Abstract
Currently, the development of new effective drugs for cancer therapy is not only hindered by development costs, drug efficacy, and drug safety but also by the rapid occurrence of drug resistance in cancer. Hence, new tools are needed to study the underlying mechanisms in cancer. Here, we discuss the current use of metabolic modelling approaches to identify cancer-specific metabolism and find possible new drug targets and drugs for repurposing. Furthermore, we list valuable resources that are needed for the reconstruction of cancer-specific models by integrating various available datasets with genome-scale metabolic reconstructions using model-building algorithms. We also discuss how new drug targets can be determined by using gene essentiality analysis, an in silico method to predict essential genes in a given condition such as cancer and how synthetic lethality studies could greatly benefit cancer patients by suggesting drug combinations with reduced side effects.
Keywords: cancer, drug repurposing, drug target discovery, metabolic modelling, personalized medicine, systems biology
Introduction
Since Otto Warburg, it is known that some cancer cells have an altered metabolism such as preferring the production of ATP from aerobic glycolysis over oxidative phosphorylation [1]. What was believed to be the consequence of high mutation rates in cancer cells, is now regarded as required rewiring of metabolism, tailored by mutations and selection, to meet the high need for energy and cellular building blocks to sustain rapid proliferation rates [2]. This altered metabolism in cancer cells is an important research topic, as it potentially allows identifying cancer-specific vulnerabilities that could be targeted without harming healthy cells and hence are expected to have fewer side effects.
A wide catalogue of mutations across different tumours was gathered by the COSMIC database [3], as well as large transcriptomic datasets from thousands of cancer cell lines (CCLE [4], NCI-60 [5], 1000 Genomes Project [6,7]) and cancer patients such as TCGA [8] or Metabric [9]. The bottleneck in the understanding of metabolic rewiring is the integration of huge amounts of cancer data gathered from different experimental settings and literature. Genome-scale and context-specific models [10], that have been successfully used for the integration of -omics data, are very promising approaches that allow, among others, to understand how mutations affect cancer metabolism by mapping them onto context-specific models to study their metabolism [11] and to determine if the phenotype can be rescued by alternative pathways.
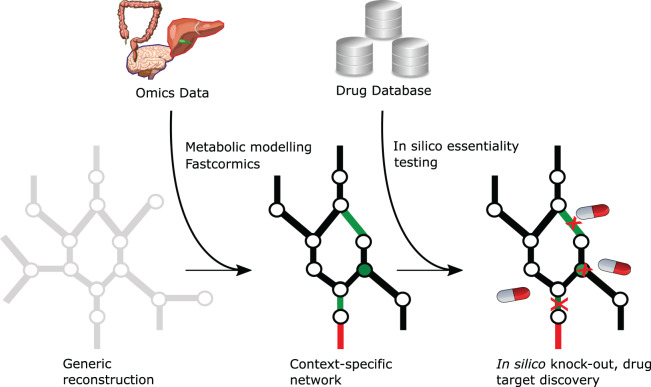
More interesting applications of genome-scale metabolic models are in silico knockout studies to discover cancer-specific essential genes [12] that could serve as potential drug targets or to identify oncometabolites by blocking the flux of the enzyme that consumes these metabolites [13]. A workflow using these approaches has previously been published [14] and is depicted in Figure 1. Because the in vitro identification of drug targets and drug screenings is a meticulous task, with drug combination screenings having endless possibilities, metabolic modelling can be used to narrow down the number of targets, therefore reducing the time and costs of experiments.
Fig. 1.
Drug repurposing workflow using metabolic modelling and public databases. A context-specific reconstruction (black network) can be extracted from a generic reconstruction (grey network) using -omics data and context-specific model reconstruction algorithms such as FASTCORMICS. Circles and lines represent metabolites and reactions, respectively. Genes that can be targeted by existing drugs and oncometabolites are mapped to the model to obtain a set of targetable reactions (green lines) or metabolites (green circles). To identify essential genes, reactions, or metabolites, one or more objective function(s) (red line) can be set and the effect of a drug-induced knockout on the objective function(s) can be simulated by preventing the targeted reactions to carry a flux. Depending on the network topology, the knockout can either have no effect on the flux through the objective function(s), or the flux is reduced if alternative pathways are present, or the knockout can cause a loss of all the flux through the objective function.
Furthermore, context-specific models can be used to identify cancer-specific flux distributions using random sampling [15,16], flux variability analysis [17], FBA [18], parsimonious FBA [19], or cancer-specific sub-pathway activation patterns by combining metabolic models with machine-learning approaches [14].
In this review article, we will discuss the current advances in metabolic modelling in regards to analysing cancer metabolic rewiring as well as possible applications in drug discovery.
Cancer and metabolic modelling
Metabolic alteration in cancer and their potential role as drug target
Oncogenes and tumour suppressor genes have, besides their iconic targets, metabolic targets that have been shown to act as metabolic regulators. For example, the constitutive expression of MYC affects glycolysis [20] and glutamine metabolism [21], whereas mutations or inactivation of the tumour protein p53 can lead to an increase in glycolysis while inhibiting gluconeogenesis [22]. Furthermore, mutations in the metabolic enzymes themselves can be a driving force for cancer such as mutations in the fumarate hydratase [23] and succinate dehydrogenase [24] have previously been associated with cancer by increasing cellular vascularization, invasion, and metastasis through the action of HIF-1 [25]. Loss-of-function mutations can cause the accumulation of fumarate and succinate, that are competitive inhibitors of -ketoglutarate-dependent dioxygenases, perturbing histone and DNA demethylation [26]. Other enzymes (IDH1 and IDH2), when mutated, can also affect the activity of HIF-1 subunits by accumulating 2-hydroxyglutarate, a product of the conversion of -ketoglutarate, mainly taking place in cancer cells [27].
Even though metabolic alterations play a lesser role in the contribution to cancer morphology and progression than mutations, these enzymes, whose deregulation causes the accumulation of sub-products of metabolism, are potential drug targets [28–31]. Other alterations with no transforming power themselves can facilitate the production of building blocks or maintain the redox state. These enabling alterations are often under the control of tumour suppressors or oncogenes and could be considered as potential drug targets.
Metabolic models of cancer and context-specific models
Metabolic models are powerful tools to identify metabolic alterations and mechanisms in human diseases such as Alzheimer’s [32,33], to predict biomarkers for inborn errors of metabolism [34,35], liver metabolism [36,37], pathogen infection of alveolar macrophages [38], obesity [39], Leigh syndrome fibroblasts [40], diabetes [41], co-morbidity [42], obesity and diabetes application have been reviewed in [43] as well as drug target prediction in cancer [12,14,18,44–52].
Two different strategies are being used to study cancer metabolism:
The first approach, a bottom-up approach, aims to reconstruct a cancer core metabolic model, which only contains reactions present in all cancer samples. The small size of these models enable them to be manually curated and thoroughly analysed. For example, a model of ATP production showed that the Warburg effect is dependent on glucose uptake [53], a core model including the main metabolic pathway showed that the blackcancer phenotype induces metabolic changes [45], or the presence of metabolic differences in three cancer core models [18].
In the second approach, cancer genome-scale models are reconstructed using context-specific building algorithms such as MBA [37], iMAT [54], INIT [46,47], GIMME [55], PRIME [49], mCADRE [56], RegrEx [57], CORDA [58], FASTCORE family [14,59,60] using patient transcriptomic data as input. An overview of the model-building algorithms can be found in Supplementary Table S1. Data from various patients and samples can be pooled to reconstruct a single cancer type model or subgroup model of different patients. Even though pooling samples allows building models that are more robust to noise while displaying common alterations, the creation of patient-specific models without pooling allows detecting less common rewiring strategies [61].
Because metabolic rewiring strategies are tightly related to the identification of novel anticancer drugs, metabolic models were often used to identify potential drug targets.
The first genome-scale metabolic model of cancer was presented by [12] in order to study common metabolic alterations in cancer. The model is based on the manual selection of highly expressed core genes from the NCI-60 cancer cell lines as well as a minimal set of reactions needed to activate the core genes via an MBA. A total of 52 cytostatic metabolic drug targets were successfully predicted using in silico gene deletion and validated using sh-RNA screening data.
In a follow-up paper, [44] further investigated the effects of synthetic lethality in FH1-deficient cells, a deficiency that can lead to renal-cell cancer. The same model building approach was used as in [12] to create one deficient and one control model for FH1 that showed that the inhibition of Hmox is synthetically lethal in FH1 deficient cells.
