Abstract
Background
Sex chromosomes have evolved independently multiple times in eukaryotes and are therefore considered a prime example of convergent genome evolution. Sex chromosomes are known to emerge after recombination is halted between a homologous pair of chromosomes, and this leads to a range of non-adaptive modifications causing gradual degeneration and gene loss on the sex-limited chromosome. However, the proximal causes of recombination suppression and the pace at which degeneration subsequently occurs remain unclear.
Results
Here, we use long- and short-read single-molecule sequencing approaches to assemble and annotate a draft genome of the basket willow, Salix viminalis, a species with a female heterogametic system at the earliest stages of sex chromosome emergence. Our single-molecule approach allowed us to phase the emerging Z and W haplotypes in a female, and we detected very low levels of Z/W single-nucleotide divergence in the non-recombining region. Linked-read sequencing of the same female and an additional male (ZZ) revealed the presence of two evolutionary strata supported by both divergence between the Z and W haplotypes and by haplotype phylogenetic trees. Gene order is still largely conserved between the Z and W homologs, although the W-linked region contains genes involved in cytokinin signaling regulation that are not syntenic with the Z homolog. Furthermore, we find no support across multiple lines of evidence for inversions, which have long been assumed to halt recombination between the sex chromosomes.
Conclusions
Our data suggest that selection against recombination is a more gradual process at the earliest stages of sex chromosome formation than would be expected from an inversion and may result instead from the accumulation of transposable elements. Our results present a cohesive understanding of the earliest genomic consequences of recombination suppression as well as valuable insights into the initial stages of sex chromosome formation and regulation of sex differentiation.
Keywords: Sex chromosomes, W-chromosome, Recombination suppression, Willow, Salix
Background
Sex chromosomes, genomic regions associated with either males or females, have evolved independently many times in the eukaryotes [1, 2]. Sex chromosomes come in two general forms in organisms where sex is expressed in the diploid phase of the life cycle. X-Y sex chromosomes form where the sex-specific Y chromosome is associated with males (male heterogamety), and Z-W sex chromosomes form where the sex-specific W chromosome is associated with females (female heterogamety). Both of these sex chromosome types emerge after recombination is halted between a homologous pair of chromosomes [3, 4], which allows the X and Y or Z and W chromosomes to diverge from each other. Studies in systems with highly diverged, independently evolved sex chromosomes have revealed many shared genomic properties across a broad array of taxa [1, 2, 5], and sex chromosomes therefore represent an important example of convergent genome evolution.
In addition to promoting sex chromosomes’ divergence from one another, recombination arrest in the sex-determining region (SDR), the region harboring the sex-determining locus, leads to a range of non-adaptive consequences for the sex-limited Y or W chromosome. These include the build-up of deleterious variation and repetitive elements, as well as the loss of gene activity [6–8]. Due to the longstanding focus on systems with highly divergent sex chromosomes, the speed and order at which these processes occur after recombination suppression remain largely unclear.
Additionally, over evolutionary time, the non-recombining region can expand, resulting in strata or regions with differing levels of divergence between the X and Y or Z and W chromosomes [9–13]. Broadly defined, a stratum is a contiguous region along the sex chromosome where recombination suppression occurred at roughly the same time. In a nascent stratum, recombination may still occur occasionally, but the sex chromosomes are nonetheless in the earliest stages of divergence. Most importantly, although there will be substantial variance in divergence, repetitive element accumulation, and transcriptional loss, there will be more similarity for these traits within a stratum than among different strata or the pseudo-autosomal regions. Multiple strata can occur, resulting in regions along the sex chromosome with different levels of divergence [10, 14, 15].
Expansion of the non-recombining region and the emergence of new strata may occur gradually, in which case we might expect only partial recombination suppression in the youngest stratum, in conjunction with substantial heterogeneity in X-Y or Z-W divergence [16–19]. Alternatively, some have suggested that strata form instantaneously, via large-scale inversions on the Y or W chromosome [20], which prevent recombination between the sex chromosomes along the entirety of the reversed region.
The answers to these questions have important implications beyond sex chromosomes. Halting recombination permanently links co-adapted gene complexes [21–24], also referred to as supergenes. Y and W chromosomes are thought to represent sex-specific supergenes, linking loci with sex-benefit alleles to the sex-determining locus [25–28]. Supergenes have resurfaced recently as a major potential adaptive mechanism [29–33], and in so doing have implicated recombination suppression as a crucial component of complex phenotypic adaptation. However, it is important to note that alternative, non-adaptive mechanisms have been suggested for recombination suppression on sex chromosomes, which can occur simply through the shifting of a sex-specific recombination hotspot away from the sex-determining locus [34–37].
Sex chromosomes are therefore a powerful system to understand the evolutionary consequences of recombination suppression. Furthermore, detailed studies of nascent sex chromosomes are critical if we want to understand the initial causes of recombination suppression, as well as the order and rate of the evolutionary processes that follow it. For example, recent studies of young sex chromosome systems have revealed substantial intra-specific variation in the degree of recombination suppression across populations [38–41], suggesting that the boundaries of recombination suppression are not fixed within a species.
Plants in particular are useful in the study of the earliest stages of sex chromosome formation, as many plant sex chromosomes emerged only very recently in evolutionary time [42–45]. Recent studies based on next-generation sequencing of plant sex chromosomes have shown important patterns in the earliest stages of sex determination [46–51]. Studies on plant sex chromosomes have also revealed the importance of haploid selection in maintaining gene activity in the non-recombining region [17, 52] in the face of rapid loss of gene expression following recombination suppression [8, 53].
Recent work in Salix viminalis, the basket willow, has revealed the presence of nascent Z-W sex chromosomes, with a highly restricted SDR [54, 55]. The sex chromosomes of Salix have evolved independently from the X-Y system in the sister genus Populus [54, 56], which also exhibits very low levels of divergence between the sex chromosomes [43]. The Salicaceae family, which includes willows and poplars, therefore presents a powerful system for studying the earliest stages of sex chromosome formation. Here, we use long- and short-read single-molecule sequencing (PacBio and 10× Genomics Chromium linked-reads approaches) in S. viminalis to assemble a female reference genome. Importantly, our approach allowed us to obtain phased male and female haplotypes using large, continuous haplotype scaffolds. This allows us to transcend the current limitations of short-read next-generation sequencing, which hinder the assembly of repetitive regions, common in SDRs, as well as complicate accurate phasing. Our results shed unprecedented detail on the earliest stages of sex chromosome formation and reveal that the initial stages of recombination suppression are incomplete, as would be expected from gradual selection against recombination rather than from the build-up of inversions in the SDR.
Results and discussion
Assembly and annotation of the basket willow reference genome
In order to gain a better understanding of the evolution and genomic architecture of the recently formed sex chromosomes in Salix viminalis, we sequenced and assembled the complete genome of a single diploid heterogametic female (ZW) which was previously part of a large association mapping population [57]. To this end, we used a combination of long- and short-read single-molecule sequencing strategies and generated ~ 19 Gb of Pacific Biosciences (PacBio) long reads in a female and ~ 58 Gb of 10× Genomics linked-reads in the same female and a male (Additional File 1: Table S1). The full assembled genome has ~ 357 Mb of sequence spanning 2372 scaffolds above 1 kb in length, a scaffold N50 of ~ 1.3 Mb, and 92% of the genome in scaffolds longer than 50 kb. With this estimated genome size, our sequencing constitutes > 50× PacBio and > 160× 10× Genomics coverage for autosomes, and > 25× and > 40× coverage of the W chromosome accounting for the hemizygous nature of the female-limited region.
Assembly quality, as assessed by whole-genome DNA and transcriptome short-read mapping, suggests a high completeness and contiguity with ~ 98% and ~ 84% of the reads, respectively, aligned to the assembled sequence (Additional File 1: Table S2). Importantly, we obtained a high proportion of properly paired reads (Additional File 1: Table S2). An initial assessment with BUSCO also identified more than 94% of complete core Embryophyta genes in the assembly (Additional File 1: Table S2). We also mapped 1987 genotype by sequencing (GBS) [54, 58] markers in order to verify their presence and order. Consequently, our reference genome of the basket willow S. viminalis is essentially complete and properly assembled. Given the inherent difficulties in assembling an ancient polyploid genome such as S. viminalis [59], the relative completeness of our assembly reveals the benefits of incorporating single-molecule and long-read sequencing.
Annotation of the basket willow genome followed an in-house pipeline based on MAKER v3.00.0 [60] that combined transcriptome data [55, 61], reference proteins, and ab initio predictors. We identified 36,490 gene models, with 28,212 (77.3%) of them having functional annotation, and predicted 3469 ncRNA and 1139 tRNAs (Additional File 1: Table S3). Finally, we also identified several families of repetitive elements which together represent ~ 35% of the assembly. The basket willow genome is publicly available for the community through the PopGenIE Integrative Explorer (http://popgenie.org) [62].
Delimitation of the SDR in the female assembly
Differences between male and female genomes in read depth or single nucleotide polymorphim (SNP) density can be used to identify different forms of sex chromosome divergence [12, 63]. In nascent sex chromosome systems, this method is particularly useful when combined with genetic mapping studies of sex-determining regions [41, 55]. These methods are based on the different patterns of divergence and gene coverage differences between males and females on the sex chromosomes. In female heterogametic systems, W-specific reads are present only in females, resulting in higher female read coverage for W scaffolds. Conversely, as the W degrades, we expect a greater male read depth for the corresponding region of the Z chromosome, as females retain only one copy of the Z. Additionally, in the earliest stages of recombination suppression, we expect W regions to retain significant similarity to the Z chromosome, and therefore, females may show similar read coverage for these regions as males. However, once recombination is halted, the W is expected to accumulate polymorphisms that are not shared with the Z, and so we might expect a greater density of SNPs in females compared to males in these regions even before significant divergence lowers mapping efficiency.
In order to assess these different degrees of sex chromosome divergence, we mapped male and female short-read DNA-seq data (~ 69× and ~ 66× average sequencing coverage for females and males, respectively) to our female assembly. Because we assembled the genome of a heterogametic Z-W female, and given the relatively high levels of heterozygosity across the genome (~ 0.5% or 1 SNP per 200 bp), we expect a proportion of divergent regions in the genome, including Z and W haplotypes, to assemble separately in different scaffolds. As this would likely bias our SNP density estimates, where regions with elevated numbers of polymorphisms would appear to be homozygous, we first constructed a non-redundant assembly by removing smaller scaffolds that showed strong evidence of sequence overlap with longer scaffolds. We then aligned our non-redundant scaffolds to the Populus trichocarpa genome [64], revealing broad synteny as expected between these sister genera (Additional File 1: Fig. S1, Fig. 1a). In total, we anchored ~ 272 Mb (76.4% of the full assembly) to P. trichocarpa chromosomes.
