Abstract
Red flour beetle (Tribolium castaneum) is one of the most destructive pests of stored cereals worldwide. The essential oil (EO) of Artemisia vulgaris (mugwort) is known to be a strong toxicant that inhibits the growth, development, and reproduction of T. castaneum. However, the molecular mechanisms underlying the toxic effects of A. vulgaris EO on T. castaneum remain unclear. Here, two detoxifying enzymes, carboxylesterase (CarEs) and cytochrome oxidase P450 (CYPs), were dramatically increased in red flour beetle larvae when they were exposed to A. vulgaris EO. Further, 758 genes were differentially expressed between EO treated and control samples. Based on Gene Ontology (GO) analysis, numerous differentially expressed genes (DEGs) were enriched for terms related to the regulation of biological processes, response to stimulus, and antigen processing and presentation. Our results indicated that A. vulgaris EO disturbed the antioxidant activity in larvae and partially inhibited serine protease (SP), cathepsin (CAT), and lipase signaling pathways, thus disrupting larval development and reproduction as well as down-regulating the stress response. Moreover, these DEGs showed that A. vulgaris indirectly affected the development and reproduction of beetles by inducing the expression of genes encoding copper-zinc-superoxide dismutase (CuZnSOD), heme peroxidase (HPX), antioxidant enzymes, and transcription factors. Moreover, the majority of DEGs were mapped to the drug metabolism pathway in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Notably, the following genes were detected: 6 odorant binding proteins (OBPs), 5 chemosensory proteins (CSPs), 14 CYPs, 3 esterases (ESTs), 5 glutathione S-transferases (GSTs), 6 UDP-glucuronosyltransferases (UGTs), and 2 multidrug resistance proteins (MRPs), of which 8 CYPs, 2 ESTs, 2 GSTs, and 3 UGTs were up-regulated dramatically after exposure to A. vulgaris EO. The residual DEGs were significantly down-regulated in EO exposed larvae, implying that partial compensation of metabolism detoxification existed in treated beetles. Furthermore, A. vulgaris EO induced overexpression of OBP/CYP, and RNAi against these genes significantly increased mortality of larvae exposed to EO, providing further evidence for the involvement of OBP/CYP in EO metabolic detoxification in T. castaneum. Our results provide an overview of the transcriptomic changes in T. castaneum in response to A. vulgaris EO.
Keywords: Tribolium castaneum, Artemisia vulgaris, insecticidal activity, development, reproduction, metabolism system
Introduction
Insects are serious pests of stored products such as oilseeds, pulses, and cereals (Phillips and Throne, 2010; Wakil et al., 2010). These pests are distributed worldwide and cause huge economic losses (Flinn et al., 2010). The red flour beetle (Tribolium castaneum; Tenebrionidae) is one of the most economically crucial insect pests of stored-products (Salem et al., 2018) and a common pest of indoor food storage facilities (Wang Y. et al., 2015). The use of chemical fumigants (methyl bromide and phosphine) is currently one of the most effective methods for controlling stored-product insect pests (Athanassiou et al., 2015; Thompson and Reddy, 2016). Chemical insecticides, such as pyrethroids, phosphine, and dichlorvos, are also used to control these pests (Nayak et al., 2003; Hori and Kasaishi, 2005; Bomzan et al., 2018). However, improper and indiscriminate application of these insecticides can result in long-term undesirable effects on human health, non-targeted animals, and the environment (Dey, 2016). Because of the underlying ozone-depleting property of methyl bromide and the potential genotoxicity of phosphine to warm-blooded animals, the use of these insecticides has been restricted (Danse et al., 1984; Bell and Wilson, 1995; Kalsi and Palli, 2015). Therefore, the search for natural products of botanical-origin that have insecticidal activity has intensified in the scientific community (Isman, 2006).
Essential oils (EOs) of plants are mixtures of volatile compounds that have been widely used as bioactive agents. EOs are effective as insecticides (Qing et al., 2010), ovicides (Tunç et al., 2000), antifeedants (Huang et al., 2000), oviposition inhibitors (Ho et al., 1996), and repellents (Ogendo et al., 2008). Moreover, these chemicals affect certain biological parameters such as the life span, growth rate, and reproduction of insects (Naseri et al., 2017). The EO of Ocimum basilicum (Labiatae) exhibits highly toxic and repellent activity against T. castaneum adults when applied topically or impregnated on filter paper, grains, or glass pebbles (Obeng-Ofori and Reichmuth, 1997). Similar activities have been pointed out for EOs from Artemisia annua (Asteraceae) and Baccharis salicifolia (Asteraceae) (García et al., 2005; Goel et al., 2007), Trachyspermum ammi (Umbelliferae), Nigella sativa (Ranunculaceae), and Anethum graveolens (Umbelliferae) (Jana and Shekhawat, 2010; Khani and Asghari, 2012; Soni et al., 2016).
Numerous studies have reported the insecticidal proprieties of EOs made from Artemisia species against a wide range of insect pests. Artemisia is a genus of fragrant annual herb species belonging to the Compositae family, with a wide distribution in Asia, Europe, and North America (Kordali et al., 2006; Jiang et al., 2019). Previous studies show that Artemisia EOs exhibit antifeedant, repellent, and insecticidal activities against various insect pests including T. castaneum, Callosobruchus maculatuss (Bruchidae), Rhyzopertha dominica (Bostrichidae), and Plodia interpunctella (Phycitinae) (Tripathi et al., 2000; Sharifian et al., 2012; Borzoui et al., 2016). Furthermore, Artemisia EOs and their constituents also exhibit sublethal effects on the survival, fecundity, development, and life table parameters of Sitophilus granaries (Curculionidae), Tribolium confusum (Tenebrionidae), C. maculatuss, P. interpunctella, Trogoderma granarium (Dermestidae), and T. castaneum (Kordali et al., 2006; Wang et al., 2006; Stamopoulos et al., 2007; Izakmehri et al., 2013; Borzoui et al., 2016; Nouri-Ganbalani and Borzoui, 2017). The repellent and fumigant activities of Artemisia vulgaris (Asteraceae) EO has been demonstrated against Musca domestica (Muscidae) and the stored-product insect pest T. castaneum (Wang et al., 2006; Alizadeh et al., 2012). Furthermore, the insecticidal activity of A. vulgaris EO has been reported against C. maculatus, R. dominica, and T. castaneum (Sharifian et al., 2013). The insecticidal and larvicidal properties of A. vulgaris EO have been attributed to the presence of camphene, a chloro derivative of camphene, and α-Thujone (Pandey and Singh, 2017). Camphor has also been reported as one of the active components of A. vulgaris EO, which possesses moth repellent properties, and therefore has been used as a preservative in pharmaceuticals and cosmetics (Corrêa-Ferreira et al., 2014).
Currently, three modes of action of plant EOs have been identified against insect pests: (1) action on the nervous system of insects, thus suppressing normal growth, development, metamorphosis, and reproduction; (2) suppression of mitochondrial membrane respiratory enzymes; (3) regulation of oxygen consumption and carbon dioxide released (Pare and Tumlinson, 1999; Kostyukovsky et al., 2002; Mansour and Abdel-Hamid, 2015; Nascimento et al., 2015; Oboh et al., 2017). Insect genomes encode a variety of detoxification enzymes, including carboxylesterase (CarE, also called CCE/EST/CES), cytochrome oxidase P450 (CYP), and glutathione S-transferase (GST), to cope with xenobiotic compounds (Li et al., 2013; Zhang et al., 2013; Xiong et al., 2019b). Determination of the activity of these metabolic enzymes in insects after insecticide application has been widely used to try to understand the insecticidal mechanism of xenobiotic compounds (Francis et al., 2005). On the other hand, transcriptional regulation of gene expression in insects plays an significant role in insect response to various stresses (Chen et al., 2016).
However, molecular mechanisms underlying the insecticidal effects of A. vulgaris EO on stored-product insect pests remains unclear. In this paper, we aimed to assess the lethal effects of A. vulgaris EO against T. castaneum under laboratory conditions. Furthermore, we examined roles of the three most important detoxification enzymes (CarEs, CYPs, and GSTs) in the response of T. castaneum to A. vulgaris EO. To further explore how A. vulgaris affects the physiological activity of T. castaneum, we performed comparative transcriptome analyses of T. castaneum larvae treated with or without A. vulgaris EO. To validate the sequencing results and survey expression variation of response time-course for key responding genes, we provided bioassay analysis for a group of differentially expressed genes (DEGs) selected randomly and key responding genes. This paper provides the first overview of the molecular events underlying the response of T. castaneum to A. vulgaris EO. Our results provide a strong foundation for the development of plant EOs as novel, natural, and environmentally friendly insecticides.
Materials and Methods
Insect Culturing, Extraction, and Insecticidal Efficacy Assay of A. vulgaris EO
The T. castaneum Georgia-1 (GA-1) strain was used in this study. Insects were reared in whole wheat flour containing 5% brewer’s yeast in a growth chamber maintained at 30°C under a 14-h light/10-h dark cycle as described previously (Xiong et al., 2019b).
Artemisia vulgaris EO was extracted using the subcritical butane extraction apparatus (Henan Subcritical Biological Technology Co., Ltd., Anyang, China) with the following parameters: liquid:solid ratio = 30:1, temperature = 45°C, extraction time = 34 min, and particle size = 0.26 mm. The extract was subjected to hydrodistillation in a Clevenger-type apparatus for 2 h. To separate the EO from residual water, the oil/water emulsion was collected and stored at 4°C overnight. The EO was then transferred and stored in a glass bottle at room temperature until further use (Jiang et al., 2019).
To test the insecticidal efficacy, A. vulgaris EO was dissolved in acetone at seven different concentrations (1, 1.25, 1.7, 2.5, 5, 10, and 100%). Twenty-day-old T. castaneum larvae were used to determine the contact activity of A. vulgaris EO, as described by Lu et al. (2012). Fifteen larvae were treated with approximately 50 μL of each EO solution or acetone (control) for 1 min and then placed on Whatman filter paper for air drying for approximately 2 min. Subsequently, the larvae were transferred into an 8-ml glass vial and maintained under standard conditions. Mortality was recorded at 24, 48, and 72 h after treatment. Larvae were considered dead if they were unable to move or show response when disturbed with a pair of tweezers or a brush. Each experiment was repeated at least three times.
Assessment of Enzyme Activities
During enzyme preparation, we treated another set of test insects with A. vulgaris EO at 5% concentration (approximately median lethal concentration, LC50) or acetone (control); larvae were sampled at 12, 24, 48, 60, and 72 h, when they were weighed and washed twice or three times with pre-cooled saline. Then, larvae were dried using a piece of filter paper and ground in liquid nitrogen using a mortar and pestle. The resultant powder was transferred to a centrifuge tube to obtain 10% tissue homogenate in physiological saline and the issue suspension was centrifuged at 3,500 rpm for 10 min at 4°C. The supernatant was taken into a new tube for the enzyme activity test. Enzyme activity estimation and calculation method were refereed as previously described (Guo et al., 2012; Xiong et al., 2019b).
CarE enzyme activity was quantified using a spectrophotometric assay kit (catalog no. A133-1-1; Nanjing Jiancheng Bioengineering Institute, Nanjing, China), according to the manufacturer’s protocol, from proteins in the tissue samples that were extracted using a protein extraction kit (catalog no. A045-4-2; Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The extracted protein samples were incubated with 1-naphthalenol, and following incubation at room temperature for 30 min with agitation, absorbance was measured at 450 nm.
Total CYP activity was measured using a linked immunosorbent assay (ELISA) method using a commercially available kit (catalog no. H190-1, Nanjing Jiancheng Bioengineering Institute, Nanjing, China), by adding 50 μL of standards (7.5, 15, 30, 60, and 120 ng/mL) to the wells and drawing an A 4-parameter calibration curve (CurveExpert 1.4) for protein quantification. Then, 50 μL of the sample to be tested was added to the sample wells, with blank standards as a control, and washed, to which horseradish peroxidase (HRP) labeled antibody was also added. Samples were washed again, and HRP substrate was added and reacted for 5 min in the dark at 37°C, before absorbance at 450 nm was recorded.
Glutathione S-transferase activity test was conducted using a GST detection kit (catalog no. A004-1-1, Nanjing Jiancheng Bioengineering Institute, Nanjing, China), where one unit of activity was defined as a 1 μmol decrease in glutathione concentration in 1 mg of tissue protein for 1 min at 37°C, when the effects of non-enzymatic reactions are eliminated; absorbance was measured at 412 nm. Total protein concentration was determined using a BCA protein assay kit (catalog no. A045-4-2; Nanjing Jiancheng Bioengineering Institute, Nanjing, China), according to the manufacturer’s instructions.
RNA Isolation, cDNA Synthesis, and RNA-Seq
Because preliminary biochemical assays showed that CarE and CYP enzyme activities were significantly enhanced in larvae treated with 5% EO at 36 h, six samples (including three control and three EO treated samples harvested at the 36-h time point; each sample contained six larvae) were selected for RNA-Seq analysis. Total RNA was isolated from T. castaneum larvae using RNAisoTM Plus (TaKaRa, Kyoto, Japan), according to the manufacturer’s instructions, and treated with DNaseI at 37°C for 20 min to remove residual DNA contamination. The absorbance ratio at 260 and 280 nm (A260/A280) of total RNA was measured to assess the yield and purity of the sample, and the integrity of total RNA was assessed by electrophoresis on 1.5% agarose gel using an Agilent 2100 Bioanalyzer. Intact total RNA samples (1 μg) with an A260/A280 ratio > 1.8 were used for mRNA purification with Oligo(dT) magnetic beads. Then, the purified mRNA was fragmented and reverse transcribed to synthesize first-strand cDNA using Moloney murine leukemia virus reverse transcriptase and Oligo(dT)18 primer (TaKaRa, Kyoto, Japan). Double-stranded cDNA (dscDNA) was generated using the N6 random primer. The dscDNAs were end-repaired by the addition of phosphate at the 5′ end and nucleotide A at the 3′ end, and then ligated to an adaptor at the 3′ end. Two sequence-specific primers were used to amplify the ligation products. The PCR products were heat-denatured and circularized using splint oligo and DNA ligase. The resulting products were sequenced at Beijing Genomics Institute (BGI; Shenzhen, China) on the BGISEQ-500 platform1. Raw sequence reads were deposited in the NCBI Sequence Read Archive (SRA) database under the following accession numbers: control: SRR7646221, SRR7646223, and SRR7646224; EO treatment: SRR7646222, SRR7646225, and SRR7646226.