More cancer-specific models shortly followed by integrating cancer data with different genome-scale reconstructions and model-building algorithms for data integration (see Supplementary Table S2) such as models for each of the cell lines in the NCI-60 to identify metabolic sub-pathways that provide energy and lipids for cancer growth [62] or HCC models that allow stratifying patients according to acetate utilization [63]
Recently, due to the decrease in computational demands and reconstruction times of context-specific metabolic models, initiated by the publication of FASTCORE [59] and due to the number of published cancer metabolic models, metabolic modelling could be combined with machine learning.
In a pioneer study, Christian Diener and colleagues [19] used regression approaches to predict cancer growth rates from the TCGA dataset while using the NCI-60 cancer cell line panel and TCGA as a training set. They showed that patients with a high predicted growth rate have a worse survival expectancy. Furthermore, they used the predicted growth rates to obtain the flux distributions via parsimonious FBA for more than 3000 samples using already published cancer models and identified pathways that are up-regulated in cancer such as the pentose phosphate pathway, retinol, branched-chain amino acid metabolism, and ROS detoxification.
In a second study, 10 005 context-specific models for the TCGA dataset were built using an extension of the FASTCORMICS workflow [60] for RNA-seq data [14]. A reverse feature selection approach was used to extract gene and reactions signatures that allow segregating between cancerous and control samples for 13 different cancer types. Furthermore, cancer models were shown to be smaller than their healthy counterparts and reactions from the cancer core metabolism were enriched for essential genes. Generic cancer-type models were also reconstructed to predict drug targets and propose drugs for repurposing in cancer. For colorectal cancer, three of the predicted drugs have been successfully validated in vitro.
Personalized modelling and stratification of cancer patients
A future application of context-specific algorithms is the reconstruction and analysis of patient-specific metabolic models towards personalized treatment. This calls for algorithms that are robust to noise but nevertheless able to capture metabolic variations between different patients that result from inter-tumour variability. In some cancers, such as colon or breast cancer, numerous cancer subtypes were identified, each showing a different prognosis and drug response [64,65]. Being able to accurately model the inter-tumour heterogeneity would allow identifying subtype or even patient-specific drugs and biomarkers. The challenge resides in the distinction between real metabolic variations and noise or algorithm-related bias.
In recent years, benchmarking methods have been proposed to increase the quality of context-specific algorithms [66–68] and their generic reconstructions from which context-specific models are extracted from [69]. Standardizing the benchmarking workflows as well as eliminating any heuristic thresholds during the model reconstruction will improve the quality of the context-specific models so that they could eventually be used in personalised medicine.
Another hurdle that needs to be overcome is the intra-tumour heterogeneity. As tumours can be composed of different clones carrying different mutations, the reconstruction of models based on these biopsies might miss some of the clones and modellers risk to predict drugs that only select for clones that were captured by the biopsy. Furthermore, the use of bulk RNA-seq data might mask the intra-cellular variation. The next logical step would be to take biopsies at different locations of a tumour and to build single-cell models.
In silico gene deletions are used to predict drug targets
The possibility to reconstruct context-specific models based on genomic and transcriptomic data allows for the exploration and comparison between the metabolism of different tissues, conditions, and patients. Thus, the metabolism of cancer cells can be compared with their healthy counterpart tissue (structural analysis) and alternative pathways can be elucidated. Furthermore, new potential drug targets with low toxicity can be predicted using in silico gene deletions or essentiality analysis [70] by focussing on cancer-specific vulnerabilities.
During gene essentiality analysis, the flux controlled by the knockout genes are set to zero (according to the gene–protein–reaction rules) and flux balance analysis [71] is run to determine the maximum flux through an objective function before and after the gene knockout [72]. In general, essential genes are defined as genes whose knockout affects the growth or survival of a cell, therefore, they are often used as a surrogate for potential drug targets. Conventionally, the objective function is defined as biomass production and often used to determine the growth rate of a cell [73]. This might be true for fast proliferating cells such as cancer but not for non-proliferating cells such as neurons. It is therefore important to choose the correct objective function (which differs between cell types, tissues and species) for the model [74] and to define a cancer and tissue-specific biomass instead of relying on the Escherichia coli biomass, currently used in most metabolic models. Even though one could take the ATP demand reaction as an objective for these cells, it would be more suitable to have a well-defined set of metabolic tasks that a cell needs to fulfil [46].
Similar to gene essentiality analysis, synthetic lethality analysis knocks down two genes simultaneously and the flux through the objective function is measured. Whereas the knockout of one gene might not have a significant effect on the cell, the knock-down of two genes can result in lethality or significantly reduced cell functioning. Synthetic lethality studies have already shown promising results in E. coli [72,75] and can be used in anticancer therapy [12,76]. Because cancer cells have high mutation rates and thus some genes are shut down a priori, synthetic lethality takes advantage of these non-lethal mutations in cancer to specifically kill malignant cells without harming healthy cells. Therefore, patients can also benefit from synthetic lethality studies because drug combinations that target multiple genes of a synergistic lethal couple are less likely to cause resistance as it is more difficult for cancer cells to simultaneously develop resistance to two targets [77]. Moreover, the use of drug combinations allows reducing the dosage which is in turn likely to reduce the toxicity of each compound [78–80].
Several algorithms can perform single, double, and multiple knockouts on genes as well as on reactions and metabolites to simulate the effect of oncometabolites (Table 1). Oncometabolites are competitors for the access to the catalytic site of an enzyme, therefore inhibiting the normal conversion of a metabolite. Reactions consuming these metabolites are regarded as inactive during the simulations. Besides the usual brute force approaches, several algorithms were proposed that used a more targeted approach to reduce computational demands. Notably, an algorithm for the study of synergistic lethality was proposed that allows impairing undesired functions while guaranteeing the production of key metabolites [81].
Table 1. Knockout tools.
| Deletion type | Tools and algorithms |
|---|---|
| Single gene deletion | singleGeneDeletion of the Cobra toolbox [83]) (Flux Balance Analysis, MOMA, linear MOMA) |
| Fast-SL [84] | |
| FastMM_singleGeneKO_multi [85] (Flux Balance Analysis) | |
| Double gene deletion | doubleGeneDeletion of the Cobra toolbox (Flux Balance Analysis, MOMA, linear MOMA) |
| Fast-SL | |
| FastMM_doubleGeneKO_multi (Flux Balance Analysis) | |
| gMCSs [82] | |
| OptKnock [86] | |
| Multiple gene deletion | Fast-SL |
| gMCSs [82] | |
| OptKnock | |
| Single reaction deletion | singleRxnDeletion of the Cobra toolbox (Flux Balance Analysis, MOMA, linear MOMA) |
| Single metabolite deletion | singleMetKO from fastMM toolbox |
| Double metabolite deletion | doubleMetKO, from the fastMM toolbox |
Another strategy to identify synergistic lethality has been proposed in 2017 and is based on genetic minimal cut sets or gMCSs [82]. Their framework finds the minimal number of genes that have to be knocked out in order to block a metabolic task such as the biomass production. The analysis is performed on the generic reconstruction to avoid any bias linked to heuristic thresholds in the -omics data during the context-specific model reconstruction. The -omics data is only used to drive the selection.
Consequently, in silico knockouts are promising to find drug targets in cancer as has already been demonstrated in several publications [12,14,44–49,87]. Notably, [87] and [14] performed in silico drug predictions and found Ifenprodil as a potential repurposed drug for the prostate cancer and Naftifine, Mimosine and Ketoconazole for colon cancer, respectively. Both groups validated their respective targets in vitro.
However, robust validation methods need to be established for the predicted drug targets. One possibility is to compare the predictions to an essential gene screenings [88]. Even though there exist different high-throughput screenings that used shRNA [89], RNAi [90], or CRISPR/Cas9 on cell lines [91–93] and patient-derived glioblastoma cells [94] to experimentally determine essential genes, their application is still limited: Screenings cannot be performed for every condition and cell type and they only allow targeting one gene at the time. Thus, complete (synthetic) lethality screenings for all cancer and cell types targeting two genes at the same time would be practically impossible due to the sheer number of possibilities.
