Abstract
The BCOR family of tumors includes a number of undifferentiated sarcomas, occurring in various age groups and anatomic sites, characterized by a spindle and round cell phenotype and diffuse immunoreactivity for BCOR. Prior RNA sequencing data revealed that NTRK3 was a top upregulated gene in BCOR-CCNB3 sarcomas. In this study we investigate a large cohort of tumors harboring BCOR/YWHAE genetic alterations for NTRK3 upregulation at both the mRNA and protein levels, compared to other sarcoma types. Pan-Trk immunohistochemistry was assessed for intensity and extent. A correlation between NTRK3 expression and the type of BCOR alteration and BCOR immunoreactivity was also performed. Most soft tissue undifferentiated round cell sarcomas with YWHAE or BCOR rearrangements or BCOR internal tandem duplications (ITD) showed NTRK3, but not NTRK1 or NTRK2, up-regulation by RNA sequencing data analysis. Cytoplasmic pan-Trk immunoreactivity was also observed in most soft tissue round cell sarcomas with YWHAE rearrangements (100%), BCOR ITD (80%), and BCOR-CCNB3 fusions (67%), as well as clear cell sarcomas of kidney (75%), another BCOR family tumor, and ossifying fibromyxoid tumors with ZC3H7B-BCOR fusion (100%), with variable staining intensity and extent. Pan-Trk staining was also seen in solitary fibrous tumors (100%) and less frequently in synovial sarcoma and Ewing sarcoma, but rarely in other sarcomas tested. Tumors harboring rare fusion variants of BCOR, such as BCOR-CHD9, a novel fusion identified by targeted RNA sequencing, and KMT2D-BCOR, were also positive for pan-Trk staining and NTRK3 overexpression. In conclusion, NTRK3 upregulation resulting in pan-Trk overexpression is common in the BCOR family of tumors as well as in subsets of BCOR expressing sarcomas through alternative mechanisms. The therapeutic implication of this finding awaits further investigation.
Keywords: NTRK3, BCOR, YWHAE, round cell sarcoma, PMMTI, CCSK
INTRODUCTION
Sarcomas with BCOR and YWHAE gene alterations encompass different pathologic entities sharing an undifferentiated round to spindle morphology and BCOR upregulation. Two categories of BCOR genetic events have been described including BCOR internal tandem duplications (ITD)[1] and BCOR fusions, often involving the CCNB3 gene and occasionally less common partners (MAML3, ZC3H7B)[2, 3]. In addition to this genetic variability, there is also a significant clinical heterogeneity. Tumors with BCOR ITD and YWHAE gene rearrangements generally have overlapping clinicopathologic presentations: in children, they may represent one of the following entities: undifferentiated round cell sarcoma in infants, primitive myxoid mesenchymal tumor of infancy (PMMTI), clear cell sarcoma of the kidney (CCSK), central nervous system high-grade neuroepithelial tumor[1, 4, 5]; in adults the diagnosis varies from uterine high grade endometrial stromal sarcomas to other rare visceral round cell sarcomas[6-8]. A similar scenario of genetic promiscuity is seen with BCOR fusions: tumors may represent bone or soft tissue high grade undifferentiated sarcomas, ossifying fibromyxoid tumors, or high grade endometrial stromal sarcomas[2, 3, 9, 10]. To complicate things further, other sarcoma types, such as a subset of synovial sarcoma and solitary fibrous tumors (SFT) [11, 12], may also show overexpression of BCOR, through a yet undetermined mechanism and represent diagnostic pitfalls.
In a previous study, we identified NTRK3 as one of the top-upregulated genes in BCOR-CCNB3 sarcomas.[2] As specific NTRK inhibitors, such as larotrectinib, have been recently FDA approved for NTRK-fusion positive tumors[13], it is crucial to understand the specificity of NTRK1/2/3 expression in diagnosing these tumors and possible diagnostic pitfalls. In this study we investigate a large spectrum of tumors with BCOR genetic alterations and other sarcoma types for expression of NTRK3 at both the mRNA and protein levels.
MATERIALS AND METHODS
Case Selection for pan-Trk immunohistochemistry.
The study group included 35 cases of sarcomas with YWHAE or BCOR gene abnormalities. There were 24 soft tissue and bone round cell sarcomas with various genetic alterations, including: YWHAE rearrangement (n=3), BCOR ITD (n=5), BCOR-CCNB3 (n=12), KMT2D-BCOR (n=2), BCOR-CHD9 (n=1), and ZC3H7B-BCOR (n=1) fusions (Table 1). In addition, we examined 8 CCSK and 3 ossifying fibromyxoid tumors (OFMT) with ZC3H7B-BCOR fusions.
Table 1.
