Abstract
We integrated genomic and bioinformatic analyses, using data from the largest genome-wide association study of cocaine dependence (CD; n = 6546; 82.37% with CD; 57.39% male) and the largest postmortem gene-expression sample of individuals with cocaine use disorder (CUD; n = 36; 51.35% with CUD; 100% male). Our genome-wide analyses identified one novel gene (NDUFB9) associated with the genetic predisposition to CD in African-Americans. The genetic architecture of CD was similar across ancestries. Individual genes associated with CD demonstrated modest overlap across European-Americans and African-Americans, but the genetic liability for CD converged on many similar tissue types (brain, heart, blood, liver) across ancestries. In a separate sample, we investigated the neuronal gene expression associated with CUD by using RNA sequencing of dorsal–lateral prefrontal cortex neurons. We identified 133 genes differentially expressed between CUD case patients and cocaine-free control subjects, including previously implicated candidates for cocaine use/addiction (FOSB, ARC, KCNJ9/GIRK3, NR4A2, JUNB, and MECP2). Differential expression analyses significantly correlated across European-Americans and African-Americans. While genes significantly associated with CD via genome-wide methods were not differentially expressed, two of these genes (NDUFB9 and C1qL2) were part of a robust gene coexpression network associated with CUD involved in neurotransmission (GABA, acetylcholine, serotonin, and dopamine) and drug addiction. We then used a “guilt-by-association” approach to unravel the biological relevance of NDUFB9 and C1qL2 in the context of CD. In sum, our study furthers the understanding of the genetic architecture and molecular neuropathology of human cocaine addiction and provides a framework for translating biological meaning into otherwise obscure genome-wide associations.
SIGNIFICANCE STATEMENT Our study further clarifies the genetic and neurobiological contributions to cocaine addiction, provides a rapid approach for generating testable hypotheses for specific candidates identified by genome-wide research, and investigates the cross-ancestral biological contributions to cocaine use disorder/dependence for individuals of European-American and African-American ancestries.
Keywords: cocaine dependence, cocaine use disorder, genome-wide association study (GWAS), RNA sequencing, multiancestry, GWAS follow-up
Introduction
Neuroscience research has facilitated the identification of genes studied in hypothesis-driven human genetic research, often called “candidate gene studies.” The candidate gene literature proposes numerous associations between genetic variants within neurotransmitter system genes and cocaine use/addiction. However, some experts question the validity of candidate gene research due to a lack of reproducibility (Colhoun et al., 2003; Munafò and Flint, 2009) and encourage the use of hypothesis-free genome-wide methods.
Genome-wide association studies (GWASs) have identified thousands of genetic variants associated with human traits. However, linking molecular mechanisms to GWAS findings is challenging. Significant GWAS results do not generally conform to a priori candidate genes and often reside in non-protein-coding genomic regions (Maurano et al., 2012). Therefore, individual gene variants from GWASs are rarely interpreted with concrete mechanisms. Experimental laboratory studies have unraveled mechanisms for a few GWAS findings (Claussnitzer et al., 2015; Sekar et al., 2016), but these studies are expensive and time intensive, so it is not feasible to apply this line of research for all GWAS findings. Systematic approaches are needed to prioritize individual genes from GWASs for follow-up investigation in specific tissues or cell types. Another important caveat of GWAS research is that most findings are based on individuals of European ancestry/ethnicity (Martin et al., 2019), highlighting a priority to investigate the genetic basis of traits among non-Europeans.
GWASs have discovered four significant genes contributing to predisposition to cocaine dependence [CD; see Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV)]: FAM53B, KCTD20, STK38, and C1qL2 (Gelernter et al., 2014; Huggett and Stallings, 2020). The relevance of these genes to CD is not fully understood. Follow-up investigation in mice revealed that Fam53b might influence cocaine self-administration via midbrain coexpression with Cyfip2 (Dickson et al., 2016), a gene that influences cocaine-induced sensitization (Kumar et al., 2013). Similarly, our previous work found that KCTD20 was associated with human cocaine abuse/dependence through a hippocampal gene coexpression network implicated in synaptic plasticity (Huggett and Stallings, 2020). This work provides a “guilt-by-association” approach to infer the role of newly associated disease genes and helps to contextualize and interpret otherwise ambiguous genetic associations. Given the surplus of publically available bioinformatic data, systems-based computational follow-up may be a fruitful line of inquiry that could help to translate biological meaning to obscure genetic associations.
Despite the rising rates of cocaine and drug-related overdoses in the United States (NIDA, 2020) postmortem brain data on substance use disorders remain limited. The largest cocaine-related human brain sample used RNA-sequencing (RNA-seq) on dorsolateral PFC (dlPFC) neurons from individuals of mixed ancestries (Ribeiro et al., 2017). The PFC is a critical region for the neuropathology of cocaine addiction and plays a role in decision-making and salience attribution, and promotes inhibitory control over drug addiction (Goldstein and Volkow, 2011). Rodent models suggest that PFC glutamate neurons provide “top-down” control of reward circuitry and increase motivation to seek/use cocaine (Kalivas et al., 2005), but little is known regarding the neuroadaptations underlying PFC dysfunction in human cocaine addicts. Ribeiro et al. (2017) identified associations of various immediate early genes (FOS, JUN, and JUNB) with dlPFC neuroadaptations of cocaine use disorder (CUD; see DSM-V) and found one gene coexpression network associated with CUD that was enriched for neuroplasticity processes and GWAS associations for body mass index and obesity. Notably, while, genome-wide research has begun to disentangle the genetic architecture of human traits across ancestries (Peterson et al., 2019), we are aware of no transcriptome-wide studies characterizing potential similarities/differences across ancestries/ethnicities. Future research is warranted to clarify the links between the genetic risk for substance abuse and the neurobiological characteristics of the addicted brain, while also investigating how gene expression generalizes across ethnicities.
This study aimed to unravel the genetic architecture and molecular neuropathology of human cocaine addiction. Integrating genomic and bioinformatic methods, we identified specific genes and tissues associated with the predisposition to CD and characterized PFC neuroadaptations associated with CUD. We translated findings across ancestries and methods, and sought to make human genetic findings more relevant for neuroscientists.
Materials and Methods
Genome-wide analyses
Sample.
We used case-control GWAS summary statistics from the study by Gelernter et al. (2014), which were based on data from 3370 African-Americans (44.18% female; mean age = 41.71 years) and 3176 European-Americans (40.96% female; mean age = 37.35 years). Participants were a part of the Study of Addiction: Genetics and Environment (SAGE) or were recruited via clinical settings in the northeastern United States. Genome-wide analyses were performed separately by ancestry to account for population stratification. GWAS summary statistics corrected for relatedness via generalized estimating equations and adjusted for three ancestral principal components, age, and sex, but not co-occurring substance abuse or other psychiatric comorbidities. All participants reported trying cocaine and 90.39% of African-Americans and 73.96% of European-Americans had a lifetime diagnosis of CD (3+ Symptoms using DSM-IV criteria). In a portion of this sample, measurements of CD yielded high internal reliability (k > 0.80; Pierucci-Lagha et al., 2005), indicating reliable trait measurement. Stringent quality control was applied to the genotypic data of all subjects and imputation was performed using the 1000 Genomes Project reference panel.
Experimental design and analysis—gene-based associations.
To detect specific protein-coding genes underlying the predisposition of CD, we conducted Multi-marker Analysis of GenoMic Annotation (MAGMA, version 1.06; de Leeuw et al., 2015) gene-based association tests by submitting summary statistics to the Functional Mapping and Annotation (FUMA, version 1.1.2) GWAS pipeline (Watanabe et al., 2017). Contrary to GWAS, which performs millions of regressions for all common gene variants across the genome, gene-based associations perform one regression per protein-coding gene and therefore reduce the multiple testing burdens of GWAS and offer more interpretable results. Most protein-coding genes have a multitude of gene variants. MAGMA gene-based tests use a principal components analysis to reduce the numerous variants for a certain gene into a single signal, which is then associated with the trait (de Leeuw et al., 2015). Our gene-based analyses included all single nucleotide polymorphisms (SNPs) within protein-coding regions of the genome (Ensembl version 85) that had a minor allele frequency >1%. In total, our gene-based tests included 18,122 genes for the African-American sample and 18,220 genes for the European-American sample (18,903 shared genes). We compared the results of our previously published gene-based test of CD in European-Americans (Huggett and Stallings, 2020) to the African-American sample and used a Bonferroni correction for multiple testing to determine genome-wide significance (p < 2.7e-6). Note that this standard Bonferroni p value correction (FUMA default) demarks a less significant threshold than the original GWAS (p < 5.0e-8; Gelernter et al., 2014) due to the reduction of tests performed (∼18,000 vs ∼9 million).
To interrogate specific alleles underlying the genetic predisposition to CD, we investigated specific SNPs driving genome-wide significant associations. First, we reported the lead SNP from each genomic region, the total SNPs within each gene, as well as the number of parameters for each gene, which reflects independent linkage disequilibrium blocks within genes. “Causal” SNPs are more likely to confer a biological consequence in protein or transcript function. Leveraging DNA sequencing data from 71,702 individuals, we queried the Genome Aggregation Database (version 3; Karczewski et al., 2019; https://gnomad.broadinstitute.org/) for missense mutations, or SNPs that code for an amino acid substitution, among genome-wide significant gene-based test results. To determine whether a missense mutation was significantly associated with CD, we used a Bonferroni correction for all missense variants within each gene. We also estimated the relationship between particular missense mutations and the lead SNP of a gene using LDlink (Machiela and Chanock, 2015; https://ldlink.nci.nih.gov/), which computes linkage disequilibrium between loci by ancestry.
