Abstract
The metazoan nucleus is equipped with a meshwork of intermediate filament proteins called the A- and B-type lamins. Lamins lie beneath the inner nuclear membrane and serve as a nexus to maintain the architectural integrity of the nucleus, chromatin organization, DNA repair and replication and to regulate nucleocytoplasmic transport. Perturbations or mutations in various components of the nuclear lamina result in a large spectrum of human diseases collectively called laminopathies. One of the most well-characterized laminopathies is Hutchinson–Gilford progeria (HGPS), a rare segmental premature aging syndrome that resembles many features of normal human aging. HGPS patients exhibit alopecia, skin abnormalities, osteoporosis and succumb to cardiovascular complications in their teens. HGPS is caused by a mutation in LMNA, resulting in a mutated form of lamin A, termed progerin. Progerin expression results in a myriad of cellular phenotypes including abnormal nuclear morphology, loss of peripheral heterochromatin, transcriptional changes, DNA replication defects, DNA damage and premature cellular senescence. A key challenge is to elucidate how these different phenotypes are causally and mechanistically linked. In this mini-review, we highlight some key findings and present a model on how progerin-induced phenotypes may be temporally and mechanistically linked.
Keywords: lamin A, progeria, senescence
Introduction
Aging can be defined as a gradual deterioration of cell and tissue function, resulting in an elevated risk of developing a large number of chronic illnesses. These include cardiovascular disease, diabetes, decreased cognitive and hearing ability, neurodegenerative diseases, and loss of bone and muscle mass. Aging is also correlated with an elevated cancer risk. On a cellular level, increased levels of DNA damage, genomic instability, epigenetic alterations including loss of heterochromatin, telomere shortening, mitochondrial dysfunction, impaired nutrient sensing, loss of proteostasis and permanent growth arrest (senescence) are all aging-associated phenomena [1]. However, given the complexity of the aging phenotype, and the ethical and practical limitations of longitudinal studies, little is known about how all of these ‘hallmarks of aging' are causally connected. Progeroid syndromes, which are rare premature aging disorders, may, however, offer a unique window on to the mechanisms that contribute to human physiological aging [2–5]. Progeria patients exhibit characteristic features of normal aging. Typically, these are variously represented by cardiovascular disease, osteoporosis, lipodystrophy, alopecia and aberrant skin pigmentation. Depending upon the type and severity of the particular syndrome, patients may succumb as early as in their mid-teens or may survive until their fifth decade. Progeroid syndromes can be classified into two main groups: The first group is caused by defects in DNA repair pathways or impaired telomere maintenance and includes Werner syndrome (WS), xeroderma pigmentosum (XP), Bloom syndrome (BS), Nijmegen breakage syndrome (NBS), Cockayne Syndrome (CS), Rothmund–Thomson syndrome (RTS), ataxia telangiectasia (AT), Fanconi anemia (FA), dyskeratosis congenita (DC) and Hoyeraal–Hreidarsson syndrome (HHS). The second group is caused by mutations in components of the nuclear lamina and is represented by Hutchinson–Gilford progeria syndrome (HGPS), restrictive dermopathy (RD), Néstor–Guillermo progeria syndrome (NGPS) and mandibuloacral dysplasia (MAD) [6–8]. In this mini-review, we will focus mainly on HGPS as it is arguably the best-studied progeroid syndrome.
The nuclear lamina
A defining feature of all eukaryotic cells is the nucleus, a compartment encapsulated by a pair of membranes that are separated by the perinuclear space. The outer nuclear membrane (ONM) faces the cytoplasm, is contiguous with the endoplasmic reticulum and serves as an anchor point for the cytoskeleton, whilst the inner nuclear membrane (INM) faces the nuclear lamina; a ∼15 nm thick proteinaceous meshwork consisting of type V intermediate filament proteins [9–12] known as A- and B-type lamins. The ONM and INM are connected where they are spanned by nuclear pore complexes, massive channels that facilitate the nuclear-cytoplasmic transport of macromolecular cargoes. The INM and the lamina are connected through interactions with multiple integral membrane proteins. Prominent among these are a family of INM proteins, that share a 40 amino acid LAP2-emerin-MAN1 (LEM) domain, that binds both the nuclear lamina and repressed chromatin [13]. Most LEM-family proteins, including LAP2β, LEMD1 and Emerin are tethered to the INM via a single C-terminal transmembrane domain; they interact with each other as well as with the A- and B-type lamin proteins of the nuclear lamina.
The basic structure of the nuclear lamins features a ∼30 amino acid non-helical amino terminal head domain and a central α-helical coiled-coil rod domain of ∼45 nm. The rod domain is followed by a highly conserved immunoglobulin-like fold (Ig fold). Sequences downstream of the Ig fold domain are largely unstructured and, with the exception of lamins C and C2, terminate in a CaaX motif (C, cysteine; a, aliphatic amino acid; X, any amino acid but usually methionine) [9,11,14,15], the function of which is described in the next section. In contrast with their cytoplasmic intermediate filament counterparts, all members of the lamin family possess a classical SV-40-type nuclear localization sequence that is situated between the central alpha-helical rod and Ig fold domains.
As members of the intermediate filament family, both the A- and B-type lamins will assemble to form tetrameric filaments. These in turn may associate to form the higher-order nuclear lamina structure that is essential for the maintenance of nuclear architecture and stability. The A-type lamins are encoded by a single gene, LMNA, and include the major isoforms lamin A and lamin C, as well as minor isoforms such as CΔ10 and C2 [16]. The B-type lamins, lamin B1 and lamin B2, are encoded by two separate genes, LMNB1 and LMNB2, respectively. The latter also encodes the germ cell-specific lamin B3 as a splice variant of lamin B2. The fact that lamins are notoriously insoluble, form filaments and lack any catalytic activity led to the initial notion that lamins function primarily to maintain nuclear shape and integrity. However, research in the past 20 years demonstrated that lamins play a crucial role in various cellular processes including mechanotransduction, modulation of nuclear stiffness and elasticity, chromatin organization, DNA replication and repair, and transcriptional regulation [4,17,18].
