Env-specific Abs served as discriminatory features forming a robust signature of reduced SHIV infection risk.
Figure 6. CH505-Specific Ab Binding to FcγRIIIa Robustly Predicts Challenge Resistance after the Final Vaccination.
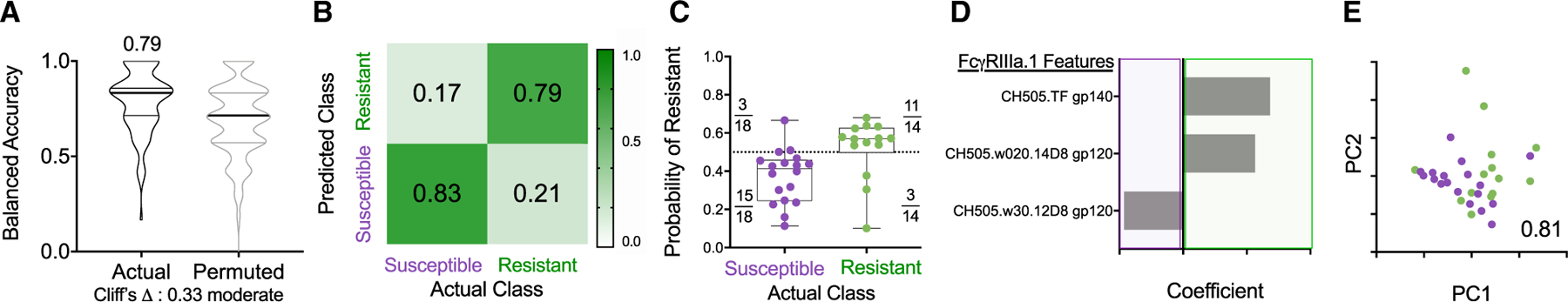
(A) Balanced accuracy of models learned on training data when applied to test data. Models learned from actual data (left) perform moderately better (Cliff’s delta, 0.33) than those learned from permuted data (right). Average accuracy across cross-validation runs is reported in inset.
(B and C) Confusion matrix confidence (B) and model confidence (C) as described in Figure 5.
(D and E) Coefficients of features (D) and PC biplot of features (D) contributing to the final model with accuracy reported in inset. Resistant RMs are indicated in green; susceptible RMs are indicated in purple.
Both Vaccine Regimens Induced Immune Responses Associated with Reduction in Viremia
Plasma viral load (VL) was monitored in the SHIV-infected RMs over 18 weeks (Figure 7A). Significant VL differences (Kruskal-Wallis tests of differences over the three groups at the individual times) were found at several time points (weeks 2–9 post-infection). RMs from both vaccine groups showed lower acute viremia during the first 4 weeks post-infection, defined as area under the curve (AUC, weeks 0–4), compared to controls (Figure 7B). These data indicate that both vaccine regimens induced immune responses able to control viremia. RMs in the separate administration group decreased acute viremia to lower levels, although we cannot exclude that the different number of infected RMs (9 versus 15 vaccinees) may have contributed to this outcome.
Figure 7. Both Vaccine Regimens Induce Immune Responses Able to Reduce Viremia.
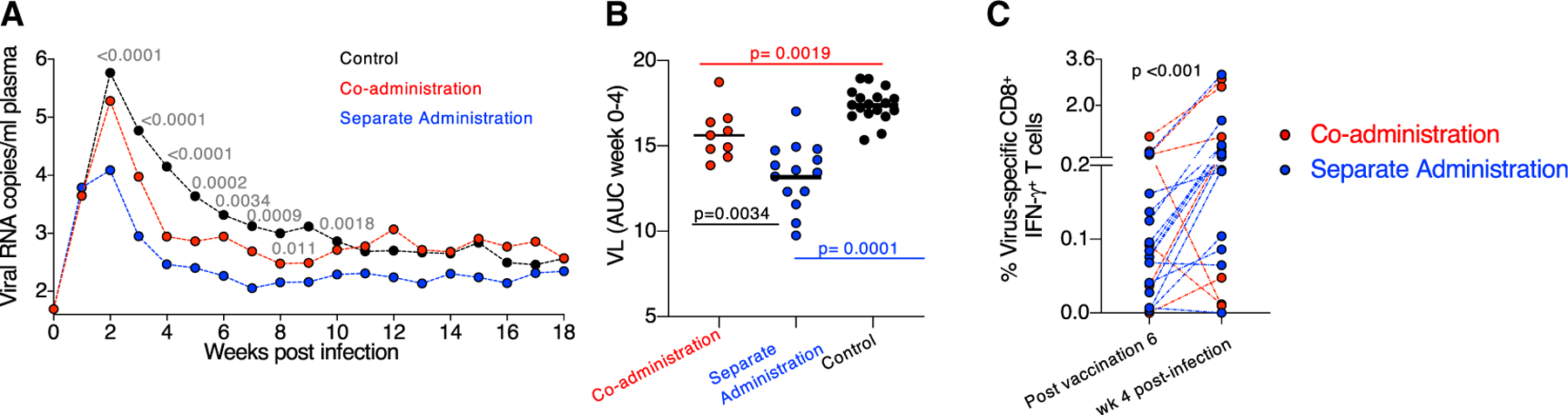
(A) VL of all infected animals monitored over 18 weeks of follow-up showing the geometric mean VL. The p values (Kruskal-Wallis tests of differences over the three groups at the individual times) weeks 2–9 are given, with wide fluctuations in some of the individual levels at weeks 6–9.
(B) Comparison of early viremia (weeks 0–4) as AUC with p values of pairwise comparisons (Kruskal-Wallis test).
(C) Changes in the level of virus-specific (Env and Gag) CD8+ T cells measured after sixth vaccination (3 weeks before challenge start) and 4 weeks after infection (expressed as a percentage of total CD3+ T cells). p value, Wilcoxon signed rank test.
Comparison of T cell responses measured 2 weeks after the last vaccination and 4-weeks post infection showed a robust anamnestic increase of virus-specific (Env and Gag) IFN-γ+ CD8+ T cells (Figure 7C) in the infected vaccinees, mediated by cytotoxic (GrzB+ and/or CD107a+) virus-specific T cells that likely contributed to control of viremia. Taken together, the co-administration regimen elicited humoral responses protecting against viral acquisition, and both vaccine regimens induced cellular immunity able to control viremia.
DISCUSSION
The co-administration regimen induced higher Env-Ab titers and more Env-specific CD4+ T cells than the separate administration regimen, and this anatomic correlation of decreased transmission risk points to a contribution of the CD4+ T cell help in the development of humoral immune responses. Plasma Env Abs, peak ADCC activity at early time points (after the third and fourth vaccinations), and development of CH505 Env Abs with high binding to FcγRIIIa (after the sixth vaccination) were identified as correlates of reduced risk of infection. Some of the correlations waned with subsequent immunizations, but the underlying reason(s) are unclear. It is possible that some functional properties and/or epitope specificities present in the mucosal Abs, but not properly addressed by the current assays or by the time points analyzed, are also responsible for the potent protection against the vaginal challenge observed in the co-administration group. Although single Ab features at the time of challenge failed to explain the mechanism mediating protection in the animals from the co-administration group, a multicomponent approach combining different Ab binding specificities and functional properties was successful in distinguishing protected from non-protected immunized RMs, clearly indicating that the vaccine-induced humoral responses are responsible for these different outcomes.
In addition to the ADCC peak activity, Abs with elevated binding to FcγRIIIa—the main receptor expressed on NK cells and directly responsible for the ADCC activity mediated by these cells—were also identified as a correlate for reduced risk of infection. Although high binding affinity of Env Abs to FcγRIII was identified as a correlate of protection, it is not the cause of sterilizing immunity. High affinity to FcγRIII provides the Ab with the ability to mediate ADCC, which could contribute to limit the systemic viral dissemination, but this does not exclude other IgG-functional properties.
Taken together, we observed that non-neutralizing Env Abs, Env-specific CD4+ T cell responses, ADCC, and Env-Ab binding to FcγRIIIa contribute to the protective signature. Thus, our study provides another example of the potent role of non-neutralizing HIV Abs in protection from infection (Ackerman et al., 2018; Barouch et al., 2018; Bradley et al., 2017; Ehrenberg et al., 2019; Haynes et al., 2012; Neidich et al., 2019; reviewed in Ackerman et al., 2017; Alter and Barouch, 2018)).
