Abstract
Viral factories are intracellular compartments of the host cell that contain viral replication organelles and necessary elements for assembly and maturation of new infectious viral particles. In this article we revise the methods used to study viral factories and the current knowledge on the structure, functions and biogenesis of these structures. We also describe some of the most emblematic examples of viral factories characterized so far. Finally, we describe how the identification of mechanisms involved in the biogenesis and functional architecture of viral factories will provide new means for antiviral intervention.
Keywords: Viral morphogenesis, Viral replication complex, Virus factory, Virus replication organelle
Abbreviations: CLEM, Correlative light and electron microscopy; ER, Endoplasmic reticulum; ET, Electron tomography; LM, Light microscopy; RO, Replication organelle; TEM, Transmission electron microscopy; VF, Virus factory; VRC, Viral replication complex
Glossary
- Aggresome
Perinuclear inclusion body assembled around the microtubule organizing center (MTOC) and surrounded by a cage of vimentin where the cell encloses misfolded proteins. Some DNA viruses hijack this pathway to concentrate materials and build their factories.
- Autophagy
Mechanism for degradation and recycling of cellular components. Expendable constituents are isolated inside a double-membrane organelle known as autophagosome that later fuses with a lysosome where materials are degraded. Some RNA viruses induce and use components of this pathway for building their factories.
- Correlative light and electron microscopy
Group of methods that combine light and electron microscopy imaging of the same specimen. Light microscopy offers a large field of view and imaging the dynamics of living cells. Particular cells with interesting features are selected for high resolution electron microscopy (EM). This approach is very useful to study the biogenesis of virus factories.
- Replication organelle
Membranous structure that harbors viral replication complexes inside the viral factory. It is usually made of single-membrane vesicles or spherules, double-membrane vesicles, multi-membraned vesicles, tubule-vesicular cubic membranes or planar oligomeric arrays.
- Viral replication complex
Macromolecular structure constituted by viral polymerases, other viral non-structural proteins and cell co-factors that makes multiple copies of the viral genome and transfers them to the assembly sites.
- Virus factory or viral factory
Intracellular compartment that contains the viral replication organelles and the sites for viral particle assembly and maturation.
Introduction
Viruses are obligate intracellular parasites. In the extracellular environment, viral particles exist as rather simple macromolecular entities made of protein shells also known as capsids, and/or membranes, also known as envelopes, that protect and transport the viral genome from cell to cell. It is only during the intracellular phase of their life that viruses synthesize distinct molecules and perform activities that are characteristic of living entities. One of these activities is the construction of a platform for genome replication and morphogenesis, also known as virus factory (VF). The central element of the factory is the replication organelle (RO) where the viral replication complexes (VRC) make multiple copies of the viral genome. In many cases, factories are made of remodeled cell membranes and contain functional compartments with specific roles in virus replication, assembly and egress. Other cell elements such as mitochondria and the cytoskeleton are often recruited to viral factories and make contacts with the RO. In most cases, the specific role of these contacts is not known. VFs can be rather big, with a diameter of several microns and occupy large intracellular regions. VFs are dynamic structures that maintain their communication with intracellular transport pathways to incorporate essential components and to facilitate the exit of newly formed viral particles (Fig. 1 ). Furthermore, VFs change over time with the assembly and disassembly of particular structures needed for specific steps of the virus life. In some cases, VFs or only the ROs are mobile compartments that move inside the infected cell, like in the case of the early ROs of Vaccinia Virus, Hepatitis C Virus, rotaviruses and the mouse hepatitis coronavirus. In the case of some plant viruses, ROs are transported from cell to cell through intercellular channels known as plasmodesmata.
Fig. 1.