pan-Trk immunohistochemical staining in BCOR family of tumors and controls
| Positive cases |
Intensity | Extent | |||
|---|---|---|---|---|---|
| Strong | Moderate | Weak | |||
| BCOR family sarcoma | |||||
| YWHAE-rearranged soft tissue sarcoma | 3/3 (100%) |
-- | 3/3 (100%) |
-- | 90~95% |
| BCOR ITD sarcoma of soft tissue | 4/5 (80%) |
-- | 2/5 (40%) |
2/5 (40%) | 30~90% |
| Clear cell sarcoma of kidney | 6/8 (75%) |
1/8 (13%) |
2/8 (25%) |
3/8 (38%) |
10~90% |
| BCOR-CCNB3 sarcoma | 8/12 (67%) |
1/12 (8%) | 4/12 (33%) |
3/12 (25%) | 10%~95% |
| KMT2D-BCOR sarcoma | 2/2 (100%) |
1/2 (50%) |
1/2 (50%) |
-- | 60%~95% |
| BCOR-CHD9 sarcoma | 1/1 (100%) |
1/1 (100%) | -- | -- | 100% |
| Round cell sarcoma with ZC3H7B-BCOR | 0/1 | -- | -- | -- | -- |
| OFMT with ZC3H7B-BCOR | 3/3 (100%) |
-- | 3/3 (100%) |
-- | 80~90% |
| Control group | |||||
| 1. Other soft tissue tumors known to express BCOR | |||||
| Solitary fibrous tumor | 15/15 (100%) |
13/15* (87%) |
2/15 (13%) |
-- | 35~100% |
| Synovial sarcoma (whole section) | 2/7 (29%) |
-- | 2/7 (29%) |
-- | 20~90% |
| Synovial sarcoma (TMA) | 1/50 (2%) |
-- | 1/50 (2%) |
-- | 90% |
| 2. Other round cell sarcomas | |||||
| Ewing sarcoma | 2/6 (33%) |
1/6# (17%) | -- | 1/6 (17%) | 10~85% |
| CIC-DUX4 sarcoma | 0/4 | -- | -- | -- | -- |
| 3. Other sarcomas | |||||
| Chondrosarcoma (TMA) | 0/20 | -- | -- | -- | -- |
| Chordoma (TMA) | 0/20 | -- | -- | -- | -- |
| Myxofibrosarcoma (TMA) | 0/20 | -- | -- | -- | -- |
| Angiosarcoma (TMA) | 0/40 | -- | -- | -- | -- |
| MPNST (TMA) | 0/20 | -- | -- | -- | -- |
| Myxoid liposarcoma (TMA) | 0/20 | -- | -- | -- | -- |
| Low-grade fibromyxoid sarcoma (TMA) | 1/10 (10%) |
-- | -- | 1/10 (10%) | 90% |
ITD indicates internal tandem duplication. OFMT, ossifying fibromyxoid tumor. MPNST, malignant peripheral nerve sheath tumor. TMA, tissue microarray.
One case with cytoplasmic and nuclear staining.
Membranous staining pattern. All other cases showed cytoplasmic staining.
Among the control group we included 3 categories: 1. sarcomas with various gene fusions known to express BCOR (synovial sarcoma, SFT); 2. undifferentiated round cell sarcomas with various gene fusions; and 3. other sarcoma types.
In the first control cohort we included 57 synovial sarcomas (7 cases with whole sections and 50 cases with tissue microarray) and 15 SFT (10 with BCOR expression; 8 malignant; including soft tissue, pleural, and meningeal origins). The second group included sarcomas with a primitive round cell morphology: 6 Ewing sarcomas (5 with EWSR1-FLI1, 1 with EWSR1-ERG fusion) and 4 CIC-DUX4 fusion positive sarcomas. The last cohort of the control group comprised a broad range of bone and soft tissue sarcoma types, including chondrosarcoma (n=20), chordoma (n=20), myxofibrosarcoma (n=20), angiosarcoma (n=40), malignant peripheral nerve sheath tumor (n=20), myxoid liposarcoma (n=20), and low-grade fibromyxoid sarcoma (n=10), using tissue microarrays. Whole sections of formalin-fixed paraffin-embedded tissues were used for pan-Trk staining except for cases using tissue microarrays mentioned above. The study was approved by the Institutional Review Board.
Immunohistochemistry
Immunohistochemical staining was performed using a pan-Trk antibody (EPR17341; Abcam, Cambridge, MA)[14], which recognizes Trk proteins including Trk-A, Trk-B, and Trk-C, encoded by the NTRK1, NTRK2, and NTRK3 genes, respectively. Immunostaining for BCOR using clone C-10 (sc-514576; Santa Cruz, Dallas, TX), was performed and/or reviewed, as previously described.[11] In a small subset of cases additional immunostains were applied including NTRK1 (Ab76291, 1:1,500, ABCAM), H3K27me3 (C36B11 (1:200 dilution; Cell Signaling Technology, Danvers, MA) and TLE1 (Santa Cruz Biotech, clone Poly; sc-9121; 1:100 dilution) .
The staining patterns of pan-Trk were recorded as cytoplasmic, membranous, and/or nuclear. The intensities were recorded as weak, moderate, or strong, and the percentage of tumor cells positive for pan-Trk staining was also assessed in each case. Cases with >5% tumor cells staining for pan-Trk were considered positive. The staining results of BCOR were also recorded whenever available.
RNA sequencing data analysis
The mRNA levels of BCOR, NTRK1/2/3, and NTF3 expression were evaluated in round cell sarcomas with BCOR ITD, BCOR fusions, or YWHAE fusions using RNA sequencing (RNAseq) data and compared to other sarcoma types available on the same platforms. Datasets from 2 RNAseq platforms were analyzed including 10 cases studied on whole transcriptome sequencing (6 BCOR ITD, 2 BCOR-CCNB3, 1 BCOR-MAML3, and 1 YWHAE-NUTM2B) and 7 cases tested on targeted RNAseq using the TruSight RNA Fusion Panel (Illumina, San Diego, CA) (1 BCOR ITD, 3 BCOR-CCNB3, 2 KMT2D-BCOR, and 1 BCOR-CHD9), as previously described.[1, 2] Whole transcriptome sequencing data was also analyzed for NTRK3 expression at the exon level. Control groups of other sarcomas with available data in each dataset included: Ewing sarcoma (n=1), CIC-DUX4 sarcomas (n=9), and OFMT with ZC3H7B-BCOR fusion (n=1) in whole transcriptome sequencing and synovial sarcoma (n=1) and SFT (n=4) in the targeted TruSight RNA Fusion Panel. In addition, one infantile fibrosarcoma with an EML4-NTRK3 fusion with targeted RNAseq data was also included as reference for the enhanced level of NTRK3 mRNA upregulation.