Our data suggest that synchronized uptake and processing of the vaccine components (DNA and adjuvanted protein) in the draining LNs triggers a primary immune response with qualitatively different properties than the responses induced by the same components processed individually in separated anatomical sites. Of note, all vaccinees received the same number of vaccinations, the same doses of each DNA and protein component, the same kind and amount of adjuvant, and the same delivery routes and number of anatomical sites (left and right inner thighs). The only difference was the delivery of the two components either together in the co-administration protocol or separately (DNA in the left and protein in the right thigh). In the co-administration vaccine group, the draining LNs were exposed to DNA + protein, triggering simultaneous T and B cell recognition within the same follicles. In contrast, the separate administration vaccine group likely elicited independent primary responses triggered by the protein alone (Ab responses upon B cell recognition) or the DNA (mainly T cell responses triggered by the processed peptides from the endogenously synthesized proteins). Analysis of cellular processes within the draining LNs early after priming will be critical to dissect and understand the sequence and topology of immune events associated with either of the two vaccine regimens (Havenar-Daughton et al., 2016; Liang et al., 2017). In fact, it was reported that priming of high numbers of Env-specific CD4+ T cells in the draining LNs, directly correlates with increased T-follicular-helper-cell differentiation and germinal center formation (Liang et al., 2017).
Both vaccine regimens rapidly induced high Ab levels, as we previously noted for SIV-Env-based co-administered DNA + protein vaccines (Patel et al., 2013; Singh et al., 2018). Thus, inclusion of protein in the priming vaccination resulted in more efficient Ab production. Interestingly, similar conclusions were reported in two recently completed clinical trials (Pantaleo et al., 2019; Rouphael et al., 2019), which also included protein in the prime. In these trials, canarypox (NYVAC) or DNA vaccine vectors were combined with gp120 Env and administered separately in contralateral sites of the body. The human trials and our RM studies share the key finding of rapid induction of humoral immune responses when protein is included in the priming vaccination and further show that this outcome is found using different vector platforms (DNA, NYVAC). Furthermore, our results show that co-administration of the vaccine components induces higher Env-specific humoral and cellular immune response and provides significant protection from low-dose pathogenic tier-2 SHIV.CH505 challenge. Both vaccine regimens (co-administration and separate administration) induced tier-1A NAb, but neither of the protocols led to the development of tier-2 autologous or heterologous bNAbs, a difficult task (Cirelli and Crotty, 2017) that, despite robust early Ab development, was not achieved. We found potent prevention of vaginal infection, and we cannot exclude an Ab contribution to protection that is not readily measured by canonical NAb assays.
Although one aim of the study has been to explore whether the set of sequentially isolated CH505 Envs is able to induce bNAbs, our results do not indicate that this was accomplished. This could be the result of the selected immunogens used in this study, the vaccine regimens, or the restricted ability of the macaque immune system to initiate bNAb development efficiently, due to the low frequency of specific B cell precursors. Although the present study failed to show an advantage of sequential vaccination with these serially isolated CH505 Envs to direct development of bNAbs, improved immunogen designs such as altered immunogens including the stabilized SOSIP (soluble cleaved HIV Env trimer) and stronger adjuvants should be tested before a final conclusion about this concept is drawn. In support of this notion, two other immunogenicity studies using a different series of sequentially isolated CH505 Envs resulted in induction of some autologous NAbs (McCurley et al., 2017; Saunders et al., 2017b): cleavage-deficient CH505 gp140 immunogens induced sporadic tier-2 NAbs (Saunders et al., 2017b); and CH505 gp160 immunogens in a DNA/MVA regimen induced very low autolo gous CD4bs NAbs in a few RMs (McCurley et al., 2017). Native-like BG505 Env SOSIP trimers (Pauthner et al., 2017, 2019) induced autologous NAbs contributing to control of infection. An epitope-based vaccine induced bNAbs in RMs (Xu et al., 2018). Thus, we hypothesize that immunogen design could further improve our vaccine platform. Unlike current vaccination efforts, HIV infection leads to bNAb development in ~50% of HIV-infected persons (Hraber et al., 2014), likely due to persistent exposure to constantly evolving Env immunogen. Induction of such bNAbs is complicated perhaps due to the limited immunogen diversity and the lack of sequence evolution associated with this vaccine regimen, as well as the low frequency of B cell precursors necessary to induce the B cell lineages targeting conserved HIV-Env-neutralizing epitopes with sufficient affinity and selectivity (Haynes and Mascola, 2017; Kelsoe and Haynes, 2017, 2018; Kwong and Mascola, 2018; Williams et al., 2017).
Finally, it should be noted that all RMs were female and the challenges were intravaginal. Females have been reported to have better Env Ab responses compared to males (Gilbert et al., 2010) and we cannot rule out a role of gender in the protection seen in this study. Thus, effective delivery of immunogens (DNA, adjuvanted protein) to the same draining LNs may result in stronger immune activation and improved Ab responses, thereby enhancing vaccine efficacy. We hypothesize that optimization of immunogens to better target the rare B cell precursor, combined with the co-administration of vaccine vector and protein in the same draining LNs, could provide an immunological advantage over current protocols, resulting in significantly improved protection.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, BarbaraK. Felber (Barbara.felber@nih.gov).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The RStudio and Python code generated during this study are available at https://github.com/dAbL-Dartmouth/p186.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All RMs used in this study were colony-bred female Indian rhesus macaques (Macaca mulatta) obtained from Tulane National Primate Research Center (TNPRC) (Covington, LA, USA). Studies involving RMs were conducted at the Tulane National Primate Research Center in accordance with the laws, regulations, and guidelines promulgated by the United States Department of Agriculture (e.g., the Animal Welfare Act and its regulations, and the Animal Care Policy Manual), Institute for Laboratory Animal Research (e.g., Guide for the Care and Use of Laboratory Animals, 8th edition), Public Health Service, National Research Council, Centers for Disease Control and Prevention, and the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International. The study (SVEU P186) was approved by the TNPRC Institutional Animal Care and Use Committee (IACUC) (Protocol # P0284 and P0284R). RM housing rooms were maintained on a 12–12 hour light:dark cycle with a relative humidity of 30%–70%, and a temperature of 64 to 84° F. All RMs were fed a commercially formulated nonhuman primate diet with fruit offered at least 3 times weekly as part of the enrichment program. RMs were observed twice daily for general behavior, abnormal clinical conditions, and local reactions after injections. Vaccinations were performed under anesthesia with ketamine hydrochloride (10mg/kg IM, KetaVed, Vedko). No adverse effects were found following any of the vaccination time points. RMs were anesthetized as described above for blood sample collections for clinical chemistry panels, complete blood counts, and other hematology parameters, body weight measurements, and temperature recordings. Regularly scheduled physical exams were performed by a veterinarian. Prior to all biopsies, the RMs were anesthetized with tiletamine hydrochloride and zolazepan hydrochloride (5–8 mg/kg IM; Tiletamine-Zolazepan, Zoetis, Kalamazoo, MI). Sustained release buprenorphine hydrochloride (0.2 mg/kg SQ; Buprenorphine SR, ZooPharm, Laramie, WY) was administered for analgesia.
At study initiation, the RMs were on average 9 years of age and were seronegative for SIV, STLV and SRV antibodies. The groups included 3 (Co-administration and control) or 4 (Separate Administration) Mamu A*01+ animals, no Mamu B*08 and 1 Mamu B*017+ animal per group (Table S1). Macaque (FE35, Separate Administration group) developed health issues and had to be euthanized after the 2nd vaccination. Two RMs from each vaccine group (FG06, JE14, GH91, GM52) were subjected to elected necropsy at 2 weeks after the 6th vaccination, prior to challenge start; animal GH35 had elected necropsy at week 3 PI before peak VL was established. Animal (EI21) from the control group developed AIDS-related illness and had to be sacrificed 9 weeks post-infection.
METHOD DETAILS
Plasmid DNA encoding sequentially isolated CH505 Env gp145 and the matching gp120 recombinant proteins were used for the vaccination. HIV Env sequences were derived from patient CH505 at different times post-infection (Liao et al., 2013). CH0505s TF, CH505.M5 and M11 represent inferred Env sequences (Bonsignori et al., 2017; Bonsignori et al., 2016; Gao et al., 2014; Liao et al., 2013; Saunders et al., 2017b). Endotoxin-free DNA expressing 6 sequentially isolated HIV CH505 gp145 Env [M5 (HV1300656), M11 (HV1300662), w020.14 (HV1300635), w030.20D8 (HV1300688), w30.12D8 (HV1300646), w136.B18D8 (HV1300724)], SIV gag [p57Gag (206S) and p27CE Gag (286S) (Hu et al., 2016)], macaque IL-12 (AG157;(Jalah et al., 2012; Jalah et al., 2013) and sham DNA CMVkan were used. The matching gp120 Env protein was purified from HEK293F cells and purified by Galanthus nivalis lectin (Saunders et al., 2017a). The proteins: CH505.M5D8gp120/293F/MON, CH505.M11D8gp120/293F/MON, CH505w020.14D8gp120/293F/MON, CH505w030.20D8gp120/293F/MON, CH505.w30.12D8gp120/293F/MON, CH505.w136.B18D8gp120/293F/MON are referred to as M5, M11, w20.14, w30.20, w30.12 and w136.B18 in this paper. The TLR-4 agonist (GLA-SE) (Kramer et al., 2018) adjuvant was obtained from Infectious Disease Research Institute (IDRI), Seattle, Washington.