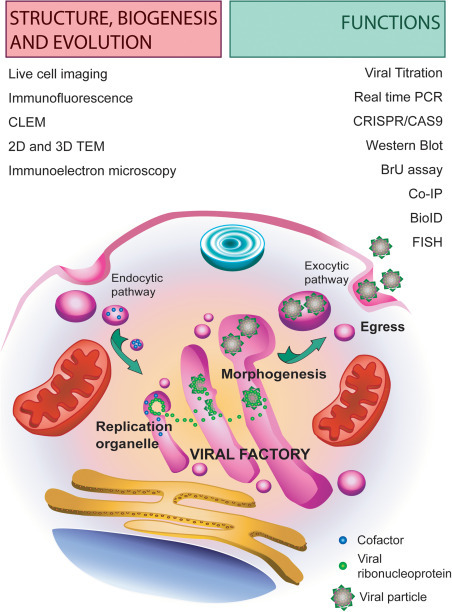
Main components and functions of virus factories and methods for their characterization. Techniques for characterizing the structure, biogenesis and evolution of VFs are listed on the left, those for studying their functions are listed on the right.
To build their replication neo-organelles and factories, viruses make use of cellular materials and signaling pathways. The study of VFs and the way they hijack cellular pathways give also insight in the pathway itself. Researchers have identified two main pathways hijacked by viruses to build their factories: The aggresome formation for DNA viruses and autophagy for RNA viruses. Moreover, viruses manipulate lipid synthesis and flows to create membranes with particular lipid composition and biophysical properties that are ideal for the activation and efficient performance of the macromolecular complexes that work in viral genome replication and assembly.
Although present in many VFs, it is not known why and how mitochondria are recruited to viral factories. It has been proposed that mitochondria would provide the energy storage molecule adenosine triphosphate (ATP) necessary for virus replication, but direct evidence is lacking. Alternatively, viruses may need mitochondrial components such as particular proteins that incorporate into the replication organelles where they would enhance viral genome replication. Another possibility is that viruses recruit mitochondria around VFs to block the innate antiviral immunity response mediated by these organelles.
Methods to Study Virus Factories
The study of virus factories includes the characterization of their structure, biogenesis, functions, and evolution over time (Fig. 1). Virus factories were identified with light and electron microscopy imaging techniques. The first evidences of the existence of VFs came from studies of thin-sections of infected cells visualized by transmission electron microscopy (TEM). These images showed viral particles associated with macro-structures made of organelles that had been remodeled and transported to particular intracellular locations. Light microscopy (LM) then showed how viruses change the organization of whole infected cells to build their VFs. Nowadays, advances in fluorescent probes and LM methods are providing new insights into the biogenesis and functional architecture of VFs. Time-course assays combined with immunofluorescence and confocal microscopy provide a general view of the progression of infection. For these studies, cells at different times post-infection (tpi) are processed for immunolabeling with primary antibodies specific for viral proteins and secondary antibodies conjugated with fluorophores. When fluorescent viruses are available, real time, live cell microscopy shows an accurate view of all steps of the viral infection, including the early stages of VF assembly (Fig. 2(a)–(c) ). Moreover, simultaneous labeling of cell compartments with specific markers allows the identification of particular organelles subverted by the virus and provides a dynamic view of their transformation during VF assembly.
Fig. 2.