RESULTS
Most YWHAE and BCOR-altered round cell sarcomas express cytoplasmic pan-Trk – regardless of the genetic alterations
Among the tumors with YWHAE and BCOR ITD abnormalities, pan-Trk immuno-staining was positive in all 3 tumors with YWHAE rearrangements (2 infantile soft tissue tumors and 1 adult soft tissue) with a diffuse (≥90%) and moderate cytoplasmic pattern (Fig 1A, Table 1); 80% of soft tissue round cell sarcomas with BCOR ITD (Fig. 1B) and 75% of the CCSK (Fig. 1C) with a more variable extent and intensity.
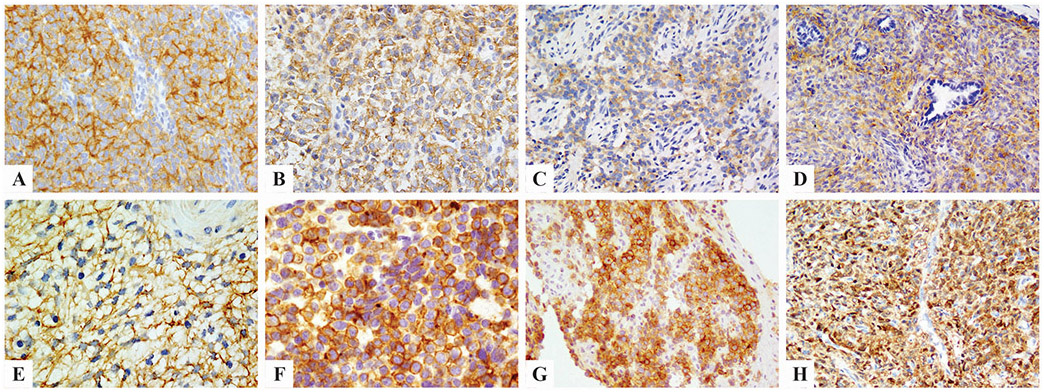
Figure 1. Immunohistochemical stains for pan-Trk.
showed diffuse moderate staining in YWHAE-rearranged sarcomas (A), variable staining in soft tissue round cell sarcoma with BCOR ITD (B), clear cell sarcoma of kidney with BCOR ITD (C), and BCOR-CCNB3 fused sarcomas (D). Ossifying fibromyxoid tumor harboring ZC3H7B-BCOR fusion showed diffuse but moderate cytoplasmic pan-Trk staining (E). One Ewing sarcoma with EWSR1-FLI1 fusion had diffuse membranous staining for both pan-Trk (F) and NTRK1 (G). A BCOR-negative, NAB2-STAT6 positive solitary fibrous tumor showed diffuse and strong pan-Trk staining (H).
Eight of 12 (67%) BCOR-CCNB3 sarcomas were positive for pan-Trk, with a cytoplasmic pattern; ranging from focal (10%) to diffuse (>90%) and variable intensity (Fig. 1D). Three cases of rare soft tissue tumors with alternative BCOR fusions, BCOR-CHD9 (n=1) and KMT2D-BCOR (n=2), were all diffusely positive for pan-Trk, while the only ZC3H7B-BCOR soft tissue round cell sarcoma tested was negative for pan-Trk staining.
All except 4 cases with BCOR/YWHAE genetic alterations and available BCOR IHC results (2 infantile YWHAE sarcomas, 3 BCOR ITD soft tissue sarcomas, 7 CCSK, 8 BCOR-CCNB3 sarcoma, 2 KMT2D-BCOR sarcoma) were strongly and diffusely immunoreactive to BCOR. The only 4 cases which were BCOR negative, included a renal sarcoma with BCOR-CHD9 fusion and 3 OFMTs with ZC3H7B-BCOR fusions. The latter 3 OFMT showed diffuse, moderate cytoplasmic staining with pan-Trk (80~90% of tumor cells) (Fig. 1E). No association between BCOR and pan-Trk staining was found.
Pan-Trk expression is seen in solitary fibrous tumor, small subsets of Ewing sarcoma and synovial sarcoma, but rarely in other sarcoma types
All 15 SFTs showed pan-Trk immunoreactivity, regardless of location (soft tissue, pleural, or meningeal), risk of malignancy, and BCOR immunoreactivity. The staining pattern was diffuse, strong, and cytoplasmic in most cases (Fig. 1H), except one case with multifocal moderate staining in 35% of tumor area and another one with diffuse moderate expression.
Two of 7 (29%) synovial sarcomas tested using whole tissue sections for pan-Trk stain also showed moderate cytoplasmic staining, in 90% and 20% tumor cells, respectively. Pan-Trk staining was observed at a much lower rate (1/50, 2%) when using tissue microarray sections of synovial sarcomas. A subset of Ewing sarcoma (2/6, 33%) also showed immunoreactivity to pan-Trk. One Ewing sarcoma showed strong membranous staining in 85% of tumor cells (Fig. 1F), in contrast to the cytoplasmic staining observed in most other soft tissue tumors. This particular case was also diffusely positive for NTRK1 (Fig. 1G), while being negative for BCOR. We tested the pretreatment biopsy and posttreatment resection specimens of this patient, and the results were similar. The other Ewing sarcoma case positive for pan-Trk had only weak cytoplasmic staining in 10% cells. All other sarcomas tested, including 4 CIC-DUX4 sarcomas, were negative for pan-Trk staining, except for one low grade fibromyxoid sarcoma which showed diffuse weak staining.
