Abstract
Epigenetic mechanisms, including DNA methylation, histone post-translational modifications, and chromatin structure regulation, are critical for the interactions between tumor and immune cells. Emerging evidence shows that tumors commonly hijack various epigenetic mechanisms to escape immune restriction. As a result, the pharmaceutical modulation of epigenetic regulators, including ‘writers’, ‘readers’, ‘erasers’, and ‘remodelers’, is able to normalize the impaired immunosurveillance and/or trigger antitumor immune responses. Thus, epigenetic targeting agents are attractive immunomodulatory drugs and will have major impacts on immuno-oncology. Here, we discuss epigenetic regulators of the cancer–immunity cycle and current advances in developing epigenetic therapies to boost anticancer immune responses, either alone or in combination with current immunotherapies.
Epigenetics and Cancer
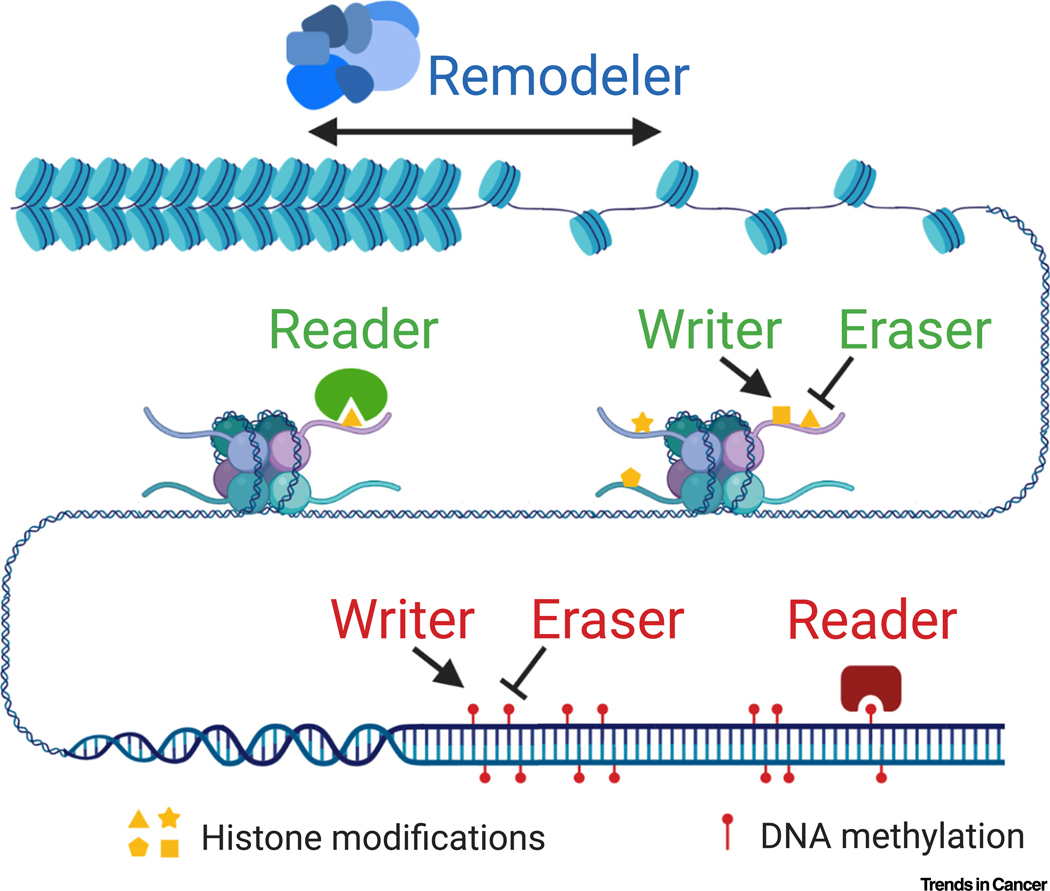
Epigenetics is defined as the DNA sequence-independent inheritance of phenotype or gene expression [1]. By modulating which, when, and where genes are expressed, epigenetic machinery determines cell fates during differentiation and maintains cell identities during and after cell division [2]. There are four major mechanisms of epigenetic regulation: DNA methylation, histone post-translational modifications, chromatin structure regulation, and noncoding RNA regulation [1]. Here, we mainly focus on the first three mechanisms, which are chromatin-based mechanisms (Figure 1). These epigenetic mechanisms usually work in a coordinated manner to provide precise and durable gene regulation. DNA and histone marks and chromatin structure are dynamically regulated by four classes of epigenetic regulators. These regulators are commonly known as ‘writers’, which add the epigenetic marks; ‘erasers’, which remove the epigenetic marks; ‘readers’, which recognize specific epigenetic marks to mediate downstream effects; and ‘remodelers’, which modulate chromatin status (Figure 1). There are ~1000 epigenetic regulators in mammals, forming one of the largest protein groups (Table 1).
Figure 1. Molecular Basis of Chromatin-Based Epigenetic Mechanisms and their Regulators.
Genomic DNA is wrapped around histone octamers to form nucleosomes, which are further packaged into the human nucleus in a highly organized manner. The packaging states of chromatin are dynamically regulated by chromatin-remodeling complexes (‘remodelers’) to allow or deny access of selected cis-elements by their trans-factors. Core histones can be modified at multiple residues through covalent bonds by methylation, acetylation, phosphorylation, ubiquitination, and many other modifications. DNA can be methylated (or hydroxymethylated) at the 5th position on the pyrimidine ring in cytosines, and less commonly in mammals at the nitrogen in the 6th position on the adenine in adenosines. Histone post-translational modifications and DNA methylation are added or removed by specific enzymes (‘writers’ and ‘erasers’, respectively) and recognized by their binding proteins (‘readers’).
Table 1.
Major Groups of Epigenetic Regulators
| Epigenetic features | Regulator | Gene family |
|---|---|---|
| DNA methylation | Writer | DNA methyltransferases (DNMTs) |
| Reader | 5-methylcytosine-binding domain proteins (MeCP2 and MBDs) | |
| Eraser | Ten-eleven translocation dioxygenases (TETs), ALKBH1 | |
| Histone modifications | Writer | Lysine methyltransferases (KMTs), protein arginine methyltransferases (PRMTs), lysine acetyltransferases (KATs or HATs), histone ubiquitin ligases, histone kinases, and others |
| Reader | Chromodomain, Tudor domain, MBT domain, PhD finger, bromodomain-containing proteins | |
| Eraser | Lysine demethylases (KDMs), histone deacetylases (HDACs and SIRTs), histone deubiquitinating enzymes, histone phosphatases, and others | |
| Chromatin structure | Remodeler | SWI/SNF, ISWI, CHD, and INO80/SWR complexes |
Epigenetic features are commonly dysregulated in cancer. Genome-wide DNA hypomethylation in cancer cells was first observed during the 1980s, while tumor suppressor genes are usually silenced by DNA hypermethylation at their promoters [3]. Similarly, loss of histone H4K16 acetylation and H4K20 trimethylation was reported to be a common hallmark of human cancer [4]. One of the surprising findings from cancer genome-sequencing studies is the high rate of alterations in many epigenetic regulator genes [5], such as loss-of-function mutations of genes encoding the SWI/SNF chromatin remodeling complex in ~20% of all cancers [6].