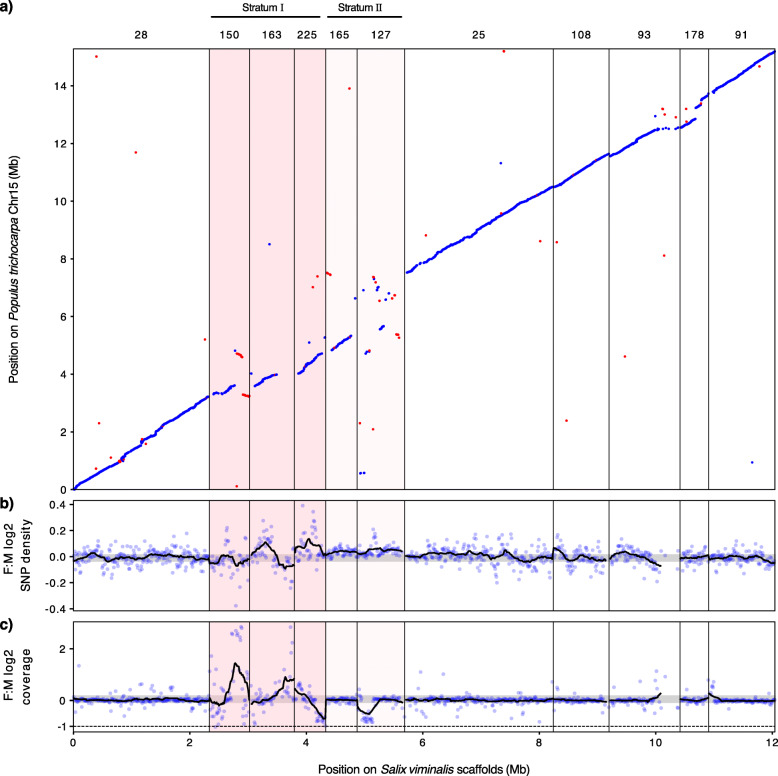
Fig. 1.
Identification of two evolutionary strata in the sex-determining region (SDR) of S. viminalis. Chromosome positions for S. viminalis and P. trichocarpa are shown in Mb with the S. viminalis scaffold names shown on the top. The two identified strata are shown with different hues of pink and labeled above the plot. a Anchoring of S. viminalis scaffolds to the autosomal chromosome 15 of P. trichocarpa. Forward alignments are drawn in blue and reverse alignments are drawn in red. b Log2 differences of normalized SNP density between S. viminalis females and males in non-overlapping windows of 10 kb. A moving average of 25 windows is shown in the black line. The gray shaded area corresponds to the bootstrap 95% confidence interval of the autosomal data. c Log2 differences of normalized read coverage between females and males in non-overlapping windows of 10 kb. Moving average and bootstrap statistics are as in b. Values close to −1 indicate twice the coverage in males in comparison with females, thus potentially Z-linked
We previously identified chromosome 15 as the sex chromosome [54, 55] and mapped the extent of the SDR on this chromosome (highlighted in pink, Fig. 1). Our results show that the five scaffolds within the SDR show significant deviations relative to the autosomal or pseudo-autosomal scaffolds of both female:male SNP density (p < 0.0001, one-sided p value from 10,000 permutations), indicative of the build-up of female-specific SNPs on the W, and/or female:male read coverage differences, suggesting regions of significant divergence between the Z and W chromosomes (Fig. 1). It is important to note that because S. viminalis exhibits only a limited divergence between the Z and W, and our long-read assembly was based on a female sample, the assembly of the sex chromosome regions likely represents Z-W chimeras. Chimeras such as this can obscure signals of Z-W divergence, particularly if sliding windows are very large. This chimerism is evident in scaffolds 150 and 163, which both show a region of similar coverage in males and females and a region of strong female bias that likely represents W-specific genetic material (Fig. 1). These scaffolds, in addition to scaffold 225, show the greatest deviations in the read depth between males and females and likely represent a region where recombination was first suppressed between the emerging Z and W chromosomes (stratum I). Our previous linkage mapping of the same population with GBS markers [54, 58] also placed scaffold 127 together with scaffolds 163 and 225 in the SDR (Additional File 1: Fig. S2). However, the former scaffold shows far fewer differences in female:male read depth while having higher polymorphism in females relative to males. As a result, this likely represents a region where recombination has been suppressed very recently or remains partially incomplete (stratum II).
The SDR region spans a total of ~ 3.4 Mb, or ~ 3.1 Mb when excluding the putatively chimeric regions, and this estimate is somewhat smaller than that of our previous estimation of ~ 5.3 Mb [55]. This difference is likely due to the fact that our previous estimate was based on a male assembly and included non-aligned regions on chromosome 15 of P. trichocarpa. In Salix purpurea, a close relative of S. viminalis with a divergence time of ~ 10 million years [65], the SDR is also located on chromosome 15; however, it is much larger (> 10 Mb) [66]. It has been suggested that these sex chromosomes share a common origin [66], although it remains unclear whether the SDR in these two species is in the same syntenic region. In order to test whether the SDR regions overlapped between the lineages leading to these the two species, we aligned our S. viminalis genome assembly to the S. purpurea assembly. We found that all scaffolds inferred to be part of the S. viminalis SDR aligned to the SDR region in S. purpurea (Fig. 2), suggesting a shared origin, albeit with several potential rearrangements between them.
Fig. 2.
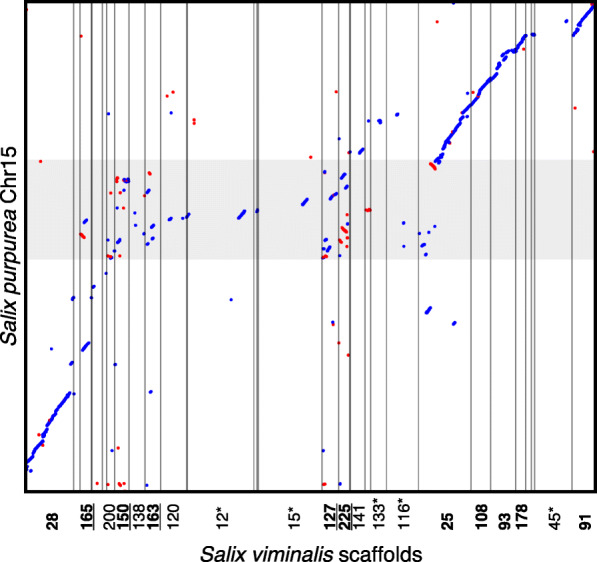
Alignment between S. viminalis and S. purpurea SDR regions. One-to-one orthologous alignments between S. viminalis scaffolds and chromosome 15 of S. purpurea, with forward alignments drawn in blue and reverse alignments drawn in red. The SDR region of S. purpurea is delimited by the gray shaded area (10.7–15.3 Mb, from Zhou et al. [66]). S. viminalis scaffolds anchored to chromosome 15 of P. trichocarpa are highlighted in bold, those inferred to be part of the S. viminalis SDR are underlined, and scaffolds well anchored with several colinear markers on other chromosomes are marked with an asterisk
Two evolutionary strata on the S. viminalis sex chromosomes
It is possible to quantify divergence between the sex chromosomes by comparing dN (a measure of non-synonymous divergence) and dS (a measure of synonymous divergence) between males and females in the sex-linked region. To accurately estimate this divergence, we constructed 10× Genomics Chromium de novo assemblies using one individual of each sex. Fully phased diploid genotypes were obtained for 65.8% and 61.6% of the genome in our female and the male samples, respectively. Similar phasing efficiency was also achieved for chromosome 15 (Additional File 1: Fig. S3) and for genes on the SDR, resulting in an average of 137 (63.7%) and 113 (52.5%) of genes phased in the female and male diploid assemblies, respectively. Our results show significantly greater dN and dS between stratum I and the genomic average in our female sample, but not in our male sample (genomic averages in female dS 0.007460, female dN 0.002080, male dS 0.008151, male dN 0.002261; stratum I female dS: mean = 0.012286, p = 0.00072; female dN: mean = 0.007036, p = 0.000077; stratum I male dS: mean = 0.008700, p = 0.65; male dN: mean = 0.003456, p = 0.25, based on Mann-Whitney U test relative to the genome, Fig. 3), indicating low but detectable divergence between the Z and W in this region. When stratum II is also included, the SDR shows a marginally non-significant divergence in the female (female dS: mean = 0.006402, p = 0.89; female dN: mean = 0.004020, p = 0.061; male dS: mean = 0.005052, p = 0.99; male dN: mean = 0.002084, p = 0.94, Mann-Whitney U test relative to the genome) despite the presence of sex-linked markers in this region (Additional File 1: Fig. S2), reinforcing the conclusion that either recombination was suppressed very recently in this region, or is not yet entirely complete. These estimates are comparatively lower than those obtained in many other plant systems [67, 68]. With the exception of the higher dS in stratum I, they are also similar to those estimated in S. purpurea [66]. dN and dS were also marginally significantly higher between the pseudo-autosomal region (PAR) and the genome in females (dS: mean = 0.009345, p = 0.0019; dN: mean = 0.002308, p = 0.0133, Mann-Whitney U test), but not in males (dS: mean = 0.007991, p = 0.93; dN: mean = 0.002176, p = 0.94, Mann-Whitney U test).
Fig. 3.
