Abstract
The electrochemical DNA biosensor has been developed for the detection of Listeria monocytogenes in raw milk samples. The electrochemical studies of the developed biosensor was recorded by cyclic voltammetry (CV) and electrochemical impedance (EI) using methylene blue (MB) and potassium ferricyanide K3Fe(CN)−6 as redox indicators. The selectivity of the developed biosensor was demonstrated using complementary and mismatch oligonucleotide sequences. The sensitivity (S) of the developed sensor was recorded as 3461 (μA/cm2)/ng and limit of detection (LOD) was found to be 82 fg/6 µl with the regression coefficient (R2) 0.941 using CV. The sensor was characterized by field emission scanning electron microscopy (FE-SEM). The electrode was found to be stable for six months, with only 10% loss in the initial CV current.
Keywords: DNA biosensor, plcA gene, CNF, AuNP, Foodborne pathogens, listeria monocytogenes, Selectivity
Introduction
Nowadays, the consumption of ready to eat (RTE) food is a leading cause of majority of problems related to human health. Listeria monocytogenes, a gram-positive, facultative anaerobic bacterium that grows at temperatures between 0.4 and 45 °C is one of the most virulent pathogen found in both unprocessed and processed foods (Valimaa et al. 2015). Listeria monocytogenes is the common cause of Zoonosis (Vizzini et al. 2019). The genus Listeria includes seven species, out of which only L. monocytogenes and L. ivanovii are pathogenic, facultative intracellular bacteria which can cross intestinal barrier and can proliferate within macrophages, epithelial, and endothelial cells.
The bacterium contains different virulent factors that disrupt the vacuolar membrane and may cause listeriosis in humans. Phosphatidylinositol-specific phospholipase C gene (PlcA) of L. monocytogenes is a virulent gene and encodes a 33-kDa protein responsible for the lysis of primary single-membraned vacuoles (Vazquez et al. 2001). L. monocytogenes causes listeria infection especially in pregnant women, newborn babies, and immunocompromised patients (Soni et al. 2014). This pathogen is responsible for mild fever to gastroenteritis infection, meningitis, and septicemia. The infection rate of this pathogen is high in neonates and in people above 65 years of age, and regarded as one of the factor for abortions and still birth in India (Jamshidi and Zeinali 2019). The reported rate of hospitalization with infection by listeriosis is found to be 94%, with a fatality rate of 12–17% (Zhao et al. 2017), and mortality rate of 20–30% (Soni et al. 2018).
In past decade, various techniques have been used for the detection of L. monocytogenes and include bacterial culturing methods, sugar fermentation, haemolytic properties, immunological methods such as enzyme-linked immunosorbent assay (ELISA), enzyme-linked fluorescent assay (ELFA), (Jaakohuhta et al. 2007) and molecular methods viz, polymerase chain reaction (PCR), and microarray study (Ingianni et al. 2001; Palumbo et al. 2003; Volokhov et al. 2002; Sergeev et al. 2004; Chemburu et al. 2005). The colony morphology, sugar fermentation, and haemolytic properties are considered as a gold standard methods for identification of L. monocytogenes. All the methods mentioned above have some limitations of being expensive and less sensitive, and some of these methods also lead to false-positive results (Valimaa et al. 2015). Culture-based methods, however, are simple, but time-consuming (require up to 5–7 days), labor-intensive, and may not be appropriate to test foods with short shelf life (Jadhav et al. 2012; Datta et al. 2013). To overcome all above problems, there is a need for the development of rapid and sensitive method of detection. In present study, an electrochemical DNA biosensor with high sensitivity and specificity was developed for the detection of L. monocytogenes with short time of assay. Also, this is the first plcA-based electrochemical DNA biosensor developed for the detection of L. monocytogenes in raw milk samples using CNF/AuNp-based electrode.
