Abstract
Melanoma, originating from epidermal melanocytes, is a heterogeneous disease that has the highest mortality rate among all types of skin cancers. Numerous studies have revealed the cause of this cancer as related to various somatic driver mutations, including alterations in KIT—a proto-oncogene encoding for a transmembrane receptor tyrosine kinase. Although accounting for only 3% of all melanomas, mutations in c-KIT are mostly derived from acral, mucosal, and chronically sun-damaged melanomas. As an important factor for cell differentiation, proliferation, and survival, inhibition of c-KIT has been exploited for clinical trials in advanced melanoma. Here, apart from the molecular background of c-KIT and its cellular functions, we will review the wide distribution of alterations in KIT with a catalogue of more than 40 mutations reported in various articles and case studies. Additionally, we will summarize the association of KIT mutations with clinicopathologic features (age, sex, melanoma subtypes, anatomic location, etc.), and the differences of mutation rate among subgroups. Finally, several therapeutic trials of c-KIT inhibitors, including imatinib, dasatinib, nilotinib, and sunitinib, will be analyzed for their success rates and limitations in advanced melanoma treatment. These not only emphasize c-KIT as an attractive target for personalized melanoma therapy but also propose the requirement for additional investigational studies to develop novel therapeutic trials co-targeting c-KIT and other cytokines such as members of signaling pathways and immune systems.
Keywords: c-KIT protein, melanoma, mutation, therapeutics, clinical trial
OVERVIEW OF c-KIT AND ITS RTKs FAMILY
c-KIT or CD117 is a member of class III transmembrane receptor tyrosine kinases (RTKs) along with platelet-derived growth factor receptors (PDGFRs), fms like tyrosine kinase 3 (FLT3)/CD135, and macrophage colony stimulating factor receptors (M-CSFRs). It was discovered in 1987 as a cellular homologue of viral oncogene v-kit, which was isolated from a feline retrovirus.1,2 A variety of cell types were identified to express c-KIT including hematopoietic cells, germ cells, gastrointestinal (GI) tract Cajal cells, melanoma cells, B cell progenitors, and mast cells.
Wild-type c-KIT protein contains 976 amino acids (aa) divided into three main regions including an extracellular ligand-binding domain with 519 aa, a hydrophobic transmembrane domain with 23 aa, and an intracellular tail (Fig. 1).3,4 The extracellular domain consists of five immunoglobulin-like domains D1–D5, in which D1–D3 is responsible for stem cell factor (SCF) binding while D4–D5 contains motif for receptor dimerization. The 433 aa cytoplasmic region includes a juxta-membrane domain and a tyrosine kinase domain with an insertion of approximately 80 residues. Most of phosphorylation sites on c-KIT are located at the juxta-membrane region, the kinase insertion domain, and the C-terminal tail. Human c-KIT is encoded by a proto-oncogene located on the chromosome 4 at position of q11–12.5 This 90 kb gene is transcribed and translated into a core protein of 110 kDa, which is subsequently heterogeneously N-linked glycosylated, mainly in the extracellular domain closest to the plasma membrane, before maturing to a size of 145–160 kDa.2,6 c-KIT has four isoforms generated by alternative splicing mechanism.7,8,9 The first two isoforms differ in the presence of a glycine-asparagine-asparagine-lysine (GNNK) tetra-peptide adjacent to the extracellular transmembrane domain. The other two relate to the presence or absence of a serine amino acid at position 715 (Ser715) in the kinase insertion region.
Fig. 1. Structure of c-KIT receptor tyrosine kinase.
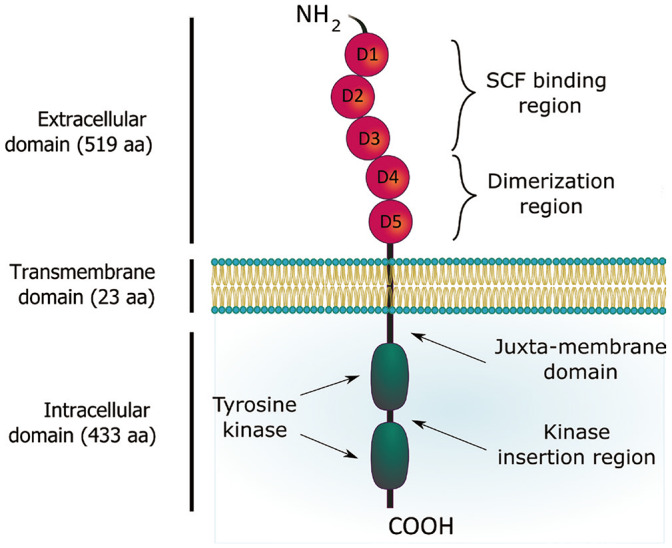
The ligand SCF of c-KIT is a hematopoietic cytokine, which signals to maintain survival of hematopoietic cells as well as to promote cell proliferation, differentiation and regulation of growth and development.10 Upon binding to D1–D3 region of c-KIT, a homodimer of SCF induces a conformational change that enables a homotypic interaction between D4–D5 regions of two adjacent c-KIT molecules.11,12 This dimerization allows a trans-phosphorylation of tyrosine residues in the intracellular region of each c-KIT monomer by the other, leading to signal transduction through plasma membrane. Many mutations in c-KIT have been found to perturb these characteristics. For example, a mutation in D4 at key residues disrupts the transmembrane regions of each monomer, thereby blocking the subsequent trans-phosphorylation of tyrosine kinase domains.13 However, this kind of mutation dramatically reduces tyrosine phosphorylation but does not influence the dimerization. A comprehensive summary of KIT mutations and their respective clinical implications will be further discussed in detail later.
Downstream signaling pathways of c-KIT
Many studies have been done on various cell lines to describe different downstream signaling cascades of c-KIT, including mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) pathway, PI3K/AKT pathway, Src family kinases pathway, phospholipases PLC-γ pathway, and JAK/STAT pathway.14,15,16,17,18 These signaling pathways can be activated independently or concomitantly by c-KIT and they are integrated into a signaling circuit. Since different pathways relate to different cell types as well as cancer types, we will review in detail only pathways that are mostly well-known and researched on in c-KIT derived melanoma, which are MAPK/ERK pathway and PI3K/AKT pathway (Fig. 2).
Fig. 2. c-KIT mediated signaling pathways.