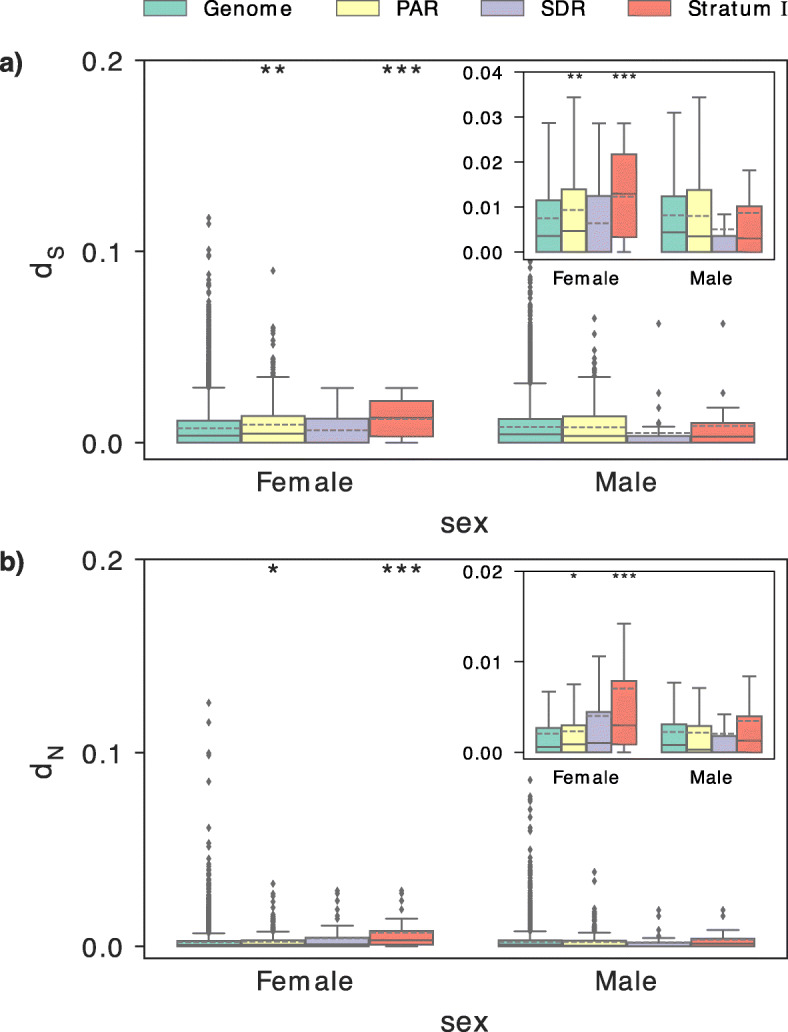
Comparison of polymorphisms at synonymous (dS) and non-synonymous (dN) sites. a Boxplots of dS estimates. b Boxplots of dN estimates. dS and dN were calculated based on the coding sequence alignment of phased diploid haplotypes from one female and one male individuals in the genome (excluding chromosome 15), the pseudo-autosomal region (PAR), the sex-determining region (SDR), and the more divergent stratum I. The inset plots show the quartile distributions of dS and dN estimates without outliers. Significant values from the Mann-Whitney U test relative to the genome are indicated with asterisks: *p < 0.05; **p < 0.01; ***p < 0.001
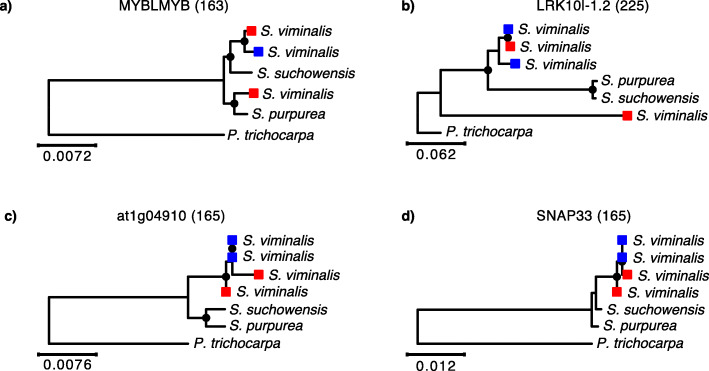
Phylogenetic analysis of Z-W orthologs in conjunction with outgroup species can reveal the relative timing of recombination suppression [13]. We therefore used our phased male and female haplotypes in the SDR together with orthologous genes from two closely related Salix species (S. suchowensis and S. purpurea) and poplar (P. trichocarpa). Our phylogenetic analyses provide further support for two evolutionary strata with different times since recombination suppression (Fig. 4, Additional File 1: Fig. S4). Phylogenies based on genes located in stratum I tend to show one female haplotype, corresponding to the W haplotype, clustering as an outgroup from the other three S. viminalis haplotypes (two male Z haplotypes and the female Z haplotype). This phylogenetic structure indicates that recombination ceased in stratum I prior to S. viminalis speciation. The phylogenetic structure in stratum II shows most female haplotypes clustered together with the male haplotypes, in line with more recent, or possibly partially incomplete, recombination suppression.
Fig. 4.
Examples of phylogenetic trees between gametologous gene pairs in the basket willow SDR. a, b The W-linked copy of the female gametolog is more divergent and does not cluster with the other S. viminalis haplotypes, indicating that suppression of recombination in stratum I occurred prior to S. viminalis speciation. c, d The female W-linked copy clusters within the species’ branch suggesting that recombination has been halted more recently. Female S. viminalis gametologs are indicated with red squares, and male haplotypes are in blue. Trees were estimated by maximum likelihood. Bootstrap values > 75% are indicated with black dots on the respective nodes. The poplar (P. trichocarpa) ortholog was used to root the trees
Distinct evolutionary strata are evident in many sex chromosome systems [9–13], and the mechanism behind recombination suppression, whether it is a large-scale inversion on the sex-limited chromosome [20] or a more gradual suppression of recombination [16–19], remains unclear. Crucially, males and females can differ substantially both in frequency and in location of recombination hotspots [69–72], referred to as heterochiasmy. Local sex-specific recombination rates within the genome may be important in both initial sex chromosome divergence and subsequent expansion of the non-recombining region [28]. Importantly, once recombination has been halted around the SDR in the heterogametic sex, selection to maintain gene order is abolished [73], and selection against inversions is greatly reduced. This suggests that inversions might follow recombination suppression, as has been recently observed [35], even if they are not the cause of recombination suppression initially.
If inversions are the cause of recombination suppression between the Z and W, we would expect our female assembly to be heterozygous for inversions between the Z and W chromosomes in the SDR. To identify potential structural variations between the Z and W chromosomes, we mapped the female 10× Genomics sequencing reads to the reference genome with the barcode-aware Long Ranger pipeline from 10× Genomics. With this approach, we could detect several heterozygous deletions that largely overlap with the differences in coverage between females and males, but, consistent with the conserved synteny between the homologous Z- and W-linked scaffolds (Fig. 5, see below), we observe no evidence for inversions associated with either stratum I or stratum II (Additional File 1: Table S4).
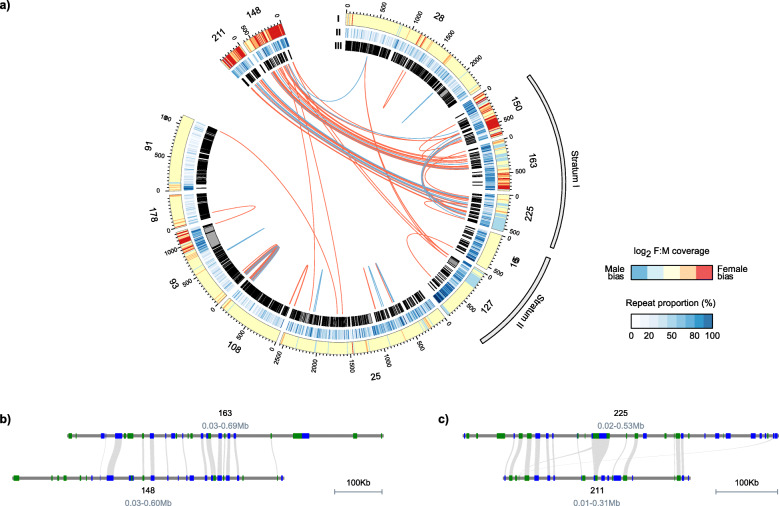
Fig. 5.
Synteny analyses of Z- and W-linked resolved haplotypes. a Circular plots showing that scaffolds 148 and 211 are W-linked and align to the SDR of chromosome 15. From the outside to the center, (I) depicts the heatmap of log2 females:males read depth in non-overlapping windows of 5 kb, (II) shows the repeat proportion in non-overlapping windows of 10 kb, and (III) indicates the location of annotated genes. Links between genes were computed from the best BLASTP hits and are color coded relative to the BLASTP alignment percent identity, with percent identity > 80% in blue and > 90% in red. Positions are shown in kb. b, c A highly conserved synteny between Z- and W-linked scaffolds
It is still possible that inversions formed within the few remaining breakpoints in between our scaffolds, which we would not be able to detect. However, it is worth noting that we observed considerable overlap in both dS and dN estimates (Fig. 3) between the two strata and also the incomplete segregation of some female stratum I Z and W haplotypes (Fig. 4, Additional File 1: Fig. S4), suggesting a gradual divergence with residual or ongoing recombination in the sex chromosomes of S. viminalis. This gradual divergence is not consistent with a major inversion, which would result in a more similar phylogenetic signal for all Z-W orthologs within the inversion as recombination would be suppressed at the same time. Older sex chromosomes also show substantial variation in divergence within perceived strata [10, 13]; however, the limited number of loci remaining on the oldest regions of the sex-limited chromosome complicates these analyses. In these older systems, strata may also have formed through shifts in sex-specific recombination hotspots, resulting in gradual expansions rather than large-scale inversion events.
Together, our evidence suggests that at the earliest stages of sex chromosome formation and expansion, recombination suppression is a gradual process and may result from changes in sex-specific recombination hotspots or from epigenetic variation [74, 75]. Therefore, theoretical models about local changes in heterochiasmy as a result of sexually antagonistic alleles [71, 72] may prove to be key to sex chromosome evolution. Alternatively, recent evidence from fungal mating-type chromosomes, analogous to sex chromosomes in many ways, has suggested non-adaptive explanations for the origin and expansion of the non-recombining region due to neutral rearrangements [36, 76] or shifts in recombination hotspots [35].
These non-adaptive models may also explain some of the curious intra-specific heterogeneity in the extent of sex chromosome divergence in younger systems [38–41]. If recombination suppression occurs more gradually, population-level differences in sex-specific recombination hotspots, often observed [70], will drive different levels of divergence in the earliest stages of sex chromosomes, leading to inter-population differences in sex chromosome divergence.
Degeneration of the W chromosome
Although studies of old, highly degenerate Y and W chromosomes have revealed the accumulation of significant repetitive DNA [77, 78], it remains unclear how quickly this material accumulates after recombination suppression. Additionally, the build-up of repetitive elements on the W chromosome may in itself act as a mechanism to suppress recombination with the corresponding region of the Z [79–81]. Repetitive sequences can also trigger the recruitment of the DNA methylation and histone modification machine, and by an increase in heterochromatinization indirectly promote recombination suppression in the sex chromosomes [75, 82–84]. However, the difficulty associated with phasing short-read data has previously hampered efforts to study the earliest stages of sex chromosome divergence. Although it is possible to identify sex-specific transcripts from pedigrees based on inheritance through familial pedigrees [67, 85–87], this method misses non-coding sequence, making it difficult to assess whether non-coding repetitive elements are associated with the earliest stages of recombination suppression.
