Abstract
Methanol production by co-culture of methanotrophs Methylocystis bryophila and Methyloferula stellata was examined from methane, a greenhouse gas. Co-culture exhibited higher methanol yield of 4.72 mM at optimum ratio of M. bryophila and M. stellata (3:2) compared to individual cultures. The immobilized co-culture within polyvinyl alcohol (PVA) showed relative efficiency of 90.1% for methanol production at polymer concentration of 10% (v/v). The immobilized co-culture cells within PVA resulted in higher bioprocess stability over free cells at different pH, and temperatures. Free and encapsulated co-cultures showed maximum methanol production of 4.81 and 5.37 mM under optimum conditions, respectively. After five cycles of reusage under batch conditions, free and encapsulated co-cultures retained methanol production efficiency of 23.8 and 61.9%, respectively. The present investigation successfully revealed the useful influence of co-culture on the methanol production over pure culture. Further, encapsulation within the polymeric matrix proved to be a better approach for the enhanced stability of the bioprocess.
Keywords: Co-culture, Encapsulation, Greenhouse gas, Methanol production, Methylocystis bryophila, Methyloferula stellata
Introduction
A substantial increase in the emission of greenhouse gases (GHGs) especially methane (CH4) has been noted by growing anthropogenic activities in commercial and industrial sectors [1–4]. CH4 has been suggested as potential feedstock to produce value-added products to counter its harmful environmental impact [1, 5–7]. Methanotrophs use CH4 (carbon source) as feed to produce various economically important bioproducts such as biofuels, biopolymers, carotenoids, lactic acid, single cell protein, and vitamin B12 [8–10]. The production of biofuel such as methanol is reported widely by methanotrophs including Methylosinus sporium and Methylosinus tricosporium [11–13]. Methanol is an industrially important compound, employed for producing chemical compounds such as acetic acid, alcohols, formaldehyde, paints, and resins [1, 2, 14]. Production of methanol over CH4 is pondered more valuable in terms of safety, transportation and storage concerns [1, 15, 16].
Biotransformation approaches have been recognized to be more beneficial compared to chemical processes that requires high energy and financial investments. In addition, there are issues of low selectivity and non-ecofriendly nature [17–24]. The conversion of CH4 to methanol is catalyzed by CH4 monooxygenases (MMOs). Thereafter, methanol is converted to formaldehyde followed by formate, and finally to CO2 by the action of methanol dehydrogenase (MDH), formaldehyde- and formate-dehydrogenases, respectively [1, 3]. A few limiting factors hindering efficient bioconversion are lower conversion yield of methanol, fewer methanotroph screenings, low stability of free cells under varying physiological conditions and repeated batch production. A few strategies have been proposed to enhance production of methanol through: (i) screening microbes, (ii) optimization of process parameters, (iii) supplementation of metal ions such as copper (Cu2+) and iron (Fe+2), (iv) addition of formate, and MDH inhibitors, and (v) immobilization of whole-cells. The biocatalytic activity of MMOs is significantly modulated in the presence of Cu2+ and Fe+2 metal ions during the methanol production [2, 3, 12]. Also, the partial inhibition of MDH activity by ethylenediaminetetraacetate, magnesium chloride (MgCl2), phosphate buffer, and sodium chloride and exhibited diverse influences on methanol production seems necessary for increasing it [2, 12]. It may be remarked that higher inhibition may not lead to increase in methanol production due to lowering of co-factor regeneration in the case of soluble MMOs (sMMOs) activity. Further, the immobilization of biocatalyst has demonstrated to improve their stability [25–32]. A few reports on the immobilization of methanotrophs through encapsulation have demonstrated the enhancement of methanol production [13, 33]. In addition, only few investigations have established the improvement in methanol production with the help of co-cultures of methanotrophs, including i) Methylococcus capsulatus, M. sporium NCIMB 11,126 and M. trichosporium OB3b enriched cultures from landfill soil [34], and ii) Methylomonas methanica and Methylocella tundrae [16]. In the present investigation, co-culture of Methylocystis bryophila and Methyloferula stellata was used for producing methanol from CH4. Further, influence of co-culture immobilization within polyvinyl alcohol (PVA) through encapsulation was evaluated to improve the stability on the methanol production process. These findings imply that methanol production by co-culture is more beneficial over pure culture and encapsulation of co-culture proved more effective in improving methanol production stability under repeated batch culture.