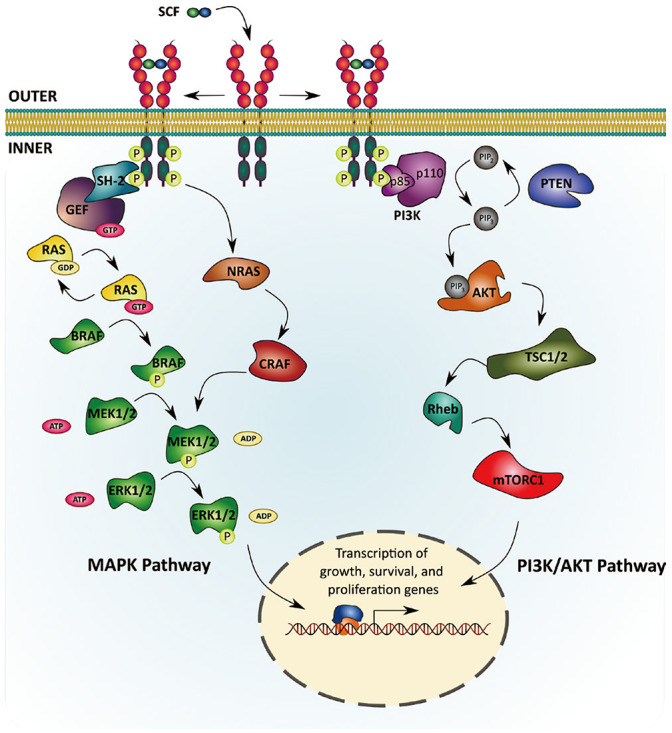
MAPK/ERK pathway
Through trans-phosphorylation by SCF binding, c-KIT is activated and recruits adaptor proteins containing a Src homology 2 (SH-2) domain. This SH-2 protein will then associate with a guanine nucleotide exchange factor (GEF) that exchanges guanosine triphosphate (GTP) and guanosine diphosphate (GDP). This SH-2/GEF complex activates the G-protein RAS by transferring its GTP.19 Activation of RAS leads to the activation of a serine/threonine protein kinase BRAF, which subsequently phosphorylates mitogen-activated protein kinase kinase1/2 (MAP2K1/2 or MEK1/2). MEK1/2, in turn, phosphorylates and activates ERK1/2. Several ERK1/2-activated transcription factors (TFs) induce the expression of genes related to cell proliferation, apoptosis, differentiation, adhesion, and mobility.20,21
PI3K/AKT pathway
PI3K/AKT pathway is responsible for cell survival and regulation of apoptosis. This signaling pathway can be activated via two mechanisms: either directly through interaction with c-KIT at position Tyr721 or indirectly through scaffold protein Gab2 and adapter protein Grb2.22,23 The p85 subunit of PI3K interacts with autophosphorylated c-KIT through its SH2 domain, changes conformation, and associates with the enzymatic p110 subunit to fully activate PI3K. That activation step also recruits PI3K to plasma membrane, placing it in close proximity of its lipid substrate, phosphatidylinositol 4,5-biphosphate (PIP2), which converts PIP2 to phosphatidylinositol 3,4,5-triphosphate (PIP3). The PIP3 then activates the pleckstrin homology (PH) domain, which contains proteins such as the serine/threonine kinase AKT.
AKT can activate several downstream proteins such as Bad, Foxo, and nuclear factor kappa-light chain enhancer of activated B cells (NF-κB) to interfere with the initiation of apoptosis and promote cell survival.24,25,26,27 AKT also activates mTORC1 or mTor complex 1 (mTor, Raptor, GβL, PRAS40, and Deptor) through downstream pathway members like TSC1/2 and Rheb. This mTORC1 relates to melanocyte proliferation and migration.28 The PI3K/AKT pathway can be inhibited by the phosphatase and tensin homolog (PTEN) gene. The PTEN protein removes one inorganic phosphate group from PIP3 to regenerate PIP2, which prevents the activation of AKT.
KIT interactions with other cytokines
It is well established that c-KIT interacts with various families of adaptor proteins, which contain multiple interaction domains, including SH2 and PH. Among growth factor receptorbound proteins, c-KIT was shown to interact with Grb7 at Tyr-936 position in the downstream signaling of cell migration.29,30 Another member in this family, Grb10, was found to interact with c-KIT to facilitate its PI3K-kinase-dependent activation and following association with AKT.31 CrkL, a member of Crk family, has its phosphorylation induced by c-KIT and can interact with the ubiquitin E3 ligase c-Cbl.32 Activation of c-KIT can also trigger the binding to its juxta-membrane region of Dok1, an adaptor that can interact with many signaling proteins such as Abl, SHIP, PLCγ1, and CrkL.33 Another interactor of c-KIT, Lnk, may negatively regulate function of c-KIT as lnk−/− mice had an enhanced hematopoiesis.34
Apart from adaptor proteins, c-KIT can interact with and facilitate functions of many different cytokines. For examples, in primary mast cells, SCF-induced activation of c-KIT is required to evoke optimal IL-33-induced cytokine production.35 c-KIT can also affect erythropoiesis as it can replace erythropoietin (Epo) to activate its receptor (Epo-R) by tyrosine phosphorylation and induce maturation of progenitors.36 Similar with the interaction between c-KIT and Epo-R, the interaction with IL-7 of c-KIT can indirectly stimulate Jak-Stat pathway in T-lymphoid cells under the absence of Stat5 activation.37 In myeloid cell line, interestingly, members of the transmembrane 4 superfamily (TM4SF), including CD9, CD63, and CD81, show their physical association and serve as negative modulators of c-KIT, thus, regulating its sensitivity to Steel factor (SLF) in hematopoietic progenitors.38
KIT mutations in melanoma
Dysregulation of c-KIT can affect cell proliferation, tumor growth, and metastasis in various cancer types such as gastrointestinal stromal tumors (GIST), leukemia (the first tumor found linked to KIT mutation), lung cancer, acute myeloid leukemia, and melanoma.3,7,39,40 In fact, KIT mutations (mainly in-frame deletions of exon 11) are found in 80% of GIST tumors.41,42 Apart from gene amplifications, KIT mutations in melanoma are almost all missense substitutions and widely distributed (Fig. 3). Table 1 shows a catalogue of 47 recorded KIT mutations based on data collected from a pooled analysis of 1635 patients samples from 12 recent melanoma genomics studies using cBioPortal (www.cbioportal.org) and several separate studies.43,44,45,46,47,48,49,50,51,52,53,54 KIT mutations are identified in 3% of all melanomas and more specifically in 36% of acral melanomas, 39% of mucosal melanomas, and 28% of melanomas on chronically sun-damaged (CSD) skin but none in melanomas on skin without CSD or non-CSD (NCSD).46 About 70% of KIT mutations in melanoma are localized to exon 11, most often a lysine-to-proline mutation at codon 576 (L576P), and to exon 13, most often a methionine-to-glutamic acid mutation at codon 642 (K642E). L576P affects the juxta-membrane domain and K642E affects a kinase domain. Both mutations lead to constitutive activation of c-KIT tyrosine kinase activity and subsequent induction of both MAPK and PI3K/AKT pathways.55 Interestingly, mutations in KIT almost never occur in conjunction with BRAF (V600E) and NRAS (G12/Q61) mutations thereby suggesting an epistatic relationship.56 Melanomas without these recurrent alterations in BRAF and NRAS have a significant enrichment for either KIT mutations or alterations in NF1, a downstream modulator in c-KIT/MITF signaling axis.48
Fig. 3. Wide distribution of KIT genetic alterations in melanoma (from cBioPortal).43,44,45,46,47,48,49,50,51,52,53,54.