In order to identify a W-specific sequence, we mapped female and male sequencing reads to our female assembly. We were able to identify an additional subset of 35 scaffolds spanning ~ 3.3 Mb and with 119 protein-coding genes (Additional File 1: Table S5), which likely represent W-specific sequence, i.e., with a significant excess of female:male read coverage over the entire scaffold length based on genomic confidence intervals. Despite the recent origin of recombination suppression, these scaffolds show a significant enrichment of repetitive sequences in comparison with both the corresponding Z-linked portion of the SDR and the genomic average (Additional File 1: Fig. S5, W genome p < 1 × 10−46; W-SDR p = 0.00058, Mann-Whitney U test). These results suggest that either repetitive sequence can accumulate very quickly following the arrest of recombination, or alternatively repetitive elements may in fact act to halt recombination in the absence of inversions.
The loss of recombination on the sex-limited SDR has important evolutionary effects, namely the build-up of deleterious variation and repetitive elements, as well as the loss of gene activity [6–8]. The latter effect in particular can lead to profound differences in gene content between X and Y or Z and W chromosomes in older sex chromosome systems [6]. Studies in other plant sex chromosomes have indicated that gene loss occurs in the SDR [8, 53], however, it remains unclear how quickly this occurs. Additionally, the extended haploid phase in plants may prevent loss of SDR genes expressed in the haploid phase [17, 52].
In order to identify gene content differences between the Z and the W chromosome, we used two of the W-linked scaffolds identified above, scaffolds 148 and 211. These scaffolds align almost entirely to the SDR where read mapping coverage is male-biased (Z-linked), as would be expected for sex-linked homologous regions (Fig. 5a). In both cases, we observed a high degree of synteny in the aligned regions, indicating that both gene content and gene order are still largely conserved between Z and W homologs, even in the most divergent region of the SDR (Fig. 5b, c). This is likely a function of both the recent divergence of this sex chromosome system [55], as well as the preservative effects of haploid selection on genes expressed in plant reproductive tissues. Nevertheless, seven protein-coding genes on the corresponding Z-linked scaffolds with known products are missing from the W assembly. Using a translated BLAST search of these proteins to the corresponding Z-linked scaffolds and considering a minimum query coverage of 80%, we inferred that at least two of them (os02g0180000 on scaffold 163 and TIR on scaffold 225) have likely been pseudogenized on the W. These results suggest that gene loss can occur very quickly, even in nascent sex chromosome systems.
Candidate sex determination genes in S. viminalis
We scanned for genes unique to the S. viminalis W chromosome, or without preserved synteny to the Z homolog, as potential candidates for sex-determining loci in S. viminalis. We recovered several genes located on the W-linked scaffold 148 that could possibly be involved in this role, including WOX1, two genes in tandem of the two-component response regulator implicated in phytohormone signaling, ARR5 and ARR17, and three pre-mRNA splicing factor RNA helicases of the ESP3 gene family (Table 1). We could not recover ortholog copies of ARR17 and ESP3_2 in the genome, or evidence for a pseudogene in the Z chromosome, suggesting that they could have originated through either a translocation to the W or gene loss on the Z.
Table 1.
Genes on W chromosome scaffolds 148 and 211 with non-preserved synteny relative to the homologous region on the Z chromosome. Orthologs were searched with BLASTP using an e value threshold of 1 × 10−3 and 75% minimum sequence identity
| Scaffold | Gene | Product | Scaffold of the best ortholog (location in P. trichocarpa) |
|---|---|---|---|
| 211 | ADT2 | Arogenate dehydratase/prephenate dehydratase 2, chloroplastic | 100 (Chr08) |
| 211 | 30220 | Hypothetical protein | |
| 211 | 30217 | Hypothetical protein | |
| 211 | POPTR_0012s05040g | l-Ala-d/l-amino acid epimerase | |
| 211 | 30210 | Hypothetical protein | 71 (Chr18) |
| 211 | FBA | Fructose-bisphosphate aldolase | 402* (Chr15) |
| 148 | KP1_5 | Kinesin KP1 | 150 (Chr15) |
| 148 | ESP3_4 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DEAH1 | 127 (Chr15) |
| 148 | CDC48MEE29 | Cell division cycle protein 48 homolog | 47 (Chr12) |
| 148 | ESP3_2 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DEAH1 | |
| 148 | ESP3_6 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DEAH1 | 127 (Chr15) |
| 148 | ARR5_2 | Two-component response regulator ARR5 | 25 (Chr15) |
| 148 | ARR17 | Two-component response regulator ARR17 | |
| 148 | WOX1_4 | WUSCHEL-related homeobox 1 | 150 (Chr15) |
| 148 | ATM_6 | Serine/threonine-protein kinase ATM | 25 (Chr15) |
| 148 | BADH4_2 | Betaine aldehyde dehydrogenase, chloroplastic | 326 (Chr12) |
| 148 | ZDS_7 | Zeta-carotene desaturase, chloroplastic/chromoplastic | 593 (Chr15) |
| 148 | At4g28100 | Uncharacterized GPI-anchored protein | 12 (Chr12) |
| 148 | 27648 | Hypothetical protein | |
| 148 | CDKE-1_12 | Cyclin-dependent kinase E-1 | 4 (Chr01) |
| 148 | 27660 | Hypothetical protein |
*Scaffold 402 was inferred as an allelic variation of scaffold 150
DEAD-box ATP-dependent RNA helicase homologs are known to regulate the formation of male gametophytes in rice [88] and have been recently identified in the candidate sex-determining region of grapevine [89]. WOX1 is a WUSCHEL-related homeobox protein, involved in the cytokinin regulatory pathway that coordinates stem cell proliferation with differentiation [90]. The Silene latifolia homolog of WOX1, SlWUS1, is also sex-linked on the X chromosome with the homologous copy in the Y chromosome likely lost [91]. ARR proteins are one of the final targets of the cytokinin signaling system, which is known to play important roles in flower development and floral sex differentiation in several plant species [92]. In particular, a C-type cytokinin response regulator that acts as a dominant suppressor of carpel development, resulting in female lethality, was specifically identified as the possible male sex-determining gene in kiwifruit [93, 94], and a regulator of the cytokinin metabolism is also a major potential candidate for sex determination in grapevine [89, 95]. Interestingly, within the Salicaceae family, sex-linked polymorphisms in the poplar genome map to a small region on chromosome 19 that includes ARR17 [43, 96]. Similarly, the sex-determining region of the purple willow, S. purpurea, appears to contain palindromic sequences of ARR17 that could be involved in sex determination [97].
Taken together, these observations suggest that cytokinin regulators are likely candidates for major sex-determining genes in S. viminalis, and it is possible that phytohormone signaling mediated by cytokinin regulators plays an important role in the sex determination cascade of this species. Additionally, it is reasonable to speculate on the evolutionary convergence in the control of genetic sex determination in plants as similar genes are likely being independently recruited in different plant families for these functions. Further functional and comparative studies will help to elucidate if these mechanisms are shared among the Salicaceae or, alternatively, evolved independently in the different groups.
It is worth noting that dioecy evolved early in the Salicaceae lineage in which S. viminalis is embedded and is shared by most members of the clade [98]. This means that the standard model for the evolution of sex chromosomes in plants, which assumes an immediate hermaphrodite ancestor, may not be applicable. The model posits two linked mutations encoding male and female sterility [99] as the progenitor of sex chromosomes, and this model has received some empirical support [47]. However, the ancient dioecy found in Salicaceae and the observation of small and heterogeneous levels of divergence in the basket willow [55] and poplar [43] sex chromosomes are difficult to reconcile with this two-gene model, although it is of course still possible. Indeed, recent work has pointed out alternative sex determination mechanisms in flowering plants, either determined by a single gene as in the case of Diospyros [46] or, as in Cucumis and Mercurialis, as a polygenic trait controlled by several genes distributed across different chromosomes [100, 101]. The Salicaceae family with its young sex chromosomes derived from ancient dioecy therefore provides a valuable comparative system to elucidate this process.
Conclusions
Here, we use multiple types of single-molecule sequencing to assemble the genome of the basket willow, S. viminalis, and used this to reveal the earliest stages of sex chromosome evolution. This approach allows us unprecedented power to phase our data, allowing us to resolve Z and W haplotypes at this early stage of divergence. Our results suggest that the SDR is of limited size and divergence, and we recover no evidence that recombination suppression is due to inversions in this region. Even at this early stage of divergence, we see evidence of pseudogenization and the accumulation of repetitive elements in the SDR, suggesting that these processes occur very swiftly after recombination ceases. Furthermore, we found W-linked genes involved in cytokinin regulation, suggesting that phytohormone signaling could be important in the sex determination cascade in S. viminalis. In total, our results shed new light on the fundamental process of sex chromosome formation.
Methods
Plant material and DNA extraction
Fresh young leaves (approximately 200 mg) were sampled from a female and a male S. viminalis (accession 78183 and 81084, respectively), described in [57, 102], and DNA was extracted following a CTAB protocol described in [55]. In brief, approximately 200 mg fresh leaves were snap-frozen and pulverized. To every sample, 950 μl of extraction buffer (100 mM Tris-HCl pH 7.5–8, 25 mM EDTA, 2 M NaCl, 2% (w/v) CTAB, 2% (w/v) PVP K30, 5% (w/v) PVPP, 50 μg/ml RNAse) was added, and the sample was thoroughly mixed before incubation for 30 min at 65 °C. Subsequently, 300 μl chloroform to isoamyl alcohol 24:1 was added, the sample mixed and centrifuged for 10 min at 13,000 rpm, the supernatant was transferred to a new tube, and the process was repeated. 1.5 volumes of ice-cold isopropanol were added to the supernatant followed by an incubation overnight at − 20 °C. After centrifugation for 10 min at 13,000 rpm at 4 °C, the supernatant was removed and the pellet rinsed with chilled 100% EtOH followed by another centrifugation of 5 min at 13,000 rpm at 4 °C. The supernatant was then removed and the DNA was air-dried before it was dissolved in 100 μl TE buffer (10 mM Tris-HCl, 1 mM EDTA). DNA concentration was assessed by Qubit 3.0 Fluorometer (Thermo Fisher Scientific).