Materials and Methods
Materials
M. bryophila (DSM21852), and M. stellata (DSM22108) were acquired from German Collection of Microorganisms and Cell Cultures (DSMZ). Copper sulfate (CuSO4), MgCl2, ferrous sulfate (FeSO4), and PVA were obtained from Sigma-Aldrich, USA. Pure CH4 was acquired from NK Co., Busan, Republic of Korea. All other chemicals employed were of analytical grade.
Growth Conditions
Strains were cultured on nitrate mineral salt medium (200 mL in 1-L Erlenmeyer flask) with CH4 (20%) as feed (added on alternate days) at 30 °C and shaking of 200 rpm and incubated up to 7 days [2, 35]. Further, fully-grown cells were recovered through centrifugation (4000 rpm for 30 min at 4 °C) followed by washing twice with phosphate buffer (20 mM, pH 7.0). Finally, the harvested cells were stored at 4 °C [36, 37].
Co-culture Preparation and Optimization of Pure Cultures Ratio
Pure strains of M. bryophila and M. stellata were used for the preparation of co-culture by mixing them at a final inoculum concentration of 3 mg of dry cell mass (mg-DCM)/mL. The effective strain ratios of (M. bryophila: M. stellate) 4:1, 3:2, 2:1; 1:1, 1:2, 2:3, and 1:4 in the co-culture were examined for optimum conditions to produce methanol.
Biomethanol Production
The methanol production by pure or co-culture was carried out in serum bottles of 120 mL capacity. Reaction mixture of 20 mL was prepared containing Fe2+ (10 µM), Cu2+ (5 µM), MgCl2 (20 mM), formate (100 mM) and 3 mg-DCM/mL of inoculum in phosphate buffer (100 mM, pH 7.0). This mixture was incubated at 30 °C for 24 h and stirred at 150 rpm after saturating the headspace with CH4 (30%) as a feed [2, 35].
Encapsulation of Co-culture Within PVA
Co-culture immobilization was achieved by encapsulating them within PVA at various concentrations (5–15%) using 3 mg-DCM/mL cells [33, 36]. Loosely attached cells were removed by washing (twice) with distilled water, which was followed by buffer (20 mM, pH 7.0). Encapsulated bacterial cells were stored at 4 °C [16]. The relative efficiency (RE) of methanol production was calculated as ratio of methanol produced by encapsulated and free co-culture × 100.
Optimization of Encapsulated Co-culture
The optimization of methanol production conditions for free or encapsulated co-culture was examined by altering buffer pH (6.0–8.0) and incubation temperatures (25–40 °C). Further, methanol production patterns of the cultures were monitored up to 72 h.
Reusability
The encapsulated co-culture within PVA reusability was assessed for methanol production from CH4 (30%) under repeated-batch conditions up to five cycles as reported previously. During each cycle (24 h) of operation, encapsulated cells were separated by filtration followed by washing with buffer and used as inoculum. The methanol production efficiency of the zero cycle was taken as 100% [16].
Instrumental Analysis
Methanol concentration was analyzed by gas chromatography (GC, Agilent 7890A, USA) system equipped with HP-5 column (Agilent 19091 J-413, USA) and flame ionization detector as reported earlier [3, 38]. Absorbance was assessed spectrophotometrycally (6705 UV/Vis. Spectrophotometer, Jenway Scientific, UK) [39, 40]. The experimental data are given as mean values ± standard deviations of three replicates.
Results and Discussion
Production of Methanol by Co-culture
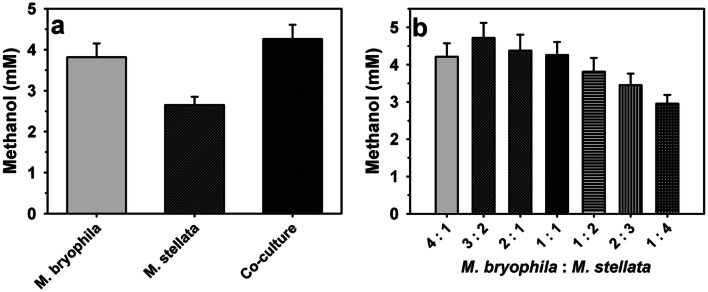
The co-culture of M. bryophila and M. stellata (1:1) led to higher methanol production of 4.24 mM in comparison to 3.82 and 2.65 mM by respective free cells (Fig. 1a). Here, an increase in the methanol production can be associated with the syntrophic behavior between the two organisms. This influence might be helpful to improve physiological stability of methanol production process. Previously, enhancement in methanol production has been reported by enrichment of mixed culture (M. capsulatus, M. sporium NCIMB 11,126, and M. trichosporium OB3b) from landfill soil [34], methanotrophs consortium from anaerobic digestion [41], type I methanotrophs mixed culture from activated sludge [42] and mixed methanotrophic species from landfill soil [43]. The use of undefined methanotrophic consortium is unreliable as methanotrophic population may be strongly influenced even by slight variations in enrichment conditions or sampling period. Therefore, the use of defined co-culture for methanol production can be expected to overcome such issues, and also for improving the process efficiency and operation stability than the pure cultures [11, 14, 16, 34, 41].