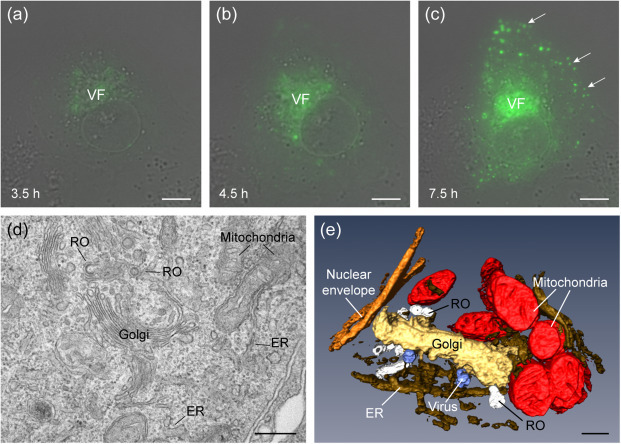
Visualization of a viral factory by live cell imaging, 2D TEM and 3D TEM. (a, b, c) Snapshots from a live cell microscopy movie showing the biogenesis and evolution of a bunyavirus factory (VF). BHK-21 cells were infected with the fluorescent bunyavirus eGFP-Gc-BUNV and transferred to a TCS-SP5 microscope (Leica Microsystems). Images were recorded every 15 min from 1 to 18 h post-infection (hpi). (a) Initial stage of factory assembly. (b) The viral factory grows over time with perinuclear localization. (c) Late stage of infection with the large factory and some secretory vesicles (arrows) with viruses at the plasma membrane. (d) Ultrathin section of viral factories observed after conventional embedding and TEM. BHK-21 cells were adsorbed with bunyavirus and fixed at 10 hpi; Golgi stacks and replication organelles (RO) that contain the replication complexes are shown with mitochondria and endoplasmic reticulum (ER) membranes surrounding the factories. (e) Factory as visualized by TEM of serial sections, 3D reconstruction and image processing. Vero cells were adsorbed with bunyavirus for 10 h. The modified Golgi stack (gold) with spherules/ROs (white) and newly formed viral particles (blue) is surrounded by mitochondria (red) and ER (brown), adjacent to the nuclear envelope (orange). Scale bars = 10 µm in (a, b, c); 500 nm in (d); and 200 nm in (e).
For a detailed analysis of VFs and their components, the high resolution of transmission electron microscopy is needed. The size of most viruses is beyond the resolution limit of optical microscopy and EM provides the necessary resolution to study viruses and their VFs. The most common approach is the study of thin sections of virus-infected cells. These sections must be thin enough (~50–100 nm) for electrons to go through and generate a projection image in the TEM. With advances in sample preparation methods for TEM we are getting close to the visualization of the VF native-like structure. Cryo-preparation techniques such as high-pressure freezing followed by freeze-substitution or frozen-hydrated sections are some of the options to visualize VFs close to their natural state. In addition, ultrastructural imaging can be done either in two dimensions (2D) or three dimensions (3D). The images a TEM generates from a thin section are 2D projections of three-dimensional, much larger structures. In these projections, we can observe considerable details about the elements that build the factory (Fig. 2(d)) but the spatial information within the volume of the structure is lost. Different 3D imaging techniques for biological samples are now available. They are unveiling how different elements compose the VF macro-structure inside cells. Methods such as 3D reconstruction of serial sections (Fig. 2(e)) and electron tomography (ET) are currently applied to study the three-dimensional architecture of viral factories. 3D studies of VFs discovered inter-organelle contacts that do not exist in non-infected cells.
The group of techniques known as correlative light and electron microscopy (CLEM) combines both types of microscopy and allows the selection of particular cells with interesting features detected by (live) light microscopy imaging to study them at higher resolution by TEM. This strategy is very useful to study the different steps of the infection, such as the assembly of the VF and the heterogeneous response of different cells to the virus.
Microscopy studies show when, where and how the VF is built and therefore provide useful information about the most adequate tpi for other studies, such as the quantification of viral genome synthesis with real-time polymerase chain reaction (RT-PCR), viral protein production by western-blot and the production of intracellular and extracellular infectious viruses with titration assays. All these methods work together to identify VF functions in the infected cell. However, to assign particular activities to VF sub-compartments, an in situ molecular mapping of the structures is necessary. Detection of viral and cellular macromolecular complexes in VFs can be done with fluorescent probes and super-resolution light microscopy techniques. Stimulation emission depletion (STED), stochastic optical reconstruction (STORM) and total internal reflection fluorescence (TIRF) microscopy are some of these techniques that produce images at higher resolution than the one imposed by the diffraction limit. Their application in virology is still limited but they have a great potential to study the macromolecular architecture of VFs. At the ultrastructural level, molecular mapping is done with specific primary antibodies and secondary antibodies conjugated with colloidal gold particles in immunogold labeling assays. Recently, clonable tags for EM have been developed. The method known as metal-tagging TEM (METTEM) uses the metal-binding protein metallothionein (MT) coupled with a gold nano-cluster as an electron-dense tag. MT has unveiled the 3D organization of the Tombusvirus polymerase molecules in ROs and the movement of newly synthesized influenza virus ribonucleoproteins from factories to the plasma membrane before viral particle assembly and egress.
