Abstract
Adeno-associated virus (AAV) vectors have been successfully used in patients with bleeding disorders and blindness. For human liver targeting, two major factors restrict effective AAV transduction after systemic administration of AAV vectors: human hepatocyte tropism and neutralizing antibodies (Nabs). In this study, we attempted to isolate AAV variants with the ability to transduce human hepatocytes and escape Nabs using a directed evolution approach in vivo. After four cycles of selection, 14 AAV capsid mutants were identified from a capsid shuffling library selected in the presence of human Intravenous Immunoglobulin (IVIG) and isolated from human hepatocytes xenografted into chimeric mice. AAV neutralization assays using IVIG showed that most of the mutants showed the Nab escape pattern in a manner similar to that of AAV8 or AAV9 and better than that of other AAV serotypes. Different mutants displayed varying capacities to escape Nab activity from individual serum samples collected from healthy subjects or hemophilia patients. The mutant AAV LP2-10 was found in 12 colonies out of 25, which was composed of capsids from AAV serotypes 2, 6, 8, and 9, with VP3 subunits derived from AAV8 swapped with AAV6 from residues 261 to 272. The mutant AAV LP2-10 manifested a higher ability than that of other serotypes to escape Nabs in IVIG and most human serum samples. After injection of AAV vectors encoding a self-complementary GFP cassette into chimeric mice, LP2-10 transduced human hepatocytes with efficiency similar to that of AAV8. In summary, AAV mutants can be isolated in humanized mice with both human hepatocyte tropism and the ability to evade Nab activity.
Keywords: AAV, human hepatocyte, tropism, Nabs, chimeric mice
Graphical Abstract
AAV mutant vectors were identified from a capsid shuffling library selected in the presence of human IVIG and isolated from human hepatocytes xenografted into chimeric mice. LP2-10, whose VP3 subunits were derived from AAV8 and AAV6, manifested the highest ability to escape Nabs in IVIG and most human serum samples.
Introduction
The adeno-associated virus (AAV) vector is a popular and effective transgene delivery vehicle and has been successfully applied in clinical trials for many diseases, such as Leber congenital amaurosis1 and hemophilia B.2, 3, 4 AAV is a single-stranded (ss) DNA virus that is composed of the Rep gene and the Cap gene flanked by the inverted terminal repeats. AAV Rep proteins are responsible for the viral replication life cycle and perform virus genome packaging. The AAV Cap gene encodes three alternatively spliced capsid proteins, which play a major role for virus tissue tropism. AAV vectors are generated by replacement of the AAV Rep and Cap genes with a therapeutic transgene cassette, and the Cap gene provided in trans may be modified to influence specific aspects of transgene delivery.
Although a therapeutic level of clotting factors has been achieved after liver targeting of AAV vectors encoding clotting factors VIII (FVIII) or IX (FIX) via systemic administration, two major concerns have been raised: low transduction efficiency of AAV vectors for human hepatocytes and the high prevalence of AAV neutralizing antibodies (Nabs) in the population. In pre-clinical trials for hemophilia therapy with AAV vectors, much more efficient hepatocyte transduction and protein expression were observed in animal models. Compared to patients in clinical trials using a similar amount of particles per kilogram of AAV vector, FIX levels were approximately 100-fold higher in mice and 10-fold higher in primates.5 These results indicate that AAV transduction efficiency in current animal models may not be predictive for clinical studies. It is imperative to develop an authentic animal model to examine AAV transduction efficiency in order to guide future clinical trials. Recently, chimeric mice xenografted with human hepatocytes have been used to study AAV vector tropism in human hepatocytes.6, 7, 8 To explore novel AAV vectors for enhanced transduction, genetic modification of the AAV capsid via rational design or directed evolution is a popular strategy. These approaches have been used to develop novel AAV mutants with high human hepatocyte tropism in chimeric mice,8 as well as primary human islet cells and human embryonic-stem-cell-derived β cells.9
To overcome AAV Nabs, several lab-based and clinical approaches have been investigated, including coating the AAV virion surface to avoid Nab recognition,10 elimination of Nabs by plasmapheresis, elimination of B cells with antibodies,11 utilization of AAV empty virions as a decoy,12,13 and genetic modification of AAV capsids to modify epitopes recognized by Nabs.14,15 Engineering of AAV capsids represents a very promising strategy to develop novel AAV vectors with the ability to evade Nabs. Similar to transduction enhancement with genetic modification of AAV capsid, the approaches with rational design and directed evolution have also been used to exploit AAV variants for Nab evasion.14 In order to isolate AAV mutants with the ability to both evade Nabs and transduce human hepatocytes, we have used a combined strategy of directed evolution with an AAV shuffling library in chimeric mice xenografted with human hepatocytes in the presence of human Nabs. After 4 cycles of selection in mice in the presence of human IVIG, one mutant (AAV LP2-10) was the dominant isolate, which was composed of capsids derived from AAV2, -6, -8, and -9. Nab analysis showed that the mutant AAV LP2-10 had an increased ability to escape Nab activity not only from IVIG but also from sera of healthy subjects or hemophilia patients when compared to that of AAV serotypes. However, the mutant AAV LP2-10 did not show a higher human hepatocyte tropism than AAV8 in chimeric mice.