Our study then refined the focus of gene-based associations with CD from a genome-wide perspective to a candidate systems approach, selecting genes from typically studied neurotransmitter systems. In total, these analyses included 130 genes from GABA, glutamate, acetylcholine, endocannabinoid, dopamine, epinephrine/norepinephrine, and serotonin systems encompassing synthesis, vesicular transport, receptors, degradation, and reuptake genes. Since these classical genes rarely surpass conservative genome-wide significance thresholds, we assessed whether these hypothesis-driven neurotransmitter genes surpassed a nominally significant p value threshold (p < 0.05), as is typically used in the candidate gene literature. Collapsing across ancestries, we tested whether these candidate neurotransmitter genes were enriched for being nominally associated with CD using a Fisher's exact test.
Tissue enrichment.
To identify tissues underlying the genetic pathophysiology of CD, we performed tissue enrichment analyses. These analyses assess where genes underlying the predisposition of a trait might be exerting a functional role. Tissue enrichment analyses identified which tissues a list of input genes demonstrated differential expression (upregulated or downregulated). We assessed tissue enrichment in 53 tissues from hundreds of healthy human samples (GTEx Consortium, 2013) and performed analyses separately by ethnicity- including genes nominally associated with CD (unadjusted p < 0.05; 901 genes in African-Americans and 1008 genes in European-Americans). Tissue enrichment analyses used competitive hypergeometric tests to compare a specific tissue type versus all other tissues and incorporated a Bonferroni multiple-testing correction to ascertain significantly enriched tissues (p < 0.05/53).
Neuron-specific RNA-seq analyses
Sample.
Next, we performed neuron-specific RNA-seq analyses using publically available data from the largest postmortem human brain study on cocaine use (Gene Expression Omnibus #GSE99349). Cocaine users (n = 19; 100% male; 6 African-Americans, 6 European-Americans, and 7 Hispanics or Latinos) died from the toxic effects of chronic cocaine abuse and met the criteria for CUD (see DSM-V criteria). Age-matched and race-matched cocaine-free control subjects (n = 17; 7 African-Americans, 4 European-Americans, and 6 Hispanics or Latinos) were selected from homicides and accidental or cardiac-related deaths and had negative urine screening results for common drugs before death. Case patients and control subjects did not significantly differ on postmortem index (PMI), RNA integrity (RIN), age, or brain pH level, (all | t | > 1.69, all p > 0.100).
Data preparation.
For more details on the sample, tissue preparation, RNA extraction, library construction, and RNA-seq protocol, see the study by Ribeiro et al. (2017). Briefly, dlPFC tissue was extracted from the middle frontal gyrus at the lateral portion of Brodmann's area 46. Fluorescence-activated cell sorting dissociated dlPFC cell types, and neuronal nuclei were isolated/extracted via the mouse anti-NeuN antibody. RNA isolation was conducted via a Direct-zol RNA Miniprep Kit (catalog #R2050, Zymo Research). Indexed libraries were constructed using 10 ng of nuclear RNA from each sample with the Clontech SMARTer Stranded Total RNA-Seq Library Preparation Kit (catalog #634839, Takara). Paired end (2 × 125) RNA-seq was performed using the HiSeq-2000 Sequencing System (Illumina) and resulted in an average of 50,925,315 read pairs per sample.
We preprocessed the RNA-seq data from the study by Ribeiro et al. (2017), via Trimmomatic version 0.36 to eliminate short and low-quality reads (Phred score < 20 or < 100 bases) as well as Illumina adapters, which resulted in an average of 30,486,006 read pairs per sample. We then aligned the RNA-seq data to the hg19 reference genome via the Spliced Transcripts Alignment to a Reference (STAR; Dobin et al., 2013). On average, we had 26,476,583 (SD = 6,173,119) uniquely mapped read pairs per sample, with a mean alignment rate of 86.84% (SD = 5.86%) and observed no significant differences in read alignment between case patients and control subjects (t = 0.668, p = 0.509). Our study used HTSeq software (Anders et al., 2015) to transfer mapped reads into discrete genes/transcripts.
Our reanalysis of the data from the study by Ribeiro et al. (2017) differed in two ways. First, we defined differentially expressed genes with an adjusted p value threshold of Benjamini–Hochberg false discovery rate (FDR) < 0.05. Second, we normalized RNA-seq data with SCnorm, a method that uses quantile regression and seems to properly handle data derived from single-cell types (Bacher et al., 2017). RNA-seq approaches from a single cell (type) differ from regular RNA-seq due to the presence of technical noise (i.e., zero-inflated read counts of genes not expressed in sequenced cells) and may require sensitive statistical care. To test whether SCnorm increased power, we assessed the number of differentially expressed genes identified from this technique compared with a standard normalization method (DESeq2 scale factors). Without covariates, we found just six differentially expressed genes/transcripts (padj < 0.05) using the standard scale factor approach, but identified 250 differentially expressed genes/transcripts (padj < 0.05) via the SCnorm technique. Additionally, we found appreciable evidence for zero-inflated read counts and discovered that SCnorm successfully accommodated for this noise (data are available on request), perhaps stemming from non-neuronal genes/transcripts. Accordingly, the lowest decile of normalized read counts was enriched for cortical astrocytes (padj = 6.92e-4) and oligodendrocytes (padj = 0.002), but not for cortical neuronal cell types (all padj > 0.999), as observed from a cell-specific expression analysis (Dougherty et al., 2010). Thus, we normalized the RNA-seq data with SCnorm (for our differential expression analyses) as it appeared to properly account for technical artifacts and afforded increased statistical power.
Experimental design and analysis—differential expression.
We used DESeq2 (Love et al., 2014) to assess differentially expressed genes/transcripts and to investigate the association of differential expression analyses across ancestries. We used the full sample to identify differentially expressed genes/transcripts (49,496 total genes/transcripts), which controlled for RIN, PMI, age, race (European-American = −1; Hispanic = 0; African-American = 1), blood alcohol content, smoking status (smokers = 1; nonsmokers = 0), and hidden batch effects (two surrogate variables via the svaseq package; Leek, 2014).
To complement our genome-wide analyses, we investigated the association between ancestry-specific differential expression results from African-American (n = 13) and European-American (n = 10) subsamples. Because of low sample size for ancestry-specific differential expression analyses, we did not control for all possible confounds, but adjusted for two common and salient covariates (PMI and age). Log fold change estimates from differential expression analyses are estimated with noise, especially among lowly expressed genes/transcripts. To accommodate for this error/noise and enable transcriptome-wide investigation (e.g., low and high expressed genes/transcripts), our cross-ancestry RNA-seq analysis focused on test statistics from differential expression analyses (DESeq2 Wald statistics), which account for log fold change effect size and standard error (SE) for individual genes/transcripts. Additionally, we selected the genes/transcripts with a differential expression Wald-statistic > |2| in either European-American- or African-American-specific analyses (705 genes/transcripts) and investigated the cross-ancestry correlation of cocaine-related gene/transcripts.
Gene coexpression networks.
Next, our study used a systems-genetics approach to model clusters of genes derived from correlated RNA expression (gene coexpression networks/gene networks). The reader should note that these analyses do not determine gene coexpression networks a priori, but rather create gene networks from the observed RNA-seq data. Specifically, we conducted a signed weighted gene coexpression network analysis (WGCNA; Langfelder and Horvath, 2008), using the same input parameters as our previous work (Huggett and Stallings, 2020). Briefly, we filtered genes/transcripts based on expression level, such that we only included genes/transcripts with an average baseline expression >1 read count per sample, which resulted in a total of 15,178 genes/transcripts for WGCNA modeling. Our WGCNA approach computed Pearson Product-Moment Correlations of normalized RNA expression (log2-counts per million) of all WGCNA genes/transcripts with themselves and weighted these correlations by raising them to the (default) power of 12, which satisfied WGCNA distribution assumptions (scale-free topology = 0.84). Then, using a dynamic tree-cutting algorithm, we split clusters of correlated/coexpressed genes into defined WGCNA gene coexpression networks (minimum module size = 50).
To validate our WGCNA gene networks, we used a Z summary module preservation statistic (Langfelder et al., 2011). Z summary statistics >10 indicate that gene networks are highly robust and reproducible, and Z summary statistics >2 suggest that WGCNA gene networks are weak to moderately reproducible. Our validation approach was based on previous work (Vanderlinden et al., 2013) that incorporates a within-sample and out-of-sample gene network validation technique. Our within-sample gene network validation analysis is indicative of WGCNA network stability and compared the WGCNA networks from the current study to 100 bootstrapped samples from the same dataset (human dlPFC neurons; n = 37). Then to assess whether gene networks were robust in a separate sample, we tested whether our constructed WGCNA gene networks were reproducible in an independent sample using RNA-seq data of hippocampal tissue from human cocaine users/addicts and control subjects (Zhou et al., 2011).
