Abstract
Many eukaryotic genomes harbour large numbers of duplicated sequences, of diverse biotypes, resulting from several mechanisms including recombination, whole genome duplication and retro-transposition. Such repeated sequences complicate gene/transcript quantification during RNA-seq analysis due to reads mapping to more than one locus, sometimes involving genes embedded in other genes. Genes of different biotypes have dissimilar levels of sequence duplication, with long-noncoding RNAs and messenger RNAs sharing less sequence similarity to other genes than biotypes encoding shorter RNAs. Many strategies have been elaborated to handle these multi-mapped reads, resulting in increased accuracy in gene/transcript quantification, although separate tools are typically used to estimate the abundance of short and long genes due to their dissimilar characteristics. This review discusses the mechanisms leading to sequence duplication, the biotypes affected, the computational strategies employed to deal with multi-mapped reads and the challenges that still remain to be overcome.
Keywords: RNA-seq, Multi-mapped reads, Duplicated genes, Noncoding RNAs, Gene isoforms, Expectation–maximization algorithm
1. Introduction
Gene duplications are common genomic occurrences resulting in the addition of new genetic material in a genome and representing one of the mechanisms enabling molecular evolution [1], [2]. With time, the two identical sequences will often gradually diverge through the acquisition of mutations. Different molecular mechanisms can cause gene duplications, with different genomic regions presenting a different susceptibility to these mechanisms, as described in section 2. Duplicated genomic regions represent a challenge when analysing high-throughput RNA sequencing datasets as the reads align equally well at more than one such genomic location. When reads cannot be unambiguously aligned to a reference, genes cannot be accurately quantified [3]. The short reads from technologies such as Illumina compound the problem, although many repeated sequences correspond to short genes for which longer reads would not solve the problem. Fractions of RNA sequencing (RNA-seq) reads that cannot be uniquely aligned vary depending on the organism, the sample, the type of molecule enriched for in the experiment, the aligner and the reference annotation used. Proportions of multi-mapped reads typically range from 5 to 40% of total reads mapped [4], [5], representing a substantial subset of reads. Diverse strategies have been devised to deal with multi-mapping reads, including ignoring them, splitting them equally between the multi-mapped genes, distributing them between the multi-mapped genes or gene portions proportionally to their uniquely mapped reads or based on a statistical model of mapping uncertainty, and providing quantifications for gene groups rather than individual genes. Section 3 of the review describes these strategies. In addition to duplicated genomic sequences, duplicated transcriptomic sequences, due to alternative splicing for example, cause similar problems during RNA-seq read alignment to transcriptomic sequences. This mini-review explores the mechanisms by which genomic and transcriptomic sequences can be duplicated, the classes of transcripts with repeated sequences and the computational strategies to manage the mapping of reads to duplicated sequences.
2. Genomic and transcriptomic sequence duplication mechanisms and affected RNA biotypes
2.1. Recombination and whole genome duplication
Recombination is a molecular mechanism enabling genetic sequence exchange that can be reciprocal or not. Recombination usually results from a sequence exchange between DNA regions from the same locus on homologous chromosomes. In some cases, the crossing-over of the homologous chromosomes can be unequal, leading to tandem duplication of genes [1], [2], [6]. In addition, recombination can also occur ectopically between non-homologous loci, leading to the insertion of different genetic material in one of the loci involved and resulting in the duplication of sequence [6], [7]. Such duplication events can generate new functional genes termed paralogs of the original copy. However, if they lack gene expression capability or acquire mutations affecting their capacity to code for proteins, they are referred to as pseudogenes. Though under different selective pressure, paralogs but even more so pseudogenes progressively acquire mutations compared to their parental copy, the age of the copy reflecting the extent of sequence divergence with the parent gene [8]. Most duplicated genes are thought to eventually become pseudogenes or be lost [9].
Whole genome duplication is another mechanism resulting in sequence duplication. Strong evidence points to the occurrence of whole genome duplication in diverse ancestral organisms including in the lineage leading to the baker’s yeast Saccharomyces cerevisiae, in early chordate evolution and in diverse plant lineages [10], [11], [12], [13]. And while such events are typically followed by widespread gene loss, a subset of genes remain present in more than one copy as a consequence. For example, many gene families in vertebrates are believed to have been formed or expanded by a whole genome duplication event in early chordate evolution [11].