Results
Characterization of Isolated AAV Mutants from Human Hepatocytes
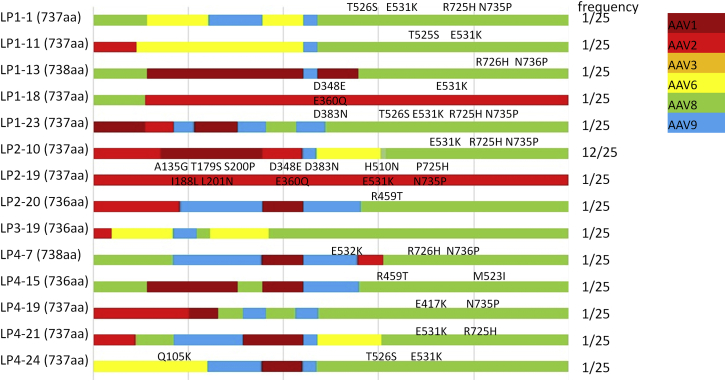
To isolate AAV variants with the ability to escape Nabs and maintain human hepatocyte tropism, we performed an in vivo selection in the presence of Nabs in chimeric humanized mice xenografted with human hepatocytes (Figure 1). After passively transferring 30 mg human IVIG containing anti-AAV Nabs into naive mice via retro-orbital injection, mice were systemically administered 2 × 1011 particles of the AAV capsid shuffling library; then adenovirus dl309 was applied to increase AAV genome replication and virion assembly in human hepatocytes. Three days later, human chimeric mouse livers were collected for the generation of cell lysate. The cell lysate was used to infect mice with pre-IVIG treatment for a second cycle of AAV amplification in human hepatocytes, as described earlier. Such amplification was repeated for two more cycles, and then livers were harvested for DNA extraction. PCR for AAV capsid DNA was performed, and its products were cloned into pXR plasmids. The capsid DNA in the different clones was sequenced. A total of 25 validated clones were recovered, where one capsid AAV LP2-10 was found in 12 clones, and the other 13 capsids were each detected in only a single clone. Except for two capsids (AAV LP2-20 and AAV LP4-15) with the C terminus from AAV2, all other capsid mutants had the C terminus derived from AAV8 with different point mutations (Figure 2). Seven mutants had AAV2 N-terminal sequences, 5 mutants had AAV8 N- terminal sequences, and the N-termini of the other 2 mutants were derived from AAV1 and AAV3. Specifically, the mutant AAV LP2-10 capsid was composed of domains from AAV2, AAV6, AAV8, and AAV9. Its VP3 subunit was from AAV8, with a swap of AAV6 from residue 261 to residue 272 (Figure 2). The mutation E531K in the AAV8 capsid was reserved in most clones.
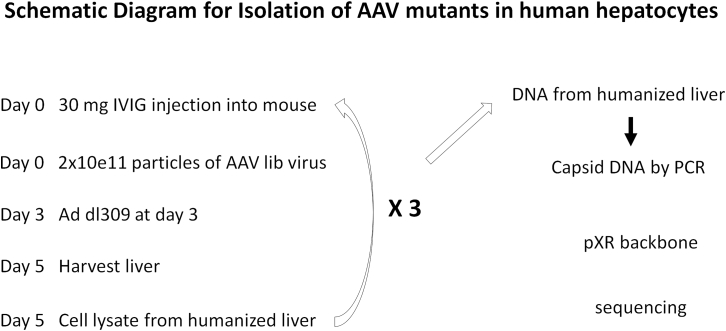
Figure 1.
Schematic Diagram for Isolation of AAV Mutants from Hepatocytes of Humanized Mice
Humanized mice were first pretreated by injecting 30 mg IVIG via retro-orbital vein and then administered 2 × 1011 particles of AAV capsid shuffling library viruses. Three days later, adenovirus dl309 was injected into the mice, and at day 5, hepatocytes in the mice were harvested and lysed. The AAV in the cell lysate from humanized liver was re-injected into another group of IVIG-pretreated mice for the next round of AAV amplification as described above. The AAV amplification was repeated three times more for a total of four rounds of selection. After the last round of AAV amplification, DNA was extracted from the humanized liver, and the AAV capsid DNA was amplified using capsid specific primers and then reconstructed into the pXR backbone for sequencing and AAV vector production.
Figure 2.
Schematic Capsid Diagram for AAV Mutants Isolated from the Livers of Humanized Mice
Different colors are used to indicate the composition of capsids from different AAV serotypes, and point mutations are indicated on the top of each clone diagram. The frequency representation indicates the number of capsids isolated from a total of 25 available clones.
Next, we studied the vector production efficiency from these mutants, as shown in Figure 3. Four mutants (AAV LP1-11, LP1-13, LP4-15, and LP4-21) generated low-yield AAV vectors and were not pursued for further experimentation. When compared to AAV2 vector yields, the titers of most recovered AAV mutant vectors, including AAV LP2-10, were a little lower but generally comparable to the yields of AAV1, AAV8, and AAV9 (Figure 3A). Next, these vectors were used to determine transduction efficiency in culture using Huh7 cells. Most of the selected vectors had lower transduction efficiency in cell culture than AAV2, -3, and 6 but performed similarly to or better than AAV8. The exceptions were 3 mutants (AAV LP1-18, LP2-19, and LP4-7), which induced transduction at a rate similar to or less than that of AAV9 (Figure 3B).
Figure 3.
AAV Packaging and Transduction Efficiency of Isolated AAV Capsid Mutants
(A) Packaging efficiency of individual AAV mutants. The production efficiency of mutants and wild-type AAV vectors was normalized to AAV2. (B) Transduction efficiency in Huh 7 cells from mutant AAV vectors. Huh7 cells were infected with 1 × 104 particles per cell of AAV vectors encoding the luciferase gene in 96-well plates, and 48 h later, the transduction efficiency was measured by luciferase assay.
