Abstract
Comprehensive characterization of the genomic landscape of epidermal growth factor receptor (EGFR)-mutated lung cancers have identified patterns of secondary mutations beyond the primary oncogenic EGFR mutation. These include concurrent pathogenic alterations affecting p53 (60–65%), RTKs (5–10%), PIK3CA/KRAS (3–23%), Wnt (5–10%), and cell cycle (7–25%) pathways as well as transcription factors such as MYC and NKX2-1 (10–15%). The majority of these co-occurring alterations were detected or enriched in samples collected from patients at resistance to tyrosine kinase inhibitor (TKI) treatment, indicating a potential functional role in driving resistance to therapy. Of note, these co-occurring tumor genomic alterations are not necessarily mutually exclusive, and evidence suggests that multiple clonal and sub-clonal cancer cell populations can co-exist and contribute to EGFR TKI resistance. Computational tools aimed to classify, track and predict the evolution of cancer clonal populations during therapy are being investigated in pre-clinical models to guide the selection of combination therapy switching strategies that may delay the development of treatment resistance. Here we review the most frequently identified tumor genomic alterations that co-occur with mutated EGFR and the evidence that these alterations effect responsiveness to EGFR TKI treatment.
Keywords: Lung cancer, oncogenes, mutations, therapy
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide (1). Research into the complex genetic basis of lung cancer initiation and progression has yielded novel treatments that have revolutionized lung cancer management (2). The expansion of clinical sequencing efforts has led to the identification of several novel oncogenic driver alterations (3,4), and has spurred the development of small molecule tyrosine kinase inhibitors (TKIs) aimed at suppressing signaling pathways downstream (2). Activating mutations in the epidermal growth factor receptor (EGFR) gene have been identified as oncogenic driver mutations in approximately 15% of lung adenocarcinoma (LUAD) patient tumors in Western populations and in over 50% of patients of East Asian descent and represent the most frequent targetable genetic alterations identified in lung cancer (4-7). The two most common EGFR-activating alterations are in-frame deletions in exon 19 (E746-A750del) and amino acid substitution in exon 21 (L858R), which together account for >90% of known EGFR driver mutations (6,8).
With the identification of these driver alterations, a new paradigm for lung cancer treatment emerged. Tumor biopsy and sequencing to identify a driver mutations followed by TKI treatment (e.g., erlotinib, gefitinib, afatinib, osimertinib in the case of EGFR) replaced first line platinum-based chemotherapy, which had limited clinical efficacy with a median overall survival (OS) <12 months and a 5-year survival rate <1% (9). Unfortunately, these practice changes resulted in only modest improvements in progression-free survival (PFS) over platinum therapy for patients with EGFR mutations, ultimately limiting long-term OS (10). Even with a targeted therapy approach, the clinical success of treatment with TKIs is almost uniformly limited by the development of drug resistance and progression emerges after a median of 8–18 months (11,12).
The EGFR T790M on-target resistance mutation was first identified as a common mechanism of resistance to first- and second-generation EGFR TKIs (13,14). Similarly, although less frequent, the EGFR C797S mutation has emerged as an on-target resistance mutation to the 3rd generation EGFR TKI osimertinib (15). Additional studies profiled and identified tumor genetic alterations that are pre-existing or acquired after TKI treatment and can bypass EGFR signaling pathway inhibition, promoting resistance to EGFR-directed therapy (5,16-20). Beyond direct tumor sequencing, liquid biopsies can capture the heterogenous mutational landscape of metastatic tumors at different sites. Analysis of circulating tumor DNA (ctDNA) from patients with advanced EGFR-mutated non-small cell lung cancer (NSCLC) point to co-occurring tumor genomic alterations and tumor mutational heterogeneity as a common feature of EGFR-mutant lung cancers (5,21). This may ultimately contribute to the limited depth and duration of response to EGFR inhibitors. Herein, we review our current understanding of how concurrent tumor genetic alterations limit response and lead to resistance to EGFR-targeted therapy.
Types of concurrent resistance mutations by pathway in lung cancer
EGFR-dependent co-alterations
Acquired resistance to EGFR TKI treatment can occur through the acquisition or selection of pre-existing TKI resistance mutations (Table 1). In 60% of EGFR-mutant NSCLC patients treated with 1st (erlotinib, gefitinib) and 2nd (afatinib) generation EGFR TKIs, acquisition of resistance is triggered by the substitution of threonine to methionine at position 790 in exon 20 (T790M) which impedes drug binding and increases ATP affinity in the EGFR ATP-binding pocket (11,22). Third generation EGFR TKIs have been developed to overcome T790M-mediated resistance. Osimertinib (AZD9291) (68) selectively targets both canonical EGFR activating mutations as well as the T790M resistance mutations by covalently binding the C797 residue in the ATP pocket of mutant EGFR (69). Patients whose tumors harbored the EGFR T790M mutation and were treated with osimertinib in the second line setting had a median PFS of 9.6 months (68). Furthermore, osimertinib resulted in significantly improved PFS compared to platinum-based chemotherapy (23). In the FLAURA study, osimertinib was tested as first-line therapy in comparison to 1st generation EGFR TKIs (gefitinib or erlotinib), and demonstrated significantly improved PFS (18.9 vs. 10.2 months) and OS (38.6 vs. 31.8 months), as well as a better safety profile (10,23). Although these results supported the use of osimertinib as first-line therapy in advanced-stage EGFR-mutated NSCLC patients, intrinsic and acquired osimertinib resistance can occur through tertiary EGFR mutations including C797 (7% of patients treated with front-line osimertinib), or the more rare G796, L792, L718, G719, G724 residue substitutions and additional exon 20 mutations (23-34,70). These mutations can co-exist at low frequency with sensitizing EGFR mutations and sterically interfere with the binding of the drug to the active site (23).
Table 1. Concurrent functional gene alterations in EGFR-mutant NSCLC.