Table 1. Catalogue of KIT Mutations in Melanomas.
| No | KIT mutation | Mutation type | Variation type | Copy number | Exon location | Corresponding region | Cancer type |
|---|---|---|---|---|---|---|---|
| 1 | G32V | Missense | G → T | ShallowDel | 2 | Extracellular domain | Cutaneous melanoma |
| 2 | P36Q | Missense | C → A | Gain | 2 | Extracellular domain | Cutaneous melanoma |
| 3 | V50L | Missense | G → C | - | 2 | Extracellular domain | Desmoplastic melanoma |
| 4 | S106F | Missense | C → T | Diploid | 2 | Extracellular domain | Cutaneous melanoma |
| 5 | S123F | Missense | C → T | Diploid | 3 | Extracellular domain | Cutaneous melanoma |
| 6 | L160V | Missense | T → G | - | 3 | Extracellular domain | Cutaneous melanoma |
| 7 | A207S | Missense | G → T | Diploid | 3 | Extracellular domain | Cutaneous melanoma |
| 8 | G226W | Missense | G → T | Gain | 4 | Extracellular domain | Cutaneous melanoma |
| 9 | T245M | Missense | C → T | Diploid | 4 | Extracellular domain | Cutaneous melanoma |
| 10 | P363S | Missense | C → T | Diploid | 6 | Extracellular domain | Cutaneous melanoma |
| 11 | G445E | Missense | G → A | - | 8 | Extracellular dimerization motif | Cutaneous melanoma |
| 12 | S451C | Missense | C → G | Gain | 9 | Extracellular dimerization motif | Cutaneous melanoma |
| 13 | P456Q | Missense | C → A | - | 9 | Extracellular dimerization motif | Cutaneous melanoma |
| 14 | P467Q | Missense | C → A | Diploid | 9 | Extracellular dimerization motif | Cutaneous melanoma |
| 15 | N463S | Missense | - | - | 9 | Extracellular dimerization motif | Mucosal melanoma |
| 16 | Y503del_insFAH | In-frame Ins | - → TTGCCC | Amp | 9 | Extracellular dimerization motif | Cutaneous melanoma |
| 17 | V532I | Missense | G → A | Diploid | 10 | Transmembrane domain | Melanoma |
| 18 | M541L | Missense | A → C | ShallowDel | 10 | Transmembrane domain | Melanoma of unknown primary |
| 19 | W557R | Missense | T → A/C | Amp | 11 | Juxta-membrane domain | Acral, mucosal, cutaneous melanoma |
| 20 | V559A | Missense | T → C | Diploid/ShallowDel | 11 | Juxta-membrane domain | Acral, mucosal, cutaneous melanoma |
| 21 | V559D | Missense | T → A | - | 11 | Juxta-membrane domain | Acral, mucosal melanoma |
| 22 | V569A | Missense | T → C | - | 11 | Juxta-membrane domain | Cutaneous melanoma |
| 23 | Y570H | Missense | - | - | 11 | Juxta-membrane domain | CSD melanoma |
| 24 | Y570_L576del | In-frame Del | TT → - | - | 11 | Juxta-membrane domain | Cutaneous melanoma |
| 25 | L576P | Missense | T → C | Amp | 11 | Juxta-membrane domain | Acral, mucosal melanoma |
| 26 | W582L | Missense | G → T | ShallowDel | 11 | Juxta-membrane domain | Cutaneous melanoma |
| 27 | K642E | Missense | A → G | Amp/Gain/Diploid | 13 | TKI domain/ATP-binding pocket | Acral, mucosal, cutaneous melanoma |
| 28 | V654A | Missense | - | - | 13 | TKI domain/ATP-binding pocket | Mucosal melanoma |
| 29 | T666I | Missense | C → T | - | 14 | Kinase insertion domain | Cutaneous melanoma, lentigo maligna melanoma |
| 30 | F681I | Missense | T → A | - | 14 | Kinase insertion domain | Desmoplastic Melanoma |
| 31 | L706F | Missense | C → T | Diploid | 14 | Kinase insertion domain | Cutaneous melanoma |
| 32 | M722I | Missense | G → T | Diploid | 15 | Kinase insertion domain | Cutaneous melanoma |
| 33 | Q775K | Missense | C → A | Diploid | 16 | Kinase domain | Cutaneous melanoma |
| 34 | G803C | Missense | G → T | Diploid | 17 | Kinase domain | Cutaneous melanoma |
| 35 | R815_D816delinsS | IF del | Diploid | 17 | Kinase domain | Cutaneous melanoma | |
| 36 | D816N | Missense | G → A | Amp | 17 | Kinase domain | Cutaneous melanoma |
| 37 | D820Y | Missense | - | - | 17 | Kinase domain | Mucosal melanoma |
| 38 | N822I | Missense | A → T | Amp | 17 | Kinase domain | Cutaneous melanoma |
| 39 | N822K | Missense | T → G | Gain | 17 | Kinase domain | Cutaneous melanoma |
| 40 | N822Y | Missense | A → T | Amp | 17 | Kinase domain | Cutaneous melanoma |
| 41 | A829P | Missense | - | - | 18 | Kinase domain | Mucosal melanoma |
| 42 | P838L | Missense | - | - | 18 | Kinase domain | Acral melanoma |
| 43 | V8521 | Missense | - | - | 18 | Kinase domain | Mucosal melanoma |
| 44 | A895T | Missense | G → A | - | 19 | Kinase domain | Cutaneous melanoma |
| 45 | P911L | Missense | C → T | - | 20 | Kinase domain | Cutaneous melanoma |
| 46 | R956Q | Missense | G → A | - | 21 | C-terminal tail | Melanoma |
| 47 | S967F | Missense | CC → TT | - | 21 | C-terminal tail | Cutaneous melanoma |
CSD, chronically sun-damaged.
CLINICAL IMPLICATIONS OF KIT MUTATIONS
Correlations between clinical features and KIT mutations
The KIT mutation rate and its association with various clinicopathological features remains controversial as results of different studies are inconsistent. Here, we summarize the data from Gong, et al's 2018 meta-analysis57 of the clinical implications of KIT mutations (Table 2). In this analysis, selected studies must satisfy three inclusion criteria: 1) KIT mutations detected in the tissue samples of human melanoma, but not in cell lines, 2) the incidence of KIT mutations according to clinicopathologic parameters was described in detail, and 3) studies were carried out on humans and published in English. This study collected data from 497 out of 5224 patients harboring KIT mutations, comprising 360 Asian patients and 137 White patients.
Table 2. Associations between KIT Mutations and Various Clinicopathological Features/Races of Melanomas.
| Clinicopathologic characteristics | OR | 95% CI | p value | Association with KIT mutation |
|---|---|---|---|---|
| Age (≥60 yr) | ||||
| Asian | 1.349 | 1.056–1.723 | 0.017 | Positive |
| White | 0.795 | 0.337–1.879 | 0.602 | None |
| Overall | 1.296 | 1.025–1.641 | 0.031 | Positive |
| Sex | ||||
| Asian | 1.134 | 0.910–1.412 | 0.264 | None |
| White | 0.860 | 0.426–1.735 | 0.674 | None |
| Overall | 1.106 | 0.897–1.364 | 0.347 | None |
| Mucosal melanoma | ||||
| Asian | 1.080 | 0.842–1.386 | 0.545 | None |
| White | 3.003 | 1.895–4.758 | <0.001 | Positive |
| Overall | 1.363 | 1.094–1.697 | 0.006 | Positive |
| Acral melanoma | ||||
| Asian | 1.361 | 1.087–1.702 | 0.007 | Positive |
| White | 1.435 | 0.901–2.286 | 0.128 | None |
| Overall | 1.374 | 1.123–1.682 | 0.002 | Positive |
| Cutaneous melanoma with NCSD | ||||
| Asian | 0.613 | 0.424–0.886 | 0.009 | Negative |
| White | 0.094 | 0.018–0.500 | 0.006 | Negative |
| Overall | 0.562 | 0.392–0.805 | 0.002 | Negative |
| Cutaneous melanoma with CSD | ||||
| Asian | 1.643 | 0.962–2.806 | 0.069 | None |
| White | 7.791 | 1.370–44.291 | 0.021 | Positive |
| Overall | 1.880 | 1.127–3.136 | 0.016 | Positive |
| Melanoma on the extremities | 0.294 | 0.105–0.820 | 0.019 | Negative |
| Breslow thickness | ||||
| >1 mm | 0.910 | 0.586–1.413 | 0.674 | None |
| >4 mm | 1.177 | 0.928–1.492 | 0.179 | None |
| Ulceration | 0.968 | 0.772–1.215 | 0.781 | None |
CSD, chronically sun-damaged; NCSD, non-CSD; OR, odds ratio; CI, confidence interval.