Filtering and Mapping of RNA-Seq Data
Raw sequence reads were filtered using FASTX2 to remove adaptor sequences, reads with >10% unknown nucleotides, low-quality reads (percentage of low-quality nucleotides >50% or nucleotides with sequence quality ≤5), and short reads (<20 bp). The T. castaneum genome sequence and corresponding annotations were downloaded from the BeetleBase database3 (version: T. castaneum 5.2), and clean reads were mapped to the reference gene sequences using Bowtie2 (Stephen et al., 2008) and to the T. castaneum reference genome using hierarchical indexing for spliced alignment of transcripts (HISAT) (Daehwan et al., 2015).
Determination of Gene Expression and Identification of DEGs
Gene expression levels were quantified using the RNA-Seq of the Expectation Maximization (RSEM) software package (Bo and Dewey, 2011). The relative transcript level of each gene was normalized and evaluated using fragments per kilobase of exons per million fragments mapped (FPKM) values. Repeatability among the three biological replicates was assessed using cluster analysis calculated by the Euclidean method. The mean FPKM value of three biological replicates of each gene was calculated to determine the expression level of each gene, and then genes with FPKM values ≥ 0.5 were analyzed further. The DEGs identified between the control and EO treatment groups were screened using the NOISeq package4 (Tarazona et al., 2011) of R. DEGs with a fold-change (FC) ≥ 2(| log2 ratio| ≥ 1 and divergence probability ≥ 0.8 were selected for further analysis.
Functional Annotations of DEGs
To understand the main biological and molecular functions of DEGs, annotations were performed using the Gene Ontology (GO) database5, according to the annotation information in the non-redundant (NR) database, and categorized into three standardized gene functional classifications (molecular functions, cellular components, and biological processes) using the Blast2GO pipeline (Ana et al., 2005). The Kyoto Encyclopedia of Genes and Genomes (KEGG6) database is the main public repository of known pathways. KEGG annotations were performed using the NCBI BLAST tool for all T. castaneum genes, followed by KEGG enrichment analysis of DEGs using the KOBAS 2.0 software7. Significance levels of all KEGG and GO terms were corrected by controlling the false discovery rate (FDR) of multiple pairwise comparisons, and terms with significantly enriched values (FDR < 0.05) were determined.
Validation of RNA-Seq Data by Quantitative Real-Time PCR (qRT-PCR)
To validate RNA-Seq results, the expression of 16 randomly selected DEGs was analyzed by qRT-PCR using the same RNA samples as those used for RNA-Seq analysis. The qRT-PCR was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) using SYBR Green Master Mix (Roche, Indianapolis, IN, United States) as a fluorescent dye, according to the manufacturer’s instructions. Three biological replications were performed for each experiment. Relative transcript levels of genes were determined using T. castaneum ribosomal protein S3 (rps3; GenBank accession number CB335975) as the internal reference gene, and gene expression levels were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001). Primers used for qRT-PCR are listed in Supplementary Table S1.
Induction of TcCYP4BN6 and TcOBPC11 Induction in Response to the A. vulgaris EO
Two genes (TcCYP4BN6 and TcOBPC11) that were significantly induced by A. vulgaris EO were selected for further study. To measure TcCYP4BN6 and TcOBPC11 induction patterns after EO treatments, 20-day-old larvae were first collected and then separated into three groups. Briefly, approximately 36 synchronous individuals in each group were loaded into 1.5-mL EP tubes and exposed to 120 μL of 5% A. vulgaris EO or acetone (3 beetles/10 μL). After soaking for 1 min, the treated larvae in each group were placed on filter paper and allowed to air dry for 2 min. Each group was then transferred to an 8-mL glass vial and maintained under standard conditions as previously described (Wei et al., 2019). The acetone exposure group served as the negative control group in this study.
RNAi to Evaluate the Roles of TcCYP4BN6 and TcOBPC11 Response to the A. vulgaris EO
To further investigate the functions of TcCYP4BN6 and TcOBPC11 in response to the A. vulgaris EO, RNA interference (RNAi) was used. Here, primers tailed with T7 promoters on the 5′ side were used to synthesize isoform-specific double stranded RNAs (dsRNAs), which target specific region for each isoform (Supplementary Table S1). dsRNAs are synthesized as previously described (Bi et al., 2019). A total of 200 ng of dsRNAs were injected into each larva of T. castaneum. Further, the collected 20-day-old larvae were separated into three groups and then used for ds-TcCYP4BN6 injection, ds-TcOBPC11 injection, and ds-GFP injection. The insects were further treated with 5% A. vulgaris EO to evaluate mortality. Prior to A. vulgaris EO exposure, RNAi efficiency was assessed by determining the transcript levels of the target genes after RNAi application. Further, at fifth day following dsRNA injection, three larvae were randomly selected for RNA extraction, and the RNA samples were employed in qRT-PCR. Briefly, approximately 50 μL of A. vulgaris EO was applied to the 15 ds-TcCYP4BN6 injection, ds-TcOBPC11 injection, and ds-GFP injection larvae. The mortalities of the larvae in the different treatment groups were recorded from 24 to 72 h after EO exposure. In this assay, the beetles were considered dead if they were unable to move and failed to respond when disturbed with a tweezer or brush. Each bioassay was replicated three times.
Statistical Analysis
Mortality data from the insecticidal efficacy assay of A. vulgaris EO were analyzed for the LC50 values and their 95% confidence intervals (95% CIs) by probit analysis using the SPSS program (SPSS Inc., Chicago, IL, United States). The effect of essential oils on enzymatic activities, the gene expression data, the mean values of the RNAi-treated versus the mean values of the control insects were compared by Student’s t-test and one-way analysis of variance (ANOVA) in combination with a Fisher’s least significant difference (LSD) multiple comparison tests, respectively, by using the SPSS statistics program (Chicago, IL, United States). All data are presented as the mean ± standard error (SE). Differences were considered significant at P-value < 0.05.
Results
Insecticidal Activity of A. vulgaris EO
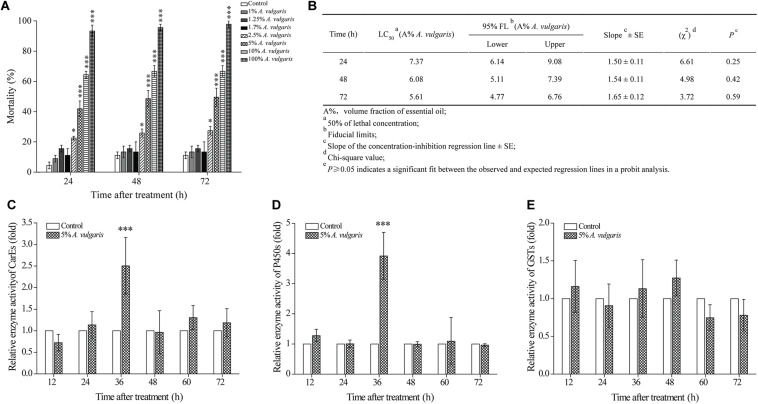
To investigate the insecticidal activity of A. vulgaris EO against T. castaneum larvae, we performed contact assays. Larvae treated with 1–1.7% A. vulgaris EO showed a slight increase in the mortality rate at 48, 60, and 72 h. By contrast, in larvae treated with EO concentrations of 2.5, 5, 10, and 100%, the cumulative mortality reached 27.41% ± 2.82%, 49.52% ± 5.97%, 66.67% ± 3.85%, and 99.78% ± 2.22%, respectively, at 72 h (Figure 1A). At the same dose, slightly increased effect of exposure was observed over the time course of 24–48–72 h. The largest concentration of essential oils (100% A. vulgaris EO) caused the mortality of 93.33% ± 3.84%, 95.56% ± 2.22%, and 97.78% ± 2.22% in T. castaneum after 24, 48, and 72 h of EO treatment, respectively. The corresponding LC50 values were 6.14, 5.11, and 4.77% A. vulgaris EO, respectively (Figure 1B). These results demonstrate that the EO caused dose-dependent increase in larval mortality, while there were not apparent time-dependent effects.
FIGURE 1.
(A) The effect of Artemisia vulgaris essential oil (EO) on Tribolium castaneum larvae. Larvae were treated with eight different concentrations (0.1, 1, 1.25, 1.7, 2.5, 5, 10, and 100%) of A. vulgaris EO or with acetone (control). Larval mortality was observed at 24, 48, and 72 h post-treatment. (B) Contact toxicity of A. vulgaris EO against T. castaneum larvae. At different times with 5% A. vulgaris EO on carboxylesterase (CarEs) (C), cytochrome P450 (CYPs) (D), and glutathione S-transferase (GSTs) (E) in larval T. castaneum in vivo. Fold change of the total enzyme activity was calculated by dividing the enzyme activity of each treatment by that of the acetone only control, which had been ascribed an arbitrary value of one. The times of 12, 24, 36, 48, 60, and 72 h were six different test points after insecticide treatment. Data represent mean ± SE of three independent experiments. Asterisks indicate significant differences between the control and EO treatment groups (∗P < 0.05, ∗∗∗P < 0.001; Student’s t-test).
Effect of A. vulgaris EO on Enzyme Activities in T. castaneum
Next, we determined the effect of A. vulgaris EO on the activities of CarE, CYP, and GST enzymes in T. castaneum larvae. In larvae treated with 5% EO (approximately LC50 for 72 h), activities of CarE and CYP enzymes were increased in a similar pattern from 12 to 72 h (Figures 1C,D). Activities of both CarE and CYP enzymes in EO treated larvae reached a peak at 36 h (2.50- and 3.92-fold, respectively) and declined thereafter (Figures 1C,D). By contrast, GST activity in EO treated larvae showed no significant change from 12 to 72 h (Figure 1E).
RNA-Seq and Read Alignment
To explore the gene expression profiles of T. castaneum in response to A. vulgaris EO treatment, six RNA-Seq libraries (three control and three EO treatment groups) were sequenced. An average of 23,647,023 raw reads were generated, from which 23,271,263 clean reads were obtained after filtering out low-quality sequences (Table 1). The ratio of clean reads to raw reads for the six libraries ranged from 99.66 to 99.85%. On average, 53.578% of the control reads mapped either to a unique location (52.16%) or to multiple locations (1.61%) in the T. castaneum genome. A similar proportion (54.74%) of the EO treatment group reads mapped to the T. castaneum genome, most of which mapped to a unique location (53.34%), and a small proportion mapped to multiple locations (1.40%; Table 1). These data demonstrate that all six libraries were of high quality (Kim et al., 2016).
TABLE 1.
Summary of read numbers based on the RNA-sequencing data.
| Category | Control |
Treatment |
||||
| Control_rep1 | Control_rep2 | Control_rep3 | 5% A. vulgaris_rep1 | 5% A. vulgaris_rep2 | 5% A. vulgaris_rep3 | |
| Total reads number | 23203638 | 23649862 | 24087570 | 24054117 | 24071256 | 21688417 |
| Total mapped reads | 13245867 | 10272901 | 14644323 | 15017197 | 10780866 | 12359145 |
| (57.09%) | (43.44%) | (60.80%) | (62.43%) | (44.79%) | (56.99%) | |
| Perfect match | 10762958 | 8295964 | 11945631 | 12317982 | 8834002 | 10034020 |
| (46.38%) | (35.08%) | (49.59%) | (51.21%) | (36.70%) | (46.26%) | |
| Mismatch | 2482909 | 1976937 | 2698692 | 2699215 | 1946864 | 2325125 |
| (10.70%) | (8.36%) | (11.20%) | (11.22%) | (8.09%) | (10.72%) | |
| Unique match | 12892239 | 9909736 | 14218684 | 14604563 | 10479759 | 12094489 |
| (55.56%) | (41.90%) | (59.03%) | (60.72%) | (43.54%) | (55.76%) | |
| Multi-position match | 353628 | 363165 | 425639 | 412634 | 301107 | 264656 |
| (1.52%) | (1.54%) | (1.77%) | (1.72%) | (1.25%) | (1.22%) | |
| Total unmapped reads | 9957771 | 13376961 | 9443247 | 9036920 | 13290390 | 9329272 |
| (42.91%) | (56.56%) | (39.20%) | (37.57%) | (55.21%) | (43.01%) | |
An average of 12,156 and 12,239 genes (80 to ≥2,000 bp in length) were detected in the control and EO treatment samples, respectively. Based on their length, these genes were divided into five categories. Approximately 50% of the genes were 500–1,500 bp in length, 20.19% ranged from 500 to 1,000 bp, and 22.47% ranged from 1,000 to 1,500 bp (Supplementary Figure S1A). The distribution of genes in each category was similar between the control and treatment samples (Supplementary Figure S1B). The majority of genes showed FPKM values ranging from 10 to 100 in both control (6,424 genes, 52.84%) and EO treatment groups (6,382 genes, 52.15%; Supplementary Figure S1B). A few genes showed FPKM values >1,000, indicating that only a small proportion of genes were expressed to high levels. No significant differences in FPKM values were detected between the control and EO treatment groups (Supplementary Figure S1B).
Next, we performed sequence saturation analysis to determine whether the number of detected genes increased proportionally with the number of sequence reads. The results indicated that the number of detected genes stopped increasing beyond 1.5 million sequence reads (Supplementary Figures S2G–L). To determine mRNA expression, redundancy and heterogeneity were considered as two key characteristics. Further, a small proportion of mRNAs were expressed at high levels, while the majority was expressed at low levels. Thus, we used the distribution of tag expression to assess the normality of RNA-seq data. In addition, the distribution of distinct tags over different tag abundance categories showed that the proportions of the control and EO treatment libraries were nearly equal (Supplementary Figures S2G–L). Both types of libraries exhibited similar distribution patterns, with more than 50% of detected genes showing a coverage of >80% (Supplementary Figures S2A–F). In total, the control and EO treatment libraries yielded 12,684 genes, thus providing abundant data for analyzing the influence of A. vulgaris EO on the physiological characteristics of T. castaneum larvae.