| Co-alteration | Gene | Type | Frequency | Clinical context | Stage | refs | |
|---|---|---|---|---|---|---|---|
| Alteration | Result | ||||||
| EGFR dependent | EGFR | SNVs | T790M | 50% | 1st, 2nd gen. TKIs, > post-TKIs | Met. | (11,22,23) |
| C797X | 10–26% | 3rd gen. TKI, 1st/2nd line | Met. | (23-25) | |||
| G796X | Rare | 3rd gen. TKI, 2nd line | Met. | (23,25-27) | |||
| L792X | Rare | 3rd gen. TKI, 2nd line | Met. | (23,25) | |||
| L718X | Rare | 3rd gen. TKI, 1st/2nd line | Met. | (23,25,27,28) | |||
| G719A | Rare | 3rd gen. TKI, 2nd line | Met. | (23,25,27) | |||
| G724S | Rare | 3rd gen. TKI, 1st line | Met. | (23,26,29-32) | |||
| SNVs ins |
Exon 20 | Rare | 3rd gen. TKI, 1st/2nd line | Met. | (23,28,32,33) | ||
| CNV | Amp | Under study | 3rd gen. TKI, 1st line | Met. | (23,34) | ||
| EGFR independent | |||||||
| P53 pathway | TP53 | SNVs | Mts | 60–65% | > post-TKI, 1st, 2nd gen. TKIs | Early/met. | (5,16,35-39) |
| MDM2 | CNV | Amp | 12% | > pre-TKI | Met. | (16,40) | |
| RTKs | MET | CNV | Amp | 9.9% | > post-TKI, 1st, 2nd, 3rd (1st/2nd line) gen. TKIs | Met. | (5,16,17,23,28,41-46) |
| ERBB2 | CNV | Amp | 8.6–12% | Pre/post-TKI, 1st, 2nd, 3rd (1st/2nd line) gen.TKIs | Met. | (5,16,19,23,28,34) | |
| PDGFRA | SNVs, CNV | Mts, amp | 4.7% | post-TKI? | Met. | (5) | |
| PIK3CA/KRAS pathways | PIK3CA | SNVs, CNV | Mts, amp | 13% | > post-TKI, 1st, 3rd (1st/2nd line) gen. TKIs | >met. | (5,16,17,23,28,34,47-49) |
| KRAS | SNVs, CNV | Mts, amp | 4.7% | > post-TKI? 1st, 2nd, 3rd (1st/2nd line) gen. TKIs | Met. | (5,16,17,23,46,47,50-54) | |
| BRAF | SNV | V600E | 3% | > post-TKI, 1st, 3rd (2nd line) gen. TKIs | Met. | (5,16,23,28,34,55) | |
| NF1 | SNVs | Mts | 16–23% | Pre/post-TKI | Met. | (5,56) | |
| mTOR | SNV | E2419K | 1 pt | Post-TKI, 1st gen. TKI | Met. | (16) | |
| Wnt pathway | CTNNB1 | SNVs | Mts | 5.3–9.6% | Pre/post-TKI, 1st, 2nd gen. TKIs | >met. | (5,16,39,57-60) |
| TF | MYC | CNV | Amp | 10.6% | Pre/post-TKI, 1st gen TKI | Early/met. | (5,61) |
| NKX2-1 | CNV | Amp | 15%–11% | > pre-TKI | Early/met. | (16,39,62,63) | |
| Gene fusions | FGFR3-TACC3 | Fusion | 3–10% | Post-TKI, 1st, 3rd (2nd line) gen. TKIs | Met. | (16,23,34,47) | |
| RET-ERC1 | Fusion | 3–10% | Post-TKI, 3rd (2nd line) gen. TKI | Met. | (17,23,34) | ||
| CCDC6-RET | Fusion | 3–10% | Post-TKI, 3rd (2nd line) gen. TKI | Met. | (17,23,47) | ||
| NTRK1-TPM3 | Fusion | 3–10% | Post-TKI, 3rd (2nd line) gen. TKI | Met. | (17,23) | ||
| NCOA4-RET | Fusion | 3–10% | Post-TKI, 3rd (2nd line) gen. TKI | Met. | (17,23) | ||
| GOPC-ROS1 | Fusion | 3–10% | Post-TKI, 3rd (2nd line) gen. TKI | Met. | (23,64) | ||
| AGK-BRAF | Fusion | 3–10% | Post-TKI, 1st, 3rd (2nd line) gen. TKI | Met. | (16,23,65) | ||
| ESYT2-BRAF | Fusion | 3–10% | Post-TKI, 3rd (2nd line) gen. TKI | Met. | (23,47) | ||
| SPTBN1-ALK | Fusion | 1 pt | Post-TKI, 3rd (2nd line) gen. TKI | Met. | (23) | ||
| PLEKHA7-ALK | Fusion | Rare | Post-TKI, 3rd (2nd line) gen. TKI | Met. | (23,66) | ||
| EML4-ALK | Fusion | Rare | Post-TKI, 3rd (2nd line) gen. TKI | Met. | (23,67) | ||
| Cell cycle | Rb1 | SNVs CNV | Mts, del | 9.5–10.3% | > post-TKI, 1st gen. TKI | Early/met. | (16,39) |
| CDK4 | CNV | Amp | 7–10% | Pre/post-TKI, 3rd (1st line) gen. TKI | Met. | (5,23,39) | |
| CDK6 | CNV | Amp | 7% | Pre/post-TKI, 3rd (1st/2nd line) gen. TKI | Met. | (5,23,39) | |
| CCNE1 | CNV | Amp | 6.9% | Pre/post-TKI, 3rd (1st/2nd line) gen. TKI | Early/met. | (5,23,39) | |
| CDKN2A | CNV | Del | 24.6% | > post-TKI, 1st, 3rd (2nd line) gen. TKIs | Early/met. | (16,39) | |
| CDKN2B | CNV | Del | 20.2% | > post-TKI, 1st gen. TKI | Early/met. | (16,39) | |
TKI, tyrosine kinase inhibitor; NSCLC, non-small cell lung cancer; TF, transcription factors; SNVs, single nucleotide variants; CNV, copy number variation; mts, mutations; gen., generation; early, early stage; met., metastatic.
EGFR-independent co-alterations
A. P53 pathway
TP53 is the most frequently altered gene in human cancers (71). TP53 is a tumor suppressor whose gene product is responsible for the induction of cell-cycle-arrest or apoptosis programs in response to cellular stress, including stress induced by oncogene activation (72). When TP53 is mutated and this crucial cell programming is lost, unchecked cell proliferation can occur leading to carcinogenesis (73). TP53 codes for a 393-aa protein composed of three domains: one trans-activation domain, which is a target of post-translational regulation, one DNA-binding domain (DBD) (spanning exons 5–8, aa 102–292) accounting for most of its tumor suppressor activity, and a C-terminal domain responsible for negative protein regulation through oligomerization (74). p53-DNA interactions are mediated by loops L2 (aa 163–195) and L3 (aa 236–251) in the DBD (75). Many TP53 mutations affect the DBD and generate a mutated protein with a dominant negative effect (76). In addition, TP53 mutations with gain of function activities have been described and may contribute to cancer progression and drug resistance (77-79).
TP53 mutations are classified as “disruptive” or “nondisruptive”. Disruptive mutations lead to complete or near complete loss of p53 activity and include nonsense and missense mutations in L2 or L3 loops affecting residue polarity and in-frame deletions within L2 or L3 loops (74). All other types of TP53 mutations are classified as nondisruptive mutations, which cause partial loss of p53 protein function and often associate with gain of function activity (74,80,81).