The accumulation of genetic and epigenetic alterations is a key characteristic of cancer cells [7]. Some genetic mutations create neoantigens, while epigenetic alterations may lead to the reactivation of genes, the expression of which is normally limited to immune-privileged stages or organs, such as cancer/testis antigens (CTAs) [8]. Both tumor neoantigens and autoantigens can be immunogenic [9].
Recent studies revealed that epigenetic regulation is critical for anticancer immune response and the evasion of immunosurveillance by tumor cells, nominating epigenetic targeting agents a new category of immune modulators. In this review, we will discuss the impact of epigenetic regulation on the interactions between tumor cells and immune cells and the emerging strategies to target the epigenetic machinery to boost anti-tumor immune responses.
Cancer–Immunity Cycle and Cancer Immunotherapy
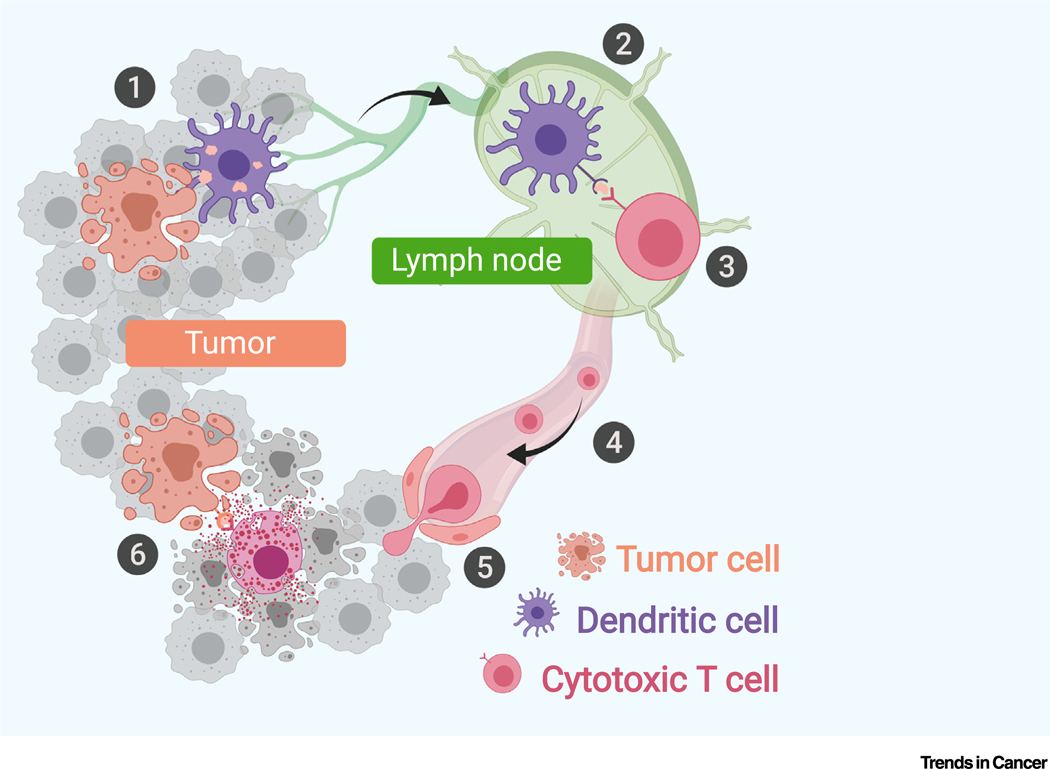
The human immune system should theoretically be capable of eradicating cancer cells through an acquired immune response executed by T cells. A series of stepwise events, called the ‘cancer–immunity cycle’, is required for tumor cell clearance by the immune system [10] (Figure 2). This self-amplifying process supposedly will end with complete clearance of nascent tumors. However, clinically detectable tumors often develop due to failed immunosurveillance through various mechanisms. For example, dendritic cell (DC)-mediated T cell priming and activation can be prevented by the lack of DC cells, DC-suppressive mechanisms, or activation of an immune check-point, such as CTLA-4 [11,12]. Lack of proper chemokines and an immunosuppressive tumor microenvironment (TME) may block the migration or infiltration of T cells into tumor tissue [13,14]. Even if tumor antigen-specific T cells infiltrate tumor tissue, their tumor-killing activity can be blocked by regulatory cells in the TME, such as regulatory T cells, macrophages, myeloid-derived suppressor cells, and cancer-associated fibroblasts, or by activation of immune checkpoints, such as PD-L1, on tumor cells or macrophages [15].
Figure 2. The Cancer–Immunity Cycle.
The cancer–immunity cycle comprises six major steps: (1) releasing: tumor-associated antigens are released by tumor cells into the microenvironment, mostly due to cell death; (2) presenting: released antigens are captured by dendritic cells (DCs) in the tumor microenvironment (TME). Antigen-loaded DCs then process and present the antigens on the cell surface with major histocompatibility complex (MHC) complexes and travel to lymphoid organs; (3) priming: naïve T cells in lymphoid organs recognize selected peptide–MHC complexes through T cell receptors (TCRs), which triggers the priming and activation of effector T cells; (4) trafficking: differentiated effector T cells leave lymphoid organs, and travel along blood vessels to scan peripheral tissues until they find their antigens in tumors; (5) infiltrating: T cells enter the tumor bed and migrate into the TME to become tumor-infiltrating lymphocytes (TILs); and (6) attacking: T cells recognize cancer cells carrying the matched antigen through interaction between the TCR and peptide–MHC complex and kill cancer cells by direct or indirect immune attack. Immune attack leads to the release of additional antigens from the dying tumor cells, which triggers a new round of antitumor immune response.
An understanding of these immune escape mechanisms has provided therapeutic opportunities by lifting immune suppression and restoring antitumor immune responses. For example, the discovery of immune checkpoint-mediated immune suppression led to the development of immune checkpoint blockade therapies (ICBTs). Antibody-based therapies targeting CTLA-4, PD-1, or PD-L1 have achieved lasting responses in some patients against a range of cancer types, especially those with considerable immunogenicity [16–18]. The success of ICBTs is, arguably, the most significant advance in cancer treatment over the past decade.
