Abstract
Background
Multiple synchronous lung tumors (MSLT), particularly within a single lobe, represent a diagnostic and treatment challenge. While histologic assessment was once the only method to possibly distinguish multiple primary lung cancers, there is a growing interest in identifying unique genomic features or mutations to best characterize these processes.
Methods
In order to differentiate multiple primary lung malignancies from intrapulmonary metastases in patients with MSLT, we performed whole exome sequencing (WES) on 10 tumor samples from 4 patients with MSLT.
Results
Shared mutations between tumors from the same patient varied from 0–91%. Patient 3 shared no common mutations; however, in Patients 2 and 4, identical mutations were identified among all tumors from each patient, suggesting that the three tumors identified in Patient 3 represent separate primary lung cancers, while those of Patients 1, 2 and 4 signify hematogenous and lymphatic spread.
Conclusions
A high proportion of shared mutations between different lung tumors is likely indicative of intrapulmonary metastatic disease, while tumors with distinct genomic profiles likely represent multiple primary malignancies driven by distinct molecular events. Application of genomic profiling in the clinical setting may prove to be important to precise management of patients with MSLT.
Keywords: Non-small cell lung cancer, multiple synchronous lung cancers, genomic heterogeneity, gene sequencing
Introduction
Lung cancer is a leading cause of cancer death worldwide. Approximately, 222,500 new cases were diagnosed in the United States in 2017 (1). It has been estimated that 0.2–8% of lung cancer patients present with multiple synchronous lung tumors (MSLT) at the time of diagnosis. These tumors are distinct in that they are anatomically separate, with unclear etiologic and genomic relationships to one another (2-5). It has been noted that the incidence of MSLT has been increasing as lung cancer screening has become more widely implemented. MSLT raise a clinical dilemma, as they may represent metastatic disease from a single primary cancer, either by hematogenous spread or regional extension, or multiple primary cancers (5,6).
Determination of the etiology of MSLT is important to appropriate staging, treatment, and prognosis of these unique disease processes. Over recent years, research groups and medical communities have established guidelines to classify MSLT (5,7). Although these criteria have been widely adopted into clinical practice and patient management, they are rather empirical with little to no underwriting with molecular evidence. Thus, the classification of these tumors remains difficult.
Genome-scale sequencing studies have revealed that lung cancer is a heterogeneous disease with a complex genomic landscape. Each lung cancer may have a unique genomic profile that may be attributed to a distinct genetic background among individual patients or differing levels of carcinogen exposure (8,9). On the other hand, studies from our group and others have demonstrated that the majority of somatic mutations are universally present in different regions within a single tumor, suggesting limited intratumoral heterogeneity (10,11).
More recently, we have performed comprehensive genomic analysis from six Chinese patients with MSLT and demonstrated distinct genomic profiles in different MSLT from the same patients (12). In spite of the fact that MSLT from a single patient share identical constitutional genetic backgrounds and exposure histories, these tumors are no more similar to each other than tumors from different patients. Since metastatic lesions usually retain a significant fraction of genomic aberrations from the founding primary tumors, these data suggest that genomic profiling may be used as a complementary approach to clinicopathological analyses to accurately distinguish simultaneous primary lung cancers from intrapulmonary metastases (13,14).
In our previous study, all 6 patients were suggested to have multiple primary lung cancers by genomic analyses, and we were not able to demonstrate how close intrapulmonary metastases resemble their primary tumors (12). In addition, since all 6 patients were Asian patients, how these findings apply to Caucasian lung cancer patients is unknown. The importance of ethnicity upon the unique genomic alterations, diagnosis, and management of lung cancer are increasingly appreciated, and thus the relevance of such an investigation among an exclusively Caucasian population is understood (15-17).
Herein, in this study, we performed whole exome sequencing (WES) of ten tumors (seven adenocarcinomas, two squamous cell carcinomas and one regional lymph node metastasis) from four Caucasian patients with MSLT in order to assess the clonal relationship between different tumors from within the same patient. We aimed to determine whether an assessment of genomic heterogeneity could distinguish synchronous lung cancers from intrapulmonary metastases.