The composition of the lamina changes during embryonic development and, even in adults, there are obvious differences according to tissue type. Pioneering studies 30 years ago revealed that lamins are differentially expressed. B-type lamins are expressed in embryonic stem cells, throughout embryonic development and in all somatic cell types. In contrast, expression of A-type lamins commences midway through gestation and may vary significantly between different somatic cell types [19–22]. However, we still lack a complete picture of the precise composition of the nuclear lamina, as well as the stoichiometry of its components, in different human tissues. This is important as the differential expression of lamins—or lamina-associated factors—may explain how mutations or perturbations of the lamina give rise to such a large spectrum of different diseases, and why certain tissues are not affected [21–23].
Processing of lamins
Soon after translation, and prior to assembly into the lamina, the C-terminus of lamins A, B1 and B2 undergoes many sequential modification steps. The first of these is the addition of a farnesyl group onto the cysteine residue of the C-terminal CaaX motif by a farnesyltransferase (FTase), the linkage consisting of a thioether bond. Farnesylation is followed by cleavage of the terminal-aaX tripeptide domain by the Zinc metallopeptidase FACE1/ZMPSTE24 or FACE2/Rce1. The newly exposed farnesylcysteine is then carboxymethylated by the isoprenyl-cysteine-carboxy-methyltransferase (ICMT) [24,25]. The hydrophobic farnesyl tail facilitates the interaction of newly synthesized lamins to the INM [26]. Finally, in the case of lamin A only, the terminal 15 amino acids, including the farnesylcysteine α-methyl ester are cleaved again by ZMPSTE24 resulting in the generation of mature non-farnesylated lamin A. This non-membrane bound mature lamin A has both lamina-associated and nucleoplasmic populations [27]. Neither lamin B1 nor B2 undergo this final processing step and instead both remain permanently farnesylated and tethered to the INM. Blocking lamin A farnesylation by mutating its C-terminal cysteine residue to a serine, and removal of its nuclear localization signal result in cytosolic accumulation of lamin A [28]. Inadequate processing of lamin A results in many devastating human diseases including HGPS, RD and MAD [29].
Hutchinson–Gilford progeria syndrome
HGPS is a rare, autosomal dominant premature aging syndrome that affects roughly 1 in 4–8 million newborns. Patients are born without any overt clinical features, but develop signs of aging 18–24 months after birth. These include failure to thrive, alopecia, thin and wrinkled skin, aberrant pigmentation, loss of subcutaneous fat, stiffening of joints and weakened bone structure [8]. HGPS is fatal and patients generally die in their mid-teens as a result of cardiovascular complications [2,30]. Clinical examination of individuals with HGPS revealed a loss of vascular smooth muscle cells, aortic calcification, accumulation of atherosclerotic plaques and adventitial thickening. Collectively, these cardiovascular alterations result in myocardial infarction and stroke and are the main causes of death in progeria patients [2,8,30]. Two independent groups discovered in 2003 that HGPS is caused by a de novo heterozygous point mutation (1824C > T) in the LMNA gene [31,32]. This point mutation activates a cryptic splice donor site that results in the deletion of a 50 amino acid segment, including the second ZMPSTE24 cleavage recognition site, and results in the formation of a truncated and permanently farnesylated and carboxymethylated mutant form of lamin A, termed progerin.
Atypical progeroid syndromes: restrictive dermopathy and mandibuloacral dysplasia
RD and MAD are caused by mutations in ZMPSTE24 that result in partial or complete loss of enzymatic function, or mutations in the ZMPSTE24 proteolytic cleavage site in lamin A, thereby resulting in the accumulation of aberrantly processed and permanently farnesylated lamin A [33]. RD is characterized by clinical features before birth, including severe intrauterine growth defects, reduced fetal movement and perinatal lethality. The skin of RD patients is thin, translucent, tightly adherent, lacks elastic fibers and exhibits disorganized collagen organization. MAD can be classified into type A and B: mutations in lamin A that affect processing by ZMPSTE24 belong to type A, whilst mutations that impair the proteolytic activity of ZMPSTE24 are classified as type B. Both types share clinical features that resemble progeria but vary in severity; whilst patients with progeria display signs of aging 1–2 years after birth, patients with MAD generally develop abnormalities around the age of ∼4–5 years. These include skeletal abnormalities, partial alopecia, insulin resistance, lipodystrophy and abnormal skin pigmentation [34]. In conclusion, RD, HGPS and MAD are all caused by an accumulation of permanently farnesylated lamin A.
Cellular phenotypes
HGPS patient-derived fibroblasts and various cell-based experimental systems have greatly contributed to our understanding how lamin A mutants perturb cell physiology and function. Original studies revealed that HGPS patient cells and various human cells expressing progerin develop nuclear abnormalities, loss of peripheral heterochromatin and a thickening of the nuclear lamina [35–37]. In addition, progeric cells display a persistent activation of the DNA damage response (DDR) checkpoints, telomere shortening, increased reactive oxygen species (ROS), genomic instability, impaired proliferation and premature cellular senescence [23,38–42]. What remains unknown is how these different phenotypes are mechanistically and/or causally linked, and whether they are a cause or a consequence of senescence.
Nuclear abnormalities
The presence of the permanently farnesylated and INM-anchored progerin protein results in a thickening of the nuclear lamina, nuclear shape alterations such as invaginations and prominent lobulations, and clustering of nuclear pore complexes [35]. As a result of these nuclear envelop perturbations, HGPS nuclei are stiffer and more resistant to mechanical forces [43]. Contrasting these observations, nuclei from Lmna-deficient mice are softer and less resistant to mechanical stress [44]. Taken together, progeric nuclei may respond differently to physical stress than their normal counterparts. This may be particularly significant in those tissues that are subjected to constant mechanical stress, such as the vasculature, bones and joints [17,45,46].