Fig. 1.
Methanol production by co-culture of Methylocystis bryophila and Methyloferula stellata: a production yield and b effect of strains ratio in co-culture
Since, the ratio of population of strains in the co-culture can significantly influence the methanol yield [16], therefore, various ratios of M. bryophila and M. stellata were examined (Fig. 1b). The various ratios of these two organisms resulted in methanol production ranging from 2.95 to 4.72 mM. The ratio of 3:2 was observed to be the most efficient for methanol production. On the other hand, the ratio of 1:4 showed lowest methanol production. These results suggest that higher concentration M. bryophila is more beneficial over M. stellata in the co-culture preparation. Figure 2 reflects that pure and co-cultures followed quite similar production trends. Initially, methanol production was observed to increase with longer incubation period up to 24 h. Thereafter, a decline in production was noted that might be a consequence of subsequent methanol utilization due to partial inhibition of MDH activity [13]. The optimum incubation period was 24 h, where a maximum production of 2.65, 3.82 and 4.72 mM by M. stellata, M. bryophila and co-culture under similar experimental conditions, respectively were recorded. Here, co-culture of M. bryophila and M. stellata exhibited higher methanol production than that of 0.02–0.71 mM by pure cultures of M. sporium KCTC 22,312, and M. trichosporium strains OB3b and IMV [12, 44, 45].
Fig. 2.
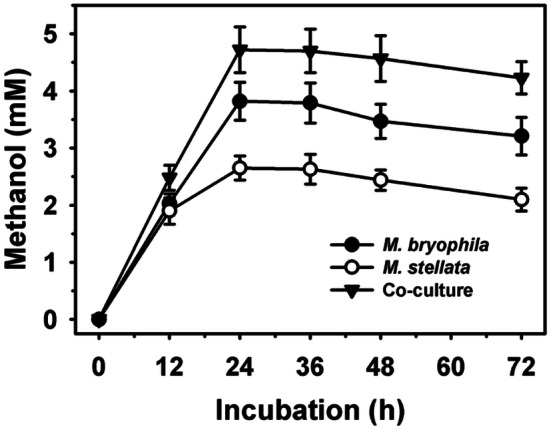
Methanol production profile of pure and co-cultures of Methylocystis bryophila and Methyloferula stellata
Encapsulation and Characterization of Co-culture
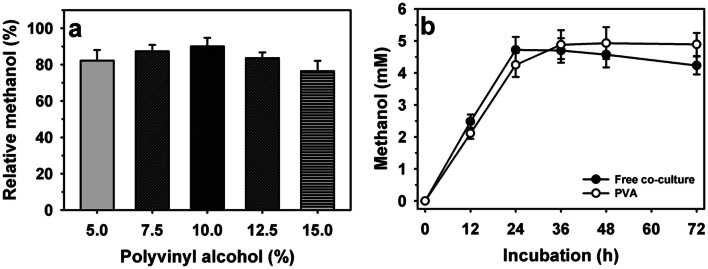
The encapsulation of co-culture (M. bryophila: M. stellata, 3:2) was evaluated using different concentrations of PVA (5–15%, v/v) to enhance the stability of the methanol production process (Fig. 3a). The concentrations of PVA showed variable influence on the methanol production with the RE in range of 82.2–90.1%. Here, a variation in the RE might be correlated with either rigidity of polymers or mass transfer limitation [13, 14]. PVA proved more efficient as support for co-culture to achieve higher RE than 72.4% reported for alginate immobilized M. tundrae [14]. The concentration of PVA (10%, v/v) was noted as an optimum for retaining maximum RE of 90.1%. A comparison of the methanol production stability of free and encapsulated co-cultures revealed a remarkable increase in the methanol production of 4.72 with an increase in incubation up to 24 h by free co-culture, which declined to 4.23 mM at longer incubation period of 72 h (Fig. 3b). Encapsulated co-culture showed higher maximum methanol production of 4.93 mM at the incubation of 48 h compared to free cells (4.72 mM). Also, immobilized cells exhibited better stability to retain 16% higher methanol production at longer incubation (72 h) than those of free cells. Previously, the higher stability of pure cultures immobilized within PVA was demonstrated for M. sporium B-2121 [33].