However, the Cancer Dependency Map Project made an effort to gather information about gene and drug screenings while combining the data in a comprehensive and regularly updated website (https://depmap.org). The aim of this project is to characterize as many cell lines as possible and identify potential genetic vulnerabilities and drug targets in cancer. The Cancer Dependency Map was created by combining genetics screens, cell line characterization data and drug sensitivity data from Achilles [89,95], DRIVE [96], Score [97], CCLE [4], CCLF [98], PRISM [99], CTRP [100–102], GDSC [103], and CTD2 [104].
From potential candidate gene to drug target validation
Even though the process of finding appropriate drugs for candidate genes is straightforward, some pitfalls will need to be overcome. This can be achieved by using the databases and tools described in the following section.
Challenges
The first challenge is the inconsistency of nomenclature used by the creators of metabolic models and the second is database updates that might significantly alter results between two releases.
Nomenclature
In the metabolic modelling community, there exist different genome-scale metabolic reconstructions for humans (and other organisms) that can in themselves already be seen as a primary database [105–112]. A genome-scale metabolic reconstruction is a collection of all the known genes, reactions, metabolites, and their interactions that are present or can take place in any given cell at the time of reconstruction.
First off, there is no consensus for the identifiers that should be used in a reconstruction. For example, Recon 1 [105] and Recon 2 [108] use Entrez Gene identifiers [113], whereas HMR [110] and Recon 2.2 [111] use Ensembl gene identifiers [114] and HGNC identifiers [115], respectively. The same goes for metabolite identifiers which can be in the BiGG [116], SEED [117], or BioCyc [118] format, but might also have more common identifiers such as the CAS number, ChEBI ID, PubChem ID, or KEGG ID associated. In some versions of some reconstruction, there is sometimes a mix of different identifiers, which makes matching identifiers between the models difficult. Similar problems arise with the names of the proteins, interacting drugs, chemicals, and diseases as many databases associate internal identifiers to them.
The problem of a non-standardized vocabulary is well known in the scientific community [119] and makes data retrieval and integration challenging [120].
Data retrieval and update intervals
Even though many databases offer online tools to the user that are useful for looking up a few genes or drugs but with big data appearing, more holistic approaches are being used and the user wants a whole overview of the data. Unfortunately, not all databases offer direct and free access to downloadable files, making data retrieval unnecessarily difficult.
Another challenge with online databases is maintenance and update intervals. Some databases have scheduled updates, which is per se good practice, but it also requires re-downloading, updating, or adjusting a user-defined script. With updates, besides the addition of content, some entries in a database might be changes or be withdrawn such as for some genes. Unfortunately, other databases are not updated on a regular basis and they are left with outdated information that needs to be revised. It is therefore important to mark the version number of a database.
Resources and databases
In general, for drug target prediction in cancer (and other diseases), the most important databases are interaction databases, which link a gene or mutation to a specific disease, or a drug to a protein. Interaction databases are vital to interconnect the different pieces of information and to create a more global and focussed view on the disease and its treatment strategies.
In their publication, [121], described different approaches for data integration at a systems level and some of the available data repositories. In the following subsection, we will shortly describe the most important databases for gene information, proteins, interactions and simply list others for further information. More information can also be found in the review from [122] that describe drug-related data types, and web-based drug repositioning tools.
Gene databases
After retrieving a list of essential genes, more information about these genes needs to be gathered.
The best-known databases for gene information are Ensembl [114], NCBI Entrez Gene [113], and HGNC [115]. It is useful to download a text file to convert the different identifiers (http://www.genenames.org/cgi-bin/download), Biomart (https://www.ensembl.org/biomart/martview/), David (https://david.ncifcrf.gov/conversion.jsp), or bioDBnet (https://biodbnet-abcc.ncifcrf.gov/db/db2db.php) to give some examples. If one is working with microarray data, the probe IDs should also be converted to the correct genes by downloading the gene annotation files for the used platform (https://www.ncbi.nlm.nih.gov/ for example).
Whereas some databases focus more on the expression of genes across specific tissues or conditions, such as the Human Protein Atlas [123] or CCLE [4], other databases collect information about gene mutations and their associated diseases such as ClinVar [124] or COSMIC [125], which can be useful tools for model validation.
A brief overview of these databases can be found in the Supplementary Tables S3, 43, and S5.
Essential gene screenings
To this date, there exist several large-scale collections of essential gene screenings for human and cancer cell lines that can be used to validate predicted essential genes (Supplementary Table S6). As different methods are used to determine essential genes in a given cell line or tissue, the results are not always comparable and finding a core set of essential genes in cancer is still ongoing. The Cancer Dependency Map (https://depmap.org) is currently gathering and harmonizing several essential gene screenings into one comprehensible platform.
As stated in the beginning, gene essentiality analysis is often performed in metabolic modelling studies in order to predict essential genes that could be considered as drug targets. Here, in vitro performed essential gene screenings can be used to validate the predicted essential genes using statistical test, e.g. a hypergeometric test. This is especially useful to predict drug targets because one could directly assess the effect of the gene deletion in a cancer cell compared with a healthy cell.
Proteins, drug targets, and protein-drug interactions
Besides genes, protein databases have become increasingly important as they include information on the protein sequence, structure, and biological function(s), which are relevant for drug target prediction and validation.
The first available protein database was The Protein Data Bank [126] (https://www.wwpdb.org/), which currently stores more than 150 000 entries on the protein structure. Whereas some databases, such as PDB, focus more on the 3D structure of a protein, other databases such as UniProt [127] focus more on the sequence of a protein. These can be useful to determine new drug binding sites. There also exist databases that focus more on the biological functions and pathways of a protein such as Gene Ontology [128] and KEGG [129]. Extensive lists on protein databases with their advantages and drawbacks have already been discussed elsewhere [130], here we will focus more on protein interaction databases for drug discovery.
By linking the predicted essential genes with their respective proteins, protein interaction or protein binding databases can be used to retrieve a list of known drugs or chemicals that interact with these proteins/genes. Examples of such databases are the Binding Database [131], which mainly gives information on the binding affinity between a protein and a ligand but also pharmacokinetics, 3D structures, and links to other databases, the DrugBank [132], which focusses more on the drugs themselves and its pharmacokinetics but also provides information on the protein targets and the type of interaction (i.e. inhibitor, activator, substrate,…) and the Stitch [133] database, which is a collection of chemical and protein interaction networks with biological evidence that also focuses on the interactions between chemicals. There also exist other protein and drug interaction databases which are listed in Table 2.
Table 2. Drug and interaction databases.