Methods
Patients
Surgical specimens were collected from four patients who had MSLT present upon initial evaluation at the University of Texas MD Anderson Cancer Center (MDACC) in Houston, Texas. Two patients (Patients 1 and 2) were found to have two tumors each; Patient 3 was observed to have three tumors; and Patient 4 had two anatomically distinct tumors with additional nodal involvement (Table 1). Patients 1 and 4 were never smokers, while Patients 2 and 3 were former smokers. Tumors from Patients 1, 2, and 3 were adenocarcinomas, and tumors from Patient 4 were squamous cell carcinoma. No patients had preoperative chemotherapy or radiation therapy. All patients were free of extrathoracic metastases. Upon pathologic examination, tumor sizes ranged from 0.5 to 5.1 centimeters. Tumor characteristics are shown in Table S1 and Figure S1. According to the American College of Chest Physicians (ACCP) criteria, Patients 1 and 2 had satellite nodules, while Patients 3 and 4 had multiple primaries (18). The collection and analysis of patient samples was approved by the MDACC Institutional Review Board [Cancer Prevention & Research Institute of Texas Multi-Investigator Research Awards (CPRIT-MIRA), RP160668]. Informed consents were obtained from all patients.
Table 1. Clinical characteristics and sequencing information of the four patients with multiple synchronous lung cancers.
| Patient ID | Tumor ID |
Sequencing depth | Histology | Nodal stage | ACCP category | Histopathologic analysis | Genomic profile | Adjuvant therapy | Follow-up (months) | Recurrence | Smoking status |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pa1 | Pa1T1 | 177× | ADC | N0 | Satellite nodules | Metastatic | Metastatic | CT | 32 | Yes | Non-smoker |
| Pa1T2 | 188× | ADC | |||||||||
| Pa2 | Pa2T1 | 30× | ADC | N0 | Satellite nodules | Metastatic | Metastatic | CT | 73 | No | Former smoker |
| Pa2T2 | 205× | ADC | |||||||||
| Pa3 | Pa3T1 | 173× | ADC | N2 | Multiple primaries | Metastatic | Primary | CRT | 73 | No | Former smoker |
| Pa3T2 | 87× | ADC | |||||||||
| Pa3T3 | 195× | ADC | |||||||||
| Pa4 | Pa4T1 | 193× | SCC | N1 | Multiple primaries | Metastatic | Metastatic | None | 59 | No | Non-smoker |
| Pa4T2 | 161× | SCC | |||||||||
| Pa4LN | 210× | SCC |
ACCP, American College of Chest Physicians; Pa, patient; T, tumor; ADC, adenocarcinoma; N, node; CT, chemotherapy; CRT, chemoradiotherapy; SCC, squamous cell carcinoma; LN, lymph node.
Table S1. Histomorphological subtypesa and their percentages in the 10 intrathoracic lesions.
| Patient ID | Sample ID | Histology | Location | Tumor (HE assessment) | Malignant cell (HE assessment) |
|---|---|---|---|---|---|
| Pa1 | Pa1T1 | ADC | LLL | 80 | 80 |
| Pa1T2 | ADC | LLL | 80 | 80 | |
| Pa2 | Pa2T1 | ADC | LUL | 80 | 80 |
| Pa2T2 | ADC | LUL | >10 | >10 | |
| Pa3 | Pa3T1 | ADC | RUL | 10 | 10 |
| Pa3T2 | ADC | RUL | 90 | 90 | |
| Pa3T3 | ADC | RUL | 30 | 30 | |
| Pa4 | Pa4T1 | SCC | RUL | 90 | 60 |
| Pa4T2 | SCC | RUL | 90 | 80 | |
| Pa4LN | SCC | Right hilar | 10 | 10 |
a, according to the multidiscipline classification criteria for adenocarcinoma in 2011. HE, hematoxylin-eosin; T, tumor; LN, lymph node; ADC, adenocarcinoma; SCC, squamous cell carcinoma; LLL, left lower lobe; LUL, left upper lobe; RUL, right upper lobe.
Figure S1.
Representative computed tomography images of 10 intra-thoracic MSLC lesions (red arrow). Pa, patient; T, tumor; LN, lymph node metastasis; MSLC, multiple synchronous lung cancer.
Sample collection and processing
Patients underwent resection of all tumors, including nodal disease. After resection, ten 10-µm formalin-fixed paraffin-embedded (FFPE) sections from each tumor sample were collected. Hematoxylin-eosin (HE)-stained slides were reviewed by experienced lung cancer pathologists to determine the histomorphological subtype, as well as the proportion of malignant cells relative to nonmalignant stromal (inflammatory, vascular, and fibroblast) cells. In addition, tumor cell viability was addressed by examining the presence of necrosis in the tissues. Tumor cells were enriched by having a pathologist scrape tumor tissue from each slide. Genomic DNA was then extracted from all samples.
