Abstract
Hereditary genetic disorders, cancer, and infectious diseases of the liver affect millions of people around the globe and are a major public health burden. Most contemporary treatments offer limited relief as they generally aim to alleviate disease symptoms. Targeting the root cause of diseases originating in the liver by regulating malfunctioning genes with nucleic acid-based drugs holds great promise as a therapeutic approach. However, employing nucleic acid therapeutics in vivo is challenging due to their unfavorable characteristics. Lipid nanoparticle (LNP) delivery technology is a revolutionary development that has enabled clinical translation of gene therapies. LNPs can deliver siRNA, mRNA, DNA, or gene-editing complexes, providing opportunities to treat hepatic diseases by silencing pathogenic genes, expressing therapeutic proteins, or correcting genetic defects. Here we discuss the state-of-the-art LNP technology for hepatic gene therapy including formulation design parameters, production methods, preclinical development and clinical translation.
Keywords: Gene therapy, liver, lipid nanoparticle (LNP), lipids, hepatocyte, small interfering RNA (siRNA), messenger RNA (mRNA), DNA, guide RNA (gRNA), CRISPR/Cas9, gene silencing, gene expression, gene editing
Graphical abstract
1. Introduction
“Survival rates have improved for almost every disease of every organ in the last few decades, with one notable exception: liver disease” [1]. This statement by The Lancet Commission clearly illustrates the global burden of liver disorders and the need for more effective therapeutic strategies [2]. The most frequently occurring liver diseases include hepatitis, liver cancer, alcoholic liver disease, fatty liver disease, and hereditary diseases. In addition to direct harmful effects, these diseases can significantly affect the liver’s carbohydrate, fat, and protein metabolism. The increase in lifestyle-related incidence rates and the limited therapeutic efficacy of currently available treatments have resulted in substantial drug development efforts targeting the liver [2]. Our ability to treat hepatic diseases by targeting their genetic background is increasingly becoming a clinical reality owing to the development of nucleic acid-based therapeutics. In contrast to small molecule drugs and biologics which target gene products (i.e. proteins), nucleic acid therapeutics have the potential to therapeutically regulate essentially any gene of interest at the DNA or RNA level. Their versatility in treating inherited or acquired disorders originating in the liver stems from the ability to induce efficient gene silencing (inhibiting pathological/mutant protein production), gene expression (producing therapeutic proteins) or gene editing (correcting dysfunctional/mutated genes). Several nucleic acid therapeutics have been approved by the U.S. Food and Drug Administration (FDA) and the European Medicines Agency (EMA) with many more in various stages of clinical evaluation. These therapeutics include antisense oligonucleotides (ASO) [3], small interfering RNA (siRNA) [4,5], plasmid DNA (pDNA) [6,7], messenger RNA (mRNA) [8,9], and complexes containing guide RNA (gRNA) as part of gene editing approaches [10,11].
Using nucleic acids therapeutically in vivo is challenging because of their unfavorable physicochemical characteristics, such as negative charge and relatively large size, which prevents their efficient uptake into cells [12]. In addition, nucleic acids are susceptible to degradation by nucleases in the circulation, suffer from rapid renal clearance, and induce immunostimulatory effects via pattern recognition receptors, resulting in adverse effects [13]. Therefore, the clinical translation of nucleic acid therapeutics has been dependent on chemical modifications and advanced delivery technologies to improve nucleic acids’ stability, promote their target tissue accumulation, enable their cellular internalization, and increase their target affinity [14].
Lipid nanoparticle (LNP) systems are currently one of the most sophisticated non-viral gene delivery technologies enabling gene therapies [15]. Decades of designing lipid-based delivery systems for small molecule therapeutics [16] has driven efforts in adapting LNP technology for nucleic acid delivery [17,18], particularly following the discovery of RNA interference (RNAi) [19,20]. These efforts included systematically optimizing all LNP components for efficient gene silencing and incorporating siRNA payload modification and chemistry [21,22], polyethylene glycol (PEG) lipids [[23], [24], [25], [26]], helper lipids [27,28], and, particularly, ionizable cationic lipids [[29], [30], [31]].
In 2018, these developments culminated in the approval of Onpattro® (patisiran), the first RNAi drug, for treating hereditary amyloidogenic transthyretin (ATTRv) amyloidosis [32,33]. This systemic disease, which generally presents as progressive neuropathy, is caused by mutations in the gene encoding the transthyretin (TTR) protein, resulting in amyloid fibril deposition in multiple organs [34]. Onpattro® relies on LNP technology for efficient TTR siRNA delivery to hepatocytes following systemic infusion, inhibiting mutant TTR protein production and subsequent fibril formation.
In this review, we provide an overview of the lipid nanotechnology-mediated gene regulation approaches in the liver for treating various diseases. First, we describe the liver’s microanatomy and how its cell subtypes affect LNP accumulation and clearance. Second, we discuss design criteria and production methods [35,36] for intravenously-administered LNPs delivering nucleic acid therapeutics to the liver. Finally, we highlight the (pre)clinical development of LNP-based genetic drugs for treating genetic liver diseases, hepatocellular carcinoma (HCC), and infections.
Of note, readers are referred to several excellent reviews covering other clinically relevant liver-targeted nucleic acid therapeutics, such as ASOs [3], N-acetylgalactosamine (GalNAc)-siRNA conjugates [4], or adenovirus-associated vectors [6,7].
2. Liver microanatomy
With more than 500 functions ranging from metabolism (e.g. of lipids, carbohydrates, or amino acids) and protein secretion (e.g. hemostasis factors, plasma proteins, or hormones) to immune responses, the liver is one of the most vital organs [37]. Hundreds of hepatic disorders affect millions of people globally, with significant personal and systemic costs [2,38]. In order to treat such diseases and develop relevant therapeutics, it is crucial to understand the liver’s microanatomical and subcellular features.
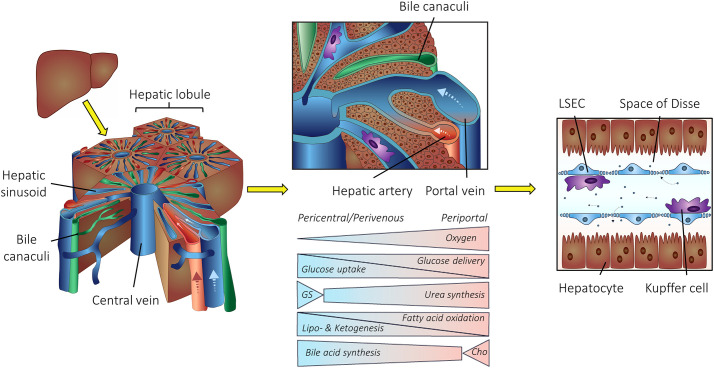
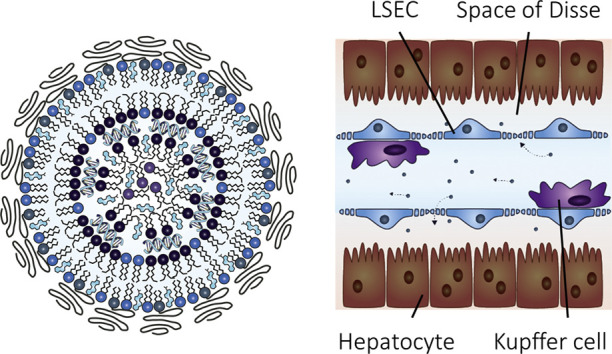
The liver is divided into functional subunits called hepatic lobules. Nutrient and oxygen-rich blood from the portal vein and the hepatic artery traverse the lobules to the central vein (Fig. 1 ), resulting in LNP exposure to scavenger cells within the liver. Liver sinusoidal endothelial cells (LSECs) line the sinusoidal vessels, while liver-resident macrophages, i.e. Kupffer cells, are localized within the hepatic sinusoids. Hepatocytes, the most prominent cell type and a key cell type for many diseases, are stationed behind the space of Disse [39,40].
Fig. 1.
Structure of liver lobules. The hepatic lobule is the liver’s functional unit. Blood from the portal vein and the hepatic artery traverse the lobules to the central vein. Bile canaculi transport bile from the liver to the gut. Various metabolic pathways distribute along the porto-central axis of a liver lobule. GS, glutamine synthesis; Cho, cholesterol synthesis. Liver sinusoidal endothelial cells (LSECs) line the hepatic blood vessels, while liver-resident macrophages, i.e. Kupffer cells, are localized within the hepatic sinusoids. Hepatocytes are located behind the space of Disse with a sinusoidal (basolateral) membrane towards blood circulation. Figure adapted from Mosby et al. [48]
2.1. Cell types within the liver microenvironment
The term “liver gene therapy” is often used to unilaterally describe all gene therapy approaches for treating diseases originating in hepatocytes. Although hepatocytes are the most prominent cell type within the liver, several other cell types can interact with nanoparticles and affect their performance [[41], [42], [43], [44]]. It is therefore recommended that scientists expand their LNP studies to include single cell quantification rather than the whole liver. Every liver lobule comprises parenchymal (i.e. hepatocytes) and non-parenchymal liver cells (i.e. LSECs and Kupffer cells). In addition to these three major cell types, the liver consists of several other cell populations. A recent study revealed 20 discrete cell types ranging from stellate cells (also known as Ito cells) and cholangiocytes to immune cells such as B, T, or NK-like cells [45]. The human liver cell atlas revealed additional subtypes within the liver microenvironment [46]. The most relevant cell types and their implications for LNP-based gene therapy are discussed in the next sections, following the particle’s journey in the body after systemic administration [47].
Following intravenous injection, LNP-siRNA systems (composed of CLinDMA:Cholesterol:PEG-DMG; 50:44:6 mol%) accumulate in all major liver cell types (i.e. Kupffer cells, LSECs, and hepatocytes) in a time- and dose-dependent manner, as demonstrated by Shi et al. [49]. When LNP-siRNA were administered at 0.3 mg siRNA/kg, similar siRNA amounts were detected in Kupffer cells and hepatocytes. Doses of 1 mg/kg to 9 mg/kg siRNA delivered 50% to 83%, respectively, to hepatocytes. At 30 minutes post injection, LNPs were mainly localized in the space of Disse, whereas 2 hours post injection LNPs accumulated in hepatocytes. siRNAs delivered to Kupffer cells and LSECs were inactive, but delivery to hepatocytes resulted in efficient gene silencing. A similar intrahepatic distribution has recently been described for LNP-DNA barcode systems (composed of MC3 or cKK-E12) by Sago et al. [50]. However, direct comparison of datasets carried out with different LNP systems, including variations in the lipid composition, ionizable lipid, lipid-nucleic acid ratios, and nucleic acid type must be assessed with caution. At a dose of 0.3 mg DNA /kg, LNPs accumulated in all three major liver cell types, with higher doses in Kupffer cells and hepatocytes than endothelial cells. In sharp contrast, gene expression following LNP-mediated mRNA delivery demonstrated an inverse hierarchy among hepatic cells: endothelial cells > Kupffer cells > hepatocytes. Higher LNP doses corresponding to 1 mg/kg mRNA shifted expression slightly towards hepatocytes while keeping the same pattern. Transfection of all major liver cell types with equal potency was recently demonstrated for LNP-mRNA systems (composed of branched-tail 306Oi10) at a dose of 2 mg RNA/kg by Hajj et al. [51].
Indeed, targeting the right cell type with the right dose is crucial to developing effective therapeutics. It should be noted that the LNP compositions described in these preclinical studies deviate from those used in the clinic (except for MC3-based LNPs). Systematic studies are therefore needed to improve our fundamental understanding of LNPs' in vivo behavior. Rigorous control of physicochemical LNP characteristics such as size distribution, zeta potential, and entrapment will be crucial to assess the intrahepatic distribution of a single LNP composition with different payloads.
2.1.1. Kupffer cells – Main phagocytotic center within the body
Following intravenous injection, liver-resident macrophages, i.e. Kupffer cells, are the first hepatic cells to interact with LNPs (Fig. 1). These phagocytic cells are part of the mononuclear phagocyte system (MPS), also known as the reticuloendothelial system (RES). They comprise 80% of the entire macrophage population within the body, illustrating their importance in host defense and LNP elimination [52].
Three major elimination pathways have been described [56]. First, negatively charged LNP systems are recognized by class A scavenger receptors (SR-A) expressed primarily on Kupffer cells resulting in rapid clearance [41,57,58]. Second, mannose- and fucose-type receptors can be leveraged to selectively target LNP systems to Kupffer cells. Third, LNP opsonization by serum proteins results in MPS sequestration. Complement factors (e.g. C3b or C1q) and serum opsonins such as fibrinogen can coat LNPs with unfavourable characteristics including large size, high surface charge, or lack of PEGylation [[59], [60], [61]]. Several research groups have explored strategies to prevent Kupffer cell clearance in order to redirect LNPs to hepatocytes. Transient Kupffer cell depletion using clodronate-loaded liposomes or by knocking out the endocytic Caveolin1 gene are efficient methods in a research setting [41,43]. However, the clinical utility of such approaches is limited.
2.1.2. Liver sinusoidal endothelial cells – Restricting hepatocyte access
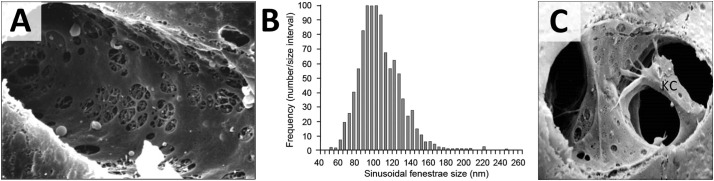
LSECs are located in close proximity to Kupffer cells and play important roles in sequestering LNPs and restricting access to hepatocytes [62]. Many structural and functional features have been elucidated by Braet and Wisse [54,[63], [64], [65], [66], [67]]. LSECs line the hepatic sinusoids and form pores, so-called fenestrations, that are clustered in sieve plates (Fig. 2 ). Endothelial fenestrae range from 50 to 200 nm in diameter and differ between species (Table 1 ). Therefore, liver fenestrae physically restrict circulating LNPs’ access to the perisinusoidal space and thus limit cellular interactions with hepatocytes according to size. Several research groups have investigated using pore-opening substances to modulate fenestrae size with limited success [64]. In addition to their structural characteristics, LSECs have high endocytic activity. A number of scavenger receptors, including stabilin-2, can efficiently sequester anionic nanoparticles [68].
Fig. 2.
Liver sinusoids. (A) Cross section of a hepatic sinusoid. Liver sinusoidal endothelial cells form clustered fenestrations also known as sieve plates [53]. Reproduced with permission. Copyright 2009 American Physiological Society (B) Distribution of sinusoidal fenestrae size in healthy humans. Average diameter of endothelial fenestrae is 107 ± 1.5 nm. Adapted from Wisse et al. [54]. (C) Kupffer cell (KC) located within the hepatic sinusoid in close proximity to endothelial cells. Adapted with permission from UCSF Office of Medical Education [55].
Table 1.
Comparison of liver fenestrations. Different species and strains have differently sized endothelial fenestrae in liver sinusoids. All studies used electron microscopy techniques to determine fenestrae diameters.
2.1.3. Stellate cells – Implications for LNP delivery
Hepatic stellate cells – also known as Ito cells, vitamin A or lipid storage cells – are localized within the perisinusoidal space of Disse [71]. In healthy human subjects, stellate cells are quiescent and function as vitamin A storage. However, liver damage and inflammatory processes induce stellate cell differentiation into a myofibroblast-like phenotype resulting in connective scar tissue production within the space of Disse [72,73]. This pathophysiological process dramatically impairs the transendothelial transport of any substance from the systemic circulation to hepatocytes with major implications for nanoparticle-based nucleic acid therapeutics [74,75].
2.1.4. Hepatocyte – The key target for hepatic gene therapy
Hepatocytes, comprising 70-80% of the total liver cell population, are the most relevant hepatic target cell type for nucleic acid therapeutics (Fig. 1). Owing to their broad range of functions, hepatocytes play a key pathogenic role in many disorders (Table 2 ). Hepatocytes are highly differentiated with a sinusoidal (basolateral) membrane towards the blood circulation and an apical membrane towards bile canaliculi. The sinusoidal membrane with its microvilli exhibits surface receptors important for LNP recognition. The most important receptors for LNP-nucleic acid are the low-density lipoprotein receptor (LDLR) and asialoglycoprotein receptor (ASGPR) [76]. Within a healthy liver, hepatocytes are postmitotic (i.e. non-dividing cells) with an average life span of up to 6 months.
Table 2.
Highlighted diseases originating in hepatocytes. Hepatocytes play major roles in various liver diseases including genetic disorders, infections, and cancer. Selected diseases are listed in order of their prevalence (from high to low), along with their pathophysiology, symptoms, current symptomatic treatments, and prevalence. Adapted with permission from Witzigmann et al. [77]
| Disease | Pathophysiology | Typical symptoms | Symptomatic treatment | Prevalence [78] | Ref. |
|---|---|---|---|---|---|
| Genetic Disease (without parenchymal damage) | |||||
| Hemophilia disorders (e.g., Hemophilia B) | Factor IX deficiency [other coagulation factor mutations A and C] |
Blood clotting disorder, hemorrhage | i.v. infusion of coagulation factor | 1:20,000 | [79,80] |
| Urea cycle disorders (e.g., OTC deficiency) | Ornithine transcarbamylase (OTC) deficiency [many other deficiencies such as Argininosuccinate synthetase (ASS; Citrullin-aemia), N-acetyl glutamate synthetase (NAGS), Carba-moylphosphate synthetase (CPS), Arginase (ARG)] |
Hyperammo-nemia; neuro-logical damage | Nitrogen scavenger therapy, hemodialysis | OTC: 1:80,000 | [81,82] |
| Familial Hyperchol-esterolemia (e.g., LDL receptor related) | LDL receptor protein mutation [also ApoB or PCSK9 mutations] | Coronary artery disease | Statins, LDL apheresis | Homozygous <1:100,000 Hetero: 1:500 |
[83,84] |
| TTR Familial amyloid polyneuropathy (FAP) | Transthyretin mutation - deposition of insoluble protein | Neurodegene-ration, poly-neuropathy | Small molecule drugs (tafamidis) | < 1:100,000, > in some countries | [85,86] |
| Thrombotic disorders (e.g., Protein C deficiency type 1) | Thrombotic disease caused by PROC gene mutation [also other inherited thrombophilias] | Risk of thrombosis | Thrombo-embolism, protein C substitution | 1:500,000–750,000 | [87,88] |
| Primary hyperoxaluria type 1 | Alanine glyoxylate aminotransferase mutation | Calcium oxalate accumulation, kidney damage | High fluid intake, kidney trans-plantation | 1:333,000–1,000,000 | [81,89] |
| Bilirubin metabolism disorders (e.g., Crigler-Najjar syndrome 1) | Uridine diphosphate glucuronosyltransferase (UGT1A1) deficiency - impairment of bilirubin conjugation | Neurological damage; kern-icterus (bilirubin encephalopathy) | Phototherapy (10–12h per day); Plasma exchange | < 1:1,000,000 | [90,91] |
| Genetic Disease (with parenchymal damage) | |||||
| α1-antitrypsin deficiency | Mutations in the SERPINA1 gene; deficiency in protease inhibitor for neutrophil elastase | Lung and liver damage | Augmentation; replacement therapy |
1–5:10,000 | [92,93] |
| Wilson´s disease | Copper-transport P-type ATPase deficiency, Copper accumulation | Liver and neuro-logical damage | Copper complexation | 1:30,000– 100,000 individuals | [94,95] |
| Tyrosinemia disorders (e.g., Tyrosinemia type 1) | Fumarylacetoacetate hydrolase (FAH) deficiency - lack of tyrosine degra-dation [other types with enzyme deficiency in tyrosine metabolism] | Hepatomegaly, liver and kidney dysfunction | Nitisinone (inhibition of tyrosine degradation) | 1:100,000 | [96,97] |
| Iron overload disorder (e.g., Hereditary hemo-chromatosis type 1) | HFE enzyme deficiency [other iron dysregulation; Type 2: HFE2 or HAMP (hepcidin); Type 3: TFR2 (transferrin receptor 2); Type 4: SLC40A1 (ferroportin)] |
Liver cirrhosis, insulin resistance | Phlebotomy, iron-chelating | Type 1: >1:1,000 Type 2/3/4: < 1 : 1,000,000 |
[98,99] |
| Glycogen storage diseases (GSD) (e.g., Pompe´s disease) | Various types of enzyme deficiencies in glycogen synthesis | Hepatomegaly, hypoglycemia | Treatments depend on type | 1:50,000–1,000,000 | [100,101] |
| Cancer | |||||
| Hepatocellular Carcinoma (HCC) | Chronic liver inflammation - cirrhosis - HCC | Liver damage, liver cancer | Curative or palliative treatment | 16:100,000 and > 700,000 new cases per year |
[[102], [103], [104]] |
| Viral Infections | |||||
| Hepatitis B | Hepatitis B Virus (HBV) infection | Liver damage, cirrhosis, HCC | Interferon α, nucleos(t)ide | 350 million chronic carriers | [[105], [106], [107], [108]] |
| Hepatitis C | Hepatitis C Virus (HCV) infection | Liver damage, cirrhosis, HCC | Interferon α, protease inhibitors | 180 million chronic carriers | [105,109] |
2.2. (Patho)physiological factors affecting hepatic gene therapy
Many factors can alter LNP accumulation and clearance. The following sections detail important (patho)physiological factors affecting intrahepatic LNP distribution.