Loss of peripheral heterochromatin
Similar to chronologically aged normal human cell, progeric cells exhibit loss of transcriptionally repressed and compacted peripheral heterochromatin, as shown by a reduction in histone H3 trimethylation at lysines 9 and 27 (H3K9me3 and H3K27me3) and electron microscopy [23,36,47,48]. Loss of heterochromatin is concomitant with the down-regulation of many proteins involved in epigenetic silencing, including the Enhancer of zeste homolog (EZH2), a member of the polycomb repressive complex (PRC) 2, heterochromatin protein 1 homolog-α (HP1α) and the histone N-methyltransferase Suppressor of variegation 3–9 homolog 1 (SUV39H1) [37,47,49]. Furthermore, a yeast two-hybrid screen using the C-terminal domain of lamin A that is deleted in progerin led to the identification of RBBP4 + 7, both components of the nucleosome remodeling and deacetylase complex (NuRD) and PRC2 complexes that are down-regulated in HGPS cells. Knockdown of both RBBP4 + 7 using siRNA recapitulated some aspects of HGPS, including a reduction in H3K9me3 and elevated levels of DNA damage [49]. Similarly, decompaction of heterochromatin by histone deacetylase inhibitors (HDACi) rendered cells more susceptible to DNA damage and senescence, although it remains unclear whether the observed phenotypes were driven directly by heterochromatin decompaction or other changes caused by HDACi treatment [50,51]. Nevertheless, the precise mechanism how progerin triggers this drastic change in chromatin compaction remains unclear. For instance, a recent transcriptional analysis of control versus progerin-expressing mouse skin found no significant changes in the expression of key epigenetic modulators [52], and expression of progerin in telomerase-positive cells did not result in any transcriptional changes [23].
What is clear is that progerin-induced heterochromatin loss is not a consequence of cells undergoing senescence; as an expression of progerin in immortalized, or telomerase-positive cells, did not prevent heterochromatin decompaction [23,36]. Furthermore, by expressing progerin during different cell cycle stages, it was recently shown that progerin triggers heterochromatin decompaction in growth-arrested (G0) cells, whilst DNA damage accumulates exclusively during DNA replication, thus separating these two phenotypes [48]. By visualizing heterochromatin levels (H3K9me3 and H3K27me3), DNA damage and progerin levels at single-cell resolution, it was also shown that cells with low levels of heterochromatin are more prone to accumulate progerin-induced DNA damage, whilst progerin removal (in G0 cells) restored heterochromatin levels and prevented DNA damage accumulation during subsequent rounds of DNA replication [48,53]. Collectively, these results suggest that chromatin structure plays an important role in causing progerin-induced DNA damage. Interestingly, cells derived from MAD or RD patients exhibit similar chromatin alterations, suggesting that this may be a conserved feature of laminopathies caused by an accumulation of incompletely processed lamin A [54,55]. Nevertheless, this conclusion is largely based on correlative data and it remains unclear whether overexpression of certain chromatin modifiers would restore, or prevent progerin-induced heterochromatin loss (and DNA damage).
Progerin-induced DNA damage
Lamins have been implicated in both non-homologous end joining and homology directed repair [38,56,57]. Indeed, cells from HGPS patients or primary human cells expressing progerin, as well as cells derived from Zmpste24-deficient mice, accumulate DNA damage foci, as judged by 53BP-1 or γH2A-X staining [38,39]. In addition, mouse embryonic fibroblasts derived from Lmna- or Zmpste24-deficient mice exhibit genomic instability including increased aneuploidy, chromosome fusions and breaks. Similarly, cells ectopically expressing progerin show an increased frequency of sister chromatid fusions, chromosomal breaks and anaphase bridges [58].
The root cause of this DNA damage has been attributed to many factors, including a delay in recruitment of the DNA repair factor p53-binding protein (53BP-1) to sites of damage, possibly as a result of impaired shuttling into the nucleus [38,59]. In contrast, we recently exposed G0-arrested human fibroblasts expressing progerin to the DNA damaging agent doxorubicin and studied the temporal dynamics of 53BP-1 and γH2A-X recruitment. Surprisingly, the timing of recruitment of these factors to sites of DNA damage were indistinguishable between control and progerin-positive fibroblasts. These results suggest that progerin expression per se may not directly affect the DDR. Nevertheless, it must be noted that these experiments were conducted in growth-arrested cells exposed to progerin for only 4 days. Thus, it is conceivable that HGPS cells that constitutively express progerin may exhibit these and other defects [48].
The nature of progerin-induced DNA damage
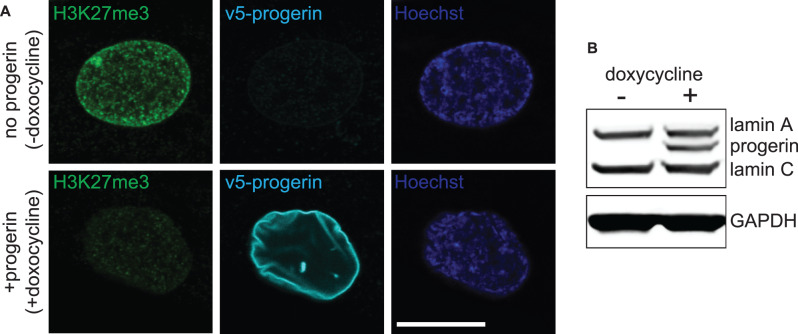
Increasing evidence suggests that the DNA damage and chronic checkpoint activation in HGPS cells are caused by progerin-induced replication defects. Lamin A/C is required to restart replication forks after hydroxyurea-induced replication stress [60] and progerin-induced DNA damage co-localizes with phospho-RPA32 (ser33) and XPA, proteins that bind to single-stranded DNA at replication forks [61]. Moreover, progerin-induced DNA damage preferentially accumulated in BrdU+ cells, suggesting that damage occurred during S-phase. In contrast, preventing the growth of progerin-expressing cells by serum starvation, or expressing progerin in contact-inhibited (G0) cells (using the doxycycline-inducible system depicted in Figure 1), reduced/prevented the accumulation of DNA damage, respectively. Collectively, these studies link progerin-induced DNA damage to replication fork collapse during S-phase [48,61–63].
Figure 1. Doxycycline-inducible expression of progerin in proliferating primary human fibroblasts.
(A) v5-progerin is visualized by anti-v5-antibody (cyan). H3K27me3 staining is indicated in green. Hoechst staining is in blue. Size marker 20 μm. (B) Western blot showing doxycyline-dependent expression of progerin. Lamin A, C, progerin and GAPDH are shown.
But how does progerin affect DNA replication? Previous studies showed that progerin or pre-lamin A expression disrupt the interaction between lamin A and the proliferating cell nuclear antigen (PCNA), a component of replisome that is essential for replication fork progression. This results in sequestration of PCNA away from the replicating fork and exposes the forks’ ds-ssDNA junction to binding by XPA [63,64], whilst reducing progerin levels restored PCNA binding to the fork [61–63]. Single-molecule replication studies using DNA fiber assays provided further evidence that progerin expression triggers replication fork stalling and deprotection. Treatment with Mirin, an inhibitor of the MRE11 nuclease, prevented fork degradation and rescued progerin-induced replication defects [65]. Lastly, by using the aforementioned inducible expression system, we determined the temporal dynamics of DNA damage accumulation during S-phase and found that progerin causes damage exclusively during the late stages of DNA replication prior to chromosome condensation and mitosis [48]. This timing coincides with the replication of telomeres that are located near the nuclear periphery, as well as with the replication of heterochromatin [66,67].