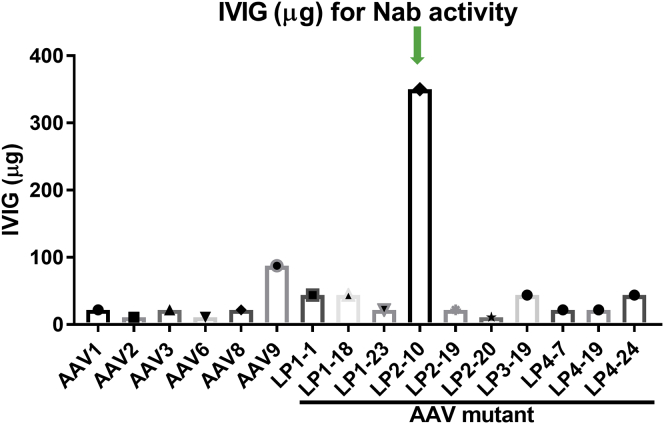
Nab Evasion Capacity of AAV Mutants for Human IVIG
Since these selected vectors were isolated from the liver of humanized mice in the presence of human IVIG, we studied the pattern of neutralizing activity of IVIG against these mutant capsids. Not surprisingly, a very low concentration of IVIG was able to inhibit transduction from AAV2 and AAV6. This was followed by increasing concentrations of IVIG needed to neutralize AAV1, AAV3, and AAV8, while more IVIG was needed to block AAV9 transduction (Figure 4). Most of the selected capsids had a Nab profile similar to that of AAV8. Mutant AAV LP2-20 had the lowest capacity to evade IVIG Nab activity, similar to AAV2. Strikingly, mutant AAV LP2-10 showed the highest ability to escape IVIG neutralizing activity, needing much more IVIG to block its transduction than AAV9 (Figure 4). This result indicates that AAV capsids can be selected on and isolated from humanized mouse livers in the presence of Nabs with the ability to escape the cognate Nab activity.
Figure 4.
AAV Neutralization Analysis of AAV Mutants against Human IVIG
1 × 109 particles of AAV/luc vectors were pre-incubated with different concentrations of IVIG for 1 h at 4°C, and then the mixtures were added into the cell culture medium of Huh7 in a 48-well plate. Forty-eight hours later, the luciferase expression was measured, and the Nab titer was determined.
The Profile of Nabs from Normal Human Serum against AAV Mutants Isolated from Chimeric Mouse Livers
The results described earlier demonstrated that the selected capsid AAV LP2-10 had a significant capability to resist the Nab activity of human IVIG. Next, we investigated whether these selected capsids isolated from the liver of humanized mice had an improved Nab profile for serum from individual healthy subjects. Twenty serum samples collected from the healthy human population were tested. Generally, the prevalence of Nabs to AAV2 and AAV3 was high (Figure 5A). The titer of Nabs to AAV9 was the lowest among the tested AAV serotypes. The pattern of Nabs for some AAV mutants was similar to those for AAV9. The Nab titer for AAV LP2-10 was lower than 1:20 dilution in a majority of subjects, and a Nab titer over 1:100 was only observed in one subject (Figure 5A). Since mutant AAV LP2-10 is composed of capsids derived from AAV2, -6, -8, and -9, we compared the prevalence of Nabs in healthy human subjects for mutant AAV LP2-10 and these four parental AAV serotypes. As shown in Figure 5B, when compared to AAV2, -6, and -8, mutant AAV LP2-10 has a significantly lower Nab titer in healthy individuals (p < 0.05). Mutant AAV LP2-10 had a low Nab titer similar to that of AAV9 (Figure 5B).
Figure 5.
The Prevalence and Titer of Nabs against AAV Mutants in Healthy Subjects and Hemophilia Patients
(A) The AAV Nab prevalence in healthy human subjects. (B) The comparison of Nab titers for mutant AAV LP2-10 and naturally occurring serotypes in healthy individuals. (C) The AAV Nab prevalence in hemophilia patients. (D) The comparison of Nab titers for mutant AAV LP2-10 and naturally occurring serotypes in hemophilia patients. ∗p < 0.05 when compared to other serotypes for Nab titer of the mutant AAV LP2-10. NS, no significance.
The Profile of Nabs from Hemophilia B Patients’ Serum against AAV Mutants Isolated from Chimeric Mouse Livers
AAV vector-mediated gene delivery has been successfully applied in patients with hemophilia B via liver targeting. To explore the profile of Nabs against AAV mutants isolated from human hepatocytes in the presence of human IVIG, we performed the Nab analysis in 26 patients with hemophilia B(Figure 5C). Similar to findings in healthy subjects, Nabs with high titers (over 1:20) were observed in AAV2 and AAV3 followed by AAV6 and AAV8. For some AAV mutants (AAV LP1-1, LP1-18, LP2-10, LP4-7, and LP4-24), no Nab titer over 1:100 was found. Specifically, only 8 patients had Nabs (>1:2) to AAV LP2-10. The case number with Nabs was increased to 8, 19, 24, and 14 for AAV1, AAV2, AAV8, and AAV9, respectively. Similar to the results against the sera from healthy subjects, a significantly lower titer of Nabs for mutant AAV LP2-10 was detected when compared to all other AAV serotypes except for AAV9 (Figure 5D). Nab analysis studies using either IVIG or sera from individual human subjects indicate that we are able to successfully isolate AAV mutants from human hepatocytes of chimeric mice with the capacity to evade Nabs.
Human Hepatocyte Transduction with Mutant AAV LP2-10 in Chimeric Mice
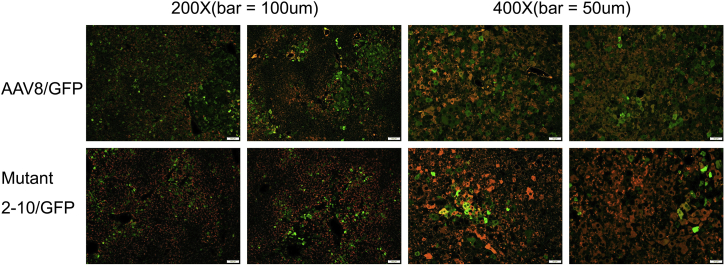
Based on the results from Nab analysis, mutant AAV LP2-10 demonstrated the ability to resist neutralization from IVIG and also from sera of individual human subjects. Next, we examined the transduction efficiency of AAV LP2-10 in human hepatocytes in vivo. Since VP3 of AAV LP2-10 is composed of capsid sequences from AAV6 and AAV8, and our previous studies have demonstrated that AAV8 had higher human hepatocyte tropism than AAV6 in mice xenografted with human hepatocytes,16 we used AAV8 as a control for direct in vivo comparison. The self-complementary (sc) AAV vectors encoding GFP at a dose of 1 × 1011 particles were injected into humanized mice via retro-orbital vein. At week 3 following AAV injection, the livers from the mice were collected. Human hepatocyte transduction was assessed by immunohistochemistry staining (Figure 6A) and flow cytometry (Table 1). Although the mutant AAV LP2-10 showed an improved ability to evade human Nabs as described earlier, human hepatocyte transduction was similar to or slightly lower than AAV8 transduction using immunohistochemistry in two separate xenografted mouse experiments with human hepatocytes of two different donors. Flow cytometry quantification of transduced cells showed that AAV LP2-10 induced human hepatocyte transduction similar to AAV8 transduction.