Similar to previous research (Ponomarev et al., 2012), we used an effect size-based approach leveraging test statistics from our full sample differential expression analysis (DESeq2 Wald statistics) to associate gene coexpression networks with CUD. That is, we calculated the average absolute value of Wald statistics for all genes/transcripts within each defined gene coexpression network. The directions of associations were determined by assessing whether mean effect sizes for gene networks were positive or negative. We ascertained significant gene networks via 100,000 permutations. That is, our permutations resampled the absolute values of Wald statistics from all WGCNA genes to approximate a null distribution. We then derived p values by determining the probability that a gene coexpression network had an average absolute Wald statistic in relation to what is expected under the null. We defined a significant association of a gene network with CUD, if it survived a Bonferroni correction (p < 0.05/number of WGCNA gene networks) and demonstrated enrichment for differentially expressed genes (FDR < 0.05).
Functional annotation.
We functionally annotated our RNA-seq results via the Database for Annotation, Visualization, and Integrated Discovery (DAVID version 6.8; Huang et al., 2009) and queried for enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, biological processes (BPs), and/or molecular functions (MFs). To control for false positive results, we required significant enrichment to survive correction for multiple testing (FDR < 0.05) and adjusted for the “background distribution” by incorporating a list of genes that were included for each analysis. We uploaded our results to GeneWeaver (https://www.geneweaver.org/; Baker et al., 2012), which can be found by searching the reported ID numbers (GS#).
Results
Genome-wide analyses
Gene-based associations
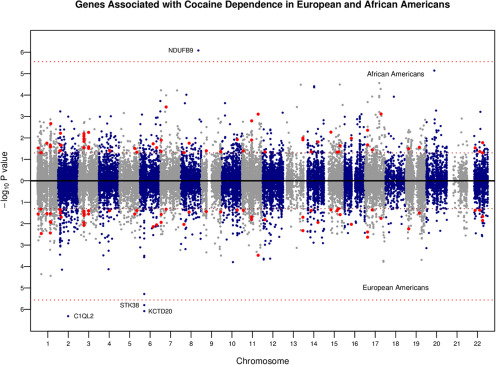
To identify specific genes underlying the predisposition to CD, we conducted gene-based association tests. Figure 1 shows the Miami plot visualizing the results of our gene-based associations with CD for African-Americans and European-Americans. Extending our previous gene-based associations with CD among European-Americans (Huggett and Stallings, 2020), we identified one novel genome-wide significant association with CD in African-Americans (p = 8.27e-07), the NADH:ubiquinone oxidoreductase subunit B9 gene (NDUFB9), but not in European-Americans (p = 0.910). The NDUFB9 gene is part of the inner mitochondrial membrane and plays a role in oxidative phosphorylation, but the relevance of this gene in the context of cocaine addiction is not known, warranting further investigation. To investigate specific loci underlying our genome-wide significant associations, we reported the most significant SNP (lead SNP) of each region and examined missense mutations for each gene associated with CD. After correction for multiple testing, we found significant associations between a missense mutation in the NDUFB9 gene (rs34095749) with CD in African-Americans and a missense mutation in the KCTD20 gene (rs2239808) with CD in European-Americans (Table 1).
Figure 1.
Miami plot visualizing results from gene-based association analyses. Each dot represents an individual protein-coding gene, the x-axis denotes chromosome number, and the y-axis shows the −log10 p value. African-American results are displayed on top and European-American results (Huggett and Stallings, 2020) are shown on the bottom. The dashed red line represents genome-wide significance, and the dashed brown line represents an unadjusted/nominal p value threshold <0.05. Red dots are genes nominally significant in both African-Americans and European-Americans.
Table 1.
SNP associations with CD
| SNPs associated with the genetic predisposition to cocaine dependence by ancestry |
||||||
|---|---|---|---|---|---|---|
| NDUFB9 | C1qL2 | KCTD20 and STK38 | ||||
| Ancestry | AA | EA | AA | EA | AA | EA |
| SNPs (n) | 174 | 205 | 10 | 74 | 215 | 174 |
| Parameters (n) | 51 | 29 | 4 | 14 | 38 | 17 |
| Lead SNP | rs77422927 | rs13020121 | rs9470273 | |||
| Minor allele | C | A | T | |||
| Minor allele frequency | 0.021 | 0.096 | 0.2247 | 0.30025 | 0.3192 | 0.21828 |
| psnp_Lead | 6.42E-06 | 0.963 | 0.902 | 2.22E-06 | 0.0253 | 1.42E-06 |
| Direction of effect | + | + | + | − | + | + |
| Missense SNP | rs34095749 | NA | rs2239808 (KCTD20) | |||
| Minor allele | T | NA | C | |||
| Minor allele frequency | 0.013 | 0.050 | NA | NA | 0.4206 | 0.21439 |
| Missense SNP | Proline_157_Serine | NA | NA | Serine_171_Threonine | ||
| psnp_Missense | 0.00568 | 0.841 | NA | NA | 0.0501 | 1.28E-05 |
| LD with lead SNP (R2) | 0.4917 | 0.5154 | NA | NA | 0.609 | 0.9629 |
| Direction of effect | + | + | NA | NA | + | + |
We collapsed KCTD20 and STK38 into a single category because they stem from the same genomic region. AA, African-American ancestry; EA, European-American ancestry. The number of parameters represents the number of independent signals tested within a protein-coding gene and differ across ancestries due to disparate LD patterns. We estimated the linkage disequilibrium patterns of missense variants with lead SNPs using LDlink, and selecting the African-American and CEU (Northern Europeans from Utah) reference panels. Note that C1qL2 only had one missense mutation but was not tested or included in the genome-wide association study on cocaine dependence due to low minor allele frequency across ancestries (<1%).
Using a nominally significant threshold (p < 0.05), our gene-based test found 901 and 1008 genes associated with CD in African-Americans (GeneWeaver ID# GS357670) and European-Americans (GeneWeaver ID# GS357669), respectively. We found a small, but significant, association between gene-based associations (Z statistics) across European-Americans and African-Americans (B = 0.017, SE = 0.008, p = 0.024; R2 = 0.0002) and observed 59 genes (p < 0.05) that were nominally associated with CD in both ancestries.
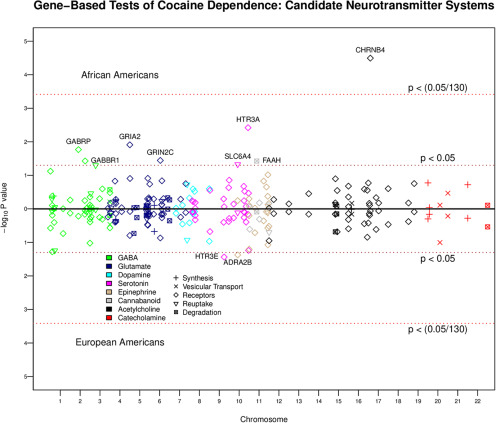
Next, we investigated gene-based associations with CD for the 130 candidate neurotransmitter system genes commonly studied with cocaine use/addiction. Of these genes, we found 10 nominally significant associations with CD from GABA, glutamate, endocannabinoid, serotonin, norepinephrine, and acetylcholine genes (Fig. 2). The most significant candidate genetic association with CD (in African-Americans) came from the CHRNB4 gene (SNPs = 422, Z = 4.00, p = 3.199e-05), which resides in a validated gene cluster for CD (Grucza et al., 2008) as well as nicotine dependence (Saccone et al., 2009). Despite the prominence of dopamine in the candidate gene literature, we found no dopamine genes to be associated with CD (all p > 0.108). Candidate neurotransmitter genes were not enriched to be (nominally) associated with CD [odds ratio (OR) = 0.73; 95% CI = 0.34, 1.40; p = 0.465]. In other words, candidate neurotransmitter genes were no more likely to be (nominally) associated with CD than we would expect by chance.
Figure 2.
Miami plot showing the associations of 130 genes from candidate neurotransmitter systems. Each gene is color coded by neurotransmitter type, and the different shapes represent the different parts of the system. The x-axis denotes chromosome number, and the y-axis shows the −log10 p value with African-Americans displayed on top and European-Americans shown on bottom. The red dashed line represents the Bonferroni correction for multiple testing (p < 0.05/130), and the brown dashed line represents the unadjusted/nominal p value threshold < 0.05.
Tissue enrichment
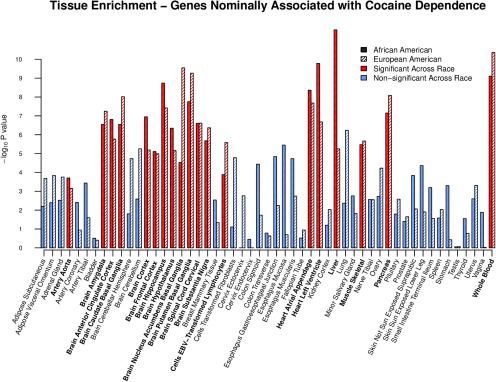
To find tissues implicated in the genetic etiology of CD, we performed tissue specificity/enrichment analyses of the genes nominally associated with CD. Genes nominally associated with CD were enriched among numerous tissue types (Fig. 3). Despite minimal overlap of individual genes associated with CD across ancestries, 70.37% of significantly enriched tissues in African-Americans were also significantly enriched in European-Americans (padj < 0.05). Tissue overlap across ancestry exceeded what we would expect by chance alone (OR = 3.65; 95% CI = 1.05, 13.70; p = 0.029). The replicated tissue types across ancestries tag plausibly implicated tissues in the genetic etiology of CD, including the heart, liver, blood, and most brain regions, and highlight various tissues for follow-up investigation.