These results imply that progerin-induced DNA damage may only occur in proliferating cells, and raises the question how relevant these cell culture-based findings may be in vivo? Although it has been thought that some adult tissues hardly turn over, there is increasing evidence that suggests otherwise [68]. Several studies demonstrated that ∼40% of cardiomyocytes are replaced during a normal lifetime [69,70]. This turnover occurs independent of ploidy and is highest in younger individuals [71]. Additionally, non-cardiomyocyte populations, such as vascular smooth muscle cells from the same hearts exhibited turnover rates of ∼15% per year [70]. Nevertheless, it is likely that DNA replication-independent factors, such as mechanical stress, contribute to progerin-induced DNA damage in some tissues.
Linking the ends to the periphery
Telomeres are nucleoprotein complexes that cap the physical ends of our linear chromosomes. They consist of 10–20 kb of tandem TTAGGG repeats and are bound by the shelterin complex. Shelterin consists of six proteins that protect the chromosome ends from nucleolytic degradation and illegitimate activation of DNA damage checkpoints. In embryonic, and some adult stem cells, germ cells and ∼85% of cancers, shelterin regulates telomere length homeostasis by recruiting telomerase, a ribonucleoprotein that can elongate telomeres, to the telomere terminus [72]. In somatic cells, telomeres shorten during each replication cycle as a result of the end replication problem. Critically shortened telomeres, or loss of shelterin components elicit a persistent DNA damage checkpoint activation and trigger, depending on cell type, senescence or apoptosis. As such, telomeres represent the Achilles’ heel of the genome and telomere shortening or dysfunction are associated with a wide spectrum of human diseases [73,74].
Telomeres are considered heterochromatic domains and are enriched near the nuclear lamina in both human and mouse cells [23,56,67]. This peripheral localization is elevated during post-mitotic nuclear reassembly [75]. What remains unclear is whether telomeres are actively tethered to the nuclear periphery, or whether they simply co-localize with heterochromatic regions of the genome. In support of a more active tethering model, lamins and associated factors such as the lamina-associated peptide-α (LAP2α) have been shown to co-localize or interact with components of the shelterin complex [23,75–77]. AKTIP, a member of the ubiquitin E2 variant (UEV) enzyme subfamily is enriched at the nuclear periphery and interacts with A- and B-type lamins, shelterin components TRF1 and 2 and PCNA. The depletion of AKTIP in primary human cells recapitulates some aspects of HGPS including replication defects and premature senescence [78].
Cells from HGPS patients have significantly shorter telomeres than their age-matched controls [23,41,42]. As a result, HGPS patient-derived fibroblasts (or cells ectopically expressing progerin) exhibit a limited proliferative capacity and prematurely undergo replicative senescence [23,41,42]. Ectopic expression of hTERT, the catalytic subunit of the telomerase, elongates telomeres in HGPS cells, prevents progerin-induced DNA damage and restores their proliferative capacity [23,58,79,80]. To investigate whether physiological levels of hTERT would be sufficient to prevent progerin-induced defects, progerin was expressed in pluripotent wild type mouse embryonic stem cells (mESC), that express TERT endogenously. Progerin-expressing mESC remained indistinguishable from their wild type counterparts, whilst differentiation of these cells into TERT-negative somatic lineages recapitulated progeric phenotypes [23,81]. Lastly, progerin expression in TERT−/− mESC resulted in massive differentiation and cell death [23]. Taken together, both ectopically expressed, or physiological levels of TERT prevent progerin-induced DNA damage and proliferation defects. Nevertheless, the ability of telomerase to prevent progerin-induced DNA damage and proliferation defects may depend on the replicative age of recipient HGPS cells; cells that have undergone senescence, or accumulated excessive genomic instability prior to hTERT transduction may not be rescued. Furthermore, hTERT is not sufficient to prevent these defects in cell that express high levels of progerin (or lamin A), and does not prevent progerin-induced heterochromatin loss or nuclear abnormalities.
What remains unclear is how hTERT prevents, or repairs, progerin-induced DNA damage and what is the physiological relevance of these findings. First, hTERT is active at telomeres only during DNA replication. Thus, the timing of progerin-induced DNA damage (late S-phase) and the ability of hTERT to prevent it coincide. Second, hTERT is expressed during embryonic development and in various adult stem cells, thus rendering these populations more resistant to the detrimental consequences of progerin expression [82]. This contrasts other premature aging syndromes, such as dyskeratosis congenita, that are caused by telomerase dysfunction and result in impaired maintenance of various stem cell compartments [83,84].
Delineating the chain of events to occur upon progerin expression
The usage of isogenic primary and hTERT-immortalized human cell types provides a tool to establish which of the above-mentioned phenotypes are a cause or a consequence of senescence. In addition, inducible expression systems facilitate the restricted expression of progerin to different cell cycle stages and provide information on how different phenotypes may be temporally connected. Based on these experiments, a picture, although incomplete, emerges that delineates the chain of events that occur upon progerin expression and ultimately result in premature senescence (Figure 2). Expression of progerin in contact-inhibited G0-arrested cells results in loss of heterochromatin marks H3K9me3 and H3K27me3, irrespective of hTERT expression, and no accumulation of DNA damage [23,48]. These results are consistent with the reduced number of DNA damage foci in serum-starved HGPS cells [61,63]. The mechanism how progerin results in loss of peripheral heterochromatin remains largely unknown. Upon reinitiating the cell cycle and entering S-phase (by plating the G0-arrested cells sparse), progerin-expressing cells accumulate DNA damage exclusively during the late stages of DNA replications, prior to chromosome condensation and mitosis, and preferentially in cells with low levels of heterochromatin [48,61–63]. Expression of hTERT, or modulation of the DDR specifically at telomeres, prevents (or rescues) progerin-induced DNA damage and senescence [23,48,58,79,85]. In somatic (hTERT-negative) cells, damaged telomeres cannot be repaired by conventional DNA repair processes and, thus, result in a permanent activation of DNA damage checkpoints and premature cellular senescence [23,40,48,58,86]. Removal of progerin in G0-arrested cells leads to a rapid restoration of heterochromatin levels. Moreover, these transiently progerin exposed cells do not exhibit any DNA damage or premature senescence after resuming proliferation. Taken together, these results suggest that progerin removal from quiescent cells or tissues, using morpholino or CRISPR-based approaches is predicted to fully restore their proliferative/regenerative capacity [48,53,87,88].