The authors reported that KIT mutations are more commonly found in older patients (≥60 years old) [odds ratio (OR)=1.296, 95% confidence interval (CI): 1.025–1.641; p=0.031] and are positively associated with both mucosal melanomas (OR=1.363, 95% CI: 1.094–1.697; p=0.006) and acral melanomas (OR=1.374, 95% CI: 1.123–1.682; p=0.002). When stratified by race, KIT was significantly associated with mucosal melanoma in White patients and acral melanoma in Asian patients. Additionally, a negative association was detected between KIT mutations and NCSD melanomas in both populations. However, there was a positive association between KIT mutations and CSD melanomas overall (OR=1.880, 95% CI: 1.127–3.136; p=0.016). KIT mutations are usually not found in melanomas that develop on the extremities and not correlated with melanoma on head/neck or trunk. Analyses also showed no significant relationship between KIT mutations and sex, Breslow thickness (either >1 mm or >4 mm), histological types, ulceration, mitotic rate, and tumor stages.
Overall, this meta-analysis documented a close association between KIT mutations and older age, acral mucosal subtypes of melanoma, and CSD sites, but did not find an association with histological subtypes or tumor stage. However, the results of this study are limited by the number of published data as well as the wide distribution of KIT mutations in various exons. The clinical implications of KIT mutations are informative clues for developing individualized therapies for patients, which will be reviewed in the next section.
Therapeutic trials of c-KIT inhibitors and success rates in melanomas
A variety of kinase inhibitors have been developed exploring c-KIT as a therapeutic target in melanoma, but not all drugs have been approved for clinical trials. In this section, we summarize data from 13 studies that reported the success rates of four widely used c-KIT inhibitors in melanoma treatments. Summary of those clinical trials in sample size, objective response rate (ORR), disease control rate (DCR), median progression-free survival (PFS), and median overall survival (OS) is shown in Table 3.
Table 3. Summary of Clinical Trials of KIT Inhibitors in KIT-Mutation Derived Melanomas.
| No. | KIT inhibitors (mg) | No. of patients | ORR (%) | DCR (%) | Median PFS (months) | Median OS (months) | Study |
|---|---|---|---|---|---|---|---|
| 1 | IMA 400 BID | 25 | 16 | 36 | 2.8 | 10.7 | Carvajal, et al.44 |
| 2 | IMA 400 QD or BID | 43 | 23.3 | 53.5 | 3.5 | 12.0 | Guo, et al.60 |
| 3 | IMA 400 QD or BID | 24 | 29.2 | 50 | 3.7 (TTP) | 12.5 | Hodi, et al.61 |
| 4 | IMA 400 QD | 78 | 21.8 | 60.3 | 4.2 | 13.1 | Wei, et al.62 |
| 5 | NIL 400 BID | 9 | 22.2 | 77.8 | 2.5 | - | Cho, et al.64 |
| 6 | NIL 400 BID | 19 | 15.8 | 52.6 | 3.3 (TTP) | 9.1 | Carvajal, et al.56 |
| 7 | NIL 400 BID | 42 | 16.7 | 57.1 | 3.3 | 11.9 | Lee, et al.67 |
| 8 | NIL 400 BID | 42 | 26.2 | 73.8 | 4.2 | 18.0 | Guo, et al.66 |
| 9 | NIL 400 BID | 25 | 16 | 64 | 6.0 | 13.2 | Delyon, et al.65 |
| 10 | DAS 70 BID | 36 | 5 | - | 2.0 | 13.8 | Kluger, et al.70 |
| 11 | DAS 70 BID | 22 | 18.2 | 50 | 2.1 | 7.5 | Kalinsky, et al.69 |
| 12 | SUN 50 QD | 10 | 40 | 50 | - | - | Minor, et al.72 |
| 13 | SUN 50 QD | 31 | 13 | 39 | 1.3 (TTP) | 4.3 | Decoster, et al.71 |
ORR, objective response rate; DCR, disease control rate; PFS, progression-free survival; OS, overall survival; BID, twice daily; QD, once daily; IMA, imatinib; NIL, nilotinib; DAS, dasatinib; SUN, sunitinib; TTP, time to progression.
Imatinib
Imatinib, or imatinib mesylate, which was initially developed as an inhibitor of the BCR-ABL fusion protein and PDGFR, was found to also inhibit c-KIT and considered an effective drug for treatment of patients with GIST.58,59 Clinical experience in four single-arm, open-label phase II trials of imatinib44,60,61,62 will be compared. The most recent study among these screened 78 patients for response after continuous treatment with 400 mg/day imatinib until intolerable toxicities or disease progression occurred. Mutations in patients were widely distributed in exons 9, 11, 13, 17, and 18, with 60.2% of mutations occurring in exon 11 or 13. L576P and K642E accounted for 24.3% of all mutations. The median PFS in the evaluable cohort was 4.2 months (95% CI: 1.9–6.4 months), and median OS was 13.1 months (95% CI: 9.6–16.7 months). A range of durability was recorded with 17 partial response (PR) patients, 30 stable disease (SD) patients, and 29 patients showing progressive disease (PD). Differences were found between patients with KIT mutations. Patients with KIT mutations in exon 11 or 13 had ORR of 24.4% and DCR of 66.7% while those for the group of other alterations had the rate of 19.4% and 54.8% respectively. This study observed generally mild to moderate adverse events (AEs), including edema (50%), rash (18%), fatigue (9%), anorexia (7%), nausea (5%), and neutropenia (2%). The additional three studies showed consistent results. These studies started with high dose of imatinib (400 mg twice a day), which was commonly associated with grades 3–4 AEs such as lymphopenia, anemia, erythema multiforme, and vomiting. Among four studies, complete responses (CRs) were only observed in the earliest study of Carvajal group with two patients durable for 95 and 94 weeks. Both of them had the exon 11 L576P mutation as well as amplification of KIT.44
Nilotinib
Nilotinib is another small kinase inhibitor with comparable or greater potency than imatinib in targeting KIT mutations.63 Five studies were conducted using this drug, in which all patients received orally nilotinib 400 mg twice a day.56,64,65,66,67 The most recent study was a phase II clinical trial conducted by the French Skin Cancer Network on 25 patients.65 At 6 months after nilotinib initiation, only four patients were responsive to nilotinib (ORR: 16%, 90% CI: 5.6–33.0%), including one CR patient and three PR patients. Out of the other 21 patients, six experienced progression and 15 died with an estimated PFS of 6.0 months and a median OS of 13.2 months (95% CI: 5.6–19.9 months). Of note, patients with CR or PR had mutations in exon 11 or 13 with overall response rate of 26%, median PFS of 6 months (95% CI: 3–46.8), and median OS of 14.4 months (95% CI: 9.97–not reached). Among patients, 56% experienced grade 3 drug-related AEs such as fatigue, rash, increased aspartate transaminase/alanine transaminase or cholestasis, and nausea. The author also observed reduced phosphorylated STAT3 in nilotinib treated KIT-mutated cell lines, suggesting that the JAK/STAT pathway can be downregulated by c-KIT inhibition and thus, associated with tumor response. However, additional mechanistic studies and molecular profiling are required, as nilotinib also targets PDGFR and ABL, which can both signal through STAT3. Other studies were conducted in patients with or without prior targeted KIT therapy. In the study of 19 patients, patients with L576P mutation showed a 25% ORR and 50% DCR while those for K642E mutation were 25% and 75% respectively.56 Of note, this study recorded two patients with reduction in size of brain metastases after nilotinib treatment by magnetic resonance imaging scanning.