Materials and methods
Strains and chemicals
The bacterial strains of L. monocytogenes (MTCC 657), Salmonella enterica (MTCC 9844), Klebsiella Pneumoniae (MTCC 39), and Escherichia coli (MTCC 50) were procured from MTCC, Institute of Microbial Technology, Chandigarh, India. Chloroform, EDTA, isoamyl alcohol, phenol, sodium chloride, sodium di-hydrogen orthophosphate and di-sodium hydrogen orthophosphate, nitric acid (HNO3), and brain heart infusion broth were purchased from Hi-media, India. 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), N-Hydroxysuccinimide (NHS), methylene blue (MB), potassium ferricyanide (K3Fe(CN)6)−, Rnase, and Proteinase K were purchased from Sigma-Aldrich, USA. All other chemicals used were of analytical grade. Screen-printed CNF/AuNP electrode was procured from DropSens, Spain, and modified in the lab.
Oligonucleotides
5´-NH2 modified plcA gene-specific single-stranded DNA (ssDNA) probe and all other oligonucleotides related to L. monocytogenes and negative control were designed and procured from Eurofins, Bengaluru, India. The stock solution of 100 pmol μl−1 of probe and oligonucleotides were prepared with autoclaved distilled water and stored at − 20 °C. The specificity of the plcA gene probe for the DNA sensing was verified by using BLAST. The sequence of oligonucleotides used in this study is shown in Table 1.
Table 1.
Oligonucleotide sequences used in the present work
| Oligonucleotides | Sequences |
|---|---|
| Probe (20 bases) | 5′- NH2-TTACTTGGTTAGGTGCGCCG-3′ |
| Negative control (20 bases) | 5′- NH2-TTACTTGGTTAGGTGCGCCG-3′ |
| Complementary sequence (20 bases) | 5′- CGGCGCACCTAACCAAGTAA-3′ |
| Complementary sequence (35 bases) | 5′-TAGCCGACGGCGCACCTAACCAAGTAAGCATCGAA-3′ |
| Single mismatch (20 bases) | 5′- CGGCGCACCTTACCAAGTAA-3′ |
Apparatus
Cyclic voltammetry (CV) and electrochemical impedance (EI) studies were carried out using the potentiostat, (Palmsens 4 Netherland) and data analysis system PSTrace 5 software. The CNF/AuNP electrode consists of a three-electrode system having working electrode (WE) and counter-electrode (CE) , made of carbon, and the reference electrode (RE), made of silver. The DNA was quantified using nanodrop spectrophotometer (QIAxpert System, Netherland). Electrode surface characterization studies were carried out using field emission scanning electron microscopy (FE-SEM), Hitachi-SU8010 microscope.
Isolation of genomic DNA from culture
The target bacterium L. monocytogenes was grown in brain heart infusion broth culture for 24 h at 37 °C and the genomic DNA (G-DNA) was isolated using phenol–chloroform method (Kaushal et al. 2012). The purity of isolated G-DNA (A 260/A280) and quantity (A260) was measured using nanodrop spectrophotometer.
Construction of the sensor
The screen-printed CNF/AuNP electrode was first cleaned with Milli Q water for 30 s and air-dried at room temperature. The working area of the electrode was further washed with 1 mM of HNO3 solution (6 μl) for 10 min, followed by washing with PBS (pH 7.4) to remove the unbound reagents. Finally, the screen-printed electrode was dried at room temperature. The working area of the electrode was further treated with a mixture of 10 mM each of 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide EDC:N-Hydroxysuccinimide NHS (1:1, v/v in PBS, pH 7.4) for 1.5 h to activate the carboxyl groups on the surface (Rahman et al. 2009). The electrode was washed with PBS (pH 7.4) and dried at room temperature. The 5′-NH2-labeled probe (6 μl) was added onto the working surface of the electrode followed by overnight incubation. This allowed the formation of an amide bond between the NH2 group of the probe and COOH group of the C-nanofibres through EDC-NHS chemistry (Rahman et al. 2009). The electrode was then washed with TE buffer (10 ml of 1 M Tris–HCl buffer and 2 ml of 0.5 M EDTA and distilled water added to make final volume upto 1000 ml and pH adjusted at 8) to remove the unbound probe and air-dried at room temperature before further electrochemical analysis. The schematic illustration of the fabrication of the sensor is shown in Scheme 1.