TP53 mutations frequently occur in NSCLC (ranging from 40–70%) and are commonly found co-occurring with EGFR mutations (35-38). A pooled analysis from 4 randomized trials by Ma et al., found TP53 mutations status to be of no prognostic value in EGFR-mutated NSCLC (82). However, when TP53 mutations were categorized by subtype, nondisruptive TP53 mutations were identified as an independent prognostic factor of reduced OS in EGFR-mutated NSCLC patients (17.8 vs. 28.4 months) (74). In another study, TP53 mutations were classified into “poor” mutations (affecting exon 4, exon 6, mutations of unknown site and multiple mutations) and “good” mutations (exon 5, exon 7, exon 8 and exon 9 mutations), with patients whose tumors harbored TP53 poor mutations having the worst prognosis in presence of concurrent exon 19/21 mutated EGFR. In contrast, patients whose tumors harbored non-exon 19/21 mutated EGFR were associated with worse prognosis when “good” TP53 mutations were identified as co-occurring with the EGFR mutation (36).
In a study of cell-free DNA from 1,122 advanced-stage EGFR-mutated lung cancer patients, 55% had a co-occurring TP53 mutation (5) (Table 1). In another study, patients with EGFR-mutant lung cancer with any co-occurring TP53 mutations had a 3-fold risk of disease progression on TKI treatment than TP53wt patients (83). Furthermore, in vitro studies demonstrated that TP53 mutant NSCLC cell lines underwent less apoptosis in response to gefitinib when compared to TP53wt controls (84).
The clinical significance of co-occurring TP53 mutations in EGFR-mutated NSCLC remains unsettled. Tsui et al. found that EGFR-mutated patients with TP53 co-mutations identified prior to TKI treatment exhibited worse OS compared to those without TP53 mutations (85). Labbé et al. found that only TP53 missense co-mutations were predictive of shorter disease-free survival after surgical resection (86). Other studies found only trends towards poorer PFS and OS when TP53 co-mutations were identified (83,87). TP53 co-mutations are frequently clonal in tumors, suggesting early occurrence during tumorigenesis, with a strong selective pressure for TP53 locus loss of heterozygosity (88). One hypothesis to explain the apparent decrease in OS associated with TP53 co-mutations is that these mutations may confer higher tolerability to genomic instability, as exemplified by high rates of tumor aneuploidy and somatic mutation burden, potentially promoting a more aggressive tumor phenotype (88).
In addition to TP53 mutations, amplification of the gene encoding the Mouse Double Minute 2 (MDM2) protein have been identified in 12% of pre-treatment metastatic EGFR-mutant lung cancers. MDM2 mediates proteasome-dependent p53 degradation, and its hyperactivation may lead to loss of p53-tumor suppressive function, potentially contributing to p53-dependent mechanisms of TKI resistance (16,40).
B. RTKs
MET
MET alterations have been found to co-occur with mutant EGFR with a frequency of ~10% (5) (Table 1). MET amplification is a classic mechanism of EGFR TKI resistance presenting in 5% to 22% of cases of acquired resistance (16,20,89). This is thought to be due to its gene location at a fragile site in chromosome 7, which facilitates its amplification by recurrent breaks within chromosomal common fragile sites (90). Subsequently, selection for clones harboring MET amplification can occur under drug pressure where MET amplification leads to resistance by maintenance of MAPK/PI3K/AKT signaling (41,91). Recent studies show that concurrent MET copy number gains (CNG) in EGFR-mutant NSCLC patients did not affect response to first and second generation of EGFR TKI in the first line setting, except when patients were classified as MET amplified (42). This highlights the importance of defining CNG thresholds in the diagnostic practice, which could guide more successful treatment combinations of MET and EGFR inhibitors (41-43). In addition, MET amplification has been identified as a common mechanism that drives resistance to first and second line osimertinib treatment in NSCLC patients. Preclinical studies and case reports suggest that this mechanism of resistance can be overcome by combination therapy with crizotinib and osimertinib (17,28,44-46).
ERBB2
ERBB2 belongs to the HER family of receptor tyrosine kinases, which activates the PI3K-AKT and MAPK downstream pathways (23). The most commonly encountered ERBB2 mutations in lung cancer are in-frame insertions in exon 20, but point mutations along the tyrosine kinase domain have also been identified (92,93). These exon 20 mutations have been demonstrated to be oncogenic drivers and tend to be mutually exclusive with the more common activating mutations in EGFR and KRAS (94). ERBB2 amplification, however, have been observed in 12% of EGFR-mutated lung cancers at acquired resistance to 1st generation EGFR TKIs gefitinib or erlotinib (19) (Table 1). Notably, all of the ERBB2 amplification positive samples tested were T790M negative, suggesting a distinct mechanism of TKI resistance attributable to ERBB2 (19). This finding was further studied in cellular models of lung cancer where ERBB2 overexpression (>50-fold above baseline, as per densitometry assessment) in EGFR-mutant NSCLCs conferred resistance to erlotinib (19). ERBB2 alterations in patients with resistance to first and second line third generation EGFR TKIs have also been described in 2% and 5% of cases respectively, and can co-exist with additional oncogenic EGFR, PIK3CA and MET alterations (28,34). The role of ERBB2 over-expression in reducing third generation EGFR TKIs sensitivity has also been confirmed in preclinical models (46).
C. PIK3CA/KRAS pathways
PIK3CA
Mutations in PIK3CA are found in approximately 7% of LUADs (4). These alterations are primarily localized to the catalytic subunit of the PI3K enzyme, in exons 9 and 20 of the helical and kinase domains of p110 alpha (4). As a result of these mutations, PI3K constitutively activates the AKT-mTOR pathway, triggering tumor cell survival and proliferation (95,96). Concurrent EGFR and PIK3CA alterations were detected in 13% of metastatic LUAD and were enriched in post-first and second EGFR TKI samples (16) (Table 1). In previous clinical investigations, the presence of these co-alterations correlated with poor prognosis but did not impact the efficacy of first-generation EGFR TKI monotherapy (97). In a recent study, longitudinal sampling of seven tumor samples from an individual patient prior to treatment and at disease progression to 1st and 3rd generation EGFR inhibitors highlighted the pre-treatment existence (PIK3CA G106V) and post-therapy enrichment (PIK3CA H1047R) of PIK3CA mutations (5). Functional characterization of PIK3CA G106V confirmed its role in promoting tumor cell invasion without affecting sensitivity to EGFR TKI treatment (5). PIK3CA mutations identified in NSCLC patients resistant to second line osimertinib treatment occur at a frequency of 4–11% and include: E545K, E542K, R88Q, N345K and E418K with the E545K functionally validated as mediator of resistance (17,34,47-49). Resistance to frontline osimertinib has also been correlated with E453K, E545K (predominant, 4% of cases) and H1047R PIK3CA mutations (28).
KRAS/BRAF/NF1
Aberrations in the RAS-MAPK pathway lead to first and second line osimertinib resistance (23,50). In patients with acquired resistance to osimertinib, concurrent KRAS G12S, G13D, Q61R, Q61K and G12D mutations have been reported (17,46,47,51,52). In preclinical models, concurrent KRAS mutations resulted in EGFR TKI resistance, which could be overcome using a combination of osimertinib and a MEK inhibitor (50,53,54), suggesting a potentially effective clinical strategy.