Despite the long-term efficacy of ICBTs for some patients, there are many patients who do not benefit from this advanced treatment for three major reasons. First, many cancers do not have strong immunogenicity, such as tumors from breast, prostate, glioblastoma, and pancreatic adenocarcinoma. The estimated percentage of patients with cancer who are eligible for any of the approved ICBTs is still <50% in the USA [19]. Second, not all patients with immunogenic tumor types respond to the treatment, due to tumor cell-intrinsic and extrinsic mechanisms [20]. A pan-cancer overall response rate is estimated to be ~25% [19]. Third, acquired resistance appears in some patients. This occurs through mechanisms that are induced or selected by the ICBTs, such as loss of antigens, inactivation of antigen-presenting machinery, desensitization to immune attack, and alternative immune suppressive pathways [20]. Intensive investigations have focused on expanding the application of current ICBTs and improve the response rate. Given that epigenetic regulation has important roles in antitumor immune responses, combining ICBTs with epigenetic drugs (epidrugs) could sensitize less-immunogenic tumors and prevent both primary and acquired resistance.
Impact of Epigenetics on the Cancer–Immunity Cycle
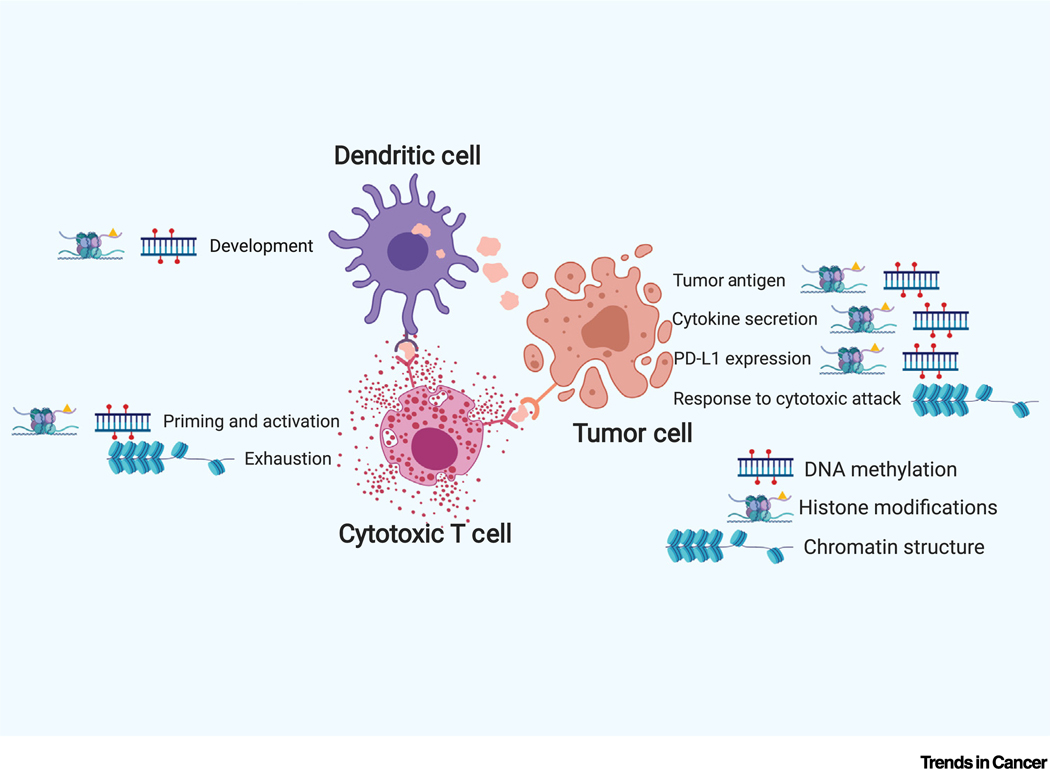
Epigenetic mechanisms are critical for many processes in the cancer–immunity cycle (Figure 3). Here, we discuss their impacts on these specific processes, either in tumor or immune cells.
Figure 3. Major Epigenetic Regulation in Tumor Immunity.
Histone post-translational modifications and DNA methylation play key roles in adaptive immune response, including dendritic cell development and T cell priming and activation. In tumor cells, histone and DNA modifications affects production of tumor antigens, silencing of anti-tumor cytokines, and induction of the PD-L1 checkpoint. Recent studies revealed the contributions of chromatin remodeling responding to cytotoxic attack in tumor cells and exhaustion phenotype in tumor infiltrating CD8 T cells. Abbreviation: PD-L1, programmed death ligand 1.
In Tumor Cells: Generation of Tumor Antigens
Deamination of 5-methyl-cytosine, either spontaneous or mutagen triggered, results in CNT transitions. Signature analysis revealed that DNA methylation-associated mutagenesis is the single most important source of genetic alterations, leading to neoantigen formation in most cancers [21]. CTAs are encoded by a group of genes, the expression of which is limited to male germ cells in healthy conditions [9]. However, demethylation of CpGs associated with these genes, as well as other epigenetic dysregulation, can cause CTA-coding genes to escape epigenetic silencing and re-express in tumors [22]. As a regenerative organ, the testis has an immune-privileged status [9]. Thus, when the protein products of these gametogenic genes are reactivated in tumor tissues, which are not immune privileged, they are capable of inducing an acquired immune response [22]. Similarly, the dysregulated epigenetic program in tumors can result in the reactivation of developmentally restricted genes, providing tumor differentiation antigens [23].
In Tumor Cells: Cytokine Production
Proinflammatory cytokines are required by effector T cells to enter the TME and execute an immune attack on tumor cells. Recent studies showed a strong connection between epigenetics and cytokine production in tumor cells. One such example is ‘viral mimicry’ as the result of DNA methyltransferase (DNMT) inhibition. Endogenous retroviruses (ERVs) represent >8% of the human genome but are predominantly silenced. DNA methylation is the major mechanism maintaining ERV silencing, and DNA demethylation in ERV promoters restores expression of ERV RNAs. ERV transcripts are mostly nonfunctional themselves. However, these transcripts can trigger the pattern-recognition receptor MDA5, which normally senses viral infection by recognizing viral double-strand (ds) RNAs. MDA5 induces signaling cascades that result in the secretion of type I interferon and eventually immune cell-induced killing. Thus, DNMT inhibitor (DNMTi) treatment tricks cancer cells into a ‘viral mimicry’ state, in which they behave as virus-infected cells, leading to activation of the interferon pathway. These changes were shown to enhance the effectiveness of immune checkpoint inhibitors [24,25]. Further studies revealed that histone deacetylases (HDACs) and KDM1A (LSD1), an ‘eraser’ of H3K4me1/2, also have similar roles in the suppression of ERVs and ERV-induced activation of the interferon pathway [26,27].