WES
All tumors, including the sampled lymph node, underwent WES. Genomic DNA was sheared into fragments with peaks of 150 to 200 base pairs (bp), and then adapters were ligated to both ends. The adapter-ligated templates were purified with AgencourtAMPure SPRI beads (Beckman Coulter, Inc., Brea, CA, USA), and fragments with an insert size of approximately 200 bp were excised. Extracted DNA was amplified by ligation-mediated PCR, purified, and hybridized to the Sure Select biotinylated RNA library (Agilent Technologies, Santa Clara, CA, USA) for enrichment according to the manufacturer’s instructions. Paired-end multiplex sequencing of samples was performed with the Illumina HiSeq 2000 System. The average sequencing depth was 162× per sample.
Sequence alignment and variant calling
Paired-end reads in FastQ format generated by the Illumina pipeline were aligned to the reference human genome (UCSC Genome Browser, Version hg19) using the Burrows-Wheeler Aligner with default settings, except for a seed length of 40, a maximum edit distance of 3, and a maximum edit distance in the seed of 2 (19). Aligned reads were further processed according to the Genome Analysis Toolkit (GATK) for duplicate removal, indel realignment, and base recalibration (20).
MuTect was employed to detect potential single nucleotide variations. In addition to the build-in filters, the following filtering criteria were applied: (I) total read count in tumor DNA ≥30; (II) total read count in germline DNA ≥15; (III) presence of variant on both strands; (IV) variant allele frequency (VAF) in tumor DNA ≥5%; (V) VAF in germline DNA ≤1% (21). Single nucleotide variants called by MuTect were used for further analysis.
Statistical analysis
Pearson’s correlation test was used to assess the association between mutational burden and various demographic and clinicopathologic factors, as well as to determine the correlation between the mutation spectra of different tumors. Fisher’s exact test was used to assess the significance of differences in mutation spectra between different tumors.
Results
Somatic point mutations
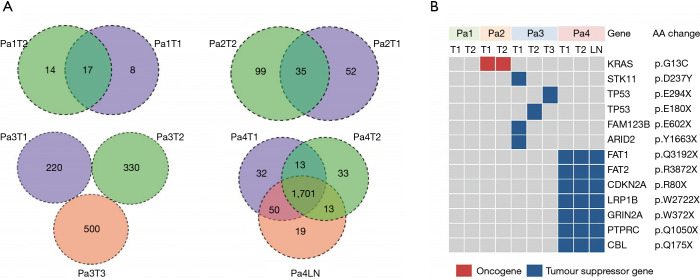
A total of 9,839 coding and splice site mutations were detected from 10 tumor samples. Of those mutations, 1,708 were detected in seven adenocarcinomas (median of 163 mutations per sample, ranging from 31–648) and 8,131 in three squamous cell carcinomas (median of 2,725 per sample, ranging from 2,678–2,728) (Table S2). All seven adenocarcinoma samples from patients 1, 2, and 3 were of an acinar pattern; the three squamous samples from patient 4 were poorly-differentiated. To determine the genomic heterogeneity of MSLT and the clonal relationships between different tumors within the same patient, we first defined functional somatic mutations as non-synonymous or stop-gain/stop-loss, frame-shift, alternative splicing mutations (Figure 1A) in exons leading to potential changes in final proteins. A total of 52 shared functional mutations were detected in the individual lung adenocarcinomas (LUAD) [17 of 39 (44%) mutations from Patient 1 and 35 of 186 (19%) from Patient 2]. No shared mutations were detected between different tumors from Patient 3. In Patient 4, two tumors (T1, T2) and a lymph node metastasis shared 1,701 (91%) of 1,861 mutations.
Table S2. Summary of somatic mutations detected in tumors obtained from whole exome sequencing.