2.2. Transposable elements
Transposons (or transposable elements) are genomic elements with the capacity to change genomic position, either by ‘cut and paste’ or ‘copy and paste’ mechanisms [14]. Approximately half to two-third of the human genome is believed to consist of transposons, although only a small proportion of these elements (<0.05%) are believed to be active today [15], [16], [17]. Transposons include both DNA transposons and retrotransposons, which move about the genome using different mechanisms. DNA transposons excise themselves from their current genomic location to integrate into another position. In contrast, retrotransposition first involves transcription of the element into RNA which is then reverse transcribed into DNA and reinserted into the genome at another locus, generating new retrotransposon copies. Transposons can be inserted in diverse genomic loci including in intergenic regions, but also within other genes, in sense or antisense, in their exons or introns, resulting in much sequence redundancy in genomes. Retrotransposition machinery can also, in addition, use cellular RNAs as substrates, reverse transcribing and inserting them in the genome, leading to new copies of existing genes [14]. Such new genes lack the genomic and thus the regulatory context of their parental copy and are often not expressed. Genes resulting from the retrotransposition of messenger RNAs are referred to as processed pseudogenes, lacking the introns of their parental copy [14]. As a consequence, they typically share sequence identity with the exons of their parental copy although since most are not expressed, they are under low selective pressure to avoid acquiring mutations, progressively losing sequence identity with their parental copy.
Many noncoding RNAs also benefit from retrotransposition, resulting in many copies in genomes and an expansion in their family member count. For example, depending on the genome, small nucleolar RNAs (snoRNAs), which are structured noncoding RNAs typically varying in length between 70 and 150 nucleotides and playing a role in ribosome biogenesis, encode dozens to thousands of copies with evidence of their propagation through retrotransposition [18], [19], [20]. Diverse other families of noncoding RNA including small nuclear RNA (involved in splicing), 7SL (the signal recognition particle RNA) and miRNA (involved in the regulation of transcript stability and translation) derive many of their members through retrotransposition [20], [21], [22], [23], [24], [25]. In the case of retrotranscribed noncoding RNAs, many copies have been shown to be expressed and functional.
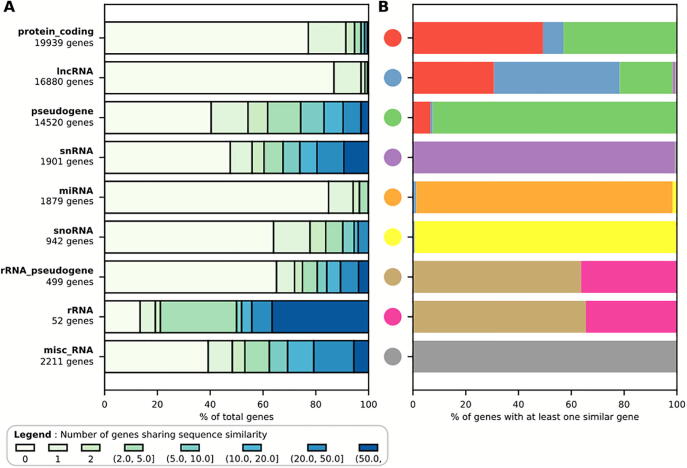
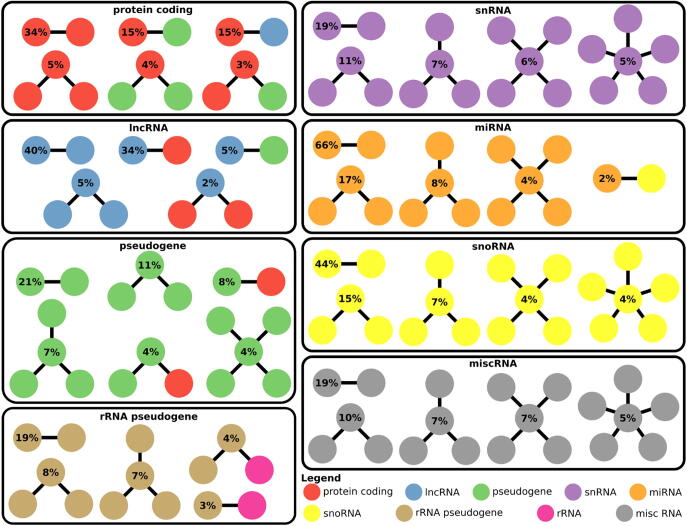
Thus as a consequence of recombination, whole genome duplication and transposition, many genomic sequences are repeated and can be distinguished in two groups: paralogous gene families resulting from the duplication of whole genes and genes containing highly repetitive elements embedded in their sequence. The former group consists of genes of any biotype and have high sequence similarity over their whole length whereas the latter group mostly affects longer genes such the protein_coding and lncRNA RNA classes and have strong sequence similarity only over a portion of the gene. Fig. 1 shows the proportion of human genes of different biotypes displaying sequence similarity to other genes, based on Ensembl annotations [26]. The gene biotypes rRNA, pseudogene, snRNA, miscellaneous RNA, snoRNA and rRNA_pseudogene show the largest proportion of members with sequence similarity to other genes (Fig. 1A). For the genes with sequence similarity to other genes, most biotypes display similarity to their own biotype (for example snRNA, miRNA, snoRNA and misc_RNA, Fig. 1B and 2). However, unsurprisingly, rRNA and rRNA pseudogenes show high levels of similarity with each other. And similarly, protein_coding, lncRNA and pseudogenes also display high levels of similarity with each other (Figs. 1B and 2). The most common similarity relationships for the main biotypes are illustrated in Fig. 2. The distribution of abundance of transcripts of different biotypes varies depending on the organism and the sample, but also the library preparation protocol and the computational pipeline used for the quantification [27], [28], which will ultimately influence the abundance distribution of each biotype and the proportion of multi-mapped reads for each biotype and overall for the sample [4], [5].