Similar to the dsRNA sensor MDA5, cyclic GMP-AMP synthase (cGAS) detects the abnormal presence of dsDNA in the cytosol, which signals infection or DNA damage [28]. cGAS then activates STING to trigger an innate immune response, especially the expression and secretion of cytokines [28]. Thus, STING agonists have been suggested as the next generation of immune-therapy agents [29]. However, robust activation of the STING pathway requires not only STING activation, but also sufficient STING protein to mediate the signaling cascade. The STING pathway is disrupted or epigenetically silenced in many tumors, enabling cancer cells to evade immunosurveillance [30]. It was found that the histone H3K4 demethylases KDM5B (JARID1B) and KDM5C (JARID1C) bind to the STING promoter and block the interferon response induced by cytosolic DNA in breast cancer cells [31]. Treatment with KDM5 inhibitors (KDM5i) induced STING expression and triggered a robust interferon response in a cytosolic DNA-dependent manner in breast cancer cells. These findings demonstrate that KDM5i act as STING inducers, representing a potential new class of cancer immunotherapeutic drugs, especially in tumors with low expression levels of STING.
Epigenetic enzymes also regulate interferons, cytokines, and chemokines through mechanisms other than through MDA5 or STING. For example, both DNMT and KMT6A (EZH2) directly suppress the expression of Th1-type chemokines, such as CXCL9 and CXCL10 [32], which are critical for T cell recruitment and infiltration. Counterintuitively, KDM6B (JMJD3), the methyltransferase with the counter role of KMT6A (EZH2), also suppresses chemokine expression [32]. Another methyltransferase, KMT3A, is required for the interferon pathway by catalyzing the methylation of STAT1, a key transcription factor of the interferon response [33].
In Tumor Cells: Tumor Antigen Presentation
Epigenetics contributes to the dysregulation of antigen-presenting machinery in tumor cells, which enables tumor cells to become invisible to T cells. To present self- and tumor-specific peptides to CD8 T cells, proteins in tumor cells need to be digested by the proteasome to generate short oligopeptides. Transporter associated with antigen processing 1 and 2 (TAP1 and TAP2) form a heterodimer to transport these peptides from the cytosol to the endoplasmic reticulum (ER). In the ER, antigen peptides are loaded onto nascent MHC-I molecules with the assistance of chaperone proteins. Antigen-loaded MHC-I, which consists of two polypeptide chains, human leukocyte antigens (HLA) and β2-microglobulin (B2M), is then delivered to the cell surface for display [34].
DNMT and HDAC both suppress MHC-I expression in tumor cells, evidenced by the re-expression of MHC-I after treating cells with DNMTi and HDACi [35,36]. In some cases, loss of MHC-I is caused by the epigenetic silencing of other genes involved in the antigen-presenting machinery, such as B2M, TAP-1, and TAP-2 [37,38]. Treating tumor cells and patients with DNMTi led to the increased expression of genes required for antigen presentation [37]. Interestingly, the deacetylation of histones was also responsible for the downregulation of MHC-I in devil facial tumor, an unusual disease that can transmit between Tasmanian devils as an infectious cell line [39].
In Tumor Cells: PD-L1 Expression
PD-L1 binds to the immune checkpoint receptor PD-1 on T cells, leading to the suppression of T cell proliferation, cytokine production, and cytotoxic activity, a phenotype described as ‘T cell exhaustion’ [40]. The upregulation of PD-L1 in some tumors is likely a result of selection caused by T cell immune responses. Epigenetic mechanisms certainly contribute to the regulation of PD-L1 expression. For example, in multiple cancers, methylation of the PD-L1 promotor was found to be negatively correlated with PD-L1 expression and prognosis [41–43]. Additionally, the histone acetylation ‘eraser’ HDAC6, methylation ‘writer’ KMT2A, and acetylation ‘reader’ bromodomain and extraterminal (BET) protein BRD4 activate PD-L1 expression in melanoma, pancreatic cancer, and ovarian cancer, respectively [44–46]. Thus, small-molecule inhibitors of HDAC6, KMT2A, and BET proteins suppressed PD-L1 expression and promoted antitumor immunity [44–46]. By contrast, ARID1A, a SWI/SNF ‘remodeler’ subunit frequently mutated in ovarian cancer, was shown to repress PD-L1 expression [47].
In Tumor Cells: Response to T Cell Attack
Besides epigenetic ‘writers’, ‘erasers’, and ‘readers’, ‘remodelers’ also alter interactions between cancer cells and immune cells in the TME. Sequencing of tumors isolated from patients with clear cell renal cell carcinoma who had received anti-PD-1 ICBT revealed that the mutation status of the PBRM1 gene was associated with clinical benefits [48]. PBRM1, ARID2, and BRD7 are signature components of the PBAF form of the SWI/SNF chromatin-remodeling complex [6]. An in vitro CRISPR/Cas9 screen identified that loss of PBRM1, ARID2, or BRD7 in melanoma cell lines sensitized cells to immune attack by CD8 T cells [49]. Increased response to IFNγ, a key cytotoxic cytokine secreted by NK and T cells, in PBAF-null cells appears to be responsible for the phenotype [49]. Thus, loss of the PBAF complex may serve as a biomarker for the response to ICBTs. However, utilization of this association to sensitize PBAF-intact tumors remains challenging, since PBRM1, ARID2, and BRD7 are not enzymes and there are no inhibitors of them available. Targeting their associated enzymes, such as BRG1, may serve as an alternative approach.
In Immune Cells: Lymphocyte Development
Epigenetic machinery has been implicated in cell-fate decisions during lymphocyte development [50]. Alterations in epigenetic regulators directly cause hematological malignancies. Key examples include translocation of KMT2A (MLL1) driving acute leukemia [51,52], and KMT6A (EZH2) gain-of-function mutations driving non-Hodgkin’s lymphoma [53].
The functional and phenotypic changes that occur during activation of the adaptive immune system also largely rely on the epigenetic machinery. Chromatin architecture and histone regulators are essential determinants of DC function [54]. For example, the histone H3K4 demethylase KDM5B negatively regulates the activation of bone marrow-derived DCs, leading to an incomplete T cell response [55]. By contrast, a ‘reader’ of methylated DNA, MBD2, is required for the phenotypic activation of DCs and their ability to initiate a T cell response [56]. Additionally, the DNA methylation ‘eraser’ TET2 and the histone acetylation ‘eraser’ HDAC2 coordinate to repress interleukin-6 expression by DCs, limiting the inflammatory response [57].