Oncogenic BRAF mutations are found in 2–3% of LUAD, among which V600E represents 50% of mutations (98-101). Concurrent BRAF alterations are present in ~11% of EGFR-mutant NSCLC patients (Table 1), with the BRAF V600E mutation identified as a mechanism of resistance to osimertinib in ~3% of cases (5,28,34). Tumor cells from patients carrying BRAF V600E resistance co-mutation showed sensitivity to combination of BRAF inhibitor and osimertinib (5,55), suggesting that this may be a viable clinical strategy to overcome resistance.
The NF1 gene encodes a GTPase-activating protein that down-regulates Ras-signaling through guanosine triphosphate (GTP) hydrolysis, thus acting as a tumor suppressor (102). Primary and acquired resistance of LUAD patients to EGFR TKI has been associated with NF1 deletion (56). Sensitivity to EGFR TKI was rescued in cells with low NF1 expression by adding a MAP-ERK kinase (MEK) inhibitor (56). In a small EGFR-mutant LUAD liquid biopsy cohort, for which treatment outcome data were available, NF1 co-alterations were present in 16–23% of clinical cases and enriched post-TKI treatment (5) (Table 1).
D. Wnt Signaling pathway
The β-catenin protein, encoded by the CTNNB1 gene, represents a key component of the WNT signaling pathway, functioning as a nuclear transcriptional activator for target genes regulating cellular proliferation and differentiation (103). When signaling through the WNT pathway is down-regulated, β-catenin is degraded by the adenomatous polyposis coli (APC)/actin/glycogen synthase kinase 3β (GSK-3β) destruction complex, whereas aberrant β-catenin activating mutations prevent GSK-3β-mediated phosphorylation and degradation, keeping β-catenin active (57).
CTNNB1 activating co-mutations show consistent co-selection with EGFR mutations in patient tumors or ctDNA (Table 1). CTNNB1 activating mutations are enriched in late stage EGFR-mutated NSCLC patients (5–10% of patients) (5,39), supporting a potential role for WNT pathway activation in promoting tumor invasion and metastasis (4,5,16,57-60). One study, for example, showed that overexpression of CTNNB1 S37F in preclinical models enhanced tumor cell invasion and reduced sensitivity to first generation EGFR TKIs through inhibition of apoptosis (5).
E. Transcription factors
MYC
MYC amplification was first reported in lung cancer in 1983 and has been correlated with primary resistance to EGFR TKI treatment as well as decreased PFS and OS (104-106) (Table 1). The MYC protein acts as a regulator of cell cycle progression and cellular transformation through its activity as a transcriptional activator (107). Concurrent MYC and EGFR alterations were observed in ~10% NSCLC patients with a trend toward enrichment in patients progressing after TKI therapy (5). Combinatorial treatments of EGFR and MYC inhibitors are currently being tested in preclinical EGFR-mutant lung cancer models as a potential approach to overcome primary TKI resistance (61).
NKX2-1
NKX2-1, also known as TTF-1, is a homeodomain transcription factor with an essential role in peripheral lung development (108). Specifically, abundant NKX2-1 expression was associated with EGFR-mutant LUADs in which NKX2-1 exerts a lineage-survival oncogenic role (62,63). Moreover, NKX2-1 gene amplification was detected in 15% and 11% of EGFR-mutant NSCLC patients at baseline and after TKI progression, respectively (16) (Table 1). Yamaguchi et al. demonstrated that NKX2-1 trans-activates ROR1 receptor which binds to EGFR favoring PI3K-AKT signaling as well as phosphorylates SRC, further enhancing AKT activity (109).
F. Gene fusions
Oncogenic fusions have been detected in 3–10% of EGFR-mutant LUADs at resistance to osimertinib treatment (23) (Table 1). The potential oncogenic fusion proteins described include: FGFR3-TACC3, RET-ERC1, CCDC6-RET, NTRK1-TPM3, NCOA4-RET, GOPC-ROS1, AGK-BRAF, ESYT2-BRAF, SPTBN1-ALK, PLEKHA7-ALK, and EML4-ALK (17,23,34,47,64-67). Of note, combination of osimertinib and a RET inhibitor, in presence of CCDC6-RET fusion, or crizotinib, in presence of EML4-ALK fusion, were effective therapies for overcoming resistance in case studies (65,67).
G. Cell cycle-G1/S regulators
The RB1 protein acts as a tumor suppressor, inhibiting G1/S progression through the cell-cycle (110). Its activity is regulated by CCND1-CDK4/6 complex-dependent phosphorylation, which tags RB1 for degradation and activates E2F transcription factors which mediates G1/S entrance (110). Inactivating RB1 mutations occur in ~10% of EGFR-mutant NSCLC, mostly co-occurring with TP53 mutations (5,16,88,111,112) (Table 1). EGFR-mutant LUAD carrying inactivated RB1, TP53 genes have higher probability of small cell transformation following EGFR TKI (112-114).
Additional frequent concurrent G1/S gene alterations detected in patients progressing to first- and second-line osimertinib are: CDK4 (7–10%), CDK6 (7%), CCNE1 (7%) amplifications and CDKN2A (25%), CDKN2B (20%) deletions, with CDKN2A/2B deletions being typically clonal alterations (5,16,88) (Table 1). Importantly, CDK4/6 amplifications correlated with resistance to first generation EGFR TKIs and reduced PFS to osimertinib (5). Preclinical studies using a combination of 1st or 2nd generation EGFR TKIs and a CDK4/6 inhibitor showed a significant delay in the onset of EGFR TKI resistance when compared to EGFR TKI monotherapy (115,116). Accordingly, clinical testing with combinatorial treatments of EGFR and CDK4/6 inhibitors are ongoing (117).
Therapeutic implications and perspectives
Intrinsic and acquired resistance to single agent EGFR-targeted therapies remains a significant challenge in the treatment of lung cancer. The impact of co-occurring genomic alterations on clinical outcomes for EGFR-mutant NSCLC patients is becoming more appreciated. Numerous EGFR-dependent and independent co-alterations are associated with reduced EGFR TKI sensitivity and shorter PFS. Additionally, tumor mutation burden increases in post-TKI treatment samples, adding complexity to the heterogeneous genomic landscape.