Figure 6.
The Transduction Profile of Mutant AAV LP2-10 in Human Hepatocytes of Chimeric Mice
1 × 1011 particles of scAAV/GFP (AAV8 or mutant AAV LP2-10) were injected into human hepatocyte xenografted mice via retro-orbital vein, and 3 weeks later, the liver was harvested and analyzed by immunohistochemistry. The antibody against human albumin was used to identify human hepatocytes (Red), and GFP expression was stained in green.
Table 1.
Human Hepatocyte Transductions by Mutant AAV LP2-10 in Xenograft Mice
| Donor No. and Mouse ID | AAV | IHC Method |
Flow Method |
|
|---|---|---|---|---|
| Transduction (%) | Transduction (%) | MFI | ||
| Donor 1 | ||||
| 2905 | 8 | 9.7 ± 2.7 | nd | nd |
| 2906 | 8 | 7.7 ± 2.7 | nd | nd |
| 2911 | LP2-10 | 4.5 ± 2.5 | nd | nd |
| 2912 | LP2-10 | 6.6 ± 5.4 | nd | nd |
| Donor 2 | ||||
| 39864 | 8 | 7.4 ± 2.7 | 15.6 | 26.1 |
| 39976 | 8 | 7.5 ± 2.0 | 4.5 | 27.1 |
| 39888 | LP2-10 | 5.0 ± 2.0 | 5.8 | 27.4 |
| 39893 | LP2-10 | 4.5 ± 2.0 | 6.4 | 41.8 |
| 39897 | LP2-10 | 4.0 ± 2.1 | 4.7 | 22.8 |
nd, not done.
Discussion
Our previous studies have demonstrated that AAV8 has a relatively high human hepatocyte tropism in xenografted mice when compared to other serotypes, as does AAV7.16 In this study, we successfully isolated several AAV mutants from the livers of humanized mice in the presence of IVIG using the directed evolution approach with an AAV capsid shuffling library. A majority of the mutants (9/10) had a C terminus derived from AAV8, which may support the high human hepatocyte transduction of AAV8. Several selected mutants demonstrated a greater ability to evade Nabs from the IVIG and sera of healthy subjects as well as hemophilia patients when compared to naturally occurring AAV serotypes. Specifically, we recovered a mutant AAV capsid LP2-10, the most highly represented isolate, which has significant identity overlap with the VP3 subunit of AAV8 and had the highest IVIG Nab escape capacity of any other isolates or natural serotypes. The human hepatocyte transduction of LP2-10 was similar to that of AAV8.
Overcoming Nab activity is a major challenge for AAV gene therapy in clinical trials, especially for patients who need systemic administration of AAV vectors. Approximately 50% of the human population is positive for AAV Nabs, and higher titer polyclonal sera demonstrate cross-reactivity between multiple AAV serotypes. The existence of Nabs can prevent patients from enrolling in clinical trials or benefitting from effective AAV-mediated gene therapy. Several approaches that have been explored to evade AAV Nabs have been used in clinical trials, including clinical methods and lab-based approaches such as the utilization of B cell depletion,17,18 plasma apheresis,19 and using empty particles as a decoy.20 However, these methods show low efficacy and cause unwanted side effects or complications in that they require either long-term administration or multiple cycles and also can create competition for AAV transduction and increase AAV capsid antigen load. More attention has been paid on genetic engineering of the AAV capsids to evade AAV Nabs, including rational design and directed evolution. Rational design requires detailed information of identified AAV immunogenic epitopes and the knowledge of AAV three-dimensional structures. However, the advantage of directed evolution is that this information and this knowledge are not required to develop immunogenically novel AAV capsids. Most studies that have isolated Nab-evading AAV mutants used either selection in animal models in the absence of AAV Nabs or selection from cell lines in vitro in the presence of Nabs. In this study, both approaches were combined to isolate Nab-escaping mutants with validated human tissue transduction using an in vivo animal model. It is well known in AAV vector gene therapy that pre-clinical results generated from mice have not directly translated to larger animals and human clinical trials. For example, in hemophilia B AAV gene therapy, over 100-fold lower FIX expression was detected in patients than in a mouse model when a similar dose of AAV vector per kilogram of body weight was used.4 Furthermore, mutant capsids that have been selected from mouse tissues lack the same transduction efficiency or tropism when tested on human or primate tissues (e.g., AAV9-PHP.B). It is, therefore, imperative to perform the selection process using humanized animal models and to isolate AAV mutants in the presence of human Nabs for human clinical applications. The mutants isolated from these systems are most likely to retain human-tissue-specific tropism and the ability to resist Nabs.