In Immune Cells: T Cell Activation
The primary T cell response in lymph nodes requires interactions between T cell receptors (TCR) on naïve T cells and MHC-peptides on antigen-presenting DCs. Upon the co-stimulation provided by mature DCs, the TCR–MHC-peptide interaction initiates an autonomous program of T cell differentiation and proliferation. This program not only increases the number of cells carrying the initial TCR sequence by clonal expansion, but also equips the lymphocytes with effector functions. During these processes, there is a global change in the epigenetic landscape in T cells, including DNA methylation [58,59], histone modifications [60,61], and genome accessibility [62], indicating the fundamental role of epigenetics in T cell activation.
The priming and activation of cytolytic T cells is accompanied by global DNA methylation remodeling [58]. Differentially methylated regions include de novo methylation on enhancers active in naïve T cells and promotor demethylation on effector genes, such as Gzmk and Gzmb [58]. Consistent with this, DNMT3A, a methyltransferase in charge of de novo DNA methylation, controls early effector CD8+ T cell fate decisions. Loss of DNMT3A leads to fewer effector cells, due to the ineffective repression of genes that are supposed to be silenced in effector cells [59].
Bivalent chromatin with the active transcriptional mark H3K4me3 and the suppressive mark H3K27me3 was found at gene loci associated with T cell proliferation and differentiation in naïve T cells. During priming and activation, most of these loci lose H3K27me3 while retaining the permissive H3K4me3 modification [60,61]. Additional analysis of enhancer marks, such as H3K4me1 and H3K27Ac, revealed a highly dynamic repertoire of enhancers during T cell activation [63].
In Immune Cells: T Cell Exhaustion
Chromatin organization has a central role in T cell exhaustion, as highlighted by recent studies. T cell exhaustion was originally discovered in a study of lymphocytic choriomeningitis mammarenavirus (LCMV), a natural pathogen of mice [64]. ATAC-seq showed that persistent LCMV infection-induced exhausted T cells have ~6000 open chromatin regions that are different from effector T cells [65]. This difference is comparable with the difference between hematopoietic lineages [66].
PD-1 blockade has the ability to reinvigorate exhausted T cells in both chronic infection and tumor settings, as shown by transcriptional, cellular, and functional changes [67–69]. However, the reinvigoration is usually not sustainable. After PD-1 blockade, reinvigorated effector T cells became re-exhausted, likely due to the failure of the blockade to reprogram the epigenetic landscape of exhausted T cells into effector T cells, shown by ATAC-seq analysis [70]. A de novo DNA methylation program in effector T cells is required for the development of fully exhausted T cells [71]. Interestingly, exhaustion-associated DNA methylation is preserved during ICBTs [71], consistent with the ATAC-seq analysis.
In addition to the chronic LCMV infection model, tumor models created by injecting antigen-carrying cancer cells into TCR transgenic mice have also been established to study the epigenetic contribution to T cell exhaustion. ATAC-seq showed that a consistent chromatin-remodeling program dominated the exhaustion of effector T cells, which was absent in the formation of memory T cells [72]. Additionally, there are two discrete chromatin states of T cell exhaustion. The first stage is plastic, because T cells can be rescued. The later stage is permanent, in which cells are resistant to reprogramming [72].
Targeting Epigenetic Regulators to Boost Antitumor Immune Responses
Targeting epigenetic aberrations is considered one of the most attractive cancer therapies for several reasons. First, recurrent mutations of epigenetic modulators and dysregulation of epigenetic features are widely observed in tumors. Second, in contrast to genetic changes, epigenetic alterations are largely reversible. Third, epigenetic features are regulated by enzymes or chromatin-binding proteins that are targetable. Thus, epidrugs can be developed to treat cancer by suppressing oncogenic epigenetic regulators and restoring normal epigenetic features.
Over the past two decades, major efforts from both academia and industry have been devoted to the development of epidrugs. Before 2020, there were only FDA-approved epidrugs for cancer treatment, including four pan-HDACi and two DNMTi (Table 2). These were approved to treat T cell lymphoma, multiple myeloma, or myelodysplastic syndromes, all of which are hematopoietic malignancies (clinical responses reviewed in [73,74]). Another HDACi, chidamide (also called HBI-8000), was approved by the Chinese Food and Drug Administration for treating peripheral T cell lymphoma (clinical responses reviewed in [75]). Several additional FDA-approved drugs, (hydralazine, procaine, and procainamide, for treating hypertension, local anesthetics, and cardiac arrhythmia, respectively) have also been shown to have DNMTi activity [76–78]. Similarly, valproic acid, an approved seizures drug, was also found to be a HDACi. These existing drugs with newly identified epigenetic modulating activities are still evaluated in the clinical trials for cancer treatment. In January 2020, tazemetostat, a KMT6A (EZH2) inhibitor, was approved for treatment of epithelioid sarcoma, making it the first approved histone ‘writer’ inhibitor and the first epidrug to treat solid tumors [79].
Table 2.
Epigenetic Drugs Approved or under Clinical Trials
| Category | Target | Approved drug and indications | Drugs under clinical trials |
|---|---|---|---|
| DNMT inhibitor | DNA methylation writers | Azacitidine (myelodysplastic syndromes), decitabine (myelodysplastic syndromes), procainamide (cardiac arrhythmias), hydralazine (essential hypertension), procaine (local anesthetics) | Tioguanine, FdCyd, TdCyd, Aza-TdCyd, fluorocyclopentenylcytosine, guadecitabine |
| HDAC inhibitor | Histone acetylation erasers | Vorinostat (cutaneous T cell lymphoma), romidepsin (cutaneous T cell lymphoma), belinostat (peripheral T cell lymphoma), panobinostat (multiple myeloma), valproic acid (seizures), chidamide (peripheral T-cell lymphoma, by CFDA) | Tacedinaline, mocetinostat, abexinostat, entinostat, pracinostat, resminostat, givinostat, quisinostat, kevetrin, tefinostat, nanatinostat, domatinostat, ricolinostat, ME-344, CG200745, CUDC-101, AR42 |
| KMT6A inhibitor | Histone methylation writer | Tazemetostat (epithelioid sarcoma) | SHR2554, CPI-1205, GSK2816126, PF-06821497, MAK683 |
| SIRT activator | Histone acetylation erasers | None | SRT2104 |
| BET inhibitor | Histone acetylation readers | None | Mivebresib, molibresib, birabresib, INCB057643, ZEN003694, FT-1101, GSK2820151, CC-90010, CPI-0610, PLX51107, ABBV-744, BAY1238097, BI 894999, BMS-986158, GS-5829 |
| PRMT5 inhibitor | Histone methylation writer | None | JNJ-64619178, PF-06939999, GSK3326595 |
| PRMT1 inhibitor | Histone methylation writer | None | GSK3368715 |
| KDM1A inhibitor | Histone methylation eraser | None | Seclidemstat, IMG-7289, tranylcypromine, GSK2879552, INCB059872, phenelzine sulfate |
| KMT4 inhibitor | Histone methylation writer | None | Pinometostat |
Interestingly, emerging evidence suggests that some epigenetics-targeting molecules, including drugs that are approved or under preclinical and clinical studies, modulate the tumor immune microenvironment and induce robust antitumor immune responses [24,25,27,31].