Sarcomas with BCOR and YWHAE alterations show NTRK3 but not NTRK1/2 mRNA up-regulation
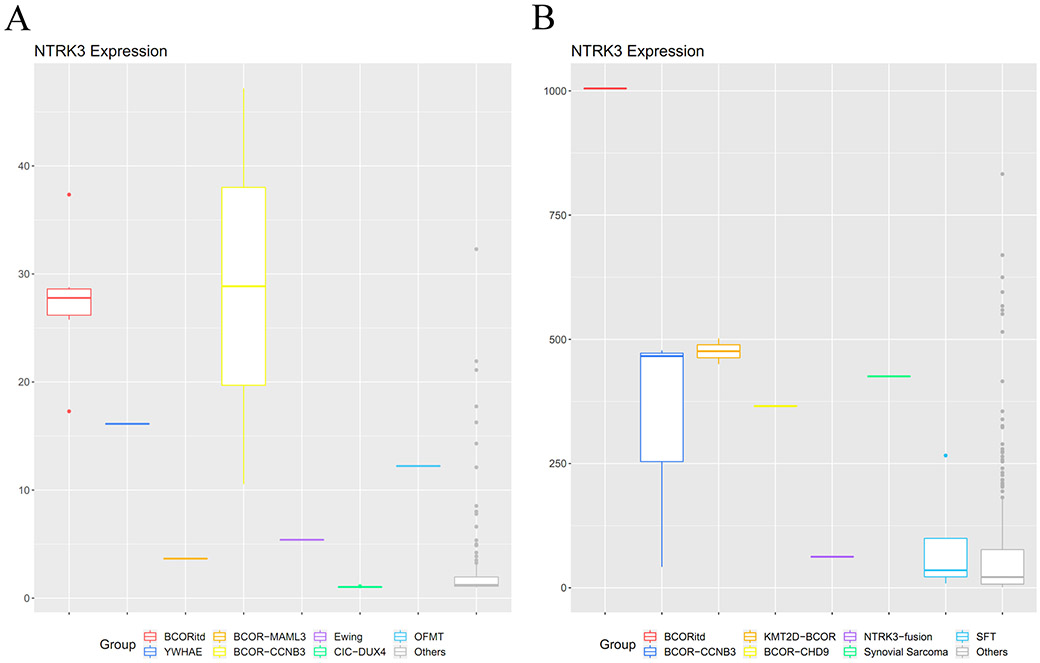
RNAseq data analysis revealed up-regulation of NTRK3 in round cell sarcomas with YWHAE rearrangement, BCOR ITD, BCOR-CCNB3, KMT2D-BCOR, and BCOR-CHD9 fusions, compared to other sarcoma subtypes in the two different molecular datasets (Fig. 2). From the targeted RNAseq data, the NTRK3 level of upregulation in BCOR family tumors was overall higher compared to the NTRK3 fusion positive control (Fig. 2B). The mRNA expressions of NTRK1 and NTRK2 were not significantly elevated in any of the BCOR family tumors (Supplementary Fig. 1), suggesting that the pan-Trk immunopositivity is the result of specific overexpression of Trk-C. The expression of NTF3 (NT-3), the gene encoding the neurotrophin-3 ligand activating Trk-C, was not elevated in BCOR family tumors. Exon level expression analysis demonstrated up-regulation of the entire coding sequence of NTRK3. One adult round cell sarcoma with a BCOR-MAML3 fusion did not show significant NTRK3 up-regulation. In other tumor types, NTRK3 expression was up-regulated in the only synovial sarcoma case available and mildly elevated in the OFMT with an ZC3H7B-BCOR fusion (Fig. 2). In contrast, one Ewing sarcoma with an EWSR1-ERG fusion and 9 CIC-DUX4 fusion sarcomas did not show NTRK3 overexpression. Moreover, SFT showed no significant NTRK3 upregulation (Fig. 2B), but instead showed NTRK1 overexpression (Supplementary Fig. 1B), which may explain the consistent pan-Trk immunoreactivity. Based on this result further immunohistochemical studies on 8 SFT cases, including BCOR-positive and BCOR-negative cases, showed diffuse and strong NTRK1 reactivity in >90% of the cells (Supplementary Fig.1E).
Figure 2. NTRK3 mRNA up-regulation in BCOR family tumors using whole transcriptome sequencing (A) and targeted RNA sequencing (B).
Up-regulated NTRK3 expression was observed in round cell sarcomas with BCOR ITD, YWHAE rearrangements, BCOR-CCNB3, KMT2D-BCOR, and BCOR-CHD9 fusions, compared to other sarcomas on each platform (expression levels in RPKM). The NTRK3 levels observed in targeted RNA sequencing were higher than an infantile fibrosarcoma with EML4-NTRK3 fusion reference (B). An ossifying fibromyxoid tumor with ZC3H7B-BCOR fusion (A) and a synovial sarcoma (B) also had relatively increased NTRK3 expressions. No significant increase of NTRK3 expression level was observed in the single case of round cell sarcoma with BCOR-MAML3 fusion, an Ewing sarcoma with EWSR1-ERG fusion, 9 CIC-DUX4 sarcomas (A), and solitary fibrous tumors (B).