Dasatinib
Dasatinib is also a c-KIT inhibitor, which also targets Src family kinases (c-Src, YES, LCK, and FYN), BCR-ABL, PDGFR-β, and EPHA2.68 Two studies of 36 and 22 evaluable patients respectively were conducted to assess clinical efficacy of dasatinib in KIT-mutated melanoma treatment.69,70 Both studies treated patients with various starting doses of dasatinib but eventually fixed at 70 mg twice a day due to toxicity. No CR case was observed in either study. In the first study, only two patients showed PR lasting 64 and 24 weeks, causing an ORR of 5%.70 Meanwhile, in the second study, four PRs and seven SDs were observed. Common grade 3 AEs were recorded, including fatigue (13%), dyspnea (12%), nausea (11%), anemia (7%), pleural effusion (5%), etc.
Sunitinib
Sunitinib, which targets both c-KIT and vascular endothelial growth factor receptors (VEGFRs), is also approved for treating melanoma. Sunitinib was examined in two studies of 10 and 31 evaluable patients harboring KIT mutations in 2012 and 2015, respectively.71,72 Both of studies started with sunitinib 50 mg daily before reducing to 37.5 and 25 mg per day. One CR with 15 months of durability was observed in the 2012 study. However, the 2015 study recorded no CR but observed 13% PR and 26% SD, which accounted for a DCR of 39%. The median PFS and median OS for the overall population in this study were 1.3 months (95% CI: 1.2–1.4) and 4.3 months (95% CI: 1.0–7.6), respectively. Most commonly recorded grade 3–4 AEs were asthenia (28%), thrombocytopenia (15%), anorexia (10%), and neutropenia (15%).
Several other kinase inhibitors targeting c-KIT were also clinically tested and recorded in case reports. For example, a patient with primary esophageal melanoma harboring KIT mutation in exon 11 showed significant response when being treated with oral masitinib.73 Masitinib treatment caused dysphagia and odynophagia disappeared within 1 week and reduced size of brain metastatic lesions and visceral lesions in the following month. Another example is a 79-year-old man at stage IV M1b metastatic anal mucosal melanoma showing CR upon sorafenib therapy.74
Epigenetic regulation of KIT
Several up/down regulations of KIT expression are caused by epigenetic changes, which includes DNA methylation and histone modifications. For example, in cardiac progenitor cells, KIT is upregulated via action of stromal cell-derived factor-1α (SDF-1α). SDF-1α, combined with CXCR4, inhibits expression and global activity of DNA methyltransferase (DNMT), which then leads to demethylation of c-KIT gene.75 In KIT-mutated mast cells, histone deacetylase inhibitors (HDACi) can decrease KIT mRNA levels, total c-KIT protein as well as cell surface c-KIT, followed later by major mast cell apoptosis.76 Some breast cancer cell lines lack the c-KIT expression due to hypermethylation of KIT promoter and treating these cell lines with methyltransferase inhibitor such as 5Aza-2dC can boost the level of KIT mRNA.77 KIT methylation is also recorded in squamous cell carcinoma of uterine cervix that overexpress c-KIT.78,79 The increased methylation of CpG islands in these skin cancer cells might interfere with the binding of CTCF repressor with KIT promoter. In GIST, a repressive complex named PRC [polycomb group (PcG) repressive complex] can reversibly suppress KIT expression via various histone modifications such as H3K27me3 and H2AK119ub1.80 In cutaneous melanoma, intriguingly, the presence of SCF leads to reduced KIT expression and increased methylation density at the KIT promoter.81 However, this epigenetic change shows no significant correlations with common genetic drivers such as BRAF, NRAS, and PTEN. This suggests that KIT may have a tumor-suppressive function in cutaneous melanoma.82,83,84 Supporting this tumorsuppressing role of KIT in melanoma, the hypermethylation of KIT is also associated with a lower OS rate, even when BRAF (V600E) is included for the survival risk prediction.85 Additionally, KIT is also found to be inhibited by microRNAs, including miR-221 and miR-222, which leads to differentiation blockade of the melanoma cells and subsequent proliferation.86 However, these microRNAs are repressed by a TF called promyelocytic leukemia zinc finger (PLZF); therefore, the silencing of PLZF can be a driver of cutaneous melanoma.
On the other hand, considering KIT as an oncogene, the epigenetic regulation of KIT is related to enhancers, those are differentially methylated regions (eDMRs) as melanomas progress from normal to primary tumors and then to metastases.87 Bell, et al.87 showed that the methylation patterns of eDMRs not only contributes to melanoma progression by overexpressing KIT but also distinguishes patient survival rates.
CONCLUSION
Overall, based on data from various studies and case reports, we created a catalogue of KIT mutations, though not sufficient, and their mostly associated melanoma subtypes. An understanding of mutational classes in melanoma will facilitate appropriate personalized treatments. Additionally, KIT mutations present distinct correlation with a number of different clinicopathologic features. KIT-mutated harboring melanomas are closely associated with older age, and acral, mucosal, or CSD sites but not with other histological parameters or tumor stage or sex. Intriguingly, no significant difference is recorded in the clinical association with KIT mutations between White populations and Asian populations although KIT mutation rate is lower in the latter one.
Upon treatment of melanoma subtypes, clinical efficacy in treating KIT-mutated melanoma has been evaluated with various small-molecular inhibitors of c-KIT, including imatinib, nilotinib, dasatinib, and sunitinib. Data collected from studies over 20 years has provided substantially critical insights into the therapeutic trials of these drugs and their success rate. However, in many cases, most patients eventually experience disease progression. One of possible explanation for this drug resistance is the frequent presence of brain and central nervous system metastases in advanced melanoma as the drug penetration is limited in these areas. With this being said, numerous additional investigational studies exploring c-KIT as a therapeutic target in combination therapy against melanoma. For instance, co-targeting c-KIT and its downstream pathways might be a plausible solution to control tumor progression. On the other hand, c-KIT inhibition showed its potential synergy with the immunological checkpoint blockade to develop antitumor effect.88,89 This is due to the ability of c-KIT inhibitors to enhance immune response such as increased T-cell activation and natural killer cell clonal expansion.90,91
ACKNOWLEDGEMENTS
This work was supported in part by a grant from the American Skin Association (to S.G.), the Melanoma Research Alliance (to H.T.) and by the generous donors to the Massachusetts General Hospital on behalf of melanoma research.
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Duc (Daniel) M. Pham and Hensin Tsao.
- Data curation: Duc (Daniel) M. Pham.
- Formal analysis: Duc (Daniel) M. Pham and Hensin Tsao.
- Funding acquisition: Hensin Tsao.
- Project administration: Hensin Tsao.
- Software: Duc (Daniel) M. Pham.
- Supervision: Hensin Tsao.