| Name | Description | URL | Citation |
|---|---|---|---|
| BindingDB | Protein binding database | https://www.bindingdb.org | [134] |
| CancerDR: Cancer Drug Resistance Database | Collection of 148 anticancer drugs, their targets and effectiveness | http://crdd.osdd.net/raghava/cancerdr/ | [135] |
| CancerResource | Drug-target interactions in cancer | http://data-analysis.charite.de/care/ | [136] |
| CGP: Cancer Genome Project | Screening of cancer cell lines with drug response data (now included in COSMIC) | http://www.sanger.ac.uk/genetics/CGP/CellLines/ | [137] |
| ChEMBL | Drug bioactivity data | https://www.ebi.ac.uk/chembl/ | [138] |
| Connectivity Map | Drug screenings | https://clue.io/ | [139] |
| CTD: Comparative Toxicogenomics Database | Gene-Drug-Disease interactions | http://ctdbase.org/ | [140] |
| CTRP: Cancer Therapeutics Response Portal | Drug Sensitivity in Cancer, 860 cell lines and 481 compounds | https://portals.broadinstitute.org/ctrp/ | [100] |
| DGIdb: The Drug Gene Interaction Database | Gene-Drug interactions | http://dgidb.genome.wustl.edu/ | [141] |
| DrugBank | Gene-Drug interactions and drug information | https://www.drugbank.ca/ | [142] |
| gCSI: The Genentech Cell Line Screening Initiative | Independent screening of 410 cancer cell lines to 16 agents of CCLE and GDSC data | http://research-pub.gene.com/gCSI-cellline-data/ | [143] |
| GDSC: Genomics of Drug Sensitivity in Cancer | Drug response data and drug sensitivity in cancer | https://www.cancerrxgene.org/ | [103] |
| Growth rate inhibition metrics | Dose-response data for breast cancer (from LINCS) | http://www.grcalculator.org/grtutorial/Home.html | |
| GSK: GlaxoSmithKline cell line collection | Response profiles of 19 compounds in 311 cell lines | [144] | |
| Hetionet | Combination of 29 public databases on genes, disease, drugs, side effects,… | https://het.io/ | [145] |
| IDG: Illuminating the Druggable Genome | Drug-targeted protein families | https://druggablegenome.net/ | [146] |
| Kegg Drug | Information on drugs and their targets | https://www.genome.jp/kegg/drug/ | [129] |
| LINCS: Library of Integrated Network-Based Cellular Signatures | Gene expression and drugs | http://www.lincsproject.org/ | [147] |
| NPC: NCGC Pharmaceutical Collection | Drug screening data & | https://tripod.nih.gov/npc/ | [148] |
| Orphanet | Rare diseases and orphan drugs | http://www.orpha.net | [149] |
| Pharmacodb | Collection of anticancer drug screenings | http://pharmacodb.ca/ | [150] |
| Pharos | Knowledgebase for the druggable genome | https://pharos.nih.gov/idg/index | [151] |
| PubChem | Chemical database | https://pubchem.ncbi.nlm.nih.gov/ | [152] |
| repoDB | Clinical trial and repositioning database | http://apps.chiragjpgroup.org/repoDB/ | [153] |
| SIDER: Side Effect Resource | Side effect database for drugs | http://sideeffects.embl.de/ | [154] |
| STITCH | Drug Target Discovery | http://stitch.embl.de/ | [133] |
| SuperTarget | Drug targets, side effects | http://insilico.charite.de/supertarget/ | [155] |
| T3DB | Gene-toxin database | http://www.t3db.ca/ | [156] |
| TCM Database | in silico drug screenings of Traditional Chinese medicine | http://tcm.cmu.edu.tw/ | [157] |
| The Drug Repurposing Hub | Drug repurposing | https://clue.io/repurposing | [158] |
| Transformer (former SuperCYP) | Cytochrome-drug interactions | http://bioinformatics.charite.de/transformer/ | [159] |
| TTD: Therapeutic Target Database | Drug targets | http://bidd.nus.edu.sg/group/cjttd/ | [160] |
| UniProt | Protein database | www.uniprot.org/ | [127] |
| YaTCM | Linking traditional Chinese medicine to targets and diseases | http://cadd.pharmacy.nankai.edu.cn/yatcm/home | [161] |
Drugs and side effects
Not only the development of new drugs is greatly hampered by drug efficacy and safety [162], severe side effects are responsible for fails during clinical trials, therefore, minimizing the toxicity of the drugs is necessary. Chemotherapeutic agents, for example, target proteins that are present in all rapidly proliferating cells, cancer cells as well as healthy cells causing the side effect. Targeted cancer drugs, on the other hand, are more selective but also come with side effects and are not always able to eradicate all cancer cells due to cancer heterogeneity. The appearance of side effects of these more targeted drugs can be explained by the drug’s affinity for similar binding sites on another protein also called off-targets [163].
Moreover, using available data on drugs and their interactions, side effects of a drug have already been predicted solely based on in silico modelling [164–166]. By combining metabolic modelling with a drug repurposing workflow, the risk of severe side effects can be reduced by suggesting a combination of two or more lower dosed drugs than one single highly dosed drug based on the metabolic modelling results. For example, one could find drug synergies between currently used anti-cancer drugs and other drugs that might allow lowering the dose of the anti-cancer drug.
Databases and datasets for cancer
There exist many different datasets of varying sizes, quality, and research focus that are stored on platforms and repositories such as NCBI Gene Expression Omnibus (GEO) [167] and ArrayExpress [168] but more specific datasets such as The Cancer Genome Atlas with more than 11000 patient samples across 33 different tumour types [8], NCI-60 [5], 1000 Genomes Project [6,7] or the Cancer Cell Line Encyclopedia [4] also exist. For a more exhaustive list of cancer datasets, see Supplementary Table S7.
Discussion
To study metabolic rewiring, two different strategies were adopted: the first focuses mostly on manually curated models of the core metabolism that were built from scratch or were extracted via model-building algorithms from already published models that were then extensively curated. The first strategy is very time consuming and only permits to capture more generic rewiring strategies. The second strategy takes advantage of the capacity of model building algorithms to reconstruct a large number of context-specific models to perform statistically relevant analysis. Whereas this approach is more subjected to noise and algorithm-related bias, it enables to capture metabolic rewiring strategies in different samples, tissues, cancers, and sub-populations.
Although context-specific metabolic models were successfully used to integrate patient data, their application to study tumour samples is still dependent of the accuracy of the model-building algorithm, the quality of the input reconstruction and the discretization/integration function used [66,67]. If the algorithm is too conservative, it will wrongly exclude lowly expressed reactions, mark alternative pathways as inactive and therefore overestimate the number of essential genes. The inverse is equally true, a model that falsely calls alternative pathways as active due to relaxed thresholds will underestimate the number of essential genes. Even though the remaining process of finding a matching drug in a database for a predicted candidate gene is straightforward, huge improvements can still be done on the modelling side, notably of the biomass composition. Currently, a very reduced number of biomass functions is published that are often used regardless of the tissue type and proliferation speed. The prediction power of a model could be improved by using more adapted biomass formulation, which considers fast and slowly proliferating cells.
Furthermore, the reaction to a drug can vary drastically from one patient to another where one might not be responding at all and another will suffer adverse effects. Therefore, patient stratification and tailored drug treatments are going to be a major challenge to find the most efficient drug or drug combination with the least side effects.
Here, metabolic modelling could be applied to predict patient-specific groups by using classifiers such as biomarkers or gene signatures that allow assigning patients into different metabolic groups or by using the metabolic variation captured by the metabolic models. These models could then be used to predict personalized drug targets and, eventually, treatments. Another important aim is to find drug combinations, which allow us to lower the overall dose and consequently reducing drug toxicity. But more importantly, cancer cells are less likely to simultaneously develop resistance to two (or more) different drugs and therefore drug combination treatments could show a higher success rate in killing cancer cells. Even though double and triple knockouts are possible, they cannot be performed experimentally for all drug combinations. As the development of a new drug is very risky and time-consuming, proposing drugs for repurposing using metabolic modelling could drastically help to develop new treatments.
Perspectives
Highlight the importance of the field: In this review, we describe a roadmap about how metabolic modelling can be used for drug discovery. We also cite the most important resources (databases and datasets) that can be used to determine novel drug targets and drugs.
A summary of the current thinking: Current cancer therapies often fail due to the appearance of resistance inside the tumour. Metabolic rewiring is a known hallmark of cancer, thus using metabolic modelling can be used to identify cancer-specific vulnerabilities and predict novel drug targets.
A comment on future directions: For the future, using personalized medicine, the creation of patient-specific models that capture inter-tumour heterogeneity as well as single-cell RNA-seq model that capture intra-tumour heterogeneity will greatly improve the drug response of a patient by increasing the effectiveness of a drug and reducing its side effect.
Competing Interests
The authors declare that there are no competing interests associated with this manuscript.