2.2.1. Metabolic and cellular liver zonation
An often-overlooked challenge in hepatic gene therapy is metabolic and cellular liver zonation, a phenomenon that separates various pathways along the porto-central axis of a liver lobule. First, some genetic disorders manifest in periportal or perivenous hepatocytes [110,111]. Second, metabolic zonation can vary among species and during development (infant versus adult). Third, different non-parenchymal cell subtypes within the liver microenvironment can affect LNP clearance [45]. All these factors impact LNP development and gene therapy outcomes.
Fig. 1 details the liver microarchitecture and its major metabolic pathways. Metabolic liver zonation for glucose homeostasis, urea synthesis, carbohydrates, bile acids, or lipid metabolism has been discussed in several excellent reviews [[110], [111], [112], [113]]. Advancements in omics and single-cell techniques are continuously elucidating new cell subtypes and improving our understanding of liver zonation [45,110]. As illustrated, ureagenesis is restricted to periportal hepatocytes, while most intravenously injected nucleic acid therapeutics predominantly target perivenous cells. Bell et al. demonstrated a zonation bias for transducing hepatocytes in different species and at different ages [114]. While transgene expression in adult mice and dogs was predominantly pericentral following viral transduction, the expression pattern in cynomolgus and rhesus macaques was mainly periportal. In contrast, newborn mice and infant rhesus macaques showed equal distribution. This bias has important implications for gene therapies, e.g. to correct ornithine transcarbamylase (OTC) deficiency [115]. Further studies are warranted to investigate such phenomena for LNPs.
Improved LNP design might facilitate gene expression within the target zone along the porto-central axis. A recent study by Sago et al. investigated whether non-parenchymal cell subtypes differentially interact with LNPs and thereby affect their clearance [50]. Interestingly, LNPs’ intra-hepatic and sub-cellular distribution varies with lipid composition. Periportal endothelial cells (CD32Low) sequestered cKK-E12-based LNPs more efficiently than central venous endothelial cells (CD32High), most likely due to the blood flow direction and the order of exposure. LNPs composed of MC3 had similar levels of delivery to both periportal and central venous endothelial cells. In contrast, clearance of MC3-based LNPs was significantly higher in tolerogenic, M2-like Kupffer cells (CD74Low) as compared to inflammatory, M1-like Kupffer cells (CD74High). Recent findings emphasize this preferential nanoparticle uptake by M2-type macrophages, with a clear hierarchy among the different phenotypes (M2c > M2 > M2a > M2b > M1) [116]. It is tempting to speculate that differences in apparent pKa values and structural LNP characteristics resulted in distinct biomolecular coronas preferentially redirecting LNPs to different cell subtypes. However, precise mechanistic studies are needed to elucidate underlying phenomena and clearance by all intermediate phenotypes [117].
Sleyster et al. demonstrated that periportal Kupffer cells are more abundant and have higher endocytic and lysosomal activity than perivenous Kupffer cells [118]. These results demonstrate that whole-tissue (entire liver) analysis should be replaced by dissociated single cell-based techniques.
2.2.2. Pathophysiological remodelling
Liver disease progression results in pathological remodelling including microanatomical or target receptor alterations that could affect nanoparticle delivery and sequestration. Firstly, liver infections or metabolic disorders can lead to chronic cell damage and cell activation. This can result in liver fenestrae re-arrangement or fibrotic material deposition by activated stellate cells within the perisinusoidal space. Thus, LNP transport to hepatocytes is inhibited, as is access to the key target cell for most gene therapies [119]. Hepatic inflammatory processes can also enhance hepatic nanoparticle sequestration by Kupffer cell activation [116]. Secondly, downregulation of surface receptors crucial for LNP binding decreases gene delivery efficiency. For example, two independent studies have demonstrated lowering of ASGPR expression during HCC progression (according to the Barcelona Clinic Liver Cancer staging) [120,121]. This has serious implications for liver cancer interventions using ASGPR-targeting approaches. Thirdly, variations in serum proteins, such as apolipoprotein E (ApoE), are known to mediate specific LNP binding and might affect efficacy. A recent study investigated the effect of ApoE polymorphisms in patients with ATTRv amyloidosis treated with Onpattro® [122]. Niemietz et al. revealed that efficacy is independent of the ApoE genotype but that ApoE downregulation reduces efficacy. All the aforementioned pathophysiological alterations affect specific cell binding and could potentially limit therapeutic outcomes. Therefore, diagnostic tools to stratify patients for LNP-based gene therapy offer interesting possibilities [123].
3. Designing LNPs for gene regulation in the liver
The fundamental LNP design parameters for nucleic acid delivery are based on those established for small molecule liposomal formulations. These parameters include appropriate particle size (for efficient terminal sterile filtration and hepatic delivery), long-term stability in storage, optimized payload release rates to produce a therapeutic effect, robust and scalable manufacturing processes, and efficient entrapment. In applying these requisites to nucleic acid delivery systems, it became obvious that additional lipid components and functionalities were required beyond those used to compose small-molecule carriers. The very first nucleic acid formulations, containing only phosphatidylcholine and cholesterol, demonstrated that nucleic acid entrapment within a particle was feasible, but the entrapment efficiency was poor [124,125]. Subsequent development of the cationic lipid 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) showed that ionic interactions between the lipids and payload can dramatically increase entrapment efficiencies and intracellular delivery. Toxicity issues, resulting from the cationic lipids’ permanent positive charge and non-biodegradable nature, plagued these initial lipoplex-like formulations [126]. Through additional formulation development, and manufacturing process optimization, it was determined that LNP systems required four components: ionizable cationic lipids, phospholipids (typically phosphatidylcholine), cholesterol, and PEG-lipids. The role of each component, the evolution of the composition, and the manufacturing processes are discussed in the following sections.
3.1. Ionizable cationic lipids
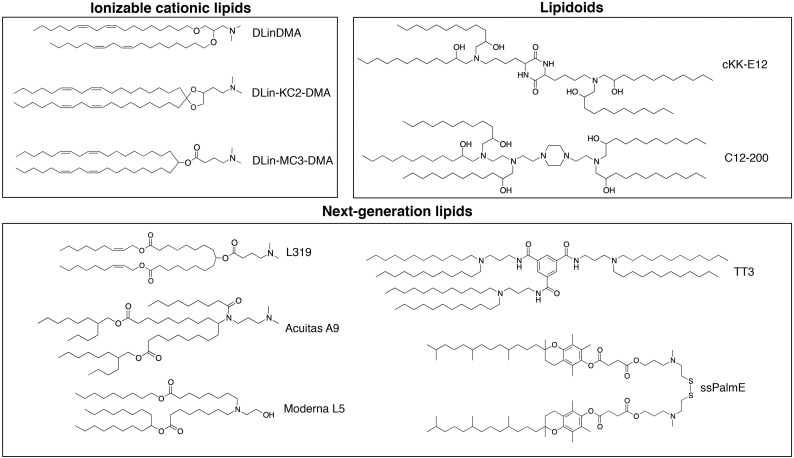
To date, a vast number of ionizable cationic lipids covering a wide range of structures have been developed (Fig. 3 ), yet they all share a few aspects: (1) The headgroups contain tertiary amines that become protonated under acidic pH and typically are uncharged (or zwitterionic) at neutral pH; (2) The lipid tails contribute to making the molecule sufficiently hydrophobic to promote incorporation into a nanoparticle during formation; and (3) The protonated lipids generate structures that help elevate propensity for membrane fusion in acidified endosomes following internalization by the target cell. In addition to these similarities, the various lipids’ performed functions are essentially identical. As the pH of the environment dictates the headgroup protonation, LNP are prepared in an acidic aqueous buffer (e.g. pH 4) that promotes the charge interaction between the ionizable cationic lipid and the anionic nucleic acid. Subsequent buffer exchange into isotonic and pH-neutral buffer generates the final LNP suspension with a near net-neutral surface charge. This uncharged state is critical to preventing immune responses upon intravenous administration and facilitates delivery to hepatocytes [126]. The next function is to maintain a positive charge in the acidified endosome and promote membrane fusion to allow cytosolic delivery of the nucleic acid. This fine balance of positive charge at acidic pH and neutral charge at physiological pH is the result of substantial efforts towards optimizing the ionizable lipid for in vivo nucleic acid delivery.
Fig. 3.
Ionizable cationic lipids or lipid-like materials (lipidoids) enabling gene therapy in the liver. Various lipid-like materials have been developed for nucleic acid delivery. The headgroups contain tertiary amines which become protonated under acidic pH and have typically no charge at neutral pH. The lipid tails contribute to making the molecule sufficiently hydrophobic to promote incorporation into LNPs while endowing either stabilizing or destabilizing properties. The above lipids are classified into three broad categories: (i) ionizable cationic lipids such as DLinDMA [133], DLin-KC2-DMA [30], and DLin-MC3-DMA [31]; (ii) lipidoids like cKK-E12 [134] and C12-200 [29]; and (iii) next- generation lipids including the biodegradable molecules L319 [130], TT3 [135], and ssPalmE [136] as well as lipids from proprietary libraries belonging to Acuitas (A9) [137] and Moderna (L5) [138].
One of the first tested ionizable lipids, known as dioleyl-dimethylaminopropane (DODMA), contained oleyl lipid tails (C18:1) conjugated to the dimethylamino-propyl headgroup through ether linkers. Using the molecular shape hypothesis as a guiding principle [127], the three components of these lipids (headgroup, linker, and tails) were systematically studied to determine optimal characteristics for each. The molecular shape hypothesis describes the macrostructure obtained upon hydration of a lipid with specific geometries. More specifically, lipids containing tails with larger cross-sectional areas than the lipid headgroups result in HII phases or inverted micelles; comparatively, when the cross-sectional area of the tails is similar to that of the head group (resulting in a cylindrical geometry), the lipids tend to from bilayers. Comparing different lipid tail-unsaturation suggested that the linoleyl chains (DLinDMA) provide optimal particle internalization and potential to generate membrane-destabilizing HII phases [128]. Subsequent studies focused on improving the headgroup and linkers. Replacing the ether linkers in DLinDMA with esters resulted in a lipid, DLin-DAP, with substantially reduced potency [30]. Further studies suggested that ester bond degradation within the acidified endosome contributed to efficacy loss [129]. Simultaneously, a series of headgroup modified lipids were tested, and DLin-KC2-DMA was designed with vastly higher potency than DLinDAP and DLinDMA (Fig. 3) [30]. Further modifications and screening led to the development of DLin-MC3-DMA (Fig. 3) [31], used in the clinical formulation, Onpattro®, and now considered the gold-standard for ionizable cationic lipids.
Although several screening methods for ionizable lipids have been devised, the critical potency test for hepatic targets was the in vivo model for hepatic gene silencing; the Factor VII (FVII) model provided a modestly high-throughput approach [29]. FVII is a serum protein produced by hepatocytes in the liver and secreted into the circulation. Its short half-life enables gene silencing assessment on the protein level within a short timeframe. It is important to stress that FVII-knockdown screens specifically identify LNPs that target hepatocytes and ignore all other hepatic cell types. LNP containing siRNA against murine FVII were intravenously administered over a dose range of 0.001-10 mg siRNA per kg body weight and circulating FVII levels were determined by chromogenic assay 24 hours later. The metric used to compare formulations was the effective dose required to achieve 50% gene silencing (ED50), and DLin-MC3-DMA (MC3) was determined to be the most potent ionizable cationic lipid for LNP-based gene silencing. The potency improvements cover the range of DLinDAP with an ED50 of ~20 mg/kg, while that for MC3 was 0.005 mg/kg in mice [30,31].
Further developments focused on lipid biodegradability to reduce potential toxicity, immunogenicity, and other adverse effects [130]. The design parameters for these lipids included high in vivo transfection efficiency, increased ability to be metabolized, and no generation or accumulation of toxic metabolites. One approach incorporates an ester linkage, which can be easily hydrolyzed by intracellular esterases or lipases, into the lipid tail. For example, Maier et al. demonstrated that including ester bonds between carbons 9 and 10 in the linoleyl chain (named L319) resulted in similar potency as the MC3-lipid but almost complete elimination over 24 hours [130]. The ester bond position was critical to the function and elimination rate. When positioned closer to the head group, these bonds affected the lipid pKa, and thus its gene silencing potency. When positioned further away from the head group, the lipids persisted in the liver for extended periods of time. In another example, Shirazi et al. detailed the synthesis of degradable multivalent cationic lipids containing a disulfide bond between the head and tail groups, resulting in improved cell viability in vitro [131]. Akita et al. also synthesized a series of disulfide bond containing lipid-like materials incorporating alpha tocopherol as the lipid tails (ssPalmE) [132].
3.2. Helper lipids - phospholipid and cholesterol
Two LNP components – phospholipids and cholesterol – have generally been seen to promote formulation stability [139]. Although that evidence is largely anecdotal in the LNP context, phospholipids such as DSPC, with strong bilayer-forming properties and high phase transition temperatures, help increase membrane rigidity and reduce membrane permeability. While the role of cholesterol remains largely unclear in the context of nucleic acid delivery systems, cholesterol-deficient particles can sequester cholesterol while in circulation, leading to potentially destabilizing effects [140]. This sequestration process is largely driven by the exchange of cholesterol away from the plasma membrane of peripheral tissues into lipoproteins in circulation followed by equilibration into circulating liposomes. Recently, Harashima and colleagues studied cholesterol-free LNP-siRNA systems (only composed of the ionizable cationic lipid CL15H6, phospholipid, and PEG-lipid) and they observed decreased potencies in the presence of serum likely due to particle instability as a result of cholesterol accumulation [141].
Two studies suggested that the amount of cholesterol typically formulated into an LNP is larger than what can be stably retained in LNPs. More recently, it was determined that ~30-40 mol% helper lipid is required to efficiently entrap siRNA within LNPs, providing additional insight into the role of these helper lipids [27]. The helper lipids serve to space-out ionizable lipids to achieve a membrane surface charge of approximately +1 per nm2 (siRNA has a surface charge of approximately -1 per nm2).
Limited information is available on the role of helper lipids for LNP activity. However, some evidence has suggested that the replacement of DSPC with DOPE in lipidoid-based LNPs improves mRNA delivery in vivo [142]. For LNP-pDNA formulations, certain unsaturated phosphatidylcholines (i.e., SOPC and DOPC) improved the LNP activity over DSPC in the presence of FBS in vitro [28]. DOPE-containing LNP-pDNA systems showed best activity in murine serum suggesting a potential role of helper lipids in modifying the LNP surface affinity to distinct apolipoprotein subtypes. An additional role of cholesterol in LNP systems was recently investigated [143]. Incorporating oxidized cholesterols such as 20α-OH redirected LNP-mRNA systems from hepatocytes to hepatic endothelial cells and Kupffer cells. Although the mechanism of modifying LNP tropism remained elusive, formation of different protein coronas and/or recognition by scavenger receptors expressed on hepatic RES (such as scavenger receptor class B type I as binding site for oxidized LDL) might have resulted in redirection of LNPs [144,145].
3.3. PEG-lipid
The final LNP component, the PEG-lipid, is engineered to perform two specific functions. First, PEG-lipids incorporate into the emerging nanoparticle during LNP formation. As LNP systems do not contain an aqueous core, PEG-lipids reside almost exclusively on the LNP surface, and their concentrations control particle size [146]. Both the PEG molecular weight as well as the molar percentage of PEG-lipid affect the characteristics of lipid-based particles [[147], [148], [149]]. Specifically, as the PEG-lipid is increased from 0.25 mol% to 5 mol%, a reduction in LNP size is observed from ~120 nm to 25 nm, but further increases to PEG-content do not modify particle size [24,147]. Second, they improve the shelf-stability by creating a steric barrier that extends away from the surface of the LNP, thereby preventing particle aggregation and improving in vivo circulation lifetimes. However, for transfection purposes, PEG-lipids have an established inhibitory effect [150,151]. Based on the hypothesized mechanism of LNP function, the nanoparticle requires an intricate balance between stability in storage and circulation, and instability within the cell to support intracellular delivery [152]. Diffusible PEG-lipids helped stabilize particles while enabling intracellular delivery [25,151]. These lipids are composed of acyl chains that are 14-carbons in length and can dissociate rapidly from the LNP in the circulation [153]. Two hours post administration, only 20% of the injected PEG-lipid is associated with the LNP. In contrast, PEG-lipids with 18-carbon acyl chains, incorporate into the LNP and do not dissociate from the particle in the circulation. At high concentrations, these PEG-lipids can contribute to extending circulation half-life (from < 30 minutes for diffusible PEG-lipid to > 2 hours) [23,153]. However, LNPs designed to target hepatic disorders do not require a prolonged circulation lifetime due to the liver’s natural ability to sequester nanoparticles. Therefore, diffusible PEG-lipids are ideal for such applications.
3.4. Manufacturing
LNP production methods have evolved over time with certain processes gaining prominence. Rapid-mixing methods have gained favor for their decreased labour requirements as they combine nanoparticle formation and nucleic acid entrapment into a single step [154], and provide more homogenous nanoparticles. The first report of rapid-mixing was by Batzri and Korn, where an ethanolic lipid solution was rapidly injected into an aqueous solution to form liposomes [155]. Applying this method to nucleic acids involved combining pre-formed cationic liposomes with nucleic acids to produce lipoplexes [156]. More recently, a T-junction mixing chamber was used for two separate mixing steps [154]. The first mixing step brought together an ethanolic lipid stream with an acidic aqueous buffer containing nucleic acid at an equal flow rate (1:1 v/v mixing). This created metastable particles that were combined with aqueous buffer in a second mixing step (through the T-mixer) to dilute the ethanol content and stabilize the nanoparticles. To simplify this process into a single step, the mixing ratio was modified to 1 part ethanol and 3 parts aqueous. These rapid-mixing methods produce homogenous nanoparticles with entrapment efficiencies > 90% and, importantly, have been proven to be fully scalable [33,146,157,158].
3.5. Optimizing LNP characteristics
A number of recent studies have demonstrated that LNP systems accumulate in various cell types within the liver [49,50,143,159]. Important physicochemical characteristics that modulate intrahepatic LNP distribution and activity are particle size, apparent pKa value (and resulting surface charge), and lipase sensitivity [159]. For clinical utility, it is also expected that these formulations display high entrapment efficiencies.
A recent study by Chen et al. investigated how LNP size (30 nm – 120 nm) influences gene silencing in hepatocytes [153]. A clear hierarchy in gene silencing potency was observed with LNP-siRNA systems around 80 nm exhibiting maximum activity (78 nm > 42 nm > 38 nm >> 27 nm > 117 nm). This LNP size optimum results from a combination of two factors: 1) smaller particles being less active (less stable and less fusogenic); and 2) larger particles (>100 nm), not being able to access hepatocytes (limited by fenestrations). Sato et al. verified these results demonstrating significant reduction in hepatocyte gene silencing for LNP-siRNA sizes above the average liver fenestrae diameters [159]. Interestingly, similar gene silencing activity in LSECs was observed for LNP-siRNA sizes up to 200 nm. This suggests that LNP sizes between 120 nm to 200 nm could be used for LSEC targeting.