Fig. 3.
Encapsulation of co-culture of Methylocystis bryophila and Methyloferula stellata using: a different concentration of polyvinyl alcohol (PVA) and b methanol production profile of immobilized cells within PVA (10%, v/v)
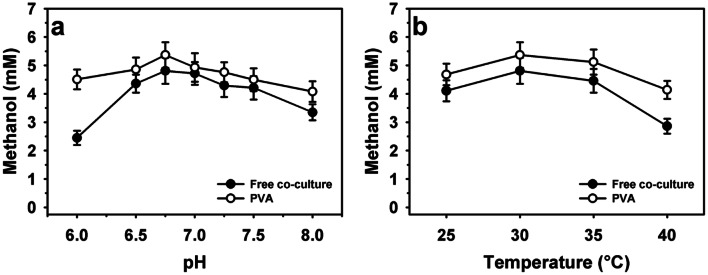
Immobilization methods of methanotrophs showed variable influence in the methanol production stability [13, 14, 46]. Therefore, the methanol production by free and encapsulated co-cultures were performed at different pH (6.0–8.0) and temperature (25–40 °C) values at optimum incubation conditions of 24 and 48 h, respectively (Fig. 4). At different pH values, encapsulated cells showed higher methanol production over free cells. The optimum pH value of 6.75 was observed for methanol production (Fig. 4a). The maximum methanol production by free and encapsulated were recorded 4.81 and 5.37 mM, respectively. Overall, encapsulated cells showed nearly 84 and 22% higher methanol production at pH 6.0 and 8.0 as compared free cells with production of 2.45 and 3.35 mM, respectively. Further, the methanol production was compared by varying incubation temperature at optimum pH and incubation period conditions for free and encapsulated co-cultures (Fig. 4b). The optimum methanol production was noted at 30 °C. The encapsulated cells showed enhancement in methanol production of 4.68 and 4.14 mM at 25 and 40 °C over free cells with the values of 4.11 and 2.86 mM, respectively. Overall, better methanol production was recorded by encapsulated cells than those of free cells, and previous reports (Table 1).
Fig. 4.
Methanol production of free and immobilized co-cultures of Methylocystis bryophila and Methyloferula stellata within polyvinyl alcohol (PVA): a at different pH and b temperature values using CH4 as a feed
Table 1.
Methanol production by encapsulated methanotrophs
| Polymer | Methanotrophs | Methanol (mM) | References |
|---|---|---|---|
| Alginate | Methylocella tundrae | 3.75 | [14] |
| Methylosinus sporium | 3.43 | [46] | |
| Methylosinus trichosporium OB3b | 3.70 | [13] | |
| Polymer matrix | M. sporium | 2.34 | [11] |
| Silica-gel | M. sporium | 3.73 | [46] |
| Polyvinyl alcohol | M. sporium | 1.94 | [33] |
| M. bryophila and M. stellata | 5.37 | This study |
Reusability
Methanol production under repeated batch conditions up to five cycles (Fig. 5) revealed that it declines in the case of free co-culture. The decline in residual methanol production was up to 69.7% within 3 cycles of reusage. Thereafter, it reduced drastically to 23.8% by the end of five cycles. Whereas, encapsulated cells showed higher residual methanol production of 83.8 and 61.9% at the end of three and five cycles, respectively. Overall, encapsulated cells showed 2.6-fold higher stability for methanol production compared to free cells in repeated batch conditions. Here, reduction in methanol production might be associated with the loss of higher biocatalytic activity by free cells as compared with immobilized cells [13, 35]. These results suggest that immobilization is a beneficial to improve the stability of co-culture. Senko et al. [11] reported a significant reduction of 90% in residual methanol production by encapsulated pure culture strains of M. sporium and M. trichosporium within polymeric matrix up to 3 cycles of reuses. Similarly, lower residual production of 57.5% after five cycles of reuses for alginate encapsulated M. tundra was also noted previously [14].
Fig. 5.