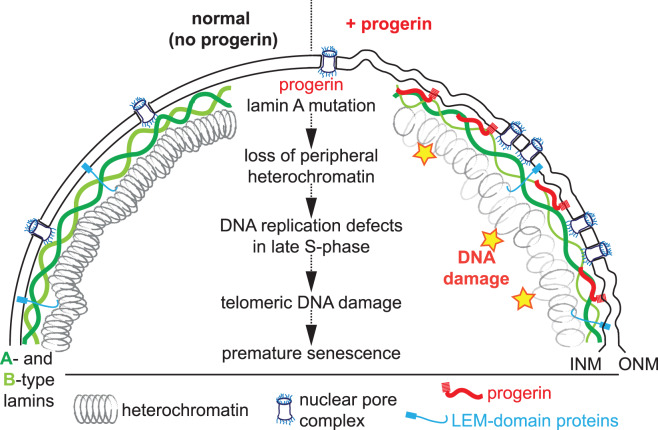
Figure 2. Schematic representation of the nuclear envelope and its associated components in normal (no progerin, left side), or progerin-expressing cells (+ progerin, right side).
The nucleus is encapsulated by the inner and outer nuclear membrane (INM, ONM). Cargo is shuttled between the nucleus and cytoplasm by nuclear pore complexes. LEM-domain proteins span the INM and connect it to A- and B-type lamins and peripheral heterochromatin. Progerin retains its farnesyl moiety and remains associated with the INM. Progerin expression results in nuclear abnormalities, heterochromatin decompaction, clustering of nuclear pore complexes, senescence-associated lamin B1 reduction and DNA damage. Center: A speculative model depicting a possible chain of events that commences with an expression of mutant lamin A (progerin), loss of heterochromatin, replication defects and an accumulation of telomeric DNA damage that results in premature cellular senescence.
Perspectives
Highlight the importance of the field: Premature aging syndromes such as Hutchinson–Gilford progeria provide a unique opportunity to identify and characterize molecular pathways and mechanisms that contribute to human aging.
A summary of the current thinking: Expression of the lamin A mutant progerin results in heterochromatin loss. Cells with low levels of heterochromatin are more prone to develop DNA replication defects and telomeric DNA damage, thereby resulting in premature cellular senescence.
A comment on future directions: Mutations and perturbations of the nuclear lamina give rise to many cellular defects and result in a plethora of human diseases (laminopathies). The key questions are to (1) elucidate the precise mechanism(s) how progerin expression perturbs heterochromatin organization and results in DNA replication defects and damage. (2) Establish how the myriad progerin-induced cellular phenotypes are causally and mechanistically linked. (3) On an organismal level, determine why some mutations exclusively affect certain tissues and how this relates to the disease etiology. Providing answers to these questions will lay the foundation for the development of novel therapeutic interventions.
Acknowledgements
Brian Burke, Mattheus Foo, Peh Fern Ong and Christopher Bong are acknowledged for critical reading of the manuscript.
Abbreviations
- HGPS
Hutchinson–Gilford progeria
- INM
inner nuclear membrane
- INM
inner nuclear membrane
- MAD
mandibuloacral dysplasia
- ONM
outer nuclear membrane
- PCNA
proliferating cell nuclear antigen
- PRC
polycomb repressive complex
Competing Interests
The author declares that there are no competing interests associated with this manuscript.
Funding
This work was supported by the Singapore Biomedical Research Council (A*STAR, Singapore) and the Skin Research Institute of Singapore.
References
- 1.López-Otín C., Blasco M.A., Partridge L., Serrano M. and Kroemer G. (2013) The hallmarks of aging. Cell 153, 1194–1217 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorado B. and Andrés V. (2017) A-type lamins and cardiovascular disease in premature aging syndromes. Curr. Opin. Cell Biol. 46, 17–25 10.1016/j.ceb.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 3.Vidak S. and Foisner R. (2016) Molecular insights into the premature aging disease progeria. Histochem. Cell Biol. 145, 401–417 10.1007/s00418-016-1411-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubben N. and Misteli T. (2017) Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat. Rev. Mol. Cell Biol. 18, 595–609 10.1038/nrm.2017.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreesen O. and Stewart C.L. (2011) Accelerated aging syndromes, are they relevant to normal human aging? Aging (Albany NY) 3, 889–895 10.18632/aging.100383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrero D., Soria-Valles C. and López-Otín C. (2016) Hallmarks of progeroid syndromes: lessons from mice and reprogrammed cells. Dis. Model Mech. 9, 719–735 10.1242/dmm.024711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foo M.X.R., Ong P.F. and Dreesen O. (2019) Premature aging syndromes: from patients to mechanism. J. Dermatol. Sci. 96, 58–65 10.1016/j.jdermsci.2019.10.003 [DOI] [PubMed] [Google Scholar]
- 8.Merideth M.A., Gordon L.B., Clauss S., Sachdev V., Smith A.C.M., Perry M.B. et al. (2008) Phenotype and course of Hutchinson–Gilford progeria syndrome. N. Engl. J. Med. 358, 592–604 10.1056/NEJMoa0706898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burke B. and Stewart C.L. (2013) The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell Biol. 14, 13–24 10.1038/nrm3488 [DOI] [PubMed] [Google Scholar]
- 10.Gerace L. and Huber M. (2012) Nuclear lamina at the crossroads of the cytoplasm and nucleus. J. Struct. Biol. 177, 24–31 10.1016/j.jsb.2011.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turgay Y., Eibauer M., Goldman A.E., Shimi T., Khayat M., Ben-Harush K. et al. (2017) The molecular architecture of lamins in somatic cells. Nature 543, 261–264 10.1038/nature21382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aebi U., Cohn J., Buhle L. and Gerace L. (1986) The nuclear lamina is a meshwork of intermediate-type filaments. Nature 323, 560–564 10.1038/323560a0 [DOI] [PubMed] [Google Scholar]
- 13.Barton L.J., Soshnev A.A. and Geyer P.K. (2015) Networking in the nucleus: a spotlight on LEM-domain proteins. Curr. Opin. Cell Biol. 34, 1–8 10.1016/j.ceb.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher D.Z., Chaudhary N. and Blobel G. (1986) cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl. Acad. Sci. U.S.A. 83, 6450–6454 10.1073/pnas.83.17.6450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKeon F.D., Kirschner M.W. and Caput D. (1986) Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature 319, 463–468 10.1038/319463a0 [DOI] [PubMed] [Google Scholar]