A key advance during the development of Onpattro® for hepatocyte gene silencing was identifying an optimized ionizable cationic lipid with an apparent pKa between 6.2 and 6.5 [17,31]. Further increasing the pKa value to 7.15 resulted in improved gene silencing in LSECs [159,160]. Incorporating ionizable cationic lipids exhibiting higher pKa values increased accumulation in the MPS, most likely due to scavenger receptor recognition [159].
The lipid sensitivity to phospholipase is another important factor modulating intrahepatic LNP distribution and activity. Three different lipases have been described including the lipoprotein and endothelial lipase in LSECs and the hepatic lipase in hepatocytes [161]. LNP-siRNA systems that incorporate ionizable cationic lipids that are sensitive to endothelial lipase (e.g. ester linkages between head and tail functions) have enhanced gene silencing in hepatocytes but exhibit significantly reduced activity in LSECs [159]. Co-treatment with lipase inhibitors or incorporating lipase-resistant ionizable cationic lipids can recover gene silencing in LSECs [159].
Based on microanatomical, subcellular, and (patho)physiological considerations, an ideal LNP for gene regulation in hepatocytes must satisfy the following design criteria: nanoparticle size < 80 nm to efficiently pass through liver fenestrae and improve LNP stability, apparent ionizable cationic lipid pKa value around 6.4, near neutral surface charge to prevent sequestration by the MPS, and lack of immune stimulation and toxic effects. Achieving these and other criteria facilitating efficient nucleic acid entrapment and LNP formulation are detailed in the following section.
It is important to mention that upon intravenous administration LNPs adsorb serum proteins on their surface. Many, if not all, of the abovementioned physiochemical characteristics impart distinct properties to the LNPs which ultimately influence protein adsorption. This “biomolecular corona” covering nanoparticles significantly impacts systemic circulation and nano-bio interactions [[162], [163], [164]]. Efficient targeting and gene regulation in hepatocytes stems from the presence of ApoE in the corona of LNPs and enabled the success of Onpattro® [33,76]. A recent publication suggested that the ionizable lipid composition plays a major role in the corona formed [165]. How the biomolecular corona can be leveraged to optimize targeting of different cell types within the liver microenvironment needs to be investigated.
4. Preclinical development and rationale for lipid nanotechnology
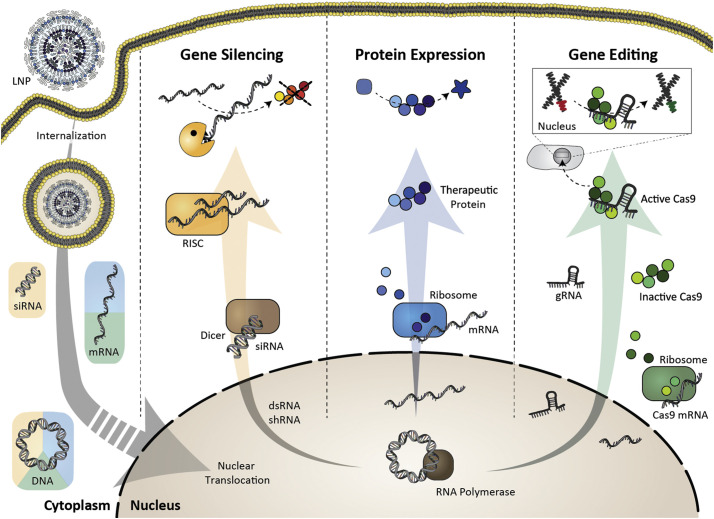
Research in the late 1980s focusing on in vivo pDNA delivery showed that in the absence of a delivery system, naked nucleic acid injected into the circulation rapidly broke down and the products accumulated in hepatic tissue [166]. As interest towards ASOs and siRNA grew, LNP compositions and production methods simply translated from plasmids to these shorter nucleic acids [167]. More recently, formulations have become sufficiently potent to support discovery and translation of mRNA therapeutics [168]. Fig. 4 illustrates the different LNP-based treatments for hepatic diseases by silencing pathogenic genes, expressing therapeutic proteins, or correcting genetic defects. Table 3 highlights preclinical LNP-based hepatic gene therapy approaches.
Fig. 4.
Therapeutic applications of LNPs enabling genetic drug delivery. LNPs can deliver siRNA, mRNA, DNA, or gene editing complexes, providing opportunities to treat hepatic diseases by silencing pathogenic genes, expressing therapeutic proteins, or correcting genetic defects. Following LNP internalization, nucleic acid therapeutics are released into the cytoplasm. DNA vectors require nuclear translocation to be active. Adapted with permission from Buck et al. [169]. Copyright 2019 American Chemical Society.
Table 3.
Selected LNP-based nucleic acid therapeutics in preclinical development. LNPs can deliver siRNA, mRNA, DNA, or gene editing complexes, providing opportunities to treat hepatic diseases by silencing pathogenic genes, expressing therapeutic proteins, or correcting genetic defects.
| Payload | Gene target/product | LNP composition | Cellular target / indication | Administration route | Model | Reference |
|---|---|---|---|---|---|---|
| Gene silencing | ||||||
| siRNA | FVII | DMAP-BLP / DSPC / cholesterol / PEG-DMG (50:10:(39.75 – x):(0.25+x)) | Hepatocytes / Screening | Subcutaneous | C57Bl/6 mice | [147] |
| siRNA | FVII | Ionizable cationic lipid / DSPC / cholesterol / PEG-DMG 40:10:40:10 |
Hepatocytes / Screening | Intravenous | C57BL/6 mice | [31] |
| siRNA | TTR | DLin-MC3-DMA / DSPC / cholesterol / PEG-DMG 50:10:38.5:1.5 |
Hepatocytes / ATTRv amyloidosis | Intravenous | Cynomolgus monkeys | [31] |
| siRNA | ApoB | DLinDMA / DSPC / cholesterol / PEG-C-DMA 40:10:48:2 |
Hepatocytes / Hypercholesterolemia | Intravenous | Cynomolgus monkeys | [133] |
| Gene expression | ||||||
| mRNA | anti-HIV-1 antibody VRC01 | Ionizable cationic lipid / PC / cholesterol / PEG-lipid (50:10:38.5:1.5) | Hepatocytes / Passive immunotherapy against HIV-1 | Intravenous | BALB/C mice | [201] |
| mRNA | Luciferase / Cre-recombinase | DOTMA/DOPE DOTMA/Chol |
Screening | Retro-orbital | NMRI mice / Reporter mice | [209] |
| pDNA | Luciferase | Cationic lipid/Chol-GALA/Malto-PEG6-C11 30:40:30 |
Screening | Intravenous | ICR mice | [174] |
| Gene editing | ||||||
| sgRNA mRNA |
TTR Cas9 |
LP01 / DSPC / cholesterol / PEG-DMG 45:9:44:2 |
Hepatocytes / ATTRv amyloidosis | CD-1 mice Sprague Dawley rats |
[206] | |
| sgRNA mRNA |
PCSK9 Cas9 |
BAMEAO16B / cholesterol / DOPE / DSPE-PEG2000 16:8:4:1 |
Hepatocytes / Hypercholesterolemia | Intravenous | C57BL/6 | [210] |
4.1. DNA delivery for long-term gene therapy and barcoding technologies
Refining lipid-DNA complexes to more advanced formulations required additional lipids, and such nanoparticles were termed stabilized plasmid lipid particles (SPLP) [169,170]. The composition of these formulations largely drew from those used for small molecule therapeutics and included about 6-8 mol% ionizable lipid at the expense of the phosphatidylcholine (i.e. DSPC). Delivery with SPLP systems showed no evidence of hepatic toxicities compared to the lipoplex-equivalent, which resulted in a 100-fold increase in serum ALT/AST levels [171]. These formulations were designed in a manner to promote accumulation at disseminated diseased sites (infection, inflammation, and solid tumors), requiring extended circulation residence times. With circulation half-lives nearing 7-8 hours, they induced reporter gene expression in tumor tissue, and substantially lower levels in the liver. However, as formulation development proceeded, it became clear that a limitation of non-viral technology was the inability to deliver nucleic acid into the nucleus of a target cell.
To address this issue, efforts have been made to increase nuclear targeting by including cell penetrating or nuclear localization sequence (NLS) peptides in the lipid formulations. These short, cationic peptides are thought to interact with the anionic DNA and enable nuclear translocation through nuclear core complexes. Initial studies using the Simian virus SV40 T antigen NLS peptide in a DOTAP/DOPE (50:50) liposome demonstrated improved nuclear targeting in vitro and up to a threefold increase in the plasmid-encoded luciferase signal [172]. Formulations utilizing the cell-penetrating peptide, octaarginine [173], or the recently identified non-peptide NLS [174], maltotriose, also demonstrated an increase in both nuclear targeting and gene expression in vitro. Maltotriose-incorporated liposomes also demonstrated higher hepatic luciferase expression levels in vivo compared to conventional DOTAP lipoplexes. Nevertheless, non-viral technology (without DNA modifications or specific nuclear targeting moieties) has generally been unsuccessful in transfecting non-dividing cells and in liver-specific applications but finds use in robust and safe transfection of neoplasms or developing tissues.
The ability to transfect dividing (liver cancer) cells was most recently highlighted in a study where LNP systems optimized for pDNA were found to yield potent transfection [28]. Starting with the formulation optimized for siRNA delivery containing MC3 and DSPC, Kulkarni et al. found that replacing the helper lipid and ionizable lipid with unsaturated lipids such as SOPC or DOPE and DLin-KC2-DMA, respectively, lead to much higher in vitro transfection of multiple liver cancer derived cell lines with little toxicity. This suggests that LNP systems, once optimized for DNA vector delivery, can not only find utility for protein expression, but potentially also in gene editing with CRISPR/Cas9-encoded plasmids for treating hepatic diseases.
While most DNA delivery applications have focused on gene therapy, a highly interesting application is employing DNA as barcodes for diagnostic and screening purposes [148]. Utilized as short fragments and each with a unique sequence, DNA barcodes allow for high-throughput, multiplexed in vivo screening to determine the biodistribution, uptake, and functional activity within the liver microenvironment (as outlined in section 2.2. and 3.5) [50,143]. Notably, as a diagnostic tool, DNA barcodes have also been used for developing personalized cancer nanomedicines by co-loading them together with anticancer drugs into lipid nanocarriers. Utilizing this strategy, multiple anticancer medicines can be administered at sub-therapeutic doses and the most effective drug can subsequently be identified in the biopsies according to their barcode [175]. As only limited therapies are available for liver cancer, this methodology could well be used towards identifying effective and novel treatments for liver cancer. Although lipid calcium phosphate nanoparticles (LCPs) are beyond the scope of this review, in the context of liver cancer, it is relevant to note the work by Leaf Huang and colleagues demonstrating that LCP-based DNA delivery enables mitigation of liver metastasis [[176], [177], [178]].
4.2. siRNA for transient gene silencing
All procedures and compositions developed for DNA delivery readily translated into effective delivery systems for other nucleic acids [167]. siRNA only requires cytoplasmic delivery as all RNA-induced silencing complex (RISC)-related machinery is located in the cytosol. This quick translation resulted in demonstrating the first robust gene silencing in non-human primates (NHP) using nanoparticles known as stable nucleic-acid lipid particles (SNALPs) containing siRNA against apolipoprotein B (ApoB) [133]. Only twelve years later, Onpattro® was approved by the FDA for treating ATTRv [179].
In the early 2000s, the concept of modifying nucleic acids was largely applied to improving their cytoplasmic persistence to enable long-term knockdown (decreased siRNA turnover). As such, modified siRNAs were entrapped into LNP systems with the rationale that a delivery system was specifically required to increase siRNA’s liver accumulation and intracellular quantity. This had to be achieved in a manner where the cost of raw materials and processing was offset by a potent formulation, i.e. a drastic reduction in material requirement made the formulation commercially viable. With LNP formulations containing DLin-MC3-DMA, murine data suggested that as little as 0.005 mg siRNA/kg body weight was required to achieve 50% gene silencing, with no observable toxicities. While alternative technologies such as siRNA-conjugates are also gaining prominence [4], the applicability of LNP technology for hepatic targets is quite clear. It should be noted that siRNA-conjugates require substantially higher doses (~1 mg/kg, weekly subcutaneous administration) in order to achieve gene knockdown [180]. LNP-siRNA systems have shown utility in decreasing viral loads and virulence, various applications in hepatic oncology, and in metabolic liver disease treatment.
RNAi finds strong support in anti-viral applications where strict adherence to treatment regimens is critical to success. LNP-siRNA treatments can provide sustained knockdown for months leading to long-term viral gene suppression with a potential to eliminate certain viruses. One example is using LNP-siRNA as a therapeutic intervention for the Ebola outbreak in 2013, which resulted in almost 28,000 cases and 11,300 deaths [181]. LNP-siRNA formulations could be rapidly adapted to provide siRNA complementarity to the specific strain and showed that a combination of three siRNAs against the viral RNA synthesis genes suppressed the infection in non-human primates (NHP) [181]. Similarly, LNP-siRNA modification with GalNAc-conjugated PEG-lipids to specifically accumulate in hepatocytes (of chimeric mice with humanized livers) reduced Hepatitis B Virus (HBV) genomic DNA and antigens [182]. Other anti-viral LNP examples include those for hepatitis delta virus (co-infected with HBV) and hepatitis C virus [183,184].
LNP-mediated siRNA delivery for hepatic oncology applications has largely focused on downregulating genes critical for cell cycle regulation, thereby inducing apoptosis. One example is LNP-siRNA against polo-like-kinase 1 (PLK1), which regulates multiple cell cycle progression stages. PLK1 is over expressed in multiple tumors including liver cancer and down-regulation has been successful as an intervention [185]. Similarly, simultaneous vascular endothelial growth factor (VEGF) and kinesin spindle protein (KSP) knockdown has been shown to inhibit proliferation in hepatocellular carcinoma and induce apoptosis [186]. Zhou et al. demonstrated that delivery of the small RNA let-7g inhibited tumor growth and dramatically extended survival in a MYC-driven genetic liver cancer tumor model [187].
Examples of LNP-siRNA delivery for liver-related metabolic disorders are plentiful. An interesting clinical observation was that loss-of-function mutations in proprotein convertase subtilisin/kexin type 9 (PCSK9) resulted in low cholesterol levels in circulation. This finding prompted the investigation into using siRNA to downregulate PCSK9 as a treatment for hypercholesterolemia. Murine and NHP studies showed that specific PCSK9 transcript lowering resulted in reversible and durable knockdown of PCSK9, apolipoprotein B (ApoB), and low-density lipoprotein associated cholesterol [188]. Similarly, in the first demonstration of RNAi in higher-order mammals, ApoB knockdown resulted in reduction of ApoB levels, serum cholesterol content, and LDL particle concentration in NHPs [133]. Other lipid-trafficking related targets include apolipoprotein C3 knockdown for hyperlipidemia [189], and angiopoietin-like 3 protein inhibition for hypertriglyceridemia [190].
Lastly, we discuss the specific case of Onpattro® (patisiran), an LNP-siRNA formulation targeting the ttr gene. TTR is a homotetrameric serum protein that is synthesized in hepatocytes and secreted into the systemic circulation (note similarity to FVII) [191]. When mutated, TTR deposits as amyloid fibrils in cardiac or peripheral nervous tissue resulting in multi-system failure including ocular, cardiovascular, nephropathy, gastrointestinal, and neuropathy (autonomic and peripheral sensorimotor) manifestations. TTR downregulation with LNP-siRNA is a powerful approach to treat this disease. Murine data suggested that at doses of 0.1 mg/kg siRNA, > 85% liver ttr mRNA knockdown and TTR protein serum concentrations could be achieved [192]. Further testing in NHPs showed that an intravenous dose of 0.3 mg/kg every 4 weeks resulted in rapid and reversible knockdown, although serum levels increased two weeks after each administration. Increasing the dosing frequency to once every three weeks resulted in sustained and robust knockdown (> 90%) following the third dose.
4.3. mRNA for gene expression and genome editing
Introducing exogenous mRNA to induce a therapeutic effect has great potential for a variety of applications. The true benefits of LNP technology for gene regulation in the liver are best highlighted with mRNA. Specifically, mRNA requires a delivery system as modifications to the nucleotides alone have not proven successful in meeting the potency requirements for clinical translation. In addition to this, the exorbitant costs of mRNA production imply that lower doses and less frequent dosing regimens are more likely to gain favourable reception. As such, dramatic advances are seen for mRNA formulations as vaccines, in protein replacement therapies, and gene editing.
LNP formulations containing mRNA are ideal vaccines. The development is conceptually straightforward and potentially very rapid. Preclinical evidence of using LNP-mRNA as vaccines against infectious diseases or cancer is extensive and several clinical trials have been initiated, including vaccines to combat the current COVID-19 pandemic. Since most vaccine applications rely on intramuscular or intradermal administration and the focus of this review is gene regulation in the liver, the reader is referred to several recent articles [[193], [194], [195], [196], [197], [198], [199], [200]]. However, several recent studies have used the “liver as a bioreactor” to produce relevant neutralizing antibodies.
Pardi et al. showed that intravenous delivery of LNP-mRNA encoding a broadly neutralizing antibody against HIV-1 resulted in sufficient expression to protect from HIV-1 challenge [201]. Similarly, another study showed that an LNP-mRNA system as prophylactic and therapeutic anti-rabies intervention protected mice from a Rabies virus challenge [202]. The prophylactic treatment involved a single dose 40 μg LNP-mRNA encoding an anti-rabies antibody intravenously administered one day prior to a 5-fold LD50 insult of Rabies virus (i.m.). The therapeutic intervention (at the same mRNA dose) was given 2 h post-Rabies virus challenge. In the same study, LNP-mRNA encoding a neutralizing antibody afforded complete protection to mice six hours following a botulinum neurotoxin challenge (4x LD50). Finally, this study also showed that LNP-mRNA encoding rituximab administered intravenously (at 10 or 50 μg mRNA) following a lethal challenge of Raji cells, resulted in either tumor growth deceleration or almost completely abolished tumor development.
LNP-based mRNA formulations have shown strong promise as therapeutics in disease states where genetic mutations result in a non-functional protein. Delivering exogenous (and functional) mRNA to generate a functional protein can alleviate stress from certain diseases. Initial studies that showed clear clinical utility are briefly highlighted here: Intravenous administration of LNP-mRNA encoding erythropoietin (EPO) resulted in increased EPO serum levels corresponding to increased reticulocyte, and elevated hematocrit in porcine and non-human primate (NHP) models [203]. Similarly, delivering mRNA encoding human clotting factor IX (FIX) to FIX-knockout mice displayed a reduction in hematocrit loss following injury, indicating FIX expression can rescue hemophilia B phenotypes [204].
Gene editing is the next major application of mRNA therapeutics. Various approaches have been explored including CRISPR/Cas9 and zinc-finger nucleases (ZFN). An initial gene editing demonstration used a combination of viral delivery (sgRNA and repair template) combined with LNP-mRNA encoding Cas9 to correct a mutation in the fumarylacetoacetate hydrolase gene [205]. The study showed approximately 6% of hepatocytes were edited and it is assumed that the limitation was the viral delivery. Comparatively, Finn et al. used LNP-mRNA formulations encoding for Cas9 protein, co-delivered with sgRNA targeting ttr. They showed sustained 12-month circulating TTR knockdown (97%) following a single administration of 3 mg/kg RNA body weight in a murine model with ~70% editing in the liver (~70% liver cells are hepatocytes) [206]. Similarly, LNP-mediated delivery of mRNA encoding ZFN targeting ttr and pcsk9 resulted in > 90% knockout at mRNA doses 10-fold lower than reported previously [137]. In the same study, co-delivery of LNP-mRNA encoding ZFN targeting the albumin gene and a viral vector for templates of promotor-less human IDS or FIX resulted in integration of those templates at the albumin locus and generated therapeutically relevant levels of those proteins in murine models. In addition to continuous efforts in optimizing ionizable cationic lipids for enhanced genome editing in the liver, a recent study by Cheng et al. demonstrated that bioengineering LNP formulations with additional lipids, so-called selective organ targeting (SORT) molecules, can tune the LNP's efficiency and biodistribution. Adding 20 mol% of an ionizable cationic lipid such as DODAP significantly enhanced the genome editing in the liver, while addition of cationic or anionic SORT molecules enabled specific gene regulation in the lung or spleen [207,208].