PacBio long-read library preparation and sequencing
A single SMRT-bell library with 20 kb insert size was constructed from 10 μg of pure high-molecular weight DNA from one S. viminalis female (accession 78183) according to the manufacturer’s protocol (Pacific Biosciences). This library was sequenced on 48 SMRT cells using P5-C3 chemistry, and 4-h movies were captured for each SMRT cell using the PacBio RSII sequencing platform (Pacific Biosciences). Primary analysis and error correction of the raw data were done using SMRT Portal (Pacific Biosciences). After filtering, the mean read length was 8924 bp (longest read was 61 kbp) and a total of ~ 19.2 Gbp of data were recovered.
10× Genomics Chromium linked-reads library preparation and sequencing
For both accessions (78183 and 81084), sequencing libraries were prepared from 0.75 ng DNA using the Chromium TM Genome Library preparation kit according to the CG00022_Chromium Genome Reagent Kit User Guide_RevA. The library preparation was performed according to the manufacturers’ instructions with the exception that 0.75 ng was used for library preparation instead of 1.25 ng recommended by the manufacturer’s instructions. This was done to account for the smaller genome size of S. viminalis compared to the human genome for which the protocol was optimized. The libraries were sequenced on an Illumina HiSeqX with a paired-end 150-bp read length using v2.5 sequencing chemistry (Illumina Inc.), resulting in ~ 58 Gb of data with a mean molecule length of ~ 40 kb.
DNA extraction and short-read Illumina sequencing
We generated additional Illumina sequencing data for the female accession 78183, the same accession used to assemble the reference genome. DNA was extracted from fresh leaves using the Fast DNA Kit (MP Biomedicals) according to the manufacturer’s instructions. Two libraries with 165 and 400 bp insert size respectively were generated with the TruSeq DNA v2 kit (manual #15005180) following the manufacturer’s protocol and sequenced on one lane each with Illumina HiSeq2000, 100- bp paired-end read length, and v3 chemistry generating ~ 28 Gb of bases (Additional File 1: Table S1).
Reference genome assembly and annotation
Falcon v0.4.2 [103] was used to assemble the sub-reads from 48 SMRT cells. The primary contigs from this first draft assembly were then polished using Quiver from the Pacific Biosciences’ SMRT suite (v2.3.0) with the PacBio reads. The resulting assembly was then corrected with Pilon v 1.17 [104] using both Illumina libraries from the same individual at 80× and 53× coverage. In addition, a 10× Genomics assembly for the same female individual was also obtained using the pseudohap-style output of Supernova v2.0.1 [105]. This 10× Genomics assembly and the PacBio assembly were then merged using Quickmerge v20160905 [106], increasing the assembly size by ~ 8 Mb. Finally, the preads (corrected PacBio reads obtained after the first step of Falcon assembly) and the Supernova pseudohap assembly were used to scaffold the merged assembly using LINKS v1.8.4 [107]. Finally, we corrected some homozygous SNPs and small insertions and deletions in the assembly using Long Ranger v2.1.2 with the 10× Genomics Chromium reads of the same female individual.
Annotation of the S. viminalis reference genome was performed with MAKER v3.00.0 [60]. The MAKER pipeline was run twice; first, based on protein and RNA sequence data only (later used to train ab initio software) and a second time combining evidence data and ab initio predictions. High-confidence protein sequences were collected from the Uniprot database [108], for proteins belonging to the Swissprot section that contain only manually annotated and reviewed curations (downloaded on August 2016), and two other specific protein sets from Salix suchowensis and Populus trichocarpa. Furthermore, to support gene predictions, we also used selected libraries of RNA-seq data from our previous studies collected from vegetative (leaf) and sex-specific reproductive tissue (catkin) from both female and male individuals [55, 61]. As the basis for the construction of gene models, we combined ab initio predictions from three sources (Augustus v2.7 [109], GeneMark_ES_ET v4.3 [110], and SNAP [111]). GeneMark_ES_ET was self-trained with the genome sequence. To train Augustus and SNAP, we first ran the MAKER pipeline the first time to create a profile using the protein evidence along with RNA-seq data. Both Augustus and SNAP were then trained with a selected set of genes from this initial evidence-based annotation. We excluded genes with an Annotation Edit Distance (AED) score equal to 1 to avoid potentially false annotations. Functional inference for genes and transcripts was performed using the translated CDS features of each coding transcript. Protein sequences were searched with BLAST in the Uniprot/Swissprot reference dataset in order to retrieve gene names and protein functions as well as in the InterProscan v5.7-48 database to retrieve additional annotations from different sources.
We created a repeat library with an in-house pipeline using RepeatModeler v1.0.8 [112]. Identification of repeat sequences in the genome was performed using RepeatMasker v4.0.3 [113] and RepeatRunner [114]. tRNAs were predicted with tRNAscan v1.3.1 [115], and broadly conserved ncRNAs were predicted with the Infernal package [116] using the RNA family database Rfam v11 [117]. The genome assembly and annotation were converted in EMBL format using EMBLmyGFF3 [118].
Identification of allelic scaffolds in single-molecule de novo assemblies
The linked-reads technology of 10× Genomics uses a large number of barcoded microdroplets (GEMs) to capture long-range information over long, single-DNA molecules, enabling the assembly through repetitive regions and to resolve heterozygous haplotypes from a diploid genome. Linked reads for the female and male accessions were assembled with Supernova v2.0.1 [105]. Fully phased heterozygous haplotypes, together with non-phased sequence (nominally homozygous), were obtained using the megabubbles-style output and a minimum sequence length of 1 kb. With this output style, Supernova generates an individual FASTA record for each homologous phased haplotype without mixing maternal and paternal alleles in the same sequence. Diploid assemblies were soft-masked with RepeatMasker v4.0.7 [113] with the “RMBlast” v2.6.0+ search engine and using our custom S. viminalis repeat library generated during genome annotation.
We used sequence alignments in order to identify homologous haplotypes in our single-molecule assemblies. A repeat-masked assembly is first aligned to itself with LAST v926 [119] using the sensitive DNA seeding MAM4 [120] and masking of repeats during alignment with the -cR11 option. To avoid false matches caused by repetitive sequences and paralogous scaffolds, orthologous alignments were generated with last-split and alignments mostly comprised of masked sequence were then discarded with last-postmask. Scaffolds were considered to represent allelic variants in the assembly if the overlap exceeded 25% of sequence length after repeat masking, and with sequence identity > 80% to other longer scaffolds.
Anchoring scaffolds to Populus trichocarpa
Pairwise alignments between P. trichocarpa v10.1 (downloaded from PopGenie v3 [62]) and our S. viminalis assembly were generated from repeat-masked genomic sequence using LAST v926 [119]. We first prepared an index of the poplar genome using the sensitive DNA seeding MAM4 [120], using the masking repeat option -cR11 during alignment. A suitable substitution and gap frequencies matrix were then determined with last-train, using parameters --revsym --matsym --gapsym -C2. Alignments were made with lastal, using the parameters -m100 -C2 followed by last-split –m1 to find 1-to-many willow-poplar orthologous matches. Finally, alignments (within scaffolds) that were composed primarily of masked sequence were ignored using last-postmask, and scaffolds with less than an overall of 10% of aligned sequence were discarded. One-to-one willow-poplar alignments were made by swapping both sequences and repeating the orthology search as above.
Neighboring alignments with < 10 kb gap lengths were linked into a single path, and the longest tiling path was used to assign scaffolds to poplar chromosomes. Forward or reverse scaffold orientation relative to poplar chromosomes was similarly obtained requiring that the total length of one alignment direction was > 70% compared to the other orientation; otherwise, the original orientation was kept. If the longest tiling path for a particular scaffold did not agree with its overall alignment path on the poplar chromosome, the scaffold was marked as unlocalized.
Preprocessing of Illumina reads
Whole-genome DNA sequencing reads were quality assessed with FastQC v0.11.5 [121] and preprocessed with BBTools v37.02 “bbduk” [122] to remove adapter sequences, to trim regions with average quality scores below Q10 from both ends of reads, and to filter out reads aligning to PhiX-174 genome (a commonly used spike-in control in Illumina sequencing runs). After filtering, read-pairs were excluded from downstream analyses if either read had an average quality score < Q20 or was < 50 bases in length. The same criteria for quality assessment and filtering were used for RNA-seq data.
Coverage and polymorphism analysis
Alignments to the genome assembly were performed with BWA v0.7.15-r1140 using the MEM [123] algorithm and default options. General processing of SAM/BAM files was performed with SAMtools v1.6 [124], and duplicated reads were flagged with biobambam v2.0.72 [125] after alignment. Per-site coverage was computed with the SAMtools depth command after filtering out reads with mapping quality ≥ Q3 that map to multiple locations, reads with secondary alignments, and duplicated reads. We then calculated the effective coverage value per scaffold and in non-overlapping windows of 10 kb, as the mean per-site coverage of every site in that class. To account for the differences in the overall coverage between individuals, the coverage data were normalized for the median coverage value of each individual in the respective class.
Polymorphism analyses were conducted using the same filters as above. Read alignments were then converted to nucleotide profiles with the sam2pro program of mlRho [126]. Only sites with a per-site coverage ≥ 5 and a SNP called for bi-allelic sites with a minor allele frequency ≥ 30% within an individual were analyzed. The average SNP density per scaffold, and window, was calculated as the number of SNPs divided by the number of sites that passed the coverage threshold of 5 for the respective class.
In order to avoid infinitely high numbers associated with log2 0 when calculating the log2 difference of coverage or SNP density between females and males, we added a small number (0.1) to each value. The 95% confidence intervals for the sliding window distributions were estimated from the mean bootstrap values with resampling of 1000 random sets of 25 windows from autosomes. We excluded the entirety of chromosome 15 (the sex chromosome), including the PAR, in the bootstrapping procedure to avoid potential linkage effects resulting from the SDR.
To identify potentially W-linked scaffolds in the assembly, we proceeded as above and calculated the log2 F:M coverage differences for each scaffold. All scaffolds where the normalized female coverage was < 10% of the normalized whole-genome coverage were excluded. This is a conservative approach because of the difficulty associated with mapping to highly repetitive potential W-linked scaffolds. These scaffolds are therefore likely to remain undetected. Scaffolds were considered W-linked if the log2 F:M coverage difference was > 95% the genome average.