Supplementary Material
References
- 1.Hecht F. (1987) On the origins of cancer genetics and cytogenetics. Cancer Genet. Cytogenet. 29, 187–190 10.1016/0165-4608(87)90050-1 [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D. and Weinberg R.A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 3.Bamford S., Dawson E., Forbes S., Clements J., Pettett R., Dogan A. et al. (2004) The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 91, 355–358 10.1038/sj.bjc.6601894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S. et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shoemaker R.H. (2006) The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 6, 813–823 10.1038/nrc1951 [DOI] [PubMed] [Google Scholar]
- 6.Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Abecasis G.R., Bentley D.R. et al. (2015) A global reference for human genetic variation. Nature 526, 68–74 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J. et al. (2015) An integrated map of structural variation in 2504 human genomes. Nature 526, 75–81 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang K., Creighton C.J., Davis C., Donehower L., Drummond J., Wheeler D. et al. (2013) The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 10.1038/ng.2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J. et al. (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsson A. and Nielsen J. (2017) Genome scale metabolic modeling of cancer. Metab. Eng. 43, 103–112 10.1016/j.ymben.2016.10.022 [DOI] [PubMed] [Google Scholar]
- 11.Wu X. and Li G. (2016) Prevalent accumulation of non-optimal codons through somatic mutations in human cancers. PLoS ONE 11, e0160463 10.1371/journal.pone.0160463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Folger O., Jerby L., Frezza C., Gottlieb E., Ruppin E. and Shlomi T. (2011) Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 7, 501 10.1038/msb.2011.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghaffari P., Mardinoglu A., Asplund A., Shoaie S., Kampf C., Uhlen M. et al. (2015) Identifying anti-growth factors for human cancer cell lines through genome-scale metabolic modeling. Sci. Rep. 5, 8183 10.1038/srep08183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pacheco M.P., Bintener T., Ternes D., Kulms D., Haan S., Letellier E. et al. (2019) Identifying and targeting cancer-specific metabolism with network-based drug target prediction. EBioMedicine 43, 98–106 10.1016/j.ebiom.2019.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Özcan E. and Çakir T. (2016) Reconstructed metabolic network models predict flux-level metabolic reprogramming in glioblastoma. Front. Neurosci. 10, 1–11 10.3389/fnins.2016.00156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turanli B., Zhang C., Kim W., Benfeitas R., Uhlen M., Arga K.Y. et al. (2019) Discovery of therapeutic agents for prostate cancer using genome-scale metabolic modeling and drug repositioning. EBioMedicine 42, 386–396 10.1016/j.ebiom.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bordbar A., Monk J.M., King Z.A. and Palsson B.O. (2014) Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 15, 107–120 10.1038/nrg3643 [DOI] [PubMed] [Google Scholar]
- 18.Di Filippo M., Colombo R., Damiani C., Pescini D., Gaglio D., Vanoni M. et al. (2016) Zooming-in on cancer metabolic rewiring with tissue specific constraint-based models. Comput. Biol. Chem. 62, 60–69 10.1016/j.compbiolchem.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 19.Diener C. and Resendis-Antonio O. (2016) Personalized prediction of proliferation rates and metabolic liabilities in cancer biopsies. Front. Physiol. 7, 1–11 10.3389/fphys.2016.00644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Connell B.C., Cheung A.F., Simkevich C.P., Tam W., Ren X., Mateyak M.K. et al. (2003) A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 278, 12563–12573 10.1074/jbc.M210462200 [DOI] [PubMed] [Google Scholar]
- 21.DeBerardinis R.J. and Cheng T. (2010) Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 29, 313–324 10.1038/onc.2009.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N.C., Nakano K., Bartrons R. et al. (2006) TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120 10.1016/j.cell.2006.05.036 [DOI] [PubMed] [Google Scholar]
- 23.Tomlinson I.P.M., Alam N.A., Rowan A.J., Barclay E., Jaeger E.E.M., Kelsell D. et al. (2002) Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 30, 406–410 10.1038/ng849 [DOI] [PubMed] [Google Scholar]
- 24.Baysal B.E., Ferrell R.E., Willett-Brozick J.E., Lawrence E.C., Myssiorek D., Bosch A. et al. (2000) Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 287, 848–851 10.1126/science.287.5454.848 [DOI] [PubMed] [Google Scholar]
- 25.Selak M.A., Armour S.M., MacKenzie E.D., Boulahbel H., Watson D.G., Mansfield K.D. et al. (2005) Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 7, 77–85 10.1016/j.ccr.2004.11.022 [DOI] [PubMed] [Google Scholar]
- 26.Xiao M., Yang H., Xu W., Ma S., Lin H., Zhu H. et al. (2012) Inhibition of -KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 26, 1326–1338 10.1101/gad.191056.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S.H. et al. (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of -ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 10.1016/j.ccr.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imming P., Sinning C. and Meyer A. (2006) Drugs, their targets and the nature and number of drug targets. Nat. Rev. Drug Discov. 5, 821–834 10.1038/nrd2132 [DOI] [PubMed] [Google Scholar]
- 29.Galluzzi L., Kepp O., Heiden M.G.V. and Kroemer G. (2013) Metabolic targets for cancer therapy. Nat. Rev. Drug Discov. 12, 829–846 10.1038/nrd4145 [DOI] [PubMed] [Google Scholar]
- 30.Frezza C., Pollard P.J. and Gottlieb E. (2011) Inborn and acquired metabolic defects in cancer. J. Mol. Med. 89, 213–220 10.1007/s00109-011-0728-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wettersten H.I., Aboud O.A., Lara P.N. and Weiss R.H. (2017) Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol. 13, 410–419 10.1038/nrneph.2017.59 [DOI] [PubMed] [Google Scholar]
- 32.Lewis N.E., Schramm G., Bordbar A., Schellenberger J., Andersen M.P., Cheng J.K. et al. (2010) Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nat. Biotechnol. 28, 1279–1285 10.1038/nbt.1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stempler S., Yizhak K. and Ruppin E. (2014) Integrating transcriptomics with metabolic modeling predicts biomarkers and drug targets for Alzheimer’s disease. PLoS ONE 9, e105383 10.1371/journal.pone.0105383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shlomi T., Cabili M.N. and Ruppin E. (2009) Predicting metabolic biomarkers of human inborn errors of metabolism. Mol. Syst. Biol. 5, 263 10.1038/msb.2009.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahoo S., Franzson L., Jonsson J.J. and Thiele I. (2012) A compendium of inborn errors of metabolism mapped onto the human metabolic network. Mol. Biosyst. 8, 2545 10.1039/c2mb25075f [DOI] [PubMed] [Google Scholar]
- 36.Gille C., Bölling C., Hoppe A., Bulik S., Hoffmann S., Hübner K. et al. (2010) HepatoNet1: a comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology. Mol. Syst. Biol. 6, 411 10.1038/msb.2010.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jerby L., Shlomi T. and Ruppin E. (2010) Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol. Syst. Biol. 6, 401 10.1038/msb.2010.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bordbar A., Lewis N.E., Schellenberger J., Palsson B.Ø. and Jamshidi N. (2010) Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions. Mol. Syst. Biol. 6, 422 10.1038/msb.2010.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mardinoglu A., Agren R., Kampf C., Asplund A., Nookaew I., Jacobson P. et al. (2013) Integration of clinical data with a genome-scale metabolic model of the human adipocyte. Mol. Syst. Biol. 9, 649 10.1038/msb.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vo T.D., Paul Lee W.N. and Palsson B.O. (2007) Systems analysis of energy metabolism elucidates the affected respiratory chain complex in Leigh’s syndrome. Mol. Genet. Metab. 91, 15–22 10.1016/j.ymgme.2007.01.012 [DOI] [PubMed] [Google Scholar]
- 41.Bordbar A., Feist A.M., Usaite-Black R., Woodcock J., Palsson B.O. and Famili I. (2011) A multi-tissue type genome-scale metabolic network for analysis of whole-body systems physiology. BMC Syst. Biol. 5, 180 10.1186/1752-0509-5-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee D.S., Park J., Kay K.A., Christakis N.A., Oltvai Z.N. and Barabasi A.L. (2008) The implications of human metabolic network topology for disease comorbidity. Proc. Natl. Acad. Sci. U.S.A. 105, 9880–9885 10.1073/pnas.0802208105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Väremo L., Nookaew I. and Nielsen J. (2013) Novel insights into obesity and diabetes through genome-scale metabolic modeling. Front. Physiol. 4, 1–7 10.3389/fphys.2013.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frezza C., Zheng L., Folger O., Rajagopalan K.N., MacKenzie E.D., Jerby L. et al. (2011) Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature 477, 225–228 10.1038/nature10363 [DOI] [PubMed] [Google Scholar]