5. Clinical translation of lipid nanotechnology
The rapid translation from lab bench to patients was primarily driven by a holistic design of LNP composition and processes to support scalability while maintaining potency. Onpattro® paved the way for the next generation of lipid-based therapeutics and its success in phase 2 trials spurred development of mRNA therapeutics. Gene therapies enabled by LNPs are under clinical development for a broad range of applications (Table 4 ) [211]. In this section we discuss the clinical data for Onpattro® and some mRNA therapeutics currently under development.
Table 4.
Highlighted LNP-based nucleic acid therapeutics in the clinic. Drug products in clinical development or approved by the U.S. Food and Drug Administration (FDA) and the European Medicines Agency (EMA). Company code names, generic (non-proprietary) names and company names for the products are given in brackets. Table adapted from Kulkarni et al. [18]
| Product | Nucleic acid / transgene | Indication | Administration route | Clinical stage | Ref. |
|---|---|---|---|---|---|
| Gene silencing | |||||
| Onpattro®, patisiran (Alnylam Pharmaceuticals) | siRNA-TTR | ATTRv amyloidosis | Intravenous | Approved (2018) | [223] |
| ALN-VSP02 (Alnylam/Ascletis) | siRNA-VSP/VEGF-A | Solid tumors (liver involvement) | Intravenous | Phase I (completed) NCT01158079 |
[224] |
| ARB-001467 (Arbutus Biopharma) | Three siRNAs against four HBV transcripts | Hepatitis B | Intravenous | Phase 2 (completed) NCT02631096 |
[225] |
| TKM-080301, TKM-PLK1 (Arbutus Biopharma) | siRNA-PLK1 | Solid tumors (NET, ACC) | Intravenous | Phase 1/2 (completed) NCT01262235 |
[226] |
| Atu027 (Silence Therapeutics) | siRNA-PKN3 (+gemcitabine) | Advanced / metastatic pancreatic cancer | Intravenous | Phase 1/2 (completed) NCT01808638 |
[227] |
| ND-L02-s0201, BMS-986263 (Nitto Denko Corporation / Bristol-Myers Squibb) | siRNA-HSP47 | Idiopathic pulmonary fibrosis | Intravenous | Phase 2 (recruiting) NCT03538301 |
[228] |
| EPHARNA (M.D. Anderson Cancer Center) | siRNA-EphA2 | Advanced or recurrent solid tumors | Intravenous | Phase 1 (recruiting) NCT01591356 |
[229] |
| Gene expression | |||||
| Lipo-MERIT (Biontech RNA Pharmaceuticals) | Four mRNAs encoding melanoma-associated antigens | Melanoma | Intravenous | Phase 1 (recruiting) NCT02410733 |
[230] |
| IVAC_W_bre1_uID and IVAC_M_uID (Biontech RNA Pharmaceuticals) | mRNAs encoding tumor-associated antigens and/or personalized neoantigens | Triple negative breast cancer | Phase 1 (recruiting) NCT02316457 |
||
| SGT-53 (SynerGene Therapeutics) | pDNA encoding wild-type p53 (+nab-paclitaxel / gemcitabine) | Metastatic pancreatic cancer | Intravenous | Phase 2 (recruiting) NCT02340117 |
[231] |
| MTL-CEBPA (Mina Alpha) | saRNA-CEBPα | Advanced liver cancer | Intravenous | Phase 1 (recruiting) NCT02716012 |
[232] |
| Gene editing | |||||
| NTLA-2001 (Intellia Therapeutics / Regeneron) | sgRNA-TTR mRNA-Cas9 |
ATTRv amyloidosis | Intravenous | Phase 1 planned |
[233] |
The Onpattro® story, while heavily reviewed in literature, makes for a compelling case to support the development of other LNP nucleic acid formulations [33]. Initial efforts laid the foundations to support further clinical development, although it was clear that improved potency was required. DLinDMA-based LNP-siRNA against ttr (ALN-TTR01) was administered once to 24 healthy subjects at doses ranging from 0.01 to 1.0 mg siRNA per kg body weight, with another eight subjects receiving placebo [212,213]. Over the period of 30 days, 38% serum TTR reduction was observed with persistent reduction for approximately one week. While the knockdown was arguably insufficient for therapeutic efficacy at the highest dose, the study validated the RNAi approach in humans. Subsequent clinical development used MC3-based LNP, named ALN-TTR02 or Onpattro® (patisiran). Another phase 1 study included 13 healthy subjects receiving Onpattro®, four subjects receiving placebo, and another six receiving a control siRNA [214]. The Onpattro® doses ranged from 0.01 to 0.5 mg/kg siRNA and TTR serum levels were measured over 70 days. At siRNA doses of 0.3 mg/kg, rapid and robust ttr knockdown was observed; this was sustained over two weeks for a period of 21 days following administration. At these doses and with promising results, further development was warranted.
In a subsequent phase 2 study, the dosing regimen for Onpattro® was established [86]. ATTRv patients received two Onpattro® infusions at doses 0.01-0.3 mg/kg every four weeks or 0.3 mg/kg every three weeks. The Q3W dosing regimen resulted in a mean 85% knockdown after the second dose. Only few mild-to-moderate infusion-related reactions were observed and one patient reported three serious adverse events. The similarity to preclinical data is quite astonishing; in NHP studies, increasing dosing frequency to Q3W (from Q4W) resulted in 96% maximal knockdown, and ~85% mean knockdown following the initial dose [192].
In the phase 3 APOLLO study, 148 patients received Onpattro® at a dose of 0.3 mg/kg once every three weeks, with 77 patients receiving placebo [32]. The primary endpoint was the modified neuropathy impairment score + 7 (mNIS + 7), which is used to measure the level polyneuropathy in ATTRv patients. The test uses highly standardized, quantitative methods to measure muscle weakness, muscle stretch reflexes, sensory loss, and autonomic impairment with higher scores corresponding to disease worsening [215]. Over a period of 18 months, ATTRv patients on placebo showed a linear increase in their mNIS+7 from 0 to 28.0. Onpattro®, with an mNIS+7 of -6.0, is the only ATTRv treatment that has been able to halt and even reverse disease progression in patients [32]. In addition to this, Onpattro® also met all secondary endpoints. This led to EMA and FDA approval in August 2018 [33].
With LNP technology validated as a safe approach for gene modulation in the liver, a wide range of applications have emerged. A substantial effort is focusing on vaccine applications without necessarily transfecting the liver. However, the potential for treating liver diseases is also clear. Translate Bio was developing a formulation for treating OTC deficiency, however disappointing preclinical toxicology data resulted in the termination of the program [216]. Moving forward, they chose to focus on developing their cystic fibrosis mRNA therapeutic. Moderna Therapeutics is advancing an LNP candidate formulation for treating methylmalonic acidemia [217]. The focus of this review is on the hepatic applications of LNP formulations, and indications for extrahepatic targets have been summarized elsewhere [218]. Highlighted LNP-based nucleic acid therapeutics in the clinic are summarized in Table 4.
5.1. Overcoming the barriers to successful clinical translation
Therapeutic development, and gene therapy in particular, requires concerted efforts from formulation developers, process developers, and clinical sponsors to allow for successful clinical translation. Specifically, the therapeutic has to be safe and effective, be producible at a large scale, and meet all regulatory requirements for the corresponding drug class. Onpattro® has shown that this is possible for systemic nucleic acid therapeutics, as it overcame barriers that typically halt the clinical translation of such nanocarrier-based therapeutics.
Intravenous administration of nanoparticulate formulations can potentially result in infusion-related reactions such as hypersensitivity manifesting as mild flu-like symptoms, or more severe cardiac anaphylaxis [219]. Both complement activation as well as complement-independent phagocytosis are involved in such reactions. The reader is referred to excellent articles on complement activation-related pseudoallergy (CARPA) and complement independent pseudoallergy (CIPA) [219,220]. Several physiochemical properties such as lamellarity, surface charge, and cholesterol content may influence hypersensitivity reactions [221]. Infusion-related reactions can be managed by pre-dosing patients with a combination of anti-histamines (H1/H2 blockers), corticosteroid immunosuppressants (e.g., dexamethasone), and oral acetaminophen in addition to reducing the rate of infusion [222]. Onpattro®’s phase 3 trial suggested that the most frequent reactions included flushing, backpain, abdominal pain, and nausea described as mild-to-moderate. The severity and frequency of these reactions decreased with repeated administration and exposure of Onpattro®. It should be noted that ASOs and GalNAc-siRNA conjugates do not require pre-medication and can be administered subcutaneously (by healthcare professionals), but the doses required to achieve equivalent gene silencing are a few orders of magnitude higher than required for LNPs and can only be limited to gene silencing applications [192].
Another substantial barrier to clinical translation is producing formulations at commercial scales. As described previously, LNP manufacturing methods rely on rapid-mixing technologies and therefore, the type and capabilities of the mixer become very important. For example, a production of 10 mL LNP-siRNA at a flow rate of 20 mL/min requires a mixing time of 30 seconds [157]. However, production of 1 L of material with the same mixer requires 50 minutes of mixing time. With an inherently unstable LNP suspension and in 25% (v/v) ethanol, this time could result in substantial changes in LNP properties negating the value of rapid-mixing technologies (homogeneity). High-throughput mixers (> 1 L/min) are required to mitigate such effects. In addition to this, the in-process volumes generated by rapid-mixing are much larger than by extrusion and can be limiting to manufacturing scales. Typical processes use low concentrations of material (0.2 mg/mL siRNA after mixing), and therefore the intermediate volumes are substantial. For example, 1 g siRNA (only 100 vials of Onpattro®) would generate 5 L of material post-mixing and would include a post-mixing dilution step to stabilize the intermediate material. This could result in up to 15-20 L of intermediate material depending on the required dilution (composition-dependent). For larger batches, even the ethanol amount handled at a facility can become limiting. Following this, the next processing steps introduce shear as buffer exchange is not done by dialysis, but rather by tangential flow filtration. Given the inherent instability of these formulations, particle size increases are observed during this step. These processes use terminal, redundant sterile filtration rather than complete aseptic processing. The impact that buffer exchange has on particle size also affects the ability to sterile filter the formulation and the yield of material. Robust process design is critical for successful and timely clinical translation of such formulations.
6. Future perspectives
Developing LNP delivery technology has enabled the clinical translation and approval of the first siRNA drug for inhibiting pathogenic protein production in hepatocytes [32,33]. Importantly, Onpattro® provides a valuable treatment for ATTRv amyloidosis patients, whose options were previously limited to TTR stabilizers or a liver transplant [34]. At the same time, LNP-siRNA development has yielded fundamental insights into optimally designing formulations for hepatocyte gene silencing, (large scale) production methods, in vivo behaviour, immunostimulatory effects, and cost-effectiveness. As these criteria and parameters are now firmly established, it is anticipated that other hepatocyte-targeted LNP-siRNA treatments will be developed, such as to knockdown proprotein convertase subtilisin/kexin type 9 for hypercholesteremia treatment [234].
While these advances in LNP development are ground-breaking, other liver-targeted nucleic acid therapeutics, such as ASOs [3] and GalNAc-siRNA conjugates [4], are also gaining momentum. For example, the ASO Tegsedi® (inotersen) was recently approved for the same indication as Onpattro® [235]. With both Onpattro® and Tegsedi® set at the same list price ($450,000 per year), it remains to be seen which treatment will prove to be most beneficial and cost-effective. Tegsedi’s® major advantage is its subcutaneous administration (versus Onpattro’s® intravenous infusion), although this advantage could be outweighed by its less favorable toxicity profile; patients require monitoring of platelet count, renal and hepatic impairment. Subcutaneous administration (and a less complex production process) is also the main advantage of GalNAc-siRNA conjugates although currently approved conjugates have to be administered by healthcare professionals. Most recently, the GalNAc-siRNA conjugate Givlaari™ (givosiran, $575,000 per year) was approved for treating acute hepatic porphyria [236,237], while New Drug Applications were filed for lumasiran for treating primary hyperoxaluria type 1, [238,239] and inclisiran for treating hypercholesteremia [[240], [241], [242]]. Vutrisiran, a GalNAc-siRNA conjugate for treating ATTRv amyloidosis, is currently undergoing phase 3 trials and has been granted Orphan Drug designation in the U.S. and the European Union [243]. Although there is preclinical evidence that LNP-siRNA can induce hepatic gene silencing following subcutaneous administration, the dose needed for effective gene silencing is considerably higher than for intravenously administered formulations [147]. Of note, while LNP-siRNA systems have been optimized for hepatic gene silencing, preclinical studies have also demonstrated their ability to induce effective gene silencing in extrahepatic target sites including the bone [244] and tumors [245,246]. A major area of interest is applying LNP-siRNA for immunotherapy, by silencing target genes in lymphocytes following intravenous administration for immunotherapy [[247], [248], [249], [250], [251]] (covered by Peer et al. in this issue [252]).
As mentioned before, LNP technology’s true benefits are currently proving to be of significant value for gene regulation approaches using large nucleic acid-based therapeutics, such as mRNA and gene editing complexes, which cannot be accomplished by nucleic acid modification or GalNAc conjugation. Intravenously administered LNP-mRNA effectively transfect hepatocytes and induce protein expression in the liver, providing opportunities for protein replacement therapy without affecting the genome. For example, An et al. demonstrated that treatment with LNP containing mRNA encoding human methylmalonyl-CoA mutase (hMUT) had sustained functional benefits in mouse models of methylmalonic acidemia, a rare, inherited, pediatric metabolic disorder [217]. Other examples include using the liver to produce coagulation factors [253], or therapeutic antibodies against HIV [201] and chikungunya virus [254]. Although this review focuses on gene therapy for diseases originating in the liver, it is worth mentioning that as with LNP-siRNA, intravenously injecting LNP-mRNA to induce protein expression in immune cells is gaining considerable traction [255,256], especially for developing (personalized) cancer immunotherapies [257,258]. In addition, LNP-mRNA systems have revealed their potential for ex vivo CAR T cell engineering [259].
Moreover, LNP-mRNA-based vaccinations following subcutaneous, intradermal, or intramuscular administration have demonstrated to effectively protect from viral challenge [[260], [261], [262], [263], [264], [265]]. Given mRNA’s relatively short optimization time from target identification to therapeutic, several companies including Moderna, BioNTech, and CureVac as well as universities around the globe have initiated LNP-mRNA vaccine programs to combat the recent SARS-CoV-2 pandemic [[266], [267], [268], [269]]. Typical vaccine production relies on isolation and large-scale virus propagation with subsequent processing to purify material (e.g. inactive virus or specific surface protein) that raises a response against a specific viral antigen. With mRNA delivery, these timelines can be dramatically reduced, and the breadth of immune coverage expanded. Additionally, mRNA vaccines leverage several aspects of LNP technology: (1) LNP systems are not completely immune-silent and can act as adjuvants [270,271], (2) few doses are required (i.e. prime and booster), and (3) the mRNA dosage is relatively low (compared to protein replacement therapies).
LNP technology, and ionizable cationic lipid development in particular, have been instrumental for translating therapeutic gene regulation in hepatocytes from bench to bedside. As LNPs are a multicomponent and modular platform, they represent a versatile toolbox with many opportunities to develop future gene therapies with more potent therapeutic effects and improved toxicity profiles. For example, incorporating lipophilic prodrugs in LNP systems has shown to be an attractive approach for reducing nucleic acid therapeutics’ immunostimulatory effects [272] or for designing combination therapies with additive therapeutic effects [273]. Generating more potent (ionizable cationic) lipids and improved understanding of nano-bio interactions in vivo are continuously fueling the optimization of LNP systems for delivering nucleic acid therapeutics. Deciphering intracellular trafficking pathways and mechanism(s) of endosomal escape will facilitate efforts to boost LNP potency [[274], [275], [276], [277]]. Elucidating the nature and dynamic of the biomolecular corona formed on LNPs (following intravenous injection) and understanding its implications for biodistribution will be crucial to develop gene therapies beyond the liver [[162], [163], [164],278,279]. Therefore, we expect that LNP-based gene therapies will be developed for indications beyond (ultra) rare diseases in the near future and increasingly become integrated in mainstream medicine.
Acknowledgments
Acknowledgements
PRC acknowledges support from the Canadian Institutes for Health Research (FDN 148469) and the NanoMedicines Innovation Network (NMIN), a Canadian Networks of Centres of Excellence (NCE) in nanomedicine. DW is supported by the Swiss National Science Foundation (#183923). JAK is supported by the NMIN Postdoctoral Fellowship Award in Gene Therapy. JL is supported by a Frederick Banting and Charles Best Canada Graduate Scholarship – Masters (6556). The work of RvdM is supported by the Netherlands Organization for Scientific Research (NWO, ZonMW Vici grant no. 016.176.622).
Declaration of Competing Interest
PRC is a co-founder of Acuitas Therapeutics and Precision Nanosystems; and Scientific Director and CEO of the NMIN.
Contributions
DW, JAK, JL, SC, PRC and RvdM conceived and co-wrote the manuscript. The final manuscript was approved by all authors.