| Single nucleotide variantsa | Pa1T1 | Pa1T2 | Pa2T1 | Pa2T2 | Pa3T1 | Pa3T2 | Pa3T3 | Pa4T1 | Pa4T2 | Pa4LN |
|---|---|---|---|---|---|---|---|---|---|---|
| Number | 107 | 132 | 189 | 430 | 684 | 698 | 1,339 | 6,250 | 6,013 | 6,385 |
| Coding | 31 | 41 | 110 | 159 | 296 | 403 | 633 | 2,691 | 2,643 | 2,689 |
| Non-synonymous | 21 | 26 | 76 | 119 | 201 | 295 | 436 | 1,634 | 1,603 | 1,622 |
| Synonymous | 6 | 10 | 28 | 30 | 82 | 80 | 149 | 948 | 934 | 958 |
| Stop-loss/stop-gain | 4 | 5 | 6 | 10 | 13 | 28 | 47 | 106 | 103 | 106 |
| Unknown | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 3 | 3 |
| Non-coding | 15 | 18 | 16 | 48 | 66 | 59 | 118 | 633 | 601 | 641 |
| UTR | 9 | 8 | 12 | 30 | 48 | 40 | 78 | 452 | 435 | 471 |
| ncRNA | 6 | 10 | 4 | 18 | 18 | 19 | 40 | 181 | 166 | 170 |
| Intronic | 41 | 45 | 61 | 171 | 268 | 216 | 496 | 2,573 | 2,458 | 2,723 |
| Splice site | 0 | 0 | 4 | 4 | 5 | 7 | 15 | 37 | 35 | 36 |
| Other | 41 | 45 | 57 | 167 | 263 | 209 | 481 | 2,536 | 2,423 | 2,687 |
| Intergenic | 20 | 28 | 2 | 52 | 54 | 20 | 92 | 353 | 311 | 332 |
a, single nucleotide variants called by MuTect. Pa, patient; T, tumor; LN, lymph node; UTR, untranslated region; ncRNA, non-coding RNA.
Figure 1.
Genomic profiling of different lesions rising from four patients (Pa) with MSLT. (A) Venn diagram illustrating the distributions of functional mutations across 10 lesions. The numbers of mutations identified in only one tumor (T) or shared by two or more lesions are as indicated. Shared mutations were defined as identical nucleotide substitutions at the same genomic coordinates. (B) Heatmap of known cancer gene mutations shared across 10 lesions.
Known cancer gene mutations
We then characterized canonical cancer gene mutations, defined as nonsynonymous mutations identical to those previously reported in known cancer genes or truncating mutations in known tumor suppressor genes (22-27). In total, 13 known cancer gene mutations were identified, of which eight cancer gene point mutations were shared between any two tumors from the same patient, including KRAS p.G13C mutation in Patient 2 (Figure 1B). Five other known cancer gene mutations (STK11 p.D237Y, TP53 p.E294X, TP53 p.E180X, FAM123B p.E602X, ARID2 p.Y1663X) were found in different tumors of Patient 3. In Patient 4, seven known tumor suppressor gene mutations (FAT1 p.Q3192X, FAT2 p.R3872X, CDKN2A p.R80X, LRP1B p.W2722X, GRIN2A p.W372X, PTPRC p.Q1050X, and CBL p.Q175X) were shared by three different tumors.
To confirm our findings, we compared the mutations in our cohort and those in 230 unrelated LUAD and 178 unrelated lung squamous cell carcinomas (LUSC) in The Cancer Genome Atlas (TCGA) study (8,9,11). Pairs of any two unrelated tumors in TCGA were less likely to have shared mutations (Figure S2). Among 230 LUAD samples (26,335 pairs): 1 pair of samples had 63 shared functional mutation, 40 pairs of samples had 2 shared functional mutations, and 1,846 pairs of samples had 1 shared functional mutation. In 178 LUSC samples (15,753 pairs): 5 pairs of samples shared 2 functional mutations and 212 pairs of samples had 1 shared functional mutation.
Figure S2.
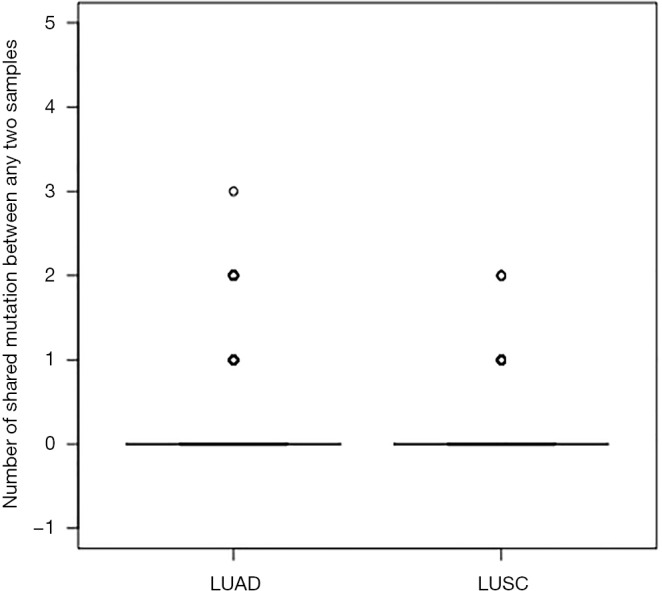
Numbers of shared mutations between any two samples in LUAD and LUSC from TCGA database. This figure shows that pairs of any two unrelated tumors in TCGA were less likely to have shared mutations. In 230 LUAD samples (i.e., 26,335 pairs), one pair of samples shared three mutations, 50 pairs of samples have two shared mutations, and 24,269 pairs of samples have one shared mutation. In 178 LUSC samples (i.e., 15,753 pairs), five pairs of samples have two shared mutations, and 301 pairs of samples shared one mutation. LUAD, lung adenocarcinomas; LUSC, lung squamous cell carcinomas; TCGA, The Cancer Genome Atlas.