- 45.Resendis-Antonio O., Checa A. and Encarnación S. (2010) Modeling core metabolism in cancer cells: surveying the topology underlying the Warburg effect. PLoS ONE 5, e12383 10.1371/journal.pone.0012383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agren R., Mardinoglu A., Asplund A., Kampf C., Uhlen M. and Nielsen J. (2014) Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol. Syst. Biol. 10, 721 10.1002/msb.v10.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agren R., Bordel S., Mardinoglu A., Pornputtapong N., Nookaew I. and Nielsen J. (2012) Reconstruction of genome-scale active metabolic networks for 69 human cell types and 16 cancer types using INIT. PLoS Comput. Biol. 8, e1002518 10.1371/journal.pcbi.1002518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L., Zhou X., Ching W.K. and Wang P. (2010) Predicting enzyme targets for cancer drugs by profiling human metabolic reactions in NCI-60 cell lines. BMC Bioinform. 11, 501 10.1186/1471-2105-11-501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yizhak K. and Gaude E., Le Dévédec S., Waldman Y.Y., Stein G.Y., van de Water B. et al. (2014) Phenotype-based cell-specific metabolic modeling reveals metabolic liabilities of cancer. eLife 3, 1–23 10.7554/eLife.03641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marín de Mas I., Aguilar E., Zodda E., Balcells C., Marin S., Dallmann G. et al. (2018) Model-driven discovery of long-chain fatty acid metabolic reprogramming in heterogeneous prostate cancer cells. PLoS Comput. Biol. 14, e1005914 10.1371/journal.pcbi.1005914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanhaiya K. and Tyagi-Tiwari D. (2019) Identification of drug targets in breast cancer metabolic network. J. Comput. Biol. 10.1089/cmb.2019.0258 [DOI] [PubMed] [Google Scholar]
- 52.Zhang C., Aldrees M., Arif M., Li X., Mardinoglu A. and Aziz M.A. (2019) Elucidating the reprograming of colorectal cancer metabolism using genome-scale metabolic modeling. Front. Oncol. 9, 681 10.3389/fonc.2019.00681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vazquez A., Liu J., Zhou Y. and Oltvai Z.N. (2010) Catabolic efficiency of aerobic glycolysis: the Warburg effect revisited. BMC Syst. Biol. 4, 58 10.1186/1752-0509-4-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shlomi T., Cabili M.N., Herrgård M.J., Palsson B.Ø. and Ruppin E. (2008) Network-based prediction of human tissue-specific metabolism. Nat. Biotechnol. 26, 1003–1010 10.1038/nbt.1487 [DOI] [PubMed] [Google Scholar]
- 55.Becker S.A. and Palsson B.O. (2008) Context-specific metabolic networks are consistent with experiments. PLoS Comput. Biol. 4, e1000082 10.1371/journal.pcbi.1000082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y., Eddy Ja. and Price N.D. (2012) Reconstruction of genome-scale metabolic models for 126 human tissues using mCADRE. BMC Syst. Biol. 6, 153 10.1186/1752-0509-6-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Estévez S.R. and Nikoloski Z. (2015) Context-specific metabolic model extraction based on regularized least squares optimization. PLoS ONE 10, e0131875 10.1371/journal.pone.0131875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schultz A. and Qutub A.A. (2016) Reconstruction of tissue-specific metabolic networks using CORDA. PLoS Comput. Biol. 12, 1–33 10.1371/journal.pcbi.1004808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vlassis N., Pacheco M.P. and Sauter T. (2014) Fast reconstruction of compact context-specific metabolic network models. PLoS Comput. Biol. 10, e1003424 10.1371/journal.pcbi.1003424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pacheco M.P., John E., Kaoma T., Heinäniemi M., Nicot N., Vallar L. et al. (2015) Integrated metabolic modelling reveals cell-type specific epigenetic control points of the macrophage metabolic network. BMC Genomics 16, 809 10.1186/s12864-015-1984-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pacheco M.P., Bintener T. and Sauter T. (2019) Towards the network-based prediction of repurposed drugs using patient-specific metabolic models. EBioMedicine 43, 26–27 10.1016/j.ebiom.2019.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feizi A. and Bordel S. (2013) Metabolic and protein interaction sub-networks controlling the proliferation rate of cancer cells and their impact on patient survival. Sci. Rep. 3, 3041 10.1038/srep03041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Björnson E., Mukhopadhyay B., Asplund A., Pristovsek N., Cinar R., Romeo S. et al. (2015) Stratification of hepatocellular carcinoma patients based on acetate utilization. Cell Rep. 13, 2014–2026 10.1016/j.celrep.2015.10.045 [DOI] [PubMed] [Google Scholar]
- 64.Guinney J., Dienstmann R., Wang X., De Reyniès A., Schlicker A., Soneson C. et al. (2015) The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 10.1038/nm.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sveen A., Bruun J., Eide P.W., Eilertsen I.A., Ramirez L., Murumägi A. et al. (2018) Colorectal cancer consensus molecular subtypes translated to preclinical models uncover potentially targetable cancer cell dependencies. Clin. Cancer Res. 24, 794–806 10.1158/1078-0432.CCR-17-1234 [DOI] [PubMed] [Google Scholar]
- 66.Pacheco M.P., Pfau T., Sauter T., Pires Pacheco M., Pfau T. and Sauter T. (2016) Benchmarking procedures for high-throughput context specific reconstruction algorithms. Front. Physiol. 6, 1–19 10.3389/fphys.2015.00410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Opdam S., Richelle A., Kellman B., Li S., Zielinski D.C. and Lewis N.E. (2017) A systematic evaluation of methods for tailoring genome-scale metabolic models. Cell Syst. 4, 318–329 10.1016/j.cels.2017.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jamialahmadi O., Hashemi-Najafabadi S., Motamedian E., Romeo S. and Bagheri F. (2019) A benchmark-driven approach to reconstruct metabolic networks for studying cancer metabolism. PLoS Comput. Biol. 15, e1006936 10.1371/journal.pcbi.1006936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lieven C., Beber M.E., Olivier B.G., Bergmann F.T., Ataman M., Babaei P. et al. (2018) Memote: a community-driven effort towards a standardized genome-scale metabolic model test suite. BioRxiv. p. 350991
- 70.Edwards J.S. and Palsson B.O. (2000) The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc. Natl. Acad. Sci. 97, 5528–5533 10.1073/pnas.97.10.5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orth J.D., Thiele I. and Palsson B.Ø. (2010) What is flux balance analysis? Nat. Biotechnol. 28, 245–248 10.1038/nbt.1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edwards J.S. and Palsson B.O. (2000) Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions. BMC. Bioinformatics. 1, 1 10.1186/1471-2105-1-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feist A.M. and Palsson B.O. (2010) The biomass objective function. Curr. Opin. Microbiol. 13, 344–349 10.1016/j.mib.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tobalina L., Pey J., Rezola A. and Planes F.J. (2016) Assessment of FBA based gene essentiality analysis in cancer with a fast context-specific network reconstruction method. PLoS ONE 11, e0154583 10.1371/journal.pone.0154583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suthers P.F., Zomorrodi A. and Maranas C.D. (2009) Genome-scale gene/reaction essentiality and synthetic lethality analysis. Mol. Syst. Biol. 5, 1–17 10.1038/msb.2009.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaelin W.G. (2005) The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 5, 689–698 10.1038/nrc1691 [DOI] [PubMed] [Google Scholar]
- 77.Lehár J., Krueger A.S., Avery W., Heilbut A.M., Johansen L.M., Price E.R. et al. (2009) Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol. 27, 659–666 10.1038/nbt.1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keith C.T., Borisy Aa. and Stockwell B.R. (2005) Innovation: multicomponent therapeutics for networked systems. Nat. Rev. Drug Discov. 4, 71–78 10.1038/nrd1609 [DOI] [PubMed] [Google Scholar]
- 79.Roth B.L., Sheffler D.J. and Kroeze W.K. (2004) Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat. Rev. Drug Discov. 3, 353–359 10.1038/nrd1346 [DOI] [PubMed] [Google Scholar]
- 80.Sharom J.R., Bellows D.S. and Tyers M. (2004) From large networks to small molecules. Curr. Opin. Chem. Biol. 8, 81–90 10.1016/j.cbpa.2003.12.007 [DOI] [PubMed] [Google Scholar]
- 81.Facchetti G., Zampieri M. and Altafini C. (2012) Predicting and characterizing selective multiple drug treatments for metabolic diseases and cancer. BMC Syst. Biol. 6, 115 10.1186/1752-0509-6-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Apaolaza I., San José-Eneriz E., Tobalina L., Miranda E.E., Garate L., Agirre X. et al. (2017) An in-silico approach to predict and exploit synthetic lethality in cancer metabolism. Nat. Commun. 8, 459 10.1038/s41467-017-00555-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heirendt L., Arreckx S., Pfau T., Mendoza S.N., Richelle A., Heinken A. et al. (2019) Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v. 3.0. Nat. Protoc. 14, 639–702 10.1038/s41596-018-0098-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pratapa A, Balachandran S and Raman K. (2015) Fast-SL: an efficient algorithm to identify synthetic lethal sets in metabolic networks. Bioinformatics 31, 3299–3305 10.1093/bioinformatics/btv352 [DOI] [PubMed] [Google Scholar]