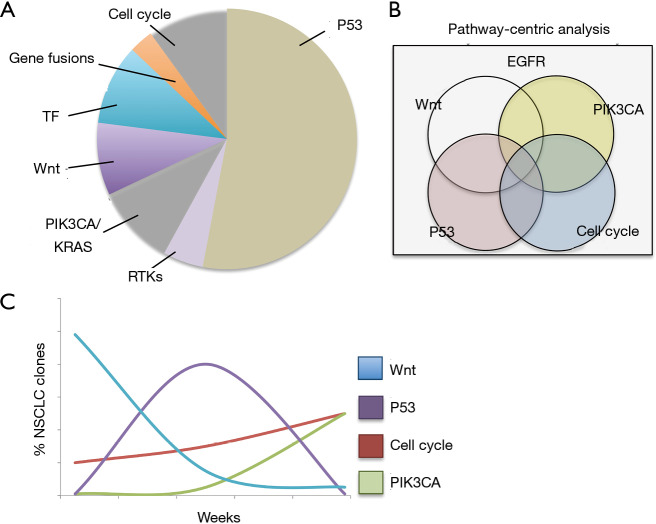
Specifically, advanced EGFR-mutated NSCLCs undergo dynamic modulation of tumor sub-clonal populations, with multiple pathogenic alterations co-existing in pre- and post-TKI specimens (Figure 1) (5). Overcoming this dynamic and complex process will require an equally adaptive treatment approach based on real-time monitoring of tumor genomic complexity. Computational frameworks that profile patterns of dynamic, actionable alterations in real-time and identify optimal therapy switching strategies have been described in preclinical models of co-occurring EGFR, BRAF, and MET alterations (Figure 1) (119). The implementation of this strategy in patients could rely on: (I) frequent liquid biopsy and profiling of ctDNA to detect emerging genomic alterations during therapy and (II) mathematical modeling to identify therapy strategies that pre-empt or overcome the outgrowth of resistant tumor subclones (Figure 1) (5,119).
Figure 1.
Classification of concurrent genomic alterations in EGFR-mutated NSCLC. (A) Most frequently co-occurring pathways altered in EGFR-mutated NSCLC are highlighted in the pie chart (not necessarily mutually exclusive), which can impact the efficacy of EGFR-targeted therapy. (B) A pathway-centric classification of tumor genomic co-alterations could guide the selection of combination therapy approaches (118). (C) Real-time monitoring of tumor clonal evolution through liquid biopsies in conjunction with mathematical modeling may inform combination therapy treatment schedules and switching strategies (hypothetical clonal evolution shown) (119). EGFR, epidermal growth factor receptor; NSCLC, non-small cell lung cancer; TF, transcription factors.
Using broad sequencing panels, it is now possible to characterize the intra-tumor heterogeneity and identify multiple co-existing, actionable oncogenic targets within individual tumors (118). Moreover, a genomic pathway-centric analysis of broad-panel next generation sequencing data has been recently proposed to identify patterns of therapeutic vulnerabilities in LUAD patients (118). The use of up-front, targeted therapy combinations and dynamic therapy switching could thus represent more durable treatments in advanced stage NSCLC and are currently investigated in pre-clinical and clinical trials (117,119).
Overall, systematic data collection and management, novel computational tools, high throughput pre-clinical testing, real-time tumor genomic assessments, and flexible clinical trial designs will be required to more effectively employ precision therapeutic approaches that address the genomic complexity and heterogeneity present in EGFR-mutant NSCLC (Figure 1).
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: The Damon Runyon Cancer Research Foundation P0528804 (to CM Blakely), Doris Duke Charitable Foundation grant # 2018110 (to CM Blakely), and V Foundation V2018-020 (to CM Blakely).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editor (Trever G. Bivona) for the series “Mechanisms of resistance to EGFR-targeted therapy” published in Journal of Thoracic Disease. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd.2020.03.78). The series “Mechanisms of Resistance to EGFR-targeted Therapy” was commissioned by the editorial office without any funding or sponsorship. Dr. Blakely reports grants from Astrazeneca, grants from Takeda, grants from Novartis, grants from Mirati, grants from Spectrum, grants from Roche, personal fees from Revolution Medicines, personal fees from Foundation Medicine, outside the submitted work. The other authors have no other conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. 10.1001/jama.2014.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014;346:256-9. 10.1126/science.1256930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network . Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. Erratum in: Nature. 2014 Oct 9;514(7521):262. Rogers, K [corrected to Rodgers, K]. Author Correction: Comprehensive molecular profiling of lung adenocarcinoma. [Nature. 2018] 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet 2017;49:1693-704. 10.1038/ng.3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. 10.1056/NEJMoa040938 [DOI] [PubMed] [Google Scholar]
- 7.Zhang XC, Wang J, Shao GG, et al. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat Commun 2019;10:1772. 10.1038/s41467-019-09762-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. 10.1126/science.1099314 [DOI] [PubMed] [Google Scholar]
- 9.Morgillo F, Della Corte CM, Fasano M, et al. Mechanisms of resistance to EGFR-targeted drugs: lung cancer. ESMO Open 2016;1:e000060. 10.1136/esmoopen-2016-000060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med 2020;382:41-50. 10.1056/NEJMoa1913662 [DOI] [PubMed] [Google Scholar]
- 11.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- 13.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73. 10.1371/journal.pmed.0020073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oxnard GR, Arcila ME, Sima CS, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res 2011;17:1616-22. 10.1158/1078-0432.CCR-10-2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. 10.1038/nm.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu HA, Suzawa K, Jordan E, et al. Concurrent Alterations in EGFR-Mutant Lung Cancers Associated with Resistance to EGFR Kinase Inhibitors and Characterization of MTOR as a Mediator of Resistance. Clin Cancer Res 2018;24:3108-18. 10.1158/1078-0432.CCR-17-2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le X, Puri S, Negrao MV, et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR-Mutant NSCLC. Clin Cancer Res 2018;24:6195-203. 10.1158/1078-0432.CCR-18-1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohashi K, Sequist LV, Arcila ME, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A 2012;109:E2127-33. 10.1073/pnas.1203530109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov 2012;2:922-33. 10.1158/2159-8290.CD-12-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 2007;104:20932-7. 10.1073/pnas.0710370104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. 10.1038/ncomms11815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oxnard GR, Arcila ME, Chmielecki J, et al. New strategies in overcoming acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer. Clin Cancer Res 2011;17:5530-7. 10.1158/1078-0432.CCR-10-2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonetti A, Sharma S, Minari R, et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121:725-37. 10.1038/s41416-019-0573-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Yang N, Ou Q, et al. Investigating Novel Resistance Mechanisms to Third-Generation EGFR Tyrosine Kinase Inhibitor Osimertinib in Non-Small Cell Lung Cancer Patients. Clin Cancer Res 2018;24:3097-107. 10.1158/1078-0432.CCR-17-2310 [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q, Zhang XC, Yang JJ, et al. EGFR L792H and G796R: Two Novel Mutations Mediating Resistance to the Third-Generation EGFR Tyrosine Kinase Inhibitor Osimertinib. J Thorac Oncol 2018;13:1415-21. 10.1016/j.jtho.2018.05.024 [DOI] [PubMed] [Google Scholar]
- 27.Ou SI, Cui J, Schrock AB, et al. Emergence of novel and dominant acquired EGFR solvent-front mutations at Gly796 (G796S/R) together with C797S/R and L792F/H mutations in one EGFR (L858R/T790M) NSCLC patient who progressed on osimertinib. Lung Cancer 2017;108:228-31. 10.1016/j.lungcan.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 28.Cho BC, Cheng Y, Zhou C, et al. Mechanisms of acquired resistance to first-line osimertinib: Preliminary data from the phase III FLAURA study. Ann Oncol 2018;29 Suppl 9:ix177. 10.1093/annonc/mdy483.007 [DOI] [Google Scholar]
- 29.Fassunke J, Muller F, Keul M, et al. Overcoming EGFR(G724S)-mediated osimertinib resistance through unique binding characteristics of second-generation EGFR inhibitors. Nat Commun 2018;9:4655. 10.1038/s41467-018-07078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peled N, Roisman LC, Miron B, et al. Subclonal Therapy by Two EGFR TKIs Guided by Sequential Plasma Cell-free DNA in EGFR-Mutated Lung Cancer. J Thorac Oncol 2017;12:e81-e84. 10.1016/j.jtho.2017.02.023 [DOI] [PubMed] [Google Scholar]
- 31.Oztan A, Fischer S, Schrock AB, et al. Emergence of EGFR G724S mutation in EGFR-mutant lung adenocarcinoma post progression on osimertinib. Lung Cancer 2017;111:84-7. 10.1016/j.lungcan.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 32.Schoenfeld AJ, Chan JM, Rizvi H, et al. Tissue-based molecular and histological landscape of acquired resistance to osimertinib given initially or at relapse in patients with EGFR-mutant lung cancers. Journal of Clinical Oncology. 2019;37:9028. 10.1200/JCO.2019.37.15_suppl.9028 [DOI] [Google Scholar]
- 33.Leventakos K, Kipp BR, Rumilla KM, et al. S768I Mutation in EGFR in Patients with Lung Cancer. J Thorac Oncol 2016;11:1798-801. 10.1016/j.jtho.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papadimitrakopoulou VA, Wu YL, Han JY, et al. Analysis of resistance mechanisms to osimertinib in patients with EGFR T790M advanced NSCLC from the AURA3 study. Ann Oncol 2018;29 Suppl 8:viii741.