Identification of structural variations
We used the linked-read data of 10× Genomics sequencing to search for large structural variations in the sex-determining region of the female assembly, as long-range information (average molecule length of ~ 40 kb) can provide accurate sequencing information spanning the region around a breakpoint, even if breakpoints are in regions that are inherently difficult to assemble, for example, in areas enriched for repetitive sequence. To facilitate the interpretation of structural variants that could span adjacent scaffolds, we concatenated all scaffolds anchored to chromosome 15 into a single pseudo-chromosome, preserving scaffold ordering, separated by runs of 100 null nucleotides (N). We then used Long Ranger (10× Genomics), which employs barcode-aware read alignments for the identification of large-scale structural variations using the 10× linked-reads library of the same female used in the assembly (accession 78183). Read mapping used the full genome assembly; however, structural variants were called specifically on the pseudo-chromosome 15.
Quantification of gene expression
Preprocessed RNA-seq reads [55, 61] were filtered for rRNA using Bowtie v2.3.2 [127], and the SILVA release 128 database of LSU and SSU NR99 rRNAs [128]. Filtered reads were then aligned to the reference assembly using HISAT2 v2.1.0 [129] with options --no-mixed --no-discordant. The resulting alignments for each library were sorted and merged by individual and by tissue (catkin and leaves) with SAMtools v1.6 [124]. Read counting per gene was performed using the count command of HTSeq [130] and reads per kilobase mapped (rpkm) expression values were calculated with edgeR [131]. Only genes with an rpkm 1 in at least one sample were considered in further analyses.
Annotation lift-over to 10× Genomics diploid assemblies
Our reference genome annotation was transferred independently for each of the inferred haplotypes derived from our 10× Genomics de novo assemblies of female and male genomes using UCSC Genome Browser’s utilities [132]. First, a pairwise alignment between each haplotype and the non-redundant reference genome was generated as described above with LAST v926 [119]. Alignments were then converted into a series of syntenic chains and nets, tuned for more divergent genomes (axtChain -linearGap=loose), using the same scoring matrix generated during the LAST alignments. Finally, annotations were moved to the haplotype assemblies using the liftOver utility with a minimum 75% ratio of mapped bases between features. Only the longest isoform of each gene was considered in the lift-over. With this approach, we transferred ~ 25,159 genes per diploid haplotype or ~ 80% of the complete annotation.
We further attempted to recover additional genes not lifted initially by aligning each gene individually back to the haplotype assemblies with BLAT v170523 [133], (-minIdentity=30 -minScore=12 -stepSize=5 -repMatch=2253 -extendThroughN), keeping the highest-scoring alignment for each query. In order to avoid potential problems caused by the BLAT alignment of paralogous sequences, we counted the average number of haplotypes aligned to each reference gene (for a fully phased diploid region we expect 2 haplotypes). These counts were then bootstrapped with 1000 iterations, and all alignments for which the haplotype coverage was below the lower bootstrap 95% confidence interval (~ 1.6× coverage) were excluded. This procedure recovered an average of 364 additional genes per haplotype.
Divergence analysis of diploid genotypes
We calculated rates of divergence at synonymous (dS) and non-synonymous (dN) sites between the coding sequences of diploid genotypes for each sex separately. In order to further increase the number of genes analyzed in the SDR, we additionally made use of the resolved haplotypes derived from the female PacBio assembly (from scaffolds 163 and 225). We identified orthologous genes with BLASTP using an e value threshold of 1 × 10−3 and only considering the top hit for each gene. This process recovered an additional set of 11 genes that have not been previously lifted-up to the 10× Genomics diploid assembly. Only sequences with a valid start codon, without internal stop codons and with a minimum sequence length of 120 bases, were analyzed. After this initial filter, pairwise alignments for the two haplotypes were obtained with PRANK v140603 [134], and dS and dN estimates were calculated using the method of Yang and Nielsen [135] as implemented in the yn00 program of PAML v4.9h [136]. Pairwise comparisons with dS > 0.2 were excluded, thereby avoiding the incorrect assignment of orthologs.
Phylogenetic analysis
We used gene trees to determine the relative age of recombination suppression for the haplotypes in each identified sex chromosome strata. In addition to our non-redundant S. viminalis genome, coding sequences for S. suchowensis v4.1 and P. trichocarpa v10.1 were obtained from PopGenie v3 [62] and sequences for S. purpurea v1.0 were obtained from Phytozome v12 [137]. Only the longest transcripts were considered. We first use the conditional reciprocal best BLAST method [138], with a BLAST e value cutoff < 1 × 10−5, to identify 14,255 one-to-one orthologs across all four species (S. viminalis, S. suchawensis, S. purpurea, and P. trichocarpa). For each ortholog group, we searched for the S. viminalis homolog in the lifted annotation of the female and male phased diploid assemblies and aligned all species’ sequences with MAFFT v7.313 [139]. Aligned columns with > 40% gaps and taxa with > 40% of missing data were removed. Maximum likelihood phylogenetic trees were obtained with RAxML v8.2.12 [140] using the rapid bootstrap algorithm with 100 bootstraps and the GTRGAMMA model of sequence evolution. Trees were rooted in the P. trichocarpa branch and were only considered if the two female haplotypes were present. Phylogenetic tree analyses were performed with ETE3 [141].
Supplementary information
Additional file 1: Table S1. Whole-genome DNA sequencing data used in this study. Table S2. Assembly statistics for the full and non-redundant assemblies of Salix viminalis. Table S3. Characterization of the different annotation classes for the Salix viminalis assembly. Table S4. Characterization of heterozygous structural variations identified in Chr15. Table S5. List of all genes found on putatively W-linked scaffolds. Figure S1. Whole genome synteny between Salix viminalis assembly and Populus trichocarpa. Figure S2. Genetic markers aligned to chromosome 15 (from Pucholt et al. 2015) on our assembly. Figure S3. Percentage of fully phased haplotypes using 10X Genomic Chromium sequence data. Figure S4. Phylogenetic trees between Z-W gene pairs in the basket willow SDR. Figure S5. Density of repetitive elements across different genomic regions.
Acknowledgements
We acknowledge the use of the University College London Legion High Performance Computing Facility (Legion@UCL), and associated support services, in the completion of this work. Computational resources were provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX). We thank A. Corral-Lopez, I Darolti, B. Furman, D. Metgzer, B. Sandkam, J. Shu, and W. van der Bijl for the helpful comments and suggestions. We thank the reviewers for their constructive suggestions that helped to improve the manuscript.
Authors’ contributions
JEM and SB conceived the study. JEM, SB, PA, BN, EP-W, AC, and LS designed the experiments. SB, A-CR-W, and PP collected the samples and DNA material. JN and TM collected the sequencing data. PA, EP-W, AC, LS, and JD analyzed the data. JEM and PA wrote the manuscript with contributions from all authors. All authors read and approved the final manuscript.
Funding
This work was funded by the European Research Council (grant agreements 260233 and 680951) to JEM. JEM also gratefully acknowledges further the support from a Royal Society Wolfson Merit Award, a Canada 150 Research Chair, and the Natural Sciences and Engineering Research Council of Canada. Sequencing and annotation were funded by the Swedish Energy Agency, grant 30599-5, and from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas) grant 2016-20031. Sequencing was performed by the National Genomics Infrastructure (NGI) Sweden at Science for Life Laboratory. EP-W, AC, LS, JD, JN, and BN were financially supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation as part of NGI and NBIS (National Bioinformatics Infrastructure Sweden) at SciLifeLab.
Availability of data and materials
Genome sequencing data and annotation generated for this study have all been deposited in EBI’s ENA (https://www.ebi.ac.uk/ena) under project number PRJEB31619 [142]. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sofia Berlin and Judith E. Mank are the shared senior authors.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12915-020-00808-1.
References
- 1.Bachtrog D, Mank JE, Peichel CL, et al. Sex determination: why so many ways of doing it. PLoS Biol. 2014;12:e1001899. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beukeboom LW, Perrin N. The evolution of sex determination. Oxford: Oxford University Press; 2014. p. 222.
- 3.Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2009;24:94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Muller HJ. Genetic variability, twin hybrids and constant hybrids, in a case of balanced lethal factors. Genetics. 1918;3:422–499. doi: 10.1093/genetics/3.5.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachtrog D, Kirkpatrick M, Mank JE, et al. Are all sex chromosomes created equal. Trends Genet. 2011;27:350–357. doi: 10.1016/j.tig.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Bachtrog D. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 2013;14:113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond Ser B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papadopulos AS, Chester M, Ridout K, Filatov DA. Rapid Y degeneration and dosage compensation in plant sex chromosomes. Proc Natl Acad Sci U S A. 2015;112:13021–13026. doi: 10.1073/pnas.1508454112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergero R, Forrest A, Kamau E, Charlesworth D. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics. 2007;175:1945–1954. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 11.Roesti M, Kueng B, Moser D, Berner D. The genomics of ecological vicariance in threespine stickleback fish. Nat Commun. 2015;6:8767. doi: 10.1038/ncomms9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013;11:e1001643. doi: 10.1371/journal.pbio.1001643.s024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright AE, Harrison PW, Montgomery SH, Pointer MA, Mank JE. Independent stratum formation on the avian sex chromosomes reveals inter-chromosomal gene conversion and predominance of purifying selection on the W chromosome. Evolution. 2014;68:3281–3295. doi: 10.1111/evo.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pandey RS, Wilson Sayres MA, Azad RK. Detecting evolutionary strata on the human X chromosome in the absence of gametologous Y-linked sequences. Genome Biol Evolution. 2013;5:1863–1871. doi: 10.1093/gbe/evt139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright AE, Moghadam HK, Mank JE. Trade-off between selection for dosage compensation and masculinization on the avian Z chromosome. Genetics. 2012;192:1433–1445. doi: 10.1534/genetics.112.145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergero R, Qiu S, Forrest A, Borthwick H, Charlesworth D. Expansion of the pseudo-autosomal region and ongoing recombination suppression in the Silene latifolia sex chromosomes. Genetics. 2013;194:673–686. doi: 10.1534/genetics.113.150755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chibalina MV, Filatov DA. Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr Biol. 2011;21:1475–1479. doi: 10.1016/j.cub.2011.07.045. [DOI] [PubMed] [Google Scholar]
- 18.Natri HM, Shikano T, Merilä J. Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes. Mol Biol Evol. 2013;30:1131–1144. doi: 10.1093/molbev/mst035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolas M, Marais G, Hykelova V, et al. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 2005;3:e4. doi: 10.1371/journal.pbio.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- 21.Dobzhansky T. Genetics of natural populations; experiments on chromosomes of Drosophila pseudoobscura from different geographic regions. Genetics. 1948;33:588–602. doi: 10.1093/genetics/33.6.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobzhansky T. Genetics of the evolutionary process. New York: Columbia University Press; 1970.