Treating tumor-bearing animals with DNMTi and/or HDACi altered the immunosuppressive TME and enhanced tumor-infiltrating lymphocytes [26,35,47,81]. These effects are the result of enhanced tumor antigen expression and/or presentation, ‘viral mimicry’ effects, activation of DC cells, suppression of T cell exhaustion, or combinations thereof. Additional epidrugs, such as inhibitors targeting KMT6A (EZH2) [82], KDM1A (LSD1) [83], PRMT5 [84], and BET proteins [85,86], were also capable of remodeling the TME in animal models. Similar changes in the TME have been observed in tumor tissues isolated from patients who received epidrugs [37,87]. Thus, epidrugs are attractive immunotherapy agents to boost antitumor immune responses, either as single agents or in combination with other anticancer agents, including ICBTs.
In addition to FDA-approved DNMTi and HDACi, new inhibitors of DNMTs and HDACs have been developed to improve efficiency and achieve stability, high specificity, and low toxicity. For example, guadecitabine (SGI-110), TdCyd, FdCyd, aza-TdCyd, and ASTX727 are cytidine analogs that inhibit DNMTs through a mechanism similar to azacitidine and decitabine. These drugs have already entered clinical trials. New categories of DNMTi, such as DNA-binding compounds, oligonucleotides, and S-adenosyl-I-methionine (SAM) competitors, are under preclinical investigation [88]. There are more than a dozen new HDACi, representing different specificities against 18 HDACs (including SIRTs, a subgroup of NAD+ dependent HDACs) in the human genome, also in clinical trials [89].
Furthermore, intensive efforts are also being dedicated to targeting other epigenetic regulators (Table 2). Many epigenetic targeting agents are under clinical investigation for the treatment of both hematological and solid tumors. For example, inhibitors that target the histone mark ‘writers’ KMT6A (EZH2), KMT4 (DOT1L), and PRMT5, ‘erasers’ KDM1A (LSD1), and ‘reader’ BET proteins, have entered clinical trials. Other inhibitors, such as KMT2, KDM4, and KDM5 inhibitors, are still at the preclinical stages for oncology indications. Since the combination of epidrugs with checkpoint inhibitors showed synergistic effects in animal experiments [71,81,85,86,90], some clinical-stage epidrugs are being evaluated in clinical trials in combination with CTLA-4, PD-1, and PD-L1 ICBTs (Table 3).
Table 3.
Clinical Trials with the Indicated Clinicaltrial.gov NCT IDs Combining Immune Checkpoint Inhibitors and Epigenetic Targeting Agentsa
| Epigenetic targeting agent | Immune checkpoint inhibitor | |||||
|---|---|---|---|---|---|---|
| Ipilimumab (anti-CTLA4) | Nivolumab (anti-PD-1) | Pembrolizumab (anti-PD-1) | Atezolizumab (anti-PD-L1) | Avelumab (anti-PD-L1) | Durvalumab (anti-PD-L1) | |
| DNMTi | ||||||
| Azacitidine | 02530463, 02397720 | 02530463, 02397720, 01928576 | 03264404, 03769532, 03094637, 02260440, 02816021, 02845297, 02546986, 02959437, 02900560, 02512172 | 02508870 | 03390296, 03699384, 02953561, 03390296, 02951156 | 02811497, 03019003, 02811497, 02775903, 03161223, 02117219, 02250326, 03019003 |
| Decitabine | 02890329 | 02664181, 03358719 | 03445858, 02996474, 03969446, 03240211, 02957968, 03233724 | 03395873 | ||
| Guadecitabine | 02608437, 02890329 | 03576963 | 02901899, 02998567, 03220477 | 02892318, 02935361, 03179943, 03206047 | 03308396, 03257761, 03085849 | |
| HDACi | ||||||
| Vorinostat | 02619253, 03150329, 03426891, 02395627 | |||||
| Romidepsin | 02393794 | 03278782, 02512172 | 03161223 | |||
| Panobinostat | 02032810 | |||||
| Chidamide | 02718066 | |||||
| Mocetinostat | 03565406 | 03565406, 02954991 | 03220477 | 02805660, 02993991 | ||
| Valproic acid | 02648633 | 03357757 | ||||
| Abexinostat | 03590054 | |||||
| Entinostat | 03552380, 02453620 | 03838042, 03552380, 02453620, 03250273, 01928576 | 03179930, 02936752, 02909452, 03765229, 02437136, 02697630, 03978624 | 02708680, 03280563 | 02915523 | |
| Domatinostat | 03812796 | |||||
| BETi | ||||||
| INCB057643 | 02959437 | |||||
| BMS-986158 | 024-19417 | |||||
| KMT6Ai | ||||||
| Tazemetostat | 03854474 | 02220842 | ||||
| CPI-1205 | 03525795 | |||||
| KDM1Ai | ||||||
| INCB059872 | 02712905 | 02959437 | ||||
In addition, sintilimab (anti-PD-1) is being tested in combination with chidamide (HDACi) (NCT03820596), and tremelimumab (anti-CTLA-4) is being tested in combination with azacitidine (DNMTi) (NCT03019003) and guadecitabine (DNMTi) (NCT03085849).
Concluding Remarks
Cancer epigenetics and cancer immunology are both fast-moving fields, attracting major investigational efforts. Recent studies have shown that epigenetic regulation affects all cancer hall-marks, including all aspects of the interaction between tumor cells and the immune system. As a result, epigenetic modulation can elicit robust antitumor immune responses. Although some changes are broad, others are more restricted to certain cells and/or tissues. Here, we suggest that epigenetic therapies are novel immunotherapies by themselves. These findings provide unique opportunities to combine epidrug-based therapies with other cancer treatment strategies, including current and upcoming immunotherapies. These combinations could not only have synergistic effects, but could also reduce adverse effects and prevent drug resistance. Identifying the most effective epigenetic targeting strategies to boost anti-tumor immune responses, especially in solid tumors, and developing rationale-based combinational strategies will have major impacts on our practice of immuno-oncology in the future (see Outstanding Questions).