- 35.Chiba I, Takahashi T, Nau MM, et al. Mutations in the p53 gene are frequent in primary, resected non-small cell lung cancer. Lung Cancer Study Group. Oncogene 1990;5:1603-10. [PubMed] [Google Scholar]
- 36.Jiao XD, Qin BD, You P, et al. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non-small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer 2018;123:70-5. 10.1016/j.lungcan.2018.07.003 [DOI] [PubMed] [Google Scholar]
- 37.Jin Y, Shi X, Zhao J, et al. Mechanisms of primary resistance to EGFR targeted therapy in advanced lung adenocarcinomas. Lung Cancer 2018;124:110-6. 10.1016/j.lungcan.2018.07.039 [DOI] [PubMed] [Google Scholar]
- 38.Shajani-Yi Z, de Abreu FB, Peterson JD, et al. Frequency of Somatic TP53 Mutations in Combination with Known Pathogenic Mutations in Colon Adenocarcinoma, Non-Small Cell Lung Carcinoma, and Gliomas as Identified by Next-Generation Sequencing. Neoplasia 2018;20:256-62. 10.1016/j.neo.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer 2019;19:495-509. 10.1038/s41568-019-0179-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou H, Sun D, Zhang X. The role of MDM2 amplification and overexpression in therapeutic resistance of malignant tumors. Cancer Cell Int 2019;19:216. 10.1186/s12935-019-0937-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. 10.1126/science.1141478 [DOI] [PubMed] [Google Scholar]
- 42.Lai GGY, Lim TH, Lim J, et al. Clonal MET Amplification as a Determinant of Tyrosine Kinase Inhibitor Resistance in Epidermal Growth Factor Receptor-Mutant Non-Small-Cell Lung Cancer. J Clin Oncol 2019;37:876-84. 10.1200/JCO.18.00177 [DOI] [PubMed] [Google Scholar]
- 43.Gainor JF, Niederst MJ, Lennerz JK, et al. Dramatic Response to Combination Erlotinib and Crizotinib in a Patient with Advanced, EGFR-Mutant Lung Cancer Harboring De Novo MET Amplification. J Thorac Oncol 2016;11:e83-5. 10.1016/j.jtho.2016.02.021 [DOI] [PubMed] [Google Scholar]
- 44.Wang Q, Yang S, Wang K, et al. MET inhibitors for targeted therapy of EGFR TKI-resistant lung cancer. J Hematol Oncol 2019;12:63. 10.1186/s13045-019-0759-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi P, Oh YT, Zhang G, et al. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett 2016;380:494-504. 10.1016/j.canlet.2016.07.021 [DOI] [PubMed] [Google Scholar]
- 46.Ortiz-Cuaran S, Scheffler M, Plenker D, et al. Heterogeneous Mechanisms of Primary and Acquired Resistance to Third-Generation EGFR Inhibitors. Clin Cancer Res 2016;22:4837-47. 10.1158/1078-0432.CCR-15-1915 [DOI] [PubMed] [Google Scholar]
- 47.Oxnard GR, Hu Y, Mileham KF, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients with EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 2018;4:1527-34. 10.1001/jamaoncol.2018.2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong MH, Kim MH, Kim SY, et al. Molecular landscape of osimertinib resistance revealed by targeted panel sequencing and patient-derived cancer models in non-small cell lung cancer patients. Ann Oncol 2018;29 Suppl 8:viii516.