- 23.Dobzhansky T, Pavlovsky O. Indeterminate outcome of certain experiments on Drosophila populations. Evolution. 1953;7:198–210. doi: 10.1111/j.1558-5646.1953.tb00082.x. [DOI] [Google Scholar]
- 24.Dobzhansky T, Pavlovsky O. Interracial hybridization and breakdown of coadapted gene complexes in Drosophila paulistorum and Drosophila willistoni. Proc Natl Acad Sci U S A. 1958;44:622–629. doi: 10.1073/pnas.44.6.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bull JJ. Evolution of sex determining mechanisms. San Francisco: Benjamin-Cummings Publishing Company; 1983. p. 316. [Google Scholar]
- 26.Fisher RA. The evolution of dominance. Genetics. 1931;6:345–368. [Google Scholar]
- 27.Rice WR. Evolution of the Y sex chromosome in animals. BioScience. 1996;46:331–343. doi: 10.2307/1312947. [DOI] [Google Scholar]
- 28.Wright AE, Dean R, Zimmer F, Mank JE. How to make a sex chromosome. Nat Commun. 2016;7:12087. doi: 10.1038/ncomms12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iijima T, Kajitani R, Komata S, et al. Parallel evolution of Batesian mimicry supergene in two Papilio butterflies, P. polytes and P. memnon. Science Advances. 2018;4:eaao5416. doi: 10.1126/sciadv.aao5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joron M, Frezal L, Jones RT, et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature. 2011;477:203–206. doi: 10.1038/nature10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Küpper C, Stocks M, Risse JE, et al. A supergene determines highly divergent male reproductive morphs in the ruff. Nat Genet. 2016;48:79–83. doi: 10.1038/ng.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamichhaney S, Fan G, Widemo F, et al. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax) Nat Genet. 2016;48:84–88. doi: 10.1038/ng.3430. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Wurm Y, Nipitwattanaphon M, et al. A Y-like social chromosome causes alternative colony organization in fire ants. Nature. 2013;493:664–668. doi: 10.1038/nature11832. [DOI] [PubMed] [Google Scholar]
- 34.Ironside JE. No amicable divorce? Challenging the notion that sexual antagonism drives sex chromosome evolution. BioEssays. 2010;32:718–726. doi: 10.1002/bies.200900124. [DOI] [PubMed] [Google Scholar]
- 35.Sun Y, Svedberg J, Hiltunen M, Corcoran P, Johannesson H. Large-scale suppression of recombination predates genomic rearrangements in Neurospora tetrasperma. Nat Commun. 2017;8:1140. doi: 10.1038/s41467-017-01317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Branco S, Badouin H, Rodríguez de la Vega RC et al. Evolutionary strata on young mating-type chromosomes despite the lack of sexual antagonism. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:7067–7072. doi:10.1073/pnas.1701658114. [DOI] [PMC free article] [PubMed]
- 37.Kent TV, Uzunović J, Wright SI. Coevolution between transposable elements and recombination. Philos Trans R Soc Lond Ser B Biol Sci. 2017;372. 10.1098/rstb.2016.0458. [DOI] [PMC free article] [PubMed]
- 38.Dufresnes C, Bertholet Y, Wassef J, et al. Sex-chromosome differentiation parallels postglacial range expansion in European tree frogs (Hyla arborea) Evolution. 2014;68:3445–3456. doi: 10.1111/evo.12525. [DOI] [PubMed] [Google Scholar]
- 39.Reichwald K, Petzold A, Koch P, et al. Insights into sex chromosome evolution and aging from the genome of a short-lived fish. Cell. 2015;163:1527–1538. doi: 10.1016/j.cell.2015.10.071. [DOI] [PubMed] [Google Scholar]
- 40.Rodrigues N, Vuille Y, Loman J, Perrin N. Sex-chromosome differentiation and ‘sex races’ in the common frog (Rana temporaria) Proceedings Biological Sciences. 2015;282:20142726. doi: 10.1098/rspb.2014.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright AE, Darolti I, Bloch NI, et al. Convergent recombination suppression suggests role of sexual selection in guppy sex chromosome formation. Nat Commun. 2017;8:14251. doi: 10.1038/ncomms14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Filatov DA. Homomorphic plant sex chromosomes are coming of age. Mol Ecol. 2015;24:3217–3219. doi: 10.1111/mec.13268. [DOI] [PubMed] [Google Scholar]
- 43.Geraldes A, Hefer CA, Capron A, et al. Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus) Mol Ecol. 2015;24:3243–3256. doi: 10.1111/mec.13126. [DOI] [PubMed] [Google Scholar]
- 44.Ming R, Bendahmane A, Renner SS. Sex chromosomes in land plants. Annu Rev Plant Biol. 2011;62:485–514. doi: 10.1146/annurev-arplant-042110-103914. [DOI] [PubMed] [Google Scholar]
- 45.Veltsos P, Cossard G, Beaudoing E, et al. Size and content of the sex-determining region of the Y chromosome in dioecious Mercurialis annua, a plant with homomorphic sex chromosomes. Genes. 2018;9. 10.3390/genes9060277. [DOI] [PMC free article] [PubMed]
- 46.Akagi T, Henry IM, Tao R, Comai L. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science. 2014;346:646–650. doi: 10.1126/science.1257225. [DOI] [PubMed] [Google Scholar]
- 47.Harkess A, Zhou J, Xu C, et al. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat Commun. 2017;8:1279. doi: 10.1038/s41467-017-01064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell JR, Pannell JR. Sex determination in dioecious Mercurialis annua and its close diploid and polyploid relatives. Heredity. 2015;114:262–271. doi: 10.1038/hdy.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tennessen JA, Wei N, Straub SCK, Govindarajulu R, Liston A, Ashman TL. Repeated translocation of a gene cassette drives sex-chromosome turnover in strawberries. PLoS Biol. 2018;16:e2006062. doi: 10.1371/journal.pbio.2006062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torres MF, Mathew LS, Ahmed I, et al. Genus-wide sequencing supports a two-locus model for sex-determination in Phoenix. Nat Commun. 2018;9:3969. doi: 10.1038/s41467-018-06375-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Na JK, Yu Q, et al. Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci U S A. 2012;109:13710–13715. doi: 10.1073/pnas.1207833109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandler G, Beaudry FEG, Barrett SCH, Wright SI. The effects of haploid selection on Y chromosome evolution in two closely related dioecious plants. Evolution Letters. 2018;2:368–377. doi: 10.1002/evl3.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridout KE, Veltsos P, Muyle A, et al. Hallmarks of early sex-chromosome evolution in the dioecious plant Mercurialis annua revealed by de novo genome assembly, genetic mapping and transcriptome analysis. bioRxiv. 2017. 10.1101/106120.
- 54.Pucholt P, Rönnberg-Wästljung AC, Berlin S. Single locus sex determination and female heterogamety in the basket willow (Salix viminalis L.) Heredity. 2015;114:575–583. doi: 10.1038/hdy.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pucholt P, Wright AE, Conze LL, Mank JE, Berlin S. Recent sex chromosome divergence despite ancient dioecy in the willow Salix viminalis. Mol Biol Evol. 2017;34:1991–2001. doi: 10.1093/molbev/msx144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hou J, Ye N, Zhang D, et al. Different autosomes evolved into sex chromosomes in the sister genera of Salix and Populus. Sci Rep. 2015;5:9076. doi: 10.1038/srep09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berlin S, Trybush SO, Fogelqvist J, et al. Genetic diversity, population structure and phenotypic variation in European Salix viminalis L. (Salicaceae) Tree Genet Genomes. 2014;10:1595–1610. doi: 10.1007/s11295-014-0782-5. [DOI] [Google Scholar]
- 58.Pucholt P, Hallingbäck HR, Berlin S. Allelic incompatibility can explain female biased sex ratios in dioecious plants. BMC Genomics. 2017;18:251. doi: 10.1186/s12864-017-3634-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berlin S, Fogelqvist J, Lascoux M, Lagercrantz U, Rönnberg-Wästljung AC. Polymorphism and divergence in two willow species, Salix viminalis L. and Salix schwerinii E. Wolf. G3. 2011;1:387–400. doi:10.1534/g3.111.000539. [DOI] [PMC free article] [PubMed]
- 60.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Darolti I, Wright AE, Pucholt P, Berlin S, Mank JE. Slow evolution of sex-biased genes in the reproductive tissue of the dioecious plant Salix viminalis. Mol Ecol. 2018;27:694–708. doi: 10.1111/mec.14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sjödin A, Street NR, Sandberg G, Gustafsson P, Jansson S. The Populus Genome Integrative Explorer (PopGenIE): a new resource for exploring the Populus genome. New Phytologist. 2009;182:1013–1025. doi: 10.1111/j.1469-8137.2009.02807.x. [DOI] [PubMed] [Google Scholar]
- 63.Vicoso B, Bachtrog D. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature. 2013;499:332–335. doi: 10.1038/nature12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tuskan GA, Difazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 65.Zhao YJ, Liu XY, Guo R, Hu KR, Cao Y, Dai F. Comparative genomics and transcriptomics analysis reveals evolution patterns of selection in the Salix phylogeny. BMC Genomics. 2019;20:253. doi: 10.1186/s12864-019-5627-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou R, Macaya-Sanz D, Rodgers-Melnick E, et al. Characterization of a large sex determination region in Salix purpurea L. (Salicaceae) Mol Gen Genomics. 2018;293:1437–1452. doi: 10.1007/s00438-018-1473-y. [DOI] [PubMed] [Google Scholar]
- 67.Martin H, Carpentier F, Gallina S, et al. Evolution of young sex chromosomes in two dioecious sister plant species with distinct sex determination systems. Genome Biol Evolution. 2019;11:350–361. doi: 10.1093/gbe/evz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Veltsos P, Ridout KE, Toups MA, et al. Early sex-chromosome evolution in the diploid dioecious plant Mercurialis annua. Genetics. 2019;212:815–835. doi: 10.1534/genetics.119.302045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burt A, Bell G, Harvey PH. Sex differences in recombination. J Evol Biol. 1991;4:259–277. doi: 10.1046/j.1420-9101.1991.4020259.x. [DOI] [Google Scholar]
- 70.Kong A, Thorleifsson G, Gudbjartsson DF, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 71.Lenormand T. The evolution of sex dimorphism in recombination. Genetics. 2003;163:811–822. doi: 10.1093/genetics/163.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lenormand T, Dutheil J. Recombination difference between sexes: a role for haploid selection. PLoS Biol. 2005;3:e63. doi: 10.1371/journal.pbio.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flot JF, Hespeels B, Li X, et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature. 2013;500:453–457. doi: 10.1038/nature12326. [DOI] [PubMed] [Google Scholar]