Figure 3.
The implicated tissue types based on genes nominally associated with cocaine dependence (CD) separately by ancestry are shown. The x-axis shows all tissue types (GTEx) sorted alphabetically, and the y-axis represents the –log10 p value. Solid boxes denote results from the African-American analysis, and dashed boxes show European-American results from the study by Huggett and Stallings (2020). Red bars show replicated tissue types that were significantly enriched (padj < 0.05) across both ancestries. The labels of replicated tissues are emphasized in bold text on the x-axis.
Neuron-specific RNA-seq analyses
Differential expression
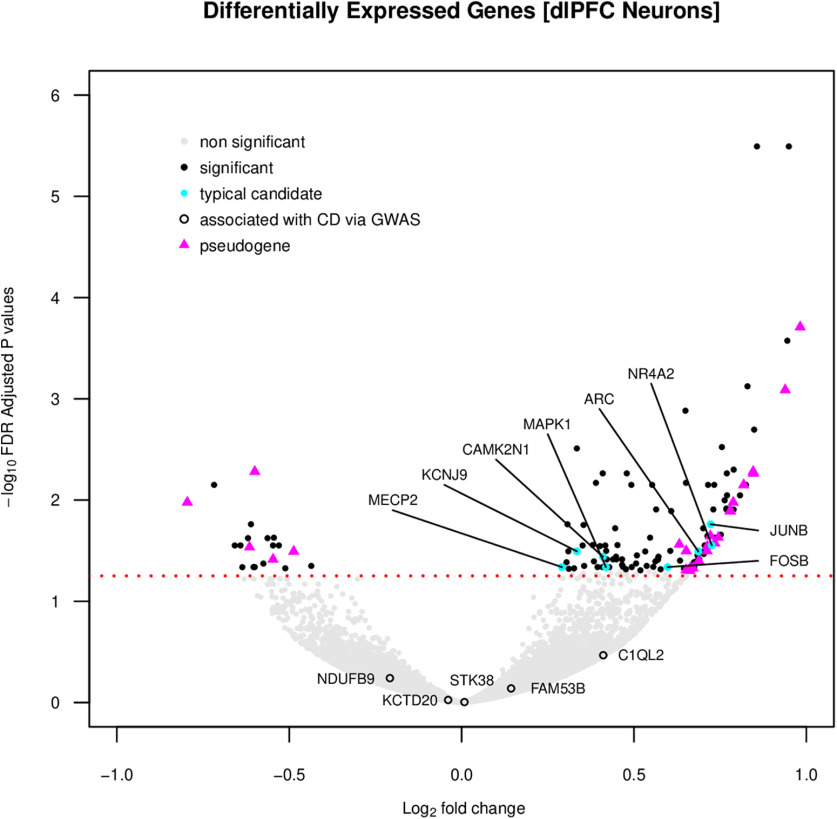
To follow-up genome-wide associations with CD, we used publically available data from dlPFC neurons from individuals with CUD (n = 19) and cocaine-free control subjects (n = 17; Ribeiro et al., 2017). After successful data normalization and adjustment for covariates, we found 133 differentially expressed genes/transcripts (all padj < 0.05; Fig. 4; GeneWeaver ID# GS357661). Similar to Ribeiro et al. (2017), 42.86% of differentially expressed genes/transcripts were noncoding, and, of these, pseudogenes were the most abundant, including 15 pseudogenes derived from inner mitochondrial membrane parent genes. Given that most noncoding transcripts lack detailed functional characterization, perhaps it is not surprising that differentially expressed genes/transcripts were not enriched for any KEGG pathways, BPs, or MFs (all padj > 0.089), although we did identify some typical candidates for cocaine use/dependence. That is, consistent with previous research, we found increased expression of FOSB (Larson et al., 2010), JUNB (Guez-Barber et al., 2011), ARC (Zavala et al., 2008; Salery et al., 2017), MECP2 (Im et al., 2010; Deng et al., 2014), NR4A2 (López et al., 2019), KCNJ9/GIRK3 (Rifkin et al., 2018; McCAll et al., 2019), MAPK1 (Cahill et al., 2016), and CAMK2N1 (Ribeiro et al., 2018). These genes represent various “immediate early genes” whose expression is induced by cocaine, intracellular signaling cascades that modulate neural responsiveness, and nuclear epigenetic transcripts that perturb the expression of numerous genes. No genome-wide significant association with CD (FAM53B, C1qL2, KCTD20, STK38, or NDUFB9) was significantly differentially expressed in dlPFC neurons (all | log2 fold change | < 0.411, all p > 0.026, all padj > 0.341).
Figure 4.
Volcano plot showing genes/transcripts that are expressed differently in human PFC neurons between control subjects (n = 17) and individuals with CUD (n = 19). Each dot represents a gene/transcript. The x-axis denotes the log2 fold change with positive values corresponding to increased expression in those with CUD. The y-axis shows the –log10 FDR-adjusted p value, and all genes above the red dashed line survive correction for multiple testing (133 gene/transcripts; padj < 0.05). We labeled all genes significantly associated with the genetic predisposition to CD and highlighted significantly differentially expressed genes/transcripts previously implicated in cocaine use and the most abundant noncoding transcripts (pseudogenes).
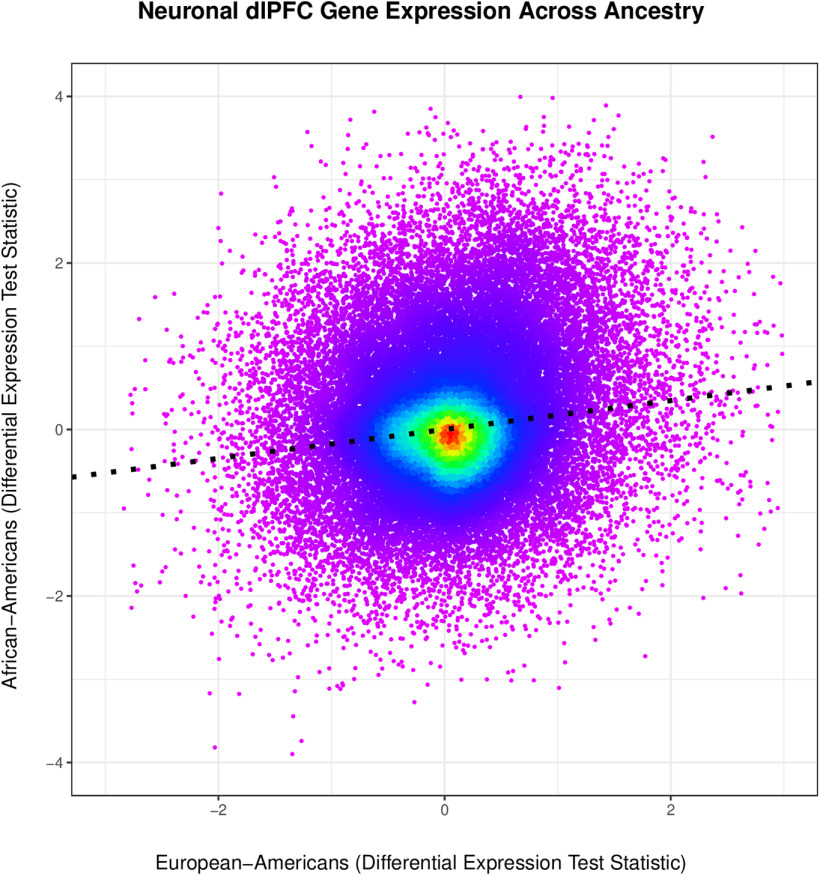
To complement our genome-wide analyses, we explored whether neurotranscriptomic associations with CUD generalized across European-Americans and African-Americans. After covariate adjustment, we found one gene that was significantly differentially expressed in European-Americans (PAX8-AS1: log2 fold change = −7.90, padj = 4.36e-5). In African-Americans, we found 37 significant differentially expressed genes; the most significant was the CHRNG gene (log2 fold change = 30.00, padj = 4.29e-30). While the top associations were different across ancestry, we found that the transcriptome-wide differential expression results significantly correlated across ancestries (r = 0.174, p < 2.e-16; Fig. 5). This association persisted after selecting cocaine-related genes/transcripts (r = 0.332, p < 2.e-16; 705 genes/transcripts).
Figure 5.
Heat scatter plot depicting the correlation of neuronal dlPFC gene expression associated with CUD from African-Americans (n = 13) and European-Americans (n = 10). The x-axis shows the Wald statistics from the European-American differential expression analysis, and the y-axis represents the Wald statistics from the African-American differential expression analysis. Each dot represents a specific gene/transcript, and the bright red color shows the highest frequency, whereas the light purple/pink indicates the lowest frequency of genes/transcripts. The dashed black line highlights the Pearson product correlation of gene expression across ethnicities (r = 0.174, p < 2e-16).