GO and KEGG Enrichment Analyses of DEGs
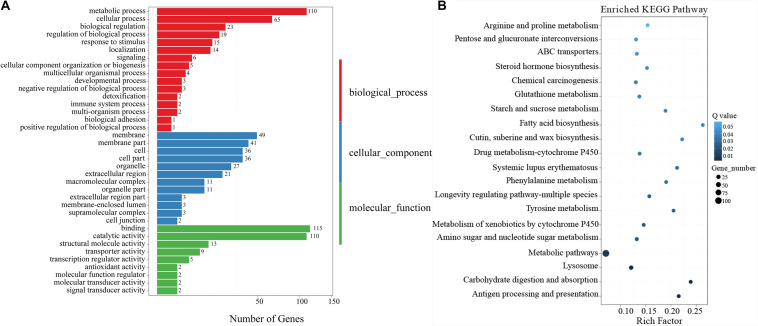
Functional annotations of DEGs were performed by GO enrichment analysis. A total of 142, 99, and 198 DEGs (P ≤ 0.05) were assigned to the cellular component, molecular function, and biological process categories, respectively, and further divided into 37 sub-categories (16 in biological processes, 12 in cellular components, and 9 in molecular function categories; Figure 2A). In these categories, regulation of biological processes (GO:0050789), developmental processes (GO: 0032502), and response to stimulus (GO: 0050896) were closely associated with the influence of A. vulgaris EO on T. castaneum larvae. A total of 19 DEGs encoding Wnt 8a, netrin receptor UNC5C, and myosin 9, which play significant roles in insect eclosion and reproduction, were associated with the regulation of biological processes (GO:0050789; Table 2). The GO term response to stimulus (GO: 0050896) contained 15 DEGs, including Peroxidase, Defensin1, and Defensin2 (Supplementary Table S4), which were involved in immunity and stress responses simultaneously. Moreover, the GO terms binding (GO: 0005488), catalytic activity (GO: 0003824) with 109 DEGs, and metabolic processes (GO: 0008152) contained 115, 109, and 100 DEGs, respectively (Figure 2A).
FIGURE 2.
(A) Gene Ontology (GO) enriched terms of differentially expressed genes (DEGs) of T. castaneum after exposure of its larvae to 5% A. vulgaris essential oil (EO) versus the control group. The x-axis is the number of DEGs involved in each term. The y-axis lists the sub-GO terms under categories of biological processes, cellular components, and molecular functions. (B) The most enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of T. castaneum after exposure of its larvae to 5% A. vulgaris EO. Pathway significance is shown together with Q-value (color), rich factor (vertical ordinate), and a number of involved genes (size of circles).
TABLE 2.
Differentially expressed genes (DEGs) involved in regulation of biological process.
| Gene ID | Log2 ratio (T/C) | Regulation (T/C) | Protein | P-value | Phenotype after RNAi of DEGs in T. castaneuma |
| LOC664422 | 2.50 | Up | Gadd45 | 0 | / |
| LOC663364 | 2.10 | Up | DETS 6 | 5.37E-218 | / |
| LOC662363 | 2.05 | Up | ADAMTS7 | 0 | / |
| LOC100141722 | 1.58 | Up | Octopamine receptor 1 | 7.54E-05 | / |
| LOC103314939 | 1.37 | Up | Transcription factor GATA 4 | 7.94E-35 | / |
| LOC663803 | 1.10 | Up | Eukaryotic translation initiation factor 4E-binding protein 2 | 3.44E-205 | / |
| LOC660084 | −1.00 | Down | Wnt 8a | 7.50E-06 | / |
| LOC664175 | −1.02 | Down | Netrin receptor UNC5C | 5.56E-156 | / |
| LOC663899 | −1.12 | Down | Myosin 9 | 0 | / |
| LOC658656 | −1.13 | Down | Apterous | 9.30E-31 | / |
| LOC656430 | −1.15 | Down | Fork head domain-containing protein FD4 | 4.07E-19 | / |
| LOC100142549 | −1.17 | Down | Targeting protein for xklp2 | 9.34E-48 | 80% embryo/egg not developed/not fertilizedc |
| LOC641604 | −1.22 | Down | Orthodenticle 2 | 2.50E-05 | / |
| LOC656829 | −1.27 | Down | Transmembrane protein 80 | 6.07E-10 | / |
| LOC641600 | −1.29 | Down | Scr | 2.78E-87 | 30%b; 30 ∼ 50% embryo/egg with embryonic tissue, no cuticlec |
| LOC662160 | −1.31 | Down | TIMELESS-interacting protein | 9.24E-22 | / |
| LOC662392 | −1.47 | Down | Histone H2A | 0 | / |
| LOC103314024 | −1.81 | Down | Enhancer of split malpha protein | 5.57E-43 | / |
| LOC659737 | −2.04 | Down | Neuropeptide Y receptor | 5.35E-05 | / |
C, control; T, 5% A. vulgaris treatment; Gadd45, growth arrest and DNA damage-indueible genes; DETS 6, DNA-binding protein DETS-6; ADAMTS7, A disintegrin and metalloproteinase with thrombospondin motifs 7; Scr, Sex combs reduced; ametamorphosis and survival after RNAi were analyzed (http://ibeetle-base.uni-goettingen.de); bLethalities 11 days after larval injection (includes death as larva, prepupa, and pupa); c14 days after female pupal injection.
The results of KEGG analysis showed that 511 DEGs were assigned to 262 biological pathways (Supplementary Table S2). A total of 19 pathways including antigen processing and presentation (ko04612), carbohydrate digestion and absorption (ko04973), lysosome (ko04142), and metabolism of xenobiotics by cytochrome P450 (ko00980) were significantly enriched in DEGs compared with the whole transcriptome background, with a Q-value ≤ 0.05 (Figure 2B and Supplementary Table S2). A total of 28 DEGs were involved in the lysosome pathway (ko04142) including lysosomal alpha-mannosidase like, beta-glucuronidase (BG), cathepsin L precursor (CatL precursor), alpha-N-acetylgalactosaminidase, cathepsin B1 (CatB1), arylsulfatase B-like, serine protease P38 (SPP38), MD-2-related lipid-recognition (ML), Lipase 1, Lipase 3-like, and Lipase 4 (Table 3), which probably contributed to the development and reproduction of T. castaneum. Moreover, 14 DEGs participated in antigen processing and presentation (Table 3 and Supplementary Table S5), of which six included members of the lipase family that overlapped with the lysosome pathway. These six genes are likely involved in the stress response and innate immunity as well as in the reproduction and development of larvae. Notably, 44 DEGs were responsible for three drug metabolism pathways (ko00980, ko00982, and ko00983), which contained genes encoding flavin-containing monooxygenase (FMO), esterase (EST), UDP-glucuronosyltransferase (Ugt), BG, xanthine dehydrogenase (XDH), GST, and CYP enzymes (Table 4). The DEGs also encoded odorant binding proteins (OBPs) and chemosensory proteins (CSPs), which have been suggested to be involved in drug metabolism in Plutella xylostella (Bakhetia et al., 2005). Together, our results demonstrate considerable differences in physiological processes between EO treatment and control samples.
TABLE 3.
Differentially expressed genes associated with the lysosome pathway.
| Gene ID | Log2 ratio (T/C) | Regulation (T/C) | P-value | Protein | Phenotype after RNAi of DEGs in T. castaneuma |
| LOC658343 | 2.92 | Up | 1.06E-24 | CP 1 | / |
| LOC660669 | 2.62 | Up | 8.30E-75 | Cat L precursor | / |
| LOC659367 | 2.08 | Up | 1.84E-77 | Cat L precursor | / |
| LOC664569 | 2.04 | Up | 0 | Beta-hexosaminidase subunit alpha | / |
| LOC663215 | 1.96 | Up | 3.22E-17 | MFS15 | / |
| LOC657117 | 1.81 | Up | 1.26E-06 | Cat B1 like | / |
| LOC107398158 | 1.79 | Up | 7.41E-05 | Cat L1 like | / |
| LOC100141821 | 1.48 | Up | 4.30E-12 | Snmp1 | / |
| LOC659039 | 1.41 | Up | 3.36E-28 | Lipase member K | / |
| LOC103312286 | 1.30 | Up | 5.53E-295 | Lipopolysaccharide | / |
| LOC664564 | 1.28 | Up | 3.89E-07 | Glucosylceramidase like | / |
| LOC660551 | 1.26 | Up | 1.04E-307 | Cat L | / |
| LOC103313498 | 1.10 | Up | 0 | Crammer | / |
| LOC103314303 | 1.03 | Up | 1.29E-19 | CD63 antigen like | / |
| LOC661588 | 1.03 | Up | 4.25E-67 | Arylsulfatase B | / |
| LOC656698 | −1.11 | Down | 2.40E-18 | Lysosomal alpha-mannosidase like | / |
| LOC657825 | −1.12 | Down | 1.27E-15 | BG | / |
| LOC660368 | −1.16 | Down | 3.69E-175 | Cat L precursor | 100%b, 40% vitellogenic egg chamber not presentc |
| LOC662197 | −1.28 | Down | 9.74E-90 | Alpha-N-acetylgalactosaminidase | / |
| LOC663117 | −1.30 | Down | 7.73E-28 | Cat B1 | / |
| LOC659525 | −1.32 | Down | 5.41E-84 | Arylsulfatase B like | / |
| LOC655752 | −1.36 | Down | 9.57E-12 | Serine protease P38 | / |
| LOC103312725 | −1.63 | Down | 3.19E-11 | ML | / |
| LOC661631 | −1.75 | Down | 8.99E-81 | Lipase 1 | / |
| LOC659587 | −1.78 | Down | 0 | Arylsulfatase B like | / |
| LOC659121 | −1.82 | Down | 2.14E-132 | Lipase 1 like | 100% previtellogenic egg chamber orientation irregularc |
| LOC661718 | −2.14 | Down | 6.36E-24 | Lipase 3 like | / |
| LOC661966 | −2.97 | Down | 2.16E-296 | Lipase 4 | 20%b |
C, control; T, 5% A. vulgaris treatment; CP, cysteine proteinase; Cat, cathepsin; Snmp1, sensory neuron membrane protein 1; MFS15, major facilitator superfamily transporter 15; BG, beta-glucuronidase; ML, MD-2-related lipid-recognition; ametamorphosis and survival after RNAi were analyzed (http://ibeetle-base.uni-goettingen.de); bLethalities 11 days after larval injection (includes death as larva, prepupa, pupa); c13 days after female pupal injection.
TABLE 4.
Differentially expressed genes (DEGs) involved in drug metabolism.
| Gene ID | Log2 ratio (T/C) | Regulation (T/C) | P-value | Protein | Metabolism detoxification process |
| LOC656161 | 4.28 | Up | 2.26E-65 | OBPC11 | Phase 0 |
| LOC660270 | 4.03 | Up | 0 | CYP4BN6 | Phase I |
| LOC100240681 | 2.12 | Up | 6.93E-09 | OBPC17 | Phase 0 |
| LOC664471 | 2.04 | Up | 5.36E-52 | CYP6BQ7 | Phase I |
| LOC658048 | 2.03 | Up | 2.33E-37 | CYP6A2 | Phase I |
| LOC107398166 | 1.91 | Up | 1.98E-07 | Ugt86Dg | Phase II |
| LOC664595 | 1.91 | Up | 2.23E-217 | OBP10 | Phase 0 |
| LOC662506 | 1.87 | Up | 0 | α-EST5 | Phase I |
| LOC657602 | 1.76 | Up | 3.47E-76 | GSTs6 | Phase II |
| LOC103313315 | 1.63 | Up | 6.44E-23 | CYP351A2 | Phase I |
| LOC100141953 | 1.58 | Up | 2.82E-06 | Ugt2B7-like | Phase II |
| LOC656937 | 1.47 | Up | 1.65E-114 | MRP4 | Phase III |
| LOC661102 | 1.43 | Up | 1.12E-09 | α-EST2 | Phase I |
| LOC664599 | 1.41 | Up | 5.71E-155 | OBPC02 | Phase 0 |
| LOC656243 | 1.40 | Up | 0 | OBPC12 | Phase 0 |
| LOC661621 | 1.37 | Up | 0 | α-EST6 | Phase I |
| LOC661270 | 1.36 | Up | 2.72E-31 | CSP12 | Phase 0 |
| LOC657697 | 1.33 | Up | 0 | CSP20 | Phase 0 |
| LOC664285 | 1.31 | Up | 2.63E-37 | CYP9Z2 | Phase I |
| LOC657454 | 1.27 | Up | 2.98E-30 | CYP9AC1 | Phase I |
| LOC661469 | 1.24 | Up | 0.000982 | CSP17 | Phase 0 |
| LOC656120 | 1.22 | Up | 1.19E-27 | Ugt2B16 | Phase II |
| LOC659847 | 1.15 | Up | 4.96E-63 | MRP49 | Phase III |
| LOC659009 | 1.13 | Up | 4.50E-45 | GSTs7 | Phase II |
| LOC656770 | 1.12 | Up | 6.93E-55 | CYP6BK11 | Phase I |
| LOC661219 | 1.12 | Up | 1.22E-37 | CSP11 | Phase 0 |
| LOC657560 | 1.07 | Up | 3.78E-13 | CYP6A14 | Phase I |
| LOC657825 | −1.12 | Down | 1.27E-15 | BG | Phase II |
| LOC100142486 | −1.17 | Down | 1.98E-46 | Ugt86Dc | Phase II |
| LOC656306 | −1.22 | Down | 3.25E-57 | CYP4BN5 | Phase I |
| LOC655331 | −1.24 | Down | 5.47E-242 | GSTe7 | Phase II |
| LOC655415 | −1.33 | Down | 3.03E-92 | GSTe8 | Phase II |
| LOC658886 | −1.42 | Down | 1.80E-08 | XDH | Phase I |
| LOC661967 | −1.48 | Down | 7.35E-79 | Ugt2B20 | Phase II |
| LOC659878 | −1.51 | Down | 1.31E-71 | CYP4Q1 | Phase I |
| LOC662300 | −1.99 | Down | 3.03E-22 | CYP351A3 | Phase I |
| LOC661799 | −2.15 | Down | 9.46E-10 | CSP8 | Phase 0 |
| LOC103313826 | −2.22 | Down | 8.77E-05 | FMO 2 | Phase I |
| LOC664598 | −2.25 | Down | 0 | OBPC01 | Phase 0 |
| LOC660846 | −2.44 | Down | 7.10E-103 | Ugt2C1 | Phase II |
| LOC662337 | −2.49 | Down | 6.97E-127 | CYP4C1 | Phase I |
| LOC107398495 | −2.77 | Down | 7.86E-25 | GSTe4 like | Phase II |
| LOC664475 | −2.87 | Down | 5.91E-05 | CYP6A20 | Phase I |
| LOC661930 | −3.09 | Down | 5.08E-05 | CYP349A1 | Phase I |
C, control; T, 5% A. vulgaris treatment; OBP, odorant binding protein; CYP, cytochrome P450; Ugt, UDP-glucuronosyltransferase; EST, esterase; GST, glutathione S-transferase; MRP, multidrug resistance protein; BG, beta-glucuronidase; XDH, xanthine dehydrogenase; FMO, flavin-containing monooxygenase.