The vaccine comprised 2 mg of gp145 Env DNA (vaccination 1 used 2 mg each of M5 and M11; vaccination 6 used 0.5 mg each of the 4 Env) and 0.2 mg matching gp120 proteins (vaccination 1, 0.2 mg each of M5 and M11; vaccination 6, 0.05 mg each of the 4 Env). The gp120 protein was adjuvanted with GLA-SE using 25 microG for vaccinations 1 and 2 and using 10 microG for vaccinations 3–6. The DNA mixture also contained gag DNA [(vaccination 1–2, 2 mg; vaccination 3, 3 mg Gag CE DNA; vaccinations 4–6: 2 mg Gag CE+2 mg p57Gag DNA). All DNA formulations, including the sham DNA (CMVkan), contained optimized 0.2 mg of macaque IL-12 DNA per injection. The same vaccine dose was administered in both groups. The DNA vaccine (1 ml) was delivered via IM injection in the Co-administration group (0.5 mL each in left and right inner thighs) or Separate Administration group (1 mL in the left inner thigh) followed by in vivo electroporation using the Cellectra® 5P device (Inovio Pharmaceuticals, Inc, Plymouth Meeting, PA). The GLA-SE-adjuvanted recombinant CH505 gp120 protein was administered by needle and syringe immediately following the DNA electroporation at same anatomical locations (0.25 mL each in left and right inner thighs) in the Co-administration group, and in the right inner thigh (0.5 ml) for animals in the Separate Administration group. The same anatomical sites for DNA and protein immunization were kept constant throughout the study. Twenty female RMs served as controls: ten received sham vaccine (sham DNA, IL-12 DNA and GLA-SE) and ten were treatment-naive.
SHIV CH505 virus and challenge
The RMs were challenged weekly intravaginally with the SHIV.CH505 challenge stock (SHIV.C.CH505.375H.dCT, lot 12/22/2015) grown in primary, activated, purified rhesus CD4+ T cells from naive RMs (Li et al., 2016) at 1:4 dilution with 1x PBS. This SHIV stock was shown to be genetically homogeneous and to retain the tier-2 neutralization phenotype and antigenic profile of the original HIV-1 CH505 transmitted/founder virus of the human. For groups of 4 animals, two vials of 0.5 mL of the viral stock were mixed and diluted with 3 mL of 1x PBS prior to inoculation and 1 mL of virus was used per challenge. Plasma virus load (VL) was measured over the course of the study using the NASBA assay with a threshold of detection of 50 SIV gag RNA copies per milliliter (Advanced BioScience Laboratories) (Romano et al., 2000). VL was measured weekly to monitor virus acquisition and infection was defined with two consecutively positive VL.
Antigen-specific T cell responses in PBMC
Isolated PBMC were cultured in 96-well plates and stimulated for 12 hours with peptide pools (HIV Env, SIV Gag; 15-mer peptides overlapping by 11 AA), at a final concentration of 1 mg/ml for each individual peptide. To prevent cytokine secretion, monensin (Golgi-stop, BD PharMingen, San Jose, CA) was added to the wells 60 minutes after addition of the peptides (Singh et al., 2018). Two peptide pools (120 and 115 peptides each) were designed to cover the 6 CH505 Env variants. Ten minutes after addition of peptides, the CD107a-PE Ab (Cat#12-1079-42, ThermoFisher) was added. Antigen-specific T cells were measured by intracellular cytokine staining followed by polychromatic flow cytometry using the following cocktail of cell surface antibodies: CD3-APC Cy7 (clone SP34–2, Cat#557757, BD PharMingen), CD4-V500 (clone L200, Cat#561488, BD), CD8-AlexaFluor-405 (clone 3B5, Cat#MHCD0826, Invitrogen), CD95FITC (clone DX2, Cat#556640, BD), CD28-PerCPCy5.5 (clone CD28.2, Cat#302922, Biolegend). After cell permeabilization, intracellular staining was performed using IFN-γ-PE Cy7 (clone B27, Cat#557643, BD PharMingen), Ki67-AF700 (clone B56, Cat#561277, BD) and Granzyme B-APC antibodies (clone GB12, Cat#MHGB05, Invitrogen). As negative and positive controls, PBMCs were cultured in medium without peptide stimulation or with a commercial mixture of PMA and calcium ionophore (Cat#00-4970-93, Invitrogen), respectively. Samples were acquired on a Fortessa or LSRII flow cytometers (BD Biosciences). The data were analyzed using FlowJo software (Tree Star, Inc.). Samples were considered positive if the frequency of IFN-γ+ T cells was 2-fold higher than that of unstimulated medium only control and greater than 0.01 after subtracting the medium control value.
Determination of antibody levels
Plasma Ab to a series of CH505 Env, including CD4bs competition assays (sCD4, CH106, DH235), and Gag were performed by direct-binding ELISA (Bonsignori et al., 2011). Briefly, antigens coated 384-well plates were incubated with 3x serial dilution of plasma samples starting at 1:30. An HRP-conjugated anti-macaque IgG Ab was added in assay diluent and developed using TMB substrate; plates were read at 450 nm in a SpectraMax 384 PLUS reader (Molecular Devices, Sunnyvale, CA); results are reported as logarithm 10 of area under the curve (AUC log10).
Plasma and vaginal IgG Ab binding to a panel of Env antigens (primary Env variants, consensus Env proteins, gp70-V1V2) and plasma IgG Ab to linear peptides (15-mer peptides overlapping by 12 amino acids) covering Env from CH505 and different clades were quantified by the multiplex binding antibody assay (BAMA) at the Immunology Core (Duke University and Medical Center, Tomaras lab) (Tomaras et al., 2008) as listed in Table S3. Briefly, antigens were coupled to carboxylated fluorescent beads and incubated with diluted test samples. Antigen-specific IgG were detected with biotinylated goat anti-monkey IgG, followed by incubation with streptavidin-phycoerythrin (PE). After washing the beads, the samples were acquired on a Bio-Plex instrument to measure florescence intensity. Plasma samples were tested in serial dilutions, and the mean MFI of the area under plasma dilution curve (AUC) was calculated and reported using the trapezoidal curve fit method. The array data were processed using pepStat (Huang et al., 2009). Mucosal samples were tested at a 1:2 dilution. Specific binding activity (SA) values were calculated as MFI*dilution/total IgG concentration (micrograms per ml). The total IgG concentration in mucosal samples was measured by a custom ELISA after sample elution and preparation for Ab assays. The BAMA was run under good clinical laboratory practice (GCLP)-compliant conditions, including tracking of positive controls by Levy-Jennings charts, using 21 CFR part 11-compliant software. Magnitude-Breadth (MB) scores for individual animals was determined from the linear peptide responses (He and Fong, 2019) The vaginal samples were assayed at a dilution of 1:2, and the binding magnitude is reported as specific activity (MFI*dilution/total IgG concentration; microG/ml). SIV Gag Ab levels were also evaluated by ELISA using serial dilutions plasma along with serial dilutions of pooled normal RM serum containing known amounts of IgG (for standard curve generation). Plates were developed by consecutive treatment with biotin conjugated goat anti-human IgG (Southern Biotech, AL), streptavidin-peroxidase (Sigma) and tetramethylbenzidine substrate (Sigma) with 1M H2SO4 stop solution. Absorbance at 450nm was measured and standard curves were generated using SoftMax Pro (Molecular Devices) to calculate total IgG levels in samples. Linear peptide array binding assay as previously described (Imholte and Gottardo, 2016; Karasawas et al., 2012). Microarray binding was performed using the HS4800 Pro Hybridization Station (Tecan, Männedorf, Switzerland) and scanned using an Axon Genepix 4300 Scanner (Molecular Devices, Sunnyvale, CA, USA). Images were analyzed using Genepix Pro software (Molecular Devices). Binding magnitude was calculated as log2-transformed signal intensity after pre-immunization baseline subtraction.
Binding avidity measurements
Surface plasmon resonance (SPR) tests were performed (Haynes et al., 2012; Lynch et al., 2014) using Biacore 4000 instruments. Env proteins were immobilized and the avidity score was calculated by determining the RU/Kd value (Haynes et al., 2012). The binding magnitude (in response units [RU]) and dissociation rate constant (kd, s-1) were measured for duplicate samples of purified serum IgG (at 200 μg/ml) against a panel of HIV-1 Env. Pre time-point values were subtracted and mean values from replicate measurements are used. The dissociation rate and avidity score (RU/kd) were measured.