References
- 1.Williams R., Aspinall R., Bellis M., Camps-Walsh G., Cramp M., Dhawan A., Ferguson J., Forton D., Foster G., Gilmore I., Hickman M., Hudson M., Kelly D., Langford A., Lombard M., Longworth L., Martin N., Moriarty K., Newsome P., O’Grady J., Pryke R., Rutter H., Ryder S., Sheron N., Smith T. Addressing liver disease in the UK: a blueprint for attaining excellence in health care and reducing premature mortality from lifestyle issues of excess consumption of alcohol, obesity, and viral hepatitis. Lancet Lond. Engl. 2014;384:1953–1997. doi: 10.1016/S0140-6736(14)61838-9. [DOI] [PubMed] [Google Scholar]
- 2.Asrani S.K., Devarbhavi H., Eaton J., Kamath P.S. Burden of liver diseases in the world. J. Hepatol. 2019;70:151–171. doi: 10.1016/j.jhep.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Crooke S.T., Witztum J.L., Bennett C.F., Baker B.F. RNA-targeted therapeutics. Cell Metab. 2018;27:714–739. doi: 10.1016/j.cmet.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Springer A.D., Dowdy S.F. GalNAc-siRNA conjugates: leading the way for delivery of RNAi therapeutics. Nucleic Acid Ther. 2018;28:109–118. doi: 10.1089/nat.2018.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulkarni J.A., Witzigmann D., Chen S., Cullis P.R., Meel R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Acc. Chem. Res. 2019;52:2435–2444. doi: 10.1021/acs.accounts.9b00368. [DOI] [PubMed] [Google Scholar]
- 6.Dunbar C.E., High K.A., Joung J.K., Kohn D.B., Ozawa K., Sadelain M. Gene therapy comes of age. Science. 2018;359 doi: 10.1126/science.aan4672. [DOI] [PubMed] [Google Scholar]
- 7.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines-a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahin U., Türeci Ö. Personalized vaccines for cancer immunotherapy. Science. 2018;359:1355–1360. doi: 10.1126/science.aar7112. [DOI] [PubMed] [Google Scholar]
- 10.Xu C.F., Chen G.J., Luo Y.L., Zhang Y., Zhao G., Lu Z.D., Czarna A., Gu Z., Wang J. Rational designs of in vivo CRISPR-Cas delivery systems. Adv. Drug Deliv. Rev. 2019 doi: 10.1016/j.addr.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Wilbie D., Walther J., Mastrobattista E. Delivery aspects of CRISPR/Cas for in vivo genome editing. Acc. Chem. Res. 2019;52:1555–1564. doi: 10.1021/acs.accounts.9b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowdy S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017;35:222–229. doi: 10.1038/nbt.3802. [DOI] [PubMed] [Google Scholar]
- 13.Barros S.A., Gollob J.A. Safety profile of RNAi nanomedicines. Adv. Drug Deliv. Rev. 2012;64:1730–1737. doi: 10.1016/j.addr.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Wittrup A., Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Miao L., Satterlee A., Huang L. Delivery of oligonucleotides with lipid nanoparticles. Adv. Drug Deliv. Rev. 2015;87:68–80. doi: 10.1016/j.addr.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen T.M., Cullis P.R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 17.Cullis P.R., Hope M.J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 2017;25:1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulkarni J.A., Cullis P.R., Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. 2018;28:146–157. doi: 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
- 19.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 20.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 21.Dong Y., Siegwart D.J., Anderson D.G. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev. 2019 doi: 10.1016/J.ADDR.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ku S.H., Jo S.D., Lee Y.K., Kim K., Kim S.H. Chemical and structural modifications of RNAi therapeutics. Adv. Drug Deliv. Rev. 2016;104:16–28. doi: 10.1016/j.addr.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 23.Mui B.L., Tam Y.K., Jayaraman M., Ansell S.M., Du X., Tam Y.Y.C., Lin P.J., Chen S., Narayanannair J.K., Rajeev K.G., Manoharan M., Akinc A., Maier M.A., Cullis P., Madden T.D., Hope M.J. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol. Ther. Nucleic Acids. 2013;2 doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar V., Qin J., Jiang Y., Duncan R.G., Brigham B., Fishman S., Nair J.K., Akinc A., Barros S.A., Kasperkovitz P.V. Shielding of lipid nanoparticles for siRNA delivery: impact on physicochemical properties, cytokine induction, and efficacy. Mol. Ther. Nucleic Acids. 2014;3 doi: 10.1038/mtna.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Judge A., McClintock K., Phelps J.R., MacLachlan I. Hypersensitivity and loss of disease site targeting caused by antibody responses to PEGylated liposomes. Mol. Ther. 2006;13:328–337. doi: 10.1016/j.ymthe.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Heyes J., Hall K., Tailor V., Lenz R., MacLachlan I. Synthesis and characterization of novel poly(ethylene glycol)-lipid conjugates suitable for use in drug delivery. J. Control. Release. 2006;112:280–290. doi: 10.1016/j.jconrel.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 27.Kulkarni J.A., Witzigmann D., Leung J., Tam Y.Y.C., Cullis P.R. On the role of helper lipids in lipid nanoparticle formulations of siRNA. Nanoscale. 2019;11:21733–21739. doi: 10.1039/c9nr09347h. [DOI] [PubMed] [Google Scholar]
- 28.Kulkarni J.A., Myhre J.L., Chen S., Tam Y.Y.C., Danescu A., Richman J.M., Cullis P.R. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomed. Nanotechnol. Biol. Med. 2017;13:1377–1387. doi: 10.1016/j.nano.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 29.Akinc A., Zumbuehl A., Goldberg M., Leshchiner E.S., Busini V., Hossain N., Bacallado S.A., Nguyen D.N., Fuller J., Alvarez R., Borodovsky A., Borland T., Constien R., Fougerolles A., Dorkin J.R., Jayaprakash K.N., Jayaraman M., John M., Koteliansky V., Manoharan M., Nechev L., Qin J., Racie T., Raitcheva D., Rajeev K.G., Sah D.W.Y.Y., Soutschek J., Toudjarska I., Vornlocher H.-P.P., Zimmermann T.S., Langer R., Anderson D.G. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W., Stebbing D., Crosley E.J., Yaworski E., Hafez I.M., Dorkin J.R., Qin J., Lam K., Rajeev K.G., Wong K.F., Jeffs L.B., Nechev L., Eisenhardt M.L., Jayaraman M., Kazem M., Maier M.A., Srinivasulu M., Weinstein M.J., Chen Q., Alvarez R., Barros S.A., De S., Klimuk S.K., Borland T., Kosovrasti V., Cantley W.L., Tam Y.K., Manoharan M., Ciufolini M.A., Tracy M.A., Fougerolles A., MacLachlan I., Cullis P.R., Madden T.D., Hope M.J. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 31.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., Rajeev K.G., Hafez I.M., Akinc A., Maier M.A., Tracy M.A., Cullis P.R., Madden T.D., Manoharan M., Hope M.J. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.-C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L., Lin K.-P., Vita G., Attarian S., Planté-Bordeneuve V., Mezei M.M., Campistol J.M., Buades J., Brannagan T.H., Kim B.J., Oh J., Parman Y., Sekijima Y., Hawkins P.N., Solomon S.D., Polydefkis M., Dyck P.J., Gandhi P.J., Goyal S., Chen J., Strahs A.L., Nochur S.V., Sweetser M.T., Garg P.P., Vaishnaw A.K., Gollob J.A., Suhr O.B. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 33.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M.J., Madden T.D., Mui B.L., Semple S.C., Tam Y.K., Ciufolini M., Witzigmann D., Kulkarni J.A., Meel R., Cullis P.R. The Onpattro® story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 34.Adams D., Koike H., Slama M., Coelho T. Hereditary transthyretin amyloidosis: a model of medical progress for a fatal disease. Nat Rev Neurol. 2019;15 doi: 10.1038/s41582-019-0210-4. [DOI] [PubMed] [Google Scholar]
- 35.Maeki M., Kimura N., Sato Y., Harashima H., Tokeshi M. Advances in microfluidics for lipid nanoparticles and extracellular vesicles and applications in drug delivery systems. Adv. Drug Deliv. Rev. 2018;128:84–100. doi: 10.1016/j.addr.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 36.Evers M.J.W., Kulkarni J.A., van der Meel R., Cullis P.R., Vader P., Schiffelers R.M. State-of-the-Art design and rapid-mixing production techniques of lipid nanoparticles for nucleic acid delivery. Small Methods. 2018;2:1700375. doi: 10.1002/smtd.201700375. [DOI] [Google Scholar]
- 37.Trefts E., Gannon M., Wasserman D.H. The liver. Curr. Biol. 2017;27:R1147–R1151. doi: 10.1016/j.cub.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F.-S., Fan J.-G., Zhang Z., Gao B., Wang H.-Y. The global burden of liver disease: the major impact of China. Hepatol. Baltim. Md. 2014;60:2099–2108. doi: 10.1002/hep.27406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gissen P., Arias I.M. Structural and functional hepatocyte polarity and liver disease. J. Hepatol. 2015;63:1023–1037. doi: 10.1016/j.jhep.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Z., Xu M.-J., Gao B. Hepatocytes: a key cell type for innate immunity. Cell. Mol. Immunol. 2016;13:301–315. doi: 10.1038/cmi.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y.-N., Poon W., Tavares A.J., McGilvray I.D., Chan W.C.W. Nanoparticle–liver interactions: cellular uptake and hepatobiliary elimination. J. Control. Release. 2016;240:332–348. doi: 10.1016/j.jconrel.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 42.Poon W., Zhang Y.-N., Ouyang B., Kingston B.R., Wu J.L.Y., Wilhelm S., Chan W.C.W. Elimination pathways of nanoparticles. ACS Nano. 2019;13:5785–5798. doi: 10.1021/acsnano.9b01383. [DOI] [PubMed] [Google Scholar]
- 43.Sago C.D., Lokugamage M.P., Lando G.N., Djeddar N., Shah N.N., Syed C., Bryksin A.V., Dahlman J.E. Modifying a commonly expressed endocytic receptor retargets nanoparticles in vivo. Nano Lett. 2018;18:7590–7600. doi: 10.1021/acs.nanolett.8b03149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dolina J.S., Sung S.-S.J., Novobrantseva T.I., Nguyen T.M., Hahn Y.S. Lipidoid nanoparticles containing PD-L1 siRNA delivered in vivo enter kupffer Cells and enhance NK and CD8(+) T cell-mediated hepatic antiviral immunity. Mol. Ther. Nucleic Acids. 2013;2 doi: 10.1038/mtna.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.S.A. MacParland, J.C. Liu, X.-Z. Ma, B.T. Innes, A.M. Bartczak, B.K. Gage, J. Manuel, N. Khuu, J. Echeverri, I. Linares, R. Gupta, M.L. Cheng, L.Y. Liu, D. Camat, S.W. Chung, R.K. Seliga, Z. Shao, E. Lee, S. Ogawa, M. Ogawa, M.D. Wilson, J.E. Fish, M. Selzner, A. Ghanekar, D. Grant, P. Greig, G. Sapisochin, N. Selzner, N. Winegarden, O. Adeyi, G. Keller, G.D. Bader, I.D. McGilvray, Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations, Nat. Commun. 9 (2018) 4383. 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed]
- 46.Aizarani N., Saviano A., Sagar L. Mailly, Durand S., Herman J.S., Pessaux P., Baumert T.F., Grün D. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertrand N., Leroux J.-C. The journey of a drug-carrier in the body: An anatomo-physiological perspective. J. Controlled Release. 2012;161:152–163. doi: 10.1016/j.jconrel.2011.09.098. [DOI] [PubMed] [Google Scholar]
- 48.Mosby . 8th edition. 2020. Mosby’s Medical Dictionary. [Google Scholar]
- 49.Shi B., Keough E., Matter A., Leander K., Young S., Carlini E., Sachs A.B., Tao W., Abrams M., Howell B., Sepp-Lorenzino L. Biodistribution of small interfering RNA at the organ and cellular levels after lipid nanoparticle-mediated delivery. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2011;59:727–740. doi: 10.1369/0022155411410885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sago C.D., Krupczak B.R., Lokugamage M.P., Gan Z., Dahlman J.E. Cell subtypes within the liver microenvironment differentially interact with lipid nanoparticles. Cell. Mol. Bioeng. 2019;12:389–397. doi: 10.1007/s12195-019-00573-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hajj K.A., Melamed J.R., Chaudhary N., Lamson N.G., Ball R.L., Yerneni S.S., Whitehead K.A. A potent branched-tail lipid nanoparticle enables multiplexed mrna delivery and gene editing in vivo. Nano Lett. 2020 doi: 10.1021/acs.nanolett.0c00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bilzer M., Roggel F., Gerbes A.L. Role of Kupffer cells in host defense and liver disease. Liver Int. Off. J. Int. Assoc. Study Liver. 2006;26:1175–1186. doi: 10.1111/j.1478-3231.2006.01342.x. [DOI] [PubMed] [Google Scholar]
- 53.Vollmar B., Menger M.D. The hepatic microcirculation: mechanistic contributions and therapeutic targets in liver injury and repair. Physiol. Rev. 2009;89:1269–1339. doi: 10.1152/physrev.00027.2008. [DOI] [PubMed] [Google Scholar]
- 54.Wisse E., Jacobs F., Topal B., Frederik P., Geest B.D. The size of endothelial fenestrae in human liver sinusoids: implications for hepatocyte-directed gene transfer. Gene Ther. 2008;15:1193–1199. doi: 10.1038/gt.2008.60. [DOI] [PubMed] [Google Scholar]
- 55.Electron Miscroscopy 2020. https://meded.ucsf.edu (accessed February 16, 2016)
- 56.Toth C.A., Thomas P. Liver endocytosis and Kupffer cells. Hepatol. Baltim. Md. 1992;16:255–266. doi: 10.1002/hep.1840160137. [DOI] [PubMed] [Google Scholar]
- 57.PrabhuDas M.R., Baldwin C.L., Bollyky P.L., Bowdish D.M.E., Drickamer K., Febbraio M., Herz J., Kobzik L., Krieger M., Loike J., McVicker B., Means T.K., Moestrup S.K., Post S.R., Sawamura T., Silverstein S., Speth R.C., Telfer J.C., Thiele G.M., Wang X.-Y., Wright S.D., Khoury J.E. A consensus definitive classification of scavenger receptors and their roles in health and disease. J. Immunol. 2017;198:3775–3789. doi: 10.4049/jimmunol.1700373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peiser L., Gordon S. The function of scavenger receptorsexpressed by macrophages and their rolein the regulation of inflammation. Microbes Infect. 2001;3:149–159. doi: 10.1016/S1286-4579(00)01362-9. [DOI] [PubMed] [Google Scholar]
- 59.Dobrovolskaia M.A., Aggarwal P., Hall J.B., McNeil S.E. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol. Pharm. 2008;5:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aggarwal P., Hall J.B., McLeland C.B., Dobrovolskaia M.A., McNeil S.E. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv. Drug Deliv. Rev. 2009;61:428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen D., Ganesh S., Wang W., Amiji M. The role of surface chemistry in serum protein corona-mediated cellular delivery and gene silencing with lipid nanoparticles. Nanoscale. 2019;11:8760–8775. doi: 10.1039/C8NR09855G. [DOI] [PubMed] [Google Scholar]
- 62.Poisson J., Lemoinne S., Boulanger C., Durand F., Moreau R., Valla D., Rautou P.-E. Liver sinusoidal endothelial cells: Physiology and role in liver diseases. J. Hepatol. 2017;66:212–227. doi: 10.1016/j.jhep.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 63.Wisse E. An electron microscopic study of the fenestrated endothelial lining of rat liver sinusoids. J. Ultrastruct. Res. 1970;31:125–150. doi: 10.1016/S0022-5320(70)90150-4. [DOI] [PubMed] [Google Scholar]
- 64.Braet F., Wisse E. Structural and functional aspects of liver sinusoidal endothelial cell fenestrae: a review. Comp. Hepatol. 2002;1:1. doi: 10.1186/1476-5926-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wisse E., de Zanger R.B., Charels K., van der Smissen P., McCuskey R.S. The liver sieve: considerations concerning the structure and function of endothelial fenestrae, the sinusoidal wall and the space of disse. Hepatology. 1985;5:683–692. doi: 10.1002/hep.1840050427. [DOI] [PubMed] [Google Scholar]
- 66.Braet F., Fraser R., McCuskey R.S. Thirty-five years of liver sinusoidal cells: Eddie wisse in retirement. Hepatology. 2003;38:1056–1058. doi: 10.1002/hep.1840380434. [DOI] [PubMed] [Google Scholar]
- 67.Braet F., Riches J., Geerts W., Jahn K.A., Wisse E., Frederik P. Three-dimensional organization of fenestrae labyrinths in liver sinusoidal endothelial cells. Liver Int. Off. J. Int. Assoc. Study Liver. 2009;29:603–613. doi: 10.1111/j.1478-3231.2008.01836.x. [DOI] [PubMed] [Google Scholar]
- 68.Campbell F., Bos F.L., Sieber S., Arias-Alpizar G., Koch B.E., Huwyler J., Kros A., Bussmann J. Directing Nanoparticle biodistribution through evasion and exploitation of stab2-dependent nanoparticle uptake. ACS Nano. 2018;12:2138–2150. doi: 10.1021/acsnano.7b06995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Charels K., De Zanger R., Van Bossuyt H., Van Der Smissen P., Wisse E. Influence of acute alcohol administration on endothelial fenestrae of rat livers: an in vivo and in vitro scanning electron microscopic study. Cells Hepatic Sinusoid. 1986;1:497–502. [Google Scholar]
- 70.Snoeys J., Lievens J., Wisse E., Jacobs F., Duimel H., Collen D., Frederik P., Geest B.D. Species differences in transgene DNA uptake in hepatocytes after adenoviral transfer correlate with the size of endothelial fenestrae. Gene Ther. 2007;14:604–612. doi: 10.1038/sj.gt.3302899. [DOI] [PubMed] [Google Scholar]
- 71.Blomhoff R., Wake K. Perisinusoidal stellate cells of the liver: important roles in retinol metabolism and fibrosis. FASEB J. 1991;5:271–277. doi: 10.1096/fasebj.5.3.2001786. [DOI] [PubMed] [Google Scholar]
- 72.Koyama Y., Brenner D.A. Liver inflammation and fibrosis. J. Clin. Invest. 2017;127:55–64. doi: 10.1172/JCI88881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsuchida T., Friedman S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017;14:397–411. doi: 10.1038/nrgastro.2017.38. [DOI] [PubMed] [Google Scholar]
- 74.Hernandez-Gea V., Friedman S.L. Pathogenesis of liver fibrosis. Annu. Rev. Pathol. Mech. Dis. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 75.Iredale J.P. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J. Clin. Invest. 2007;117:539–548. doi: 10.1172/JCI30542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R., Butler J.S., Qin L., Racie T., Sprague A., Fava E., Zeigerer A., Hope M.J., Zerial M., Sah D.W.Y., Fitzgerald K., Tracy M.A., Manoharan M., Koteliansky V., de Fougerolles A., Maier M.A. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. J. Am. Soc. Gene Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Witzigmann D. 2020. Hepatocyte-Specific Drug Delivery using Active Targeted Nanomedicines - Evaluation of Targeting Strategies in Vitro and in Vivo. Thesis, University_of_Basel, 2016. doi:info:doi/10.5451/unibas-006699992. [Google Scholar]
- 78.Orphanet 2020. http://www.orpha.net/consor/cgi-bin/index.php (accessed February 20, 2016)
- 79.Kay M.A. State-of-the-art gene-based therapies: the road ahead. Nat. Rev. Genet. 2011;12:316–328. doi: 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- 80.High K.A. Gene therapy for haemophilia: a long and winding road. J. Thromb. Haemost. 2011;9:2–11. doi: 10.1111/j.1538-7836.2011.04369.x. [DOI] [PubMed] [Google Scholar]
- 81.Fagiuoli S., Daina E., D’Antiga L., Colledan M., Remuzzi G. Monogenic diseases that can be cured by liver transplantation. J. Hepatol. 2013;59:595–612. doi: 10.1016/j.jhep.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 82.Bergmann K.R., McCabe J., Smith T.R., Guillaume D.J., Sarafoglou K., Gupta S. Late-onset ornithine transcarbamylase deficiency: treatment and outcome of hyperammonemic crisis. Pediatrics. 2014;133:E1072–E1076. doi: 10.1542/peds.2013-1324. [DOI] [PubMed] [Google Scholar]