- 16.Broers J.L., Machiels B.M., Kuijpers H.J., Smedts F., van den Kieboom R., Raymond Y. et al. (1997) A- and B-type lamins are differentially expressed in normal human tissues. Histochem. Cell Biol. 107, 505–517 10.1007/s004180050138 [DOI] [PubMed] [Google Scholar]
- 17.Dahl K.N., Ribeiro A.J.S. and Lammerding J. (2008) Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 102, 1307–1318 10.1161/CIRCRESAHA.108.173989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikegami K., Secchia S., Almakki O., Lieb J.D. and Moskowitz I.P. (2020) Phosphorylated lamin A/C in the nuclear interior binds active enhancers associated with abnormal transcription in progeria. Dev. Cell 52, 699–713.e11 10.1016/j.devcel.2020.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rober R.A., Weber K. and Osborn M. (1989) Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development 105, 365–378 PMID: [DOI] [PubMed] [Google Scholar]
- 20.Lehner C.F., Stick R., Eppenberger H.M. and Nigg E.A. (1987) Differential expression of nuclear lamin proteins during chicken development. J. Cell Biol. 105, 577–587 10.1083/jcb.105.1.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swift J., Ivanovska I.L., Buxboim A., Harada T., Dingal P.C., Pinter J. et al. (2013) Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341, 1240104 10.1126/science.1240104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J., Lian Q., Zhu G., Zhou F., Sui L., Tan C. et al. (2011) A human iPSC model of hutchinson Gilford progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell 8, 31–45 10.1016/j.stem.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 23.Chojnowski A., Ong P.F., Wong E.S.M., Lim J.S.Y., Mutalif R.A., Navasankari R. et al. (2015) Progerin reduces LAP2a-telomere association in Hutchinson–Gilford progeria. eLife 4, e07759 10.7554/eLife.07759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young S.G., Fong L.G., Michaelis S. and Prelamin A. (2005) Zmpste24, misshapen cell nuclei, and progeria–new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J. Lipid Res. 46, 2531–2558 10.1194/jlr.R500011-JLR200 [DOI] [PubMed] [Google Scholar]
- 25.Rusiñol A.E. and Sinensky M.S. (2006) Farnesylated lamins, progeroid syndromes and farnesyl transferase inhibitors. J. Cell Sci. 119, 3265–3272 10.1242/jcs.03156 [DOI] [PubMed] [Google Scholar]
- 26.Holtz D., Tanaka R.A., Hartwig J. and McKeon F. (1989) The CaaX motif of lamin A functions in conjunction with the nuclear localization signal to target assembly to the nuclear envelope. Cell 59, 969–977 10.1016/0092-8674(89)90753-8 [DOI] [PubMed] [Google Scholar]
- 27.Dechat T., Gesson K. and Foisner R. (2010) Lamina-independent lamins in the nuclear interior serve important functions. Cold Spring. Harb. Symp. Quant. Biol. 75, 533–543 10.1101/sqb.2010.75.018 [DOI] [PubMed] [Google Scholar]
- 28.Wu D., Flannery A.R., Cai H., Ko E. and Cao K. (2014) Nuclear localization signal deletion mutants of lamin A and progerin reveal insights into lamin A processing and emerin targeting. Nucleus 5, 66–74 10.4161/nucl.28068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreiber K.H. and Kennedy B.K. (2013) When lamins go bad: nuclear structure and disease. Cell 152, 1365–1375 10.1016/j.cell.2013.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamczyk M.R., del Campo L. and Andrés V. (2018) Aging in the cardiovascular system: lessons from Hutchinson–Gilford progeria syndrome. Annu. Rev. Physiol. 80, 27–48 10.1146/annurev-physiol-021317-121454 [DOI] [PubMed] [Google Scholar]
- 31.Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L. et al. (2003) Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature 423, 293–298 10.1038/nature01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I. et al. (2003) Lamin A truncation in Hutchinson–Gilford progeria. Science 300, 2055 10.1126/science.1084125 [DOI] [PubMed] [Google Scholar]
- 33.Navarro C.L., Cadinanos J., De Sandre-Giovannoli A., Bernard R., Courrier S., Boccaccio I. et al. (2005) Loss of ZMPSTE24 (FACE-1) causes autosomal recessive restrictive dermopathy and accumulation of lamin A precursors. Hum. Mol. Genet. 14, 1503–1513 10.1093/hmg/ddi159 [DOI] [PubMed] [Google Scholar]
- 34.Cenni V., D'Apice M.R., Garagnani P., Columbaro M., Novelli G., Franceschi C. et al. (2018) Mandibuloacral dysplasia: a premature ageing disease with aspects of physiological ageing. Ageing Res. Rev. 42, 1–13 10.1016/j.arr.2017.12.001 [DOI] [PubMed] [Google Scholar]
- 35.Goldman R.D., Shumaker D.K., Erdos M.R., Eriksson M., Goldman A.E., Gordon L.B. et al. (2004) Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proc. Natl. Acad. Sci. U.S.A. 101, 8963–8968 10.1073/pnas.0402943101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shumaker D.K., Dechat T., Kohlmaier A., Adam S A., Bozovsky M.R., Erdos M.R. et al. (2006) Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. U.S.A. 103, 8703–8708 10.1073/pnas.0602569103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scaffidi P. and Misteli T. (2006) Lamin A-dependent nuclear defects in human aging. Science 312, 1059–1063 10.1126/science.1127168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu B., Wang J., Chan K.M., Tjia W.M., Deng W., Guan X. et al. (2005) Genomic instability in laminopathy-based premature aging. Nat. Med. 11, 780–785 10.1038/nm1266 [DOI] [PubMed] [Google Scholar]
- 39.Liu Y., Rusinol A., Sinensky M., Wang Y. and Zou Y. (2006) DNA damage responses in progeroid syndromes arise from defective maturation of prelamin A. J. Cell Sci. 119, 4644–4649 10.1242/jcs.03263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bridger J.M. and Kill I.R. (2004) Aging of Hutchinson–Gilford progeria syndrome fibroblasts is characterised by hyperproliferation and increased apoptosis. Exp. Gerontol. 39, 717–724 10.1016/j.exger.2004.02.002 [DOI] [PubMed] [Google Scholar]