Cyclic V2 measurements
Cyclic V2 Ab were measured in heat inactivated monkey plasma (1:50 dilution; in triplicate) by surface plasmon Resonance (SPR, Biacore) using N-linked biotinylated cyclic HIV-1 V2 peptides (JPT) captured onto the Streptavidin-immobilized CM7 sensor chips followed by secondary anti-monkey IgG antibodies. The HIV cyclic biotin V2 peptide sequences used were as follows:
V2.CH505: CSFNITTELRDKREKKNALFYKLDIVQLDGNSSQYRLINC;
V2.AE.92TH023: CSFNMTTELRDKKQKVHALFYKLDIVPIEDNTSSSEYRLINC;
V2.B.MN: CSFNITTSIGDKMQKEYALLYKLDIEPIDNDSTSYRLISC;
V2.C.1086 CSFKATTELKDKKHKVHALFYKLDVVPLNGNSSSSGEYRLINC.
For each plasma sample or controls, 4 replicates for each peptide were collected at rate of 1 Hz, with an analysis temperature at 25° C. All sample injections were conducted at flow rate of 10 μl/min. Data analysis was performed using Biacore 4000 Evaluation software 4.1 with double subtractions for unmodified surface and buffer for blank (Barouch et al., 2012; Singh et al., 2018; Vaccari et al., 2016).
Systems serology assays
ADCC (antibody-dependent cellular cytotoxicity) activity was measured as described with the flow-based ADCC-GranToxiLux assay (Lai et al., 2012; Pollara et al., 2011) using HIV CH505 gp120 protein coated CEM.NKRCCR5 target cells (Pollara et al., 2011). ADCC endpoint titers were defined as the reciprocal of the highest dilution indicating a positive response, and as the maximum % of GrzB within the target cell population (ADCC peak) at any plasma dilution. Specific killing is defined as percentage of gp120-coated target cells taking up granzyme B with a positivity cut-off at 8%. NK functions, antibody-dependent complement deposition (ADCD), antibody-dependent cellular phagocytosis (ADCP), antibody-dependent neutrophil phagocytosis (ADNP) effector functions, and antibody glycosylation were analyzed as previously described (Barouch et al., 2015; Vaccari et al., 2016) using purified HIV CH505 gp120. ADCP and ADNP of antigen-coated beads was performed using gp120-coated fluorescent beads. Beads were incubated with plasma from vaccinated RMs, washed and then placed in co-culture with THP1 cells or primary human neutrophils, respectively, as effector cells. The level of phagocytosis was quantified as the composite of the percent and mean fluorescence intensity of bead uptake. Antibody-dependent NK cell activation was performed using gp120-coated 96-well plates. Plasma was added to antigen-coated plates, after which non-binding antibodies were washed away, and purified human NK cells were added in the presence of the Golgi-blocking Brefeldin A. The level of degranulation, IFN-γ and macrophage inflammatory protein-1β (MIP-1β) were measured. ADCD was measured on beads using gp120-coated beads. Antigen-coated beads were cultured with plasma, washed to remove all non-binding Ab, and then guinea pig complement was added. The level of C3 deposition was then detected by flow cytometry. All assays were repeated in duplicate, non-infected NHP plasma background were subtracted for each assay. The Fc and Fab characteristics of virus-specific antibodies were simultaneously probed using the multiplexed Fc Array (Brown et al., 2017, 2015). Systemic vaccine effects were monitored in plasma samples collected pre vaccination and day 7 post 1st vaccination using the NHP Meso Scale Discovery multiplex assay.
Infected Cell Antibody Binding Assay (ICABA)
Measurement of plasma Ab binding to HIV-1 Env expressed on the surface of infected cells was conducted by indirect surface staining followed by flow cytometry (Ferrari et al., 2011). Briefly, mock infected and HIV-1 CH505 T/F-infected CEM.NKRCCR5 cells were incubated for 2 hr at 37° C with 1:100 dilutions of plasma samples from the vaccinated RMs. The cells were stained with a vital dye (Live/Dead Aqua) to exclude dead cells, and subsequently washed and permeabilized using BD Cytofix/Cytoperm solution. After permeabilization, cells were stained with FITC-conjugated goat-anti-Rhesus polyclonal antisera to detect binding of the plasma Ab, and RD1-conjugated anti-HIV-1 p24Gag to identify infected cells. Cells positive for NHP plasma binding were defined as viable, p24Gag positive, and FITC positive. Final results are reported as percent of FITC positive cells and FITC MFI among the viable p24Gag positive events after subtracting the background of the staining of the secondary Ab only and, in addition, to the subtraction of the background observed using the mock-infected cells. Baseline correction for the FITC MFI was performed by subtracting the MFI obtained using pre-immunization plasma samples, wherever applicable.
Neutralizing antibody assay
NAb activity in serum samples was measured in 96-well culture plates by using Tat-regulated luciferase (Luc) reporter gene expression to quantify reductions in virus infection in TZM-bl cells. TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program, as contributed by John Kappes and Xiaoyun Wu (Derdeyn et al., 2000; Platt et al., 2009, 1998; Takeuchi et al., 2008; Wei et al., 2002). Assays were performed with HIV-1 Env-pseudotyped viruses (CH0505.TF and variants), replication competent viruses produced in 293T cells (SHIV CH505.375H) or activated human PBMCs (SHIV SF162P3), and CH0505 and 426c glycan mutants produced in 293T or 293S/GnT1- cells essentially as previously described (Montefiori, 2009). Serum samples were heat-inactivated at 56° C for 1 hr, then diluted over a range of 1:20 to 1:43740 in cell culture medium and pre-incubated with virus (~150,000 relative light unit equivalents) for 1 hr at 37° C before addition of cells. Following a 48-hr incubation, cells were lysed and Luc activity determined using a microtiter plate luminometer and BriteLite Plus Reagent (Perkin Elmer). Neutralization titers are the sample dilution at which relative luminescence units (RLU) were reduced by 50% compared to RLU in virus control wells after subtraction of background RLU in cell control wells.
Fc array
The Fc and Fab characteristics of virus-specific antibodies were simultaneously probed using the multiplexed Fc Array, as described (Brown et al., 2017, 2015) for samples at baseline and weeks 42, 58, 77 (post vaccinations 4, 5, and 6) (Figure S5). Briefly, uniquely fluorescently coded antigen-coupled beads were incubated with serum samples, and the Fc regions of bound, antigen-specific antibodies were subsequently probed using phycoerythrin-labeled detection reagents including human and rhesus FcγRs, human C1q, MBL, and anti-IgG (Tables S2 and S3). Prior to modeling, features were filtered at each time point by excluding measurements that were not significantly different than baseline as defined by Student’s t test at p < 0.05 (unadjusted).
Classification
Least absolute shrinkage and selection operator (Tibshirani, 1996) (LASSO)-regularized binomial logistic regression (Cox, 1958) was used to learn models combining relatively sparse sets of features linearly in order to distinguish between RMs in different vaccine groups, as previously described (Pittala et al., 2019). Briefly, the ‘glmnet’(Friedman et al., 2010) package in R was employed with default options to develop models, and the penalty parameter (lambda) for regularization that achieved the lowest classification error was used in final models. Modeling performance was assessed in terms of the balanced accuracy over 100 repetitions of 10-fold cross-validation. Robustness was evaluated with permutation tests (Ojala and Garriga, 2010), in which the same modeling approach was applied across each of 100 different random shufflings of group labels. Differences in the accuracy of models learned from actual versus permuted data were characterized by effect size (Cliff’s delta). Visual inspection of predicted classes was based on a single run of 10-fold cross-validation. Simplified final models were developed by training on all the RMs but retaining only the two features making the greatest contribution to class predictions. These simplified final models were used for visual inspection of regression coefficients and distinctions between subjects in principal component biplots. To evaluate the robustness of this simplified feature set over time, these models (features and coefficients) were then applied to data collected at other time points and used to predict class across the longitudinal series.
For classification of challenge resistance, RMs were defined as relatively susceptible or resistant to infection based on their infection status as of the tenth challenge. This boundary was selected as it reasonably reflected the bimodal distribution of challenge outcomes that was observed overall (20 susceptible: 15 resistant) and among the Co-administration animals (9 susceptible: 11 resistant) only. For modeling protection among the animals in the Co-administration group only, the same classification approach described above for discriminating between Co-administration and Separate Administration was employed. However, given the reduced number of features to be considered, modeling on the restricted set of FcγRIIIa-associated features was performed with Sequential Forward Floating Selection (SFFS). The SFFS implementation from the mlxtend library in Python was used alongside logistic regression with L2 regularization from the default scikit-learn Python library with the number of features contributing capped at 30. Modeling performance was assessed in terms of balanced accuracy over 20 repetitions of 5-fold cross-validation. Training/testing splits were constructed using stratified folds in order to ensure proportional representation of the vaccine groups across testing sets. The feature set that achieved the highest mean classification accuracy was used in the final model. Visualizations of predicted classes and regression coefficients were based on a sparse final model relying on three features that was trained using all the animals. Robustness was evaluated with permutation tests, in which the same modeling approach, relying on the same number of features, was applied across 100 different random shufflings of group labels and accuracy of classification compared for effect size (Cliff’s delta).