Fig. 1.
Proportion of human genes with sequence similarity to other genes, per biotype. (A) Stacked bar chart displaying the percentage of genes per biotype with specified number of genes sharing sequence similarity. (B) For each biotype and for all genes of that biotype that share similarity with another human gene, the distribution of the biotypes of their similar genes is shown. To calculate gene similarity, human genes were obtained from the Ensembl annotation (version 99). Pairwise sequence similarity was measured with BLAST (version 2.9.0 from bioconda). The BLAST database was composed of the genomic sequence from each gene, and the spliced sequence of their transcript having the highest number of exons. The spliced transcript is used to identify processed pseudogenes by reducing gap bias. The blastn algorithm was run for all pairs of sequences in the database, with 1e-20 as a minimum e-value, and keeping only the best hit for each pairwise comparison. Each BLAST hit was scored as the average of the alignment length divided by the whole length of the sequence and multiplied by the percentage of identical matches for each sequence in the pair. Results were then parsed, eliminating self-hits (a gene with itself or its transcript), and analysed using a BLAST pairwise score threshold of 60%.
Fig. 2.
Most common sequence similarity relationships between human genes, per biotype. The network of sequence similarity relationships was measured for all human genes as described in Fig. 1. The most common sequence similarity patterns are illustrated here, per biotype.
2.3. Alternative splicing as a source of duplicated sequences
Alternative splicing, as well as the use of alternative promoters, increase the number of different transcripts produced by a gene [29], [30], [31]. While such isoforms contain unique sequences, all common exons included in these transcripts will have identical sequences, increasing the amount of sequence duplication when one considers the entire transcript content of an RNA sample (ie its transcriptome) [3]. While alternative splicing does not result in sequence duplication in a genome, it does in the context of a transcriptome (ie in the context of a transcriptome annotation, alternative splicing results in more than one transcript from the same gene, usually with overlapping sequences). Thus if RNA-seq reads are aligned to a transcriptome rather than to a genome, multiple overlapping transcripts annotated for a given gene will appear as duplicated sequences in the reference.
3. Strategies to deal with repeated sequences when analysing RNA-seq
RNA-seq is a powerful high-throughput approach which enables the quantification of RNAs in a sample. As extensively reviewed in [32], [33], the RNA-seq methodology involves isolating the RNA of interest with optional fragmentation depending on the technology and the RNAs considered, construction of the sequencing library, typically implicating reverse transcription to DNA, generating the inserts that will be sequenced, to which adapters are added and then amplification, followed by the sequencing itself. Many variations of the protocol exist to sequence specific RNAs or portions of RNAs of interest [32]. The analysis of RNA-seq involves evaluating the quality of the individual sequencing reads and alignment of the reads to the genome or transcriptome of the organism from which the RNA was obtained, if available, followed by attribution of the aligned reads to specific genes or transcripts and their quantification [34]. In this context, all sequence repetition, whether genomic or transcriptomic, adds complexity to RNA-seq analysis because all the reads aligning to one copy of a repeated sequence will also align to most or all of its copies. The RNA-seq reads mapping to more than one locus, or multireads, are thus difficult to quantify and add uncertainty to downstream analysis. Both types of RNA-seq analyses, either gene- or transcript-level, have different types of multireads and strategies to quantify them, although recent methods deal with them simultaneously within the same model (summarized in Table 1). Here, we discuss the methods that deal with both multireads mapping to more than one genomic location and to more than one transcript (in the case of transcript-level analyses).
Table 1.
Computational strategies and methods that handle multi-mapped reads.