Gene coexpression networks
Next, we modeled systems of coexpressed genes (gene networks) using WGCNA. Similar to previous WGCNA results with these data (Ribeiro et al., 2017), we constructed 12 gene coexpression networks, each of which is arbitrarily assigned to a color. To evaluate the stability and validity of our gene coexpression networks, we used a standard network preservation technique (Langfelder et al., 2011) to assess within-sample and out-of-sample gene network reproducibility. Our analyses suggest that our WGCNA networks were highly reproducible/valid within sample (e.g., stable; all Z summary > 16.19) and, except for the tan (Z summary = 9.75) and yellow (Z summary = 0.47) gene networks, were valid in an independent RNA-seq sample (e.g., robust; all Z summary > 12.67) of hippocampal tissue from human cocaine users and control subjects (Huggett and Stallings, 2020).
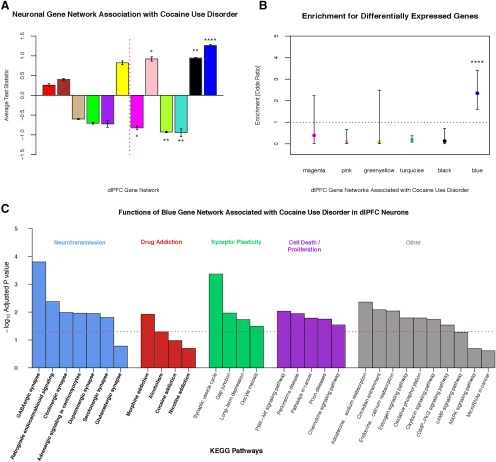
After validating our WGCNA gene coexpression networks, we associated these networks with CUD. Using an effect size-based approach and permuting p values, we found six gene networks associated with CUD (all padj < 0.049; Fig. 6A). We subsequently tested significantly associated WGCNA gene networks for the enrichment of differentially expressed genes/transcripts (133 genes). We found significant enrichment of differentially expressed genes among one WGCNA network, the blue gene network (2735 genes; Fig. 6B; GeneWeaver ID# GS357662). Thus, the blue gene network was robustly associated with CUD and selected for follow-up investigation.
Figure 6.
A, The x-axis shows the twelve WGCNA gene coexpression networks. The y-axis shows the absolute value of Wald statistics (from whole-sample differential expression analysis) of all genes within a defined/discrete WGCNA network. The directions of associations were determined by assessing whether mean effect sizes for gene networks were positive or negative. All WGCNA gene networks to the right of the dashed red line are significantly associated with cocaine use disorder (*padj < 0.05; **padj < 0.01; ***padj < 0.001; ****padj < 0.0001). B, The six associated WGCNA gene networks were subsequently tested for the enrichment of the 133 differentially expressed genes in dlPFC neurons. The y-axis represents the odds ratio calculated by a two-sided Fisher's exact test. Only the blue gene network demonstrated significant enrichment and was selected for follow-up investigation. ****p < 0.0001. C, Potential functions of blue gene network via functional annotation analysis of pathways from KEGG. We picked 30 representative functions/pathways and grouped them into five domains, which are labeled by colors.
The blue gene network recapitulated many molecular processes and was significantly enriched for 31 KEGG pathways (padj < 0.05; Fig. 6C). Similar to the study by Ribeiro et al. (2017), our blue gene network was enriched for neuroplasticity processes and also overrepresented for various neurotransmitter signaling pathways, morphine addiction, intracellular signaling, and circadian entrainment. Note that other KEGG drug addiction pathways (nicotine addiction, alcoholism, and cocaine addiction) approached significant enrichment (all p values = 0.007–0.054; all padj values = 0.051–0.201). Additionally, the blue gene network was enriched for the 130 candidate neurotransmitter system genes (OR = 2.51; 95% CI = 1.61, 3.83; p = 3.175e-05).
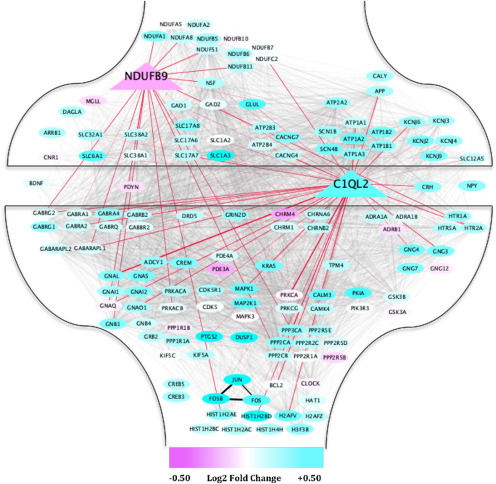
We then assessed the overlap between genetic predispositions to CD and the blue gene network robustly associated with CUD. Of the five genome-wide significant associations with CD, our analyses identified the NDUFB9 and C1qL2 genes to be central entities [>50th percentile of module membership (kME); kME > 0.58] of the blue gene network. To better understand the role of NDUFB9 and C1qL2 in the context of cocaine addiction, we visualized their coexpression patterns with the blue network genes annotated for neurotransmitter signaling and drug addictions (Fig. 7). Of particular note, we found that our data-derived blue gene network recapitulated previously established connections between FOSB and JUN genes, which are thought to perpetuate chronic cocaine/drug-seeking behavior (Nestler et al., 2001), and further highlights the validity of the coexpression patterns from this gene network.
Figure 7.
The genes from the blue gene network significantly enriched for drug addiction and neurotransmission from KEGG (2019) and their relation to the genes associated with the predisposition to CD (in triangles) are shown. Coexpression patterns with NDUFB9 and C1qL2 are highlighted in red. Only coexpression patterns above a weighted r > 0.05 are shown. Genes shown in cyan represent increased expression in dlPFC neurons for those with CUD, and those in magenta represent decreased expression.
We used coexpression patterns in the blue gene network to better understand biological functions of NDUFB9 and C1qL2 with cocaine use via a guilt-by-association approach (Oliver, 2000). Guilt-by-association analyses are commonly used to unravel the biological role of new disease genes and are based on the principle that if genes are highly associated with each other (e.g., coexpressed), they are more likely to share a function (van Dam et al., 2018). In our guilt-by-association technique, we selected the most highly coexpressed genes (weighted r > 0.05 or raw r > 0.78) separately for NDUFB9 and C1qL2 within the blue gene network and then investigated enrichment for biological processes, molecular functions, and KEGG pathways. Our guilt-by-association analyses indicated that the NDUFB9 gene might play a role in cell death, synaptic plasticity, and cell adhesion (Table 2; all padj < 0.037). Highly coexpressed genes with C1qL2 were significantly enriched for neurotransmitter signaling, drug response, synaptic plasticity, cell proliferation, and neurodevelopment (Table 2; all padj < 0.045).
Table 2.
Guilt-by-association analyses: inferring function of NDUFB9 and C1qL2 with CUD
| Potential functions of NDUFB9 and C1qL2 | |||
|---|---|---|---|
| NDUFB9 | C1qL2 | ||
| Biological processes, molecular function or KEGG pathways | padj | Biological processes, molecular function or KEGG pathways | padj |
| Neurodegeneration/cell death | Neurotransmitter signaling | ||
| Phagosome acidification | 0.0056 | GABAergic synapse | 2.4e-7 |
| Parkinson's disease | 0.0156 | Serotonergic synapse | 4.5e-4 |
| Alzheimer's disease | 0.0299 | Cholinergic synapse | 6.2e-4 |
| Huntington's disease | 0.0320 | Glutamatergic synapse | 0.0441 |
| Synaptic plasticity | Ion channels and drug addiction | ||
| Synaptic vesicle cycle | 6.3e-4 | Aldosterone-regulated sodium reabsorption | 0.0013 |
| Cadherin binding involved in cell-cell adhesion | 0.0010 | Alcoholism | 0.0032 |
| GTPase activity | 0.0016 | Response to drug | 0.0041 |
| GTP binding | 0.0067 | Nicotine addiction | 0.0265 |
| Cell-to-cell adhesion | 0.0170 | Potassium ion import | 0.0428 |
| Other processes | Neurodevelopment and synaptic plasticity | ||
| Oxidative phosphorylation | 2.6e-5 | Small GTPase mediated signal transduction | 2.2e-5 |
| Protein binding | 7.9e-4 | Positive regulation of cell proliferation | 0.0177 |
| Endocrine-regulated calcium reabsorption | 0.0367 | Nervous system development | 0.0416 |
Our guilt-by-association approach assesses the function of genes/transcripts that are highly coexpressed with NDUFB9 and C1qL2 in the blue gene network associated with CUD and assesses their enrichment for biological processes, molecular functions, and KEGG pathways using DAVID (Huang et al., 2009). We selected the most highly coexpressed genes with NDUFB9 (300 genes/transcripts) and C1qL2 (694 genes/transcripts) in the blue gene network by using an arbitrary coexpression threshold (weighted r > 0.05; raw r > 0.78).