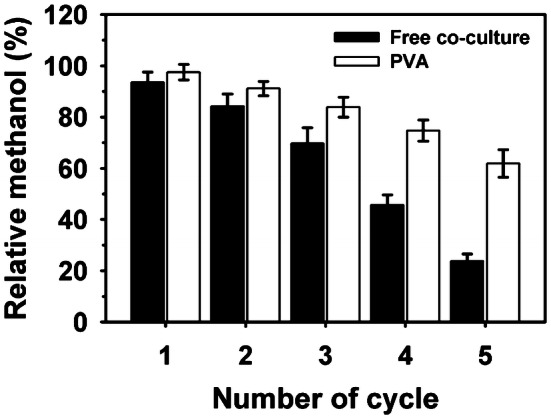
Reusability of free and immobilized co-cultures of Methylocystis bryophila and Methyloferula stellata within polyvinyl alcohol (PVA) using CH4 as a feed
Conclusion
Biotransformation of GHG such as CH4 to value-added products by methanotrophs to methanol have been recognized as a suitable tactic for its mitigation. This investigation demonstrated that the methanotrophs based co-culture approach improves the methanol production from CH4. Co-culture consisting of M. bryophila and M. stellata showed better methanol production over their pure cultures. The immobilization of co-cultures within PVA retained high RE for methanol production and led to better stability over free cells. Further, encapsulated cells retained significantly improved residual methanol production efficiency in repeated batch culture conditions than those of free cells. This study revealed the beneficial influence of using co-culture and immobilized cells for better methanol production using GHGs over pure cultures and free cells, respectively. Further, utilization of biogas originated form anaerobic digestion process of biowaste as a feedstock by co-cultures can be more useful for sustainable development.
Acknowledgements
This work was supported by Brain Pool grant (NRF-2019H1D3A201060226) by National Research Foundation of Korea (NRF) to work at Konkuk University. This research was supported by Basic Science Research Program through the NRF funded by the Ministry of Science, ICT & Future Planning (NRF-2020H1D3A2A01060467, NRF-2019R1C1C11009766). This work was also supported by KU Research Professor program of Konkuk University. This paper was supported by Konkuk University Researcher Fund in 2018.
Compliance with Ethical Standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/21/2021
A Correction to this paper has been published: 10.1007/s12088-021-00946-2
Contributor Information
Vipin Chandra Kalia, Email: vckaliaku@gmail.com.
Jung-Kul Lee, Email: jkrhee@konkuk.ac.kr.
References
- 1.Fei Q, Guarnieri MT, Tao L, Laurens LML, Dowe N, Pienkos PT. Bioconversion of natural gas to liquid fuel: opportunities and challenges. Biotechnol Adv. 2014;32:596–614. doi: 10.1016/j.biotechadv.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Patel SKS, Mardina P, Kim S-Y, Lee J-K, Kim I-W. Biological methanol production by a type II methanotroph Methylocystis bryophila. J Microbiol Biotechnol. 2016;26:717–724. doi: 10.4014/jmb.1601.01013. [DOI] [PubMed] [Google Scholar]
- 3.Patel SKS, Selvaraj C, Mardina P, Jeong JH, Kalia VC, Kang YC, Lee J-K. Enhancement of methanol production from synthetic gas mixture by Methylosinus sporium through covalent immobilization. Appl Energy. 2016;171:383–391. doi: 10.1016/j.apenergy.2016.03.022. [DOI] [Google Scholar]
- 4.Kondaveeti S, Patel SKS, Pagolu R, Li J, Kalia VC, Choi M-S, Lee J-K. Conversion of simulated biogas to electricity: sequential operation of methanotrophic reactor effluents in microbial fuel cell. Energy. 2019;189:116309. doi: 10.1016/j.energy.2019.116309. [DOI] [Google Scholar]
- 5.Pieja AJ, Morse MC, Cal AJ. Methane to bioproducts: the future of the bioeconomy? Curr Opin Chem Biol. 2017;41:123–131. doi: 10.1016/j.cbpa.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 6.Patel SKS, Kondaveeti S, Otari SV, Pagolu RT, Jeong SH, Kim SC, Cho BK, Kang YC, Lee J-K. Repeated batch methanol production from a simulated biogas mixture using immobilized Methylocystis bryophila. Energy. 2018;145:477–485. doi: 10.1016/j.energy.2017.12.142. [DOI] [Google Scholar]