- 74.Metzger DCH, Schulte PM. The DNA methylation landscape of stickleback reveals patterns of sex chromosome evolution and effects of environmental salinity. Genome Biol Evol. 2018;10:775–785. doi: 10.1093/gbe/evy034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang W, Wang X, Yu Q, Ming R, Jiang J. DNA methylation and heterochromatinization in the male-specific region of the primitive Y chromosome of papaya. Genome Res. 2008;18:1938–1943. doi: 10.1101/gr.078808.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Branco S, Carpentier F, Rodríguez de la Vega RC, et al. Multiple convergent supergene evolution events in mating-type chromosomes. Nature Communications. 2018;9:2000. doi: 10.1038/s41467-018-04380-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Soh YQ, Alföldi J, Pyntikova T, et al. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell. 2014;159:800–813. doi: 10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tomaszkiewicz M, Rangavittal S, Cechova M, et al. A time- and cost-effective strategy to sequence mammalian Y chromosomes: an application to the de novo assembly of gorilla Y. Genome Res. 2016;26:530–540. doi: 10.1101/gr.199448.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ben-Aroya S, Mieczkowski PA, Petes TD, Kupiec M. The compact chromatin structure of a Ty repeated sequence suppresses recombination hotspot activity in Saccharomyces cerevisiae. Mol Cell. 2004;15:221–231. doi: 10.1016/j.molcel.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 80.He L, Dooner HK. Haplotype structure strongly affects recombination in a maize genetic interval polymorphic for Helitron and retrotransposon insertions. Proc Natl Acad Sci U S A. 2009;106:8410–8416. doi: 10.1073/pnas.0902972106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sasaki M, Tischfield SE, van Overbeek M, Keeney S. Meiotic recombination initiation in and around retrotransposable elements in Saccharomyces cerevisiae. PLoS Genet. 2013;9:e1003732. doi: 10.1371/journal.pgen.1003732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li SF, Zhang GJ, Yuan JH, Deng CL, Gao WJ. Repetitive sequences and epigenetic modification: inseparable partners play important roles in the evolution of plant sex chromosomes. Planta. 2016;243:1083–1095. doi: 10.1007/s00425-016-2485-7. [DOI] [PubMed] [Google Scholar]
- 83.Zhou Q, Ellison CE, Kaiser VB, Alekseyenko AA, Gorchakov AA, Bachtrog D. The epigenome of evolving Drosophila neo-sex chromosomes: dosage compensation and heterochromatin formation. PLoS Biol. 2013;11:e1001711. doi: 10.1371/journal.pbio.1001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 85.Hough J, Hollister JD, Wang W, Barrett SC, Wright SI. Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. Proc Natl Acad Sci U S A. 2014;111:7713–7718. doi: 10.1073/pnas.1319227111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muyle A, Käfer J, Zemp N, Mousset S, Picard F, Marais GA. SEX-DETector: a probabilistic approach to study sex chromosomes in non-model organisms. Genome Biol Evol. 2016;8:2530–2543. doi: 10.1093/gbe/evw172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Muyle A, Zemp N, Fruchard C, et al. Genomic imprinting mediates dosage compensation in a young plant XY system. Nature Plants. 2018;4:677–680. doi: 10.1038/s41477-018-0221-y. [DOI] [PubMed] [Google Scholar]
- 88.Li X, Gao X, Wei Y, et al. Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell. 2011;23:1416–1434. doi: 10.1105/tpc.110.082636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Badouin H, Velt A, Gindraud F et al. The wild grape genome sequence provides insights into the transition from dioecy to hermaphroditism during grape domestication. bioRxiv. 20202020.01.07.897082. doi:10.1101/2020.01.07.897082. [DOI] [PMC free article] [PubMed]
- 90.Somssich M, Je BI, Simon R, Jackson D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 2016;143:3238–3248. doi: 10.1242/dev.133645. [DOI] [PubMed] [Google Scholar]
- 91.Kazama Y, Nishihara K, Bergero R et al. SlWUS1; an X-linked gene having no homologous Y-linked copy in Silene latifolia. G3. 2012;2:1269–1278. doi:10.1534/g3.112.003749. [DOI] [PMC free article] [PubMed]
- 92.Wybouw B, De Rybel B. Cytokinin - a developing story. Trends Plant Sci. 2019;24:177–185. doi: 10.1016/j.tplants.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 93.Akagi T, Henry IM, Ohtani H, et al. A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. Plant Cell. 2018;30:780–795. doi: 10.1105/tpc.17.00787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Akagi T, Pilkington SM, Varkonyi-Gasic E, et al. Two Y-chromosome-encoded genes determine sex in kiwifruit. Nature Plants. 2019;5:801–809. doi: 10.1038/s41477-019-0489-6. [DOI] [PubMed] [Google Scholar]
- 95.Coito JL, Ramos MJ, Cunha J, et al. VviAPRT3 and VviFSEX: two genes involved in sex specification able to distinguish different flower types in Vitis. Front Plant Sci. 2017;8:98. doi: 10.3389/fpls.2017.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McKown AD, Klápště J, Guy RD, et al. Sexual homomorphism in dioecious trees: extensive tests fail to detect sexual dimorphism in Populus. Sci Rep. 2017;7:1831. doi: 10.1038/s41598-017-01893-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou R, Macaya-Sanz D, Carlson CH, et al. A willow sex chromosome reveals convergent evolution of complex palindromic repeats. Genome Biol. 2020;21:38. doi: 10.1186/s13059-020-1952-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cronk QC, Needham I, Rudall PJ. Evolution of catkins: inflorescence morphology of selected Salicaceae in an evolutionary and developmental context. Front Plant Sci. 2015;6:1030. doi: 10.3389/fpls.2015.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Charlesworth B, Charlesworth D. A model for the evolution of dioecy and gynodioecy. Am Nat. 1978;112:975–997. doi: 10.2307/2460344. [DOI] [Google Scholar]
- 100.Boualem A, Troadec C, Camps C, et al. A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science. 2015;350:688–691. doi: 10.1126/science.aac8370. [DOI] [PubMed] [Google Scholar]
- 101.Cossard GG, Pannell JR. Enhanced leaky sex expression is an adaptive plastic response to pollen limitation in the dioecious plant Mercurialis annua. bioRxiv. 2019703009. doi:10.1101/703009. [DOI] [PMC free article] [PubMed]
- 102.Hallingbäck HR, Fogelqvist J, Powers SJ, et al. Association mapping in Salix viminalis L. (Salicaceae) - identification of candidate genes associated with growth and phenology. Global Change Biol Bioenergy. 2016;8:670–685. doi: 10.1111/gcbb.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chin CS, Peluso P, Sedlazeck FJ, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB. Direct determination of diploid genome sequences. Genome Res. 2017;27:757–767. doi: 10.1101/gr.214874.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chakraborty M, Baldwin-Brown JG, Long AD, Emerson JJ. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 2016;44:e147. doi: 10.1093/nar/gkw654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Warren RL, Yang C, Vandervalk BP, et al. LINKS: scalable, alignment-free scaffolding of draft genomes with long reads. GigaScience. 2015;4:35. doi: 10.1186/s13742-015-0076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Magrane M, UniProt C. UniProt Knowledgebase: a hub of integrated protein data. Database. 2011;2011:bar009. doi:10.1093/database/bar009. [DOI] [PMC free article] [PubMed]
- 109.Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smit AFA, Hubley R. RepeatModeler Open-1.0. 2015. http://www.repeatmasker.org.
- 113.Smit AFA, Hubley R, Green P. RepeatMasker Open-4.0. 2015. http://www.repeatmasker.org.
- 114.Yandell M. Comparative genomics library - RepeatRunner. 2006. [Google Scholar]
- 115.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nawrocki EP, Burge SW, Bateman A, et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015;43:D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Norling M, Jareborg N, Dainat J. EMBLmyGFF3: a converter facilitating genome annotation submission to European Nucleotide Archive. BMC Research Notes. 2018;11:584. doi: 10.1186/s13104-018-3686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Frith MC, Noé L. Improved search heuristics find 20,000 new alignments between human and mouse genomes. Nucleic Acids Res. 2014;42:e59. doi: 10.1093/nar/gku104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2016. [Google Scholar]
- 122.Bushnell B. BBMap short read aligner, and other bioinformatic tools. 2016. [Google Scholar]
- 123.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Nat Genet. 2013;1303(3997).
- 124.Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tischler G, Leonard S. biobambam: tools for read pair collation based algorithms on BAM files. Source Code for Biol Med. 2014;9:2078. doi: 10.1186/1751-0473-9-13. [DOI] [Google Scholar]
- 126.Haubold B, Pfaffelhuber P, Lynch M. mlRho - a program for estimating the population mutation and recombination rates from shotgun-sequenced diploid genomes. Molecular Ecol. 2010;19(Suppl 1):277–284. doi: 10.1111/j.1365-294X.2009.04482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci U S A. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Löytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 136.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 137.Goodstein DM, Shu S, Howson R, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Aubry S, Kelly S, Kümpers BM, Smith-Unna RD, Hibberd JM. Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 2014;10:e1004365. doi: 10.1371/journal.pgen.1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huerta-Cepas J, Serra F, Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Almeida P, Proux-Wera E, Churcher A et al. Genome assembly of the basket willow, Salix viminalis, reveals earliest stages of sex chromosome expansion. Supplementary Datasets. 2020. ENA accesion: PRJEB31619. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Whole-genome DNA sequencing data used in this study. Table S2. Assembly statistics for the full and non-redundant assemblies of Salix viminalis. Table S3. Characterization of the different annotation classes for the Salix viminalis assembly. Table S4. Characterization of heterozygous structural variations identified in Chr15. Table S5. List of all genes found on putatively W-linked scaffolds. Figure S1. Whole genome synteny between Salix viminalis assembly and Populus trichocarpa. Figure S2. Genetic markers aligned to chromosome 15 (from Pucholt et al. 2015) on our assembly. Figure S3. Percentage of fully phased haplotypes using 10X Genomic Chromium sequence data. Figure S4. Phylogenetic trees between Z-W gene pairs in the basket willow SDR. Figure S5. Density of repetitive elements across different genomic regions.
Data Availability Statement
Genome sequencing data and annotation generated for this study have all been deposited in EBI’s ENA (https://www.ebi.ac.uk/ena) under project number PRJEB31619 [142]. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.