| Tool | Quantification level | Input | Strandedness can be specified | Count type | Strategy | Paired end | Confidence level | Focus | Reference |
|---|---|---|---|---|---|---|---|---|---|
| HTSeq-count | Gene | BAM | Y | Counts | Ignore | Y | N | Long RNA | [35] |
| STAR geneCounts | Gene | Fastq | Y | Counts | Ignore | Y | N | Long RNA | [36] |
| Cufflinks | Transcript | BAM | Y | RPKM | Split equally, Rescue | Y | N | Long RNA | [38] |
| featureCounts | Gene | BAM | Y | Counts | Ignore, count all, split equally | Y | N | Long RNA | [37] |
| CoCo | Gene | BAM | Y | Counts, CPM, TPM | Rescue | Y | N | Small RNA Long RNA |
[28] |
| ERANGE | Transcript | BAM | N | RPKM | Rescue | Y | N | Long RNA | [39] |
| EMASE | Transcript | BAM | N | Counts, TPM | EM | Y | N | Long RNA | [48] |
| IsoEM2 | Both | SAM | Y | FPKM, TPM | EM | Y | Confidence intervals | Long RNA | [56] |
| Kallisto | Transcript | Fastq | Y | TPM | EM | Y | Bootstrap values | Long RNA | [49] |
| RSEM | Both | Fastq, BAM | Y | Counts, TPM, FPKM | EM | Y | 95% credibility intervals | Long RNA | [45] |
| Salmon | Transcript | Fastq | Y | Counts, TPM | EM | Y | Bootstrap values | Long RNA | [50] |
| MMR | N/A | BAM | Y | N/A | Read coverage | Y | N/A | Long RNA | [44] |
| MuMRescueLite | Genomic loci | Custom format | N | Counts | Read coverage | N | N | Short sequence tags | [41] |
| Rcount | Gene | BAM | Y | Counts | Read coverage | N | N | Long RNA | [42] |
| ShortStack | Gene | Fastq, BAM | N | Counts, RPM | Read coverage | N | N | Small RNA | [43] |
| mmquant | Gene | BAM | Y | Counts | Gene Clustering | Y | N | Small RNA Long RNA |
[51] |
| SeqCluster | Gene | BAM | N | Counts | Gene clustering | N | N | Small RNA | [53] |
| Fuzzy method | Gene | Custom format | N | Fuzzy counts | Fuzzy sets | N | Fuzzy counts | Small RNA Long RNA |
[54] |
| geneQC | Gene | SAM | Y | NA | ML | Y | Mapping uncertainty level | Small RNA Long RNA |
[5] |
3.1. Simple methods
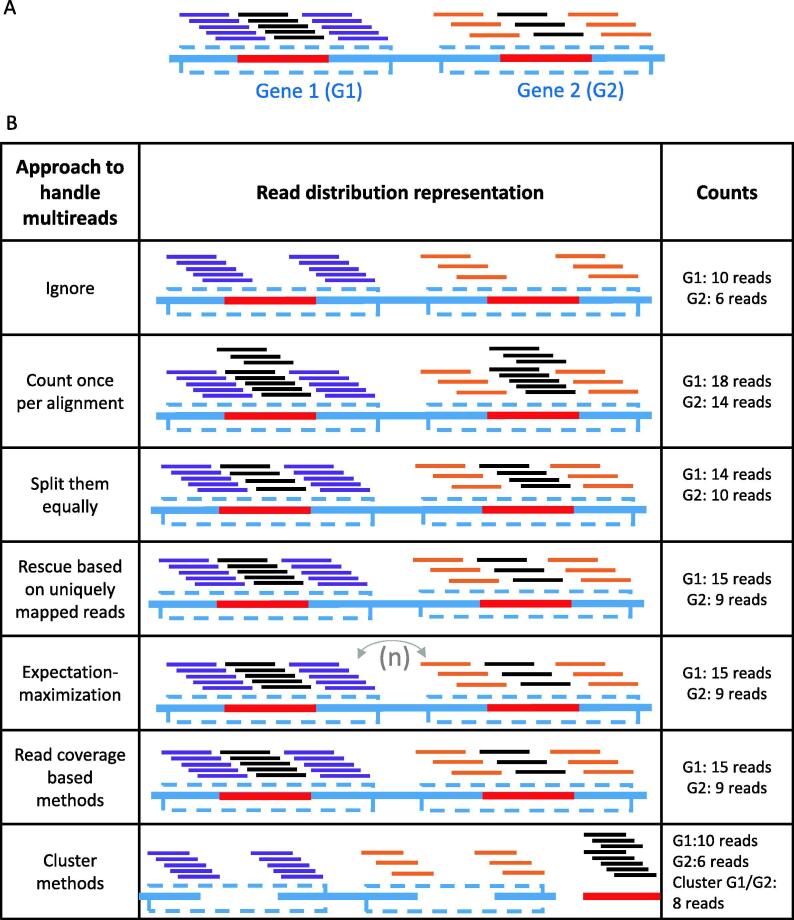
The simplest ways of handling multireads are to ignore them all together, count them once for each alignment, randomly assign them to one of the best alignments or split them equally between each alignment (Fig. 3). The first method consisting in discarding all the multi-mapped reads is often used in gene-level quantification. This approach is employed by popular tools such as HTSeq-count [35], STAR geneCounts [36] and Subreads’s featureCounts [37] using their default parameters. This strategy reduces the uncertainty of the multi-copy gene quantification, but will also lead to an underestimation of certain gene sets and biotypes (Fig. 3). This is not an option in transcript-level analyses, since complete exons would be ignored. The second option, which is to count all valid alignments for a read will have the opposite effect of systematically overestimating these gene sets and RNA biotypes. Using this strategy, a read may be counted more than once, inflating the number of molecules that appear to have been sequenced, misrepresenting the studied sample (Fig. 3). This option is available using Subread’s featureCounts with –M option. The last simple strategy is to equally split the multi-mapped reads between all their alignments (Fig. 3). This can be achieved by using featureCounts –M --fraction options and Cufflinks [38]. Uniformly distributing the multireads, by either keeping a single random alignment or by splitting the count between each alignment, will ensure that every read is counted only once, and will give a better representation of the proportion of each RNA biotype in the sequenced sample. The downfall of these approaches is the dilution of the effect on a single active gene copy between all the other inactive ones. For instance, if a protein coding gene having multiple inactive copies is expressed (for example, in Fig. 2, one of the protein_coding genes with similarity to one or several pseudogenes could be in this situation), the reads that align to the active and inactive copies will be shared, and the region of the inactive copies that diverge from the parent gene won’t have any expression. Thus, the truly expressed gene will be underestimated and the inactive copies overestimated.