- 83.Raal F.J., Santos R.D. Homozygous familial hypercholesterolemia: Current perspectives on diagnosis and treatment. Atherosclerosis. 2012;223:262–268. doi: 10.1016/j.atherosclerosis.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 84.Seidah N.G., Awan Z., Chretien M., Mbikay M. PCSK9: A key modulator of cardiovascular health. Circ. Res. 2014;114:1022–1036. doi: 10.1161/CIRCRESAHA.114.301621. [DOI] [PubMed] [Google Scholar]
- 85.Hawkins P.N., Ando Y., Dispenzeri A., Gonzalez-Duarte A., Adams D., Suhr O.B. Evolving landscape in the management of transthyretin amyloidosis. Ann. Med. 2015;47:625–638. doi: 10.3109/07853890.2015.1068949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suhr O.B., Coelho T., Buades J., Pouget J., Conceicao I., Berk J., Schmidt H., Waddington-Cruz M., Campistol J.M., Bettencourt B.R., Vaishnaw A., Gollob J., Adams D. Efficacy and safety of patisiran for familial amyloidotic polyneuropathy: a phase II multi-dose study. Orphanet J. Rare Dis. 2015;10:109. doi: 10.1186/s13023-015-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bereczky Z., Kovacs K.B., Muszbek L. Protein C and protein S deficiencies: similarities and differences between two brothers playing in the same game. Clin. Chem. Lab. Med. 2010;48:S53–S66. doi: 10.1515/CCLM.2010.369. [DOI] [PubMed] [Google Scholar]
- 88.Qi X., De Stefano V., Wang J., Bai M., Yang Z., Han G., Fan D. Prevalence of inherited antithrombin, protein C, and protein S deficiencies in portal vein system thrombosis and Budd-Chiari syndrome: a systematic review and meta-analysis of observational studies. J. Gastroenterol. Hepatol. 2013;28:432–442. doi: 10.1111/jgh.12085. [DOI] [PubMed] [Google Scholar]
- 89.Hoppe B. An update on primary hyperoxaluria. Nat. Rev. Nephrol. 2012;8:467–475. doi: 10.1038/nrneph.2012.113. [DOI] [PubMed] [Google Scholar]
- 90.Bosma P.J. Inherited disorders of bilirubin metabolism. J. Hepatol. 2003;38:107–117. doi: 10.1016/s0168-8278(02)00359-8. [DOI] [PubMed] [Google Scholar]
- 91.van Dijk R., Beuers U., Bosma P.J. Gene replacement therapy for genetic hepatocellular jaundice. Clin. Rev. Allergy Immunol. 2015;48:243–253. doi: 10.1007/s12016-014-8454-7. [DOI] [PubMed] [Google Scholar]
- 92.Flotte T.R., Mueller C. Gene therapy for alpha-1 antitrypsin deficiency. Hum. Mol. Genet. 2011;20:R87–R92. doi: 10.1093/hmg/ddr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stoller J.K., Aboussouan L.S. A review of alpha(1)-antitrypsin deficiency. Am. J. Respir. Crit. Care Med. 2012;185:246–259. doi: 10.1164/rccm.201108-1428CI. [DOI] [PubMed] [Google Scholar]
- 94.Rosencrantz R., Schilsky M. Wilson disease: pathogenesis and clinical considerations in diagnosis and treatment. Semin. Liver Dis. 2011;31:245–259. doi: 10.1055/s-0031-1286056. [DOI] [PubMed] [Google Scholar]
- 95.Purchase R. The treatment of Wilson’s disease, a rare genetic disorder of copper metabolism. Sci. Prog. 2013;96:19–32. doi: 10.3184/003685013X13587771579987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nobili V., Jenkner A., Francalanci P., Castellano A., Holme E., Callea F., Dionisi-Vici C. Tyrosinemia type 1: metastatic hepatoblastoma with a favorable outcome. Pediatrics. 2010;126:E235–E238. doi: 10.1542/peds.2009-1639. [DOI] [PubMed] [Google Scholar]
- 97.Kitagawa T. Hepatorenal tyrosinemia. Proc. Jpn. Acad. Ser. B-Phys. Biol. Sci. 2012;88:192–200. doi: 10.2183/pjab.88.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pietrangelo A. Hereditary hemochromatosis: pathogenesis, diagnosis, and treatment. Gastroenterology. 2010;139:393–408. doi: 10.1053/j.gastro.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 99.Babitt J.L., Lin H.Y. The molecular pathogenesis of hereditary hemochromatosis. Semin. Liver Dis. 2011;31:280–292. doi: 10.1055/s-0031-1286059. [DOI] [PubMed] [Google Scholar]
- 100.Ozen H. Glycogen storage diseases: new perspectives. World J. Gastroenterol. 2007;13:2541–2553. doi: 10.3748/wjg.v13.i18.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Byrne B.J., Falk D.J., Pacak C.A., Nayak S., Herzog R.W., Elder M.E., Collins S.W., Conlon T.J., Clement N., Cleaver B.D., Cloutier D.A., Porvasnik S.L., Islam S., Elmallah M.K., Martin A., Smith B.K., Fuller D.D., Lawson L.A., Mah C.S. Pompe disease gene therapy. Hum. Mol. Genet. 2011;20:R61–R68. doi: 10.1093/hmg/ddr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.El-Serag H.B., Rudolph K.L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 103.El-Serag H.B. Hepatocellular carcinoma. N. Engl. J. Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 104.Forner A., Llovet J.M., Bruix J. Hepatocellular carcinoma. Lancet Lond. Engl. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 105.Bouchard M.J., Navas-Martin S. Hepatitis B and C virus hepatocarcinogenesis: lessons learned and future challenges. Cancer Lett. 2011;305:123–143. doi: 10.1016/j.canlet.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chisari F.V., Isogawa M., Wieland S.F. Pathogenesis of hepatitis B virus infection. Pathol. Biol. (Paris). 2010;58:258–266. doi: 10.1016/j.patbio.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liaw Y.-F., Chu C.-M. Hepatitis B virus infection. Lancet Lond. Engl. 2009;373:582–592. doi: 10.1016/S0140-6736(09)60207-5. [DOI] [PubMed] [Google Scholar]
- 108.Trépo C., Chan H.L.Y., Lok A. Hepatitis B virus infection. Lancet Lond. Engl. 2014;384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 109.Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin. Microbiol. Infect. 2011;17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 110.Gebhardt R., Matz-Soja M. Liver zonation: Novel aspects of its regulation and its impact on homeostasis. World J. Gastroenterol. WJG. 2014;20:8491–8504. doi: 10.3748/wjg.v20.i26.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kietzmann T. Metabolic zonation of the liver: the oxygen gradient revisited. Redox Biol. 2017;11:622–630. doi: 10.1016/j.redox.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jungermann K., Katz N. Functional specialization of different hepatocyte populations. Physiol. Rev. 1989;69:708–764. doi: 10.1152/physrev.1989.69.3.708. [DOI] [PubMed] [Google Scholar]
- 113.Gebhardt R. Metabolic zonation of the liver: regulation and implications for liver function. Pharmacol. Ther. 1992;53:275–354. doi: 10.1016/0163-7258(92)90055-5. [DOI] [PubMed] [Google Scholar]
- 114.Bell P., Wang L., Gao G., Haskins M.E., Tarantal A.F., McCarter R.J., Zhu Y., Yu H., Wilson J.M. Inverse zonation of hepatocyte transduction with AAV vectors between mice and non-human primates. Mol. Genet. Metab. 2011;104:395–403. doi: 10.1016/j.ymgme.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baruteau J., Waddington S.N., Alexander I.E., Gissen P. Gene therapy for monogenic liver diseases: clinical successes, current challenges and future prospects. J. Inherit. Metab. Dis. 2017;40:497–517. doi: 10.1007/s10545-017-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.MacParland S.A., Tsoi K.M., Ouyang B., Ma X.-Z., Manuel J., Fawaz A., Ostrowski M.A., Alman B.A., Zilman A., Chan W.C.W., McGilvray I.D. Phenotype determines nanoparticle uptake by human macrophages from liver and blood. ACS Nano. 2017;11:2428–2443. doi: 10.1021/acsnano.6b06245. [DOI] [PubMed] [Google Scholar]
- 117.Guillot A., Tacke F. Liver macrophages: old dogmas and new insights. Hepatol. Commun. 2019;3:730–743. doi: 10.1002/hep4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sleyster E.C., Knook D.L. Relation between localization and function of rat liver Kupffer cells. Lab. Investig. J. Tech. Methods Pathol. 1982;47:484–490. [PubMed] [Google Scholar]
- 119.Bartneck M., Warzecha K.T., Tacke F. Therapeutic targeting of liver inflammation and fibrosis by nanomedicine. Hepatobiliary Surg. Nutr. 2014;3:364–376. doi: 10.3978/j.issn.2304-3881.2014.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shi B., Abrams M., Sepp-Lorenzino L. Expression of asialoglycoprotein receptor 1 in human hepatocellular carcinoma. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2013;61:901–909. doi: 10.1369/0022155413503662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Witzigmann D., Quagliata L., Schenk S.H., Quintavalle C., Terracciano L.M., Huwyler J. Variable asialoglycoprotein receptor 1 expression in liver disease: Implications for therapeutic intervention. Hepatol. Res. 2016;46:686–696. doi: 10.1111/hepr.12599. [DOI] [PubMed] [Google Scholar]
- 122.Niemietz C., Nadzemova O., Zibert A., Schmidt H.H.-J. APOE polymorphism in ATTR amyloidosis patients treated with lipid nanoparticle siRNA. Amyloid. 2020;27:45–51. doi: 10.1080/13506129.2019.1681392. [DOI] [PubMed] [Google Scholar]
- 123.van der Meel R., Sulheim E., Shi Y., Kiessling F., Mulder W.J.M., Lammers T. Smart cancer nanomedicine. Nat. Nanotechnol. 2019;14:1007–1017. doi: 10.1038/s41565-019-0567-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fraley R.T., Fornari C.S., Kaplan S. Entrapment of a bacterial plasmid in phospholipid vesicles: potential for gene transfer. Proc. Natl. Acad. Sci. U. S. A. 1979;76:3348–3352. doi: 10.1073/pnas.76.7.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fraley R., Subramani S., Berg P., Papahadjopoulos D. Introduction of liposome-encapsulated SV40 DNA into cells. J. Biol. Chem. 1980;255:10431–10435. [PubMed] [Google Scholar]
- 126.Guo X., Wang H., Li Y., Leng X., Huang W., Ma Y., Xu T., Qi X. Transfection reagent Lipofectamine triggers type I interferon signaling activation in macrophages. Immunol. Cell Biol. 2019;97:92–96. doi: 10.1111/imcb.12194. [DOI] [PubMed] [Google Scholar]
- 127.Gruner S.M., Cullis P.R., Hope M.J., Tilcock C.P.S. Lipid polymorphism:the molecular basis of nonbilayer phases. Annu. Rev. Biophys. Biophys. Chem. 1985;14:211–238. doi: 10.1146/annurev.bb.14.060185.001235. [DOI] [PubMed] [Google Scholar]
- 128.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release Off. J. Control. Release Soc. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 129.Lin P.J.C., Tam Y.Y.C., Hafez I., Sandhu A., Chen S., Ciufolini M.A., Nabi I.R., Cullis P.R. Influence of cationic lipid composition on uptake and intracellular processing of lipid nanoparticle formulations of siRNA. Nanomedicine Nanotechnol. Biol. Med. 2013;9:233–246. doi: 10.1016/j.nano.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 130.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., Zhang X., Wang Q., Panesar S., Hutabarat R., Carioto M., Hettinger J., Kandasamy P., Butler D., Rajeev K.G., Pang B., Charisse K., Fitzgerald K., Mui B.L., Du X., Cullis P., Madden T.D., Hope M.J., Manoharan M., Akinc A. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shirazi R.S., Ewert K.K., Leal C., Majzoub R.N., Bouxsein N.F., Safinya C.R. Synthesis and characterization of degradable multivalent cationic lipids with disulfide-bond spacers for gene delivery. Biochim. Biophys. Acta. 2011;1808:2156–2166. doi: 10.1016/j.bbamem.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Akita H., Noguchi Y., Hatakeyama H., Sato Y., Tange K., Nakai Y., Harashima H. Molecular tuning of a vitamin E-scaffold pH-sensitive and reductive cleavable lipid-like material for accelerated in vivo hepatic siRNA delivery. ACS Biomater. Sci. Eng. 2015;1:834–844. doi: 10.1021/acsbiomaterials.5b00203. [DOI] [PubMed] [Google Scholar]
- 133.Zimmermann T.S., Lee A.C.H., Akinc A., Bramlage B., Bumcrot D., Fedoruk M.N., Harborth J., Heyes J.A., Jeffs L.B., John M., Judge A.D., Lam K., McClintock K., Nechev L.V., Palmer L.R., Racie T., Röhl I., Seiffert S., Shanmugam S., Sood V., Soutschek J., Toudjarska I., Wheat A.J., Yaworski E., Zedalis W., Koteliansky V., Manoharan M., Vornlocher H.P., MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 134.Dong Y., Love K.T., Dorkin J.R., Sirirungruang S., Zhang Y., Chen D., Bogorad R.L., Yin H., Chen Y., Vegas A.J., Alabi C.A., Sahay G., Olejnik K.T., Wang W., Schroeder A., Lytton-Jean A.K.R., Siegwart D.J., Akinc A., Barnes C., Barros S.A., Carioto M., Fitzgerald K., Hettinger J., Kumar V., Novobrantseva T.I., Qin J., Querbes W., Koteliansky V., Langer R., Anderson D.G. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl. Acad. Sci. U. S. A. 2014;111:3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li B., Luo X., Deng B., Wang J., McComb D.W., Shi Y., Gaensler K.M.L., Tan X., Dunn A.L., Kerlin B.A., Dong Y. An orthogonal array optimization of lipid-like nanoparticles for mRNA delivery in vivo. Nano Lett. 2015;15:8099–8107. doi: 10.1021/acs.nanolett.5b03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Akita H., Ishiba R., Togashi R., Tange K., Nakai Y., Hatakeyama H., Harashima H. A neutral lipid envelope-type nanoparticle composed of a pH-activated and vitamin E-scaffold lipid-like material as a platform for a gene carrier targeting renal cell carcinoma. J. Control. Release Off. J. Control. Release Soc. 2015;200:97–105. doi: 10.1016/j.jconrel.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 137.Conway A., Mendel M., Kim K., McGovern K., Boyko A., Zhang L., Miller J.C., DeKelver R.C., Paschon D.E., Mui B.L., Lin P.J.C., Tam Y.K., Barbosa C., Redelmeier T., Holmes M.C., Lee G. Non-viral delivery of zinc finger nuclease mRNA enables highly efficient in vivo genome editing of multiple therapeutic gene targets. Mol. Ther. J. Am. Soc. Gene Ther. 2019;27:866–877. doi: 10.1016/j.ymthe.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., Almarsson Ö., Stanton M.G., Benenato K.E. A novel amino lipid series for mrna delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. J. Am. Soc. Gene Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cheng X., Lee R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016;99:129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 140.Rodrigueza W.V., Wheeler J.J., Klimuk S.K., Kitson C.N., Hope M.J. Transbilayer movement and net flux of cholesterol and cholesterol sulfate between liposomal membranes. Biochemistry. 1995;34:6208–6217. doi: 10.1021/bi00018a025. [DOI] [PubMed] [Google Scholar]
- 141.Sato Y., Okabe N., Note Y., Hashiba K., Maeki M., Tokeshi M., Harashima H. Hydrophobic scaffolds of pH-sensitive cationic lipids contribute to miscibility with phospholipids and improve the efficiency of delivering short interfering RNA by small-sized lipid nanoparticles. Acta Biomater. 2020;102:341–350. doi: 10.1016/j.actbio.2019.11.022. [DOI] [PubMed] [Google Scholar]
- 142.Kauffman K.J., Dorkin J.R., Yang J.H., Heartlein M.W., DeRosa F., Mir F.F., Fenton O.S., Anderson D.G. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 143.Paunovska K., Da Silva Sanchez A.J., Sago C.D., Gan Z., Lokugamage M.P., Islam F.Z., Kalathoor S., Krupczak B.R., Dahlman J.E. Nanoparticles containing oxidized cholesterol deliver mRNA to the liver microenvironment at clinically relevant doses. Adv. Mater. Deerfield Beach Fla. 2019;31 doi: 10.1002/adma.201807748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.De Rijke Y.B., Biessen E.A., Vogelezang C.J., van Berkel T.J. Binding characteristics of scavenger receptors on liver endothelial and Kupffer cells for modified low-density lipoproteins. Biochem. J. 1994;304:69–73. doi: 10.1042/bj3040069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gillotte-Taylor K., Boullier A., Witztum J.L., Steinberg D., Quehenberger O. Scavenger receptor class B type I as a receptor for oxidized low density lipoprotein. J. Lipid Res. 2001;42:1474–1482. [PubMed] [Google Scholar]
- 146.Belliveau N.M., Huft J., Lin P.J., Chen S., Leung A.K., Leaver T.J., Wild A.W., Lee J.B., Taylor R.J., Tam Y.K., Hansen C.L., Cullis P.R. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol. Ther. Nucleic Acids. 2012;1 doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen S., Tam Y.Y.C., Lin P.J.C., Leung A.K.K., Tam Y.K., Cullis P.R. Development of lipid nanoparticle formulations of siRNA for hepatocyte gene silencing following subcutaneous administration. J Control Release. 2014;196:106–112. doi: 10.1016/j.jconrel.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 148.Dahlman J.E., Kauffman K.J., Xing Y., Shaw T.E., Mir F.F., Dlott C.C., Langer R., Anderson D.G., Wang E.T. Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2060–2065. doi: 10.1073/pnas.1620874114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sieber S., Grossen P., Uhl P., Detampel P., Mier W., Witzigmann D., Huwyler J. Zebrafish as a predictive screening model to assess macrophage clearance of liposomes in vivo. Nanomed. Nanotechnol. Biol. Med. 2019;17:82–93. doi: 10.1016/j.nano.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 150.Holland J.W., Hui C., Cullis P.R., Madden T.D. Poly(ethylene glycol)--lipid conjugates regulate the calcium-induced fusion of liposomes composed of phosphatidylethanolamine and phosphatidylserine. Biochemistry. 1996;35:2618–2624. doi: 10.1021/bi952000v. [DOI] [PubMed] [Google Scholar]
- 151.Harvie P., Wong F.M.P., Bally M.B. Use of poly(ethylene glycol)–lipid conjugates to regulate the surface attributes and transfection activity of lipid–DNA particles. J. Pharm. Sci. 2000;89:652–663. doi: 10.1002/(SICI)1520-6017(200005)89:5<652::AID-JPS11>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 152.Hafez I.M., Maurer N., Cullis P.R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 153.Chen S., Tam Y.Y.C., Lin P.J.C., Sung M.M.H., Tam Y.K., Cullis P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Control. Release Off. J. Control. Release Soc. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 154.Jeffs L.B., Palmer L.R., Ambegia E.G., Giesbrecht C., Ewanick S., MacLachlan I. A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm. Res. 2005;22:362–372. doi: 10.1007/s11095-004-1873-z. [DOI] [PubMed] [Google Scholar]
- 155.Batzri S., Korn E.D. Single bilayer liposomes prepared without sonication. Biochim. Biophys. Acta. 1973;298:1015–1019. doi: 10.1016/0005-2736(73)90408-2. [DOI] [PubMed] [Google Scholar]
- 156.Hirota S., de Ilarduya C.T., Barron L.G., Szoka F.C. Simple mixing device to reproducibly prepare cationic lipid-DNA complexes (lipoplexes) BioTechniques. 1999;27:286–290. doi: 10.2144/99272bm16. [DOI] [PubMed] [Google Scholar]
- 157.Kulkarni J.A., Darjuan M.M., Mercer J.E., Chen S., van der Meel R., Thewalt J.L., Tam Y.Y.C., Cullis P.R. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano. 2018;12:4787–4795. doi: 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- 158.Kulkarni J.A., Tam Y.Y.C., Chen S., Tam Y.K., Zaifman J., Cullis P.R., Biswas S. Rapid synthesis of lipid nanoparticles containing hydrophobic inorganic nanoparticles. Nanoscale. 2017;9:13600–13609. doi: 10.1039/c7nr03272b. [DOI] [PubMed] [Google Scholar]