Scheme 1.
Fabrication of electrochemical sensor using immobilization of 5′-NH2-labeled ssDNA probe of plcA gene on CNF/AuNp and hybridization with ssG-DNA of L. monocytogenes. EDC:NHS chemistry was used in fabrication of biosensor
Hybridization with ssDNA
Different concentrations of isolated genomic DNA of L. monocytogenes were prepared and denatured at 95 C for 5 min. The single-stranded DNA probe (ssDNA) was hybridized on working electrode surface using different concentrations of single-stranded genomic DNA (ssG-DNA). After hybridization, the electrode surface was washed with TE buffer (pH 8). Further electrochemical analysis of electrode with CV and EI was done by potentiostat using 1 mM MB and 2.5 mM K3Fe(CN)−6 that works as an artificial redox indicator (Zhu et al. 2008; Nasef et al. 2010). Electrode surface was regenerated after every measurement using PBS buffer (pH 7.4) that removes the hybridized DNA strands from the electrode surface except immobilized probe and TE (pH 8) was used to provide favorable electrochemical conditions.
Surface characterization of DNA biosensor
Surface morphology characterization of CNF/AuNP electrodes at different stages of fabrication viz. CNF/AuNP-based electrode, CNF/AuNP immobilized with NH2-labeled probe ssDNA, and CNF/AuNP/dsDNA after hybridization with 50 ng/6 µl ssG-DNA of L. monocytogenes were carried out using field emission scanning electron microscopy (FE-SEM).
Specificity, selectivity, and stability of DNA biosensor
The specificity of the DNA biosensor was studied by hybridization of ssG-DNA of L. monocytogenes and other selected foodborne pathogens (E. coli, K. pneumoniae, and S. enterica) with ssDNA probe specific to plcA gene of L. monocytogenes. The relative peak current (Ip) in each case was calculated with respect to the immobilized probe.
The selectivity of the sensor was evaluated using different oligonucleotides mentioned in Table 1. The hybridization efficiency of the negative control, complementary, and mismatch oligonucleotides with respect to the immobilized probe DNA was investigated using CV analysis. The stability of the developed sensor on storage at 4 °C was studied using CV analysis at regular interval of 30 days each for upto 6 months.
Validation of DNA biosensor
The validation study of developed biosensor was done using 20 raw milk samples and 3 milk samples artificially spiked with L. monocytogenes. The peak current (Ip) values of CV after hybridization with ssG-DNA of raw milk samples were compared with the probe, standard positive (samples artificially spiked with L. monocytogenes), and negative control (milk sample without L. monocytogenes), respectively.