There are a few examples of humanized mice that have been used to test AAV transduction efficiency and isolate AAV variants with human-tissue-specific tropism. Specifically, mice xenografted with human hepatocytes have been used to develop novel AAV mutants with higher transduction efficiency. Some mutants isolated from the humanized mice showed higher human hepatocyte transduction as well as a substantially greater ability to evade Nabs when compared to the parental serotypes using AAV shuffling library.8 In this study, we successfully isolated several AAV variants from the liver of chimeric mice xenografted with human hepatocytes in the presence of human Nabs. A few of these selected mutants demonstrate better IVIG evasion ability than AAV8; however, among natural serotypes, AAV9 has the greatest potential to resist Nab activity from IVIG. Only one mutant isolate, AAV LP2-10, showed a higher capacity for IVIG evasion when compared to AAV9. It is interesting to note that AAV LP2-10, which was the most highly represented isolate in the pool, needs 4-fold more IVIG than AAV9 for neutralization. This result strongly suggests that it is possible to isolate AAV mutants with greater potential to evade pooled Nabs than parental AAV serotypes using directed evolution strategy combined with an AAV capsid shuffling library. However, when individual human samples from healthy subjects or hemophilia patients were tested, the Nab escape profile of isolated mutants differs between individuals. Mutants including AAV LP2-10 did not show significantly increased Nab evasion potential in most individual patient samples when compared to AAV9 (Tables S1 and S2). This is consistent with our previous findings that AAV mutants isolated from muscles in the presence of individual patient serum had a better ability to escape the Nabs from cognate serum but not necessarily from sera of other subjects.21 This consistent result provides insight about the potential limitation of selecting AAV mutants for Nab evasion using a directed evolution approach in the presence of Nabs.
Based on the interaction of the monoclonal antibody with the AAV virion, it has been identified that residue 265 on AAV2 can be recognized by the A20 antibody, which is able to bind to intact AAV2 virions and block AAV2 transduction.22,23 Our previous studies have demonstrated that modification of the residue 265 demonstrated a decreased ability to bind to A20 and resisted A20 neutralizing activity.23 In support of this finding, this study shows most selected mutants demonstrated swapping of the domain covering residue 265 from other serotypes while retaining the C terminus from AAV8. For example, the mutant LP2-10 has the domain from aa residues 261–272 of AAV6, which may suggest that the domain replacement with AAV6 involving the residue 265 may offer higher resistance of mutant AAV LP2-10 to IVIG when compared to AAV8 and AAV6.
The point mutation E531K on the AAV8 C terminus may also play a helping role for the mutants to resist Nabs.24 Among 14 selected AAV mutants, only 5 mutants, including AAV LP3-19 and AAV LP4-19, did not have the E531K point mutation. Compared to AAV8, we found that a similar Nab profile was observed in both mutants AAV LP3-19 and AAV LP4-19 in human sera, especially from hemophilia B patients. According to previous reports, the E531K mutation is responsible for the heparin binding ability of AAV6, which might have influenced the virus titer, transduction efficiency, and penetration of the blood-brain barrier (BBB).25 Additionally, the E531K mutation in AAV capsids could also affect Nab resistance due to E531K being located in a basic cluster near the spikes that surround the icosahedral 3-fold axes of the AAV capsid.23 We also identified several novel mutations on the AAV8 capsid in isolated mutants, including E417K, H510N, T526S, R725H, and N735P. Their function for Nab resistance is unknown and worth further investigation. Most of the AAV mutants isolated from this study contained a highly variable composition of the N terminus from different capsid serotypes; thus, it is impossible to exclude the role of a chimeric N terminus on these mutants in Nab resistance. Further experiments are needed to warrant the potential role of the N terminus in Nab evasion.
Similar to our previous finding that the mutants isolated from mouse muscle in the presence of Nabs did not induce higher transduction than AAV6,26 which is the best serotype for mouse muscle tropism, the mutant AAV LP2-10 induced similar or slightly lower human hepatocyte transduction than AAV8 in a xenograft mouse model. Again, the slightly lower transduction of human hepatocytes from mutant AAV LP2-10 may result from the substitution of the AAV8 domain with an AAV6 domain in the VP3 subunits. Our previous studies have demonstrated that modification of the amino acid (aa) residue 265 (using VP1 numbering) impacts AAV transduction,27 and a recent study showed that the swap of the 265 domain from AAVrh10, which crosses the BBB in mice, into AAV1 enabled AAV1 to transport across the BBB.28 These studies demonstrated that the domain involving residue 265 plays a fundamental role in AAV function in addition to Nab recognition. It has been demonstrated that AAV6 induces much lower human hepatocyte transduction than AAV8 in chimeric humanized mice.16 The combination of these observations and slightly lower transduction of mutant AAV LP2-10 in human hepatocytes may allow us to conclude that the domain around residue 265 confers high AAV8 human hepatocyte tropism.
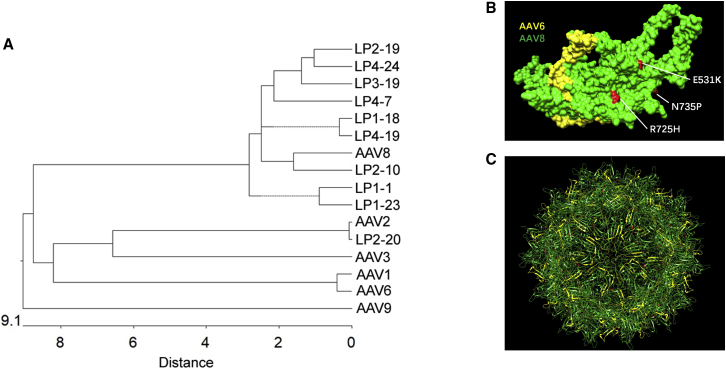
Structural studies have demonstrated the role of different domains on the AAV surface in AAV transduction efficiency and tissue tropism. These domains are also the sites for Nab recognition. We performed phylogenetic analysis of AAV mutants isolated from the liver of humanized mice and their parental serotypes and found that most mutants were located in the same clade as AAV8. It is interesting to note that the most popular and effective Nab evasion mutant, AAV LP2-10, is phylogenetically closer to AAV8 than to other mutants (Figure 7A). Finally, we performed structural modeling of mutant AAV LP2-10 to determine the structure-function relationship (Figure 7B). The domain of mutant AAV LP2-10 varied from the AAV8 capsid sequence between aa residues 261 and 272, which is derived from AAV1/AAV6. This domain is included in variable region (VR)I of the AAV virion, and it has been implicated that VRI is involved in AAV transduction and Nab recognition.29 The results from this study support that VRI on the AAV8 capsid plays a role in high human hepatocyte transduction and Nab recognition, while genetic modification of VRI may change transduction efficiency and Nab evasion.