- 159.Sato Y., Hatakeyama H., Hyodo M., Harashima H. Relationship between the physicochemical properties of lipid nanoparticles and the quality of siRNA delivery to liver cells. Mol. Ther. 2016;24:788–795. doi: 10.1038/mt.2015.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shobaki N., Sato Y., Harashima H. Mixing lipids to manipulate the ionization status of lipid nanoparticles for specific tissue targeting. Int. J. Nanomedicine. 2018;13:8395–8410. doi: 10.2147/IJN.S188016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yu K.C.-W., David C., Kadambi S., Stahl A., Hirata K.-I., Ishida T., Quertermous T., Cooper A.D., Choi S.Y. Endothelial lipase is synthesized by hepatic and aorta endothelial cells and its expression is altered in apoE-deficient mice. J. Lipid Res. 2004;45:1614–1623. doi: 10.1194/jlr.M400069-JLR200. [DOI] [PubMed] [Google Scholar]
- 162.Monopoli M.P., Åberg C., Salvati A., Dawson K.A. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 163.Francia V., Yang K., Deville S., Reker-Smit C., Nelissen I., Salvati A. Corona composition can affect the mechanisms cells use to internalize nanoparticles. ACS Nano. 2019;13:11107–11121. doi: 10.1021/acsnano.9b03824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Albanese A., Walkey C.D., Olsen J.B., Guo H., Emili A., Chan W.C.W. Secreted biomolecules alter the biological identity and cellular interactions of nanoparticles. ACS Nano. 2014;8:5515–5526. doi: 10.1021/nn4061012. [DOI] [PubMed] [Google Scholar]
- 165.Miao L., Lin J., Huang Y., Li L., Delcassian D., Ge Y., Shi Y., Anderson D.G. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 2020;11:2424. doi: 10.1038/s41467-020-16248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kawabata K., Takakura Y., Hashida M. The fate of plasmid DNA after intravenous injection in mice: involvement of scavenger receptors in its hepatic uptake. Pharm. Res. 1995;12:825–830. doi: 10.1023/a:1016248701505. [DOI] [PubMed] [Google Scholar]
- 167.Maurer N., Wong K.F., Stark H., Louie L., McIntosh D., Wong T., Scherrer P., Semple S.C., Cullis P.R. Spontaneous entrapment of polynucleotides upon electrostatic interaction with ethanol-destabilized cationic liposomes. Biophys. J. 2001;80:2310–2326. doi: 10.1016/S0006-3495(01)76202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Main Acuitas. 2020. https://acuitastx.com/ (accessed January 19, 2020)
- 169.Buck J., Grossen P., Cullis P.R., Huwyler J., Witzigmann D. Lipid-based DNA therapeutics: hallmarks of non-viral gene delivery. ACS Nano. 2019;13:3754–3782. doi: 10.1021/acsnano.8b07858. [DOI] [PubMed] [Google Scholar]
- 170.Wheeler J.J., Palmer L., Ossanlou M., MacLachlan I., Graham R.W., Zhang Y.P., Hope M.J., Scherrer P., Cullis P.R. Stabilized plasmid-lipid particles: construction and characterization. Gene Ther. 1999;6:271–281. doi: 10.1038/sj.gt.3300821. [DOI] [PubMed] [Google Scholar]
- 171.Tam P., Monck M., Lee D., Ludkovski O., Leng E.C., Clow K., Stark H., Scherrer P., Graham R.W., Cullis P.R. Stabilized plasmid-lipid particles for systemic gene therapy. Gene Ther. 2000;7:1867–1874. doi: 10.1038/sj.gt.3301308. [DOI] [PubMed] [Google Scholar]
- 172.Aronsohn A.I., Hughes J.A. Nuclear localization signal peptides enhance cationic liposome-mediated gene therapy. J. Drug Target. 1998;5:163–169. doi: 10.3109/10611869808995871. [DOI] [PubMed] [Google Scholar]
- 173.Khalil I.A., Kimura S., Sato Y., Harashima H. Synergism between a cell penetrating peptide and a pH-sensitive cationic lipid in efficient gene delivery based on double-coated nanoparticles. J. Control. Release Off. J. Control. Release Soc. 2018;275:107–116. doi: 10.1016/j.jconrel.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 174.Akita H., Masuda T., Nishio T., Niikura K., Ijiro K., Harashima H. Improving in vivo hepatic transfection activity by controlling intracellular trafficking: the function of GALA and maltotriose. Mol. Pharm. 2011;8:1436–1442. doi: 10.1021/mp200189s. [DOI] [PubMed] [Google Scholar]
- 175.Yaari Z., da Silva D., Zinger A., Goldman E., Kajal A., Tshuva R., Barak E., Dahan N., Hershkovitz D., Goldfeder M., Roitman J.S., Schroeder A. Theranostic barcoded nanoparticles for personalized cancer medicine. Nat. Commun. 2016;7 doi: 10.1038/ncomms13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang Y., Miao L., Satterlee A., Huang L. Delivery of oligonucleotides with lipid nanoparticles, Adv. Drug Deliv Rev. 2015;87:68–80. doi: 10.1016/j.addr.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Goodwin T.J., Shen L., Hu M., Li J., Feng R., Dorosheva O., Liu R., Huang L. Liver specific gene immunotherapies resolve immune suppressive ectopic lymphoid structures of liver metastases and prolong survival. Biomaterials. 2017;141:260–271. doi: 10.1016/j.biomaterials.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hu M., Wang Y., Xu L., An S., Tang Y., Zhou X., Li J., Liu R., Huang L. Relaxin gene delivery mitigates liver metastasis and synergizes with check point therapy. Nat. Commun. 2019;10:2993. doi: 10.1038/s41467-019-10893-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.O. of the Commissioner FDA Approves First-of-its kind Targeted RNA-Based therapy to treat a Rare Disease, FDA. 2019. http://www.fda.gov/news-events/press-announcements/fda-approves-first-its-kind-targeted-rna-based-therapy-treat-rare-disease (accessed January 19, 2020)
- 180.Foster D.J., Brown C.R., Shaikh S., Trapp C., Schlegel M.K., Qian K., Sehgal A., Rajeev K.G., Jadhav V., Manoharan M., Kuchimanchi S., Maier M.A., Milstein S. Advanced siRNA designs further improve in vivo performance of GalNAc-siRNA conjugates. Mol. Ther. 2018;26:708–717. doi: 10.1016/j.ymthe.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Thi E.P., Mire C.E., Lee A.C.H., Geisbert J.B., Zhou J.Z., Agans K.N., Snead N.M., Deer D.J., Barnard T.R., Fenton K.A., MacLachlan I., Geisbert T.W. Lipid nanoparticle siRNA treatment of Ebola-virus-Makona-infected nonhuman primates. Nature. 2015;521:362–365. doi: 10.1038/nature14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sato Y., Matsui H., Yamamoto N., Sato R., Munakata T., Kohara M., Harashima H. Highly specific delivery of siRNA to hepatocytes circumvents endothelial cell-mediated lipid nanoparticle-associated toxicity leading to the safe and efficacious decrease in the hepatitis B virus. J. Control. Release Off. J. Control. Release Soc. 2017;266:216–225. doi: 10.1016/j.jconrel.2017.09.044. [DOI] [PubMed] [Google Scholar]
- 183.Ye X., Tateno C., Thi E.P., Kakuni M., Snead N.M., Ishida Y., Barnard T.R., Sofia M.J., Shimada T., Lee A.C.H. Hepatitis B virus therapeutic agent ARB-1740 Has inhibitory effect on hepatitis delta virus in a new dually-infected humanized mouse model. ACS Infect. Dis. 2019;5:738–749. doi: 10.1021/acsinfecdis.8b00192. [DOI] [PubMed] [Google Scholar]
- 184.Moon J.-S., Lee S.-H., Han S.-H., Kim E.-J., Cho H., Lee W., Kim M.-K., Kim T.-E., Park H.-J., Rhee J.-K., Kim S.-J., Cho S.-W., Han S.H., Oh J.-W. Inhibition of hepatitis C virus in mouse models by lipidoid nanoparticle-mediated systemic delivery of siRNA against PRK2. Nanomed. Nanotechnol. Biol. Med. 2016;12:1489–1498. doi: 10.1016/j.nano.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 185.Semple S.C., Judge A.D., Robbins M., Klimuk S., Eisenhardt M., Crosley E., Leung A., Kwok R., Ambegia E., McClintock K., MacLachlan I. Abstract 2829: preclinical characterization of TKM-080301, a lipid nanoparticle formulation of a small interfering RNA directed against polo-like kinase 1. Cancer Res. 2011;71:2829. doi: 10.1158/1538-7445.AM2011-2829. [DOI] [Google Scholar]
- 186.Doan C.C., Le L.T., Hoang S.N., Do S.M., Le D.V. Simultaneous silencing of VEGF and KSP by siRNA cocktail inhibits proliferation and induces apoptosis of hepatocellular carcinoma Hep3B cells. Biol. Res. 2014;47:70. doi: 10.1186/0717-6287-47-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhou K., Nguyen L.H., Miller J.B., Yan Y., Kos P., Xiong H., Li L., Hao J., Minnig J.T., Zhu H., Siegwart D.J. Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model. Proc. Natl. Acad. Sci. 2016;113:520–525. doi: 10.1073/pnas.1520756113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Frank-Kamenetsky M., Grefhorst A., Anderson N.N., Racie T.S., Bramlage B., Akinc A., Butler D., Charisse K., Dorkin R., Fan Y., Gamba-Vitalo C., Hadwiger P., Jayaraman M., John M., Jayaprakash K.N., Maier M., Nechev L., Rajeev K.G., Read T., Röhl I., Soutschek J., Tan P., Wong J., Wang G., Zimmermann T., de Fougerolles A., Vornlocher H.-P., Langer R., Anderson D.G., Manoharan M., Koteliansky V., Horton J.D., Fitzgerald K. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Jørgensen A.B., Frikke-Schmidt R., Nordestgaard B.G., Tybjærg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N. Engl. J. Med. 2014;371:32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 190.Tikka A., Soronen J., Laurila P.-P., Metso J., Ehnholm C., Jauhiainen M. Silencing of ANGPTL 3 (angiopoietin-like protein 3) in human hepatocytes results in decreased expression of gluconeogenic genes and reduced triacylglycerol-rich VLDL secretion upon insulin stimulation. Biosci. Rep. 2014;34 doi: 10.1042/BSR20140115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hou X., Aguilar M.-I., Small D.H. Transthyretin and familial amyloidotic polyneuropathy. Recent progress in understanding the molecular mechanism of neurodegeneration. FEBS J. 2007;274:1637–1650. doi: 10.1111/j.1742-4658.2007.05712.x. [DOI] [PubMed] [Google Scholar]
- 192.Butler J.S., Chan A., Costelha S., Fishman S., Willoughby J.L.S., Borland T.D., Milstein S., Foster D.J., Gonçalves P., Chen Q., Qin J., Bettencourt B.R., Sah D.W., Alvarez R., Rajeev K.G., Manoharan M., Fitzgerald K., Meyers R.E., Nochur S.V., Saraiva M.J., Zimmermann T.S. Preclinical evaluation of RNAi as a treatment for transthyretin-mediated amyloidosis. Amyloid Int. J. Exp. Clin. Investig. Off. J. Int. Soc. Amyloidosis. 2016;23:109–118. doi: 10.3109/13506129.2016.1160882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pardi N., LaBranche C.C., Ferrari G., Cain D.W., Tombácz I., Parks R.J., Muramatsu H., Mui B.L., Tam Y.K., Karikó K., Polacino P., Barbosa C.J., Madden T.D., Hope M.J., Haynes B.F., Montefiori D.C., Hu S.-L., Weissman D. Characterization of HIV-1 nucleoside-modified mRNA vaccines in rabbits and rhesus macaques. Mol. Ther. Nucleic Acids. 2019;15:36–47. doi: 10.1016/j.omtn.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines-a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pardi N., Weissman D. Nucleoside modified mRNA vaccines for infectious diseases. Methods Mol. Biol. Clifton NJ. 2017;1499:109–121. doi: 10.1007/978-1-4939-6481-9_6. [DOI] [PubMed] [Google Scholar]
- 196.Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S.J., Mui B.L., Tam Y.K., Karikó K., Barbosa C.J., Madden T.D., Hope M.J., Krammer F., Hensley S.E., Weissman D. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E., LaBranche C.C., Montefiori D.C., Lobby J.L., Saunders K.O., Liao H.-X., Korber B.T., Sutherland L.L., Scearce R.M., Hraber P.T., Tombácz I., Muramatsu H., Ni H., Balikov D.A., Li C., Mui B.L., Tam Y.K., Krammer F., Karikó K., Polacino P., Eisenlohr L.C., Madden T.D., Hope M.J., Lewis M.G., Lee K.K., Hu S.-L., Hensley S.E., Cancro M.P., Haynes B.F., Weissman D. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R., Wagner W., Granados A., Greenhouse J., Walker M., Willis E., Yu J.-S., McGee C.E., Sempowski G.D., Mui B.L., Tam Y.K., Huang Y.-J., Vanlandingham D., Holmes V.M., Balachandran H., Sahu S., Lifton M., Higgs S., Hensley S.E., Madden T.D., Hope M.J., Karikó K., Santra S., Graham B.S., Lewis M.G., Pierson T.C., Haynes B.F., Weissman D. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., Mihai C., Lynn A., McFadyen I., Moore M.J., Senn J.J., Stanton M.G., Almarsson Ö., Ciaramella G., Brito L.A. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pardi N., Secreto A.J., Shan X., Debonera F., Glover J., Yi Y., Muramatsu H., Ni H., Mui B.L., Tam Y.K., Shaheen F., Collman R.G., Kariko K., Danet-Desnoyers G.A., Madden T.D., Hope M.J., Weissman D. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat Commun. 2017;8:14630. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Thran M., Mukherjee J., Pönisch M., Fiedler K., Thess A., Mui B.L., Hope M.J., Tam Y.K., Horscroft N., Heidenreich R., Fotin-Mleczek M., Shoemaker C.B., Schlake T. mRNA mediates passive vaccination against infectious agents, toxins, and tumors. EMBO Mol. Med. 2017;9:1434–1447. doi: 10.15252/emmm.201707678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. J. Am. Soc. Gene Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.DeRosa F., Guild B., Karve S., Smith L., Love K., Dorkin J.R., Kauffman K.J., Zhang J., Yahalom B., Anderson D.G., Heartlein M.W. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 2016;23:699–707. doi: 10.1038/gt.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Finn J.D., Smith A.R., Patel M.C., Shaw L., Youniss M.R., Heteren J., Dirstine T., Ciullo C., Lescarbeau R., Seitzer J., Shah R.R., Shah A., Ling D., Growe J., Pink M., Rohde E., Wood K.M., Salomon W.E., Harrington W.F., Dombrowski C., Strapps W.R., Chang Y., Morrissey D.V. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 207.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.van der Meel R. Nanotechnology for organ-tunable gene editing. Nat. Nanotechnol. 2020;15:253–255. doi: 10.1038/s41565-020-0666-9. [DOI] [PubMed] [Google Scholar]
- 209.Rosigkeit S., Meng M., Grunwitz C., Gomes P., Kreft A., Hayduk N., Heck R., Pickert G., Ziegler K., Abassi Y., Röder J., Kaps L., Vascotto F., Beissert T., Witzel S., Kuhn A., Diken M., Schuppan D., Sahin U., Haas H., Bockamp E. Monitoring translation activity of mRNA-loaded nanoparticles in mice. Mol. Pharm. 2018;15:3909–3919. doi: 10.1021/acs.molpharmaceut.8b00370. [DOI] [PubMed] [Google Scholar]
- 210.Liu J., Chang J., Jiang Y., Meng X., Sun T., Mao L., Xu Q., Wang M. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 2019;31:1902575. doi: 10.1002/adma.201902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Trial to Evaluate Safety and Tolerability of ALN-TTR01 in Transthyretin (TTR) Amyloidosis - Full Text View - ClinicalTrials.gov. 2020. https://clinicaltrials.gov/ct2/show/NCT01148953 (accessed January 19, 2020)
- 213.Coelho T., Adams D., Silva A., Lozeron P., Hawkins P.N., Mant T., Perez J., Chiesa J., Warrington S., Tranter E., Munisamy M., Falzone R., Harrop J., Cehelsky J., Bettencourt B.R., Geissler M., Butler J.S., Sehgal A., Meyers R.E., Chen Q., Borland T., Hutabarat R.M., Clausen V.A., Alvarez R., Fitzgerald K., Gamba-Vitalo C., Nochur S.V., Vaishnaw A.K., Sah D.W.Y., Gollob J.A., Suhr O.B. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 214.Trial to Evaluate Safety, Tolerability, and Parmacokinetics of ALN-TTR02 in Healthy Volunteer Subjects - Full Text View - ClinicalTrials.gov. 2020. https://clinicaltrials.gov/ct2/show/NCT01559077 (accessed January 19, 2020)
- 215.Dyck P.J.B., González-Duarte A., Obici L., Polydefkis M., Wiesman J.F., Antonino I., Litchy W.J., Dyck P.J. Development of measures of polyneuropathy impairment in hATTR amyloidosis: from NIS to mNIS + 7. J. Neurol. Sci. 2019;405:116424. doi: 10.1016/j.jns.2019.116424. [DOI] [PubMed] [Google Scholar]
- 216.T.B. Inc Translate Bio Announces FDA Clearance to Proceed with a Single-ascending Dose (SAD) Phase 1/2 Clinical Trial for Ornithine Transcarbamylase (OTC) Deficiency, GlobeNewswire News Room. 2019. http://www.globenewswire.com/news-release/2019/06/26/1874354/0/en/Translate-Bio-Announces-FDA-Clearance-to-Proceed-with-a-Single-ascending-Dose-SAD-Phase-1-2-Clinical-Trial-for-Ornithine-Transcarbamylase-OTC-Deficiency.html (accessed January 25, 2020)
- 217.An D., Frassetto A., Jacquinet E., Eybye M., Milano J., DeAntonis C., Nguyen V., Laureano R., Milton J., Sabnis S., Lukacs C.M., Guey L.T. Long-term efficacy and safety of mRNA therapy in two murine models of methylmalonic acidemia. EBioMedicine. 2019;45:519–528. doi: 10.1016/j.ebiom.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Haussecker D. Current issues of RNAi therapeutics delivery and development. J. Controlled Release. 2014;195:49–54. doi: 10.1016/j.jconrel.2014.07.056. [DOI] [PubMed] [Google Scholar]
- 219.Szebeni J. Mechanism of nanoparticle-induced hypersensitivity in pigs: complement or not complement? Drug Discov. Today. 2018;23:487–492. doi: 10.1016/j.drudis.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 220.Szebeni J., Muggia F., Gabizon A., Barenholz Y. Activation of complement by therapeutic liposomes and other lipid excipient-based therapeutic products: prediction and prevention. Adv. Drug Deliv. Rev. 2011;63:1020–1030. doi: 10.1016/j.addr.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 221.Szebeni J., Bedocs P., Rozsnyay Z., Weiszhár Z., Urbanics R., Rosivall L., Cohen R., Garbuzenko O., Báthori G., Tóth M., Bünger R., Barenholz Y. Liposome-induced complement activation and related cardiopulmonary distress in pigs: factors promoting reactogenicity of Doxil and AmBisome. Nanomed. Nanotechnol. Biol. Med. 2012;8:176–184. doi: 10.1016/j.nano.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 222.Adams D., Suhr O.B., Dyck P.J., Litchy W.J., Leahy R.G., Chen J., Gollob J., Coelho T. Trial design and rationale for APOLLO, a Phase 3, placebo-controlled study of patisiran in patients with hereditary ATTR amyloidosis with polyneuropathy. BMC Neurol. 2017;17:181. doi: 10.1186/s12883-017-0948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L., Lin K.P., Vita G., Attarian S., Plante-Bordeneuve V., Mezei M.M., Campistol J.M., Buades J., Brannagan T.H., 3rd, Kim B.J., Oh J., Parman Y., Sekijima Y., Hawkins P.N., Solomon S.D., Polydefkis M., Dyck P.J., Gandhi P.J., Goyal S., Chen J., Strahs A.L., Nochur S.V., Sweetser M.T., Garg P.P., Vaishnaw A.K., Gollob J.A., Suhr O.B., Brannagan T.H., 3rd, Kim B.J., Oh J., Parman Y., Sekijima Y., Hawkins P.N., Solomon S.D., Polydefkis M., Dyck P.J., Gandhi P.J., Goyal S., Chen J., Strahs A.L., Nochur S.V., Sweetser M.T., Garg P.P., Vaishnaw A.K., Gollob J.A., Suhr O.B. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 224.Tabernero J., Shapiro G.I., LoRusso P.M., Cervantes A., Schwartz G.K., Weiss G.J., Paz-Ares L., Cho D.C., Infante J.R., Alsina M., Gounder M.M., Falzone R., Harrop J., White A.C.S., Toudjarska I., Bumcrot D., Meyers R.E., Hinkle G., Svrzikapa N., Hutabarat R.M., Clausen V.A., Cehelsky J., Nochur S.V., Gamba-Vitalo C., Vaishnaw A.K., Sah D.W.Y., Gollob J.A., Burris H.A., Burris H.A., 3rd First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- 225.Eley T. Pharmacokinetics and exploratory exposure-response of siRNAs administered monthly as ARB-001467 (ARB-1467) in a Phase 2a study in HBeAg positive and negative virally suppressed subjects with chronic hepatitis B. Hepatology. 2017;66 [Google Scholar]
