Abstract
Human Endogenous Retroviruses are a class of genomic elements that are the result of ancient retroviral infection of the human germline. Many are biologically active elements that have been implicated in multiple diseases including cancer. The most recent class to invade the human genome is the HERV-K(HML-2) (HERV-K) family. Approximately 90 HERV-K proviruses and many smaller elements have been identified to date in the human genome. Additional proviruses are continually being discovered with the rapid advancement of deep-sequencing and long-read sequencing technologies. HERV-K proviruses are poorly annotated in human transcriptome databases making their analysis in RNA-seq data difficult. To enable analysis, we compiled the sequences of 91 HERV-K proviruses identified in NCBI GenBank (ID JN675007-JN675097) and created a proviral alignment tool for visualizing RNA-seq reads aligned across individual proviruses. This allowed us to analyse publicly available RNA-seq data from 10 hepatoblastoma samples and 3 normal liver controls (GEO Accession ID: GSE89775). This data report includes the raw FASTA sequence files of the HERV-K proviruses from NCBI, a differential gene expression list between hepatoblastoma samples, and genomic alignment figures from 5 HERV-K proviruses identified as differentially expressed in the companion research article “Upregulation of Human Endogenous Retrovirus-K (HML-2) mRNAs in hepatoblastoma: Identification of potential new immunotherapeutic targets and biomarkers [1]. The data provided here are available for other research groups interested in evaluating individual HERV-K proviral expression using RNA-seq data. Furthermore, the data analysis is highly flexible and will accommodate the addition of other HERV-K proviruses.
Keywords: Human endogenous retrovirus-K, Hepatoblastoma, Transcriptome analysis, Genomic alignment
Specifications table
| Subject | Immunology and Microbiology: Virology |
| Specific subject area | Human Endogenous Retroviruses and Oncology |
| Type of data | Tables Figures Additional Data- text file (FASTA) of genomic sequences |
| How data were acquired | Bioinformatic analysis of HERV-K elements in RNA-seq data (Salmon, HISAT2, DESeq2) |
| Data format | Raw and Analysed |
| Parameters for data collection | 91 HERV-K proviral sequences contained in the NCBI Data Repository (GenBank ID JN675007-JN675097) were concatenated into a single FASTA file. The HERV-K FASTA file was used to analyze a publically available RNA-seq dataset of Hepatoblastoma and Normal Liver Controls. |
| Description of data collection | HERV-K FASTA file was used to perform a standard differential gene expression analysis across conditions with ensuing gene enrichment analysis (GO & KEGG) as well as a positional alignment analysis of RNA-seq reads across individual proviruses. |
| Data source location | University of Virginia School of Medicine Charlottesville, Virginia United States |
| Data accessibility | With the article |
| Related research article | David F Grabski, Aakrosh Ratan, Laurie R Gray, Stefan Bekiranov, David Rekosh, Marie-Louise Hammarskjold, Sara K Rasmussen; Upregulation of Human Endogenous Retrovirus-K (HML-2) mRNAs in hepatoblastoma: Identification of potential new immunotherapeutic targets and biomarkers; Jounral of Pediatric Surgery; Submitted. |
Value of the data
-
•
Human Endogenous Retrovirus-K are biologically active genomic elements in many cancers and during fetal development, making evaluation in fetal malignancy especially interesting. These data support one of the first investigations of HERV-K mRNA expression in fetal tumors.
-
•
The data presented in this investigation will assist virologists and immunologists investigating HERV-K mRNA expression in human systems. It will also assist translational oncologists interested in studying the development of HERV-K as a potential neoantigen and therapeutic target for immune therapy.
-
•
These data characterize HERV-K mRNA expression in hepatoblastoma. Additional experimental validation will determine a potential role for this expression as either a tumor marker or as a immunotherapeutic target.
-
•
The data in this investigation are presented in a flexible, easy to modify format making reproducible analyses in other experimental conditions (e.g. other cancers or biological conditions) quickly feasible.
1. Data description
The data in this investigation relates to the expression of Human Endogenous Retrovirus-K in Hepatoblastoma. It is a companion data manuscript to the research article “Upregulation of Human Endogenous Retrovirus-K (HML-2) mRNAs in hepatoblastoma: Identification of potential new immunotherapeutic targets and biomarkers [1].” Human Endogenous Retroviruses are a class of genomic elements that resulted from ancient retroviral infection of the human germline. Though often transcriptionally silent, HERV's are biologically active in many cancers [2] as well as during fetal development [3]. We developed an approach to measure HERV-K mRNA using RNA-seq data and examined HERV-K mRNA expression in 10 Hepatoblastoma (HB) and 3 normal liver controls (NC) using a publicly available RNA-seq dataset (NCBI Biorepository: GEO accession ID GSE89). We report data on the differential gene expression of Hepatoblastomas with high and low HERV-K expression and ensuing Gene Enrichment Analysis. We also report data on RNA-seq read alignment across specific HERV-K proviruses that were found to be differentially expressed.