Figure 7.
Phylogeny of AAV Mutants and Structural Modeling of Mutant AAV LP2-10
(A) A phylogenetic tree was used to analyze the genetic distance between wild-type AAVs and Nab-escaping AAV mutants isolated from the livers of humanized mice based on DNA sequence. (B) The fragments and point mutations on LP2-10 capsid subunit were structurally analyzed. Capsid domains derived from different AAV serotypes in the VP3 subunit of the mutant AAV LP2-10 and point mutations are highlighted as follows (according to VP1 numbering): yellow (AAV6), green (AAV8), and red (point mutations at residues 531, 725, and 735 of AAV8). (C) The fully assembled mutant AAV LP2-10 virion (60-mer) was displayed based on the AAV8 VP3 subunit from (B).
In summary, we were able to isolate AAV capsid mutants with the ability to evade AAV Nabs in humanized mice, while the transduction efficiency of some isolates was slightly comprised when compared to the best serotypes in the AAV shuffling library. In the future, more effective strategies need to be developed to isolate AAV mutants with the potential to evade Nab activity while maintaining high human hepatocyte transduction efficiency.
Materials and Methods
Human Sera
Normal human serum samples were purchased from Valley Biomedical (Winchester, VA, USA). The sera from hemophilia patients were collected in The Hemophilia Treatment Center of the School of Medicine at UNC at Chapel Hill (UNC IRB#15-2126). Human IVIG was purchased from Grifols Therapeutics (Research Triangle Park, NC, USA). All sera and IVIG solutions were aliquoted and stored in −80°C.
Cells and Mice
HEK293 and Huh7 cell lines were incubated at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin.
The chimeric mice xenografted with human hepatocytes with 70% repopulation were purchased from Yecuris (Yecuris, Tualatin, OR, USA). Mice were maintained in a specific pathogen-free facility at the University of North Carolina (UNC) at Chapel Hill. All procedures were approved by the UNC Institutional Animal Care and Use Committee. 4- to 6-month-old female mice were used.
AAV Vector Production
AAV vector was produced using the triple plasmid transfection. Briefly, HEK293 cells were transfected with an AAV transgene plasmid (ss pTR-CBA-Luciferase or sc pTR-CBh-GFP), an AAV helper plasmid (the Rep and Cap genes), and the adenovirus helper plasmid pXX6-80. Forty-eight hours later after transfection, HEK293 cells were harvested and lysed, and AAV vector was purified by cesium chloride (CsCl) gradient density centrifugation. The virus titer was determined by qPCR.
Isolation of AAV Mutants in Human Hepatocytes
The AAV library, which consisted of shuffled capsids from serotypes 1–6, 8, 9, and an AAV8 with the E531K mutation was produced as described previously.24,26,30 The humanized mice were administered with 30 mg human IVIG via retro-orbital injection. Four hours later, 2 × 1011 particles of AAV shuffling library were intravenously injected. At 3 days after AAV injection, adenovirus dl309 was intravenously injected at 2 × 109 of MOI. Two days later, mice were euthanized, and the livers were harvested and lysed. The cell lysate was generated by three freeze-thaw cycles, heated to 56°C for 30 min to inactivate adenovirus, and then clarified by centrifugation to remove cellular debris. The pooled supernatant of cell lysate was used to infect human hepatocytes in other humanized mice treated with 30 mg human IVIG, and then adenovirus dl309 was supplied for further amplification of AAV capsids. This process was repeated for two more cycles (Figure 1). Following the final cycle, capsid DNA sequences from isolated liver DNA were amplified through PCR and cloned into the pXR shuttle vector. Individual clones, each potentially representing a chimeric capsid DNA sequence, were sequenced, and recombinant vectors packaged with the GFP and luciferase were generated for in vitro and in vivo characterization.
Nab Assay
Huh7 cells were plated with 1 × 105 cells per well in a 48-well plate and cultured for overnight. 1 × 109 particles of AAV/LUC vectors were incubated with IVIG or sera from healthy subjects or hemophilia patients at different dilutions for 1 h at 4°C and then added into Huh7 cell culture medium. After culturing for 48 h, the luciferase activity in the cell lysate was measured with a Wallac-1420 Victor 2 automated plate reader.
Human Hepatocyte Transduction with AAV Vectors in Chimeric Mice
All mice received retro-orbital injections of 1 × 1011 particles of sc AAV/GFP vectors. Three weeks later, mice were euthanized, and livers were harvested. Part of the liver tissues was fixed in 10% formalin for immunohistochemistry staining. Another part of the liver tissues was used to make single hepatocytes for flow cytometry analysis.
Immunohistochemistry
As described earlier,31 after fixation, liver tissues were embedded in paraffin and sectioned. The liver sections were rehydrated with a serial concentration of ethanol, and then antigen was retrieved. After blocking, liver tissues were incubated with a primary antibody goat anti-human albumin and then with the Alexa Fluor 488-conjugated secondary antibody. After washing, liver tissues were stained with chicken anti-GFP primary antibody followed by the Alexa Fluor 594-conjugated secondary antibody. Finally, the tissues were stained with DAPI.
Flow Cytometry Analysis
Single-cell suspensions from liver tissues were fixed and permeabilized with Fixation/Permeabilization Solution (BD Biosciences) and then incubated with a goat anti-human albumin antibody (Bethyl Laboratories) for 30 min. After being washed with cold PBS, cells were stained with Allophycocyanin (APC)-conjugated donkey anti-goat immunoglobulin G (IgG) antibody (R&D Systems). After washing with PBS, cells were analyzed by flow cytometry.