Results and discussion
In this study, an electrochemical biosensor based on CNF/AuNp electrode was developed to detect L. monocytogenes in milk samples with high specificity and sensitivity. A 5′- NH2-labeled single-stranded probe DNA (ssDNA) specific to plcA gene was immobilized on the working surface of nanofabricated electrode. The complementary ssG-DNA of L. monocytogenes was hybridized with immobilized probe and was detected using CV and EIS. The sensitivity and limit of detection (LOD) of the sensors was improved by using nanomaterials, i.e., carbon nanofibers fabricated with gold nanoparticles on screen-printed electrode. These CNF/AuNP screen-printed electrode have a high surface area-to-volume ratio and excellent dimensional stability with high thermal and high electrical conductivity (Zhu et al. 2013, 2015). The results of this study are discussed as follows:
Cyclic voltammetry (CV) studies of CNF/AuNp-based biosensor
Cyclic voltammetry studies were performed to analyze the immobilization of 5′-NH2-labeled ssDNA probe and hybridization of different concentrations of the ssG-DNA on CNF/AuNp electrode as shown in Fig. 1. The CV peak current of ssDNA probe was found higher, i.e., 114 µA (curve a) as compared to bare electrode (not shown), and on further increase in the concentration of DNA, an increase in CV peak current was observed. The peak current for CV after hybridization with different DNA concentration was found as 177 µA (curve b), 209 µA (curve c), 248 µA (curve d), 297 µA (curve e), 330 µA (curve f), 362 µA (curve g), 401 µA (curve h), 427 µA (curve i), and 445 µA (curve j), respectively. This increase in electrode peak current was attributed to the presence of more unhybridized nitrogenous base pairs that lead to oxidation of methylene blue molecules at electrode surface. The plot between the relative Ip of probe and the ssG-DNA was found to be hyperbolic (Fig. 1 inset [A]) with a regression coefficient (R2) of 0.941 (Fig. 1 inset [B]). The sensitivity (S) of DNA sensor was calculated using the formula S = m/A and was found to be 3461 (μA/cm2/ng−1) where m is the slope of the linear equation and A is the area of the working electrode (0.126 cm2). The LOD of the DNA sensor was calculated using the formula LOD = 3 (σ/S), where σ is the standard deviation and S is the sensitivity, and was found to be 82 fg/6 µl (Fig. 1 inset [B]).
Fig. 1.
CV study of developed CNF/AuNp electrochemical biosensor at different stages of fabrication (a) ssDNA-NH2/CNF/AuNp electrode and (b–j) hybridization with 0.11–30 ng/6 μl of L. monocytogenes ssG-DNA at 50 mVs−1 using 1 mM MB in 50 mM PBS, pH 7.4. The inset-A shows hyperbolic curve from 0 to 30 ng/6 μl with linear peak current (Ip) up to 0.234 ng/6 μl of ssG-DNA of L. monocytogenes. Inset-B shows the linear plot from 0–0.234 ng/6 μl ssG-DNA for the calculation of sensitivity and LOD
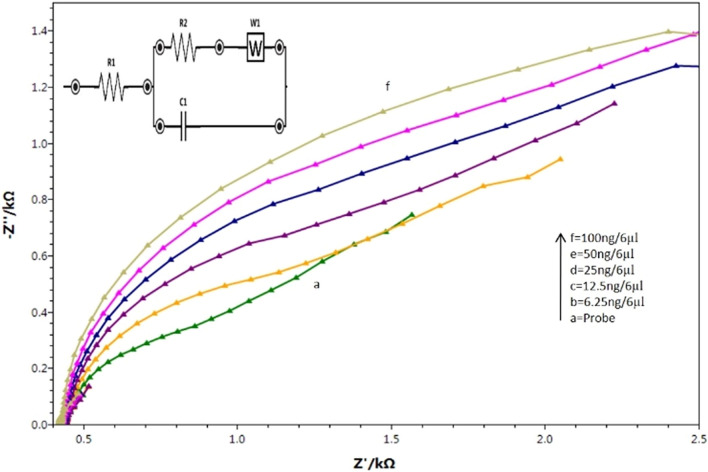
The electrochemical impedance (EIS) of CNF/AuNp-based biosensor
The Nyquist plot for electrochemical impedance studies of the immobilized probe and the hybridization of different concentration (6.25, 12.5, 25, 50, and 100 ng/6 μl) of ssG-DNA using 1 mM K3Fe(CN)−6 is shown in Fig. 2. In the Nyquist plots, the diameter of the semicircle reflected the charge transfer resistance (Rct) value (Fan et al. 2019). The Rct value of immobilized probe was found higher (0.9 × 103 Ω, curve a) than the bare electrode (not shown in the figure). This may be attributed to the negatively charged phosphate backbone of ssDNA, which prevented the K3Fe(CN)−6 ions from reaching the electrode surface. The Rct values for different concentrations of ssG-DNA after hybridization with the immobilized probe were found to be 1.1 × 103 Ω (curve b), 1.3 × 103 Ω (curve c), 1.6 × 103 Ω (curve d), 1.8 × 103 Ω (curve e), and 2.0 × 103 Ω (curve f), respectively. This may be due to the increase in the number of negatively charged phosphate groups after hybridization on the electrode surface that causes an increase in surface thickness which in turn resulted in an increase in the Rct value (Park and Park 2009). Total bacterial detection time of developed sensor was found to be 30 min that includes DNA processing, immobilization, and hybridization with immobilized probe.