- 49.Shi Y, Xing P, Han X, et al. P1.13-18 Exploring the Resistance Mechanism of Osimertinib and Monitoring the Treatment Response Using Plasma ctDNA in Chinese NSCLC Patients. J Thorac Oncol 2018;13:S589. 10.1016/j.jtho.2018.08.875 [DOI] [Google Scholar]
- 50.Eberlein CA, Stetson D, Markovets AA, et al. Acquired Resistance to the Mutant-Selective EGFR Inhibitor AZD9291 Is Associated with Increased Dependence on RAS Signaling in Preclinical Models. Cancer Res 2015;75:2489-500. 10.1158/0008-5472.CAN-14-3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nukaga S, Yasuda H, Tsuchihara K, et al. Amplification of EGFR Wild-Type Alleles in Non-Small Cell Lung Cancer Cells Confers Acquired Resistance to Mutation-Selective EGFR Tyrosine Kinase Inhibitors. Cancer Res 2017;77:2078-89. 10.1158/0008-5472.CAN-16-2359 [DOI] [PubMed] [Google Scholar]
- 52.Del Re M, Rofi E, Restante G, et al. Implications of KRAS mutations in acquired resistance to treatment in NSCLC. Oncotarget 2017;9:6630-43. 10.18632/oncotarget.23553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Del Re M, Tiseo M, Bordi P, et al. Contribution of KRAS mutations and c.2369C > T (p.T790M) EGFR to acquired resistance to EGFR-TKIs in EGFR mutant NSCLC: a study on circulating tumor DNA. Oncotarget 2017;8:13611-9. 10.18632/oncotarget.6957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guibert N, Pradines A, Farella M, et al. Monitoring KRAS mutations in circulating DNA and tumor cells using digital droplet PCR during treatment of KRAS-mutated lung adenocarcinoma. Lung Cancer 2016;100:1-4. 10.1016/j.lungcan.2016.07.021 [DOI] [PubMed] [Google Scholar]
- 55.Ho CC, Liao WY, Lin CA, et al. Acquired BRAF V600E Mutation as Resistant Mechanism after Treatment with Osimertinib. J Thorac Oncol 2017;12:567-72. 10.1016/j.jtho.2016.11.2231 [DOI] [PubMed] [Google Scholar]
- 56.de Bruin EC, Cowell C, Warne PH, et al. Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer Discov 2014;4:606-19. 10.1158/2159-8290.CD-13-0741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakayama S, Sng N, Carretero J, et al. beta-catenin contributes to lung tumor development induced by EGFR mutations. Cancer Res 2014;74:5891-902. 10.1158/0008-5472.CAN-14-0184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015;5:850-9. 10.1158/2159-8290.CD-15-0285 [DOI] [PubMed] [Google Scholar]
- 59.Malladi S, Macalinao DG, Jin X, et al. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell 2016;165:45-60. 10.1016/j.cell.2016.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. 10.1126/scitranslmed.3002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong J, Li L, Wang Z, et al. Potential Resistance Mechanisms Revealed by Targeted Sequencing from Lung Adenocarcinoma Patients with Primary Resistance to Epidermal Growth Factor Receptor (EGFR) Tyrosine Kinase Inhibitors (TKIs). J Thorac Oncol 2017;12:1766-78. 10.1016/j.jtho.2017.07.032 [DOI] [PubMed] [Google Scholar]
- 62.Takeuchi T, Tomida S, Yatabe Y, et al. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol 2006;24:1679-88. 10.1200/JCO.2005.03.8224 [DOI] [PubMed] [Google Scholar]
- 63.Tanaka H, Yanagisawa K, Shinjo K, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res 2007;67:6007-11. 10.1158/0008-5472.CAN-06-4774 [DOI] [PubMed] [Google Scholar]
- 64.Zeng L, Yang N, Zhang Y, et al. GOPC-ROS1 Rearrangement as an Acquired Resistance Mechanism to Osimertinib and Responding to Crizotinib Combined Treatments in Lung Adenocarcinoma. J Thorac Oncol 2018;13:e114-e116. 10.1016/j.jtho.2018.02.005 [DOI] [PubMed] [Google Scholar]
- 65.Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of Acquired Resistance to Osimertinib in EGFR-Mutant NSCLC and Clinical Validation of Combined EGFR and RET Inhibition with Osimertinib and BLU-667 for Acquired RET Fusion. Cancer Discov 2018;8:1529-39. 10.1158/2159-8290.CD-18-1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schrock AB, Zhu VW, Hsieh WS, et al. Receptor Tyrosine Kinase Fusions and BRAF Kinase Fusions are Rare but Actionable Resistance Mechanisms to EGFR Tyrosine Kinase Inhibitors. J Thorac Oncol 2018;13:1312-23. 10.1016/j.jtho.2018.05.027 [DOI] [PubMed] [Google Scholar]
- 67.Offin M, Somwar R, Rekhtman N, et al. Acquired ALK and RET Gene Fusions as Mechanisms of Resistance to Osimertinib in EGFR-Mutant Lung Cancers. JCO Precis Oncol 2018. doi: 10.1200/PO.18.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 69.Cross DA, Ashton SE, Ghiorghiu S, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 2014;4:1046-61. 10.1158/2159-8290.CD-14-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, He B, Zhou D, et al. Newly emergent acquired EGFR exon 18 G724S mutation after resistance of a T790M specific EGFR inhibitor osimertinib in non-small-cell lung cancer: a case report. Onco Targets Ther 2018;12:51-6. 10.2147/OTT.S188612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hollstein M, Sidransky D, Vogelstein B, et al. p53 mutations in human cancers. Science 1991;253:49-53. 10.1126/science.1905840 [DOI] [PubMed] [Google Scholar]
- 72.Aubrey BJ, Strasser A, Kelly GL. Tumor-Suppressor Functions of the TP53 Pathway. Cold Spring Harb Perspect Med 2016. doi: . 10.1101/cshperspect.a026062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2010;2:a001008. 10.1101/cshperspect.a001008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Molina-Vila MA, Bertran-Alamillo J, Gasco A, et al. Nondisruptive p53 mutations are associated with shorter survival in patients with advanced non-small cell lung cancer. Clin Cancer Res 2014;20:4647-59. 10.1158/1078-0432.CCR-13-2391 [DOI] [PubMed] [Google Scholar]
- 75.Cho Y, Gorina S, Jeffrey PD, et al. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 1994;265:346-55. 10.1126/science.8023157 [DOI] [PubMed] [Google Scholar]
- 76.Lehmann BD, Pietenpol JA. Targeting mutant p53 in human tumors. J Clin Oncol 2012;30:3648-50. 10.1200/JCO.2012.44.0412 [DOI] [PubMed] [Google Scholar]
- 77.Xu J, Reumers J, Couceiro JR, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol 2011;7:285-95. 10.1038/nchembio.546 [DOI] [PubMed] [Google Scholar]
- 78.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009;9:701-13. 10.1038/nrc2693 [DOI] [PubMed] [Google Scholar]
- 79.Leroy B, Fournier JL, Ishioka C, et al. The TP53 website: an integrative resource centre for the TP53 mutation database and TP53 mutant analysis. Nucleic Acids Res 2013;41:D962-9. 10.1093/nar/gks1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scian MJ, Stagliano KE, Deb D, et al. Tumor-derived p53 mutants induce oncogenesis by transactivating growth-promoting genes. Oncogene 2004;23:4430-43. 10.1038/sj.onc.1207553 [DOI] [PubMed] [Google Scholar]
- 81.Vikhanskaya F, Lee MK, Mazzoletti M, et al. Cancer-derived p53 mutants suppress p53-target gene expression--potential mechanism for gain of function of mutant p53. Nucleic Acids Res 2007;35:2093-104. 10.1093/nar/gkm099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma X, Le Teuff G, Lacas B, et al. Prognostic and Predictive Effect of TP53 Mutations in Patients with Non-Small Cell Lung Cancer from Adjuvant Cisplatin-Based Therapy Randomized Trials: A LACE-Bio Pooled Analysis. J Thorac Oncol 2016;11:850-61. 10.1016/j.jtho.2016.02.002 [DOI] [PubMed] [Google Scholar]
- 83.Canale M, Petracci E, Delmonte A, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin Cancer Res 2017;23:2195-202. 10.1158/1078-0432.CCR-16-0966 [DOI] [PubMed] [Google Scholar]