QUANTIFICATION AND STATISTICAL ANALYSIS
These data were analyzed using GraphPad Prism version 8.0 for MacOS X (GraphPad Software, Inc, La Jolla, CA) and SAS Software Version [9.4]. Correlation coefficients and significance levels were calculated using Spearman rank correlation. Two-group test significance levels were calculated using Mann-Whitney analysis. Due to the exploratory nature of this research, the alpha level was set at 0.05 with no adjustment for multiple comparisons. Percentages uninfected as a function of challenge number were estimated by the Kaplan-Meier method, and p values of two-group comparisons were calculated by the exact log-rank test. The Wilcoxon signed rank test was used to compare measurement of individual RMs over time. Kruskal-Wallis tests were used to assess differences at individual time points across individual groups. Following previously established approaches (Pittala et al., 2019) models were trained in the setting of repeated cross-validation to use humoral and cellular response data to discriminate between RMs in the different immunization groups or with different resistance to infection following viral challenge. Cross-validated classifier performance was evaluated for accuracy and robustness by comparison to predictions learned from permuted data. For robust predictions, simplified final models comprised of minimal feature sets and learned on complete data are reported, and the ability of principal components derived from these features to differentiate between classes are provided for visualization.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-CD3 Monoclonal Antibody, APC-Cy7 Conjugated, Clone SP34–2 | BD Biosciences | Cat# 557757; RRID:AB_396863 |
| Mouse Anti-CD4 Monoclonal Antibody, V500 Conjugated, Clone L200 | BD Biosciences | Cat# 561488; RRID;AB_10693557 |
| Mouse Anti-CD95 Monoclonal Antibody, FITC Conjugated, Clone DX2 | BD Biosciences | Cat# 556640; RRID:AB_396506 |
| CD8 Monoclonal Antibody (3B5), Alexa Fluor 405 | Thermo Fisher | Cat# MHCD0826; RRID:AB_10372951 |
| PerCP/Cyanine5.5 anti-human CD28 antibody | BioLegend | Cat# 302922; RRID:AB_2073718 |
| CD107a (LAMP-1) Monoclonal Antibody (eBioH4A3), PE, eBioscience | Thermo Fisher | Cat# 12-1079-42; RRID:AB_10853326 |
| Mouse Anti-IFN-gamma Monoclonal Antibody, PE-Cy7 Conjugated, Clone B27 | BD Biosciences | Cat# 557643; RRID:AB_396760 |
| AlexaFluor 700 Mouse anti-Ki-67, clone B56 | BD Biosciences | Cat# 561277; RRID:AB_10611571 |
| Granzyme B Monoclonal Antibody (GB12), APC | Thermo Fisher | Cat#MHGB05; RRID:AB_10373420 |
| Biotinylated goat anti-monkey IgG | Rockland Immunochemicals | Cat# 617-106-006 |
| Streptavidin-phycoerythrin (PE) | Rockland Immunochemicals | Cat# S000-08 |
| Biotin conjugated goat anti-human IgG | Southern Biotech | Cat# 2040-08 |
| Streptavidin-peroxidase | Sigma | Cat# S5512 |
| Tetramethylbenzidine substrate | Sigma | Cat# T0440 |
| Streptavidin-immobilized CM7 Sensor Chips | GE Healthcare | Cat# 28–953828 |
| BD Golgi Stop (containing Monensin) | BD Biosciences | Cat# 554724 |
| Live/Dead Aqua | Thermo Fisher Scientific | Cat# L34957 |
| eBioscience Intracellular Fixation & Permeabilization Buffer Set | Invitrogen | Cat# 88-8824-00 |
| FITC-conjugated goat anti-Rhesus polyclonal antisera | Southern Biotech | Cat# 6200-02; RRID:AB_2796265 |
| RD1-conjugated anti-HIV-1 p24Gag | KC57, Beckman Coulter | Cat# 6604667; RRID:AB_1575989 |
| BriteLite Plus Reagent | Perkin Elmer | Cat# 6066761 |
| HRP-conjugated anti-macaque IgG antibody | Rockland Immunochemicals | Cat# 617-103-012 |
| Bacterial and Virus Strains | ||
| SHIV.C.CH505.375H (lot # 12/22/2015) | Shaw Lab | Lietal., 2016 |
| CH505.TF and variants HIV-1 Env-pseudotyped | See Table S3 | N/A |
| Biological Samples | ||
| Plasma and PBMCs from macaques in study | this study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cell Stimulator cocktail (500x) | Invitrogen | Cat#00-4970-93 |
| CH505 Env peptides | Infinity Biotech Research and Resource | N/A |
| SIV p57Gag peptides | NIH AIDS Reagent Program | Cat#12364 |
| TLR-4 agonist (GLA-SE) | Infectious Disease Research Institute Seattle, WA | Lot# QH079 |
| CH505.M5D8gp120/293F/MON | Haynes, B. | Lot# 226HC |
| CH505.M11D8gp120/293F/MON | Haynes, B. | Lot #150505 and Lot# 150506 |
| CH505.w020.14D8gp120/293F/MON | Haynes, B. | Lot# 1505132, Lot# 150513 and Lot# 150924 |
| CH505.w030.20D8gp120/293F/MON | Haynes, B. | Lot#150910 and Lot# 150813 |
| CH505.w30.12D8gp120/293F/MON | Haynes, B. | Lot# 150917 and Lot# 150814 |
| CH505.w136.B18D8gp120/293F/MON | Haynes, B. | Lot# 150901, Lot# 150902 and Lot# 150824 |
| Env peptides for Ab mapping | JPT Peptide Technologies GmbH (Germany) | N/A |
| N-linked biotinylated cyclic V2.CH505 | JPT Peptide Technologies GmbH (Germany) | N/A |
| N-linked biotinylated cyclic V2.AE.92TH023 | JPT Peptide Technologies GmbH (Germany) | N/A |
| N-linked biotinylated cyclic V2.B.MN | JPT Peptide Technologies GmbH (Germany) | N/A |
| N-linked biotinylated cyclic V2.C.1086 | JPT Peptide Technologies GmbH (Germany) | N/A |
| Critical Commercial Assays | ||
| NHP U-plex Biomarker | Meso Scale Discovery | Cat# K15082K |
| Experimental Models: Cell Lines | ||
| HEK293F (Expi293TM GnTI-Expression System Kit | Thermo Fisher Scientific | Cat# A39250 |
| Human cell line TZM-bl | NIH AIDS Reagent Program | Cat# 8129 |
| HEK293T | Montefiori, D. | N/A |
| Monocytic THP-1 cell line | ATCC | Cat# TIB-202 |
| Primary human neutrophils | Ferrari, G. | N/A |
| HIV-1 CH505 T/F-infected CEM.NKRCCR5 Cells | Ferrari, G. | N/A |
| HIV-1 CH505 T/F- gp120 coated CEM.NKRCCR5 Cells | Ferrari, G. | N/A |
| Experimental Models: Organisms/Strains | ||
| M. mulatta-Indian Rhesus macaques | Tulane National Primate Research Center | N/A |
| Recombinant DNA | ||
| CH505.M5gp145 | Haynes, B. | HV1300656 |
| CH505.M11 gp145 | Haynes, B. | HV1300662 |
| CH505.w020.14gp145 | Haynes, B. | HV1300635 |
| CH505.w030.20D8gp145 | Haynes, B. | HV1300688 |
| CH505.w30.12D8gp145 | Haynes, B. | HV1300646 |
| CH505.w136.B18D8gp145 | Haynes, B. | HV1300724 |
| p27CE Gag | Felber, B.K. | 286S |
| p57Gag | Felber, B.K. | 206S |
| macaque IL-12 | Felber, B.K. | AG157 |
| CMVkan | Felber, B.K. | N/A |
| Software and Algorithms | ||
| GraphPad Prism v8.0 | GraphPad Software | N/A |
| SAS Software v9.4 | SAS Institute | N/A |
| FlowJo Software v10 | BD | N/A |
| Biacore 4000 Evaluation Software v4.1 | GE Healthcare Life Sciences | N/A |
| SoftMax Pro | Molecular Devices, LLC | N/A |
| Genepix Pro Software | Molecular Devices, LLC | N/A |
| (LASSO)-regularized binomial logistic regression | Cox, 1958 | N/A |
| RStudio and Python Code | Ackerman, M | https://github.com/dAbL-Dartmouth/p186 |
| Other | ||
| Cellectra 5P | Inovio Pharmaceuticals, Inc | N/A |
| Fortessa and LSRII flow cytometers | BD Biosciences | N/A |
| Bio-Plex Instrument | N/A | |
| HS4800 Pro Hybridization Station | Tecan | N/A |
| Axon Genepix 4300 | Molecular Devices, LLC | N/A |
| Biacore 4000 | Biacore | N/A |
| Infected Cell Antibody Binding Assay (ICABA) | Ferrari, G. | N/A |
| Env-specific humoral immune assays | Tomaras, G., Montefiori, D. | N/A |
| NASBA Assay | Advanced Biosciences Laboratory | N/A |
| SpectraMax Plus 384 Microplate Spectrophotometer | Molecular Devices, LLC | N/A |
| Luminometer | Perkin Elmer | N/A |
| MHC Class I Assay | Refer to Table S1 | N/A |
| Flow-based ADCC-GranToxiLux Assay | Pollaraetal. (2011) | N/A |
| Multiplexed Fc Array | Brown et al. (2017, 2015); See Tables S2 and S3 | N/A |
Highlights.