Outstanding Questions.
Why are current epigenetic drugs ineffective against most solid tumors?
Which epigenetic targeting strategies can achieve specific immunomodulatory effects?
Which epidrugs are synergistic with current and upcoming cancer immunotherapies?
Highlights.
Epigenetic mechanisms affect all aspects of the cancer–immunity cycle.
Some epigenetic targeting strategies are novel immunotherapies.
Epigenetic drugs could have synergistic effects with current cancer therapies, including immunotherapies, and prevent resistance to current cancer therapies.
Acknowledgments
This work was supported by a start-up fund and a New Investigator Award (both to J.C.) provided by Rutgers Cancer Institute of New Jersey (State of NJ appropriation and National Institutes of Health grant P30CA072720), a Melanoma Research Foundation Career Development Award (to J.C.), a Breast Cancer Alliance Exceptional Project Grant (to Q.Y.), and National Institutes of Health grants R01CA237586 and P50CA121974 (both to Q.Y.). We thank for Sarah Radford for her critical reading of the manuscript and valuable comments. We apologize to our colleagues whose works are not cited because of space limitations.
Footnotes
Uncited reference
[80]
References
- 1.Cavalli G and Heard E. (2019) Advances in epigenetics link genetics to the environment and disease. Nature 571, 489–499 [DOI] [PubMed] [Google Scholar]
- 2.John RM and Rougeulle C. (2018) Developmental epigenetics: phenotype and the flexible epigenome. Front. Cell Dev. Biol 6, 130–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fardi M. et al. (2018) Epigenetic mechanisms as a new approach in cancer treatment: an updated review. Genes Dis. 5, 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraga MF. et al. (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet 37, 391–400 [DOI] [PubMed] [Google Scholar]
- 5.Lawrence MS. et al. (2014) Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadoch C. et al. (2013) Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet 45, 592–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D and Weinberg RA. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 8.Schumacher TN and Schreiber RD. (2015) Neoantigens in cancer immunotherapy. Science 348, 69–74 [DOI] [PubMed] [Google Scholar]
- 9.Simpson AJ. et al. (2005) Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer 5, 615–625 [DOI] [PubMed] [Google Scholar]
- 10.Chen DS and Mellman I. (2013) Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10 [DOI] [PubMed] [Google Scholar]
- 11.Fu C and Jiang A. (2018) Dendritic cells and CD8 T cell immunity in tumor microenvironment. Front. Immunol 9, 3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y. et al. (2019) Hijacking antibody-induced CTLA-4 lysosomal degradation for safer and more effective cancer immunotherapy. Cell Res. 29, 609–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krummel MF. et al. (2016) T cell migration, search strategies and mechanisms. Nat. Rev. Immunol 16, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joyce JA and Fearon DT. (2015) T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80 [DOI] [PubMed] [Google Scholar]
- 15.Rabinovich GA. et al. (2007) Immunosuppressive strategies that are mediated by tumor cells. Annu. Rev. Immunol 25, 267–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodi FS. et al. (2010) Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med 363, 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Topalian SL. et al. (2012) Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med 366, 2443–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brahmer JR. et al. (2012) Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med . 366, 2455–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haslam A and Prasad V. (2019) Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw. Open 2, e192535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma P. et al. (2017) Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandrov LB. et al. (2013) Signatures of mutational processes in human cancer. Nature 500, 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitehurst AW. (2014) Cause and consequence of cancer/testis antigen activation in cancer. Annu. Rev. Pharmacol. Toxicol 54, 251–272 [DOI] [PubMed] [Google Scholar]
- 23.Ilyas S and Yang JC. (2015) Landscape of tumor antigens in T cell immunotherapy. J. Immunol 195, 5117–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roulois D. et al. (2015) DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous Transcripts. Cell 162, 961–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiappinelli KB. et al. (2015) Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Topper MJ. et al. (2017) Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 171, 1284–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheng W. et al. (2018) LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan X. et al. (2018) Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol 72, 447–478 [DOI] [PubMed] [Google Scholar]
- 29.Ramanjulu JM. et al. (2018) Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature 564, 439–443 [DOI] [PubMed] [Google Scholar]
- 30.Xia T. et al. (2016) Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 14, 282–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu L. et al. (2018) KDM5 histone demethylases repress immune response via suppression of STING. PLoS Biol. 16, e2006134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng D. et al. (2015) Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 527, 249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen K. et al. (2017) Methyltransferase SETD2-mediated methylation of STAT1 is critical for interferon antiviral activity. Cell 170, 492–506 [DOI] [PubMed] [Google Scholar]
- 34.Leone P. et al. (2013) MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J. Natl. Cancer Inst 105, 1172–1187 [DOI] [PubMed] [Google Scholar]
- 35.Luo N. et al. (2018) DNA methyltransferase inhibition upregulates MHC-I to potentiate cytotoxic T lymphocyte responses in breast cancer. Nat. Commun 9, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magner WJ. et al. (2000) Activation of MHC class I, II, and CD40 gene expression by histone deacetylase inhibitors. J. Immunol 165, 7017–7024 [DOI] [PubMed] [Google Scholar]
- 37.Li H. et al. (2014) Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epi- thelial cancers. Oncotarget 5, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kortenhorst MS. et al. (2013) Analysis of the genomic response of human prostate cancer cells to histone deacetylase inhibitors. Epigenetics 8, 907–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddle HV. et al. (2013) Reversible epigenetic down-regulation of MHC molecules by devil facial tumour disease illustrates immune escape by a contagious cancer. Proc. Natl. Acad. Sci. U. S. A 110, 5103–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong H. et al. (2002) Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med 8, 793–800 [DOI] [PubMed] [Google Scholar]
- 41.Goltz D. et al. (2017) PD-L1 (CD274) promoter methylation predicts survival in patients with acute myeloid leukemia. Leukemia 31, 738–743 [DOI] [PubMed] [Google Scholar]