Molecular Modeling
Wild-type AAV cryo-structures were obtained from the Protein Data Bank (PDB). The identification numbers for AAV2, -6, -8, and -9 are (PDB: 6IHB, 3SHM, 3RA2, and 6NXE, respectively. After alignment with the wild-type capsids, the capsid aa sequence of AAV LP2-10, which was composed of the fragments from AAV2, -6, -8, and -9 was re-assembled according to the DNA sequencing result by Chimera (UCSF). Capsid aa sequences are listed in reference to the VP1 start site.
Phylogenetic Analysis
ClustalW was used to align the entire capsid aa sequences of all AAV mutants and wild serotypes,32 and MEGAv5.05 was applied to generate phylogenetic trees.33
Statistics
The Student’s t test (paired) was used to perform statistical analysis. p <0.05 was considered a statistically significant difference (∗p < 0.05).
Author Contributions
X.P. completed the main experiments. W.S. and A.X. established the Nab detection system. C.A., X.C., and C.C. helped to prepare the AAV mutant library. Y.L.A., D.A.G., E.P.M., and T.C.N. collected the sera samples. W.L. helped to construct the mutant plasmids. R.J.S and C.L. designed this project.
Conflicts of Interest
R.J.S. is the founder and a shareholder at Asklepios BioPharmaceutical and Bamboo Therapeutics. He holds patents that have been licensed by the UNC to Asklepios Biopharmaceutical, for which he receives royalties. He has consulted for Baxter Healthcare and has received payment for speaking. C.L. is a cofounder of Bedrock Therapeutics. He has licensed patents by UNC and has received royalties from Bedrock Therapeutics and Asklepios BioPharmaceutical.
Acknowledgments
We would like to thank the UNC Vector Core for AAV vector production. The authors acknowledge the UNC Histological Research Core and Flow Cytometry Core Facilities for their assistance in liver immunohistochemistry and flow cytometry analysis. This work was supported by National Institutes of Health grants R01HL125749 and R01HL144661 (to T.C.N. and C.L.), R01AI117408 and P01HL112761 (to R.J.S. and C.L.), N01_75N92019D00041 (to T.C.N.), and NCBC 2015-CFG-8009 (to C.L.). Animal Studies were performed within the UNC Lineberger Animal Studies Core Facility at the UNC at Chapel Hill. The UNC Lineberger Animal Studies Core is supported in part by an NCI Center Core support grant (CA16086) to the UNC Lineberger Comprehensive Cancer Center.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.06.003.
Supplemental Information
References
- 1.Cwerman-Thibault H., Augustin S., Ellouze S., Sahel J.-A., Corral-Debrinski M. Gene therapy for mitochondrial diseases: Leber Hereditary Optic Neuropathy as the first candidate for a clinical trial. C R Biol. 2014;337:193–206. doi: 10.1016/j.crvi.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuddenham E. Gene therapy for haemophilia B. Haemophilia. 2012;18(Suppl. 4):13–17. doi: 10.1111/j.1365-2516.2012.02823.x. [DOI] [PubMed] [Google Scholar]
- 4.George L.A., Sullivan S.K., Giermasz A., Rasko J.E.J., Samelson-Jones B.J., Ducore J., Cuker A., Sullivan L.M., Majumdar S., Teitel J. Hemophilia B Gene Therapy with a High-Specific-Activity Factor IX Variant. N. Engl. J. Med. 2017;377:2215–2227. doi: 10.1056/NEJMoa1708538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurlbut G.D., Ziegler R.J., Nietupski J.B., Foley J.W., Woodworth L.A., Meyers E., Bercury S.D., Pande N.N., Souza D.W., Bree M.P. Preexisting immunity and low expression in primates highlight translational challenges for liver-directed AAV8-mediated gene therapy. Mol. Ther. 2010;18:1983–1994. doi: 10.1038/mt.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vercauteren K., Hoffman B.E., Zolotukhin I., Keeler G.D., Xiao J.W., Basner-Tschakarjan E., High K.A., Ertl H.C.J., Rice C.M., Srivastava A. Superior In vivo Transduction of Human Hepatocytes Using Engineered AAV3 Capsid. Mol. Ther. 2016;24:1042–1049. doi: 10.1038/mt.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L., Bell P., Somanathan S., Wang Q., He Z., Yu H., McMenamin D., Goode T., Calcedo R., Wilson J.M. Comparative Study of Liver Gene Transfer With AAV Vectors Based on Natural and Engineered AAV Capsids. Mol. Ther. 2015;23:1877–1887. doi: 10.1038/mt.2015.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulk N.K., Pekrun K., Zhu E., Nygaard S., Li B., Xu J., Chu K., Leborgne C., Dane A.P., Haft A. Bioengineered AAV Capsids with Combined High Human Liver Transduction In Vivo and Unique Humoral Seroreactivity. Mol. Ther. 2018;26:289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pekrun K., De Alencastro G., Luo Q.J., Liu J., Kim Y., Nygaard S., Galivo F., Zhang F., Song R., Tiffany M.R. Using a barcoded AAV capsid library to select for clinically relevant gene therapy vectors. JCI Insight. 2019;4:131610. doi: 10.1172/jci.insight.131610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee G.K., Maheshri N., Kaspar B., Schaffer D.V. PEG conjugation moderately protects adeno-associated viral vectors against antibody neutralization. Biotechnol. Bioeng. 2005;92:24–34. doi: 10.1002/bit.20562. [DOI] [PubMed] [Google Scholar]