Expression Profiles of DEGs
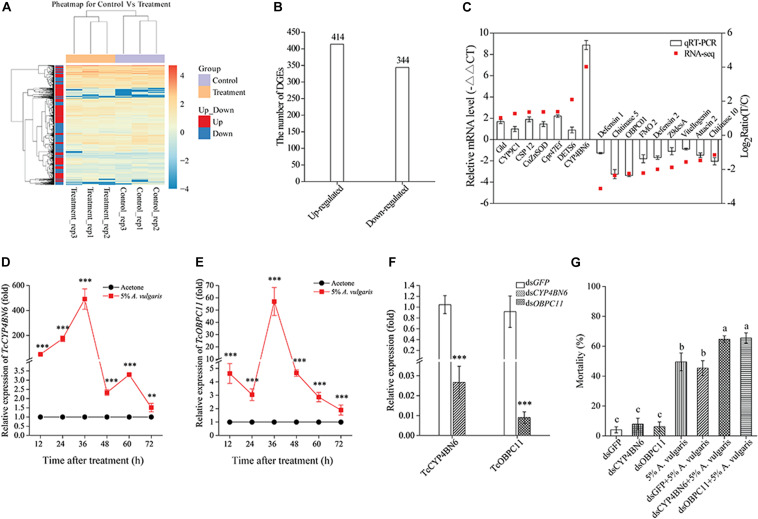
To confirm the significant differences in gene expression between the EO treatment and control groups, a likelihood ratio test was conducted based on the FPKM-derived read count. A reliable statistical analysis was applied to genes with an FPKM value ≥ 2 in at least one of the two biological replicates to minimize the identification of false positives and negatives. It is worth noting that these statistical significance tests were based on expected sampling distributions. To identify unigenes potentially involved in the response of T. castaneum larvae to A. vulgaris EO, the FPKM values of unigenes were compared between libraries. All three biological replicates of both control and EO treatment groups showed a high correlation, based on cluster analysis (Figure 3A), and A. vulgaris EO treated samples formed a distinct cluster separate from the control samples (Supplementary Figure S1C). Furthermore, to confirm which of the genes were differentially expressed to a significant degree between the treatment and control groups, thresholds of FC ≥ 2 and FDR ≤ 10–3 were used. Based on these criteria, 758 DEGs were identified. Of these, 344 DEGs were up-regulated and 414 DEGs were down-regulated in EO treated samples compared with control samples (Figure 3B and Supplementary Table S3). To validate the quality of RNA-Seq results, we verified the expression of 16 randomly selected DEGs by qRT-PCR. The expression profiles of these DEGs were similar between qRT-PCR and BGISEQ data, thus confirming the validity of RNA-Seq data (Figure 3C).
FIGURE 3.
(A) Heat map of 758 genes differentially expressed in larvae exposed to 5% A. vulgaris essential oil (EO) and control. Rows represent a single gene and columns are comparisons between genes in the exposure treatment and the control. The left-hand column clusters samples (N = 6) based on similarity of log10 transformed gene expression. Lighter colors indicate lower levels of differential expression. (B) Changes in the gene expression profile between the control and 5% A. vulgaris EO exposure groups. (C) A comparison of the expression profiles of the selected genes as determined by RNA-sequencing and qRT-PCR. OBP, Odorant binding protein; CYP, Cytochrome P450; Ugt, UDP-glucuronosyltransferase; EST, Esterase; GST, Glutathione S-transferase; MRP, Multidrug resistance protein; BG, Beta-glucuronidase; XDH, Xanthine dehydrogenase; FMO, Flavin-containing monooxygenase. Effects of 5% A. vulgaris EO exposure on the accumulation of TcCYP4BN6 (D) and TcOBPC11 (E) in 20-day-old larvae of T. castaneum. Acetone treatment represents the negative control group, which has been ascribed an arbitrary value of 1. The 12, 24, 36, 48, 60, and 72 h are six different test time points following EO treatment. (F) RNAi-mediated gene silencing. Twenty-day-old larvae were used for dsRNA injection. RNA was extracted and quantified by qRT-PCR at fifth day. Control beetles were injected with the same amount of green fluorescent protein (GFP) dsRNA. (G) Effect of TcCYP4BN6/TcOBPC11 silencing by injection of ds-TcCYP4BN6/ds-TcOBPC11 on toxicity of EO to 20-day-old larvae. Larvae injected with ds-TcCYP4BN6/ds-TcOBPC11/ds-GFP only served as untreated controls. Following injection with ds-TcCYP4BN6/ds-TcOBPC11/ds-GFP, larvae were treated with EO for 72 h. Data shown are mean ± SE (n = 3). An asterisk above bars indicated significant differences in the mRNA expression among the control and treatments (**P < 0.01, ***P < 0.001; Student’s t-test). Different letters above bars indicate significant differences (P < 0.05) according to LSD multiple comparison tests.
Validation of RNA-Seq Data by qRT- PCR
To confirm the quality of the transcriptome data and DEG results, we selected and evaluated seven up-regulated and nine down-regulated genes with qRT-PCR. The results of qRT-PCR were consistent with RNA-Seq data but with some quantitative variation in the regulation range. In short, qRT-PCR analysis confirmed the directional change in gene expression detected in DEG analysis (Figure 3C).
Induction of TcCYP4BN6 and TcOBPC11 Expression by A. vulgaris EO
To determine whether the expression of TcCYP4BN6 and TcOBPC11 could be induced by 5% A. vulgaris EO, the transcript levels of these two genes were detected by using qRT-PCR after 20-day-old larvae treated with EO at different time points. Interestingly, the transcript levels of TcCYP4BN6 and TcOBPC11 were both upregulated in response to 5% A. vulgaris EO exposure from 12 to 72 h, and they displayed similarly upregulation patterns (Figures 3D,E). Compared with the acetone controls, TcCYP4BN6 and TcOBPC11 were significantly induced by EO, both reaching maximum expression at 36 h with transcript levels of 491.49 ± 81.43- and 57.00 ± 11.38-fold, respectively, their expression then gradually returned to normal levels at 72 h (Figures 3D,E).
Effect of TcCYP4BN6 and TcOBPC11 Silencing on T. castaneum Response to A. vulgaris EO
To further determine the effects of TcCYP4BN6 and TcOBPC11 on insect response to 5% A. vulgaris EO, RNAi was used to knock down these two genes. On the fifth day after injection of dsRNAs, qRT-PCR was performed to investigate the gene expression of TcCYP4BN6 and TcOBPC11 in T. castaneum (Figure 3F). Compared to the control, transcript levels of TcCYP4BN6 and TcOBPC11 were significantly decreased by 97.3 and 99.1% in T. castaneum, respectively, whereas injection of ds-GFP had no significant effect. Hence, RNAi effectively suppressed the expression of TcCYP4BN6 and TcOBPC11 in T. castaneum. Mortality of the 20-day-old larvae injected with ds-GFP, ds-TcCYP4BN6, or ds-TcOBPC11 and exposed to the EO was shown in Figure 3G. Obviously, RNAi against TcCYP4BN6 or TcOBPC11 both significantly enhanced insecticidal activity of EO, whereas injection of ds-GFP did not (Figure 3G). After exposure to 5% A. vulgaris EO, mortality of T. castaneum was 45.33% ± 4.98%, 64.67% ± 2.33%, and 65.50% ± 3.50% when injected with ds-GFP, ds-TcCYP4BN6, or ds-TcOBPC11, respectively. Compared to the treatment with 5% A. vulgaris EO alone, mortality of T. castaneum was significantly increased by 16% or 17% after injection with ds-TcCYP4BN6 or ds-TcOBPC11 and treated with EO (Figure 3G). Our results clearly indicated that injection with ds-TcCYP4BN6 or ds-TcOBPC11 and exposure to the 5% A. vulgaris EO resulted in higher mortality than treatment with the EO alone. These results suggest that TcCYP4BN6 or TcOBPC11 may play a vital role in response to A. vulgaris EO in T. castaneum.
Discussion
In this paper, the contact activity and insecticidal effect of A. vulgaris EO on T. castaneum larvae, as assessed following biochemical and comparative RNA-Seq analyses, were characterized and investigated. A pattern of distinct dose-dependence contact activity of A. vulgaris EO were detected against T. castaneum larvae, consistent with previous studies (Wang et al., 2006). However, we found that the LC50 values were 6.14, 5.11, and 4.77% A. vulgaris EO at 24, 48, and 72 h, respectively, revealing there wasn’t apparent time-dependent effect. Actually, as shown in Figures 1C,D, the enzyme activities of CYP and CarE in the larva showed a corresponding pattern of increase at 36 h, but the mortality did not appear time-dependent during this period even at a later time, which may suggest that this degree of induced enzyme activity was not sufficient to cause the change in mortality. This similar phenomenon was also reported in Apis mellifera and Myzus persicae (Carvalho et al., 2013; Czerniewicz et al., 2018). Meanwhile, to prevent oxidative damage, insects employ detoxifying enzymes, such as CYPs and CarEs, for the metabolism of plant secondary metabolites (Pan et al., 2016). This suggests that the CYPs and CarEs participate in the response of T. castaneum larvae to A. vulgaris EO. It is possible that the death of T. castaneum larvae after A. vulgaris EO treatment is caused by the disruption of CYP and CarE homeostasis in insects.
RNA-Seq results showed significant up-regulation of 414 DEGs and significant down-regulation of 344 DEGs in A. vulgaris EO treated larvae (Supplementary Table S3). GO enrichment analysis revealed that 142, 99, and 198 DEGs were classified into 37 functional sub-categories in the cellular component, molecular function, and biological process categories, respectively (Figure 2A). KEGG analysis revealed that 511 DEGs were enriched in 262 biological pathways (Supplementary Table S2). Among these groups, we discussed the regulation of biological processes, response to stimuli, antigen processing and presentation, and drug metabolism in further detail, as these are most likely associated with the influence of A. vulgaris EO on T. castaneum larvae. Exposure to A. vulgaris EO promoted the expression of copper-and zinc-containing superoxide dismutase (CuZnSOD) and heme peroxidase (HPX); both these genes were simultaneously assigned to the response to stimulus, regulation of biological processes, antioxidant activity, developmental processes, and reproduction GO terms (Table 2 and Supplementary Tables S3, S4). Typically, reactive oxygen species (ROS), including superoxide anion and hydrogen peroxide (H2O2), produced in cells during metabolism, play pivotal roles in the innate immunity of insects as potent pathogen-killing agents (Bahia et al., 2013; Deng and Zhao, 2014). In addition, the expression of CuZnSOD is up-regulated immediately after the exposure of Brachionus calyciflorus to H2O2 treatment, and this increased expression promotes resistance to oxidative stress (Van Raamsdonk et al., 2009; Zhang et al., 2013). Under normal growth conditions, the dual oxidase (Duox) gene is responsible for ROS generation, which maintains microorganism homeostasis in the fruit fly gut (Kim and Lee, 2014). Nevertheless, higher ROS levels caused by overexpression of Duox disrupt cellular function and structure, leading to severe oxidative damage to DNA, RNA, proteins, and lipids (Zhu et al., 2016). These data indicate that the up-regulation of the CuZnSOD in EO treated larvae was accompanied by a response to excessive ROS disturbance, further suggesting that A. vulgaris EO affects antioxidative stress responses through crosstalk with antioxidant activities of T. castaneum larvae. Interestingly, antioxidant activities are also referred to development and reproduction (Taracena et al., 2015; Deng et al., 2016). In Anopheles gambiae, the expression of HPX15, which encodes as an active peroxidase that functions in reproductive organs to limit oxidative stress, was highly up-regulated. Peroxidases prevent cellular damage caused by free hydroxyl radicals through catalyzing the reduction of H2O2 to water, implying that HPX15 is required to maintain fertility (Shaw et al., 2014). These results indicate that the up-regulation of HPX in T. castaneum larvae after A. vulgaris EO treatment is a protective mechanism that prevents damage to reproductive organs or cells involved in fertility. Furthermore, in the transcriptome data, we found 28 DEGs associated with the lysosome pathway, which is correlated with the likely mode of action of A. vulgaris EO in insects. Among these 28 DEGs, 12 were involved in development and reproduction in T. castaneum (Table 3). The Cat and Lipase genes, which play a significant role in development and reproduction, were significantly down-regulated. For example, CatB/CatB-like and CatL/CatL precursor participate in metamorphosis by controlling tissue remolding and larval fat body decomposition in Delia radicum and Sarcophaga peregrina, respectively (Takahashi et al., 1993; Hegedus et al., 2002). Additionally, both CatB/CatB-like and CatL/CatL precursors are thought to be vital for embryogenesis in mosquito via the degradation of yolk protein (Cho et al., 1999; Uchida et al., 2001). Furthermore, tissue immunohistochemistry indicates that CatB participates in the embryonic degradation of yolk protein and fat body histolysis in Helicoverpa armigera (Zhao et al., 2005; Yang et al., 2006). In this study, the CatL precursor gene was significantly down-regulated after A. vulgaris EO treatment. Silencing of CatL precursor by RNAi leads to 100% immortality in insects and the absence of vitellogenic egg chambers in 40% of T. castaneum eggs (Dönitz et al., 2014). It is possible that EO treatment decreases the activity of CAT enzymes, along with low spawning and insect death. Furthermore, in this paper, the expression level of Lipase-1 like was dramatically down-regulated after A. vulgaris EO treatment. Previously, knockdown of Lipase-1 like resulted in 100% previtellogenic egg chamber orientation in red flour beetle (Dönitz et al., 2014). In addition, Lipase-3 and Lipase-3 like were dramatically down-regulated in the EO treatment group. Furthermore, knockdown of Lipase-3 reduced the hemolymph lipid concentration, which most likely disturbed lipolysis, leading to eclosion defects in beetles (data not shown). These consequences suggest that A. vulgaris EO directly affected lipase biosynthesis in T. castaneum. In addition to Spp38, two members of the serine protease (Sp) gene family were notably down-regulated, and 10 members were observably up-regulated in response to A. vulgaris EO treatment (Q-value ≤ 0.05; Table 3 and Supplementary Table S3). The SP genes play a significant role in multiple physiological processes including innate immunity, stress responses, reproduction, and development (Gorman et al., 2000; Zou et al., 2006; Wang et al., 2016). In a previous study, transcript levels of Sp3, Sph42, and Sp49 expression levels were elevated after saline or Escherichia coli injection, and pathogen challenge further enhanced the expression of Sp1, Sp2, Sp6, and Sp41 genes in Apis mellifera (Zou et al., 2006). Ulteriorly, RNAi against larvae Sph115 causes developmental arrest with 60% lethality in T. castaneum, and knockdown of Spp2c in silkworm embryos dramatically reduces the degradation rate of residual yolk proteins on embryonic day 10, further resulting in the suspension of embryogenesis (Dönitz et al., 2014). Moreover, knockdown of Spp163 or Spp123 in T. castaneum pupae leads to the formation of embryonic tissues without a cuticle, whereas RNAi of Sph111 causes musculature defects in 30% of larvae (Dönitz et al., 2014). These outcomes indicate that the SP signaling pathway is activated and amplified by A. vulgaris EO, which affects stress and immune responses, development, and reproduction in T. castaneum. Notably, the loss of activity of these SP proteins in T. castaneum larvae was at least partially compensated by up-regulation of the expression of the other primary Sp genes (Table 3 and Supplementary Table S4). This result was similar to the SP inhibitor feeding experiment, in which the expression of some Sp genes was significantly down-regulated, whereas that of other Sp genes was up-regulated, as part of an integrated compensation response in T. castaneum (Oppert et al., 2010; Perkin et al., 2017). In total, these data imply that large-scale Sp gene expression patterns were dramatically increased when the red flour beetle larvae were exposed to A. vulgaris EO, which amplified the SP signaling pathways, ultimately regulating multiple physiological functions in T. castaneum. Furthermore, 14 of the DEGs involved were identified in the antigen processing and presentation pathway (Supplementary Table S5). In addition to the seven members of Lipases discussed above, these DEGs included four HSPs (HSP68a, HSP68b, HSP70a, and HSP70b), which were up-regulated after A. vulgaris treatment. HSPs are rapidly synthesized in response to various environmental stressors, including heat shock, cold shock, pesticide application, and heavy metal exposure (Huang and Kang, 2007; Rinehart et al., 2007; Shu et al., 2011; Sun et al., 2014; Yang et al., 2016). The expression of HSP70 is significantly induced in Drosophila melanogaster, Nilaparvata lugens, Lymantria dispar, and Apolygus lucorum to increase resistance against applied pesticides (Mrdaković et al., 2016; Sun et al., 2016; Lu et al., 2017). Thus, we speculate that DEGs encoding HSPs participate in the response of T. castaneum to A. vulgaris EO.