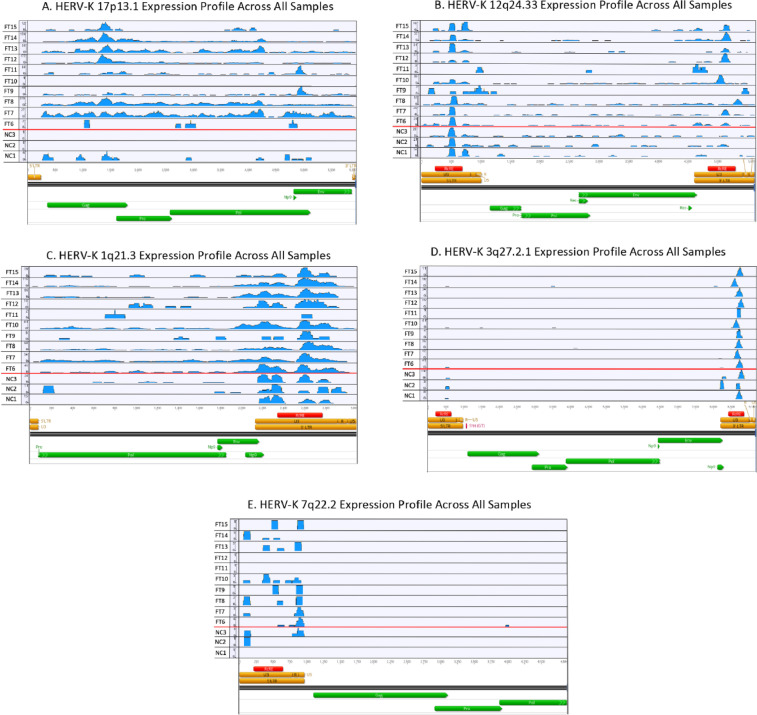
Supplemental File 1 represents the raw HERV-K FASTA file used to create our transcriptome alignments. Supplemental File 2 represents the full differential gene expression list (775 genes), which includes log 2-fold change and p-adjusted values following analysis and comparison of high HERV-K expressing tumors to low HERV-K expressing tumors. A Gene Enrichment analysis of the differential gene expression list was conducted using both Gene Otology (GO) terms as well as a Kyoto Encyclopedia of Genes and Genomes (KEGG). Table 1 includes the GO Molecular Function analysis. Table 2 includes the GO Cellular Localization analysis. Table 3 includes the KEGG fucntional analysis. For the HERV-K proviruses that were differentially expressed between HB and NC (1q21.3, 3q27.2, 7q22.2, 12q24.33 and 17p13.1), we plotted the read distribution of each sample across the respective provirus. Fig. 1 represents the RNA-seq read alignments from all samples across provirus 17p13.1 (panel A), 12q24.33 (panel B), 1q21.3 (panel C), 3q27.2 (panel D) and 7q22.2 (Panel E). Larger images for each individual provirus in Fig. 1 are provided in Supplemental File 3.
Table 1.
Gene Ontology (GO) molecular function analysis following differential gene expression analysis of high HERV-K expressing Hepatoblastoma vs low HERVK expressing Hepatoblastoma.
| Functional Category | Genes in list | Total genes | Enrichment False Discovery Rate (Adjusted-p-value) |
|---|---|---|---|
| Phospholipid binding | 32 | 441 | 0.021578265 |
| Collagen binding | 10 | 72 | 0.023567822 |
| Lipid binding | 45 | 761 | 0.023567822 |
| Identical protein binding | 92 | 1871 | 0.023567822 |
| Extracellular matrix structural constituent | 16 | 179 | 0.040987689 |
| Growth factor binding | 14 | 150 | 0.043859833 |
| Extracellular matrix binding | 8 | 56 | 0.043859833 |
| Protein kinase binding | 39 | 673 | 0.046968545 |
Table 2.