- 7.Patel SKS, Jeon MS, Gupta RK, Jeon Y, Kalia VC, Kim SC, Cho B-K, Kim DR, Lee J-K. Hierarchical macro-porous particles for efficient whole-cell immobilization: application in bioconversion of greenhouse gases to methanol. ACS Appl Mater Interfaces. 2019;11:18968–18977. doi: 10.1021/acsami.9b03420. [DOI] [PubMed] [Google Scholar]
- 8.Strong PJ, Kalyuzhnaya M, Silverman J, Clarke WP. A methanotrophs-based biorefinery: potential scenarios for generating multiple products from a single fermentation. Bioresour Technol. 2016;215:314–323. doi: 10.1016/j.biortech.2016.04.099. [DOI] [PubMed] [Google Scholar]
- 9.Patel SKS, Mardina P, Kim D, Kim S-Y, Kalia VC, Kim I-W, Lee J-K. Improvement in methanol production by regulating the composition of synthetic gas mixture and raw biogas. Bioresour Technol. 2016;218:202–208. doi: 10.1016/j.biortech.2016.06.065. [DOI] [PubMed] [Google Scholar]
- 10.Cantera S, Sánchez-Andrea I, Lebrero R, García-Encina PA, Stams AJM, Muñoz R. Multi-production of high added market value metabolites from diluted methane emissions via methanotrophic extremophiles. Bioresour Technol. 2018;267:401–407. doi: 10.1016/j.biortech.2018.07.057. [DOI] [PubMed] [Google Scholar]
- 11.Senko O, Makhlis T, Bihovsky M, Podmasterev V, Efremenko E, Razumovsky S, Varfolomeyev S (2007) Methanol production in the flow system with immobilized cells Methylosinus sporium, XV International Workshop on Bioencapsulation. Vienna, Austria. September 6–8, pp. 2–16, 1–4. https://impascience.eu/bioencapsulation/340_contribution_texts/2007-09-06_P2-16.pdf
- 12.Yoo Y-S, Hana J-S, Ahn C-M, Kim C-G. Comparative enzyme inhibitive methanol production by Methylosinus sporium from simulated biogas. Environ Technol. 2015;36:983–991. doi: 10.1080/09593330.2014.971059. [DOI] [PubMed] [Google Scholar]
- 13.Taylor A, Molzahn P, Bushnell T, Cheney C, LaJeunesse M, Azizian M, Semprni L. Immobilization of Methylosinus trichosporium OB3b for methanol production. J Ind Microbiol Biotechnol. 2018;45:201–211. doi: 10.1007/s10295-018-2010-z. [DOI] [PubMed] [Google Scholar]
- 14.Mardina P, Li J, Patel SKS, Kim I-W, Lee J-K, Selvaraj C. Potential of immobilized whole-cell Methylocella tundrae as a biocatalyst for methanol production from methane. J Microbiol Biotechnol. 2016;26:1234–1241. doi: 10.4014/jmb.1602.02074. [DOI] [PubMed] [Google Scholar]
- 15.Patel SKS, Singh R, Kumar A, Jeong JH, Jeong SH, Kalia VC, Kim I-W, Lee J-K. Biological methanol production by immobilized Methylocella tundrae using simulated biohythane as a feed. Bioresour Technol. 2017;241:922–927. doi: 10.1016/j.biortech.2017.05.160. [DOI] [PubMed] [Google Scholar]
- 16.Patel SKS, Kumar V, Mardina P, Li J, Lestari R, Kalia VC, Lee J-K. Methanol production from simulated biogas mixtures by co-immobilized Methylomonas methanica and Methylocella tundrae. Bioresour Technol. 2018;263:25–32. doi: 10.1016/j.biortech.2018.04.096. [DOI] [PubMed] [Google Scholar]
- 17.Kumar V, Patel SKS, Gupta RK, Otari SV, Gao H, Lee JK, Zhang L. Enhanced saccharification and fermentation of rice straw by reducing the concentration of phenolic compounds using an immobilized enzyme cocktail. Biotechnol J. 2019;14:1800468. doi: 10.1002/biot.201800468. [DOI] [PubMed] [Google Scholar]
- 18.Kumar A, Park GD, Patel SKS, Kondaveeti S, Otari S, Anwar MZ, Kalia VC, Singh Y, Kim SC, Cho B-K, Sohn J-H, Kim DR, Kang YC, Lee J-K. SiO2 microparticles with carbon nanotube-derived mesopores as an efficient support for enzyme immobilization. Chem Eng J. 2019;359:1252–1264. doi: 10.1016/j.cej.2018.11.052. [DOI] [Google Scholar]