To identify the active ROs in infected cells, we count with assays of brome-uridine or brome-deoxiuridine incorporation for RNA or DNA viruses, respectively. These assays can be combined with immunofluorescence and confocal microscopy or immunogold labeling and electron microscopy to localize the sites where the viral polymerases are making new copies of the viral genome. In addition, probes for imaging specific viral RNA molecules are powerful tools to study viral replication and assembly in live cells. For example, fluorescence in situ hybridization (FISH) shows where the viral genome localizes and assembles to form new viral particles. Live FISH is a new technique under development. Never used for VF studies, it might show a dynamic view of viral genome synthesis, transport and assembly in virions.
A very active field of research is the identification of cell factors used by viruses to remodel compartments and build their factories. Gene expression microarrays can give us some clues by showing the genes that are over- or under-expressed in infected cells. Also, because viral non-structural (NS) proteins are usually involved in the biogenesis of ROs and VFs, techniques that detect protein-protein interactions such as yeast two-hybrid (Y2H), co-immunoprecipitation and proximity-dependent biotin identification (Bio-ID) followed by proteomics can catch relevant cell factors that interact with NS viral proteins early in infection. Validation of candidates can be laborious and involves methods such as gene silencing with small interference RNA (siRNA) or gene deletion using the CRISPR-Cas9 technology. Transient expression of viral and cell proteins after transfection with plasmids together with the generation of stable cell lines to control the expression of specific (tagged)-proteins are very useful techniques for these studies. The impact of the over-expression or elimination of the selected candidates in VF assembly and function is analyzed with the morphological and functional studies of VFs described above. When a fluorescent virus is available, another strategy is possible. Cells are infected with the recombinant fluorescent virus and at different tpi fluorescent and non-fluorescent cells are separated by cell sorting. Different cell populations can be then studied by EM, Proteomics, Transcriptomics, and Lipidomics to search for cell factors participating in viral infection in general and VF biogenesis in particular.
Representative Examples of Virus Factories
With the examples of selected DNA and RNA viruses, this section revises some typical viral factories. Most currently known DNA viruses carry out replication and transcription either entirely or partially within the nucleus of the host cell. For these viruses, the nucleus provides the machinery required for particular steps of the viral life cycle. Due to the limited comprehension of the functional architecture of the cell nucleus, the organization of nuclear factories is poorly understood.
Some of the best studied nuclear VFs are the ones of Polyomaviruses (PyV). PyVs are small, non-enveloped DNA viruses that infect mammals and birds. They have also been associated with the development of cancers in their hosts. During PyV infection, viral DNA and capsid proteins concentrate in nuclear bodies, suggesting that these sites may function as virus factories. However, PyV active DNA replication has been located adjacent to these bodies, associated with the recruitment of cellular factors such as DNA repair-related proteins. By electron microscopy and electron tomography, PyV factories are seen as collections of proteinaceous tubular structures and clusters of viruses. The current model proposes that PyV assembles from the ends of these tubules by a budding process and that nuclear bodies may provide the necessary architectural foundation for PyV tubular structures.
Some DNA viruses use also the cytosol for some steps of their life cycle. This is the case of herpesvirus, which causes many different diseases and is associated with human tumors. After entering cells, viral particles are transported to the nuclear envelope and the viral DNA is injected into the nucleus. Viral DNA replication, protein expression and core assembly occur inside nuclear factories. Then, immature viral cores exit the nucleus and enter the trans-Golgi network (TGN), where they acquire their final envelope. Thus, herpesvirus needs the cytosol and Golgi complex to complete the maturation of the virus progeny.