- 41.Allsopp R.C., Vaziri H., Patterson C., Goldstein S., Younglai E V., Futcher A B. et al. (1992) Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 89, 10114–8 10.1073/pnas.89.21.10114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Decker M.L., Chavez E., Vulto I. and Lansdorp P.M. (2009) Telomere length in Hutchinson–Gilford progeria syndrome. Mech. Ageing Dev. 130, 377–383 10.1016/j.mad.2009.03.001 [DOI] [PubMed] [Google Scholar]
- 43.Dahl K.N., Scaffidi P., Islam M.F., Yodh A.G., Wilson K.L. and Misteli T. (2006) Distinct structural and mechanical properties of the nuclear lamina in Hutchinson–Gilford progeria syndrome. Proc. Natl. Acad. Sci. U.S.A. 103, 10271–6 10.1073/pnas.0601058103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lammerding J., Fong L.G., Ji J.Y., Reue K., Stewart C.L., Young S.G. et al. (2006) Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 281, 25768–25780 10.1074/jbc.M513511200 [DOI] [PubMed] [Google Scholar]
- 45.Verstraeten V.L., Ji J.Y., Cummings K.S., Lee R.T. and Lammerding J. (2008) Increased mechanosensitivity and nuclear stiffness in Hutchinson–Gilford progeria cells: effects of farnesyltransferase inhibitors. Aging Cell 7, 383–393 10.1111/j.1474-9726.2008.00382.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Booth E.A., Spagnol S.T., Alcoser T.A. and Dahl K.N. (2015) Nuclear stiffening and chromatin softening with progerin expression leads to an attenuated nuclear response to force. Soft Matter 11, 6412–6418 10.1039/C5SM00521C [DOI] [PubMed] [Google Scholar]
- 47.McCord R.P., Nazario-Toole A., Zhang H., Chines P.S., Zhan Y., Erdos M.R. et al. (2013) Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson–Gilford progeria syndrome. Genome Res. 23, 260–269 10.1101/gr.138032.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chojnowski A., Ong P.F., Foo M.X.R., Liebl D., Hor L.-P., Stewart C.L. et al. (2020) Heterochromatin loss as a determinant of progerin-induced DNA damage in Hutchinson–Gilford progeria. Aging Cell 19, e13108 10.1111/acel.13108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pegoraro G., Kubben N., Wickert U., Göhler H., Hoffmann K. and Misteli T. (2009) Ageing-related chromatin defects through loss of the NURD complex. Nat. Cell Biol. 11, 1261–1267 10.1038/ncb1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Micco R., Sulli G., Dobreva M., Liontos M., Botrugno O.A., Gargiulo G. et al. (2011) Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat. Cell Biol. 13, 292–302 10.1038/ncb2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H., Gu X., Su I., Bottino R., Contreras J.L. and Tarakhovsky A. (2009) Polycomb protein Ezh2 regulates pancreatic β-cell Ink4a/Arf expression and proliferation in aging. Genes Dev. 23, 975–985 10.1101/gad.1742509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sola-Carvajal A., Revêchon G., Helgadottir H.T., Whisenant D., Hagblom R., Döhla J. et al. (2019) Accumulation of progerin affects the symmetry of cell division and is associated with impaired Wnt signaling and the mislocalization of nuclear envelope proteins. J. Invest. Dermatol. 139, 2272–2280 10.1016/j.jid.2019.05.005 [DOI] [PubMed] [Google Scholar]
- 53.Scaffidi P. and Misteli T. (2005) Reversal of the cellular phenotype in the premature aging disease Hutchinson–Gilford progeria syndrome. Nat. Med. 11, 440–445 10.1038/nm1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filesi I., Gullotta F., Lattanzi G., D'Apice M.R., Capanni C., Nardone A.M. et al. (2005) Alterations of nuclear envelope and chromatin organization in mandibuloacral dysplasia, a rare form of laminopathy. Physiol. Genom. 23, 150–158 10.1152/physiolgenomics.00060.2005 [DOI] [PubMed] [Google Scholar]
- 55.Lattanzi G., Columbaro M., Mattioli E., Cenni V., Camozzi D., Wehnert M. et al. (2007) Pre-lamin A processing is linked to heterochromatin organization. J. Cell Biochem. 102, 1149–1159 10.1002/jcb.21467 [DOI] [PubMed] [Google Scholar]
- 56.Gonzalez-Suarez I., Redwood A.B., Perkins S.M., Vermolen B., Lichtensztejin D., Grotsky D.A. et al. (2009) Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 28, 2414–2427 10.1038/emboj.2009.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graziano S., Kreienkamp R., Coll-Bonfill N. and Gonzalo S. (2018) Causes and consequences of genomic instability in laminopathies: Replication stress and interferon response. Nucleus 9, 289–306 10.1080/19491034.2018.1454168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benson E.K., Lee S.W. and Aaronson S.A. (2010) Role of progerin-induced telomere dysfunction in HGPS premature cellular senescence. J. Cell Sci. 123, 2605–2612 10.1242/jcs.067306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cobb A.M., Larrieu D., Warren D.T., Liu Y., Srivastava S., Smith A.J.O. et al. (2016) Prelamin A impairs 53BP1 nuclear entry by mislocalizing NUP153 and disrupting the Ran gradient. Aging Cell 15, 1039–1050 10.1111/acel.12506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh M., Hunt C.R., Pandita R.K., Kumar R., Yang C.-R., Horikoshi N. et al. (2013) Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest. Mol. Cell. Biol. 33, 1210–1222 10.1128/MCB.01676-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wheaton K., Campuzano D., Weili M., Sheinis M., Ho B., Brown G.W. et al. (2017) Progerin-induced replication stress facilitates premature senescence in Hutchinson–Gilford progeria syndrome. Mol. Cell. Biol. 37, 1–18 10.1128/MCB.00659-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cobb A.M., Murray T V., Warren D.T., Liu Y. and Shanahan C.M. (2016) Disruption of PCNA-lamins A/C interactions by prelamin A induces DNA replication fork stalling. Nucleus 7, 498–511 10.1080/19491034.2016.1239685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hilton B.A., Liu J., Cartwright B.M., Liu Y., Breitman M., Wang Y. et al. (2017) Progerin sequestration of PCNA promotes replication fork collapse and mislocalization of XPA in laminopathy-related progeroid syndromes. FASEB J. 31, 3882–3893 10.1096/fj.201700014R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Musich P.R. and Zou Y. (2011) DNA-damage accumulation and replicative arrest in Hutchinson–Gilford progeria syndrome. Biochem. Soc. Trans. 