- 19.Kondaveeti S, Patel SKS, Woo J, Wee JH, Kim S-Y, Al-Raoush RI, Kim I-W, Kalia VC, Lee J-K. Characterization of cellobiohydrolases from Schizophyllum commune KMJ820. Indian J Microbiol. 2020;60:160–166. doi: 10.1007/s12088-019-00843-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandey D, Patel SKS, Singh R, Kumar P, Thakur V, Chand D. Solvent-tolerant acyltransferase from Bacillus sp. APB-6: purification and characterization. Indian J Microbiol. 2019;59:500–507. doi: 10.1007/s12088-019-00836-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel SKS, Gupta RK, Kumar V, Mardina P, Lestari R, Kalia VC, Choi M-S, Lee J-K. Influence of metal ions on the immobilization of β-glucosidase through protein-inorganic hybrids. Indian J Microbiol. 2019;59:370–374. doi: 10.1007/s12088-019-0796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel SKS, Choi H, Lee J-K. Multimetal-based inorganic–protein hybrid system for enzyme immobilization. ACS Sustain Chem Eng. 2019;7:13633–13638. doi: 10.1021/acssuschemeng.9b02583. [DOI] [Google Scholar]
- 23.Lee J-K, Patel SKS, Sung BH, Kalia VC. Biomolecules from municipal and food industry wastes: an overview. Bioresour Technol. 2020;298:122346. doi: 10.1016/j.biortech.2029.122346. [DOI] [PubMed] [Google Scholar]
- 24.Otari SV, Patel SKS, Kalia VC, Lee J-K. One-step hydrothermal synthesis of magnetic rice straw for effective lipase immobilization and its application in esterification reaction. Bioresour Technol. 2020;302:122887. doi: 10.1016/j.biortech.2020.122887. [DOI] [PubMed] [Google Scholar]
- 25.Patel SKS, Purohit HJ, Kalia VC. Dark fermentative hydrogen production by defined mixed microbial cultures immobilized on ligno-cellulosic waste materials. Int J Hydrog Energy. 2010;35:10674–10681. doi: 10.1016/j.ijhydene.2010.03.025. [DOI] [Google Scholar]
- 26.Patel SKS, Kumar P, Mehariya S, Purohit HJ, Lee J-K, Kalia VC. Enhancement in hydrogen production by co-cultures of Bacillus and Enterobacter. Int J Hydrog Energy. 2014;39:14663–14668. doi: 10.1016/j.ijhydene.2014.07.084. [DOI] [Google Scholar]
- 27.Kumar P, Sharma R, Ray S, Mehariya S, Patel SKS, Lee J-K, Kalia VC. Dark fermentative bioconversion of glycerol to hydrogen by Bacillus thuringiensis. Bioresour Technol. 2015;182:383–388. doi: 10.1016/j.biortech.2015.01.138. [DOI] [PubMed] [Google Scholar]
- 28.Patel SKS, Kumar P, Singh S, Lee J-K, Kalia VC. Integrative approach to produce hydrogen and polyhydroxybutyrate from biowaste using defined bacterial cultures. Bioresour Technol. 2015;176:136–141. doi: 10.1016/j.biortech.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 29.Patel SKS, Lee J-K, Kalia VC. Dark-fermentative biological hydrogen production from mixed biowastes using defined mixed cultures. Indian J Microbiol. 2017;57:171–176. doi: 10.1007/s12088-017-0643-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel SKS, Ray S, Prakash J, Wee JH, Kim S-Y, Lee J-K, Kalia VC. Co-digestion of biowastes to enhance biological hydrogen process by defined mixed bacterial cultures. Indian J Microbiol. 2019;59:154–160. doi: 10.1007/s12088-018-00777-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel SKS, Kim J-H, Kalia VC, Lee J-K. Antimicrobial activity of amino-derivatized cationic polysaccharides. Indian J Microbiol. 2019;59:96–99. doi: 10.1007/s12088-018-00764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prakash J, Sharma R, Patel SKS, Kim IW, Kalia VC. Biohydrogen production by co-digestion of domestic wastewater and biodiesel industry effluent. PLoS ONE. 2018;13:e0199059. doi: 10.1371/journal.pone.0199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Razumovsky SD, Efremenko EN, Makhlis TA, Senko OV, Bikhovsky MY, Podmasterev VV, Varfolomeev SD. Effect of immobilization on the main dynamic characteristics of the enzymatic oxidation of methane to methanol by bacteria Methylosinus sporium B-2121. Russ Chem Bull Int Ed. 2008;57:1633–1636. doi: 10.1007/s11172-008-0211-8. [DOI] [Google Scholar]