Gene Ontology (GO) cellular localization analysis following differential gene expression analysis of high HERV-K expressing Hepatoblastoma vs low HERVK expressing Hepatoblastoma (Top 20 terms).
| Functional Category | Genes in list | Total genes | Enrichment FDR |
|---|---|---|---|
| Secretory granule | 80 | 946 | 9.06E−12 |
| Vesicle | 225 | 4252 | 1.39E−11 |
| Secretory vesicle | 87 | 1108 | 1.39E−11 |
| Extracellular region part | 201 | 3693 | 2.14E−11 |
| Vesicle lumen | 45 | 386 | 3.07E−11 |
| Extracellular organelle | 141 | 2326 | 6.29E−11 |
| Cytoplasmic vesicle lumen | 44 | 385 | 6.29E−11 |
| Extracellular exosome | 140 | 2300 | 6.29E−11 |
| Extracellular vesicle | 141 | 2324 | 6.29E−11 |
| Extracellular space | 188 | 3479 | 1.41E−10 |
| Secretory granule lumen | 41 | 367 | 6.38E−10 |
| Extracellular region | 228 | 4617 | 2.16E−09 |
| Cytoplasmic vesicle part | 109 | 1761 | 7.46E−09 |
| Cytoplasmic vesicle | 144 | 2625 | 2.85E−08 |
| Intracellular vesicle | 144 | 2628 | 2.88E−08 |
| Collagen-containing extracellular matrix | 38 | 425 | 1.26E−06 |
| Platelet alpha granule lumen | 14 | 70 | 1.92E−06 |
| Extracellular matrix | 44 | 551 | 2.64E−06 |
| Endomembrane system | 225 | 4988 | 6.27E−06 |
| Lysosome | 54 | 797 | 1.78E−05 |
Table 3.
Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis following differential gene expression analysis of high HERV-K expressing Hepatoblastoma vs low HERVK expressing Hepatoblastoma.
| Functional Category | Genes in list | Total genes | Enrichment False Discovery Rate (Adjusted p-value) |
|---|---|---|---|
| Amoebiasis | 14 | 96 | 0.000793149 |
| Complement and coagulation cascades | 12 | 78 | 0.001119036 |
| Fatty acid degradation | 8 | 44 | 0.005062775 |
| Legionellosis | 9 | 55 | 0.005062775 |
| Peroxisome | 10 | 82 | 0.012590092 |
| Focal adhesion | 17 | 199 | 0.012590092 |
| Human papillomavirus infection | 24 | 330 | 0.012590092 |
| PI3K-Akt signaling pathway | 24 | 353 | 0.020654396 |
| Rheumatoid arthritis | 10 | 89 | 0.020654396 |
| ECM-receptor interaction | 9 | 82 | 0.034941883 |
| AGE-RAGE signaling pathway in diabetic complications | 10 | 100 | 0.034941883 |
| Epithelial cell signaling in Helicobacter pylori infection | 8 | 68 | 0.034941883 |
| Salmonella infection | 9 | 85 | 0.036101621 |
| Regulation of actin cytoskeleton | 16 | 214 | 0.036577774 |
| Tryptophan metabolism | 6 | 42 | 0.038157306 |
| Oocyte meiosis | 11 | 124 | 0.041198024 |
| IL-17 signaling pathway | 9 | 92 | 0.047460432 |
| Toxoplasmosis | 10 | 111 | 0.04997533 |
Fig. 1.
Graphical representation of uniquely aligned reads across HERV-K provirus (A)17p13.1 (B) 12q24.33 (C) 1q21.3 (D) 3q27.2 and (E) 7q22.2 created in bioinformatics platform Geneious. The x-axis represents the genomic position along the provirus. Major annotated regions of the proviral genome at each provirus are illustrated at the bottom of the panel. Coding regions for viral proteins Gag, Pro, Pol, Env, Rec or Np9 are represented by green bars, but does not necessarily infer an open-reading frame for the protein. Individual reads from each sample are represented on the y-axis. Abbreviations: FT- fetal tumor (hepatoblastoma), NC- normal control (liver).
2. Experimental design, materials, and methods
2.1. HERV-K database
Approximately 90 HERV-K proviruses have been identified to date in the human genome. However, HERV-K proviruses are currently not well annotated in human transcriptome databases. This makes quantifying HERV-K mRNA expression difficult using standard RNA-seq pipelines which rely on gene annotation for quantification. We searched the NCBI Data Repository for HERV-K proviral sequences, excluding solo long terminal repeats (LTRs). The search resulted in 91 HERV-K proviruses (GenBank ID JN675007-JN675097) [4]. Using the sequence of each provirus we created a HERV-K FASTA file. We then employed two separate analytical pipelines for RNA-seq analysis: one for HERV-K mRNA quantification and differential gene expression, and the second for proviral alignment and visualization, both are described in detail below.
2.2. Hepatoblastoma dataset (publicly available)
For the analysis in this investigation, we utilized a publicly available RNA-seq dataset of 10 hepatoblastoma samples and 3 normal liver controls. The data was generated as part of a larger investigation to identifying activated cancer pathways in hepatoblastoma aggressive hepatoblastoma [5]. The raw sequencing data are available from the NCBI biorepository (GEO accession ID GSE89775). The raw .fastq files were downloaded using the NCBI Sequence Read Archive (SRA) Toolkit. Following download, data was analyzed with the program FASTQC and was confirmed to be from strand-specific, 100 bp paired-end libraries containing approximately 40 M reads per sample (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). Trimmomatic was used to remove illumina adaptors, low-quality reads and assure a minimum read length of 50 bp [6].