In contrast to most DNA viruses, the infection cycles of poxvirus and mimivirus take place exclusively within the host cytoplasm. Poxviruses such as vaccinia virus (VACV), are large, enveloped DNA viruses that infect vertebrate and invertebrate species. VACV replication requires rearrangement of vimentin filaments to build a vimentin cage, structure that assembles close to the microtubule organizing center (MTOC) and resembles the cellular aggresome. Before the formation of an aggresome-like structure, VACV factories are made of ER-derived membranes that build a mini-nucleus where viral genome replication takes place. Thus, VACV assembles two different factories, the first one is a mini-nucleus for viral genome replication and the second one is an aggresome-like compartment for viral morphogenesis.
Unlike DNA viruses, almost all RNA viruses form factories exclusively in the cytoplasm. These factories are in tight association with host membranes and depending on the virus, they use different cell organelles. Flaviviruses such as hepatitis C virus (HCV), dengue virus (DENV), or Zika virus (ZIKV) are positive-sense RNA viruses and important human pathogens. HCV is the major causative agent of chronic liver disease leading to cirrhosis and cancer. Early in infection, HCV induces a remodeling of ER membranes forming double-membraned vesicles (DMVs), and later on, it induces the formation of multi-membraned vesicles (MMVs) that are composed of several concentric membrane bilayers and accumulate at late tpi. These membranous rearrangements are produced by the action of viral nonstructural proteins. Viral RNA replication seems to be associated with DMVs which constitute the major component of HCV factories. DENV, the causative agent of dengue fever, is another flavivirus that induces rearrangements of ER membranes. EM has shown ER-derived structures with distinct morphologies: convoluted membranes (CMs) and vesicle packets (VPs). Using ET, it was demonstrated that these structures form part of a single ER-derived network. Furthermore, ET has allowed the study of the 3D architecture of DENV-induced ER membrane arrangements, revealing vesicles with pores that could enable the release of newly synthesized viral RNA. In addition, these studies showed the DENV particles budding into ER membranes, directly joined to vesicle pores. ZIKV is an emerging flavivirus that has been linked to abortion and birth defects associated with severe infection of the central nervous system (CNS). Like other flaviviruses, ZIKV induces rearrangements of the ER forming characteristic vesicles and other membranous structures where viral replication and particle assembly occur in a coordinated manner. In addition, newly synthesized viral particles aggregate forming arrays of viruses in the lumen of the ER.
Coronaviruses (CoV) and arteriviruses are also positive-sense RNA viruses that generate membrane alterations in the host cytoplasm. There are common features between their virus factories, despite the significant differences in their morphology. The CoV group includes important human pathogens such as the severe acute respiratory syndrome (SARS) viruses. ET was decisive to reveal that coronaviruses form a unique reticulovesicular membrane network from the ER, where viral replicase subunits and double-stranded RNA (dsRNA), a viral replication intermediate, are both located. The membranous platform is continuous with the rough ER and contains in its lumen numerous vesicles that strongly stain for anti-dsRNA antibody. VP and CM structures have been also detected in association with the ER network. Equine arteritis virus (EAV) is the prototype of the family Arteriviridae, which includes important pathogens of animals. Arteriviruses also modify ER membranes, forming a network characterized by the accumulation of double-membraned sheets and DMVs. The diameter of these DMVs is smaller compared to those induced by CoV, and CMs are not observed. In addition, ET has shown that DMVs associate with each other in the membranous web. All these studies show that flaviviruses, coronaviruses and arteriviruses induce a single ER-derived endomembrane platform where viral replication and assembly are interconnected.