Fig. 3.
Strategies to deal with multi-mapped reads. (A) Example of two genes sharing a duplicated sequence and the distribution of RNA-seq reads originating from them. The two genes are represented by boxes outlined by dashed lines and their common sequence is illutrated by a red line. The reads are represented by lines above the genes, purple for reads that are unique to Gene 1, orange for reads that are unique to Gene 2 and black for reads that are common to genes 1 and 2. (B) General classes to handle multi-mapped reads include ignoring them, counting them once per alignment, splitting them equally between the alignments, rescuing the reads based on uniquely mapped reads of the gene, expectation–maximization approaches, rescuing methods based on read coverage in flanking regions and clustering methods that group together genes/transcripts with shared sequences. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Rescue methods
To address the above issue and more accurately represent the relative abundance of the repeated genes, the rescue method was introduced. This strategy consists of distributing the multi-mapped reads between their alignments based on the uniquely mapped read ratio (Fig. 3). Taking the example cited above, if a protein coding gene has many uniquely mapped reads throughout its length, whereas the only reads aligned to the inactive copies are also aligned to the active one, all the multi-mapped reads will be assigned to the active copy. However, if two genes are completely identical, which is more frequent for short RNA genes such as snoRNAs and miRNAs, none will have uniquely mapped reads, and the multi-mapped reads will either be equally split between both copies or not counted at all using this approach, depending on the tool. This strategy was introduced by ERANGE [39], and is also used by Cufflinks with –u option and CoCo [28]. A problem with this strategy occurs when a short gene sequence is entirely contained in a longer gene. In this case, all the reads originating from the small gene will also map to the longer one, while the longer gene can have uniquely mapped reads, resulting in an overestimation of the longer gene at the expense of the shorter one. This situation is common for small noncoding RNAs such as snoRNAs and miRNAs, many of which are intronic in mammalian genomes, encoded in longer genes referred to as their host genes [40]. Such noncoding RNAs often overlap with retained introns of their host genes, resulting in reads mapping exactly to the small RNA being rather attributed to the host gene. CoCo rescues such reads by aligning them to an alternative annotation in which exonic regions overlapping an embedded small RNA are removed (these regions are typically retained introns), resulting in the attribution of the reads to the embedded small RNA. CoCo then performs a background subtraction over the whole exon to ensure that reads are not wrongly assigned to the smaller RNA [28].
3.3. Read coverage based methods
Another strategy is to weigh the multireads based on the read coverage of their different alignments surroundings (Fig. 3). MuMRescueLite [41], Rcount [42] and ShortStack [43] distribute the multireads with respect to the read coverage density in their surroundings, rather than over the whole gene as do many rescue methods. An alignment having more reads mapped in a given window upstream and downstream the repeated region will be assigned a more important portion of the multiread. MuMRescueLite and Rcount distribute the multireads based on uniquely mapped reads only, whereas ShortStack allows the user to choose between uniquely mapped reads only or both, with a lesser weight attributed to the multireads. MMR [44] starts with one alignment for every read, this alignment can either be the best, the first in the input or a random one. MMR considers the read coverage profile around every multiple alignment and keeps the one maximising the smoothness of the local coverage. While appropriate for genes consisting both of regions that are identical to other genes but also regions that are unique to them, this method may not be suited for small RNA-seq where reads accumulate in very well defined blocks and the corresponding genes do not have flanking unique sequences. MMR filters the alignments from an alignment file (BAM), and does not perform quantification. If the user keeps more than one alignment per read, the quantification tool must be chosen accordingly.