To compare the results between RNA expression analysis and immunohistochemical stains, 2 cases each with BCOR ITD and BCOR-CCNB3 were studied with both techniques. One case in each genetic category showed negative pan-Trk staining but had high NTRK3 mRNA level by RNAseq, suggesting that some technical issues with immunohistochemistry such as old slides might be at play. The other two cases showed consistent RNA expression and immunostaining results. On the other hand, the OFMT with BCOR-ZC3H7B fusion had only mild NTRK3 mRNA up-regulation but showed moderate staining in 80% of the tumor.
Rare BCOR fusion variants BCOR-CHD9 and KMT2D-BCOR express pan-Trk, regardless of BCOR expression
Most of the BCOR family tumors with RNAseq data have been characterized in our previous studies,[1, 2] except for 3 recent cases, including one BCOR ITD-positive PMMTI, and two alternative BCOR fusions (KMT2D-BCOR and BCOR-CHD9) round cell sarcomas (Fig. 3, Supplementary Table 1).
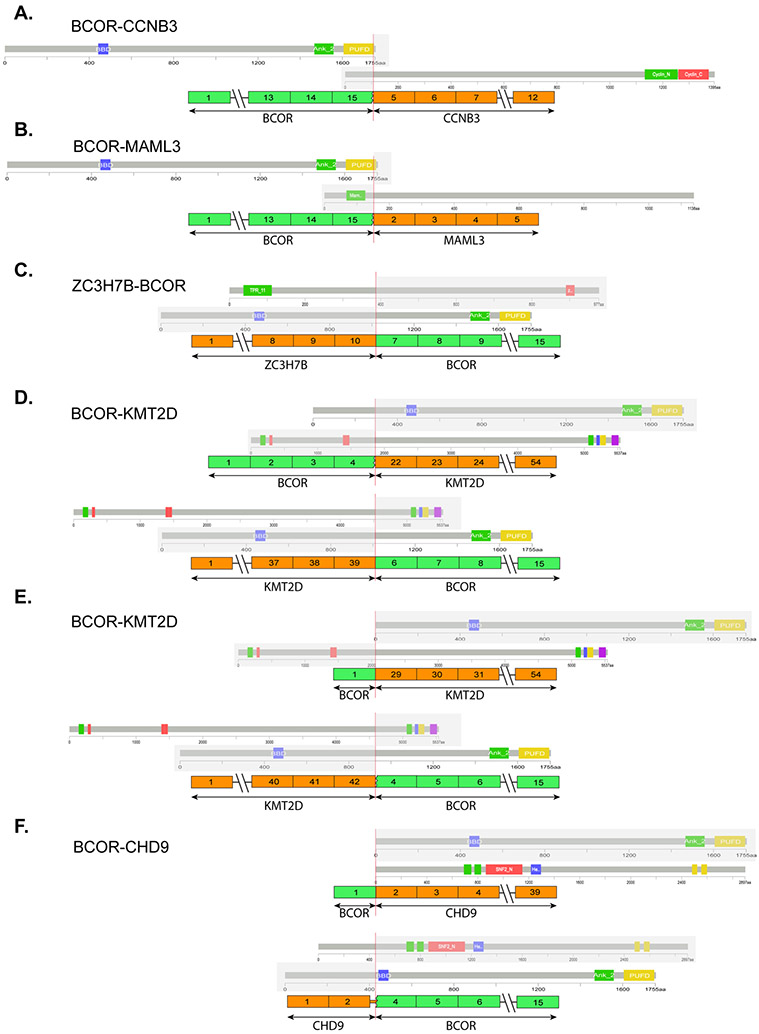
Figure 3. Genetic heterogeneity of BCOR gene fusions in soft tissue round cell sarcomas.
BCOR-CCNB3 and BCOR-MAML3 fusions involve the 3’ end of BCOR encoding the PUFD domain of BCOR protein (A-B). Other less common BCOR gene fusions showed variable breakpoints (C-F). Reciprocal fusion transcripts were identified in KMT2D-BCOR (D, recent case; E, previously reported case) and BCOR-CHD9 (F) fusions.
The novel BCOR-CHD9 gene fusion was identified in a kidney tumor of a 41-year-old woman, showing an undifferentiated round to spindle phenotype (Fig. 4A). The patient developed two subsequent intra-abdominal recurrences, one and two years after diagnosis, respectively, and showed a mixed response to Ewing sarcoma therapy. Targeted RNAseq revealed a fusion between exon 1 of BCOR to exon 2 of CHD9 and a reciprocal CHD9-BCOR fusion transcript (Fig. 3). The fusion was further confirmed by FISH showing a BCOR gene rearrangement. However, the tumor was negative for BCOR by immunohistochemistry (Fig. 4B), tested in the primary and subsequent recurrences. In contrast, pan-Trk showed diffuse reactivity with a strong staining intensity (Fig. 4C), and immunohistochemical stain for NTRK1 was negative (Fig. 4D). Other pertinent immunostains in this case revealed strong positivity for SATB2 and cyclin D1, while showing loss of H3K27me3 expression (Fig. 4E).
Figure 4. Alternative BCOR fusions overexpressing pan-Trk.