DNA + protein vaccine includes six sequentially isolated patient CH505 HIV Envs
DNA + Env protein co-administration in the same muscle reduces SHIV infection risk
Protection from infection is mediated by non-neutralizing antibody functions
Antibody binding to FcγRIIIa and ADCC activity contributes to protection
ACKNOWLEDGMENTS
We thank DAIDS/NIAID for support of SVEU P186 and N. Miller (NIAID) for discussion and support of the study, and A. Schultz (DAIDS) for continuous support. We are thankful for excellent assay performance and data analysis by H. Chung (Advanced Bioscience Laboratories), R. Parks, A. Trama,T. Gurley, K. Gronin (Duke Human Vaccine Institute); R. Mathura, S. McMillan,D. Goodman, and T. Huynh (Duke University); R. Spreng (Duke University) for statistics consultation; W. Ding (Univ. Penn) for preparation of the SHIV.CH505 challenge stock; Y. Wang (CSP, Leidos, Frederick); A. Khan (Inovio Pharmaceuticals); R.V. Blair, W. Lala, C. Mabee (Tulane National Primate Research Center, Tulane University); B.K. Hejgaard (Dartmouth) for excellent support; Hua-Xin Liao (Duke Vaccine) for discussions; and T. Jones for administrative support and editorial assistance. This work was supported in part by the Simian Virus Evaluation Unit through SVEU P186 (DAIDS, NIAID); by the Intramural Research Program of the National Cancer Institute to G.N.P. and B.K.F. with federal funds from the National Cancer Institute, National Institutes of Health; by Inovio Pharmaceuticals Plymouth Meeting, PA, under NCI CRADA#02289 to G.N.P. and B.K.F.; by the National Institute of Allergy and Infectious Disease contract no. HHSN27201100016C to D.M.; by National Institute of Allergy and Infectious Disease P01 AI120756 to G.D.T. and M.E.A.; by a NIH contract HHSN27201100016C to X.S., G.D.T., and D.M.; by a Cooperative Agreement Award (W81XWH-18-2-0040) between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the U.S. Army Medical Research and Development Command to H.V.T.; and by NIH, NIAID, Division of AIDS grants, UM1 AI100645, the Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery and the NIH, NIAID, Division of AIDS grant UM1 AI44371, and the Center for HIV/AIDS Vaccine Development, both to B.F.H.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107624.
DECLARATION OF INTERESTS
G.N.P. and B.K.F. are inventors on US Government-owned patents related to DNA vaccines and gene expression optimization. B.F.H. has submitted patent applications covering the Envs used in this study. S.G.R. is a full-time employee of Infectious Disease Research Institute and as such receives compensation in the form of salary. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The manuscript has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense. The views expressed in this article are those of the authors and do not necessarily reflect the official policy of the Department of the Army, Department of Defense, or the U.S. Government. The other authors declare no competing interests. This work was produced solely by the authors and that no other individuals or entities influenced any aspects of the work.
SUPPORTING CITATIONS
The following references appear in the Supplemental Information: He and Fong (2019); Huang et al. (2009); and Imholte and Gottardo (2016).
REFERENCES
- Ackerman ME, Barouch DH, and Alter G (2017). Systems serology for evaluation of HIV vaccine trials. Immunol. Rev 275, 262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman ME, Das J, Pittala S, Broge T, Linde C, Suscovich TJ, Brown EP, Bradley T, Natarajan H, Lin S, et al. (2018). Route of immunization defines multiple mechanisms of vaccine-mediated protection against SIV. Nat. Med 24, 1590–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G, and Barouch D (2018). Immune correlate-guided HIV vaccine design. Cell Host Microbe 24, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, Iampietro MJ, SanMiguel A, Seaman MS, Ferrari G, et al. (2012). Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature 482, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Alter G, Broge T, Linde C, Ackerman ME, Brown EP, Borducchi EN, Smith KM, Nkolola JP, Liu J, et al. (2015). Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science 349, 320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Tomaka FL, Wegmann F, Stieh DJ, Alter G, Robb ML, Michael NL, Peter L, Nkolola JP, Borducchi EN, et al. (2018). Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13–19). Lancet 392, 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, et al. (2011). Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol 85, 9998–10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang GY, Schramm CA, Wiehe K, et al. ; NISC Comparative Sequencing Program (2016). Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell 165, 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Liao HX, Gao F, Williams WB, Alam SM, Montefiori DC, and Haynes BF (2017). Antibody-virus co-evolution in HIV infection: paths for HIV vaccine development. Immunol. Rev 275, 145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley T, Pollara J, Santra S, Vandergrift N, Pittala S, Bailey-Kellogg C, Shen X, Parks R, Goodman D, Eaton A, et al. (2017). Pentavalent HIV-1 vaccine protects against simian-human immunodeficiency virus challenge. Nat. Commun 8, 15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EP, Normandin E, Osei-Owusu NY, Mahan AE, Chan YN, Lai JI, Vaccari M, Rao M, Franchini G, Alter G, and Ackerman ME (2015). Microscale purification of antigen-specific antibodies. J. Immunol. Methods 425, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EP, Dowell KG, Boesch AW, Normandin E, Mahan AE, Chu T, Barouch DH, Bailey-Kellogg C, Alter G, and Ackerman ME (2017). Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J. Immunol. Methods 443, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli KM, and Crotty S (2017). Germinal center enhancement by extended antigen availability. Curr. Opin. Immunol 47, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey L, Gilbert PB, Tomaras GD, Haynes BF, Pantaleo G, and Fauci AS (2015). Immune correlates of vaccine protection against HIV-1 acquisition. Sci. Transl. Med 7, 310rv7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR (1958). The regression-analysis of binary sequences. J. R. Stat. Soc. B 20, 215–242. [Google Scholar]
- Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O’Brien WA, Ratner L, Kappes JC, Shaw GM, and Hunter E (2000). Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol 74, 8358–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg PK, Shangguan S, Issac B, Alter G, Geretz A, Izumi T, Bryant C, Eller MA, Wegmann F, Apps R, et al. (2019). A vaccine-induced gene expression signature correlates with protection against SIV and HIV in multiple trials. Sci. Transl. Med 11, eaaw4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felber BK, Valentin A, Rosati M, Bergamaschi C, and Pavlakis GN (2014). HIV DNA vaccine: stepwise improvements make a difference. Vaccines (Basel) 2, 354–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fera D, Schmidt AG, Haynes BF, Gao F, Liao HX, Kepler TB, and Harrison SC (2014). Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains. Proc. Natl. Acad. Sci. USA 111, 10275–10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari G, Pollara J, Kozink D, Harms T, Drinker M, Freel S, Moody MA, Alam SM, Tomaras GD, Ochsenbauer C, et al. (2011). An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol 85, 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flingai S, Czerwonko M, Goodman J, Kudchodkar SB, Muthumani K, and Weiner DB (2013). Synthetic DNA vaccines: improved vaccine potency by electroporation and co-delivered genetic adjuvants. Front. Immunol 4, 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florese RH, Demberg T, Xiao P, Kuller L, Larsen K, Summers LE, Venzon D, Cafaro A, Ensoli B, and Robert-Guroff M (2009). Contribution of nonneutralizing vaccine-elicited antibody activities to improved protective efficacy in rhesus macaques immunized with Tat/Env compared with multigenic vaccines. J. Immunol 182, 3718–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, and Tibshirani R (2010). Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw 33, 1–22. [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bonsignori M, Liao HX, Kumar A, Xia SM, Lu X, Cai F, Hwang KK, Song H, Zhou T, et al. (2014). Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell 158, 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P, Wang M, Wrin T, Petropoulos C, Gurwith M, Sinangil F, D’Souza P, Rodriguez-Chavez IR, DeCamp A, Giganti M, et al. (2010). Magnitude and breadth of a nonprotective neutralizing antibody response in an efficacy trial of a candidate HIV-1 gp120 vaccine. J. Infect. Dis 202, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Carnathan DG, Torrents de la Peña A, Pauthner M, Briney B, Reiss SM, Wood JS, Kaushik K, van Gils MJ, Rosales SL, et al. (2016). Direct probing of germinal center responses reveals immunological features and bottlenecks for neutralizing antibody responses to HIV Env trimer. Cell Rep. 17, 2195–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, and Mascola JR (2017). The quest for an antibody-based HIV vaccine. Immunol. Rev 275, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. (2012). Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med 366, 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, and Fong Y (2019). Maximum diversity weighting for biomarkers with application in HIV-1 vaccine studies. Stat. Med 38, 3936–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidajat R, Xiao P, Zhou Q, Venzon D, Summers LE, Kalyanaraman VS, Montefiori DC, and Robert-Guroff M (2009). Correlation of vaccine-elicited systemic and mucosal nonneutralizing antibody activities with reduced acute viremia following intrarectal simian immunodeficiency virus SIVmac251 challenge of rhesus macaques. J. Virol 83, 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao LA, Wu L, Khan AS, Hokey DA, Yan J, Dai A, Betts MR, Draghia-Akli R, and Weiner DB (2008). Combined effects of IL-12 and electroporation enhances the potency of DNA vaccination in macaques. Vaccine 26, 3112–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, and Korber BT (2014). Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Valentin A, Dayton F, Kulkarni V, Alicea A, Rosati M, Chowdhury B, Gautam R, Broderick KE, Sardesai NY, et al. (2016). DNA prime-boost vaccine regimen to increase breadth, magnitude, and cytotoxicity of the cellular immune responses to subdominant Gag epitopes of simian immunodeficiency virus and HIV. J. Immunol 197, 3999–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gilbert PB, Montefiori DC, and Self SG (2009). Simultaneous evaluation of the magnitude and breadth of a left and right censored multivariate response, with application to HIV vaccine development. Stat. Biopharm. Res 1, 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imholte G, and Gottardo R (2016). Bayesian hierarchical modeling for subject-level response classification in peptide microarray immunoassays. Bio-metrics 72, 1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalah R, Patel V, Kulkarni V, Rosati M, Alicea C, Ganneru B, von Gegerfelt A, Huang W, Guan Y, Broderick KE, et al. (2012). IL-12 DNA as molecular vaccine adjuvant increases the cytotoxic T cell responses and breadth of humoral immune responses in SIV DNA vaccinated macaques. Hum. Vaccin. Immunother 8, 1620–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalah R, Rosati M, Ganneru B, Pilkington GR, Valentin A, Kulkarni V, Bergamaschi C, Chowdhury B, Zhang G-M, Beach RK, et al. (2013). The p40 subunit of interleukin (IL)-12 promotes stabilization and export of the p35 subunit: implications for improved IL-12 cytokine production. J. Biol. Chem 288, 6763–6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalah R, Kulkarni V, Patel V, Rosati M, Alicea C, Bear J, Yu L, Guan Y, Shen X, Tomaras GD, et al. (2014). DNA and protein co-immunization improves the magnitude and longevity of humoral immune responses in macaques. PLoS ONE 9, e91550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski JP, Krebs SJ, Trovato M, Kovarik DN, Brower Z, Sutton WF, Waagmeester G, Sartorius R, D’Apice L, Caivano A, et al. (2012). Co-immunization with multimeric scaffolds and DNA rapidly induces potent autologous HIV-1 neutralizing antibodies and CD8+ T cells. PLoS ONE 7, e31464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasawas N, Billings E, Rao M, Williams C, Zolla-Pazner S, Bailer RT, Koup RA, Madnote S, Arworn D, Shen X, et al. ; MOPH TAVEG Collaboration (2012). The Thai phase III HIV type 1 vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res. Hum. Retroviruses 28, 1444–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsoe G, and Haynes BF (2017). Host controls of HIV broadly neutralizing antibody development. Immunol. Rev 275, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsoe G, and Haynes BF (2018). What are the primary limitations in B-cell affinity maturation, and how much affinity maturation can we drive with vaccination? Breaking through immunity’s glass ceiling. Cold Spring Harb. Perspect. Biol 10, a029397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer RM, Archer MC, Orr MT, Dubois Cauwelaert N, Beebe EA, Huang PD, Dowling QM, Schwartz AM, Fedor DM, Vedvick TS, and Fox CB (2018). Development of a thermostable nanoemulsion adjuvanted vaccine against tuberculosis using a design-of-experiments approach. Int. J. Nanomedicine 13, 3689–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs SJ, McBurney SP, Kovarik DN, Waddell CD, Jaworski JP, Sutton WF, Gomes MM, Trovato M, Waagmeester G, Barnett SJ, et al. (2014). Multimeric scaffolds displaying the HIV-1 envelope MPER induce MPER-specific antibodies and cross-neutralizing antibodies when co-immunized with gp160 DNA. PLoS ONE 9, e113463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, and Mascola JR (2018). HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity 48, 855–871. [DOI] [PubMed] [Google Scholar]
- LaBranche CC, McGuire AT, Gray MD, Behrens S, Kwong PDK, Chen X, Zhou T, Sattentau QJ, Peacock J, Eaton A, et al. (2018). HIV-1 envelope glycan modifications that permit neutralization by germline-reverted VRC01-class broadly neutralizing antibodies. PLoS Pathog. 14, e1007431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBranche CC, Henderson R, Hsu A, Behrens S, Chen X, Zhou T, Wiehe K, Saunders KO, Alam SM, Bonsignori M, et al. (2019). Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancestor of CH235 lineage CD4bs broadly neutralizing antibodies. PLoS Pathog. 15, e1008026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L, Kwa SF, Kozlowski PA, Montefiori DC, Nolen TL, Hudgens MG, Johnson WE, Ferrari G, Hirsch VM, Felber BK, et al. (2012). SIVmac239 MVA vaccine with and without a DNA prime, similar prevention of infection by a repeated dose SIVsmE660 challenge despite different immune responses. Vaccine 30, 1737–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Valentin A, Kulkarni V, Rosati M, Beach RK, Alicea C, Hannaman D, Reed SG, Felber BK, and Pavlakis GN (2013). HIV/SIV DNA vaccine combined with protein in a co-immunization protocol elicits highest humoral responses to envelope in mice and macaques. Vaccine 31, 3747–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang S, Kong R, Ding W, Lee FH, Parker Z, Kim E, Learn GH, Hahn P, Policicchio B, et al. (2016). Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc. Natl. Acad. Sci. USA 113, E3413–E3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Lindgren G, Sandgren KJ, Thompson EA, Francica JR, Seu-bert A, De Gregorio E, Barnett S, O’Hagan DT, Sullivan NJ, et al. (2017). Vaccine priming is restricted to draining lymph nodes and controlled by adjuvant-mediated antigen uptake. Sci. Transl. Med 9, eaal2094. [DOI] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. ; NISC Comparative Sequencing Program (2013). Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HE, Stewart SM, Kepler TB, Sempowski GD, and Alam SM (2014). Surface plasmon resonance measurements of plasma antibody avidity during primary and secondary responses to anthrax protective antigen. J. Immunol. Methods 404, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurley NP, Domi A, Basu R, Saunders KO, LaBranche CC, Montefiori DC, Haynes BF, and Robinson HL (2017). HIV transmitted/founder vaccines elicit autologous tier 2 neutralizing antibodies for the CD4 binding site. PLoS ONE 12, e0177863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC (2009). Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol 485, 395–405. [DOI] [PubMed] [Google Scholar]
- Muthumani K, Bagarazzi M, Conway D, Hwang DS, Manson K, Ciccarelli R, Israel Z, Montefiori DC, Ugen K, Miller N, et al. (2003). A Gag-Pol/Env-Rev SIV239 DNA vaccine improves CD4 counts, and reduce viral loads after pathogenic intrarectal SIV(mac)251 challenge in rhesus macaques. Vaccine 21, 629–637. [DOI] [PubMed] [Google Scholar]
- Muthumani K, Wise MC, Broderick KE, Hutnick N, Goodman J, Flingai S, Yan J, Bian CB, Mendoza J, Tingey C, et al. (2013). HIV-1 Env DNA vaccine plus protein boost delivered by EP expands B- and T-cell responses and neutralizing phenotype in vivo. PLoS ONE 8, e84234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidich SD, Fong Y, Li SS, Geraghty DE, Williamson BD, Young WC, Goodman D, Seaton KE, Shen X, Sawant S, et al. ; HVTN 505 Team (2019). Antibody Fc effector functions and IgG3 associate with decreased HIV-1 risk. J. Clin. Invest 129, 4838–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala M, and Garriga GC (2010). Permutation tests for studying classifier performance. J. Mach. Learn. Res 11, 1833–1863. [Google Scholar]