3.4. Expectation maximization approaches
Many quantification tools use the expectation maximization (EM) algorithm to estimate the maximum likelihood value of gene or transcript abundance and can take multi-mapped reads into account (Fig. 3). The first such tool to manage mapping uncertainty with a statistical model was RSEM which can handle reads that are multi-mapped between both transcripts and genes and is capable of dealing with non-uniform read distributions [45], [46]. Several subsequent tools were proposed including IsoEM, which can also deal with multi-mapping reads between both transcripts and genes [47] and EMASE, which manages multireads between genes, transcripts and alleles [48]. Each tool has a different model usually taking into account the fragment length distribution, alignment quality, sequence bias and so on. For genes that have uniquely mapped reads in addition to their multireads, the expectation–maximization approach has been shown to be more accurate than the rescue method, which is reported as equivalent to a single iteration of the expectation maximization algorithm [45]. Some tools using this strategy such as RSEM, also report a value representing the confidence level of each gene or transcript abundance [46]. This value can be very helpful to evaluate the reliability of the differential expression analysis results. Indeed, it could indicate if a gene or transcript was deemed significantly differentially expressed mainly due to its mapping uncertainty. More recent RNA-seq quantification programs based on pseudoalignment and quasi-mapping such as Kallisto and Salmon also use the EM algorithm to attribute read counts to the most likely transcripts [49], [50]. Kallisto builds a transcriptome de Bruijn graph based on sequencing reads. If they have identical sequences, whether originating from different transcripts of the same gene or from different genes, reads will map to the same part of the de Bruijn graph and will thus be part of the same equivalence class. Kallisto samples from the equivalence classes following a multinomial distribution which are fed to the EM algorithm, resulting in transcript abundance estimates [49]. Similarly, Salmon also establishes equivalence classes, either through its own mapping procedure referred to as quasi-mapping, or from a pre-calculated alignment file, built over the initial fragment abundance estimates. An EM algorithm is then run over the equivalence classes which can span different transcripts and different genes, providing transcripts estimates [50].
3.5. Clustering methods
Instead of trying to weigh and distribute the multiread counts, some tools, such as mmquant [51], cluster genes together as a multi-mapped group (MMG) as proposed by [52]. All the genes to which a multiread aligns will be clustered as a MMG which can be used as a gene for further analysis such as differential expression analysis (Fig. 3). While this method diminishes the multiread quantification uncertainty, the handling of the MMG can be difficult. For instance, it would be difficult to evaluate normalized counts such as transcripts per million, because the shared proportion of the genes may vary inside a group, and from one group to another. In addition, MMG can span more than one biotype (Fig. 2), which can make the interpretation difficult. On the other hand, this strategy can be very useful when aligning to the genome and trying to identify multireads aligning to unannotated regions. The gene clustering can help in characterizing potential new genes as proposed by SeqCluster [53]. For example, if an unannotated region clusters with tRNA genes, it is likely that this region may encode a tRNA, and this will help refine the methodology to investigate its function. One must be careful when trying to identify new non-annotated genes if most reads are multi-mapped since it could be an inactive copy of an actual gene.
3.6. Measuring multi-mapping uncertainty
Although many tools offer different strategies for dealing with multireads, the problem is still not solved and may have an important impact in downstream analysis such as differential expression analysis and functional enrichment. Some methods were developed to help users understand or have better insights into which genes are more affected by the multiread bias. For instance, GeneQC [5] uses a machine learning approach to evaluate the uncertainty of a gene or transcript quantification based on the sequence similarity, the proportion of multireads and the number of similar genes. It can categorize a gene uncertainty as low, medium or high, which can help in selecting genes with reliable estimated counts, and raise awareness to those needing further investigation. Another paper used fuzzy sets [54] to evaluate the effect of different multiread quantification approaches on differential expression analysis and helps identify false positives. These methods offer important insights regarding the uncertainty of gene expression estimation created by multireads, and can have a crucial impact on the selection of genes for experimental validation.
4. Summary and outlook
Genomes are replete with repeated sequences, resulting from several different mechanisms that drive evolution, leading to gene duplication. RNA-seq pipelines must acknowledge this concept to ensure accuracy in gene and transcript quantification. Many strategies have been devised to deal with multi-mapping reads (Table 1). Early efforts often led to under or overestimates of repeated genes, but more recent approaches more accurately attribute multireads to gene or transcript of origin. Genes for which the whole gene sequence is repeated (typically short genes, often embedded in other genes) and those for which only a portion is duplicated do not currently benefit from the same strategies. Unified methodologies dealing well with both multi-mapped reads originating from groups of short genes and multi-mapped reads from portions of long genes will require careful design. Such methodologies will be important for the quantification of RNA-seq from low structure bias sequencing approaches such as TGIRT-seq, which detects accurately both long RNAs and short structured and often embedded RNAs simultaneously [27], [55]. A second challenge will be in the accurate quantification of RNA-seq from samples and conditions that lead to genetic sequence amplification and aberrations such as cancer, for which reference genomes or transcriptomes are not available. Accurate reference-free approaches dealing well with multi-mapping reads will be important in those cases. A third challenge will derive from technologies such as single-cell approaches which suffer from missing data, making gene and transcript estimates more difficult to evaluate, particularly for genes with large proportions of multi-mapped reads. RNA-seq is now a ubiquitous tool in molecular biology. The fast paced improvement of RNA-seq computational pipelines to deal with the widespread issue of multi-mapping reads will likely continue and these challenges be rapidly met, ensuring accurate quantification for increasingly diverse samples.