Fig. 2.
Impedance-based study of developed CNF/AuNp electrochemical biosensor at different stages of fabrication (a) ssDNA-NH2/ CNF/AuNp and (b–f) hybridization with 6.25, 12.5, 25, 50, and 100 ng/6 µl of L. monocytogenes ssG-DNA using redox indicator 1 mM K3Fe(CN)6 in 50 mM PBS, pH 7.4
FE-SEM analysis of CNF/AuNp-based biosensor
In this study, morphological changes occurring on the surface of developed sensor at different stages of fabrication were recorded. Figure 3a shows CNF/AuNP bare electrode with a smooth and uniform fibrous structure that confirms the carbon nanofibers on the electrode surface. In Fig. 3b, CNF/AuNP immobilized with NH2-labeled probe showed a change in the surface morphology. In Fig. 3c, CNF/AuNP hybridized with 50 ng/6 µl ssG-DNA of L. monocytogenes recorded a further dense surface morphology, and globular surface which confirms the binding of DNA onto the probe.
Fig. 3.
FE-SEM micrograph of developed CNF/AuNp electrode biosensor. (a) bare electrode, (b) CNF/AuNp electrode/ssDNA probe, and (c) CNF/AuNp electrode/dsDNA after hybridization with 50 ng/6 µl ssG-DNA of L monocytogenes
Specificity, selectivity, and the stability of the developed sensor
The specificity of the developed DNA biosensor was observed using CV. After CV analysis, the peak current of all the foodborne pathogens was found almost similar to the immobilized probe except with L. monocytogenes (Fig. 4) where it was found higher even at the lower concentration of genomic DNA (10 ng/6 µl). Based on these observations, the sensor is found to be highly specific for the plcA gene of L. monocytogenes.
Fig. 4.
Specificity study of DNA biosensor based on CNF/AuNp electrode for L. monocytogenes and other foodborne pathogens. The relative Ip value of CV with respect to immobilized probe as zero after hybridization with 50 ng/6 μl ssG-DNA of other pathogens and L. monocytogenes
The selectivity studies of the developed sensor were confirmed by CV analysis. The oxidation peak current of MB with ssDNA (probe) on the CNF/AuNP electrode (curve a) was found lowest as shown in Fig. 5. After hybridization with negative control, the peak current was almost equal to probe peak current (curve b). On subsequent hybridization with single base-pair mismatched ssDNA sequence (curve c), 35 mer complementary ssDNA sequence (curve d), and 20 mer complementary ssDNA sequence (curve e), a gradual increase in the peak current was recorded. This indicated the hybridization of ssDNA with complementary base-pair confirming the selectivity of the electrode surface (Sun et al. 2010). The statistical analysis was performed at 5% (p = 0.05) level of significance.
Fig. 5.
Selectivity study of developed CNF/AuNp-based electrochemical biosensor using (a) ssDNA probe, (b) negative control, (c) 20 mer single bp mismatch, (d) 35 mer complement bp, (e) 20 mer complement bp. The concentration used for the hybridized oligonucleotides was 10 µM
In the present study, the developed sensor was found stable on storage at 4 °C for up to six months with about 10% loss in CV current (Fig. 6).
Fig. 6.