When comparing the control and EO treatment group, RNA-Seq data revealed a large number of multiple metabolic detoxification enzymes in T. castaneum larvae (Figure 2B and Table 4). Cell metabolism detoxification processes are divided into four phases (phases 0–III). In phase 0, the uptake of xenobiotics is facilitated by membrane transport proteins. In phase I, CYP enzymes perform oxidation-reduction reactions, and CarE/EST enzymes are involved in the hydrolysis of ester bonds, leading to the introduction of a polar group in the toxic molecules (Montella et al., 2012; Schama et al., 2016). Meanwhile, phase II involves many cellular defense enzymes such as GSTs and Ugts mediated conjugation reactions of phase I metabolites with one of several endogenous molecules to form water-soluble products (Ando et al., 2000; Glisic et al., 2015). Finally, in phase III, reaction products of the previous step are transferred out of the cell by transport proteins (Vieira-Brock et al., 2013). In this article, genes encoding four CSP proteins (CSP11, 12, 17, and 20) and five OBPs (OBP10, 11, 12, 17, and C02) were up-regulated in EO treated larvae. The high expression of CSPs and OBPs was referred to drug metabolism because of their ability to bind lipophilic compounds, indicating that CSPs and OBPs bind to hydrophobic xenobiotics in phase 0 of the cellular detoxification process in insects (Liu et al., 2010, 2014; Bautista et al., 2015). We speculate that CSPs and OBPs are up-regulated in beetles to create resistance to A. vulgaris. In response to A. vulgaris EO treatment, CSP8 and OBPC01 were down-regulated, further disrupting the homeostasis of xenobiotics in vivo. Moreover, the expression of FMOs, ESTs, and XDHs as well as that of CYPs was also dramatically affected in EO treated larvae (Table 4). Furthermore, genes encoding CYP4BN1, CYP6BQ7, CYP6A2, CYP351A2, CYP9Z2, CYP9AC1, CYP6BK11, and CYP6A14 were up-regulated in the EO treatment group. The CYP unigenes were significantly up-regulated in the EO treated larvae, indicating that these genes are involved in A. vulgaris EO activation and detoxification pathways, which catalyze intracellular redox reactions (Janmohamed et al., 2001). However, six CYP genes (CYP4BN5 CYP4Q1, CYP351A3, CYP4C1, CYP6A20, and CYP349A1) were down-regulated, which reduced the cellular metabolic activity, thus causing insect death (Dönitz et al., 2014). Furthermore, like CYPs, the expression of FMOs, which are related to NADPH oxidase activity (N-oxidation), decreased after treatment with A. vulgaris EO. FMOs can catalyze the conversion of toxic pyrrolizidine alkaloids (PAs) into the non-toxic N-oxide form, and can also regulate the detoxification of nucleophilic nitrogen- and sulfur-containing xenobiotics in vertebrates (Hartmann, 2009). The CYP and FMO family genes, which were expressed to low levels, were involved in drug metabolism, indicating a correlation between these genes in the detoxification process (Chung et al., 2000; Zhu et al., 2015). Additionally, genes encoding CarEs (α-EST5 and α-EST2) were also up-regulated by EO treatment, suggesting that these genes could participate in catalyzing the hydrolysis of various xenobiotics in A. vulgaris EO (Table 4). In fact, our biochemical analysis showed that A. vulgaris EO caused a pronounced increase in CarE activity.
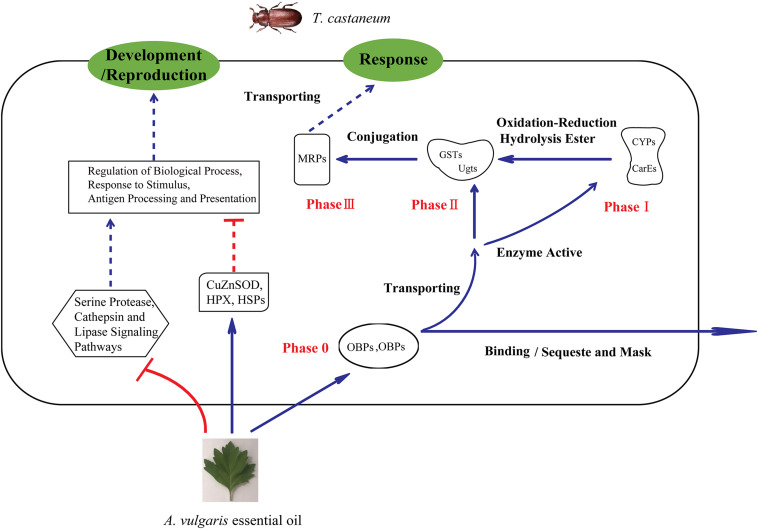
Genes encoding enzymes belonging to phase II, including BGs, GSTs, and Ugts, were also up-regulated after EO treatment. In a previous study, the activity of BGs was increased upon low-level exposure to an organophosphorus insecticide (Ueyama et al., 2010). This indicates that exposure to A. vulgaris EO caused the down-regulation of BG expression in T. castaneum larvae, suggesting that A. vulgaris EO represses phase II genes by regulating BG expression. Generally, GSTs directly detoxify organophosphates and organochlorine through the conjugation of these electrophilic compounds with the thiol group of reduced GSH, thus rendering the products more water-soluble and excretable than the non-GSH conjugated substrates (Aravindan et al., 2014). Additionally, GSTs possibly have no direct role in the metabolism of pyrethroids, but they could detoxify lipid peroxidation products induced by insecticides, which also demonstrated in coffering resistance (Vontas et al., 2001). In the current study, three GST genes (GSTs6, GSTs7, and GSTe4-like) were up-regulated in EO samples (Table 4), indicating that a growing number of toxic intermediates were transformed into innocuous substances by conjugation with GSH which also caused various endogenous molecules like sugars and glutathione pathway were expressed to conjugate xenobiotics. Moreover, two GST genes (GSTe7 and GSTe8) were down-regulated in EO samples. It is possible that redundant components could bind to the enzyme site, thus disturbing enzyme activity. The conversion of conjugated xenobiotics to innocuous substances possibly damaged the enzymes to the point of no recovery. Both induction and inhibition of GST expression in response to certain plant secondary metabolites has been previously reported (Mostofa et al., 2014; Balyan et al., 2015). Notably, Ugts participate in phase II reactions, similar to GSTs, and catalyze the transfer of glucuronic acid to lipophilic molecules to further increase the water solubility of the compound for later excretion or sequestration (Okazaki and Katayama, 2003; Crava et al., 2016). Furthermore, Ugts detoxify xenobiotics, including insecticides, and have been linked to insecticide resistance (Lv et al., 2016). In this study, we detected six Ugts in EO treated samples, implying that these genes possibly take part in the metabolic activation and detoxification of A. vulgaris EO, which accelerates the water solubility of compounds. The up-regulation of multidrug resistance proteins (MRPs) was also detected in EO treated larvae; MRPs are phase III reactions and utilize the energy derived from ATP hydrolysis to translocate a variety of physiological metabolites and xenobiotics (Table 4) (Vieira-Brock et al., 2013). It is likely that insects increase the expression of MRP genes to enhance the efficiency of xenobiotic compound excretion or degradation. This is the first report of evidence demonstrating that cellular detoxification genes of phases 0–III, including OBPs, CSPs, CYPs, ESTs, FMOs, GSTs, Ugts, and BG, are involved in the response of T. castaneum to A. vulgaris EO (Figure 4).
FIGURE 4.
Schematic of the mechanism of insect response to stimulation by A. vulgaris essential oil. OBPs, Odorant-binding proteins; CSPs, chemosensory proteins; CYPs, cytochrome P450 monooxygenases; CarE, carboxylesterase; Ugt, UDP-glucuronosyltransferase; GST, Glutathione S-transferase; MRP, multidrug resistance protein; (HPX), heme peroxidase; HSP, heat shock protein; CuZnSOD, copper-and zinc-containing superoxide dismutase.
Further, two DEGs (TcOBPC11 and TcCYP4BN6) which were likely involved in the response of T. castaneum to A. vulgaris EO were performed for EO stimulation and induction studies. Interestingly, in our study, after A. vulgaris EO treatment, the transcripts of TcOBPC11 and TcCYP4BN6 were significantly induced. Knockdown TcOBPC11 or TcCYP4BN6, the mortality of T. castaneum beetles was significantly increased compared with the controls (Figures 3D,E), indicating that TcOBPC11 or TcCYP4BN6 were involved in response to A. vulgaris EO in T. castaneum. Currently, studies examining the canonical biological functions of OBPs have focused on their activity within insect chemosensory systems, trying to understand their roles in detecting and recognizing environmental chemical stimuli (Sánchez-Gracia et al., 2009). Generally, the expression of insect OBPs associated with xenobiotic resistance is often inducible by non-toxic compounds, host allelochemicals and synthetic insecticides (Bautista et al., 2015; Xuan et al., 2015). Interestingly, such induction pattern of TcOBPC11 was reminiscent of SlituOBP9, the expression level of which was obviously promoted by chlorpyrifos in Spodoptera litura (Lin et al., 2018). Similar results have also been reported for P. xylostella PxylOBP13, T. castaneum TcOBPC01, which the transcript levels of these genes were upregulated following permethrin, carbofuran/dichlorvos treatment, respectively (Bautista et al., 2015; Xiong et al., 2019a). Thus we speculate that the activity structure of some compositions in A. vulgaris EO is similar to these insecticides. Simultaneously, in insects, CYP genes play an essential role in detoxifying exogenous compounds, including plant toxins and insecticides, and their upregulation can result in increased levels of CYP proteins and CYP enzyme activities (Zhu et al., 2010). The increased levels of TcCYP4BN6 expression observed during the response to A. vulgaris EO are ostensibly consistent with a role for TcCYP4BN6 in EO metabolism in T. castaneum (Figure 3D). The observed induction pattern of TcCYP4BN6 is reminiscent of those of SinvCYP4AB2 and SinvCYP4G15 and those of SinvCYP6A1 and SinvCYP6B1 in Solenopsis invicta, and expression of these genes has been shown to be promoted by fipronil (Zhang et al., 2016). Similar results have also been reported for M. domestica, in which the transcript levels of MuscaCYP4G2 and MuscaCYP6A38 were upregulated following permethrin treatment (Zhu et al., 2008). Furthermore, 22 CYPs unigenes in Melaleuca alternifolia EO-fumigated Sitophilus zeamais were significantly up-regulated (Liao et al., 2016). These results indicate that different types of insecticides, EOs exert some common effects on the upregulation of the expression of insecticide metabolism-related CYP genes. Notably, TcOBPC11 and TcCYP4BN6 were both significantly increased with exposure to 5% A. vulgaris EO at 12–72 h, both reaching maximum expression at 36 h, then gradually returned to normal levels at 72 h (Figures 3D,E). Perhaps it takes 72 h after exposure to EO to recruit multiple susceptibility genes in a variety of defense mechanisms (Liao et al., 2016). Moreover, a time-dependent induction of susceptibility genes is typically observed after 12 h (Zhu et al., 2008). In particular, hundreds of genes are differentially expressed during the 48–60 h period following Bemisia tabaci fungal infection (Xia et al., 2013). These studies combined with our present results (Figures 3D,E) strongly imply that this delay in response could be due to the time needed for the exogenous toxic molecules to penetrate through the cuticle of insects (Liu et al., 2014). Therefore, different CYP and OBP genes could be involved in different exposure times to protect various tissue specific physiological pathways.