CRediT authorship contribution statement
Gabrielle Deschamps-Francoeur: Investigation, Data curation, Writing - original draft, Writing - review & editing. Joël Simoneau: Methodology, Formal analysis, Visualization, Writing - review & editing. Michelle S. Scott: Conceptualization, Writing - original draft, Writing - review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The authors wish to thank members of their group for insightful discussions and to acknowledge Compute Canada as an outstanding advanced research computing resource for the Canadian scientific community.
Funding
This work was supported by the Natural Science and Engineering Research Council of Canada [NSERC grant RGPIN-2018-05412 to MSS]. The funder had no involvement in the choice of the review and its writing. GDF was supported by an Alexander-Graham-Bell Doctoral scholarship from NSERC. JS was supported by an NSERC Masters scholarship. MSS holds a Fonds de Recherche du Québec – Santé (FRQS) Research Scholar Junior 2 Career Award.
References
- 1.Ohta T. Role of gene duplication in evolution. Genome. 1989;31:304–310. doi: 10.1139/g89-048. [DOI] [PubMed] [Google Scholar]
- 2.Magadum S., Banerjee U., Murugan P., Gangapur D., Ravikesavan R. Gene duplication as a major force in evolution. J Genet. 2013;92:155–161. doi: 10.1007/s12041-013-0212-8. [DOI] [PubMed] [Google Scholar]
- 3.Treangen T.J., Salzberg S.L. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2011;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dharshini S.A.P., Taguchi Y.H., Gromiha M.M. Identifying suitable tools for variant detection and differential gene expression using RNA-seq data. Genomics. 2020;112:2166–2172. doi: 10.1016/j.ygeno.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 5.McDermaid A., Chen X., Zhang Y., Wang C., Gu S. A new machine learning-based framework for mapping uncertainty analysis in RNA-Seq read alignment and gene expression estimation. Front Genet. 2018;9:313. doi: 10.3389/fgene.2018.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benovoy D., Drouin G. Ectopic gene conversions in the human genome. Genomics. 2009;93:27–32. doi: 10.1016/j.ygeno.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Hastings P.J. Mechanisms of ectopic gene conversion. Genes (Basel) 2010;1:427–439. doi: 10.3390/genes1030427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espinosa-Cantu A., Ascencio D., Barona-Gomez F., DeLuna A. Gene duplication and the evolution of moonlighting proteins. Front Genet. 2015;6:227. doi: 10.3389/fgene.2015.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch M., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 10.Wolfe K.H., Shields D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 11.McLysaght A., Hokamp K., Wolfe K.H. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- 12.Walker J.F., Yang Y., Moore M.J., Mikenas J., Timoneda A. Widespread paleopolyploidy, gene tree conflict, and recalcitrant relationships among the carnivorous Caryophyllales. Am J Bot. 2017;104:858–867. doi: 10.3732/ajb.1700083. [DOI] [PubMed] [Google Scholar]
- 13.Xiang Y., Huang C.H., Hu Y., Wen J., Li S. Evolution of rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol Biol Evol. 2017;34:262–281. doi: 10.1093/molbev/msw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazazian H.H., Jr. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 15.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 16.Mills R.E., Bennett E.A., Iskow R.C., Devine S.E. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 17.de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitz J., Zemann A., Churakov G., Kuhl H., Grutzner F. Retroposed SNOfall–a mammalian-wide comparison of platypus snoRNAs. Genome Res. 2008;18:1005–1010. doi: 10.1101/gr.7177908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber M.J. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2006;2:e205. doi: 10.1371/journal.pgen.0020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boivin V., Faucher-Giguere L., Scott M., Abou-Elela S. The cellular landscape of mid-size noncoding RNA. Wiley Interdiscip Rev RNA. 2019;10:e1530. doi: 10.1002/wrna.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doucet A.J., Droc G., Siol O., Audoux J., Gilbert N. U6 snRNA pseudogenes: markers of retrotransposition dynamics in mammals. Mol Biol Evol. 2015;32:1815–1832. doi: 10.1093/molbev/msv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kojima K.K. Human transposable elements in Repbase: genomic footprints from fish to humans. Mob DNA. 2018;9:2. doi: 10.1186/s13100-017-0107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y.H., Bruin T., Tuzgol S., Wilson B.I., Roederer G. Two naturally occurring mutations at the first and second bases of codon aspartic acid 156 in the proposed catalytic triad of human lipoprotein lipase. In vivo evidence that aspartic acid 156 is essential for catalysis. J Biol Chem. 1992;267:1918–1923. [PubMed] [Google Scholar]