- Validation: all authors.
- Visualization: Duc (Daniel) M. Pham.
- Writing—original draft: Duc (Daniel) M. Pham.
- Writing—review & editing: all authors.
- Approval of final manuscript: all authors.
References
- 1.Besmer P, Murphy JE, George PC, Qiu FH, Bergold PJ, Lederman L, et al. A new acute transforming feline retrovirus and relationship of its oncogene v-kit with the protein kinase gene family. Nature. 1986;320:415–421. doi: 10.1038/320415a0. [DOI] [PubMed] [Google Scholar]
- 2.Yarden Y, Kuang WJ, Yang-Feng T, Coussens L, Munemitsu S, Dull TJ, et al. Human proto-oncogene c-kit: a new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987;6:3341–3351. doi: 10.1002/j.1460-2075.1987.tb02655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonescu CR. The GIST paradigm: lessons for other kinase-driven cancers. J Pathol. 2011;223:251–261. doi: 10.1002/path.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemmon MA, Ferguson KM. A new twist in the transmembrane signaling tool-kit. Cell. 2007;130:213–215. doi: 10.1016/j.cell.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 5.d'Auriol L, Mattei MG, Andre C, Galibert F. Localization of the human c-kit protooncogene on the q11-q12 region of chromosome 4. Hum Genet. 1988;78:374–376. doi: 10.1007/BF00291740. [DOI] [PubMed] [Google Scholar]
- 6.Qiu FH, Ray P, Brown K, Barker PE, Jhanwar S, Ruddle FH, et al. Primary structure of c-kit: relationship with the CSF-1/PDGF receptor kinase family--oncogenic activation of v-kit involves deletion of extracellular domain and C terminus. EMBO J. 1988;7:1003–1011. doi: 10.1002/j.1460-2075.1988.tb02907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crosier PS, Ricciardi ST, Hall LR, Vitas MR, Clark SC, Crosier KE. Expression of isoforms of the human receptor tyrosine kinase c-kit in leukemic cell lines and acute myeloid leukemia. Blood. 1993;82:1151–1158. [PubMed] [Google Scholar]
- 8.Reith AD, Ellis C, Lyman SD, Anderson DM, Williams DE, Bernstein A, et al. Signal transduction by normal isoforms and W mutant variants of the Kit receptor tyrosine kinase. EMBO J. 1991;10:2451–2459. doi: 10.1002/j.1460-2075.1991.tb07784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu WM, Dong WF, Minden M. Alternate splicing creates two forms of the human kit protein. Leuk Lymphoma. 1994;12:441–447. doi: 10.3109/10428199409073786. [DOI] [PubMed] [Google Scholar]
- 10.Chen SQ, Xiong AQ. The progress and implication of stem cell factor. Basic Med Sci Clin. 2002;22:385–390. [Google Scholar]
- 11.Philo JS, Wen J, Wypych J, Schwartz MG, Mendiaz EA, Langley KE. Human stem cell factor dimer forms a complex with two molecules of the extracellular domain of its receptor, Kit. J Biol Chem. 1996;271:6895–6902. doi: 10.1074/jbc.271.12.6895. [DOI] [PubMed] [Google Scholar]
- 12.Lemmon MA, Pinchasi D, Zhou M, Lax I, Schlessinger J. Kit receptor dimerization is driven by bivalent binding of stem cell factor. J Biol Chem. 1997;272:6311–6317. doi: 10.1074/jbc.272.10.6311. [DOI] [PubMed] [Google Scholar]
- 13.Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 14.Hendriks RW. Drug discovery: new Btk inhibitor holds promise. Nat Chem Biol. 2011;7:4–5. doi: 10.1038/nchembio.502. [DOI] [PubMed] [Google Scholar]
- 15.Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22:2954–2963. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 16.Silva CM. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene. 2004;23:8017–8023. doi: 10.1038/sj.onc.1208159. [DOI] [PubMed] [Google Scholar]
- 17.Summy JM, Gallick GE. Treatment for advanced tumors: SRC reclaims center stage. Clin Cancer Res. 2006;12:1398–1401. doi: 10.1158/1078-0432.CCR-05-2692. [DOI] [PubMed] [Google Scholar]
- 18.Yasuda T, Kurosaki T. Regulation of lymphocyte fate by Ras/ERK signals. Cell Cycle. 2008;7:3634–3640. doi: 10.4161/cc.7.23.7103. [DOI] [PubMed] [Google Scholar]
- 19.Kitamura Y, Hirotab S. Kit as a human oncogenic tyrosine kinase. Cell Mol Life Sci. 2004;61:2924–2931. doi: 10.1007/s00018-004-4273-y. [DOI] [PubMed] [Google Scholar]
- 20.Kuang D, Zhao X, Xiao G, Ni J, Feng Y, Wu R, et al. Stem cell factor/ c-kit signaling mediated cardiac stem cell migration via activation of p38 MAPK. Basic Res Cardiol. 2008;103:265–273. doi: 10.1007/s00395-007-0690-z. [DOI] [PubMed] [Google Scholar]
- 21.Liang J, Wu YL, Chen BJ, Zhang W, Tanaka Y, Sugiyama H. The C-kit receptor-mediated signal transduction and tumor-related diseases. Int J Biol Sci. 2013;9:435–443. doi: 10.7150/ijbs.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stankov K, Popovic S, Mikov M. C-KIT signaling in cancer treatment. Curr Pharm Des. 2014;20:2849–2880. doi: 10.2174/13816128113199990593. [DOI] [PubMed] [Google Scholar]
- 23.Sun J, Pedersen M, Rönnstrand L. Gab2 is involved in differential phosphoinositide 3-kinase signaling by two splice forms of c-Kit. J Biol Chem. 2008;283:27444–27451. doi: 10.1074/jbc.M709703200. [DOI] [PubMed] [Google Scholar]
- 24.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 25.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 26.Dhandapani KM, Wade FM, Wakade C, Mahesh VB, Brann DW. Neuroprotection by stem cell factor in rat cortical neurons involves AKT and NFkappaB. J Neurochem. 2005;95:9–19. doi: 10.1111/j.1471-4159.2005.03319.x. [DOI] [PubMed] [Google Scholar]
- 27.Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 28.Jeon S, Kim NH, Kim JY, Lee AY. Stem cell factor induces ERM proteins phosphorylation through PI3K activation to mediate melanocyte proliferation and migration. Pigment Cell Melanoma Res. 2009;22:77–85. doi: 10.1111/j.1755-148X.2008.00519.x. [DOI] [PubMed] [Google Scholar]
- 29.Han DC, Shen TL, Guan JL. The Grb7 family proteins: structure, interactions with other signaling molecules and potential cellular functions. Oncogene. 2001;20:6315–6321. doi: 10.1038/sj.onc.1204775. [DOI] [PubMed] [Google Scholar]
- 30.Thömmes K, Lennartsson J, Carlberg M, Rönnstrand L. Identification of Tyr-703 and Tyr-936 as the primary association sites for Grb2 and Grb7 in the c-Kit/stem cell factor receptor. Biochem J. 1999;341(Pt 1):211–216. [PMC free article] [PubMed] [Google Scholar]
- 31.Jahn T, Seipel P, Urschel S, Peschel C, Duyster J. Role for the adaptor protein Grb10 in the activation of Akt. Mol Cell Biol. 2002;22:979–991. doi: 10.1128/MCB.22.4.979-991.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sattler M, Salgia R, Shrikhande G, Verma S, Pisick E, Prasad KV, et al. Steel factor induces tyrosine phosphorylation of CRKL and binding of CRKL to a complex containing c-kit, phosphatidylinositol 3-kinase, and p120(CBL) J Biol Chem. 1997;272:10248–10253. doi: 10.1074/jbc.272.15.10248. [DOI] [PubMed] [Google Scholar]