Discussion
We extend previous genome-wide research (Gelernter et al., 2014; Huggett and Stallings, 2020) that identified four genes significantly associated with CD (C1qL2, FAM53B, KCTD20, STK38) by discovering one novel gene (NDUFB9) implicated in the genetic liability to CD for African-Americans. Our study highlights associations between two missense mutations and CD that may interfere with the product of the NDUFB9 and KCTD20 genes. Similar to other psychiatric genetic research (Johnson et al., 2017; Border et al., 2019), we found minimal evidence indicating that genes from candidate neurotransmitter systems contribute to the genetic predisposition of CD. Genome-wide significant genes associated with CD were not differentially expressed in dlPFC neurons between individuals with CUD and cocaine-free control subjects. However, NDUFB9 and C1qL2 were central parts of a gene coexpression network associated with CUD and exhibited coexpression with relevant drug addiction genes. So, while most GWAS findings tend not correspond with prehypothesized targets, they may still play a broader role in biologically relevant systems. Similarly, genome-wide associations with psychiatric traits (including alcohol dependence) demonstrated appreciable overlap with PFC gene coexpression networks associated with these traits and corresponded to neuronal, synaptic, and mitochondrial functions (Gandal et al., 2018; Kapoor et al., 2019).
Our study suggests common biological contributions to cocaine addiction across ancestries. Similar to other substance dependence research (Brick et al., 2019), we found that the individual genetic predispositions to CD demonstrated (modest) genetic overlap across African-Americans and European-Americans. Robust across ancestries, we discovered that the genetic liability of CD manifested as a multiorgan phenomenon involving the heart, liver, blood, and brain. Using RNA-seq from PFC neurons, we identified convergence of cocaine-related gene expression across African-Americans and European-Americans, albeit with small to moderate effect sizes. One potential reason for the modest magnitudes of these associations is that rates of psychiatric comorbidities between cocaine-abusing European-Americans and African-Americans seem to differ (Petry, 2003), but many other factors could be at play. To our knowledge, this is the first study to assess the cross-ancestry transcriptome-wide neurodiversity/similarity for a psychiatric trait, making interpretations difficult.
We found evidence of disrupted GABA, but not glutamate, neurotransmitter signaling in dlPFC neurons of human cocaine addicts (blue gene network; Fig. 6). PFC GABAergic signaling is sparsely studied in rodent models of cocaine use, but some evidence suggests that GABA regulates prefrontal disinhibition (Cass et al., 2013). We discovered that various GABA genes (GABBR2, GABRA1, GABRA4, GABRB2, GABARAPL1, GABARAPL2) were core elements (“hub genes”; top 10% of gene network connectivity) of PFC network function for individuals with CUD. That is, GABAergic genes demonstrated very high coexpression/connectivity patterns with other genes in the blue gene network, suggesting that GABAergic transmission plays a critical, yet unappreciated, modulatory role of PFC neurons in disordered cocaine users.
The blue gene network associated with CUD in the PFC suggests that catecholamine, acetylcholine and endocannabanoid signaling play an important role in the neuropathology of cocaine addiction. Specifically, PFC DRD5 activity may mediate executive functioning (Carr et al., 2017) and impulsive decision-making (Loos et al., 2010) and HTR1A as well as ADRA1A might regulate PFC glutamate and GABA transmission and various cocaine-related behaviors (Mitrano et al., 2012; Howell and Cunningham, 2015). Particular nicotinic (CHRNA6, CHRNB2) and muscarinic acetylcholine subunit genes (CHRM1, CHRM4) we found to be associated with CUD have previously been implicated in rodent cocaine research (Carrigan and Dykstra, 2007; Dencker et al., 2012; Sanjakdar et al., 2015) and might govern selective attention and promote incentive salience to drugs/drug-related cues (Williams and Adinoff, 2008). Cocaine has also been found to alter the expression of endocannabanoid genes/receptors in the mouse PFC (Bystrowska et al., 2019), which could facilitate the strength of connections between PFC neurons (Kasanetz et al., 2013). Overall our study utilized human brain data that corroborated specific genes and pathways studied in animal models of cocaine use and other relevant molecular endophenotypes.
The combination of genomic and bioinformatics techniques may help to contextualize and interpret nebulous genetic associations with human traits. NDUFB9 is a subunit of the inner mitochondrial complex I. Evidence indicates that cocaine inhibits complex I of the inner mitochondrial membrane (Cunha-Oliveira et al., 2013), which is similar to other genetic associations with substance use/dependence that implicate binding targets of specific drugs. Mitochondrial complex I is thought to mediate altered energy metabolism and cocaine-induced neurotoxicity (Dey and Snow, 2007; Pereira and Cunha-Oliveira, 2017) and is consistent with our guilt-by-association results, suggesting that NDUFB9 may be involved in neurodegeneration and ATP production (oxidative phosphorylation). Additionally, analogous to research highlighting the role of mitochondria in drug addiction (Sadakierska-Chudy et al., 2014), our guilt-by-association analyses suggest that NDUFB9 could be involved in cell death, synaptic plasticity, and calcium signaling. NDUFB9 is not the only mitochondrial gene implicated in cocaine addiction. We found 26 different mitochondrial inner membrane genes within the blue gene network associated with CUD, including 12 NDUF subunits, suggesting links between cocaine use and broad mitochondrial functioning. Accordingly, various mitochondrial genes have demonstrated associations with human cocaine abuse/dependence in the dlPFC (Lehrmann et al., 2003), hippocampus (Zhou et al., 2011), and midbrain (Bannon et al., 2014). Despite the mounting evidence, very little is known regarding the relation between mitochondrial genes and cocaine or drug use behavior. One study indicates that mitochondrial genes may contribute to cocaine withdrawal, as they observed differential expression of 40 mitochondrial genes in the PFCs of mice experiencing protracted abstinence after chronic high doses of cocaine use (Li et al., 2017).
The C1qL2 gene is secreted from the innate immune system and is thought to modulate trans-synaptic glutamatergic connections (Evans et al., 2019). Similar to previous work (Matsuda, 2017), we identified C1qL2 to be coexpressed with C1qL3 and found that C1qL2 may regulate glutamate receptor signaling (Table 2). Extending this research, we hypothesize and provide novel evidence that C1qL2 may be involved in broader neurotransmitter signaling (GABA, acetylcholine, and serotonin), ion transport (K+/Na+), neurodevelopment, and various drug addiction pathways. C1qL2 may be a particularly tantalizing candidate for follow-up, as it is implicated in typical biological processes underlying cocaine use, is highly conserved across species, and is differentially expressed in mouse models of cocaine use (Walker et al., 2018). Overall, we prioritize a specific cell-type for follow-up investigation (neurons) and propose specific biological roles/hypotheses for otherwise obscure genomic associations with cocaine addiction.
This study should be interpreted with the following limitations. While we used the largest GWAS of cocaine addiction to date, our (highly) selected sample had uneven case/control ratios and was not large by contemporary standards; thus, the estimates from this study were approximate. The gene-based associations we observed with CD barely surpassed genome-wide significance, warranting larger studies to replicate these findings. Although only analyzing individuals who have used cocaine may have enhanced the power to identify genes underlying the predisposition to CD (Cabana-Domínguez et al., 2019; Polimanti et al., 2020). Our tissue enrichment findings indicated plausible tissue types for cocaine addiction, suggesting the importance of follow-up among multiple tissue types; however, not all tissues seemed directly relevant for CD (e.g., muscle/skeletal) and certain genes may exert tissue-specific functions. Tissue-enrichment analyses used GTEx samples, which included mostly Caucasians and may complicate our cross-ancestry comparisons. Our RNA-seq design cannot disentangle whether findings are attributed to chronic cocaine use, acute cocaine toxicity, or psychiatric comorbidities; but it is reassuring to detect some usual suspects in the realm of cocaine addiction. While our RNA-seq results are theoretically specific to neurons, they do not distinguish between types of neurons and also included various genes/transcripts that are non-neuronal (e.g., glial genes).
In conclusion, our study translates genetic findings across methods and ancestries using independent samples. We identified significant convergence across ancestries for the genome-wide and transcriptome-wide associations with cocaine addiction. Neurotransmitter genes generally demonstrated little contribution to the genetic architecture of CD, but were prominent features underlying the neuropathology of CUD. Significant genome-wide associations with CD were linked to broad systems of genes associated with CUD in PFC neurons. Ultimately, our study represents a proof-of-principle that uses hypothesis-free methods for generating testable hypotheses regarding the role of genes detected by GWASs. We believe that this line of research provides an important alternative approach for validating genetic associations especially when no genomic replication data exist. Our study may also serve a supplemental purpose for experimental researchers to help distill lists of genes from GWASs in the context of particular tissues and cell types, while also providing molecular interpretations for otherwise obscure genetic associations.
Footnotes
This work was supported by the National Institutes of Health (Grant T32-DA-17637-14) and the National Institute on Drug Abuse (NIDA; Grant DA-035804); and in part by NIDA Grant P60-DA-11015. We thank Dr. Joel Gelernter and Dr. Henry Kranzler for giving us access to their genome-wide association summary statistics on cocaine dependence, and the Gene Expression Omnibus for providing open access to the single-cell RNA-sequencing data used in this study.
The authors declare no competing financial interests.