2.3. Proviral quantification and differential expression
We concatenated the HERV-K FASTA file onto the human cDNA transcriptome from Ensembl (GRCh38.95) (available at ftp://ftp.ensembl.org/pub/release-95/fasta/homo_sapiens/cdna/). The concatenated file allowed us to analyze HB and NC RNA-seq reads aligned to the full human transcriptome as well as HERV-K proviral loci. We used the alignment program Salmon [7] in mapping-based mode with the validateMappings flag to create a count matrix over the full human transcriptome including the concatenated HERV-K sequences (example code: salmon quant -i GRCh38_HERVK.fa -l A −1 NC_1_1.fq −2 NC_1_2.fq –validateMappings -o NC_1_quant). Transcript abundance estimates from Salmon were imported into R (version 3.5.1) using tximport [8]. Gene abundance estimates were normalized for sequencing depth using DESeq2 [9]. We then focused on the read counts assigned to HERV-K loci and performed a differential gene expression analysis also using DESeq2. A p-adjusted value less than 0.05 (calculated using Benjamini-Hochberg False Discovery Rate) and an absolute value of log2 fold change greater than 1.5 were considered significant [10].
2.4. Gene enrichment analysis
Hepatoblastoma samples demonstrated heterogeneity in overall HERV-K expression levels and were sub-classified as high HERV-K expressing tumors and low HERV-K expressing tumors. A differential gene expression analysis between the 3 highest HERV-K expressing tumors and the 3 lowest HERV-K expressing tumors was conducted in DESeq2 as described above. A Gene Enrichment analysis of the differential gene expression list was conducted using both Gene Otology (GO) as well as a Kyoto Encyclopedia of Genes and Genomes (KEGG) terms. The analysis was performed using the clusterProfiler package in R [11]. Significantly enriched terms were determined by a False Discovery Rate < 0.05.
2.5. Proviral alignment and visualization
We utilized the HERV-K FASTA file to create a positional index using the alignment program HISAT2 (example code: hisat2-build HB_Data/HERVK_Genome.FASTA HERVK_Genome_tran) [12]. We aligned the HB and NC samples to the HISAT2-HERV-K index to create .BAM files. Uniquely mapped reads were selected with SAMtools (MAPQ Score >= 50). We imported the uniquely aligned .BAM files into the bioinformatics and genomic visualization platform Geneious (Biomatters, Auckland, New Zealand). For the HERV-K proviruses that were differentially expressed between HB and NC, we plotted read distribution of each sample across the respective provirus.
Ethics statement
The RNA-sequencing data utilized in this study is publicly available genomic data from National Center for Biotechnology Information (Accession number: GSE89775). It was not generated at our institution. It is de-inidentified data that meets all criteria for exemption as described by Human Subjects Research Exemption 45 CFR 46.101(b)(4) for Existing Data, Documents, Records and Specimens.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgments
The authors kindly acknowledge Dr. Rakesh Sindhi of UPMC, who made the Hepatoblastoma RNA-seq dataset available for this analysis. Partial salary support for MLH and DR was provided by the Charles H. Ross Jr and Myles H. Thaler Professorship endowments at the University of Virginia. This work was supported by The National Cancer Institute of the National Institutes of Health (Grant numbers: T32 CA163177 and R01 CA206275).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.105895.
Appendix. Supplementary materials
References
- 1.Grabski D.F. Upregulation of Human Endogenous Retrovirus-K (HML-2) mRNAs in hepatoblastoma: identification of potential new immunotherapeutic targets and biomarkers. J. Pediatr. Surg. 2020 doi: 10.1016/j.jpedsurg.2020.05.022. [Submitted] [DOI] [PubMed] [Google Scholar]
- 2.Grabski D.F. Close to the Bedside: a Systematic Review of Endogenous Retroviruses and Their Impact in Oncology. J. Surg. Res. 2019;240:145–155. doi: 10.1016/j.jss.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grow E.J. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522(7555):221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subramanian R.P. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011;8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranganathan S. Loss of EGFR-ASAP1 signaling in metastatic and unresectable hepatoblastoma. Sci. Rep., 2016;6:38347. doi: 10.1038/srep38347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patro R. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods, 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jornsten R. DNA microarray data imputation and significance analysis of differential expression. Bioinformatics. 2005;21(22):4155–4161. doi: 10.1093/bioinformatics/bti638. [DOI] [PubMed] [Google Scholar]
- 11.Yu G. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pertea M. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.