RNAi has emerged as a powerful tool for researching gene functions within a wide range of both multicellular organisms and unicellular (Meister and Tuschl, 2004; Kim et al., 2015). Using dsRNA injection, we found that RNAi against TcOBPC11 or TcCYP4BN6 significantly increased mortality of T. castaneum when exposed to 5% A. vulgaris EO, indicating that TcOBPC11 or TcCYP4BN6 might be play a vital role during the response to A. vulgaris EO treatments (Figure 3G). Exposure to A. vulgaris EO increased the expression levels of TcOBPC11, but knocking this gene down prior to EO exposure led to even higher mortalities, which implied that the roles of this genes might be protective because TcOBPC11 is acted as buffers that have a high binding affinity with A. vulgaris EO. Increasing the expression of TcOBPC11 would decrease the toxicity of EO through high binding affinity, consequently sequestering and masking toxic molecules (Pelosi et al., 2017; Lin et al., 2018). Simultaneously, our result showed similar expression pattern and biological assay have been found in TcCYP4BN6 after A. vulgaris EO treatment, which implying that the roles of this CYP protein in T. castaneum are likely be protective. TcCYP4BN6 is known as members of the CYP family that are associated with insecticide metabolic detoxification (Schama et al., 2016), and increasing the expression of these CYP genes would be expected to decrease the toxicity of A. vulgaris EO through oxidation–reduction reactions that occur during the metabolism of toxic molecules (Wang R. L. et al., 2015). In our current study, A. vulgaris EO induced overexpression of TcOBPC11 or TcCYP4BN6 in T. castaneum. Additionally, RNAi mediated silencing of TcOBPC11 or TcCYP4BN6 significantly increased mortality of T. castaneum larvae exposed to EO. These results strongly reveal that TcOBPC11 or TcCYP4BN6 might play an important role in A. vulgaris EO metabolic detoxification in T. castaneum, which further confirms our RNA-seq result.
Conclusion
In this paper, we examined the contact toxicity of A. vulgaris EO on T. castaneum; its effects on the activities of two types of insect detoxification enzymes were also evaluated. The results suggested that A. vulgaris EO has the potential to be used as a natural insecticide. More importantly, this is the first report of a comprehensive transcriptome analysis of T. castaneum to: (1) identify genes and pathways involved in the response to A. vulgaris EO exposure, and (2) investigate the underlying molecular mechanism of insecticidal activity against T. castaneum larvae. Enzyme activities of CarEs and CYPs were dramatically increased in EO treated larvae. RNA-seq results confirmed that OBPs, CSPs, P450s, GSTs, UGTs, and MRPs participate in the metabolism of A. vulgaris EO. (3) In addition, our bioassay results also supported that OBPs and P450s might play an important role in A. vulgaris EO metabolic detoxification in T. castaneum. Thus, our results will not only accelerate studies investigating the mechanisms underlying the insecticidal effect of plant EO on insect pests but also facilitate the development of natural novel insecticides.
Data Availability Statement
Raw sequence reads were saved as FASTQ files and deposited in the NCBI Sequence Read Archive (SRA) database under the following accession numbers: control: SRR7646221, SRR7646223, and SRR7646224; EO treatment: SRR7646222, SRR7646225, and SRR7646226.
Author Contributions
SG and KZ designed the experiments. LW, GW, and WX carried out the experiments. YL, YZ, and AG analyzed the experimental results. BL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the Doctoral Scientific Research Foundation of Anyang Institute of Technology (BSJ2019009) and the National Natural Science Foundation of China (Nos. 31572326 and 31872970).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00589/full#supplementary-material
(A) Length distribution of the gene detected by RNA-sequencing. The x-axis and y-axis indicate the length of the genes and the number of genes, respectively. (B) FPKM statistic of the control and treatment. The x-axis and y-axis indicated the FPKM value of the genes and the number of genes, respectively. (C) Cluster tree. Distances between the expressed genes were calculated by the Euclidean method. The algorithm of the sum of squares of deviations was used to calculate the distance between samples to construct the cluster tree. Y-axis represented the height of the cluster tree. Samples with similar height were clustered together.
Percent of coverage representing the percentage of genes that were expressed in each of the two libraries mapped in the red flour beetle (Tribolium castaneum) genome. Panels (A–C) control groups; (D,E), 5% Artemisia vulgaris EO treatment groups. The gene coverage was the percentage of a gene covered by the reads. This value was equal to the ratio of the base number in a gene covered by unique mapping reads to the total base number of that gene. The saturation analyses of all the detected genes. Panels (G–I) control groups; (J–L) 5% Artemisia vulgaris EO treatment groups.
Primers used for qRT-PCR analysis and dsRNA synthesis in this study.
Pathway enrichment analysis of differentially expressed genes (DEGs) and all the detected genes.
A set of 758 differentially expressed genes (DEGs) between the 5% Artemisia vulgaris treatment and control groups.
Differentially expressed genes (DEGs) involved in response to stimuli.
Differentially expressed genes (DEGs) related to antigen processing and presentation.
References
- Alizadeh M., Aghaei M., Sharifian I., Saadatian M. (2012). Chemical composition of essential oil of Artemisia vulgaris from West Azerbaijan, Iran. Electron. J. Environ. Agric. Food Chem. 11 493–496. [Google Scholar]
- Ana C., Stefan G. T., Juan Miguel G. G., Javier T., Manuel T., Montserrat R. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- Ando Y., Saka H., Ando M., Sawa T., Muro K., Ueoka H., et al. (2000). Polymorphisms of UDP-glucuronosyltransferase gene and irinotecan toxicity: a pharmacogenetic analysis. Cancer Res. 60 6921–6926. 10.1002/1097-0142(20001215)89 [DOI] [PubMed] [Google Scholar]
- Aravindan V., Muthukumaravel S., Gunasekaran K. (2014). Interaction affinity of delta and epsilon class glutathione-s-transferases (GSTs) to bind with DDT for detoxification and conferring resistance in Anopheles gambiae, a malaria vector. J. Vector Borne Dis. 51 8–15. 10.1051/parasite/2014008 [DOI] [PubMed] [Google Scholar]
- Athanassiou C. G., Hasan M. M., Phillips T. W., Aikins M. J., Throne J. E. (2015). Efficacy of methyl bromide for control of different life stages of stored-product psocids. J. Econ. Entomol. 108 1422–1428. 10.1093/jee/tov069 [DOI] [PubMed] [Google Scholar]
- Bahia A. C., Oliveira J. H., Kubota M. S., Araujo H. R., Lima J. B., Rios-Velasquez C. M., et al. (2013). The role of reactive oxygen species in Anopheles aquasalis response to Plasmodium vivax infection. PLoS One 8:e57014 10.1371/journal.pone.0057014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhetia M., Charlton W., Atkinson H. J., Mcpherson M. J. (2005). RNA interference of dual oxidase in the plant nematode Meloidogyne incognita. Mol. Plant Microbe Interact. 18 1099–1106. 10.1094/MPMI-18-1099 [DOI] [PubMed] [Google Scholar]
- Balyan R., Kudugunti S. K., Hamad H. A., Yousef M. S., Moridani M. Y. (2015). Bioactivation of luteolin by tyrosinase selectively inhibits glutathione S-transferase. Chem. Biol. Interact. 240 208–218. 10.1016/j.cbi.2015.08.011 [DOI] [PubMed] [Google Scholar]
- Bautista M. A., Bhandary B., Wijeratne A. J., Michel A. P., Hoy C. W., Mittapalli O. (2015). Evidence for trade-offs in detoxification and chemosensation gene signatures in Plutella xylostella. Pest Manag. Sci. 71 423–432. 10.1002/ps.3822 [DOI] [PubMed] [Google Scholar]
- Bell C. H., Wilson S. M. (1995). Phosphine tolerance and resistance in Trogoderma granarium Everts (Coleoptera: Dermestidae). J. Stored Prod. Res. 31 199–205. 10.1016/0022-474x(95)00012-v [DOI] [Google Scholar]
- Bi J., Feng F., Li J., Mao J., Ning M., Song X., et al. (2019). A C-type lectin with a single carbohydrate-recognition domain involved in the innate immune response of Tribolium castaneum. Insect Mol. Biol. 28 649–661. 10.1111/imb.12582 [DOI] [PubMed] [Google Scholar]
- Bo L., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomzan D. P., Bhavya M. L., Chandu A., Manivannan S., Lavanya G., Ramasamy K., et al. (2018). Potential of pyrethroid-synergised pyrethrum on stored product insects and implications for use as prophylactic sprays. J. Food Sci. Technol. 55 2270–2278. 10.1007/s13197-018-3144-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borzoui E., Naseri B., Abedi Z., Karimipormehr M. S. (2016). Lethal and sublethal effects of essential oils from Artemisia khorassanica and Vitex pseudo-negundo against Plodia interpunctella (Lepidoptera: Pyralidae). Environ. Entomol. 45 1220–1226. 10.1093/ee/nvw100 [DOI] [PubMed] [Google Scholar]
- Carvalho S. M., Belzunces L. P., Carvalho G. A., Brunet J. L., Badiou-Beneteau A. (2013). Enzymatic biomarkers as tools to assess environmental quality: a case study of exposure of the honeybee Apis mellifera to insecticides. Environ. Toxicol. Chem. 32 2117–2124. 10.1002/etc.2288 [DOI] [PubMed] [Google Scholar]
- Chen Y., He M., Li Z. Q., Zhang Y. N., He P. (2016). Identification and tissue expression profile of genes from three chemoreceptor families in an urban pest, Periplaneta americana. Sci. Rep. 6:27495 10.1038/srep27495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W. L., Tsao S. M., Hays A. R., Walter R., Chen J. S., Snigirevskaya E. S., et al. (1999). Mosquito Cathepsin B-like protease involved in embryonic degradation of vitellin is produced as a latent extraovarian precursor. J. Biol. Chem. 274 13311–13321. 10.1074/jbc.274.19.13311 [DOI] [PubMed] [Google Scholar]
- Chung W. G., Park C. S., Roh H. K., Lee W. K., Cha Y. N. (2000). Oxidation of ranitidine by isozymes of flavin-containing monooxygenase and cytochrome P450. Jpn. J. Pharmacol. 84 213–220. 10.1254/jjp.84.213 [DOI] [PubMed] [Google Scholar]
- Corrêa-Ferreira M. L., Noleto G. R., Petkowicz C. L. O. (2014). Artemisia absinthium and Artemisia vulgaris: a comparative study of infusion polysaccharides. Carbohydr. Polym. 102 738–745. 10.1016/j.carbpol.2013.10.096 [DOI] [PubMed] [Google Scholar]
- Crava C. M., Brütting C., Baldwin I. T. (2016). Transcriptome profiling reveals differential gene expression of detoxification enzymes in a hemimetabolous tobacco pest after feeding on jasmonate-silenced Nicotiana attenuata plants. BMC Genomics 17:1005 10.1186/s12864-016-3348-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerniewicz P., Chrzanowski G., Sprawka I., Sytykiewicz H. (2018). Aphicidal activity of selected Asteraceae essential oils and their effect on enzyme activities of the green peach aphid, Myzus persicae (Sulzer). Pestic. Biochem. Physiol. 145 84–92. 10.1016/j.pestbp.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Daehwan K., Ben L., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danse L. H., Van Velsen F. L., Ca V. D. H. (1984). Methylbromide: carcinogenic effects in the rat forestomach. Toxicol. Appl. Pharmacol. 72 262–271. 10.1016/0041-008x(84)90311-9 [DOI] [PubMed] [Google Scholar]
- Deng F., He Q., Zhao Z. (2016). Suppressing a peroxidase gene reduces survival in the wheat aphid Sitobion avenae. Arch. Insect. Biochem. Physiol. 93 86–95. 10.1002/arch.21343 [DOI] [PubMed] [Google Scholar]
- Deng F., Zhao Z. (2014). Influence of catalase gene silencing on the survivability of Sitobion avenae. Arch. Insect. Biochem. Physiol. 86 46–57. 10.1002/arch.21161 [DOI] [PubMed] [Google Scholar]
- Dey D. (2016). Impact of indiscriminate use of insecticide on environmental pollution. Inter. J. Plant Prod. 9 264–267. 10.15740/HAS/IJPP/9.1/264-267 [DOI] [Google Scholar]
- Dönitz J., Schmitt-Engel C., Grossmann D., Gerischer L., Tech M., Schoppmeier M., et al. (2014). iBeetle-Base: a database for RNAi phenotypes in the red flour beetle Tribolium castaneum. Nucleic Acids Res. 43 D720–D725. 10.1093/nar/gku1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinn P. W., Hagstrum D. W., Reed C., Phillips T. W. (2010). Insect population dynamics in commercial grain elevators. J. Stored Prod. Res. 46 43–47. 10.1016/j.jspr.2009.09.001 [DOI] [Google Scholar]
- Francis F., Vanhaelen N., Haubruge E. (2005). Glutathione S-transferases in the adaptation to plant secondary metabolites in the Myzus persicae aphid. Arch. Insect. Biochem. Physiol. 58 166–174. 10.1002/arch.20049 [DOI] [PubMed] [Google Scholar]
- García M., Donadel O. J., Ardanaz C. E., Tonn C. E., Sosa M. E. (2005). Toxic and repellent effects of Baccharis salicifolia essential oil on Tribolium castaneum. Pest Manag. Sci. 61 612–618. 10.1002/ps.1028 [DOI] [PubMed] [Google Scholar]
- Glisic B., Mihaljevic I., Popovic M., Zaja R., Loncar J., Fent K., et al. (2015). Characterization of glutathione-S-transferases in zebrafish (Danio rerio). Aquat. Toxicol. 158 50–62. 10.1016/j.aquatox.2014.10.013 [DOI] [PubMed] [Google Scholar]
- Goel D., Goel R., Singh V., Ali M., Mallavarapu G. R., Kumar S. (2007). Composition of the essential oil from the root of Artemisia annua. J. Nat. Med. 61 458–461. 10.1007/s11418-007-0175-2 [DOI] [Google Scholar]
- Gorman M. J., Andreeva O. V., Paskewitz S. M. (2000). Sp22D: a multidomain serine protease with a putative role in insect immunity. Gene 251 9–17. 10.1016/s0378-1119(00)00181-5 [DOI] [PubMed] [Google Scholar]