References
- Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher R, Chu L-F, Leng N, Gasch AP, Thomson JA, Stewart RM, Newton M, Kendziorski C (2017) SCnorm: robust normalization of single-cell RNA-seq data. Nat Methods 14:584–586. 10.1038/nmeth.4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Jay JJ, Bubier JA, Langston MA, Chesler EJ (2012) GeneWeaver: a web-based system for integrative functional genomics. Nucleic Acids Res 40:D1067–D1076. 10.1093/nar/gkr968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon MJ, Johnson MM, Michelhaugh SK, Hartley ZJ, Halter SD, David JA, Kapatos G, Schmidt CJ (2014) A molecular profile of cocaine abuse includes the differential expression of genes that regulate transcription, chromatin, and dopamine cell phenotype. Neuropsychopharmacology 39:2191–2199. 10.1038/npp.2014.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Border R, Johnson EC, Evans LM, Smolen A, Berley N, Sullivan PF, Keller MC (2019) No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. Am J Psychiatry 176:376–387. 10.1176/appi.ajp.2018.18070881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick LA, Keller MC, Knopik VS, McGeary JE, Palmer RHC (2019) Shared additive genetic variation for alcohol dependence among subjects of African and European ancestry. Addict Biol 24:132–144. 10.1111/adb.12578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystrowska B, Frankowska M, Smaga I, Niedzielska-Andres E, Pomierny-Chamioło L, Filip M (2019) Cocaine-induced reinstatement of cocaine seeking provokes changes in the endocannabinoid and N-acylethanolamine levels in rat brain structures. Molecules 24:1125 10.3390/molecules24061125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabana-Domínguez J, Shivalikanjli A, Fernàndez-Castillo N, Cormand B (2019) Genome-wide association meta-analysis of cocaine dependence: shared genetics with comorbid conditions. Prog Neuropsychopharmacol Biol Psychiatry 94:109667. 10.1016/j.pnpbp.2019.109667 [DOI] [PubMed] [Google Scholar]
- Cahill ME, Bagot RC, Gancarz AM, Walker DM, Sun H, Wang Z-J, Heller EA, Feng J, Kennedy PJ, Koo JW, Cates HM, Neve RL, Shen L, Dietz DM, Nestler EJ (2016) Bidirectional synaptic structural plasticity after chronic cocaine administration occurs through Rap1 small GTPase signaling. Neuron 89:566–582. 10.1016/j.neuron.2016.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr GV, Maltese F, Sibley DR, Weinberger DR, Papaleo F (2017) The dopamine D5 receptor is involved in working memory. Front Pharmacol 8:666. 10.3389/fphar.2017.00666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrigan KA, Dykstra LA (2007) Behavioral effects of morphine and cocaine in M1 muscarinic acetylcholine receptor-deficient mice. Psychopharmacology (Berl) 191:985–993. 10.1007/s00213-006-0671-1 [DOI] [PubMed] [Google Scholar]
- Cass DK, Thomases DR, Caballero A, Tseng KY (2013) Developmental disruption of gamma-aminobutyric acid function in the medial prefrontal cortex by noncontingent cocaine exposure during early adolescence. Biol Psychiatry 74:490–501. 10.1016/j.biopsych.2013.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, Abdennur NA, Liu J, Svensson P-A, Hsu Y-H, Drucker DJ, Mellgren G, Hui C-C, Hauner H, Kellis M (2015) FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med 373:895–907. 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colhoun HM, McKeigue PM, Smith GD (2003) Problems of reporting genetic associations with complex outcomes. Lancet 361:865–872. 10.1016/S0140-6736(03)12715-8 [DOI] [PubMed] [Google Scholar]
- Cunha-Oliveira T, Silva L, Silva AM, Moreno AJ, Oliveira CR, Santos MS (2013) Mitochondrial complex I dysfunction induced by cocaine and cocaine plus morphine in brain and liver mitochondria. Toxicol Lett 219:298–306. 10.1016/j.toxlet.2013.03.025 [DOI] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, Posthuma D (2015) MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11:e1004219 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dencker D, Weikop P, Sørensen G, Woldbye DPD, Wörtwein G, Wess J, Fink-Jensen A (2012) An allosteric enhancer of M4 muscarinic acetylcholine receptor function inhibits behavioral and neurochemical effects of cocaine. Psychopharmacology (Berl) 224:277–287. 10.1007/s00213-012-2751-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JV, Wan Y, Wang X, Cohen S, Wetsel WC, Greenberg ME, Kenny PJ, Calakos N, West AE (2014) MeCP2 phosphorylation limits psychostimulant-induced behavioral and neuronal plasticity. J Neurosci 34:4519–4527. 10.1523/JNEUROSCI.2821-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Snow DM (2007) Cocaine exposure in vitro induces apoptosis in fetal locus coeruleus neurons through TNF-α-mediated induction of Bax and phosphorylated c-Jun NH2-terminal kinase. J Neurochem 103:542–556. 10.1111/j.1471-4159.2007.04750.x [DOI] [PubMed] [Google Scholar]
- Dickson PE, Miller MM, Calton MA, Bubier JA, Cook MN, Goldowitz D, Chesler EJ, Mittleman G (2016) Systems genetics of intravenous cocaine self-administration in the BXD recombinant inbred mouse panel. Psychopharmacology (Berl) 233:701–708. 10.1007/s00213-015-4147-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, Schmidt EF, Nakajima M, Heintz N (2010) Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res 38:4218–4230. 10.1093/nar/gkq130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AJ, Gurung S, Henley JM, Nakamura Y, Wilkinson KA (2019) Exciting times: new advances towards understanding the regulation and roles of kainate receptors. Neurochem Res 44:572–584. 10.1007/s11064-017-2450-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, Schork AJ, Appadurai V, Buil A, Werge TM, Liu C, White KP, Horvath S, Geschwind DH (2018) Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359:693–697. 10.1126/science.aad6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR, Farrer L (2014) Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry 19:717–723. 10.1038/mp.2013.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein RZ, Volkow ND (2011) Dysfunction of the prefrontal cortex in addiction: neuroimaging findings and clinical implications. Nat Rev Neurosci 12:652–669. 10.1038/nrn3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grucza RA, Wang JC, Stitzel JA, Hinrichs AL, Saccone SF, Saccone NL, Bucholz KK, Cloninger CR, Neuman RJ, Budde JP, Fox L, Bertelsen S, Kramer J, Hesselbrock V, Tischfield J, Nurnberger JI, Almasy L, Porjesz B, Kuperman S, Schuckit MA, et al. (2008) A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol Psychiatry 64:922–929. 10.1016/j.biopsych.2008.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2013) The genotype-tissue expression (GTEx) project. Nat Genet 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guez-Barber D, Fanous S, Golden SA, Schrama R, Koya E, Stern AL, Bossert JM, Harvey BK, Picciotto MR, Hope BT (2011) FACS identifies unique cocaine-induced gene regulation in selectively activated adult striatal neurons. J Neurosci 31:4251–4259. 10.1523/JNEUROSCI.6195-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell LL, Cunningham KA (2015) Serotonin 5-HT2 receptor interactions with dopamine function: implications for therapeutics in cocaine use disorder. Pharmacol Rev 67:176–197. 10.1124/pr.114.009514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huggett SB, Stallings MC (2020) Cocaine'omics: genome-wide and transcriptome-wide analyses provide biological insight into cocaine use and dependence. Addict Biol 25:e12719 10.1111/adb.12719 [DOI] [PubMed] [Google Scholar]
- Im H-I, Hollander JA, Bali P, Kenny PJ (2010) MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci 13:1120–1127. 10.1038/nn.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EC, Border R, Melroy-greif WE, Leeuw C, De Ehringer MA, Keller MC (2017) associated with schizophrenia than non-candidate genes. Biol Psychiatry 82:702–708. 10.1016/j.biopsych.2017.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW, Volkow N, Seamans J (2005) Unmanageable motivation in addiction: a pathology in prefrontal-accumbens glutamate transmission. Neuron 45:647–650. 10.1016/j.neuron.2005.02.005 [DOI] [PubMed] [Google Scholar]
- Kapoor M, Wang J-C, Farris SP, Liu Y, McClintick J, Gupta I, Meyers JL, Bertelsen S, Chao M, Nurnberger J, Tischfield J, Harari O, Zeran L, Hesselbrock V, Bauer L, Raj T, Porjesz B, Agrawal A, Foroud T, Edenberg HJ, et al. (2019) Analysis of whole genome-transcriptomic organization in brain to identify genes associated with alcoholism. Transl Psychiatry 9:89 10.1038/s41398-019-0384-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, et al. (2019) Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of- function intolerance across human protein-coding genes. bioRxiv 531210. doi: 10.1101/531210 10.1101/531210 [DOI] [Google Scholar]
- Kasanetz F, Lafourcade M, Deroche-Gamonet V, Revest J-M, Berson N, Balado E, Fiancette J-F, Renault P, Piazza P-V, Manzoni OJ (2013) Prefrontal synaptic markers of cocaine addiction-like behavior in rats. Mol Psychiatry 18:729–737. 10.1038/mp.2012.59 [DOI] [PubMed] [Google Scholar]