- 84.Rho JK, Choi YJ, Ryoo BY, et al. p53 enhances gefitinib-induced growth inhibition and apoptosis by regulation of Fas in non-small cell lung cancer. Cancer Res 2007;67:1163-9. 10.1158/0008-5472.CAN-06-2037 [DOI] [PubMed] [Google Scholar]
- 85.Tsui DWY, Murtaza M, Wong ASC, et al. Dynamics of multiple resistance mechanisms in plasma DNA during EGFR-targeted therapies in non-small cell lung cancer. EMBO Mol Med 2018. doi: . 10.15252/emmm.201707945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Labbé C, Cabanero M, Korpanty GJ, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer 2017;111:23-9. 10.1016/j.lungcan.2017.06.014 [DOI] [PubMed] [Google Scholar]
- 87.VanderLaan PA, Rangachari D, Mockus SM, et al. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer 2017;106:17-21. 10.1016/j.lungcan.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nahar R, Zhai W, Zhang T, et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun 2018;9:216. 10.1038/s41467-017-02584-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010;17:77-88. 10.1016/j.ccr.2009.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hellman A, Zlotorynski E, Scherer SW, et al. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell 2002;1:89-97. 10.1016/S1535-6108(02)00017-X [DOI] [PubMed] [Google Scholar]
- 91.Engelman JA, Jänne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res 2008;14:2895-9. 10.1158/1078-0432.CCR-07-2248 [DOI] [PubMed] [Google Scholar]
- 92.Buttitta F, Barassi F, Fresu G, et al. Mutational analysis of the HER2 gene in lung tumors from Caucasian patients: mutations are mainly present in adenocarcinomas with bronchioloalveolar features. Int J Cancer 2006;119:2586-91. 10.1002/ijc.22143 [DOI] [PubMed] [Google Scholar]
- 93.Arcila ME, Chaft JE, Nafa K, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res 2012;18:4910-8. 10.1158/1078-0432.CCR-12-0912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li C, Sun Y, Fang R, et al. Lung adenocarcinomas with HER2-activating mutations are associated with distinct clinical features and HER2/EGFR copy number gains. J Thorac Oncol 2012;7:85-9. 10.1097/JTO.0b013e318234f0a2 [DOI] [PubMed] [Google Scholar]
- 95.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 2009;9:550-62. 10.1038/nrc2664 [DOI] [PubMed] [Google Scholar]
- 96.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene 2008;27:5486-96. 10.1038/onc.2008.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eng J, Woo KM, Sima CS, et al. Impact of Concurrent PIK3CA Mutations on Response to EGFR Tyrosine Kinase Inhibition in EGFR-Mutant Lung Cancers and on Prognosis in Oncogene-Driven Lung Adenocarcinomas. J Thorac Oncol 2015;10:1713-9. 10.1097/JTO.0000000000000671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hsu KH, Ho CC, Hsia TC, et al. Identification of five driver gene mutations in patients with treatment-naive lung adenocarcinoma in Taiwan. PLoS One 2015;10:e0120852. 10.1371/journal.pone.0120852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sholl LM, Aisner DL, Varella-Garcia M, et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol 2015;10:768-77. 10.1097/JTO.0000000000000516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol 2011;29:2046-51. 10.1200/JCO.2010.33.1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Planchard D, Kim TM, Mazieres J, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:642-50. 10.1016/S1470-2045(16)00077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Le LQ, Parada LF. Tumor microenvironment and neurofibromatosis type I: connecting the GAPs. Oncogene 2007;26:4609-16. 10.1038/sj.onc.1210261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin SY, Xia W, Wang JC, et al. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A 2000;97:4262-6. 10.1073/pnas.060025397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Little CD, Nau MM, Carney DN, et al. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature 1983;306:194-6. 10.1038/306194a0 [DOI] [PubMed] [Google Scholar]
- 105.Iwakawa R, Kohno T, Kato M, et al. MYC amplification as a prognostic marker of early-stage lung adenocarcinoma identified by whole genome copy number analysis. Clin Cancer Res 2011;17:1481-9. 10.1158/1078-0432.CCR-10-2484 [DOI] [PubMed] [Google Scholar]
- 106.Seo AN, Yang JM, Kim H, et al. Clinicopathologic and prognostic significance of c-MYC copy number gain in lung adenocarcinomas. Br J Cancer 2014;110:2688-99. 10.1038/bjc.2014.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hirsch CL, Coban Akdemir Z, Wang L, et al. Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev 2015;29:803-16. 10.1101/gad.255109.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kimura S, Hara Y, Pineau T, et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev 1996;10:60-9. 10.1101/gad.10.1.60 [DOI] [PubMed] [Google Scholar]
- 109.Yamaguchi T, Yanagisawa K, Sugiyama R, et al. NKX2-1/TITF1/TTF-1-Induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell 2012;21:348-61. 10.1016/j.ccr.2012.02.008 [DOI] [PubMed] [Google Scholar]
- 110.Ekholm SV, Reed SI. Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol 2000;12:676-84. 10.1016/S0955-0674(00)00151-4 [DOI] [PubMed] [Google Scholar]
- 111.Jordan EJ, Kim HR, Arcila ME, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov 2017;7:596-609. 10.1158/2159-8290.CD-16-1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee JK, Lee J, Kim S, et al. Clonal History and Genetic Predictors of Transformation into Small-Cell Carcinomas From Lung Adenocarcinomas. J Clin Oncol 2017;35:3065-74. 10.1200/JCO.2016.71.9096 [DOI] [PubMed] [Google Scholar]
- 113.Niederst MJ, Sequist LV, Poirier JT, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun 2015;6:6377. 10.1038/ncomms7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marcoux N, Gettinger SN, O'Kane G, et al. EGFR-Mutant Adenocarcinomas That Transform to Small-Cell Lung Cancer and Other Neuroendocrine Carcinomas: Clinical Outcomes. J Clin Oncol 2019;37:278-85. 10.1200/JCO.18.01585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu M, Xu S, Wang Y, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, sensitizes lung cancer cells to treatment with epidermal growth factor receptor tyrosine kinase inhibitors. Oncotarget 2016;7:84951-64. 10.18632/oncotarget.13069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sorrentino JA, Freed DM, Bisi JE, et al. The CDK4/6 inhibitor G1T38 enhances response to targeted therapies in preclinical models of non-small cell lung cancer. Cancer Res 2018;78:abstr 1522.
- 117.Klein ME, Kovatcheva M, Davis LE, et al. CDK4/6 Inhibitors: The Mechanism of Action May Not Be as Simple as Once Thought. Cancer Cell 2018;34:9-20. 10.1016/j.ccell.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou J, Sanchez-Vega F, Caso R, et al. Analysis of Tumor Genomic Pathway Alterations Using Broad-Panel Next-Generation Sequencing in Surgically Resected Lung Adenocarcinoma. Clin Cancer Res 2019;25:7475-84. 10.1158/1078-0432.CCR-19-1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jonsson VD, Blakely CM, Lin L, et al. Novel computational method for predicting polytherapy switching strategies to overcome tumor heterogeneity and evolution. Sci Rep 2017;7:44206. 10.1038/srep44206 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as