39, 1764–1769 10.1042/BST20110687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kreienkamp R., Graziano S., Coll-Bonfill N., Bedia-Diaz G., Cybulla E., Vindigni A. et al. (2018) A cell-intrinsic interferon-like response links replication stress to cellular aging caused by progerin. Cell Rep. 22, 2006–2015 10.1016/j.celrep.2018.01.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rhind N. and Gilbert D.M. (2013) DNA replication timing. Cold Spring. Harb. Perspect. Med. 5, a010132 10.1101/cshperspect.a010132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arnoult N., Schluth-Bolard C., Letessier A., Drascovic I., Bouarich-Bourimi R., Campisi J. et al. (2010) Replication timing of human telomeres is chromosome arm-specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet. 6, e1000920 10.1371/journal.pgen.1000920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anversa P. and Nadal-Ginard B. (2002) Myocyte renewal and ventricular remodelling. Nature 415, 240–243 10.1038/415240a [DOI] [PubMed] [Google Scholar]
- 69.Bergmann O., Bhardwaj R.D., Bernard S., Zdunek S., Barnabé-Heider F., Walsh S. et al. (2009) Evidence for cardiomyocyte renewal in humans. Science 324, 98–102 10.1126/science.1164680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bergmann O., Zdunek S., Felker A., Salehpour M., Alkass K., Bernard S. et al. (2015) Dynamics of cell generation and turnover in the human heart. Cell 161, 1566–1575 10.1016/j.cell.2015.05.026 [DOI] [PubMed] [Google Scholar]
- 71.Mollova M., Bersell K., Walsh S., Savla J., Das L.T., Park S.Y. et al. (2013) Cardiomyocyte proliferation contributes to heart growth in young humans. Proc. Natl. Acad. Sci. U.S.A. 110, 1446–1451 10.1073/pnas.1214608110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Lange T. (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- 73.Armanios M. (2009) Syndromes of telomere shortening. Annu. Rev. Genom. Hum. Genet. 10, 45–61 10.1146/annurev-genom-082908-150046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Opresko P.L. and Shay J.W. (2017) Telomere-associated aging disorders. Ageing Res. Rev. 33, 52–66 10.1016/j.arr.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crabbe L., Cesare A.J., Kasuboski J.M., Fitzpatrick J.A.J. and Karlseder J. (2012) Human telomeres are tethered to the nuclear envelope during postmitotic nuclear assembly. Cell Rep. 2, 1521–1529 10.1016/j.celrep.2012.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wood A.M., Rendtlew Danielsen J.M., Lucas C.A., Rice E.L., Scalzo D., Shimi T. et al. (2014) TRF2 and lamin A/C interact to facilitate the functional organization of chromosome ends. Nat. Commun. 5, 5467 10.1038/ncomms6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dechat T., Gajewski A., Korbei B., Gerlich D., Daigle N., Haraguchi T. et al. (2004) LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J. Cell Sci. 117, 6117–6128 10.1242/jcs.01529 [DOI] [PubMed] [Google Scholar]
- 78.Burla R., Carcuro M., La Torre M., Fratini F., Crescenzi M., D'Apice M.R. et al. (2016) The telomeric protein AKTIP interacts with A- and B-type lamins and is involved in regulation of cellular senescence. Open Biol. 6, 160103 10.1098/rsob.160103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kudlow B.A., Stanfel M.N., Burtner C.R., Johnston E.D. and Kennedy B.K. (2008) Suppression of proliferative defects associated with processing-defective lamin A mutants by hTERT or inactivation of p53. Mol. Biol. Cell 19, 5238–5248 10.1091/mbc.e08-05-0492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Y., Zhou G., Bruno I.G., Zhang N., Sho S., Tedone E. et al. (2019) Transient introduction of human telomerase mRNA improves hallmarks of progeria cells. Aging Cell 18, e12979 10.1111/acel.12979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller J.D., Ganat Y.M., Kishinevsky S., Bowman R.L., Liu B., Tu E.Y. et al. (2013) Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 13, 691–705 10.1016/j.stem.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wright W.E., Piatyszek M.A., Rainey W.E., Byrd W. and Shay J.W. (1996) Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 18, 173–179 [DOI] [PubMed] [Google Scholar]
- 83.Kirwan M. and Dokal I. (2009) Dyskeratosis congenita, stem cells and telomeres. Biochim. Biophys. Acta 1792, 371–379 10.1016/j.bbadis.2009.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vulliamy T., Marrone A., Szydlo R., Walne A., Mason P.J. and Dokal I. (2004) Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat. Genet. 36, 447–449 10.1038/ng1346 [DOI] [PubMed] [Google Scholar]
- 85.Aguado J., Sola-Cavajal A., Cancila V., Revechon G., Ong P.F., Jones-Weinert C.W. et al. (2019) Sequence-specific inhibition of DNA damage response at telomeres improves the detrimental phenotypes of Hutchinson–Gilford progeria syndrome. Nat. Commun. 10, 4990 10.1038/s41467-019-13018-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.d'Adda di Fagagna, F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T. et al. (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- 87.Osorio F.G., Navarro C.L., Cadinanos J., Lopez-Mejia I.C., Quiros P.M., Bartoli C. et al. (2011) Splicing-directed therapy in a new mouse model of human accelerated aging. Sci. Transl. Med. 3, 106ra107 10.1126/scitranslmed.3002847 [DOI] [PubMed] [Google Scholar]
- 88.Santiago-Fernández O., Osorio F.G., Quesada V., Rodríguez F., Basso S., Maeso D. et al. (2019) Development of a CRISPR/Cas9-based therapy for Hutchinson–Gilford progeria syndrome. Nat. Med. 25, 423–426 10.1038/s41591-018-0338-6 [DOI] [PMC free article] [PubMed] [Google Scholar]