- Pantaleo G, Janes H, Karuna S, Grant S, Ouedraogo GL, Allen M, Tomaras GD, Frahm N, Montefiori DC, Ferrari G, et al. ; NIAID HIV Vac cine Trials Network (2019). Safety and immunogenicity of a multivalent HIV vaccine comprising envelope protein with either DNA or NYVAC vectors (HVTN 096): a phase 1b, double-blind, placebo-controlled trial. Lancet HIV 6, e737–e749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V, Jalah R, Kulkarni V, Valentin A, Rosati M, Alicea C, von Gegerfelt A, Huang W, Guan Y, Keele B, et al. (2013). DNA and virus particle vaccination protects against acquisition and confers control of viremia upon heterologous SIV challenge. Proc. Natl. Acad. Sci. USA 110, 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, et al. (2017). Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity 46, 1073–1088 e1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, Prevost J, Nedellec R, von Bredow B, Abbink P, et al. (2019). Vaccine-induced protection from homologous tier 2 SHIV challenge in nonhuman primates depends on serum-neutralizing antibody titers. Immunity 50, 241–252 e246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pissani F, Malherbe DC, Schuman JT, Robins H, Park BS, Krebs SJ, Barnett SW, and Haigwood NL (2014). Improvement of antibody responses by HIV envelope DNA and protein co-immunization. Vaccine 32, 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittala S, Bagley K, Schwartz JA, Brown EP, Weiner JA, Prado IJ, Zhang W, Xu R, Ota-Setlik A, Pal R, et al. (2019). Antibody Fab-Fc properties outperform titer in predictive models of SIV vaccine-induced protection. Mol. Syst. Biol 15, e8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, and Kabat D (1998). Effects of CCR5 and CD4 cell surface concentrations on infections by macro-phagetropic isolates of human immunodeficiency virus type 1. J. Virol 72, 2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Bilska M, Kozak SL, Kabat D, and Montefiori DC (2009). Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J. Virol 83, 8289–8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollara J, Hart L, Brewer F, Pickeral J, Packard BZ, Hoxie JA, Komoriya A, Ochsenbauer C, Kappes JC, Roederer M, et al. (2011). High-throughput quantitative analysis of HIV-1 and SIV-specific ADCC-mediating antibody responses. Cytometry A 79, 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. ; MOPH-TAVEG Investigators (2009). Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med 361, 2209–2220. [DOI] [PubMed] [Google Scholar]
- Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, deCamp AC, Carrico C, Menis S, Magaret CA, et al. (2012). Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature 490, 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano JW, Shurtliff RN, Dobratz E, Gibson A, Hickman K, Markham PD, and Pal R (2000). Quantitative evaluation of simian immunodeficiency virus infection using NASBA technology. J. Virol. Methods 86, 61–70. [DOI] [PubMed] [Google Scholar]
- Rosati M, von Gegerfelt A, Roth P, Alicea C, Valentin A, Robert-Guroff M, Venzon D, Montefiori DC, Markham P, Felber BK, and Pavlakis GN (2005). DNA vaccines expressing different forms of simian immunodeficiency virus antigens decrease viremia upon SIVmac251 challenge. J. Virol 79, 8480–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati M, Bergamaschi C, Valentin A, Kulkarni V, Jalah R, Alicea C, Patel V, von Gegerfelt AS, Montefiori DC, Venzon DJ, et al. (2009). DNA vaccination in rhesus macaques induces potent immune responses and decreases acute and chronic viremia after SIVmac251 challenge. Proc. Natl. Acad. Sci. USA 106, 15831–15836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouphael NG, Morgan C, Li SS, Jensen R, Sanchez B, Karuna S, Swann E, Sobieszczyk ME, Frank I, Wilson GJ, et al. ; HVTN 105 Protocol Team and the NIAID HIV Vaccine Trials Network (2019). DNA priming and gp120 boosting induces HIV-specific antibodies in a randomized clinical trial. J. Clin. Invest 129, 4769–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders KO, Nicely NI, Wiehe K, Bonsignori M, Meyerhoff RR, Parks R, Walkowicz WE, Aussedat B, Wu NR, Cai F, et al. (2017a). Vaccine elicitation of high mannose-dependent neutralizing antibodies against the V3-glycan broadly neutralizing epitope in nonhuman primates. Cell Rep. 18, 2175–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders KO, Verkoczy LK, Jiang C, Zhang J, Parks R, Chen H, Housman M, Bouton-Verville H, Shen X, Trama AM, et al. (2017b). Vaccine induction of heterologous tier 2 HIV-1 neutralizing antibodies in animal models. Cell Rep. 21, 3681–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Duffy R, Howington R, Cope A, Sadagopal S, Park H, Pal R, Kwa S, Ding S, Yang OO, et al. (2015). Vaccine-induced epitope specific antibodies to SIVmac239 envelope are distinct from those induced to the HIV-1 envelope in non-human primates. J. Virol 89, 8643–8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Ramírez-Salazar EG, Doueiri R, Valentin A, Rosati M, Hu X, Keele BF, Shen X, Tomaras GD, Ferrari G, et al. (2018). Control of heterologous SIVsmE660 infection by DNA and protein co-immunization regimens combined with different Toll-like receptor-4 (TLR-4) based adjuvants. J. Virol 92, e00281–e00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, McClure MO, and Pizzato M (2008). Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J. Virol 82, 12585–12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R (1996). Regression shrinkage and selection via the Lasso. J. Roy. Stat. Soc. B Met 58, 267–288. [Google Scholar]
- Tomaras GD, and Plotkin SA (2017). Complex immune correlates of protection in HIV-1 vaccine efficacy trials. Immunol. Rev 275, 245–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT, et al. (2008). Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol 82, 12449–12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari M, Gordon SN, Fourati S, Schifanella L, Liyanage NP, Cameron M, Keele BF, Shen X, Tomaras GD, Billings E, et al. (2016). Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat. Med 22, 762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin A, McKinnon K, Li J, Rosati M, Kulkarni V, Pilkington GR, Bear J, Alicea C, Vargas-Inchaustegui DA, Jean Patterson L, et al. (2014). Comparative analysis of SIV-specific cellular immune responses induced by different vaccine platforms in rhesus macaques. Clin. Immunol 155, 91–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Inchaustegui DA, Tuero I, Mohanram V, Musich T, Pegu P, Valentin A, Sui Y, Rosati M, Bear J, Venzon DJ, et al. (2014). Humoral immunity induced by mucosal and/or systemic SIV-specific vaccine platforms suggests novel combinatorial approaches for enhancing responses. Clin. Immunol 153, 308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal DO, Talbott KT, Choo DK, Shedlock DJ, and Weiner DB (2013). Synthetic DNA vaccine strategies against persistent viral infections. Expert Rev. Vaccines 12, 537–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, and Kappes JC (2002). Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother 46, 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams WB, Zhang J, Jiang C, Nicely NI, Fera D, Luo K, Moody MA, Liao HX, Alam SM, Kepler TB, et al. (2017). Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations. Nat. Commun 8, 1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao P, Zhao J, Patterson LJ, Brocca-Cofano E, Venzon D, Kozlowski PA, Hidajat R, Demberg T, and Robert-Guroff M (2010). Multiple vaccine-elicited nonneutralizing antienvelope antibody activities contribute to protective efficacy by reducing both acute and chronic viremia following simian/human immunodeficiency virus SHIV89.6P challenge in rhesus macaques. J. Virol 84, 7161–7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Acharya P, Kong R, Cheng C, Chuang GY, Liu K, Louder MK, O’Dell S, Rawi R, Sastry M, et al. (2018). Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med 24, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, deCamp AC, Cardozo T, Karasawas N, Gottardo R, Williams C, Morris DE, Tomaras G, Rao M, Billings E, et al. (2013). Analysis of V2 antibody responses induced in vaccinees in the ALVAC/AIDSVAX HIV-1 vaccine efficacy trial. PLoS ONE 8, e53629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Powell R, Yahyaei S, Williams C, Jiang X, Li W, Lu S, Wang S, Upadhyay C, Hioe CE, et al. (2016). Rationally designed vaccines targeting the V2 Region of HIV-1 gp120 induce a focused, cross-clade-reactive, biologically functional antibody response. J. Virol 90, 10993–11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RStudio and Python code generated during this study are available at https://github.com/dAbL-Dartmouth/p186.