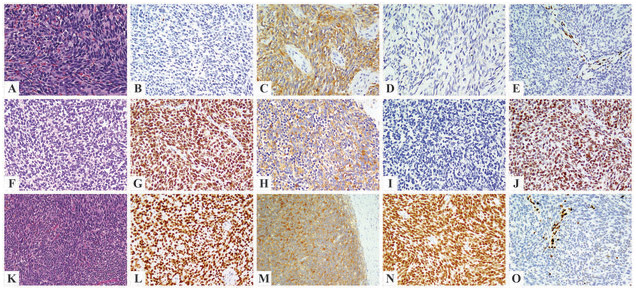
Kidney tumor with the novel BCOR-CHD9 fusion in an adult female patient (A-E) showing primitive round to short spindle cell morphology (A) and strong pan-Trk staining (C), while being negative for BCOR (B) and NTRK1 (D). Loss of H3K27me3 expression was identified (E). Two cases with KMT2D-BCOR fusion showed a monotonous round to spindle morphology (F-J, pelvic tumor of a 10-year-old female; K-O, scapular tumor of a 38-year-old male patient), positive immunostaining for BCOR (G, L), pan-Trk (H, M), and TLE1 (J, N). NTRK1 staining was negative (I). Loss of H3K27me3 expression was also identified (O).
In this study we also report a second case with KMT2D-BCOR fusion identified by targeted RNAseq and showing a fusion transcript of exon 39 of KMT2D (12q13.12) to exon 6 of BCOR (Xp11.4)(Fig. 3). Like the previously reported KMT2D-BCOR fusion sarcoma, reciprocal BCOR-KMT2D fusion transcripts were also present in this case.[2] In contrast to the previously reported case, which occurred in a 10-year-old female in the pelvic region, the recent case affected a 38-year-old male and presented as a scapular tumor. Both cases were composed of primitive round to spindle tumor cells (Fig. 4F, 4K). Immunohistochemically, both tumors showed strong positivity for BCOR (Fig. 4G, 4L), pan-Trk (Fig. 4H, 4M), SATB2, and TLE1 (Fig. 4J, 4N), while being negative for NTRK1 (Fig. 4I). H3K27me3 staining was performed on the second KMT2D-BCOR case and showed loss of expression (Fig. 4O). In both cases with KMT2D-BCOR fusions, the FISH assay could not identify the presence of BCOR gene abnormalities, likely due to cryptic gene rearrangements.
DISCUSSION
In our previous study, we identified NTRK3 as one of the top upregulated genes in BCOR-CCNB3 fusion sarcomas and infantile undifferentiated round cell sarcoma with BCOR ITD by RNAseq data analysis.[2] Significant up-regulation of NTRK3 was also recently documented in a small subset of BCOR-associated sarcomas in adults harboring BCOR-CCNB3, ZC3H7B-BCOR, CIITA-BCOR, and BCOR ITD.[8] In keeping with this finding, an earlier molecular study using Affymetrix U133A data showed high NTRK3 expression in CCSK, the renal counterpart of BCOR family tumors.[15] More recent investigations exploring the specificity and sensitivity of pan-Trk immunostaining across various tumor types have also included a few BCOR-associated sarcomas. Using the same pan-Trk antibody as in the current study, Rudzinski et al reported a lack of pan-Trk immunoexpression in one BCOR-CCNB3 sarcoma and one CCSK of unknown genetic status.[16] A few cases of synovial sarcomas have also been tested in the literature for pan-Trk, showing either focal cytoplasmic (n=2/2)[16] or negative (n=0/1) staining pattern.[14] In the present study, we further assessed the expression of NTRK3 at both the mRNA and protein levels, using a combined approach of two RNAseq platforms data and pan-Trk immunostaining, respectively. This is the largest series to date which investigates a comprehensive spectrum of BCOR genetic abnormalities and tumor types, as well as a control group spanning both look-alike round cell sarcomas or other tumors showing BCOR overexpression. Our study specifically pin-points that the pan-Trk expression in these tumors is due to NTRK3 gene upregulation and not related to NTRK1/2. Pan-Trk expression is present even in tumors with rare alternative BCOR fusions with unusual fusion breakpoints and variable BCOR protein expression. Moreover, pan-Trk expression is also seen in SFTs and a subset of synovial sarcomas. The proportion of synovial sarcomas positive for pan-Trk staining varied depending on the use of whole sections (29%) versus tissue microarrays (2%), which might be attributable to the patchy staining pattern in some cases. Therefore, the prevalence of pan-Trk expression in other sarcoma types tested using tissue microarrays may be underestimated.
With the remarkable clinical benefit of targeted therapy in tumors harboring NTRK-related fusions[13], the screening strategies and correct diagnosis of these tumors have become critical. Previous large immunohistochemical studies have suggested that pan-Trk expression has a relatively good sensitivity and specificity in most tumor types tested from various organs.[14] However, in soft tissue tumors the sensitivity and specificity of pan-Trk appears lower and the spectrum of histotypes investigated to date is rather limited.[17] Solomon et al reported pan-Trk staining, often faint cytoplasmic, in desmoplastic small round cell tumor (5/5), BCOR translocated sarcomas (3/5), and less commonly Ewing sarcoma (1/5) and rhabdomyosarcoma (1/7).[17] Our results of consistent NTRK3 upregulation in the majority of BCOR family tumors elucidate at least in part the mechanism of pan-Trk immunopositivity in certain sarcoma types lacking NTRK-related gene fusions. Moreover, these results raise caution of interpreting a positive result of pan-Trk immunohistochemistry as a surrogate for the presence of an NTRK-fusion event. Further investigation is warranted to explore if these NTRK specific inhibitors may also show clinical benefit in the tumors with BCOR and NTRK3 co-expression in the absence of NTRK fusions.