- 226.Demeure M.J., Armaghany T., Ejadi S., Ramanathan R.K., Elfiky A., Strosberg J.R., Smith D.C., Whitsett T., Liang W.S., Sekar S., Carpten J.D., Fredlund P., Niforos D., Dye A., Gahir S., Semple S.C., Kowalski M.M. A phase I/II study of TKM-080301, a PLK1-targeted RNAi in patients with adrenocortical cancer (ACC) J. Clin. Oncol. 2016;34:2547. doi: 10.1200/JCO.2016.34.15_suppl.2547. [DOI] [Google Scholar]
- 227.Schultheis B., Strumberg D., Santel A., Vank C., Gebhardt F., Keil O., Lange C., Giese K., Kaufmann J., Khan M., Drevs J. First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J. Clin. Oncol. 2014;32:4141–4148. doi: 10.1200/JCO.2013.55.0376. [DOI] [PubMed] [Google Scholar]
- 228.Soule B., Tirucherai G., Kavita U., Kundu S., Christian R. Safety, tolerability, and pharmacokinetics of BMS-986263/ND-L02-s0201, a novel targeted lipid nanoparticle delivering HSP47 siRNA, in healthy participants: A randomised, placebo-controlled, double-blind, phase 1 study. J. Hepatol. 2018;68:S112. doi: 10.1016/s0168-8278(18)30442-2. [DOI] [Google Scholar]
- 229.Wagner M.J., Mitra R., Mcarthur M.J., Baze W., Barnhart K., Wu S.Y., Rodriguez-Aguayo C., Zhang X., Coleman R.L., Lopez-Berestein G., Sood A.K. Preclinical mammalian safety studies of EPHARNA (DOPC Nanoliposomal EphA2-Targeted siRNA) Mol. Cancer Ther. 2017;16:1114–1123. doi: 10.1158/1535-7163.MCT-16-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jabulowsky R.A., Loquai C., Mitzel-Rink H., Utikal J., Gebhardt C., Hassel J.C., Kaufmann R., Pinter A., Derhovanessian E., Anft C., Attig S., Deubel A., Diken M., Gold M., Guertler C., Haas H., Heesen L., Kemmer-Brück A., Kranz L.M., Kuehlcke K., Kuhn A., Langguth P., Luxemburger U., Maurus D., Meng M., Müller F., Rae R., Sari F., Schreeb K., Schwarck-Kokarakis D., Stein M., Jäger D., Grabbe S., Kreiter S., Huber C., Türeci Ö., Sahin U. Abstract CT156: A first-in-human phase I/II clinical trial assessing novel mRNA-lipoplex nanoparticles encoding shared tumor antigens for immunotherapy of malignant melanoma. Cancer Res. 2018;78 doi: 10.1158/1538-7445.AM2018-CT156. CT156 LP-CT156. [DOI] [Google Scholar]
- 231.Pirollo K.F., Nemunaitis J., Leung P.K., Nunan R., Adams J., Chang E.H. Safety and efficacy in advanced solid tumors of a targeted nanocomplex carrying the p53 gene used in combination with docetaxel: A phase 1b study. Mol. Ther. 2016;24:1697–1706. doi: 10.1038/mt.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sarker D., Plummer R., Basu B., Meyer T., Ma Y.-T., Evans J., Palmer D.H., Huang K.-W., Chee C.E., Spalding D., Sodergren M., Habib N. 455PDFirst-in-human, first-in-class phase I study of MTL-CEBPA, a RNA oligonucleotide targeting the myeloid cell master regulator C/EBP-α, in patients with advanced hepatocellular cancer (HCC) Ann. Oncol. 2019;30 doi: 10.1093/annonc/mdz244.017. [DOI] [Google Scholar]
- 233.Intellia Therapeutics Announces Second Quarter 2019 Financial Results and Company Update, Intellia Ther 2020. https://ir.intelliatx.com/news-releases/news-release-details/intellia-therapeutics-announces-second-quarter-2019-financial (accessed January 25, 2020)
- 234.Fitzgerald K., Frank-Kamenetsky M., Shulga-Morskaya S., Liebow A., Bettencourt B.R., Sutherland J.E., Hutabarat R.M., Clausen V.A., Karsten V., Cehelsky J., Nochur S.V., Kotelianski V., Horton J., Mant T., Chiesa J., Ritter J., Munisamy M., Vaishnaw A.K., Gollob J.A., Simon A. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet Lond. Engl. 2014;383:60–68. doi: 10.1016/S0140-6736(13)61914-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Benson M.D., Waddington-Cruz M., Berk J.L., Polydefkis M., Dyck P.J., Wang A.K., Planté-Bordeneuve V., Barroso F.A., Merlini G., Obici L., Scheinberg M., Brannagan T.H., Litchy W.J., Whelan C., Drachman B.M., Adams D., Heitner S.B., Conceição I., Schmidt H.H., Vita G., Campistol J.M., Gamez J., Gorevic P.D., Gane E., Shah A.M., Solomon S.D., Monia B.P., Hughes S.G., Kwoh T.J., McEvoy B.W., Jung S.W., Baker B.F., Ackermann E.J., Gertz M.A., Coelho T. Inotersen treatment for patients with hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018;379:22–31. doi: 10.1056/NEJMoa1716793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sardh E., Harper P., Balwani M., Stein P., Rees D., Bissell D.M., Desnick R., Parker C., Phillips J., Bonkovsky H.L., Vassiliou D., Penz C., Chan-Daniels A., He Q., Querbes W., Fitzgerald K., Kim J.B., Garg P., Vaishnaw A., Simon A.R., Anderson K.E. Phase 1 trial of an RNA interference therapy for acute intermittent porphyria. N. Engl. J. Med. 2019;380:549–558. doi: 10.1056/NEJMoa1807838. [DOI] [PubMed] [Google Scholar]
- 237.Balwani M., Gouya L., Rees D., Stein P., Stölzel U., Aguilera P., Bissell D.M., Bonkovsky H., Keel S., Parker C., Phillips J., Silver S., Windyga J., D’avola D., Ross G., Stewart P., Ritchie B., Oh J., Harper P., Wang J.-D., Langendonk J., Ivanova A., Horie Y., Anderson K., Ventura P., Chan A., Penz C., Simon A., Dinh Q., Liu G., Sardh E. GS-14-ENVISION, a phase 3 study to evaluate efficacy and safety of givosiran, an investigational RNAi therapeutic targeting aminolevulinic acid synthase 1, in acute hepatic porphyria patients. J. Hepatol. 2019;70:e81–e82. doi: 10.1016/S0618-8278(19)30142-2. [DOI] [Google Scholar]
- 238.van’t Hoff W., Cochat P., Groothoff J., Harambat J., Frishberg Y., Hulton S., Magen D., Hoppe B., Lieske J., Milliner D.S., Deschenes G. Sun-325 safety and efficacy of lumasiran, an investigational rna interference (RNAi) therapeutic, in adult and pediatric patients with primary hyperoxaluria type 1. Kidney Int. Rep. 2019;4:S295. doi: 10.1016/j.ekir.2019.05.734. [DOI] [Google Scholar]
- 239.Yaacov Frishberg, Georges Deschenes, Pierre Cochat, Daniella Magen, Jaap Groothoff, Hulton Sally A., Jérôme Harambat, Hoff vant William, Bernd Hoppe, Lieske John C., Mc Gregor Tracy L., Patrick Haslett, Sandeep Talamudupula, Erbe David V., Milliner Dawn S. Mp12-14 safety and efficacy study of lumasiran, an investigational rna interference (rnai) therapeutic, in adult and pediatric patients with primary hyperoxaluria type 1 (ph1) J. Urol. 2019;201 doi: 10.1097/01.JU.0000555207.44355.e9. e174–e174. [DOI] [Google Scholar]
- 240.Iacobucci G. Inclisiran: UK to roll out new cholesterol lowering drug from next year. BMJ. 2020;368 doi: 10.1136/bmj.m139. [DOI] [PubMed] [Google Scholar]
- 241.Ray K.K., Landmesser U., Leiter L.A., Kallend D., Dufour R., Karakas M., Hall T., Troquay R.P.T., Turner T., Visseren F.L.J., Wijngaard P., Wright R.S., Kastelein J.J.P. Inclisiran in patients at high cardiovascular risk with elevated LDL cholesterol. N. Engl. J. Med. 2017;376:1430–1440. doi: 10.1056/NEJMoa1615758. [DOI] [PubMed] [Google Scholar]
- 242.Wright R.S., Collins M.G., Stoekenbroek R.M., Robson R., Wijngaard P.L.J., Landmesser U., Leiter L.A., Kastelein J.J.P., Ray K.K., Kallend D. Effects of renal impairment on the pharmacokinetics, efficacy, and safety of inclisiran: An analysis of the ORION-7 and ORION-1 studies. Mayo Clin. Proc. 2020;95:77–89. doi: 10.1016/j.mayocp.2019.08.021. [DOI] [PubMed] [Google Scholar]
- 243.Adams D., Verena K. Phase 1 study of ALN-TTRsc02, a subcutaneously administered investigational RNAi therapeutic for the treatment of transthyretin-mediated amyloidosis. Rev. Neurol. (Paris) 2019;175 doi: 10.1016/j.neurol.2019.01.339. S129. [DOI] [Google Scholar]
- 244.Basha G., Ordobadi M., Scott W.R., Cottle A., Liu Y., Wang H., Cullis P.R. Lipid nanoparticle delivery of siRNA to osteocytes leads to effective silencing of SOST and inhibition of sclerostin in vivo. Mol. Ther. Nucleic Acids. 2016;5 doi: 10.1038/mtna.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lee J.B., Zhang K., Tam Y.Y.C., Quick J., Tam Y.K., Lin P.J., Chen S., Liu Y., Nair J.K., Zlatev I., Rajeev K.G., Manoharan M., Rennie P.S., Cullis P.R. A glu-urea-lys ligand-conjugated lipid nanoparticle/siRNA system inhibits androgen receptor expression in vivo. Mol. Ther. Nucleic Acids. 2016;5 doi: 10.1038/mtna.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yamamoto Y., Lin P.J.C., Beraldi E., Zhang F., Kawai Y., Leong J., Katsumi H., Fazli L., Fraser R., Cullis P.R., Gleave M. siRNA lipid nanoparticle potently silences clusterin and delays progression when combined with androgen receptor cotargeting in enzalutamide-resistant prostate cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015;21:4845–4855. doi: 10.1158/1078-0432.CCR-15-0866. [DOI] [PubMed] [Google Scholar]
- 247.Ramishetti S., Kedmi R., Goldsmith M., Leonard F., Sprague A.G., Godin B., Gozin M., Cullis P.R., Dykxhoorn D.M., Peer D. Systemic gene silencing in primary T lymphocytes using targeted lipid nanoparticles. ACS Nano. 2015;9:6706–6716. doi: 10.1021/acsnano.5b02796. [DOI] [PubMed] [Google Scholar]
- 248.Weinstein S., Toker I.A., Emmanuel R., Ramishetti S., Hazan-Halevy I., Rosenblum D., Goldsmith M., Abraham A., Benjamini O., Bairey O., Raanani P., Nagler A., Lieberman J., Peer D. Harnessing RNAi-based nanomedicines for therapeutic gene silencing in B-cell malignancies. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E16–E22. doi: 10.1073/pnas.1519273113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Kedmi R., Veiga N., Ramishetti S., Goldsmith M., Rosenblum D., Dammes N., Hazan-Halevy I., Nahary L., Leviatan-Ben-Arye S., Harlev M., Behlke M., Benhar I., Lieberman J., Peer D. A modular platform for targeted RNAi therapeutics. Nat. Nanotechnol. 2018;13:214–219. doi: 10.1038/s41565-017-0043-5. [DOI] [PubMed] [Google Scholar]
- 250.Veiga N., Goldsmith M., Diesendruck Y., Ramishetti S., Rosenblum D., Elinav E., Behlke M.A., Benhar I., Peer D. Leukocyte-specific siRNA delivery revealing IRF8 as a potential anti-inflammatory target. J. Control. Release Off. J. Control. Release Soc. 2019;313:33–41. doi: 10.1016/j.jconrel.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 251.Novobrantseva T.I., Borodovsky A., Wong J., Klebanov B., Zafari M., Yucius K., Querbes W., Ge P., Ruda V.M., Milstein S., Speciner L., Duncan R., Barros S., Basha G., Cullis P., Akinc A., Donahoe J.S., Jayaprakash K. Narayanannair, Jayaraman M., Bogorad R.L., Love K., Whitehead K., Levins C., Manoharan M., Swirski F.K., Weissleder R., Langer R., Anderson D.G., de Fougerolles A., Nahrendorf M., Koteliansky V. Systemic RNAi-mediated gene silencing in nonhuman primate and rodent myeloid cells. Mol. Ther. Nucleic Acids. 2012;1 doi: 10.1038/mtna.2011.3. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Veiga N., Diesendruck Y., Peer D. Targeted lipid nanoparticles for RNA therapeutics and immunomodulation in leukocytes. Adv. Drug Deliv. Rev. 2020 doi: 10.1016/j.addr.2020.04.002. (In press) [DOI] [PubMed] [Google Scholar]
- 253.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kose N., Fox J.M., Sapparapu G., Bombardi R., Tennekoon R.N., de Silva A.D., Elbashir S.M., Theisen M.A., Humphris-Narayanan E., Ciaramella G., Himansu S., Diamond M.S., Crowe J.E. A lipid-encapsulated mRNA encoding a potently neutralizing human monoclonal antibody protects against chikungunya infection. Sci. Immunol. 2019;4 doi: 10.1126/sciimmunol.aaw6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Veiga N., Goldsmith M., Granot Y., Rosenblum D., Dammes N., Kedmi R., Ramishetti S., Peer D. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 2018;9:4493. doi: 10.1038/s41467-018-06936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Fenton O.S., Kauffman K.J., Kaczmarek J.C., McClellan R.L., Jhunjhunwala S., Tibbitt M.W., Zeng M.D., Appel E.A., Dorkin J.R., Mir F.F., Yang J.H., Oberli M.A., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Synthesis and biological evaluation of ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes. Adv. Mater. Deerfield Beach Fla. 2017;29 doi: 10.1002/adma.201606944. [DOI] [PubMed] [Google Scholar]
- 257.Oberli M.A., Reichmuth A.M., Dorkin J.R., Mitchell M.J., Fenton O.S., Jaklenec A., Anderson D.G., Langer R., Blankschtein D. Lipid Nanoparticle Assisted mRNA Delivery for Potent Cancer Immunotherapy. Nano Lett. 2017;17:1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H., Grunwitz C., Vormehr M., Hüsemann Y., Selmi A., Kuhn A.N., Buck J., Derhovanessian E., Rae R., Attig S., Diekmann J., Jabulowsky R.A., Heesch S., Hassel J., Langguth P., Grabbe S., Huber C., Türeci Ö., Sahin U. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 259.Billingsley M., Singh N., Ravikumar P., Zhang R., June C.H., Mitchell M.J. Ionizable lipid nanoparticle mediated mrna delivery for human CAR T cell engineering. Nano Lett. 2020 doi: 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Pardi N., LaBranche C.C., Ferrari G., Cain D.W., Tombácz I., Parks R.J., Muramatsu H., Mui B.L., Tam Y.K., Karikó K., Polacino P., Barbosa C.J., Madden T.D., Hope M.J., Haynes B.F., Montefiori D.C., Hu S.-L., Weissman D. Characterization of HIV-1 nucleoside-modified mRNA vaccines in rabbits and rhesus macaques. Mol. Ther. Nucleic Acids. 2019;15:36–47. doi: 10.1016/j.omtn.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S.J., Mui B.L., Tam Y.K., Karikó K., Barbosa C.J., Madden T.D., Hope M.J., Krammer F., Hensley S.E., Weissman D. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E., LaBranche C.C., Montefiori D.C., Lobby J.L., Saunders K.O., Liao H.-X., Korber B.T., Sutherland L.L., Scearce R.M., Hraber P.T., Tombácz I., Muramatsu H., Ni H., Balikov D.A., Li C., Mui B.L., Tam Y.K., Krammer F., Karikó K., Polacino P., Eisenlohr L.C., Madden T.D., Hope M.J., Lewis M.G., Lee K.K., Hu S.-L., Hensley S.E., Cancro M.P., Haynes B.F., Weissman D. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.N. Pardi, M.J. Hogan, R.S. Pelc, H. Muramatsu, H. Andersen, C.R. DeMaso, K.A. Dowd, L.L. Sutherland, R.M. Scearce, R. Parks, W. Wagner, A. Granados, J. Greenhouse, M. Walker, E. Willis, J.-S. Yu, C.E. McGee, G.D. Sempowski, B.L. Mui, Y.K. Tam, Y.-J. Huang, D. Vanlandingham, V.M. Holmes, H. Balachandran, S. Sahu, M. Lifton, S. Higgs, S.E. Hensley, T.D. Madden, M.J. Hope, K. Karikó, S. Santra, B.S. Graham, M.G. Lewis, T.C. Pierson, B.F. Haynes, D. Weissman, Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination, Nature. 543 (2017) 248–251. doi:10.1038/nature21428. [DOI] [PMC free article] [PubMed]
- 264.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release Off. J. Control. Release Soc. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Feldman R.A., Fuhr R., Smolenov I., Mick Ribeiro A., Panther L., Watson M., Senn J.J., Smith M., Almarsson Ӧrn, Pujar H.S., Laska M.E., Thompson J., Zaks T., Ciaramella G. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37:3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- 266.McKay P.F., Hu K., Blakney A.K., Samnuan K., Bouton C.R., Rogers P., Polra K., Lin P.J.C., Barbosa C., Tam Y., Shattock R.J. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine induces equivalent preclinical antibody titers and viral neutralization to recovered COVID-19 patients. BioRxiv. 2020 doi: 10.1101/2020.04.22.055608. 2020.04.22.055608. [DOI] [Google Scholar]
- 267.Moderna’s Work on a COVID-19 Vaccine Candidate|Moderna, Inc. 2010. https://www.modernatx.com/modernas-work-potential-vaccine-against-covid-19?utm_source=homepage&utm_medium=slider&utm_campaign=covid (accessed June 4, 2020)
- 268.BioNTech and Pfizer announce completion of dosing for first cohort of Phase 1/2 trial of COVID-19 vaccine candidates in Germany 2010. https://investors.pfizer.com/investor-news/press-release-details/2020/BioNTech-and-Pfizer-announce-completion-of-dosing-for-first-cohort-of-Phase-1-2-trial-of-COVID-19-vaccine-candidates-in-Germany/default.aspx (accessed June 4, 2020)
- 269.CureVac´s Optimized mRNA Platform Provides Positive Pre-Clinical Results at Low Dose for Coronavirus Vaccine Candidate | English, CureVac. (2020). https%3A%2F%2Fwww.curevac.com%2Fnews%2Fcurevac-s-optimized-mrna-platform-provides-positive-pre-clinical-results-at-low-dose-for-coronavirus-vaccine-candidate (accessed June 4, 2020).
- 270.Lutz J., Lazzaro S., Habbeddine M., Schmidt K.E., Baumhof P., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Heidenreich R., Fotin-Mleczek M. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. Npj Vaccines. 2017;2:1–9. doi: 10.1038/s41541-017-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Nguyen D.N., Mahon K.P., Chikh G., Kim P., Chung H., Vicari A.P., Love K.T., Goldberg M., Chen S., Krieg A.M., Chen J., Langer R., Anderson D.G. Lipid-derived nanoparticles for immunostimulatory RNA adjuvant delivery. Proc. Natl. Acad. Sci. 2012;109:E797–E803. doi: 10.1073/pnas.1121423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Chen S., Zaifman J., Kulkarni J.A., Zhigaltsev I.V., Tam Y.K., Ciufolini M.A., Tam Y.Y.C., Cullis P.R. Dexamethasone prodrugs as potent suppressors of the immunostimulatory effects of lipid nanoparticle formulations of nucleic acids. J. Control. Release Off. J. Control. Release Soc. 2018;286:46–54. doi: 10.1016/j.jconrel.2018.07.026. [DOI] [PubMed] [Google Scholar]
- 273.Modular lipid nanoparticle platform technology for siRNA and lipophilic prodrug deliverybioRxiv. 2020 doi: 10.1002/smll.202103025. https://www.biorxiv.org/content/10.1101/2020.01.16.907394v1.full [DOI] [PubMed] [Google Scholar]
- 274.Sahay G., Querbes W., Alabi C., Eltoukhy A., Sarkar S., Zurenko C., Karagiannis E., Love K., Chen D., Zoncu R., Buganim Y., Schroeder A., Langer R., Anderson D.G. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Patel S., Kim J., Herrera M., Mukherjee A., Kabanov A.V., Sahay G. Brief update on endocytosis of nanomedicines. Adv. Drug Deliv. Rev. 2019;144:90–111. doi: 10.1016/j.addr.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Patel S., Ashwanikumar N., Robinson E., DuRoss A., Sun C., Murphy-Benenato K.E., Mihai C., Almarsson Ö., Sahay G. Boosting intracellular delivery of lipid nanoparticle-encapsulated mRNA. Nano Lett. 2017;17:5711–5718. doi: 10.1021/acs.nanolett.7b02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Sayers E.J., Peel S.E., Schantz A., England R.M., Beano M., Bates S.M., Desai A.S., Puri S., Ashford M.B., Jones A.T. Endocytic profiling of cancer cell models reveals critical factors influencing LNP-mediated mRNA delivery and protein expression. Mol. Ther. 2019;27:1950–1962. doi: 10.1016/j.ymthe.2019.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Böhmert L., Voß L., Stock V., Braeuning A., Lampen A., Sieg H. Isolation methods for particle protein corona complexes from protein-rich matrices. Nanoscale Adv. 2020;2:563–582. doi: 10.1039/C9NA00537D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Zhang Y., Wu J.L.Y., Lazarovits J., Chan W.C.W. An analysis of the binding function and structural organization of the protein corona. J. Am. Chem. Soc. 2020;142:8827–8836. doi: 10.1021/jacs.0c01853. [DOI] [PubMed] [Google Scholar]