Stability of the developed CNF/AuNP sensor measured as % relative peak current with respect to probe as zero using CV for regular interval of 30 days up to 6 months of storage at 4 ºC
Validation of developed sensor with raw milk samples
The sensor was validated using 20 raw milk samples and 3 milk samples artificially spiked with L. monocytogenes and the results were recorded using CV analysis (Fig. 7). The change in relative peak current with respect to probe showed by negative control (milk sample without L. monocytogenes) and positive control (milk sample with L. monocytogenes) served as a threshold for milk samples. In Fig. 7, the y-axis denotes the relative peak current of CV with respect to the probe (as control). The negative control (without ssG-DNA in the sample to hybridize with probe) showed no increase in peak current, whereas the positive control (ssG-DNA of L. monocytogenes was hybridized with probe) showed an increase in the peak current with respect to the control (unhybridized probe). The ssG-DNA of the milk sample containing L. monocytogenes was hybridized with probe, and the relative peak current was monitored using CV. The positive and negative sample peak current were plotted against above-mentioned controls. Negative controls without L. monocytogenes (without complementary ssG-DNA) and with other pathogens (non-complementary DNA or even no DNA) showed no significant increase in the peak current, whereas in positive samples (under similar ssG-DNA concentration), the peak current showed 10% variation in the average peak current.
Fig. 7.
Validation studies of developed CNF/AuNp biosensor for the detection of L. monocytogenes by measuring the relative Ip value of CV after hybridization with ssG-DNA from raw milk samples. Twenty raw milk samples tested with developed DNA sensor using negative control (milk sample without L. monocytogenes) and positive control (milk sample with ssG-DNA of L. monocytogenes) with respect to probe (control) for validation of the sensor (significance level = 5%)
Comparative study of developed sensor with other reported sensors for detection of L. monocytogenes
The comparison of developed biosensor for the detection of L. monocytogenes causing food poisoning with other reported sensors is shown in Table 2. A similar work was carried out by Nagraik et al. (2019) in which an amperometric DNA-based biosensor was developed for the early detection of Leptospira interrogans by using MWCNT electrode. In this study the electrochemical studies before and after hybridization were done using CV and DPV in the presence of methylene blue as a redox indicator. The biosensor showed sensitivity of 264.5 (µA/cm−2/ng−1) and LOD of 0.015 ng/6 µl using CV. Due to higher sensitivity, selectivity, and lower LOD, the nanofiber-based sensor is found better than earlier-reported methods. The sensitivity of the reported sensor was found to be 3164 (µA/cm−2/ng−1) and the lower LOD was 82 fg/6 μl at 0.126 cm2 surface area of the working electrode.
Table 2.
Comparison of developed CNF/AuNp-based DNA sensor with some reported biosensors for the detection of L. monocytogenes in food samples
| Detection method | Transducer | Analyte used as marker | Target gene | Indicator used | Limit of detection (LOD) | References |
|---|---|---|---|---|---|---|
| Electrochemical Biosensor | CILE | DNA | hly gene | MB | 3.17 × 10−14 mol/L | Niu et al. 2017 |
| ElectrochemicalB osensor | Au/3DGRCILE | DNA | hly gene | MB | 3.3 × 10–15 mol L−1 | Yan et al. 2017 |
| Electrochemical Biosensor | CNF/AuNP | DNA | plcA gene | MB | 82 fg/6 µL | Present study |
SPGE screen-printed gold electrode, Au gold, 3DGR 3-dimensional graphene, CILE carbon ionic liquid electrode, MB methylene blue, ND not determined
Conclusions
The results demonstrated that the DNA biosensor could be effectively detect plcA gene of L monocytogenes. The biosensor is highly specific, rapid, has low cost, and could efficiently detect L monocytogenes specifically in raw milk. The sensitivity of biosensor was found to be 3164 (µA/cm−2/ng−1) and the lower LOD of 82 fg/6 μl at 0.126 cm2 surface area of the working electrode. This is the first-reported electrochemical biosensor based on CNF/AuNP used for the detection of plcA gene of L. monocytogenes within 30 min. The present biosensor offers lower LOD as compared to the reported biosensors for the detection of L. monocytogenes (Table 2).