- Guo Y. Q., Zhang J. Z., Yang M. L., Yan L. Z., Zhu K. Y., Guo Y. P., et al. (2012). Comparative analysis of cytochrome P450-like genes from Locusta migratoria manilensis: expression profiling and response to insecticide exposure. Insect Sci. 19 75–85. 10.1111/j.1744-7917.2011.01450.x [DOI] [Google Scholar]
- Hartmann T. (2009). Pyrrolizidine alkaloids: the successful adoption of a plant chemical defense. See Ref 17 55–81. [Google Scholar]
- Hegedus D., O’grady M., Chamankhah M., Baldwin D., Gleddie S., Braun L., et al. (2002). Changes in cysteine protease activity and localization during midgut metamorphosis in the crucifer root maggot (Delia radicum). Insect Biochem. Mol. Biol. 32 1585–1596. 10.1016/s0965-1748(02)00099-1 [DOI] [PubMed] [Google Scholar]
- Ho S. H., Koh L., Ma Y., Huang Y., Sim K. Y. (1996). The oil of garlic, Allium sativum L. (Amaryllidaceae), as a potential grain protectant against Tribolium castaneum (Herbst) and Sitophilus zeamais Motsch. Postharvest Biol. Technol. 9 41–48. 10.1016/0925-5214(96)00018-X [DOI] [Google Scholar]
- Hori M., Kasaishi Y. (2005). Estimation of the phosphine resistance level of the cigarette beetle, Lasioderma serricorne (Fabricius)(Coleoptera: Anobiidae), by the knockdown time of adult. Appl. Entomol. Zool. 40 557–561. [Google Scholar]
- Huang L. H., Kang L. (2007). Cloning and interspecific altered expression of heat shock protein genes in two leafminer species in response to thermal stress. Insect Mol. Biol. 16 491–500. 10.1111/j.1365-2583.2007.00744.x [DOI] [PubMed] [Google Scholar]
- Huang Y., Lam S. L., Ho S. H. (2000). Bioactivities of essential oil from Elettaria cardamomum (L.) Maton. to Sitophilus zeamais Motschulsky and: Tribolium castaneum (Herbst). J. Stored Prod. Res. 36 107–117. 10.1016/S0022-474X(99)00040-5 [DOI] [Google Scholar]
- Isman M. (2006). Botanical insecticides, deterrents, and repellents in modern agriculture and an increasingly regulated world. Annu. Rev. Entomol. 51 45–66. 10.1146/annurev.ento.51.110104.151146 [DOI] [PubMed] [Google Scholar]
- Izakmehri K., Saber M., Mehrvar A., Hassanpouraghdam M. B., Vojoudi S. (2013). Lethal and sublethal effects of essential oils from Eucalyptus camaldulensis and Heracleum persicum against the adults of Callosobruchus maculatus. J. Insect Sci. 13 1–10. 10.1673/031.013.15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S., Shekhawat G. S. (2010). Anethum graveolens: an Indian traditional medicinal herb and spice. Pharmacogn. Rev. 4 179–184. 10.4103/0973-7847.70915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmohamed A., Dolphin C. T., Phillips I. R., Shephard E. A. (2001). Quantification and cellular localization of expression in human skin of genes encoding flavin-containing monooxygenases and cytochromes P450. Biochem. Pharmacol. 62 777–786. 10.1016/s0006-2952(01)00718-3 [DOI] [PubMed] [Google Scholar]
- Jiang Z., Guo X., Zhang K., Sekaran G., Cao B., Zhao Q., et al. (2019). The essential oils and eucalyptol from Artemisia vulgaris L. prevent acetaminophen-induced liver injury by activating Nrf2-Keap1 and enhancing APAP clearance through non-toxic metabolic pathway. Front. Pharmacol. 10:782 10.3389/fphar.2019.00782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsi M., Palli S. R. (2015). Transcription factors, CncC and Maf, regulate expression of CYP6BQ genes responsible for deltamethrin resistance in Tribolium castaneum. Insect Biochem. Mol. Biol. 65 47–56. 10.1016/j.ibmb.2015.08.002 [DOI] [PubMed] [Google Scholar]
- Khani A., Asghari J. (2012). Insecticide activity of essential oils of Mentha longifolia, Pulicaria gnaphalodes and Achillea wilhelmsii against two stored product pests, the flour beetle, Tribolium castaneum, and the cowpea weevil, Callosobruchus maculatus. J. Insect Sci. 12:73 10.1673/031.012.7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Jang M., Shin E., Kim J., Lee S. H., Park C. G. (2016). Fumigant and contact toxicity of 22 wooden essential oils and their major components against Drosophila suzukii (Diptera: Drosophilidae). Pestic. Biochem. Physiol. 133 35–43. 10.1016/j.pestbp.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Kim S. H., Lee W. J. (2014). Role of DUOX in gut inflammation: lessons from Drosophila model of gut-microbiota interactions. Front. Cell. Infect. Microbiol. 3:116 10.3389/fcimb.2013.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. H., Soumaila Issa M., Cooper A. M. W., Zhu K. Y. (2015). RNA interference: applications and advances in insect toxicology and insect pest management. Pestic. Biochem. Physiol. 120 109–117. 10.1016/j.pestbp.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Kordali S., Aslan I., Çalmaşur O., Cakir A. (2006). Toxicity of essential oils isolated from three Artemisia species and some of their major components to granary weevil, Sitophilus granarius (L.) (Coleoptera: Curculionidae). Ind. Crop Prod. 23 162–170. 10.1016/j.indcrop.2005.05.005 [DOI] [Google Scholar]
- Kostyukovsky M., Rafaeli A., Gileadi C., Demchenko N., Shaaya E. (2002). Activation of octopaminergic receptors by essential oil constituents isolated from aromatic plants: possible mode of action against insect pests. Pest Manag. Sci. 58 1101–1106. 10.1002/ps.548 [DOI] [PubMed] [Google Scholar]
- Li S. G., Li M. Y., Huang Y. Z., Hua R. M., Lin H. F., He Y. J., et al. (2013). Fumigant activity of Illicium verum fruit extracts and their effects on the acetylcholinesterase and glutathione S -transferase activities in adult Sitophilus zeamais. J. Pest Sci. 86 677–683. 10.1007/s10340-013-0520-z [DOI] [Google Scholar]
- Liao M., Xiao J. J., Zhou L. J., Liu Y., Wu X. W., Hua R. M., et al. (2016). Insecticidal activity of Melaleuca alternifolia essential oil and RNA-Seq analysis of Sitophilus zeamais transcriptome in response to oil fumigation. PLoS One 11:e0167748 10.1371/journal.pone.0167748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Jiang Y., Zhang L., Cai Y. (2018). Effects of insecticides chlorpyrifos, emamectin benzoate and fipronil on Spodoptera litura might be mediated by OBPs and CSPs. Bull. Entomol. Res. 108 658–666. 10.1017/S0007485317001195 [DOI] [PubMed] [Google Scholar]
- Liu G. X., Xuan N., Chu D., Xie H. Y., Fan Z. X., Bi Y. P., et al. (2014). Biotype expression and insecticide response of Bemisia tabaci chemosensory protein-1. Arch. Insect Biochem. Physiol. 85 137–151. 10.1002/arch.21148 [DOI] [PubMed] [Google Scholar]
- Liu X., Luo Q., Zhong G., Rizwan-Ul-Haq M., Hu M. (2010). Molecular characterization and expression pattern of four chemosensory proteins from diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). J. Biochem. 148 189–200. 10.1093/jb/mvq050 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu K., Chen X., Liu W., Zhang Z., Wang Y., You K., et al. (2017). Characterization of heat shock protein 70 transcript from Nilaparvata lugens (Stål): its response to temperature and insecticide stresses. Pestic. Biochem. Physiol. 142 102–110. 10.1016/j.pestbp.2017.01.011 [DOI] [PubMed] [Google Scholar]
- Lu Y., Park Y., Gao X., Zhang X., Yao J., Pang Y. P., et al. (2012). Cholinergic and non-cholinergic functions of two acetylcholinesterase genes revealed by gene-silencing in Tribolium castaneum. Sci. Rep. 2:288 10.1038/srep00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y., Wang W., Hong S., Lei Z., Fang F., Guo Q., et al. (2016). Comparative transcriptome analyses of deltamethrin-susceptible and -resistant Culex pipiens pallen s by RNA-seq. Mol. Genet. Genomics 291 309–321. 10.1007/s00438-015-1109-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S. A., Abdel-Hamid N. A. (2015). Residual toxicity of bait formulations containing plant essential oils and commercial insecticides against the desert locust, Schestocerca gregaria (Forskäl). Ind. Crop Prod. 76 900–909. 10.1016/j.indcrop.2015.08.004 [DOI] [Google Scholar]
- Meister G., Tuschl T. (2004). Mechanisms of gene silencing by double-stranded RNA. Nature 431 343–349. 10.1038/nature02873 [DOI] [PubMed] [Google Scholar]
- Montella I. R., Schama R., Valle D. (2012). The classification of esterases: an important gene family involved in insecticide resistance - A review. Mem. Inst. Oswaldo Cruz 107 437–449. 10.1590/s0074-02762012000400001 [DOI] [PubMed] [Google Scholar]
- Mostofa M. G., Hossain M. A., Fujita M. (2014). Trehalose pretreatment induces salt tolerance in rice (Oryza sativa L.) seedlings: oxidative damage and co-induction of antioxidant defense and glyoxalase systems. Protoplasma 252 461–475. 10.1007/s00709-014-0691-3 [DOI] [PubMed] [Google Scholar]
- Mrdaković M., Ilijin L., Vlahović M., Matić D., Gavrilović A., Mrkonja A., et al. (2016). Acetylcholinesterase (AChE) and heat shock proteins (Hsp70) of gypsy moth (Lymantria dispar L.) larvae in response to long-term fluoranthene exposure. Chemosphere 159 565–569. 10.1016/j.chemosphere.2016.06.059 [DOI] [PubMed] [Google Scholar]
- Nascimento S. S., Araújo A. A. S., Brito R. G., Serafini M. R., Menezes P. P., Desantana J. M., et al. (2015). Cyclodextrin-complexed Ocimum basilicum leaves essential oil increases Fos protein expression in the central nervous system and produce an antihyperalgesic effect in animal models for fibromyalgia. Int. J. Mol. Sci. 16 547–563. 10.3390/ijms16010547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseri B., Abedi Z., Abdolmaleki A., Jafary-Jahed M., Borzoui E., Mozaffar Mansouri S. (2017). Fumigant toxicity and sublethal effects of Artemisia khorassanica and Artemisia sieberi on Sitotroga cerealella (Lepidoptera: Gelechiidae). J. Insect Sci. 17:100 10.1093/jisesa/iex073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak M. K., Collins P. J., Pavic H., Kopittke R. A. (2003). Inhibition of egg development by phosphine in the cosmopolitan pest of stored products Liposcelis bostrychophila (Psocoptera: Liposcelididae). Pest Manag. Sci. 59 1191–1196. 10.1002/ps.753 [DOI] [PubMed] [Google Scholar]
- Nouri-Ganbalani G., Borzoui E. (2017). Acute toxicity and sublethal effects of Artemisia sieberi Besser on digestive physiology, cold tolerance and reproduction of Trogoderma granarium Everts (Col.: Dermestidae). J. Asia Pac. Entomol. 20 285–292. 10.1016/j.aspen.2017.01.002 [DOI] [Google Scholar]
- Obeng-Ofori D., Reichmuth C. (1997). Bioactivity of eugenol, a major component of essential oil of Ocimum suave (Wild.) against four species of stored-product Coleoptera. Int. J. Pest Manag. 43 89–94. 10.1080/096708797229040 [DOI] [Google Scholar]
- Oboh G., Ademosun A. O., Olumuyiwa T. A., Olasehinde T. A., Ademiluyi A. O., Adeyemo A. C. (2017). Insecticidal activity of essential oil from orange peels (Citrus sinensis) against Tribolium confusum, Callosobruchus maculatus and Sitophilus oryzae and its inhibitory effects on acetylcholinesterase and Na+ /K+-ATPase activities. Phytoparasitica 45 501–508. 10.1007/s12600-017-0620-z [DOI] [Google Scholar]
- Ogendo J. O., Kostyukovsky M., Ravid U., Matasyoh J. C., Deng A. L., Omolo E. O., et al. (2008). Bioactivity of Ocimum gratissimum L. oil and two of its constituents against five insect pests attacking stored food products. J. Stored Prod. Res. 44 328–334. 10.1016/j.jspr.2008.02.009 [DOI] [Google Scholar]
- Okazaki Y., Katayama T. (2003). Effects of dietary carbohydrate and myo -inositol on metabolic changes in rats fed 1,1,1-trichloro-2,2-bis (p-chlorophenyl) ethane (DDT). J. Nutr. Biochem. 14 81–89. 10.1016/s0955-2863(02)00279-6 [DOI] [PubMed] [Google Scholar]
- Oppert B., Elpidina E. N., Toutges M., Mazumdar-Leighton S. (2010). Microarray analysis reveals strategies of Tribolium castaneum larvae to compensate for cysteine and serine protease inhibitors. Comp. Biochem. Physiol. Part D Genomics Proteomics 5 280–287. 10.1016/j.cbd.2010.08.001 [DOI] [PubMed] [Google Scholar]
- Pan L., Ren L., Chen F., Feng Y., Luo Y. (2016). Antifeedant activity of Ginkgo biloba secondary metabolites against Hyphantria cunea larvae: mechanisms and applications. PLoS One 11:e0155682 10.1371/journal.pone.0155682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A. K., Singh P. (2017). The genus Artemisia: a 2012-2017 literature review on chemical composition, antimicrobial, insecticidal and antioxidant activities of essential oils. Medicines 4:68 10.3390/medicines4030068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare P. W., Tumlinson J. H. (1999). Plant volatiles as a defense against insect herbivores. Plant Physiol. 121 325–332. 10.1104/pp.121.2.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P., Iovinella I., Zhu J., Wang G., Dani F. R. (2017). Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol. Rev. Camb. Philos. Soc. 93 184–200. 10.1111/brv.12339 [DOI] [PubMed] [Google Scholar]
- Perkin L. C., Elpidina E. N., Oppert B. (2017). RNA interference and dietary inhibitors induce a similar compensation response in Tribolium castaneum larvae. Insect Mol. Biol. 26 35–45. 10.1111/imb.12269 [DOI] [PubMed] [Google Scholar]
- Phillips T., Throne J. (2010). Biorational approaches to managing stored-product insects. Annu. Rev. Entomol. 55 375–397. 10.1146/annurev.ento.54.110807.090451 [DOI] [PubMed] [Google Scholar]
- Qing L. W., Hong J. C., Sha C. S., Xue Z. M., Long L. Z. (2010). Chemical composition and toxicity against Sitophilus zeamais and Tribolium castaneum of the essential oil of Murraya exotica aerial parts. Molecules 15 5831–5839. 10.3390/molecules15085831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinehart J. P., Li A., Yocum G. D., Robich R. M., Hayward S. A., Denlinger D. L. (2007). Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc. Natl. Acad. Sci. U.S.A. 104 11130–11137. 10.1073/pnas.0703538104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem N., Bachrouch O., Sriti J., Msaada K., Khammassi S., Hammami M., et al. (2018). Fumigant and repellent potentials of Ricinus communis and Mentha pulegium essential oils against Tribolium castaneum and Lasioderma serricorne. Int. J. Food Prop. 20 S2899–S2913. 10.1080/10942912.2017.1382508 [DOI] [Google Scholar]