- 85.Li G.H., Dai S, Han F, Li W, Huang J and Xiao W. (2020) FastMM: an efficient toolbox for personalized constraint-based metabolic modeling. BMC Bioinformatics 21, 1–7 10.1186/s12859-020-3410-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burgard A.P., Pharkya P and Maranas C.D. (2003) Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 84, 647–657 10.1002/bit.10803 [DOI] [PubMed] [Google Scholar]
- 87.Turanli B, Zhang C, Kim W, Benfeitas R, Uhlen M, Arga K.Y. et al. (2019) Discovery of therapeutic agents for prostate cancer using genome-scale metabolic modeling and drug repositioning. EBioMedicine 42, 386–396 10.1016/j.ebiom.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gatto F., Miess H., Schulze A. and Nielsen J. (2015) Flux balance analysis predicts essential genes in clear cell renal cell carcinoma metabolism. Sci. Rep. 5, 10738 10.1038/srep10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S. et al. (2014) Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data 1, 140035 10.1038/sdata.2014.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G. et al. (2017) A pathology atlas of the human cancer transcriptome. Science 357, eaan2507 10.1126/science.aan2507 [DOI] [PubMed] [Google Scholar]
- 91.Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K.R., MacLeod G. et al. (2015) High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 10.1016/j.cell.2015.11.015 [DOI] [PubMed] [Google Scholar]
- 92.Wang T., Birsoy K.K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J. et al. (2015) Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blomen V.A., Májek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J. et al. (2015) Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 10.1126/science.aac7557 [DOI] [PubMed] [Google Scholar]
- 94.Toledo C.M., Ding Y., Hoellerbauer P., Davis R.J., Basom R., Girard E.J. et al. (2015) Genome-wide CRISPR-Cas9 screens reveal loss of redundancy between PKMYT1 and WEE1 in glioblastoma stem-like cells. Cell Rep. 13, 2425–2439 10.1016/j.celrep.2015.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aguirre A.J., Meyers R.M., Weir B.A., Vazquez F., Zhang C.Z., Ben-David U. et al. (2016) Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. 6, 914–929 10.1158/2159-8290.CD-16-0154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McDonald E.R., de Weck A., Schlabach M.R., Billy E., Mavrakis K.J., Hoffman G.R. et al. (2017) Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 170, 577–592.e10. 10.1016/j.cell.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 97.Behan F.M., Iorio F., Picco G., Gonçalves E., Beaver cM., Migliardi G. et al. (2019) Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 568, 511–516 10.1038/s41586-019-1103-9 [DOI] [PubMed] [Google Scholar]
- 98.Boehm J.S. and Golub T.R. (2015) An ecosystem of cancer cell line factories to support a cancer dependency map. Nat. Rev. Genet. 16, 373–374 10.1038/nrg3967 [DOI] [PubMed] [Google Scholar]
- 99.Yu C., Mannan A.M., Yvone G.M., Ross K.N., Zhang Y.L., Marton M.A. et al. (2016) High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat. Biotechnol. 34, 419–423 10.1038/nbt.3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rees M.G., Seashore-Ludlow B., Cheah J.H., Adams D.J., Price E.V., Gill S. et al. (2016) Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 12, 109–116 10.1038/nchembio.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Seashore-Ludlow B., Rees M.G., Cheah J.H., Cokol M., Price E.V., Coletti M.E. et al. (2015) Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 5, 1210–1223 10.1158/2159-8290.CD-15-0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Basu A., Bodycombe N.E., Cheah J.H., Price E.V., Liu K., Schaefer G.I. et al. (2013) An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154, 1151–1161 10.1016/j.cell.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S. et al. (2013) Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961 10.1093/nar/gks1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aksoy B.A., Dančík V., Smith K., Mazerik J.N., Ji Z., Gross B. et al. (2017) CTD2 Dashboard: a searchable web interface to connect validated results from the Cancer Target Discovery and Development Network. Database 2017, 54 10.1093/database/bax054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Duarte N.C., Becker S.A., Jamshidi N., Thiele I., Mo M.L., Vo T.D. et al. (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. U.S.A. 104, 1777–1782 10.1073/pnas.0610772104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ma H., Sorokin A., Mazein A., Selkov A., Selkov E., Demin O. et al. (2007) The Edinburgh human metabolic network reconstruction and its functional analysis. Mol. Syst. Biol. 3, 135 10.1038/msb4100177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma H. and Goryanin I. (2008) Human metabolic network reconstruction and its impact on drug discovery and development. Drug Discov. Today 13, 402–408 10.1016/j.drudis.2008.02.002 [DOI] [PubMed] [Google Scholar]
- 108.Thiele I., Swainston N., Fleming R.M.T., Hoppe A., Sahoo S., Aurich M.K. et al. (2013) A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 31, 419–425 10.1038/nbt.2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mardinoglu A., Gatto F. and Nielsen J. (2013) Genome-scale modeling of human metabolism: a systems biology approach. Biotechnol. J. 8, 985–996 10.1002/biot.201200275 [DOI] [PubMed] [Google Scholar]
- 110.Mardinoglu A., Agren R., Kampf C., Asplund A., Uhlen M. and Nielsen J. (2014) Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun. 5, 3083 10.1038/ncomms4083 [DOI] [PubMed] [Google Scholar]
- 111.Swainston N., Smallbone K., Hefzi H., Dobson P.D., Brewer J., Hanscho M. et al. (2016) Recon 2.2: from reconstruction to model of human metabolism. Metabolomics 12, 1–7 10.1007/s11306-016-1051-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brunk E., Sahoo S., Zielinski D.C., Altunkaya A., Dräger A., Mih N. et al. (2018) Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. 36, 272–281 10.1038/nbt.4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maglott D. (2004) Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 33, D54–D58 10.1093/nar/gki031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J. et al. (2017) Ensembl 2017. Nucleic Acids Res. 45, D635–D642 10.1093/nar/gkw1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yates B., Braschi B., Gray K.A., Seal R.L., Tweedie S. and Bruford E.A. (2017) Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 45, D619–D625 10.1093/nar/gkw1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.King Z.A., Lu J., Dräger A., Miller P., Federowicz S., Lerman J.A. et al. (2016) BiGG models: a platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 44, D515–D522 10.1093/nar/gkv1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Henry C.S., DeJongh M., Best A.A., Frybarger P.M., Linsay B. and Stevens R.L. (2010) High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28, 977 10.1038/nbt.1672 [DOI] [PubMed] [Google Scholar]
- 118.Caspi R., Billington R., Ferrer L., Foerster H., Fulcher C.A., Keseler I.M. et al. (2016) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 44, D471–D480 10.1093/nar/gkv1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McMurry J.A., Juty N., Blomberg N., Burdett T., Conlin T., Conte N. et al. (2017) Identifiers for the 21st century: how to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data. PLoS Biol. 15, e2001414 10.1371/journal.pbio.2001414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bodenreider O. (2004) The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 32, 267D–270 10.1093/nar/gkh061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Iskar M., Zeller G., Zhao X.M., van Noort V. and Bork P. (2012) Drug discovery in the age of systems biology: the rise of computational approaches for data integration. Curr. Opin. Biotechnol. 23, 609–616 10.1016/j.copbio.2011.11.010 [DOI] [PubMed] [Google Scholar]
- 122.Turanli B., Altay O., Borén J., Turkez H., Nielsen J., Uhlen M., Arga K.Y. et al. (2019) Systems biology based drug repositioning for development of cancer therapy. Semin. Cancer Biol. 10.1016/j.semcancer.2019.09.020 [DOI] [PubMed] [Google Scholar]
- 123.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A. et al. (2015) Tissue-based map of the human proteome. Science 347, 1260419–1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 124.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S. et al. (2016) ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44, D862–D868 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J. et al. (2017) COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D777–D783 10.1093/nar/gkw1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bernstein F.C., Koetzle T.F., Williams G.J.B., Meyer E.F., Jr, Brice M.D., Rodgers J.R. et al. (1977) The protein data bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 112, 535–542 10.1016/S0022-2836(77)80200-3 [DOI] [PubMed] [Google Scholar]