- Kumar V, Kim K, Joseph C, Kourrich S, Yoo S-H, Huang HC, Vitaterna MH, de Villena FP-M, Churchill G, Bonci A, Takahashi JS (2013) C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science 342:1508–1512. 10.1126/science.1245503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Luo R, Oldham MC, Horvath S (2011) Is my network module preserved and reproducible? PLoS Comput Biol 7:e1001057 10.1371/journal.pcbi.1001057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EB, Akkentli F, Edwards S, Graham DL, Simmons DL, Alibhai IN, Nestler EJ, Self DW (2010) Striatal regulation of ΔfosB, FosB, and cFos during cocaine self-administration and withdrawal. J Neurochem 115:112–122. 10.1111/j.1471-4159.2010.06907.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT. (2014) Svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res 42:e161 10.1093/nar/gku864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrmann E, Oyler J, Vawter MP, Hyde TM, Kolachana B, Kleinman JE, Huestis MA, Becker KG, Freed WJ (2003) Transcriptional profiling in the human prefrontal cortex: evidence for two activational states associated with cocaine abuse. Pharmacogenomics J 3:27–40. 10.1038/sj.tpj.6500146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Xu P, Xu Y, Teng H, Tian W, Du Q, Zhao M (2017) Dynamic expression changes in the transcriptome of the prefrontal cortex after repeated exposure to cocaine in mice. Front Pharmacol 8:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos M, Pattij T, Janssen MCW, Counotte DS, Schoffelmeer ANM, Smit AB, Spijker S, van Gaalen MM (2010) Dopamine receptor D1/D5 gene expression in the medial prefrontal cortex predicts impulsive choice in rats. Cereb Cortex 20:1064–1070. 10.1093/cercor/bhp167 [DOI] [PubMed] [Google Scholar]
- López AJ, Hemstedt TJ, Jia Y, Hwang PH, Campbell RR, Kwapis JL, White AO, Chitnis O, Scarfone VM, Matheos DP, Lynch G, Wood MA (2019) Epigenetic regulation of immediate-early gene Nr4a2/Nurr1 in the medial habenula during reinstatement of cocaine-associated behavior. Neuropharmacology 153:13–19. 10.1016/j.neuropharm.2019.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, Chanock SJ (2015) LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31:3555–3557. 10.1093/bioinformatics/btv402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ (2019) Current clinical use of polygenic scores will risk exacerbating health disparities. Nat Genet 51:584–591. 10.1038/s41588-019-0379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K. (2017) Synapse organization and modulation via C1q family proteins and their receptors in the central nervous system. Neurosci Res 116:46–53. 10.1016/j.neures.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science 337:1190–1195. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall NM, Fernandez de Velasco EM, Wickman K (2019) GIRK channel activity in dopamine neurons of the ventral tegmental area bi-directionally regulates behavioral sensitivity to cocaine. J Neurosci 39:3600–3610. 10.1523/JNEUROSCI.3101-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrano DA, Schroeder JP, Smith Y, Cortright JJ, Bubula N, Vezina P, Weinshenker D (2012) Alpha-1 adrenergic receptors are localized on presynaptic elements in the nucleus accumbens and regulate mesolimbic dopamine transmission. Neuropsychopharmacology 37:2161–2172. 10.1038/npp.2012.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafò MR, Flint J (2009) Replication and heterogeneity in gene x environment interaction studies. Int J Neuropsychopharmacol 12:727–729. 10.1017/S1461145709000479 [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, Self DW (2001) FosB: a sustained molecular switch for addiction. Proc Natl Acad Sci U S A 98:11042–11046. 10.1073/pnas.191352698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIDA (2020) Overdose Death Rates. Available at https://www.drugabuse.gov/related-topics/trends-statistics/overdose-death-rates.
- Oliver S. (2000) Guilt-by-association goes global. Nature 403:601–603. 10.1038/35001165 [DOI] [PubMed] [Google Scholar]
- Pereira SP, Cunha-Oliveira T (2017) Role of mitochondria on the neurological effects of cocaine. In: The neuroscience of cocaine: mechanisms and treatment. Amsterdam: Elsevier. [Google Scholar]
- Peterson RE, Kuchenbaecker K, Walters RK, Chen C-Y, Popejoy AB, Periyasamy S, Lam M, Iyegbe C, Strawbridge RJ, Brick L, Carey CE, Martin AR, Meyers JL, Su J, Chen J, Edwards AC, Kalungi A, Koen N, Majara L, Schwarz E, et al. (2019) Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls and recommendations. Cell 179:589–603. 10.1016/j.cell.2019.08.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry NM. (2003) A comparison of African American and non-Hispanic Caucasian cocaine-abusing outpatients. Drug Alcohol Depend 69:43–49. 10.1016/S0376-8716(02)00255-7 [DOI] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR (2005) Diagnostic reliability of the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA). Drug Alcohol Depend 80:303–312. [DOI] [PubMed] [Google Scholar]
- Polimanti R, Gelernter J, Walters RK, Johnson EC, Bierut LJ, Bucholz KK, Fox L, Grucza R, Hartz SM, Heath AC, Madden PAF, Nelson EC, Agrawal A, McClintick JN, Edenberg HJ, Adkins AE, Adkins DE, Bacanu S-A, Riley BP, Webb BT et al. (2020) Leveraging genome-wide data to investigate differences between opioid use vs. opioid dependence in 41,176 individuals from the Psychiatric Genomics Consortium. Mol Psychiatry. Advance online publication. Retrieved May 26, 2020. doi: 10.1038/s41380-020-0677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD (2012) Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci 32:1884–1897. 10.1523/JNEUROSCI.3136-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro EA, Scarpa JR, Garamszegi SP, Kasarskis A, Mash DC, Nestler EJ (2017) Gene network dysregulation in dorsolateral prefrontal cortex neurons of humans with cocaine use disorder. Sci Rep 7:5412 10.1038/s41598-017-05720-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro EA, Salery M, Scarpa JR, Calipari ES, Hamilton PJ, Ku SM, Kronman H, Purushothaman I, Juarez B, Heshmati M, Doyle M, Lardner C, Burek D, Strat A, Pirpinias S, Mouzon E, Han M-H, Neve RL, Bagot RC, Kasarskis A, et al. (2018) Transcriptional and physiological adaptations in nucleus accumbens somatostatin interneurons that regulate behavioral responses to cocaine. Nat Commun 9:3149 10.1038/s41467-018-05657-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin RA, Huyghe D, Li X, Parakala M, Aisenberg E, Moss SJ, Slesinger PA (2018) GIRK currents in VTA dopamine neurons control the sensitivity of mice to cocaine-induced locomotor sensitization. Proc Natl Acad Sci U S A 115:E9479–E9488. 10.1073/pnas.1807788115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, Grucza RA, Sun L, Duan W, Budde J, Culverhouse RC, Fox L, Hinrichs AL, Steinbach JH, Wu M, Rice JP, Goate AM, Bierut LJ (2009) The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res 69:6848–6856. 10.1158/0008-5472.CAN-09-0786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadakierska-Chudy A, Frankowska M, Filip M (2014) Mitoepigenetics and drug addiction. Pharmacol Ther 144:226–233. 10.1016/j.pharmthera.2014.06.002 [DOI] [PubMed] [Google Scholar]
- Salery M, Dos Santos M, Saint-Jour E, Moumné L, Pagès C, Kappès V, Parnaudeau S, Caboche J, Vanhoutte P (2017) Activity-regulated cytoskeleton-associated protein accumulates in the nucleus in response to cocaine and acts as a brake on chromatin remodeling and long-term behavioral alterations. Biol Psychiatry 81:573–584. 10.1016/j.biopsych.2016.05.025 [DOI] [PubMed] [Google Scholar]
- Sanjakdar SS, Maldoon PP, Marks MJ, Brunzell DH, Maskos U, McIntosh JM, Bowers MS, Damaj MI (2015) Differential roles of α6β2* and α4β2* neuronal nicotinic receptors in nicotine- and cocaine-conditioned reward in mice. Neuropsychopharmacology 40:350–360. 10.1038/npp.2014.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, Genovese G, Rose SA, Handsaker RE, Daly MJ, Carroll MC, Stevens B, McCarroll SA (2016) Schizophrenia risk from complex variation of complement component 4. Nature 530:177–183. 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinden LA, Saba LM, Kechris K, Miles MF, Hoffman PL, Tabakoff B (2013) Whole brain and brain regional coexpression network interactions associated with predisposition to alcohol consumption. PloS one 8:e68878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam S, Võsa U, van der Graaf A, Franke L, de Magalhães JP (2018) Gene co-expression analysis for functional classification and gene-disease predictions. Brief Bioinform 19:575–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DM, Cates HM, Loh Y-HE, Purushothaman I, Ramakrishnan A, Cahill KM, Lardner CK, Godino A, Kronman HG, Rabkin J, Lorsch ZS, Mews P, Doyle MA, Feng J, Labonté B, Koo JW, Bagot RC, Logan RW, Seney ML, Calipari ES, et al. (2018) Cocaine self-administration alters transcriptome-wide responses in the brain's reward circuitry. Biol Psychiatry 84:867–880. 10.1016/j.biopsych.2018.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, van Bochoven A, Posthuma D (2017) Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8:1826. 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M, Adinoff B (2008) The role of acetylcholine in cocaine addiction. Neuropsychopharmacology 33:1779–1797. 10.1038/sj.npp.1301585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala AR, Osredkar T, Joyce JN, Neisewander JL (2008) Upregulation of Arc mRNA expression in the prefrontal cortex following cue-induced reinstatement of extinguished cocaine-seeking behavior. Synapse 62:421–431. 10.1002/syn.20502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Yuan Q, Mash DC, Goldman D (2011) Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc Natl Acad Sci U S A 108:6626–6631. 10.1073/pnas.1018514108 [DOI] [PMC free article] [PubMed] [Google Scholar]