Acknowledgements
The authors are thankful to Shoolini University, Solan, Himachal Pradesh, for providing the facility to carry out the present research work.
Compliances with ethical standards
Conflict of interest
There is no conflict of interest for authorship or related to any other context between authors.
Ethical standards
The authors have complied and worked within standard ethical norms.
References
- Chemburu S, Wilkins E, Hamid AI. Detection of pathogenic bacteria in food samples using highly-dispersed carbon particles. Biosens Bioelectron. 2005;21:491–499. doi: 10.1016/j.bios.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Datta AR, Laksanalamai P, Solomotis M. Recent developments in molecular subtyping of Listeria monocytogenes. Food Addit Contam A. 2013;30:1437–2144. doi: 10.1080/19440049.2012.728722. [DOI] [PubMed] [Google Scholar]
- Fan X, Li Z, Wang S, Liu L, Liu P, Chen F, Zheng X. Electrochemical Impedance biosensor for the determination of lipopolysaccharide using peptide as the recognition molecule. J Brazil Chem Soc. 2019;30:1762–1768. doi: 10.21577/0103-5053.20190081. [DOI] [Google Scholar]
- Ingianni A, Floris M, Palomba P, Madeddu MA, Quartuccio M, Pompei R. Rapid detection of Listeria monocytogenes in foods, by a combination of PCR and DNA probe. Mol Cell Probe. 2001;15:275–280. doi: 10.1006/mcpr.2001.0372. [DOI] [PubMed] [Google Scholar]
- Jaakohuhta S, Harma H, Tuomola M, Lovgren T. Sensitive Listeria spp. Immunoassay based on europium(III) nanoparticulate labels using time-resolved fluorescence. Int J Food Microbiol. 2007;114:288–294. doi: 10.1016/j.ijfoodmicro.2006.09.025. [DOI] [PubMed] [Google Scholar]
- Jadhav S, Bhave M, Palombo EA. Methods used for the detection and subtyping of Listeria monocytogenes. J Microbiol Methods. 2012;88:327–341. doi: 10.1016/j.mimet.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Jamshidi A, Zeinali T. Significance and characteristics of Listeria monocytogenes in poultry products. Int J Food Sci. 2019 doi: 10.1155/2019/7835253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal A, Kumar D, Khare S, Kumar A. SpeB gene as a specific genetic marker for early detection of rheumatic heart disease in human. Cell Mol Biol. 2012;58:50–54. [PubMed] [Google Scholar]
- Nagraik R, Kaushal A, Gupta S, Dhar P, Sethi S, Kumar D. Optimized DNA-based bioassay for Leptospira interrogans detection: a novel platform for leptospirosis diagnosis. 3 Biotech. 2019;9:284. doi: 10.1007/s13205-019-1815-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasef H, Beni V, O’Sullivan CK. Methylene blue as an electrochemical indicator for DF508 cystic fibrosis mutation detection. Anal Bioanal Chem. 2010;396:1423–1432. doi: 10.1007/s00216-009-3369-5. [DOI] [PubMed] [Google Scholar]
- Niu X, Zheng W, Yin C, Weng W, Li G, Sun W, Men Y. Electrochemical DNA biosensor based on gold nanoparticles and partially reduced graphene oxide modified electrode for the detection of Listeria monocytogenes hly gene sequence. J Electroanal Chem. 2017;806:116–122. doi: 10.1016/j.jelechem.2017.10.049. [DOI] [Google Scholar]
- Palumbo JD, Borucki MK, Mandrell RE, Gorski L. Serotyping of Listeria monocytogenes by enzyme-linked immunosorbent assay and identification of mixed-serotype cultures by colony immunoblotting. J Clin Microbiol. 2003;41:564–571. doi: 10.1128/jcm.41.2.564-571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Park SM. DNA hybridization sensors based on electrochemical impedance spectroscopy as a detection tool. Sensors. 2009;9:9513–9532. doi: 10.3390/s91209513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MM, Shiddiky MJ, Rahman MA, Shim YB. A lactate biosensor based on lactate dehydrogenase/nictotinamide adenine dinucleotide (oxidized form) immobilized on a conducting polymer/multiwall carbon nanotube composite film. Anal Biochem. 2009;384:159–165. doi: 10.1016/j.ab.2008.09.030. [DOI] [PubMed] [Google Scholar]