- Sánchez-Gracia A., Vieira F. G., Rozas J. (2009). Molecular evolution of the major chemosensory gene families in insects. Heredity 103 208–216. 10.1038/hdy.2009.55 [DOI] [PubMed] [Google Scholar]
- Schama R., Pedrini N., Juárez M. P., Nelson D. R., Torres A. Q., Valle D., et al. (2016). Rhodnius prolixus supergene families of enzymes potentially associated with insecticide resistance. Insect Biochem. Mol. Biol. 69 91–104. 10.1016/j.ibmb.2015.06.005 [DOI] [PubMed] [Google Scholar]
- Sharifian I., Hashemi S. M., Aghaei M., Alizadeh M. (2012). Insecticidal activity of essential oil of Artemisia herba-alba Asso against three stored product beetles. Biharean Biol. 6 90–93. [Google Scholar]
- Sharifian I., Hashemi S. M., Darvishzadeh A. (2013). Fumigant toxicity of essential oil of Mugwort (Artemisia vulgaris L.) against three major stored product beetles. Arch. Phytopathol. Plant Protect. 46 445–450. 10.1080/03235408.2012.743389 [DOI] [Google Scholar]
- Shaw W. R., Teodori E., Mitchell S. N., Baldini F., Gabrieli P., Rogers D. W., et al. (2014). Mating activates the heme peroxidase HPX15 in the sperm storage organ to ensure fertility in Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 111 5854–5859. 10.1073/pnas.1401715111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., Du Y., Wang J. (2011). Molecular characterization and expression patterns of Spodoptera litura heat shock protein 70/90, and their response to zinc stress. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 158 102–110. 10.1016/j.cbpa.2010.09.006 [DOI] [PubMed] [Google Scholar]
- Soni R., Sharma G., Jasuja N. D. (2016). Essential oil yield pattern and antibacterial and insecticidal activities of Trachyspermum ammi and Myristica fragrans. Scientifica 2016:1428194 10.1155/2016/1428194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamopoulos D. C., Damos P., Karagianidou G. (2007). Bioactivity of five monoterpenoid vapours to Tribolium confusum (du Val) (Coleoptera: Tenebrionidae). J. Stored Prod. Res. 43 571–577. 10.1016/j.jspr.2007.03.007 [DOI] [Google Scholar]
- Stephen R., Gibbs R. A., Weinstock G. M., Brown S. J., Robin D., Beeman R. W., et al. (2008). The genome of the model beetle and pest Tribolium castaneum. Nature 452 949–955. 10.1038/nature06784 [DOI] [PubMed] [Google Scholar]
- Sun Y., Sheng Y., Bai L., Zhang Y., Xiao Y., Xiao L., et al. (2014). Characterizing heat shock protein 90 gene of Apolygus lucorum (Meyer-Dür) and its expression in response to different temperature and pesticide stresses. Cell Stress Chaperones 19 725–739. 10.1007/s12192-014-0500-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zhao J., Sheng Y., Xiao Y. F., Zhang Y. J., Bai L. X., et al. (2016). Identification of heat shock cognate protein 70 gene (Alhsc70) of Apolygus lucorum and its expression in response to different temperature and pesticide stresses. Insect Sci. 23 37–49. 10.1111/1744-7917.12193 [DOI] [PubMed] [Google Scholar]
- Takahashi N., Kurata S., Natori S. (1993). Molecular cloning of cDNA for the 29 kDa proteinase participating in decomposition of the larval fat body during metamorphosis of Sarcophaga peregrina (flesh fly). FEBS Lett. 334 153–157. 10.1016/0014-5793(93)81702-2 [DOI] [PubMed] [Google Scholar]
- Taracena M. L., Oliveira P. L., Almendares O., Umana C., Lowenberger C., Dotson E. M., et al. (2015). Genetically modifying the insect gut microbiota to control Chagas disease vectors through systemic RNAi. PLoS Negl. Trop. Dis. 9:e0003358 10.1371/journal.pntd.0003358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S., Garcã-a-Alcalde F., Dopazo J., Ferrer A., Conesa A. (2011). Differential expression in RNA-seq: a matter of depth. Genome Res. 21 2213–2223. 10.1101/gr.124321.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. M., Reddy G. V. P. (2016). Effect of temperature on two bio-insecticides for the control of confused flour beetle (Coleoptera: Tenebrionidae). Fla. Entomol. 99 67–71. [Google Scholar]
- Tripathi A. K., Prajapati V., Aggarwal K. K., Khanuja S. P., Kumar S. (2000). Repellency and toxicity of oil from Artemisia annua to certain stored-product beetles. J. Econ. Entomol. 93 43–47. 10.1603/0022-0493-93.1.43 [DOI] [PubMed] [Google Scholar]
- Tunç Ý., Berger B. M., Erler F., Dağlı F. (2000). Ovicidal activity of essential oils from five plants against two stored-product insects. J. Stored Prod. Res. 36 161–168. 10.1016/S0022-474X(99)00036-3 [DOI] [Google Scholar]
- Uchida K., Ohmori D., Ueno T., Nishizuka M., Eshita Y., Fukunaga A., et al. (2001). Preoviposition activation of cathepsin-like proteinases in degenerating ovarian follicles of the mosquito Culex pipiens pallens. Dev. Biol. 237 68–78. 10.1006/dbio.2001.0357 [DOI] [PubMed] [Google Scholar]
- Ueyama J., Satoh T., Kondo T., Takagi K., Shibata E., Goto M., et al. (2010). Beta-glucuronidase activity is a sensitive biomarker to assess low-level organophosphorus insecticide exposure. Toxicol. Lett. 193 115–119. 10.1016/j.toxlet.2009.12.009 [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk J. M., Hekimi S., Kim S. K. (2009). Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 5:e1000361 10.1371/journal.pgen.1000361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira-Brock P. L., Andrenyak D. M., Nielsen S. M., Fleckenstein A. E., Wilkins D. G. (2013). Age-related differences in the disposition of nicotine and metabolites in rat brain and plasma. Nicotine Tob. Res. 15 1839–1848. 10.1093/ntr/ntt067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vontas J. G., Small G. J., Hemingway J. (2001). Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem. J. 357 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakil W., Ashfaq M., Ghazanfar M. U., Riasat T. (2010). Susceptibility of stored-product insects to enhanced diatomaceous earth. J. Stored Prod. Res. 46 248–249. 10.1016/j.jspr.2010.05.001 [DOI] [Google Scholar]
- Wang D., Zhang Y., Dong Z., Guo P., Ma S., Guo K., et al. (2016). Serine protease P-IIc is responsible for the digestion of yolk proteins at the late stage of silkworm embryogenesis. Insect Biochem. Mol. Biol. 74 42–49. 10.1016/j.ibmb.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Wang J., Zhu F., Zhou X. M., Niu C. Y., Lei C. L. (2006). Repellent and fumigant activity of essential oil from Artemisia vulgaris to Tribolium castaneum (Herbst) (Coleoptera: Tenebrionidae). J. Stored Prod. Res. 42 339–347. 10.1016/j.jspr.2005.06.001 [DOI] [Google Scholar]
- Wang R. L., Christian S., Xia Q. Q., Su Y. J., Zeng R. S. (2015). Identification and characterization of CYP9A40 from the Tobacco cutworm Moth (Spodoptera litura), a cytochrome P450 gene induced by plant allelochemicals and insecticides. Int. J. Mol. Sci. 16 22606–22620. 10.3390/ijms160922606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., You C. X., Yang K., Wu Y., Chen R., Zhang W. J., et al. (2015). Bioactivity of essential oil of Zingiber purpureum Rhizomes and its main compounds against two stored product insects. J. Econ. Entomol. 108 925–932. 10.1093/jee/tov030 [DOI] [PubMed] [Google Scholar]
- Wei L., Gao S., Xiong W., Liu J., Mao J., Lu Y., et al. (2019). Latrophilin mediates insecticides susceptibility and fecundity through two carboxylesterases, esterase4 and esterase6, in Tribolium castaneum. Bull. Entomol. Res. 109 534–543. 10.1017/S0007485318000895 [DOI] [PubMed] [Google Scholar]
- Xia J., Zhang C. R., Zhang S., Li F. F., Feng M. G., Wang X. W., et al. (2013). Analysis of whitefly transcriptional responses to Beauveria bassiana infection reveals new insights into insect-fungus interactions. PLoS One 8:e68185 10.1371/journal.pone.0068185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Gao S., Lu Y., Wei L., Mao J., Xie J., et al. (2019a). Latrophilin participates in insecticide susceptibility through positively regulating CSP10 and partially compensated by OBPC01 in Tribolium castaneum. Pestic. Biochem. Physiol. 159 107–117. 10.1016/j.pestbp.2019.06.005 [DOI] [PubMed] [Google Scholar]
- Xiong W., Gao S., Mao J., Wei L., Li B. (2019b). CYP4BN6 and CYP6BQ11 mediate insecticide susceptibility and their expression is regulated by Latrophilin in Tribolium castaneum. Pest Manag. Sci. 75 2744–2755. 10.1002/ps.5384 [DOI] [PubMed] [Google Scholar]
- Xuan N., Guo X., Xie H. Y., Lou Q. N., Lu X. B., Liu G. X., et al. (2015). Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Sci. 22 203–219. 10.1111/1744-7917.12116 [DOI] [PubMed] [Google Scholar]
- Yang X. M., Hou L. J., Dong D. J., Shao H. L., Wang J. X., Zhao X. F. (2006). Cathepsin B-like proteinase is involved in the decomposition of the adult fat body of Helicoverpa armigera. Arch. Insect Biochem. Physiol. 62 1–10. 10.1002/arch.20115 [DOI] [PubMed] [Google Scholar]
- Yang X. Q., Zhang Y. L., Wang X. Q., Dong H., Gao P., Jia L. Y. (2016). Characterization of multiple heat-shock protein transcripts from Cydia pomonella: their response to extreme temperature and insecticide exposure. J. Agric. Food Chem. 64 4288–4298. 10.1021/acs.jafc.6b01914 [DOI] [PubMed] [Google Scholar]
- Zhang B., Zhang L., Cui R., Zeng X., Gao X. (2016). Cloning and expression of multiple cytochrome P450 genes: induction by fipronil in workers of the red imported fire ant (Solenopsis invicta Buren). PLoS One 11:e0150915 10.1371/journal.pone.0150915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li D., Ge P., Yang M., Guo Y., Zhu K. Y., et al. (2013). RNA interference revealed the roles of two carboxylesterase genes in insecticide detoxification in Locusta migratoria. Chemosphere 93 1207–1215. 10.1016/j.chemosphere.2013.06.081 [DOI] [PubMed] [Google Scholar]
- Zhao X. F., An X. M., Wang J. X., Dong D. J., Du X. J., Sueda S., et al. (2005). Expression of the Helicoverpa cathepsin B-like proteinase during embryonic development. Arch. Insect Biochem. Physiol. 58 39–46. 10.1002/arch.20030 [DOI] [PubMed] [Google Scholar]
- Zhu F., Li T., Zhang L., Liu N. (2008). Co-up-regulation of three P450 genes in response to permethrin exposure in permethrin resistant house flies, Musca domestica. BMC Physiol. 8:18 10.1186/1472-6793-8-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Parthasarathy R., Bai H., Woithe K., Kaussmann M., Nauen R., et al. (2010). A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proc. Natl. Acad. Sci. U.S.A. 107 8557–8562. 10.1073/pnas.1000059107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Zhang W., Liu F., Chen X., Li H., Xu B. (2016). Characterization of an Apis cerana cerana cytochrome P450 gene (AccCYP336A1) and its roles in oxidative stresses responses. Gene 584 120–128. 10.1016/j.gene.2016.02.016 [DOI] [PubMed] [Google Scholar]
- Zhu Y. C., Blanco C. A., Portilla M., Adamczyk J., Luttrell R., Huang F. (2015). Evidence of multiple/cross resistance to Bt and organophosphate insecticides in Puerto Rico population of the fall armyworm, Spodoptera frugiperda. Pestic. Biochem. Physiol. 122 15–21. 10.1016/j.pestbp.2015.01.007 [DOI] [PubMed] [Google Scholar]
- Zou Z., Lopez D. L., Kanost M. R., Evans J. D., Jiang H. (2006). Comparative analysis of serine protease-related genes in the honey bee genome: possible involvement in embryonic development and innate immunity. Insect Mol. Biol. 15 603–614. 10.1111/j.1365-2583.2006.00684.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Length distribution of the gene detected by RNA-sequencing. The x-axis and y-axis indicate the length of the genes and the number of genes, respectively. (B) FPKM statistic of the control and treatment. The x-axis and y-axis indicated the FPKM value of the genes and the number of genes, respectively. (C) Cluster tree. Distances between the expressed genes were calculated by the Euclidean method. The algorithm of the sum of squares of deviations was used to calculate the distance between samples to construct the cluster tree. Y-axis represented the height of the cluster tree. Samples with similar height were clustered together.
Percent of coverage representing the percentage of genes that were expressed in each of the two libraries mapped in the red flour beetle (Tribolium castaneum) genome. Panels (A–C) control groups; (D,E), 5% Artemisia vulgaris EO treatment groups. The gene coverage was the percentage of a gene covered by the reads. This value was equal to the ratio of the base number in a gene covered by unique mapping reads to the total base number of that gene. The saturation analyses of all the detected genes. Panels (G–I) control groups; (J–L) 5% Artemisia vulgaris EO treatment groups.
Primers used for qRT-PCR analysis and dsRNA synthesis in this study.
Pathway enrichment analysis of differentially expressed genes (DEGs) and all the detected genes.
A set of 758 differentially expressed genes (DEGs) between the 5% Artemisia vulgaris treatment and control groups.
Differentially expressed genes (DEGs) involved in response to stimuli.
Differentially expressed genes (DEGs) related to antigen processing and presentation.
Data Availability Statement
Raw sequence reads were saved as FASTQ files and deposited in the NCBI Sequence Read Archive (SRA) database under the following accession numbers: control: SRR7646221, SRR7646223, and SRR7646224; EO treatment: SRR7646222, SRR7646225, and SRR7646226.