Mutation spectra and mutation signature
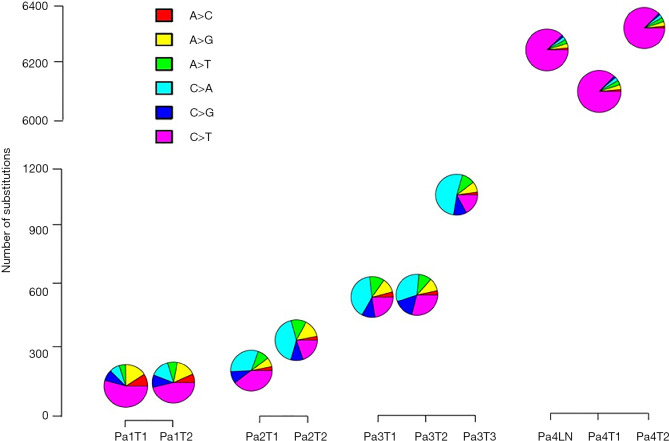
Next, we explored mutational processes in the context of independent tumors arising on a fixed genetic background and with shared exposure. The mutation spectra were different between ever smokers and never smokers (22,23,27,28). All tumors (including a metastatic lymph node) from the two never smokers (Patients 1 and 4) exhibited an enrichment for C>T mutations, while the tumors from the two smokers (Patients 2 and 3) had a higher prevalence of C>A substitutions (Figure S3). Discordant mutation spectra were observed between adenocarcinomatous tumors derived from the same patient (Patients 1, 2, and 3), while similar mutation spectra were observed between squamous cell tumors from Patient 4, indicating that different mutational processes may be involved during the development of tumors from Patients 1, 2, and 3, but not from Patient 4.
Figure S3.
Mutation spectra of all mutations across 10 MSLC lesions. An enrichment of C>T mutations were noted in all tumours of Patients 1 and 4 who were both non-smokers. The other five tumours from Patients 2 and 3 have predominantly C>A substitutions. Similar mutational spectra were observed between tumors in Patient 4. Discordant mutational spectra were observed between same-patient tumors in all adenocarcinoma lung cancer patients. MSLC, multiple synchronous lung cancer.
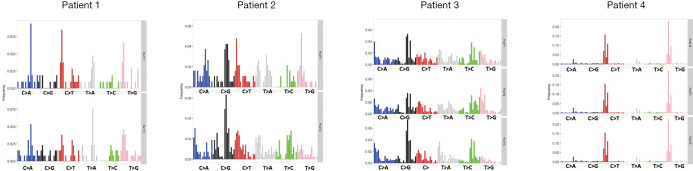
Followed by extraction of the mutational signature, we found that signature profiles differed substantially between individual tumors, except those from Patient 4. In this patient, the two tumors and associated lymph node metastasis had almost identical mutational signatures corresponding to presumptive APBOEC signature (Figure S4).
Figure S4.
Mutational signature analyses of nine MSLC tumours (T) and a lymph node metastasis (LN) across all four patients. All mutations were included in the analysis. Nucleotide substitutions in all mutations were grouped into six classes of mutations on the x-axis. In each class, mutations were grouped into 16 subclasses according to the bases immediately 5' and 3' to each mutated base. The data were the relative frequencies of the six mutation classes in each 16 tri-nucleotide contexts. MSLC, multiple synchronous lung cancer.
Discussion
MSLT are increasingly diagnosed largely as a result of improved implementation of early detection tools such as multislice spiral computed tomography, fluorescence endoscopy, and positron emission tomography (29,30). MSLT represent a clinical conundrum, as they may indicate multiple primary cancers which are potentially curable, or, on the other hand, could signify intrapulmonary metastases which would symbolize unresectable disease. As a result, many attempts have been made to distinguish these clinical entities. Although they largely represent empirical guidelines with little supporting molecular evidence, the Martini-Melamed criteria and ACCP guidelines are widely adapted clinical tools used to assess MSLT (7,31).