- 24.Mourier T., Willerslev E. Retrotransposons and non-protein coding RNAs. Brief Funct Genomic Proteomic. 2009;8:493–501. doi: 10.1093/bfgp/elp036. [DOI] [PubMed] [Google Scholar]
- 25.Smalheiser N.R., Torvik V.I. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, et al. (2019) Ensembl 2019. Nucleic Acids Res 47:D745–D51 [DOI] [PMC free article] [PubMed]
- 27.Boivin V., Deschamps-Francoeur G., Couture S., Nottingham R.M., Bouchard-Bourelle P. Simultaneous sequencing of coding and noncoding RNA reveals a human transcriptome dominated by a small number of highly expressed noncoding genes. RNA. 2018;24:950–965. doi: 10.1261/rna.064493.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deschamps-Francoeur G., Boivin V., Abou Elela S., Scott M.S. CoCo: RNA-seq read assignment correction for nested genes and multimapped reads. Bioinformatics. 2019;35:5039–5047. doi: 10.1093/bioinformatics/btz433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Dov C., Hartmann B., Lundgren J., Valcarcel J. Genome-wide analysis of alternative pre-mRNA splicing. J Biol Chem. 2008;283:1229–1233. doi: 10.1074/jbc.R700033200. [DOI] [PubMed] [Google Scholar]
- 30.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davuluri R.V., Suzuki Y., Sugano S., Plass C., Huang T.H. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–177. doi: 10.1016/j.tig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Hrdlickova R, Toloue M, Tian B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip Rev RNA 2017;8. [DOI] [PMC free article] [PubMed]
- 33.Van den Berge K., Hembach K.M., Soneson C., Tiberi S., Clement L. RNA sequencing data: Hitchhiker's guide to expression analysis. Ann Rev Biomed Data Sci. 2019;2:139–173. [Google Scholar]
- 34.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 38.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 40.Boivin V., Deschamps-Francoeur G., Scott M.S. Protein coding genes as hosts for noncoding RNA expression. Semin Cell Dev Biol. 2018;75:3–12. [Google Scholar]
- 41.Hashimoto T., de Hoon M.J., Grimmond S.M., Daub C.O., Hayashizaki Y. Probabilistic resolution of multi-mapping reads in massively parallel sequencing data using MuMRescueLite. Bioinformatics. 2009;25:2613–2614. doi: 10.1093/bioinformatics/btp438. [DOI] [PubMed] [Google Scholar]
- 42.Schmid M.W., Grossniklaus U. Rcount: simple and flexible RNA-Seq read counting. Bioinformatics. 2015;31:436–437. doi: 10.1093/bioinformatics/btu680. [DOI] [PubMed] [Google Scholar]
- 43.Johnson NR, Yeoh JM, Coruh C, Axtell MJ. Improved placement of multi-mapping small RNAs. G3 (Bethesda) 2016;6:2103–11. [DOI] [PMC free article] [PubMed]
- 44.Kahles A., Behr J., Ratsch G. MMR: a tool for read multi-mapper resolution. Bioinformatics. 2016;32:770–772. doi: 10.1093/bioinformatics/btv624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B., Ruotti V., Stewart R.M., Thomson J.A., Dewey C.N. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicolae M., Mangul S., Mandoiu I.I., Zelikovsky A. Estimation of alternative splicing isoform frequencies from RNA-Seq data. Algorithms Mol Biol. 2011;6:9. doi: 10.1186/1748-7188-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raghupathy N., Choi K., Vincent M.J., Beane G.L., Sheppard K.S. Hierarchical analysis of RNA-seq reads improves the accuracy of allele-specific expression. Bioinformatics. 2018;34:2177–2184. doi: 10.1093/bioinformatics/bty078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 50.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zytnicki M. mmquant: how to count multi-mapping reads? BMC Bioinf. 2017;18:411. doi: 10.1186/s12859-017-1816-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robert C., Watson M. Errors in RNA-Seq quantification affect genes of relevance to human disease. Genome Biol. 2015;16:177. doi: 10.1186/s13059-015-0734-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pantano L., Estivill X., Marti E. A non-biased framework for the annotation and classification of the non-miRNA small RNA transcriptome. Bioinformatics. 2011;27:3202–3203. doi: 10.1093/bioinformatics/btr527. [DOI] [PubMed] [Google Scholar]
- 54.Consiglio A., Mencar C., Grillo G., Marzano F., Caratozzolo M.F. A fuzzy method for RNA-Seq differential expression analysis in presence of multireads. BMC Bioinf. 2016;17:345. doi: 10.1186/s12859-016-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nottingham R.M., Wu D.C., Qin Y., Yao J., Hunicke-Smith S. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA. 2016;22:597–613. doi: 10.1261/rna.055558.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mandric I., Temate-Tiagueu Y., Shcheglova T., Al Seesi S., Zelikovsky A. Fast bootstrapping-based estimation of confidence intervals of expression levels and differential expression from RNA-Seq data. Bioinformatics. 2017;33:3302–3304. doi: 10.1093/bioinformatics/btx365. [DOI] [PubMed] [Google Scholar]