In the context of NTRK fusion positive tumors, the truncated TRK proteins acquire the ability of ligand-independent activation, whereas the consequence of overexpression of wild-type TRK-C as seen in these BCOR-upregulated tumors remains unclear. In a prior array-based study, infantile fibrosarcomas with ETV6-NTRK3 fusion showed up-regulation of the truncated NTRK3 as well as several other genes encoding for receptor tyrosine kinase pathway inhibitors, such as SPRY4 and DUSP6, while a group of CCSK showed only up-regulation of NTRK3 but not SPRY4 and DUSP6.[15] Up-regulation of the full-length NTRK3 transcript has also been reported in salivary gland adenoid cystic carcinoma and CYLD defective tumors of skin, such as cylindroma and spiradenoma.[18, 19] Unlike our cases, NT-3, the ligand of TRK-C, was also upregulated in adenoid cystic carcinoma and CYLD-defective cutaneous tumors. In vitro experiments using TRK targeting strategies, including RNA interference and TRK inhibitor, have shown reduced colony formation and proliferation of cultured cells from CYLD mutant tumors.[19] However, a phase 2 clinical trial exploring the usage of topical pegcantratinib, a potent TRKA inhibitor which also has activity against TRKB and TRKC, in the treatment of cutaneous CYLD defective tumors shows lack of response, which might be attributable to low drug concentration achieved in the tumors.[20]
The current study provides further evidence of genetic heterogeneity among undifferentiated sarcomas with BCOR fusions by reporting a novel BCOR-CHD9 and the second case with KMT2D-BCOR fusion. In contrast to BCOR-CCNB3 positive tumors, which occur mostly in male children and have a consistent transcript (BCOR exon 15 fused to CCNB3 exon 5) resulting in BCOR overexpression, other less frequent fusion variants, such as ZC3H7B-BCOR, KMT2D-BCOR, BCOR-CHD9, and the recently reported CIITA-BCOR, occur more commonly in middle-aged adult patients of both genders and show variable BCOR breakpoints.[3, 8] Regardless of the canonical or alternative BCOR fusions, tumors typically display a similar morphologic spectrum of primitive round to spindle cell phenotype. However, unlike in BCOR-CCNB3 and BCOR ITD positive tumors, the C-terminal PUFD domain is not consistently retained in the fusion oncoprotein of these uncommon BCOR alternative fusions (Fig. 3), which may result in more variable BCOR overexpression. In the novel BCOR-CHD9 fusion identified in this study, exon 1 of BCOR was fused to exon 2 of CHD9, which was associated with lack of BCOR expression. Intriguingly, up-regulated NTRK3 mRNA expression was demonstrated in all of the 4 tumors with KMT2D-BCOR (n=2), BCOR-CHD9 (n=1), or CIITA-BCOR (n=1) fusions.[2, 8] Three of the cases tested in this study were all positive for pan-Trk, including the BCOR-CHD9 case with negative BCOR staining, suggesting that NTRK3 expression is not affected by different exon composition of BCOR fusions. Of note, in all tumors reported to date with KMT2D-BCOR, BCOR-CHD9, or CIITA-BCOR fusions, reciprocal fusion transcripts have been identified. Based on the abundance of fusion reads or the truncated mRNA expression patterns, BCOR is likely the 3’ fusion partner gene in KMT2D-BCOR and CIITA-BCOR fusions. Diagnostic challenges might occur in diagnosing these rare BCOR fusions due to the lack of BCOR staining by immunohistochemistry, as in our BCOR-CHD9 case, as well as due to false negative FISH results for BCOR rearrangement, likely due to cryptic fusions, as seen in both our KMT2D-BCOR fusion cases and the reported CIITA-BCOR tumor. Furthermore, as these tumors typically show an undifferentiated monotonous spindle to round cell morphology, the TLE1 expression and loss of H3K27me3 expression by immunohistochemistry observed in our cases may lead to the erroneous diagnosis of synovial sarcoma and malignant peripheral nerve sheath tumor, respectively. These findings suggest the need for applying a battery of complementary ancillary tests to avoid these pitfalls.
Our results demonstrate that in the setting of an undifferentiated round to spindle cell neoplasm showing positivity for pan-Trk at immunohistochemical level, the differential diagnosis may include not only tumors with NTRK fusions, but also a large spectrum of sarcomas with BCOR genetic alterations as well as other BCOR expressing tumors, specifically SFT, and subsets of synovial sarcoma and Ewing sarcoma. Further immunohistochemical studies (e.g. BCOR, TLE1 and STAT6) and molecular tests (e.g. FISH or sequencing-based assay) are likely needed in this context for a more definitive subclassification.