Many positive-sense RNA viruses replicate in single membrane invaginations named spherules that assemble in a variety of cell organelles. For example, togaviruses assemble spherules in lysosomes. The Togaviridae family is composed of two genera which include important animal and human pathogens, such as the Semliki Forest virus (SFV) and rubella virus (RUBV). Togavirus spherules are first formed at the plasma membrane, internalized by endocytosis and finally fused with late endosomes and lysosomes. These modified lysosomes, called cytopathic vacuoles (CPVs), are the central element of the VF that harbors viral replication and morphogenesis. Mitochondria and ER are recruited around the CPVs. In the case of RUBV, the Golgi complex is also recruited and participates in virus morphogenesis.
Other viruses assemble spherules in the ER, the Golgi complex, mitochondria, chloroplasts or peroxisomes. For example, the nodavirus Flock House virus (FHV), which can replicate in insect, plant, mammalian and yeast cells, assembles spherules in mitochondria. ET and 3D visualization have shown that spherules form by invaginations of the outer mitochondrial membrane and communicate with the cytoplasm through a "neck". Tombusviruses that are frequently used as a model to study virus-host interactions, assemble their spherules in the peroxisomes of plant and yeast cells. With advanced EM techniques, it was demonstrated that a cellular protein, the ESCRT (endosomal sorting complexes required for transport) component Vps4 controls the traffic of macromolecules between the spherule and the cytosol.
Negative-sense RNA viruses also build replication factories although their structure and biogenesis are less characterized. Bunyaviruses are an exception because their large VF has been studied in detail by light and electron microscopy. Bunyaviruses constituted a large order of enveloped viruses that includes emerging pathogens of humans, animals and plants. Bunyavirus factories use the Golgi complex, where the ROs are formed and consist of typical spherules connected with atypical cylindrical structures, surrounded by mitochondria and ER membranes. Viral replication takes place in spherules but the role of the cylinders is not understood. Serial sectioning and 3D reconstruction have shown the interactions between the different elements of this complex factory. The analysis of volumes together with immunogold labeling and TEM, suggested that the cylindrical structures probably mediate the transport of essential viral and cell factors throughout the factory. The virus assembly sites locate near spherules with VRCs. Viral particles bud in cis-Golgi membranes and mature in the trans-Golgi and trans-Golgi network (TGN) subcompartments.
From the examples described in this section, we can conclude that the ER that is the largest organelle in eukaryotic cells, is also the favorite compartment for viruses to build the VFs. One of the most spectacular viral-induced ER remodeling is triggered by the human reovirus. This dsRNA virus causes disease in children and young mammals, has been involved in the etiology of the celiac disease and is currently being used as an oncolytic in anti-cancer therapies. The human reovirus uses the ER to build their VFs also known as viral inclusions. To that purpose, two non-structural viral proteins, σNS and µNS bind to the peripheral ER elements and fragment them to make a collection of small membranous tubules and vesicles that aggregate and remain attached to the ER framework. These groups of membranes concentrate and protect the viral core-associated replication complexes and possibly provide also physical support for viral particle assembly.
Viral Factories as Targets for Antiviral Therapies
Emerging and re-emerging viruses constitute a serious public health problem and the need for new broad‐spectrum antivirals is a top priority. Generally, antiviral drug development has focused on targeting specific viral components or on the modulation of the host immune response. Other strategies aim to develop molecules that target host cellular factors needed for viral replication. In this case emergence of resistant viruses is less likely. Moreover, the approaches focused on host cellular pathways used by viruses can favor the development of broad spectrum antivirals. Because different viruses use the same cell compartments and signaling pathways to build their factories, the identification of factors involved in VF biogenesis can provide new ways for antiviral design. For example, molecules targeting lipid transfer proteins (LTPs), enzymes of lipid metabolism, organelle remodeling proteins, and those involved in intracellular transport are good candidates for antiviral therapies. In addition, the current strategy known as drug repurposing is an attractive way to save time by finding molecules already approved to treat other diseases and use them to treat viral infections.