- 33.van Dijk TB, van Den Akker E, Amelsvoort MP, Mano H, Löwenberg B, von Lindern M. Stem cell factor induces phosphatidylinositol 3′-kinase-dependent Lyn/Tec/Dok-1 complex formation in hematopoietic cells. Blood. 2000;96:3406–3413. [PubMed] [Google Scholar]
- 34.Takaki S, Morita H, Tezuka Y, Takatsu K. Enhanced hematopoiesis by hematopoietic progenitor cells lacking intracellular adaptor protein, Lnk. J Exp Med. 2002;195:151–160. doi: 10.1084/jem.20011170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drube S, Heink S, Walter S, Löhn T, Grusser M, Gerbaulet A, et al. The receptor tyrosine kinase c-Kit controls IL-33 receptor signaling in mast cells. Blood. 2010;115:3899–3906. doi: 10.1182/blood-2009-10-247411. [DOI] [PubMed] [Google Scholar]
- 36.Wu H, Klingmüller U, Besmer P, Lodish HF. Interaction of the erythropoietin and stem-cell-factor receptors. Nature. 1995;377:242–246. doi: 10.1038/377242a0. [DOI] [PubMed] [Google Scholar]
- 37.Jahn T, Sindhu S, Gooch S, Seipel P, Lavori P, Leifheit E, et al. Direct interaction between Kit and the interleukin-7 receptor. Blood. 2007;110:1840–1847. doi: 10.1182/blood-2005-12-028019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anzai N, Lee Y, Youn BS, Fukuda S, Kim YJ, Mantel C, et al. C-kit associated with the transmembrane 4 superfamily proteins constitutes a functionally distinct subunit in human hematopoietic progenitors. Blood. 2002;99:4413–4421. doi: 10.1182/blood.v99.12.4413. [DOI] [PubMed] [Google Scholar]
- 39.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol. 2004;22:3813–3825. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]
- 40.Maulik G, Bharti A, Khan E, Broderick RJ, Kijima T, Salgia R. Modulation of c-Kit/SCF pathway leads to alterations in topoisomerase-I activity in small cell lung cancer. J Environ Pathol Toxicol Oncol. 2004;23:237–251. doi: 10.1615/jenvpathtoxoncol.v23.i4.10. [DOI] [PubMed] [Google Scholar]
- 41.Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 42.Joensuu H, DeMatteo RP. The management of gastrointestinal stromal tumors: a model for targeted and multidisciplinary therapy of malignancy. Annu Rev Med. 2012;63:247–258. doi: 10.1146/annurev-med-043010-091813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carvajal RD, Antonescu CR, Wolchok JD, Chapman PB, Roman RA, Teitcher J, et al. KIT as a therapeutic target in metastatic melanoma. JAMA. 2011;305:2327–2334. doi: 10.1001/jama.2011.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 47.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang WS, Hendricks W, Kiefer J, Schmidt J, Sekar S, Carpten J, et al. Integrated genomic analyses reveal frequent TERT aberrations in acral melanoma. Genome Res. 2017;27:524–532. doi: 10.1101/gr.213348.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shain AH, Garrido M, Botton T, Talevich E, Yeh I, Sanborn JZ, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat Genet. 2015;47:1194–1199. doi: 10.1038/ng.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shtivelman E, Davies MQ, Hwu P, Yang J, Lotem M, Oren M, et al. Pathways and therapeutic targets in melanoma. Oncotarget. 2014;5:1701–1752. doi: 10.18632/oncotarget.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carvajal RD, Lawrence DP, Weber JS, Gajewski TF, Gonzalez R, Lutzky J, et al. Phase II study of nilotinib in melanoma harboring KIT alterations following progression to prior KIT inhibition. Clin Cancer Res. 2015;21:2289–2296. doi: 10.1158/1078-0432.CCR-14-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gong HZ, Zheng HY, Li J. The clinical significance of KIT mutations in melanoma: a meta-analysis. Melanoma Res. 2018;28:259–270. doi: 10.1097/CMR.0000000000000454. [DOI] [PubMed] [Google Scholar]
- 58.Heinrich MC, Griffith DJ, Druker BJ, Wait CL, Ott KA, Zigler AJ. Inhibition of c-kit receptor tyrosine kinase activity by STI 571, a selective tyrosine kinase inhibitor. Blood. 2000;96:925–932. [PubMed] [Google Scholar]
- 59.Tuveson DA, Willis NA, Jacks T, Griffin JD, Singer S, Fletcher CD, et al. STI571 inactivation of the gastrointestinal stromal tumor c-KIT oncoprotein: biological and clinical implications. Oncogene. 2001;20:5054–5058. doi: 10.1038/sj.onc.1204704. [DOI] [PubMed] [Google Scholar]
- 60.Guo J, Si L, Kong Y, Flaherty KT, Xu X, Zhu Y, et al. Phase II, openlabel, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J Clin Oncol. 2011;29:2904–2909. doi: 10.1200/JCO.2010.33.9275. [DOI] [PubMed] [Google Scholar]
- 61.Hodi FS, Corless CL, Giobbie-Hurder A, Fletcher JA, Zhu M, Marino-Enriquez A, et al. Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin. J Clin Oncol. 2013;31:3182–3190. doi: 10.1200/JCO.2012.47.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei X, Mao L, Chi Z, Sheng X, Cui C, Kong Y, et al. Efficacy evaluation of imatinib for the treatment of melanoma: evidence from a retrospective study. Oncol Res. 2019;27:495–501. doi: 10.3727/096504018X15331163433914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cullinane C, Natoli A, Hui Y, Conus N, Jackson S, Brüggen J, et al. Preclinical evaluation of nilotinib efficacy in an imatinib-resistant KIT-driven tumor model. Mol Cancer Ther. 2010;9:1461–1468. doi: 10.1158/1535-7163.MCT-09-1181. [DOI] [PubMed] [Google Scholar]
- 64.Cho JH, Kim KM, Kwon M, Kim JH, Lee J. Nilotinib in patients with metastatic melanoma harboring KIT gene aberration. Invest New Drugs. 2012;30:2008–2014. doi: 10.1007/s10637-011-9763-9. [DOI] [PubMed] [Google Scholar]
- 65.Delyon J, Chevret S, Jouary T, Dalac S, Dalle S, Guillot B, et al. STAT3 mediates nilotinib response in KIT-altered melanoma: a phase II multicenter trial of the French Skin Cancer Network. J Invest Dermatol. 2018;138:58–67. doi: 10.1016/j.jid.2017.07.839. [DOI] [PubMed] [Google Scholar]