In this report, we performed genomic profiling of nine MSLT and one lymph node metastasis from four patients using WES alone. Initially, upon examination of TCGA data, it is surmised that tumors from unrelated patients are unlikely to share a high proportion of mutational similarity (12). Using this knowledge, we were able to distinguish independent primary lung cancers from intrapulmonary metastases in Patient 3, despite a shared genetic background and exposure history. These three tumors shared no mutations, similar to LUAD of different patients examined in TCGA, a majority of whom shared only a single mutation. These data from Patient 3 provide evidence that multiple molecular processes, resulting in distinct mutational profiles, may be in play during the development of independent lung cancers within the same individual, despite a shared exposure history.
Conversely, in comparison to TCGA data in which tumors from separate patients shared rare genetic markers, samples from Patients 1, 2, and 4 in our cohort shared at least 20% of the same mutations. Taken together, these results suggest that tumors from these three individuals more likely represent a single primary cancer with corresponding metastatic disease.
In addition, closer examination of the TP53 gene demonstrated disparate mutation events between the MSLT of Patient 3. Analogous intratumoral heterogeneity is observed in clear cell renal cell carcinoma, in which a single known gene implicated in carcinogenesis may have different subclones of the primary tumor. We have furthermore reported the occurrence of different mutations within a single cancer gene found in separate subclones of LUAD, which may be suggestive of convergent selection (12). This suggests a propensity for a single pathway of tumorigenesis, which is, however, driven by different biological mechanisms within a uniform biological environment (32). The data from Patient 3 suggests that, even in the context of a single genetic makeup and exposure history, the development of multiple primary lung cancers can be driven by distinct molecular events, with a possible predilection for certain pathways critical for carcinogenesis in a single patient.
Intrapulmonary metastases, either by hematogenous or local spread, are thought to derive from the same progenitor cells as the matched primary tumors (5). In this study, we profiled 4 patients, all of whom were initially clinically diagnosed with metastatic disease (either intrapulmonary metastases or satellite nodules) according to ACCP guidelines. While the MSLT from s from Patients 1, 2, and 4 shared similar molecular traits, the three tumors from Patient 3 demonstrated distinct genomic profiles. Our results suggest that the MSLT identified in Patient 3 represent independent primary lung cancers with separate progenitor cells. Conversely, the disease burden found in Patients 1, 2, and 4 most likely represents a single primary cancer with secondary metastatic disease.
During the follow-up period, only Patient 1 experienced disease recurrence after surgical resection after a follow-up duration of 20 months. The remaining patients all had a minimum postsurgical follow-up of at least 59 months (range, 59–73 months), and were without evidence of disease. Previous studies have demonstrated a slightly shorter survival in patients with satellite nodules compared with patients without satellite nodules matched for primary tumor size, lymph node and metastatic stage (33). Although our sample size was small, our data were consistent with previous studies. Our report is further novel in that we have contributed data on an exclusively Caucasian cohort, whereas our prior investigation involved sequencing of MSLT among Asian patients only (12); though we are unfortunately unable to draw direct conclusions from our assembled findings among these Asian and Caucasian patients, we feel our current report contributes to the growing body of literature on the topic of MSLT.
Of importance, all patients examined herein possessed MSLT within a single lobe. By current American Joint Committee on Cancer (AJCC) guidelines, this constellation represents T3 disease. A limitation of these criteria is demonstrated within our cohort, in which the etiology of MSLT is unclear, representing either metastatic disease or distinct primary cancers, though all patients received the benefit of surgical therapy (34). Though our investigation sheds light on this common clinical scenario of a patient with T3 disease identified imaging, our investigation is limited in that we are not able to infer the genomic profiles and molecular events which may lead to MSLT on separate lobes. Future investigations should be directed towards this additional management challenge. Additionally, though we are limited by our sample size, this work represents an exploratory investigation which we feel will still be of great importance to the literature.
To improve the diagnostic accuracy of MSLT, pioneering studies led by Travis et al have presented comprehensive histologic assessments and have shown promising results (35,36). Using a similar approach, we were able to accurately classify the molecular etiology of the 10 tumors observed, further confirming that comprehensive histologic assessment is highly valuable to distinguish multiple primary cancers from intrapulmonary metastases.