Further Reading
- Altan-Bonnet N. Lipid tales of viral replication and transmission. Trends in Cell Biology. 2017;27:201–213. doi: 10.1016/j.tcb.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson K.D., Bouchet-Marquis C., Heiser K., et al. Virion assembly factories in the nucleus of polyomavirus-infected cells. PLoS Pathogens. 2012;8 doi: 10.1371/journal.ppat.1002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández de Castro I., Volonté L., Risco C. Virus factories: Biogenesis and structural design. Cellular Microbiology. 2013;15:24–34. doi: 10.1111/cmi.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández de Castro I., Sanz-Sánchez L., Risco C. Metallothioneins for correlative light and electron microscopy. Methods in Cell Biology. 2014;124:55–70. doi: 10.1016/B978-0-12-801075-4.00003-3. [DOI] [PubMed] [Google Scholar]
- Fernández de Castro I., Tenorio R., Risco C. Virus assembly factories in a lipid world. Current Opinion in Virology. 2016;18:20–26. doi: 10.1016/j.coviro.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Fernández-Oliva A., Ortega-González P., Risco C. Targeting host lipid flows: Exploring new antiviral and antibiotic strategies. Cellular Microbiology. 2019;21 doi: 10.1111/cmi.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridmann-Sirkis Y., Milrot E., Mutsafi Y., et al. Efficiency in complexity: Composition and dynamic nature of mimivirus replication factories. Journal of Virology. 2016;90:10039–10047. doi: 10.1128/JVI.01319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harak C., Lohmann V. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology. 2015;479–480:418–433. doi: 10.1016/j.virol.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopek B.G., Perkins G., Miller D.J., Ellisman M.H., Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biology. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsafi Y., Fridmann-Sirkis Y., Milrot E., Hevroni L., Minsky A. Infection cycles of large DNA viruses: Emerging themes and underlying questions. Virology. 2014;466–467:3–14. doi: 10.1016/j.virol.2014.05.037. [DOI] [PubMed] [Google Scholar]
- Risco C., Fernández de Castro I., Sanz-Sánchez L., et al. Three-dimensional imaging of viral infections. Annual Review of Virology. 2014;1:453–473. doi: 10.1146/annurev-virology-031413-085351. [DOI] [PubMed] [Google Scholar]
- Romero-Brey I., Bartenschlager R. Membranous replication factories induced by plus-strand RNA viruses. Viruses. 2014;6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse M., de Castro I.F., Fournier G., Naffakh N., Risco C. Metal-tagging transmission electron microscopy and inmunogold labeling on Tokuyasu cryosections to image influenza A virus ribonucleoprotein transport and packaging. Methods in Molecular Biology. 2018;1836:281–301. doi: 10.1007/978-1-4939-8678-1_14. [DOI] [PubMed] [Google Scholar]
- Sachse M., Fernández de Castro I., Tenorio R., Risco C. The viral replication complexes in cells studied by electron microscopy. Adv. Virus Res. 2019;105:1–33. doi: 10.1016/bs.aivir.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strating J.R., van Kuppeveld F.J. Viral rewiring of cellular lipid metabolism to create membranous replication compartments. Current Opinion in Cell Biology. 2017;47:24–33. doi: 10.1016/j.ceb.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wileman T. Aggresomes and pericentriolar sites of virus assembly: Cellular defense or viral design? Annual Review of Microbiology. 2007;61:7905–7912. doi: 10.1146/annurev.micro.57.030502.090836. [DOI] [PubMed] [Google Scholar]
References
Relevant Websites
- https://youtu.be/NKtTv5xVJ0U –Annual Review of Virology. –Viral Manipulation of Plant Host Membranes. [DOI] [PubMed]
- https://youtu.be/57RwV2nkSkU –Flaviviridae replication organelles.
- https://youtu.be/_GrXSClpbDw –Three-dimensional imaging of viral infections.
- https://viralzone.expasy.org/1951–Viral factories. –ViralZone page.