- 11.Meliani A., Boisgerault F., Hardet R., Marmier S., Collaud F., Ronzitti G., Leborgne C., Costa Verdera H., Simon Sola M., Charles S. Antigen-selective modulation of AAV immunogenicity with tolerogenic rapamycin nanoparticles enables successful vector re-administration. Nat. Commun. 2018;9:4098. doi: 10.1038/s41467-018-06621-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mingozzi F., Anguela X.M., Pavani G., Chen Y., Davidson R.J., Hui D.J., Yazicioglu M., Elkouby L., Hinderer C.J., Faella A. Overcoming preexisting humoral immunity to AAV using capsid decoys. Sci. Transl. Med. 2013;5:194ra92. doi: 10.1126/scitranslmed.3005795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao K., Li M., Zhong L., Su Q., Li J., Li S., He R., Zhang Y., Hendricks G., Wang J., Gao G. Empty Virions In AAV8 Vector Preparations Reduce Transduction Efficiency And May Cause Total Viral Particle Dose-Limiting Side-Effects. Mol. Ther. Methods Clin. Dev. 2014;1:20139. doi: 10.1038/mtm.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tse L.V., Klinc K.A., Madigan V.J., Castellanos Rivera R.M., Wells L.F., Havlik L.P., Smith J.K., Agbandje-McKenna M., Asokan A. Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion. Proc. Natl. Acad. Sci. USA. 2017;114:E4812–E4821. doi: 10.1073/pnas.1704766114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louis Jeune V., Joergensen J.A., Hajjar R.J., Weber T. Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum. Gene Ther. Methods. 2013;24:59–67. doi: 10.1089/hgtb.2012.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao W., Pei X., Cui C., Askew C., Dobbins A., Chen X., Abajas Y.L., Gerber D.A., Samulski R.J., Nichols T.C., Li C. Superior human hepatocyte transduction with adeno-associated virus vector serotype 7. Gene Ther. 2019;26:504–514. doi: 10.1038/s41434-019-0104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velazquez V.M., Meadows A.S., Pineda R.J., Camboni M., McCarty D.M., Fu H. Effective Depletion of Pre-existing Anti-AAV Antibodies Requires Broad Immune Targeting. Mol. Ther. Methods Clin. Dev. 2017;4:159–168. doi: 10.1016/j.omtm.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corti M., Elder M., Falk D.J., Lawson L., Smith B.K., Nayak S., Conlon T.J., Clément N., Erger K., Lavassani E. B-Cell Depletion is Protective Against Anti-AAV Capsid Immune Response: A Human Subject Case Study. Mol. Ther. Methods Clin. Dev. 2014;1:14033. doi: 10.1038/mtm.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monteilhet V., Saheb S., Boutin S., Leborgne C., Veron P., Montus M.F., Moullier P., Benveniste O., Masurier C. A 10 patient case report on the impact of plasmapheresis upon neutralizing factors against adeno-associated virus (AAV) types 1, 2, 6, and 8. Mol. Ther. 2011;19:2084–2091. doi: 10.1038/mt.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 21.Li C., Wu S., Albright B., Hirsch M., Li W., Tseng Y.-S., Agbandje-McKenna M., McPhee S., Asokan A., Samulski R.J. Development of Patient-specific AAV Vectors After Neutralizing Antibody Selection for Enhanced Muscle Gene Transfer. Mol. Ther. 2016;24:53–65. doi: 10.1038/mt.2015.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lochrie M.A., Tatsuno G.P., Christie B., Wellman McDonnell J., Zhou S., Surosky R., Pierce G.F., Colosi P. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 2006;80:821–834. doi: 10.1128/JVI.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C., Diprimio N., Bowles D.E., Hirsch M.L., Monahan P.E., Asokan A., Rabinowitz J., Agbandje-McKenna M., Samulski R.J. Single amino acid modification of adeno-associated virus capsid changes transduction and humoral immune profiles. J. Virol. 2012;86:7752–7759. doi: 10.1128/JVI.00675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartel M.A., Weinstein J.R., Schaffer D.V. Directed evolution of novel adeno-associated viruses for therapeutic gene delivery. Gene Ther. 2012;19:694–700. doi: 10.1038/gt.2012.20. [DOI] [PubMed] [Google Scholar]
- 25.Boye S.L., Bennett A., Scalabrino M.L., McCullough K.T., Van Vliet K., Choudhury S., Ruan Q., Peterson J., Agbandje-McKenna M., Boye S.E. Impact of Heparan Sulfate Binding on Transduction of Retina by Recombinant Adeno-Associated Virus Vectors. J. Virol. 2016;90:4215–4231. doi: 10.1128/JVI.00200-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chai Z., Samulski R.J., Li C. Nab Escaping AAV Mutants Isolated from Mouse Muscles. Bio Protoc. 2018;8:e2841. doi: 10.21769/BioProtoc.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowles D.E., McPhee S.W., Li C., Gray S.J., Samulski J.J., Camp A.S., Li J., Wang B., Monahan P.E., Rabinowitz J.E. Phase 1 gene therapy for Duchenne muscular dystrophy using a translational optimized AAV vector. Mol. Ther. 2012;20:443–455. doi: 10.1038/mt.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albright B.H., Storey C.M., Murlidharan G., Castellanos Rivera R.M., Berry G.E., Madigan V.J., Asokan A. Mapping the Structural Determinants Required for AAVrh.10 Transport across the Blood-Brain Barrier. Mol. Ther. 2018;26:510–523. doi: 10.1016/j.ymthe.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Govindasamy L., Padron E., McKenna R., Muzyczka N., Kaludov N., Chiorini J.A., Agbandje-McKenna M. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J. Virol. 2006;80:11556–11570. doi: 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray S.J., Blake B.L., Criswell H.E., Nicolson S.E., Samulski R.J., McCown T.J., Li W. Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood-brain barrier (BBB) Mol. Ther. 2010;18:570–578. doi: 10.1038/mt.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao W., Pei X., Cui C., Askew C., Dobbins ., Chen X., Abajas Y.L., Gerber D.A., Samulski R.J., Nichols T.C., Li C. Superior human hepatocyte transduction with adeno-associated virus vector serotype 7. Gene Ther. 2019;26:504–514. doi: 10.1038/s41434-019-0104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.