- 42.Heiland DH. et al. (2017) Comprehensive analysis of PD-L1 expression in glioblastoma multiforme. Oncotarget 8, 42214–42225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gevensleben H. et al. (2016) PD-L1 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncotarget 7, 79943–79955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M, L. et al. (2016) Essential role of HDAC6 in the regulation of PD-L1 in melanoma. Mol. Oncol 10, 735–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu C. et al. (2017) The MLL1-H3K4me3 axis-mediated PD-L1 expression and pancreatic cancer immune evasion. J. Natl. Cancer Inst 109, djw283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu H. et al. (2016) BET bromodomain inhibition promotes anti-tumor immunity by suppressing PD-L1 expression. Cell Rep. 16, 2829–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukumoto T. et al. (2019) HDAC6 Inhibition Synergizes with Anti-PD-L1 Therapy in ARID1A-Inactivated Ovarian Cancer. Cancer Res. 79, 5482–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miao D. et al. (2018) Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359, 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan D. et al. (2018) A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kioussis D and Georgopoulos K. (2007) Epigenetic flexibility underlying lineage choices in the adaptive immune system. Science 317, 620–622 [DOI] [PubMed] [Google Scholar]
- 51.Milne TA. (2017) Mouse models of MLL leukemia: recapitulating the human disease. Blood 129, 2217–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krivtsov AV. et al. (2006) Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442, 818–822 [DOI] [PubMed] [Google Scholar]
- 53.Berg T. et al. (2014) A transgenic mouse model demonstrating the oncogenic role of mutations in the polycomb-group gene EZH2 in lymphomagenesis. Blood 123, 3914–3924 [DOI] [PubMed] [Google Scholar]
- 54.Boukhaled GM. et al. (2019) Chromatin architecture as an essential determinant of dendritic cell function. Front. Immunol 10, 1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ptaschinski C. et al. (2015) RSV-induced H3K4 demethylase KDM5B leads to regulation of dendritic cell-derived innate cytokines and exacerbates pathogenesis in vivo. PLoS Pathog.11, e1004978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cook PC. et al. (2015) A dominant role for the methyl-CpG-binding protein Mbd2 in controlling Th2 induction by dendritic cells. Nat. Commun 6, 6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q. et al. (2015) Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 525, 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scharer CD. et al. (2013) Global DNA methylation remodeling accompanies CD8 T cell effector function. J. Immunol 191, 3419–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ladle BH. et al. (2016) De novo DNA methylation by DNA methyltransferase 3a controls early effector CD8+ T-cell fate decisions following activation. Proc. Natl. Acad. Sci. U. S. A 113, 10631–10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Russ BE. et al. (2014) Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity 41, 853–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Araki Y. et al. (2009) Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 30, 912–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu B. et al. (2017) Epigenetic landscapes reveal transcription factors that regulate CD8(+) T cell differentiation. Nat. Immunol 18, 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He B. et al. (2016) CD8(+) T cells utilize highly dynamic enhancer repertoires and regulatory circuitry in response to infections. Immunity 45, 1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moskophidis D. et al. (1993) Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature 362, 758–761 [DOI] [PubMed] [Google Scholar]
- 65.Sen DR. et al. (2016) The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lara-Astiaso D. et al. (2014) Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bengsch B. et al. (2016) Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8(+) T cell exhaustion. Immunity 45, 358–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Staron MM. et al. (2014) The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8(+) T cells during chronic infection. Immunity 41, 802–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gubin MM. et al. (2014) Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pauken KE. et al. (2016) Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghoneim HE. et al. (2017) De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170, 142–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Philip M. et al. (2017) Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agrawal K. et al. (2018) Nucleosidic DNA demethylating epigenetic drugs - A comprehensive review from discovery to clinic. Pharmacol. Ther 188, 45–79 [DOI] [PubMed] [Google Scholar]
- 74.McClure JJ. et al. (2018) Advances and Challenges of HDAC Inhibitors in Cancer Therapeutics. Adv. Cancer Res 138, 183–211 [DOI] [PubMed] [Google Scholar]
- 75.Chan TS. et al. (2017) Chidamide in the treatment of peripheral T-cell lymphoma. Onco. Targets Ther 10, 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cornacchia E. et al. (1988) Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J. Immunol 140, 2197–2200 [PubMed] [Google Scholar]
- 77.Li YC. et al. (2018) Procaine is a specific DNA methylation inhibitor with anti-tumor effect for human gastric cancer. J. Cell. Biochem 119, 2440–2449 [DOI] [PubMed] [Google Scholar]
- 78.Lee BH. et al. (2005) Procainamide is a specific inhibitor of DNA methyltransferase 1. J. Biol. Chem 280, 40749–40756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leslie M. (2020) First EZH2 Inhibitor Approved-for Rare Sarcoma. Cancer Discov. 10.1158/2159-8290.CD-NB2020-006 [DOI] [PubMed] [Google Scholar]
- 80.Dawson MA. (2017) The cancer epigenome: concepts, challenges, and therapeutic opportunities. Science 355, 1147–1152 [DOI] [PubMed] [Google Scholar]
- 81.Knox T. et al. (2019) Selective HDAC6 inhibitors improve anti-PD-1 immune checkpoint blockade therapy by decreasing the anti-inflammatory phenotype of macrophages and down-regulation of immunosuppressive proteins in tumor cells. Sci. Rep 9, 6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang D. et al. (2018) Targeting EZH2 reprograms intratumoral regulatory T cells to enhance cancer immunity. Cell Rep. 23, 3262–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tan AHY et al. (2019) Lysine-specific histone demethylase 1A regulates macrophage polarization and checkpoint molecules in the tumor microenvironment of triple-negative breast cancer. Front. Immunol 10, 1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nagai Y. et al. (2019) PRMT5 associates with the FOXP3 homomer and when disabled enhances targeted p185(erbB2/neu) tumor immunotherapy. Front. Immunol 10, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adeegbe DO. et al. (2018) BET bromodomain inhibition cooperates with PD-1 blockade to facilitate antitumor response in Kras-mutant non-small cell lung cancer. Cancer Immunol. Res 6, 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hogg SJ. et al. (2017) BET-bromodomain inhibitors engage the host immune system and regulate expression of the immune checkpoint ligand PD-L1. Cell Rep. 18, 2162–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tomita Y. et al. (2016) The interplay of epigenetic therapy and immunity in locally recurrent or metastatic estrogen receptor-positive breast cancer: correlative analysis of ENCORE 301, a randomized, placebo-controlled phase II trial of exemestane with or without entinostat. Oncoimmunology 5, e1219008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Castillo-Aguilera O. et al. (2017) DNA methylation targeting: The DNMT/HMT crosstalk challenge. Biomolecules 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Suraweera A. et al. (2018) Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: achieving the full therapeutic potential of HDACi. Front. Oncol 8, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goswami S. et al. (2018) Modulation of EZH2 expression in T cells improves efficacy of anti-CTLA-4 therapy. J. Clin. Invest 128, 3813–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]