- Sergeev N, Distler M, Courtney S, Khaldi SF, Volokhov D, Chizhikov V. Multipathogen oligonucleotide microarray for environmental and biodefense applications. Biosens Bioelectron. 2004;20:684–698. doi: 10.1016/j.bios.2004.04.030. [DOI] [PubMed] [Google Scholar]
- Soni DK, Ahmad R, Dubey SK. Biosensor for the detection of Listeria monocytogenes: emerging trends. Crit Rev Microbiol. 2018;44:590–608. doi: 10.1080/1040841x.2018.1473331. [DOI] [PubMed] [Google Scholar]
- Soni DK, Singh M, Singh DV, Dubey SK. Virulence and genotypic characterization of Listeria monocytogenes isolated from vegetable and soil samples. BMC Microbiol. 2014;14:241. doi: 10.1186/s12866-014-0241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Qin P, Gao H, Li G, Jiao K. Electrochemical DNA biosensor based on chitosan/nano-V2O5/MWCNTs composite film modified carbon ionic liquid electrode and its application to the LAMP product of Yersinia enterocolitica gene sequence. Biosens Bioelectron. 2010;25:1264–1270. doi: 10.1016/j.bios.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Valimaa AL, Tilsala TA, Virtanen Rapid detection and identification methods for Listeria monocytogenes in the food chain—a review. Food Control. 2015;55:103–114. doi: 10.1016/j.foodcont.2015.02.037. [DOI] [Google Scholar]
- Vazquez BJ, Kuhn M, Berche P, Chakraborty T, Dominguez BG. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/cmr.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizzini P, Braidot M, Vidic J, Manzano M. Electrochemical and optical biosensors for the detection of Campylobacter and Listeria: an update look. Micromachines. 2019;10:500. doi: 10.3390/mi10080500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volokhov D, Rasooly A, Chumakov K, Chizhikov V. Identification of Listeria species by microarray-based assay. J Clin Microbiol. 2002;40:4720–4728. doi: 10.1128/jcm.40.12.4720-4728.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Zhao W, Wen Z, Li X, Niu X, Huang Y, Sun W. Electrochemical DNA Sensor for hly gene of Listeria monocytogenes by three-dimensional graphene and gold nanocomposite modified electrode. Int J Electrochem. 2017;12:4086–4095. doi: 10.20964/2017.05.04. [DOI] [Google Scholar]
- Zhao Y, Li Y, Jiang K, Wang J, White WL, Yang S, Lu J. Rapid detection of Listeria monocytogenes in food by biofunctionalized magnetic nanoparticle-based on nuclear magnetic resonance. Food Control. 2017;71:110–116. doi: 10.1016/j.foodcont.2016.06.028. [DOI] [Google Scholar]
- Zhu J, Park SW, Joh HI, Kim HC, Lee S. Preparation and characterization of isotropic pitch-based carbon fiber. Carbon Letters. 2013;14:94–98. doi: 10.5714/cl.2013.14.2.094. [DOI] [Google Scholar]
- Zhu J, Park SW, Joh HI, Kim HC, Lee S. Study on the stabilization of isotropic pitch-based fibers. Macromol Res. 2015;23:79–85. doi: 10.1007/s13233-015-3007-3. [DOI] [Google Scholar]
- Zhu L, Zhao R, Wang K, Xiang H, Shang Z, Sun W. Electrochemical behaviors of methylene blue on DNA modified electrode and its application to the detection of PCR product from NOS sequence. Sensors. 2008;8:5649–5660. doi: 10.3390/s8095649. [DOI] [PMC free article] [PubMed] [Google Scholar]