Conclusions
While tumor morphology may be controlled by complex molecular mechanisms and is furthermore subject to some degree of variable interpretation, we present a concrete molecular evidence which can help to better characterize MSLT. We demonstrate that multiple primary tumors have distinct genomic profiles, while metastatic lesions usually retain a significant fraction of genomic aberrations from the founding primary tumors (13,37). Therefore, comprehensive genomic profiling at the exome level can provide pivotal information to clinical and histologic assessment to accurately distinguish multiple primary lung cancers from intrapulmonary metastases. Application of genomic profiling in the clinical setting of staging patients with MSLT should be explored in a larger cohort to confirm the utility suggested here. If corroborated, genomic profiling may prove to be an important component of a more precise approach in managing patients presenting with MSLT.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by the MD Anderson Lung Cancer Moon Shot Program, the Cancer Prevention and Research Institute of Texas Multi-Investigator Research Award grant (RP160668), the MD Anderson Physician Scientist Program, The University of Texas (UT) Systems Stars Award (PS100149), the Welch Foundation Robert A. Welch Distinguished University Chair Award (G-0040), a Department of Defense PROSPECT grant (W81XWH-07-1-0306), the UT Lung Specialized Programs of Research Excellence Grant (P50CA70907), the MD Anderson Cancer Center Support Grant (CA016672), the T.J. Martell Foundation.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The collection and analysis of patient samples was approved by the MDACC Institutional Review Board [Cancer Prevention & Research Institute of Texas Multi-Investigator Research Awards (CPRIT-MIRA), RP160668]. Informed consents were obtained from all patients.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-1). CG has a patent named “Markers associated with ribavirin-induced anemia” that was issued in 2012. CB and IIW report grants and personal fees from Genentech/Roche, grants and personal fees from Bayer, grants and personal fees from Bristol-Myers Squibb, grants and personal fees from AstraZeneca/Medimmune, grants and personal fees from Pfizer, grants and personal fees from HTG Molecular, grants and personal fees from Merck, personal fees from GlaxoSmithKline, grants and personal fees from Guardant Health, personal fees from MSD, grants from Oncoplex, grants from DepArray, grants from Adaptive, grants from Adaptimmune, grants from EMD Serono, grants from Takeda, grants from Amgen, grants from Karus, grants from Johnson & Johnson, grants from Iovance, grants from 4D, grants from Novartis, grants from Oncocyte, grants from Akoya, outside the submitted work. JVH reports grants and other from AstraZeneca, other from Bristol-Myers Squibb, other from GlaxoSmithKline, other from Kairos Venture Investments, other from BrightPath Therapeutics, other from Hengrui Therapeutics, other from Eli Lilly, other from EMD Serono, grants and personal fees from Spectrum, other from Foundation One Medicine, grants from NIH/NCI, grants from American Cancer Society, grants from Checkmate Pharmaceuticals, outside the submitted work. Jianjun Z reports grants from Merck, grants from Johnson and Johnson, personal fees from BMS, personal fees from AZ, personal fees from GenePlus, personal fees from Innovent, outside the submitted work. MBA serves as an unpaid editorial board member of Journal of Thoracic Disease from Aug 2019 to Jul 2021. The other authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Ferguson MK, DeMeester TR, DesLauriers J, et al. Diagnosis and management of synchronous lung cancers. J Thorac Cardiovasc Surg 1985;89:378-85. 10.1016/S0022-5223(19)38787-2 [DOI] [PubMed] [Google Scholar]
- 3.Mathisen DJ, Jensik RJ, Faber LP, et al. Survival following resection for second and third primary lung cancers. J Thorac Cardiovasc Surg 1984;88:502-10. 10.1016/S0022-5223(19)38284-4 [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Wang M, MacLennan GT, et al. Evidence for common clonal origin of multifocal lung cancers. J Natl Cancer Inst 2009;101:560-70. 10.1093/jnci/djp054 [DOI] [PubMed] [Google Scholar]
- 5.Gazdar AF, Minna JD. Multifocal lung cancers--clonality vs field cancerization and does it matter? J Natl Cancer Inst 2009;101:541-3. 10.1093/jnci/djp059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Homer RJ. Pathologists' staging of multiple foci of lung cancer: poor concordance in absence of dramatic histologic or molecular differences. Am J Clin Pathol 2015;143:701-6. 10.1309/AJCPNBWF55VGKOIW [DOI] [PubMed] [Google Scholar]