- 34.Han J-S, Ahn C-M, Mahanty B, Kim C-G. Partial oxidative conversion of methane to methanol through selective inhibition of methanol dehydrogenase in methanotrophic consortium from landfill cover soil. Appl Biochem Biotechnol. 2013;171:1487–1499. doi: 10.1007/s12010-013-0410-0. [DOI] [PubMed] [Google Scholar]
- 35.Patel SKS, Kalia VC, Joo JB, Kang YC, Lee J-K. Biotransformation of methane into methanol by methanotrophs immobilized on coconut coir. Bioresour Technol. 2020;297:122433. doi: 10.1016/j.biortech.2019.122433. [DOI] [PubMed] [Google Scholar]
- 36.Patel SKS, Shanmugam R, Kalia VC, Lee J-K. Methanol production by polymer-encapsulated methanotrophs from simulated biogas in the presence of methane vector. Bioresour Technol. 2020;304:123022. doi: 10.1016/j.biortech.2020.123022. [DOI] [PubMed] [Google Scholar]
- 37.Gao H, Li J, Sivakumar D, Kim T-S, Patel SKS, Kalia VC, Kim I-W, Zhang Y-W, Lee J-K. NADH oxidase from Lactobacillus reuteri: a versatile enzyme for oxidized cofactor regeneration. Int J Biol Macromol. 2019;123:629–636. doi: 10.1016/j.ijbiomac.2018.11.096. [DOI] [PubMed] [Google Scholar]
- 38.Singh RK, Singh R, Sivakumar D, Kondaveeti S, Kim T, Li J, Sung BH, Cho B-K, Kim DR, Kim SC, Kalia VC, Zhang Y-HPJ, Zhao H, Kang YC, Lee J-K. Insights into cell-free conversion of CO2 to chemicals by a multienzyme cascade reaction. ACS Catal. 2018;8:11085–11093. doi: 10.1021/acscatal.8b02646. [DOI] [Google Scholar]
- 39.Kondaveeti S, Kim I-W, Otari S, Patel SKS, Pagolu R, Losetty V, Kalia VC, Lee J-K. Co-generation of hydrogen and electricity from biodiesel process effluents. Int J Hydrog Energy. 2019;44:27285–27296. doi: 10.1016/j.ijhydene.2019.08.258. [DOI] [Google Scholar]
- 40.Kondaveeti S, Pagolu R, Patel SKS, Kumar A, Bisht A, Dad D, Kalia VC, Kim I-W, Lee J-K. Bioelectrochemical detoxification of phenolic compounds during enzymatic pre-treatment of rice straw. J Microbiol Biotechnol. 2019;29:1760–1768. doi: 10.4014/jmb.1909.09042. [DOI] [PubMed] [Google Scholar]
- 41.Su Z, Ge X, Zhang W, Wang L, Yu Z, Li Y. Methanol production from biogas with a thermotolerant methanotrophic consortium isolated from an anaerobic digestion system. Energy Fuels. 2017;31:2970–2975. doi: 10.1021/acs.energyfuels.6b03471. [DOI] [Google Scholar]
- 42.AlSayed A, Fergala A, Khattab S, Elsharkawy A, Eldyasti A. Optimization of methane bio-hydroxylation using waste activated sludge mixed culture of type I methanotrophs as biocatalyst. Appl Energy. 2018;211:755–763. doi: 10.1016/j.apenergy.2017.11.090. [DOI] [Google Scholar]
- 43.Kim I-T, Yoo Y-S, Yoon Y-H, Lee Y-E, Jo J-H, Jeong W, Kim K-S. Bio-methanol production using treated domestic wastewater with mixed methanotroph species and anaerobic digester biogas. Water. 2018;10:1414. doi: 10.3390/w10101414. [DOI] [Google Scholar]
- 44.Markowska A, Michalkiewicz B. Biosynthesis of methanol from methane by Methylosinus trichosporium OB3b. Chem Pap. 2009;63:105–110. doi: 10.2478/s11696-008-000-5. [DOI] [Google Scholar]
- 45.Xin J-Y, Cui J-R, Niu J-Z, Hua S-F, Xia C-G, Li S-B, Zhu L-M. Biosynthesis of methanol from CO2 and CH4 by methanotrophic bacteria. Biotechnology. 2004;3:67–71. doi: 10.3923/biotech.2004.67.71. [DOI] [Google Scholar]
- 46.Patel SKS, Jeong J-H, Mehariya S, Otari SV, Madan B, Haw JR, Lee J-K, Zhang L, Kim I-W. Production of methanol from methane by encapsulated Methylosinus sporium. J Microbiol Biotechnol. 2016;26:2098–2105. doi: 10.4014/jmb.1608.08053. [DOI] [PubMed] [Google Scholar]