In conclusion, NTRK3 overexpression at both mRNA and protein levels is a consistent finding in BCOR family tumors of soft tissue and less often in synovial sarcomas. In these tumors, pan-Trk immunohistochemistry commonly shows cytoplasmic staining, with variable intensity and extent. Rare alternative BCOR fusion partners-driven adult sarcomas, such as BCOR-CHD9 and KMT2D-BCOR fusions, also show NTRK3 expression irrespective of BCOR expression status. The pan-Trk immunoreactivity observed in the majority of SFT and a subset of Ewing sarcoma might be attributable to NTRK1 up-regulation. In the context of a sarcoma with primitive round to spindle morphology, a positive pan-Trk immunoresult should not be interpreted as a surrogate for the presence of NTRK fusions without molecular confirmation. The clinical and therapeutic significance of our findings warrants further investigation
Supplementary Material
Acknowledgments
Disclosures: Supported in part by: P50 CA 140146-01 (CRA), P50 CA217694 (CRA), Cycle for Survival (CRA), Kristin Ann Carr Foundation (CRA), St Baldrick Foundation (CRA)
REFERENCES
- 1.Kao YC, Sung YS, Zhang L, Huang SC, Argani P, Chung CT, et al. Recurrent BCOR Internal Tandem Duplication and YWHAE-NUTM2B Fusions in Soft Tissue Undifferentiated Round Cell Sarcoma of Infancy: Overlapping Genetic Features With Clear Cell Sarcoma of Kidney. Am J Surg Pathol. 2016;40:1009–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kao YC, Owosho AA, Sung YS, Zhang L, Fujisawa Y, Lee JC, et al. BCOR-CCNB3 Fusion Positive Sarcomas: A Clinicopathologic and Molecular Analysis of 36 Cases With Comparison to Morphologic Spectrum and Clinical Behavior of Other Round Cell Sarcomas. Am J Surg Pathol. 2018;42:604–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Specht K, Zhang L, Sung YS, Nucci M, Dry S, Vaiyapuri S, et al. Novel BCOR-MAML3 and ZC3H7B-BCOR Gene Fusions in Undifferentiated Small Blue Round Cell Sarcomas. Am J Surg Pathol. 2016;40:433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ueno-Yokohata H, Okita H, Nakasato K, Akimoto S, Hata J, Koshinaga T, et al. Consistent in-frame internal tandem duplications of BCOR characterize clear cell sarcoma of the kidney. Nat Genet. 2015;47:861–3. [DOI] [PubMed] [Google Scholar]
- 5.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell. 2016;164:1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marino-Enriquez A, Lauria A, Przybyl J, Ng TL, Kowalewska M, Debiec-Rychter M, et al. BCOR Internal Tandem Duplication in High-grade Uterine Sarcomas. Am J Surg Pathol. 2018;42:335–41. [DOI] [PubMed] [Google Scholar]
- 7.Juckett LT, Lin DI, Madison R, Ross JS, Schrock AB, Ali S. A Pan-Cancer Landscape Analysis Reveals a Subset of Endometrial Stromal and Pediatric Tumors Defined by Internal Tandem Duplications of BCOR. Oncology. 2019;96:101–9. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida A, Arai Y, Hama N, Chikuta H, Bando Y, Nakano S, et al. Expanding the clinicopathologic and molecular spectrum of BCOR-associated sarcomas in adults. Histopathology. 2019. [DOI] [PubMed] [Google Scholar]
- 9.Antonescu CR, Sung YS, Chen CL, Richter GH, Fletcher CD, Antonescu CR. Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors--molecular characterization shows genetic overlap with endometrial stromal sarcoma. Genes Chromosomes Cancer. 2014;53:183–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis N, Soslow RA, Delair DF, Park KJ, Murali R, Hollmann TJ, et al. ZC3H7B-BCOR high-grade endometrial stromal sarcomas: a report of 17 cases of a newly defined entity. Mod Pathol. 2018;31:674–84. [DOI] [PubMed] [Google Scholar]
- 11.Kao YC, Sung YS, Zhang L, Jungbluth AA, Huang SC, Argani P, et al. BCOR Overexpression Is a Highly Sensitive Marker in Round Cell Sarcomas With BCOR Genetic Abnormalities. Am J Surg Pathol. 2016;40:1670–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Argani P, Kao YC, Zhang L, Sung YS, Alaggio R, Swanson D, et al. BCOR Overexpression in Renal Malignant Solitary Fibrous Tumors: A Close Mimic of Clear Cell Sarcoma of Kidney. Am J Surg Pathol. 2019;43:773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15:731–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D, et al. Pan-Trk Immunohistochemistry Is an Efficient and Reliable Screen for the Detection of NTRK Fusions. Am J Surg Pathol. 2017;41:1547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gadd S, Beezhold P, Jennings L, George D, Leuer K, Huang CC, et al. Mediators of receptor tyrosine kinase activation in infantile fibrosarcoma: a Children’s Oncology Group study. J Pathol. 2012;228:119–30. [DOI] [PubMed] [Google Scholar]
- 16.Rudzinski ER, Lockwood CM, Stohr BA, Vargas SO, Sheridan R, Black JO, et al. Pan-Trk Immunohistochemistry Identifies NTRK Rearrangements in Pediatric Mesenchymal Tumors. Am J Surg Pathol. 2018;42:927–35. [DOI] [PubMed] [Google Scholar]
- 17.Solomon JP, Linkov I, Rosado A, Mullaney K, Rosen EY, Frosina D, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol. 2020; 33:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov SV, Panaccione A, Brown B, Guo Y, Moskaluk CA, Wick MJ, et al. TrkC signaling is activated in adenoid cystic carcinoma and requires NT-3 to stimulate invasive behavior. Oncogene. 2013;32:3698–710. [DOI] [PubMed] [Google Scholar]
- 19.Rajan N, Elliott R, Clewes O, Mackay A, Reis-Filho JS, Burn J, et al. Dysregulated TRK signalling is a therapeutic target in CYLD defective tumours. Oncogene. 2011;30:4243–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danilenko M, Stamp E, Stocken DD, Husain A, Zangarini M, Cranston A, et al. Targeting Tropomyosin Receptor Kinase in Cutaneous CYLD Defective Tumors With Pegcantratinib: The TRAC Randomized Clinical Trial. JAMA Dermatol. 2018;154:913–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.