- 7.Martini N, Melamed MR. Multiple primary lung cancers. J Thorac Cardiovasc Surg 1975;70:606-12. 10.1016/S0022-5223(19)40289-4 [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network . Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network . Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014;346:256-9. 10.1126/science.1256930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251-6. 10.1126/science.1253462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Zhang J, Li L, et al. Genomic heterogeneity of multiple synchronous lung cancer. Nat Commun 2016;7:13200. 10.1038/ncomms13200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding L, Ellis MJ, Li S, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 2010;464:999-1005. 10.1038/nature08989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012;481:506-10. 10.1038/nature10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng L, Wu YL. Immunotherapy in the Asiatic population: any differences from Caucasian population? J Thorac Dis 2018;10:S1482-93. 10.21037/jtd.2018.05.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weksler B, Kosinski AS, Burfeind WR, et al. Racial and Ethnic Differences in Lung Cancer Surgical Stage: An STS Database Study. Thorac Cardiovasc Surg 2015;63:538-43. 10.1055/s-0035-1546295 [DOI] [PubMed] [Google Scholar]
- 17.Mitchell KA, Zingone A, Toulabi L, et al. Comparative Transcriptome Profiling Reveals Coding and Noncoding RNA Differences in NSCLC from African Americans and European Americans. Clin Cancer Res 2017;23:7412-25. 10.1158/1078-0432.CCR-17-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozower BD, Larner JM, Detterbeck FC, et al. Special treatment issues in non-small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e369S-99S. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Geest LG, Lam-Boer J, Koopman M, et al. Nationwide trends in incidence, treatment and survival of colorectal cancer patients with synchronous metastases. Clin Exp Metastasis 2015;32:457-65. 10.1007/s10585-015-9719-0 [DOI] [PubMed] [Google Scholar]
- 21.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. 10.1016/j.cell.2012.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150:1121-34. 10.1016/j.cell.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watson IR, Takahashi K, Futreal PA, et al. Emerging patterns of somatic mutations in cancer. Nat Rev Genet 2013;14:703-18. 10.1038/nrg3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 2011;39:D945-50. 10.1093/nar/gkq929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. 10.1038/nature07423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hainaut P, Pfeifer GP. Patterns of p53 G-->T transversions in lung cancers reflect the primary mutagenic signature of DNA-damage by tobacco smoke. Carcinogenesis 2001;22:367-74. 10.1093/carcin/22.3.367 [DOI] [PubMed] [Google Scholar]
- 29.Smith-Bindman R, Miglioretti DL, Johnson E, et al. Use of diagnostic imaging studies and associated radiation exposure for patients enrolled in large integrated health care systems, 1996-2010. JAMA 2012;307:2400-9. 10.1001/jama.2012.5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trousse D, Barlesi F, Loundou A, et al. Synchronous multiple primary lung cancer: an increasing clinical occurrence requiring multidisciplinary management. J Thorac Cardiovasc Surg 2007;133:1193-200. 10.1016/j.jtcvs.2007.01.012 [DOI] [PubMed] [Google Scholar]
- 31.Shen KR, Meyers BF, Larner JM, et al. Special treatment issues in lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition). Chest 2007;132:290s-305s. [DOI] [PubMed] [Google Scholar]
- 32.Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014;46:225-33. 10.1038/ng.2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deslauriers J, Brisson J, Cartier R, et al. Carcinoma of the lung. Evaluation of satellite nodules as a factor influencing prognosis after resection. J Thorac Cardiovasc Surg 1989;97:504-12. 10.1016/S0022-5223(19)34540-4 [DOI] [PubMed] [Google Scholar]
- 34.Detterbeck FC, Marom EM, Arenberg DA, et al. The IASLC Lung Cancer Staging Project: Background Data and Proposals for the Application of TNM Staging Rules to Lung Cancer Presenting as Multiple Nodules with Ground Glass or Lepidic Features or a Pneumonic Type of Involvement in the Forthcoming Eighth Edition of the TNM Classification. J Thorac Oncol 2016;11:666-80. [DOI] [PubMed] [Google Scholar]
- 35.Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. 10.1097/JTO.0000000000000630 [DOI] [PubMed] [Google Scholar]
- 36.Detterbeck FC, Franklin WA, Nicholson AG, et al. The IASLC Lung Cancer Staging Project: Background Data and Proposed Criteria to Distinguish Separate Primary Lung Cancers from Metastatic Foci in Patients with Two Lung Tumors in the Forthcoming Eighth Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:651-65. [DOI] [PubMed] [Google Scholar]
- 37.Campbell PJ, Yachida S, Mudie LJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 2010;467:1109-13. 10.1038/nature09460 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as