- 66.Guo J, Carvajal RD, Dummer R, Hauschild A, Daud A, Bastian BC, et al. Efficacy and safety of nilotinib in patients with KIT-mutated metastatic or inoperable melanoma: final results from the global, single-arm, phase II TEAM trial. Ann Oncol. 2017;28:1380–1387. doi: 10.1093/annonc/mdx079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee SJ, Kim TM, Kim YJ, Jang KT, Lee HJ, Lee SN, et al. Phase II trial of nilotinib in patients with metastatic malignant melanoma harboring KIT gene aberration: a multicenter trial of Korean Cancer Study Group (UN10-06) Oncologist. 2015;20:1312–1319. doi: 10.1634/theoncologist.2015-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, et al. Discovery of N-(2-chloro-6-methyl-phenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J Med Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 69.Kalinsky K, Lee S, Rubin KM, Lawrence DP, Iafrarte AJ, Borger DR, et al. A phase 2 trial of dasatinib in patients with locally advanced or stage IV mucosal, acral, or vulvovaginal melanoma: a trial of the ECOG-ACRIN Cancer Research Group (E2607) Cancer. 2017;123:2688–2697. doi: 10.1002/cncr.30663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kluger HM, Dudek AZ, McCann C, Ritacco J, Southard N, Jilaveanu LB, et al. A phase 2 trial of dasatinib in advanced melanoma. Cancer. 2011;117:2202–2208. doi: 10.1002/cncr.25766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Decoster L, Vande Broek I, Neyns B, Majois F, Baurain JF, Rottey S, et al. Biomarker analysis in a phase II study of sunitinib in patients with advanced melanoma. Anticancer Res. 2015;35:6893–6899. [PubMed] [Google Scholar]
- 72.Minor DR, Kashani-Sabet M, Garrido M, O'Day SJ, Hamid O, Bastian BC. Sunitinib therapy for melanoma patients with KIT mutations. Clin Cancer Res. 2012;18:1457–1463. doi: 10.1158/1078-0432.CCR-11-1987. [DOI] [PubMed] [Google Scholar]
- 73.Prosvicova J, Lukesova S, Kopecky J, Grim J, Papik Z, Kolarova R, et al. Rapid and clinically significant response to masitinib in the treatment of mucosal primary esophageal melanoma with somatic KIT exon 11 mutation involving brain metastases: a case report. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2015;159:695–697. doi: 10.5507/bp.2015.061. [DOI] [PubMed] [Google Scholar]
- 74.Quintás-Cardama A, Lazar AJ, Woodman SE, Kim K, Ross M, Hwu P. Complete response of stage IV anal mucosal melanoma expressing KIT Val560Asp to the multikinase inhibitor sorafenib. Nat Clin Pract Oncol. 2008;5:737–740. doi: 10.1038/ncponc1251. [DOI] [PubMed] [Google Scholar]
- 75.Chen Z, Pan X, Yao Y, Yan F, Chen L, Huang R, et al. Epigenetic regulation of cardiac progenitor cells marker c-kit by stromal cell derived factor-1α. PLoS One. 2013;8:e69134. doi: 10.1371/journal.pone.0069134. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Lyberg K, Ali HA, Grootens J, Kjellander M, Tirfing M, Arock M, et al. Histone deacetylase inhibitor SAHA mediates mast cell death and epigenetic silencing of constitutively active D816V KIT in systemic mastocytosis. Oncotarget. 2017;8:9647–9659. doi: 10.18632/oncotarget.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Janostiak R, Vyas M, Cicek AF, Wajapeyee N, Harigopal M. Loss of c-KIT expression in breast cancer correlates with malignant transformation of breast epithelium and is mediated by KIT gene promoter DNA hypermethylation. Exp Mol Pathol. 2018;105:41–49. doi: 10.1016/j.yexmp.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 78.Chao WR, Lin WL, Chen CK, Han LM, Lin JC, Han CP. Unusual c-KIT+squamous cell carcinoma of the uterine cervix showing paradoxical hypermethylation of the c-KIT proto-oncogene. Eur J Obstet Gynecol Reprod Biol. 2015;184:130–131. doi: 10.1016/j.ejogrb.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 79.Chang SW, Chao WR, Ruan A, Wang PH, Lin JC, Han CP. A promising hypothesis of c-KIT methylation/ expression paradox in c-KIT (+) squamous cell carcinoma of uterine cervix ----- CTCF transcriptional repressor regulates c-KIT proto-oncogene expression. Diagn Pathol. 2015;10:207. doi: 10.1186/s13000-015-0438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Syed SA, Hayashi Y, Modak MV, Tice J, Nagy RA, McGehee CD, et al. 240 Epigenetic regulation of kit expression in murine KitLow interstitial cell of cajal stem cells (ICC-SC) and KitLow gastrointestinal stromal tumor (GIST) cells by polycomb group (PcG) repressive complexes (PRC) Gastroenterology. 2013;144(5 Suppl 1):S-52 [Google Scholar]
- 81.Dahl C, Abildgaard C, Riber-Hansen R, Steiniche T, Lade-Keller J, Guldberg P. KIT is a frequent target for epigenetic silencing in cutaneous melanoma. J Invest Dermatol. 2015;135:516–524. doi: 10.1038/jid.2014.372. [DOI] [PubMed] [Google Scholar]
- 82.Isabel Zhu Y, Fitzpatrick JE. Expression of c-kit (CD117) in Spitz nevus and malignant melanoma. J Cutan Pathol. 2006;33:33–37. doi: 10.1111/j.0303-6987.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 83.Montone KT, van Belle P, Elenitsas R, Elder DE. Proto-oncogene c-kit expression in malignant melanoma: protein loss with tumor progression. Mod Pathol. 1997;10:939–944. [PubMed] [Google Scholar]
- 84.Shen SS, Zhang PS, Eton O, Prieto VG. Analysis of protein tyrosine kinase expression in melanocytic lesions by tissue array. J Cutan Pathol. 2003;30:539–547. doi: 10.1034/j.1600-0560.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- 85.Ecsedi S, Hernandez-Vargas H, Lima SC, Vizkeleti L, Toth R, Lazar V, et al. DNA methylation characteristics of primary melanomas with distinct biological behaviour. PLoS One. 2014;9:e96612. doi: 10.1371/journal.pone.0096612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 87.Bell RE, Golan T, Sheinboim D, Malcov H, Amar D, Salamon A, et al. Enhancer methylation dynamics contribute to cancer plasticity and patient mortality. Genome Res. 2016;26:601–611. doi: 10.1101/gr.197194.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Balachandran VP, Cavnar MJ, Zeng S, Bamboat ZM, Ocuin LM, Obaid H, et al. Imatinib potentiates antitumor T cell responses in gastrointestinal stromal tumor through the inhibition of Ido. Nat Med. 2011;17:1094–1100. doi: 10.1038/nm.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Seifert AM, Zeng S, Zhang JQ, Kim TS, Cohen NA, Beckman MJ, et al. PD-1/PD-L1 blockade enhances T-cell activity and antitumor efficacy of imatinib in gastrointestinal stromal tumors. Clin Cancer Res. 2017;23:454–465. doi: 10.1158/1078-0432.CCR-16-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee KC, Ouwehand I, Giannini AL, Thomas NS, Dibb NJ, Bijlmakers MJ. Lck is a key target of imatinib and dasatinib in T-cell activation. Leukemia. 2010;24:896–900. doi: 10.1038/leu.2010.11. [DOI] [PubMed] [Google Scholar]
- 91.Kreutzman A, Juvonen V, Kairisto V, Ekblom M, Stenke L, Seggewiss R, et al. Mono/oligoclonal T and NK cells are common in chronic myeloid leukemia patients at diagnosis and expand during dasatinib therapy. Blood. 2010;116:772–782. doi: 10.1182/blood-2009-12-256800. [DOI] [PubMed] [Google Scholar]