- 127.Consortium T.U. (2015) UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.The Gene Ontology Consortium (2019) The gene ontology resource: 20 years and still going strong. Nucleic Acids Res. 47, D330–D338 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kanehisa M. and Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xu D. (2012) Protein databases on the internet. In Current Protocols in Protein Science (Hoboken, N.J., ed.), vol. 1, pp. 2.6.1–2.6.17, John Wiley & Sons, Inc., USA. Available from: http://soykb.orghttp://doi.wiley.com/10.1002/0471140864.ps0206s70
- 131.Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L. and Chong J. (2016) BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 44, D1045–D1053 10.1093/nar/gkv1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R. et al. (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Szklarczyk D., Santos A., von Mering C., Jensen L.J., Bork P. and Kuhn M. (2016) STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 44, D380–D384 10.1093/nar/gkv1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kirchmair J., Göller A.H., Lang D., Kunze J., Testa B., Wilson I.D. et al. (2015) Predicting drug metabolism: experiment and/or computation? Nat. Rev. Drug Discov. 14, 387–404 10.1038/nrd4581 [DOI] [PubMed] [Google Scholar]
- 135.Kumar R., Chaudhary K., Gupta S., Singh H., Kumar S., Gautam A. et al. (2013) CancerDR: cancer drug resistance database. Sci. Rep. 3, 1445 10.1038/srep01445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gohlke B.O., Nickel J., Otto R., Dunkel M. and Preissner R. (2016) CancerResource—updated database of cancer-relevant proteins, mutations and interacting drugs. Nucleic Acids Res. 44, D932–D937 10.1093/nar/gkv1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Garnett M.J., Edelman E.J., Heidorn S.J., Greenman C.D., Dastur A., Lau K.W. et al. (2012) Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 10.1038/nature11005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bento A.P., Gaulton A., Hersey A., Bellis L.J., Chambers J., Davies M. et al. (2014) The ChEMBL bioactivity database: an update. Nucleic Acids Res. 42, D1083–D1090 10.1093/nar/gkt1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lamb J. (2007) The connectivity map: a new tool for biomedical research. Nat. Rev. Cancer 7, 54–60 10.1038/nrc2044 [DOI] [PubMed] [Google Scholar]
- 140.Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., King B.L., McMorran R. et al. (2017) The comparative toxicogenomics database: update 2017. Nucleic Acids Res. 45, D972–D978 10.1093/nar/gkw838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wagner A.H., Coffman A.C., Ainscough B.J., Spies N.C., Skidmore Z.L., Campbell K.M. et al. (2016) DGIdb 2.0: mining clinically relevant drug-gene interactions. Nucleic Acids Res. 44, D1036–D1044 10.1093/nar/gkv1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y. et al. (2014) DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 42, D1091–D1097 10.1093/nar/gkt1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Haverty P.M., Lin E., Tan J., Yu Y., Lam B., Lianoglou S. et al. (2016) Reproducible pharmacogenomic profiling of cancer cell line panels. Nature 533, 333–337 10.1038/nature17987 [DOI] [PubMed] [Google Scholar]
- 144.Greshock J., Bachman K.E., Degenhardt Y.Y., Jing J., Wen Y.H., Eastman S. et al. (2010) Molecular target class is predictive of in vitro response profile. Cancer Res. 70, 3677–3686 10.1158/0008-5472.CAN-09-3788 [DOI] [PubMed] [Google Scholar]
- 145.Himmelstein D.S., Lizee A., Hessler C., Brueggeman L., Chen S.L., Hadley D. et al. (2017) Systematic integration of biomedical knowledge prioritizes drugs for repurposing. eLife 6, 16022 10.7554/eLife.26726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rodgers G., Austin C., Anderson J., Pawlyk A., Colvis C., Margolis R. et al. (2018) Glimmers in illuminating the druggable genome. Nat. Rev. Drug Discov. 17, 301–302 10.1038/nrd.2017.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Keenan A.B., Jenkins S.L., Jagodnik K.M., Koplev S., He E., Torre D. et al. (2018) The library of integrated network-based cellular signatures NIH program: system-level cataloging of human cells response to perturbations. Cell Syst. 6, 13–24 10.1016/j.cels.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang R., Southall N., Wang Y., Yasgar A., Shinn P., Jadhav A. et al. (2011) The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci. Transl. Med. 3, 80ps16–80ps16 10.1126/scitranslmed.3001862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pavan S., Rommel K., Marquina M.E.M., Höhn S., Lanneau V. and Rath A. (2017) Clinical practice guidelines for rare diseases: the orphanet database. PLoS ONE 12, e0170365 10.1371/journal.pone.0170365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smirnov P., Kofia V., Maru A., Freeman M., Ho C., El-Hachem N. et al. (2018) PharmacoDB: an integrative database for mining in vitro anticancer drug screening studies. Nucleic Acids Res. 46, D994–D1002 10.1093/nar/gkx911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nguyen D.T., Mathias S., Bologa C., Brunak S., Fernandez N., Gaulton A. et al. (2017) Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Res. 45, D995–D1002 10.1093/nar/gkw1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A. et al. (2016) PubChem substance and compound databases. Nucleic Acids Res. 44, D1202–D1213 10.1093/nar/gkv951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Brown A.S. and Patel C.J. (2017) A standard database for drug repositioning. Sci. Data 4, 170029 10.1038/sdata.2017.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kuhn M., Letunic I., Jensen L.J. and Bork P. (2016) The SIDER database of drugs and side effects. Nucleic Acids Res. 44, D1075–D1079 10.1093/nar/gkv1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hecker N., Ahmed J., von Eichborn J., Dunkel M., Macha K., Eckert A. et al. (2012) SuperTarget goes quantitative: update on drug-target interactions. Nucleic Acids Res. 40, D1113–D1117 10.1093/nar/gkr912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wishart D., Arndt D., Pon A., Sajed T., Guo A.C., Djoumbou Y. et al. (2015) T3DB: the toxic exposome database. Nucleic Acids Res. 43, D928–D934 10.1093/nar/gku1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen C.Y.C. (2011) TCM Database@Taiwan: the world’s largest traditional Chinese medicine database for drug screening In Silico. PLoS ONE 6, e15939 10.1371/journal.pone.0015939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Corsello S.M., Bittker J.A., Liu Z., Gould J., McCarren P., Hirschman J.E. et al. (2017) The drug repurposing hub: a next-generation drug library and information resource. Nat. Med. 23, 405–408 10.1038/nm.4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hoffmann M.F., Preissner S.C., Nickel J., Dunkel M., Preissner R. and Preissner S. (2014) The transformer database: biotransformation of xenobiotics. Nucleic Acids Res. 42, D1113–D1117 10.1093/nar/gkt1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen X., Ji Z.L. and Chen Y.Z. (2002) TTD: therapeutic target database. Nucleic Acids Res. 30, 412–415 10.1093/nar/30.1.412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li B., Ma C., Zhao X., Hu Z., Du T., Xu X. et al. (2018) YaTCM: yet another traditional Chinese medicine database for drug discovery. Comput. Struct. Biotechnol. J. 16, 600–610 10.1016/j.csbj.2018.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Harrison R.K. (2016) Phase II and phase III failures: 2013–2015. Nat. Rev. Drug Discov. 15, 817–818 10.1038/nrd.2016.184 [DOI] [PubMed] [Google Scholar]
- 163.Chartier M., Morency L.P., Zylber M.I. and Najmanovich R.J. (2017) Large-scale detection of drug off-targets: hypotheses for drug repurposing and understanding side-effects. BMC Pharmacol. Toxicol. 18, 18 10.1186/s40360-017-0128-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Boland M.R., Jacunski A., Lorberbaum T., Romano J.D., Moskovitch R. and Tatonetti N.P. (2016) Systems biology approaches for identifying adverse drug reactions and elucidating their underlying biological mechanisms. Wiley Interdiscip. Rev.: Syst. Biol. Med. 8, 104–122 10.1002/wsbm.1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang Z., Clark N.R. and Ma’ayan A. (2016) Drug-induced adverse events prediction with the LINCS L1000 data. Bioinformatics 32, 2338–2345 10.1093/bioinformatics/btw168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shaked I., Oberhardt M.A., Atias N., Sharan R. and Ruppin E. (2016) Metabolic network prediction of drug side effects. Cell Syst. 2, 209–213 10.1016/j.cels.2016.03.001 [DOI] [PubMed] [Google Scholar]
- 167.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M. et al. (2013) NCBI GEO: archive for functional genomics data sets — update. Nucleic Acids Res. 41, D991–D995 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y.A., Williams E. et al. (2015) ArrayExpress update — simplifying data submissions. Nucleic Acids Res. 43, D1113–D1116 10.1093/nar/gku1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.