Abstract
Because of the unique features of spherical symmetry, angle-independency, good monodispersity, controllable components and morphologies, structural color particles (SCPs) have found great significances in various fields such as sensing, monitoring, biological assays, etc. Here, inspired by the melanosome-derived bright structural colors and the self-adhesivity of mussels, we present a kind of bioinspired SCPs assembled from polydopamine (PDA)-adhered multi-layer graphene oxide (GO) encapsulated silica nanoparticles (SNs). It is demonstrated that compared with traditional SCPs, the designed particles possess brighter and more vibrant structural colors, and no complicated modification is required during the functionalization process due to the abundant inherent functional groups of GO. The resultant SCPs are verified to be capable for direct hybridization chain reaction and multiplexed nucleic acid assays. These properties indicate the promising prospects of our designed SCPs.
Keywords: Bioinspired, Structural color, Graphene oxide, Colloidal crystal, Microfluidics
Graphical abstract
Inspired by melanosome-derived bright structural colors and self-adhesivity of mussels, the structural color particles (SCPs) assembled from the specially designed multi-layer graphene oxide (GO) and polydopamine (PDA) encapsulated silica nanoparticles (SNs) are presented. These SCPs possess brighter structural colors and require few complex modifications during functionalization process, and they are demonstrated to be capable for direct hybridization chain reaction and multiplexed nucleic acid assays.
Highlights
-
•
Structural color particles (SCPs) are assembled from specially-designed multi-layer nanoparticles.
-
•
The designed SCPs possess brighter structural colors and require few complex modifications during functionalization process.
-
•
The designed SCPs are capable for direct hybridization chain reactions and multiplexed bioassays.
1. Introduction
Structural color, a special source of colors in nature, is always vibrant and fascinating. Unlike common pigments or dyes that produce colors by molecules’ absorbing light or luminous compounds, structural colors arise through the light reflection from their complex and characteristic nanostructures, which have been widely found in natural creatures, such as peacock feathers, butterfly wings, and shells of some insects. While the pigments and dyes will degrade and fade overtime, those structural colors that rely merely on the inherent nanostructure of materials can persist a much longer time [[1], [2], [3]]. Attracted by the unique features of structural colors, numerous researchers around the world have turned their attention to various methods to understand, mimic, and recreate these natural materials [[4], [5], [6], [7], [8], [9], [10], [11], [12], [13]]. Among diversified artificial structural color materials, the development of spherical ones plays a crucial role because their symmetry property could impart the structural color particles (SCPs) with independence of the detection angle and locations [14,15]. In addition, by employing microfluidic technology into the generating process, the SCPs with excellent particle monodispersity, as well as diverse components and morphologies, can be obtained [[16], [17], [18], [19]]. These SCPs are promising in various application fields like sensing, detecting, barcoding, and anti-counterfeiting [[20], [21], [22], [23], [24], [25]]. Although great progress has been achieved in the spherical structural color materials, there are still some limitations need to be mentioned and overcome, such as the insufficient color brightness arisen from the incoherent scattering. Also, it often requires complicated modifications and operations to impart these SCPs with desired functions, which significantly limits their promotion and practical use. Thus, SCPs with excellent structural colors and inherent multiple functions are still urgently anticipated.
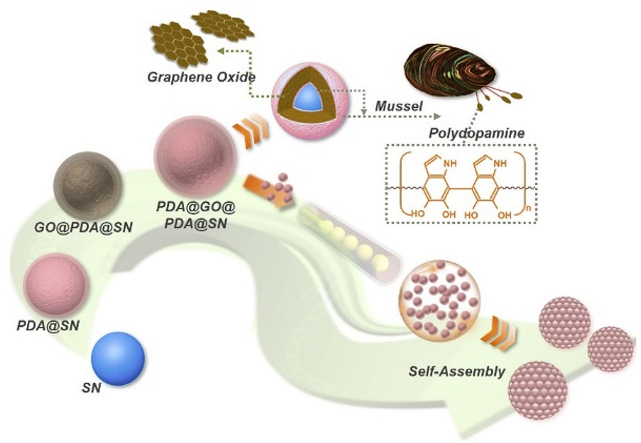
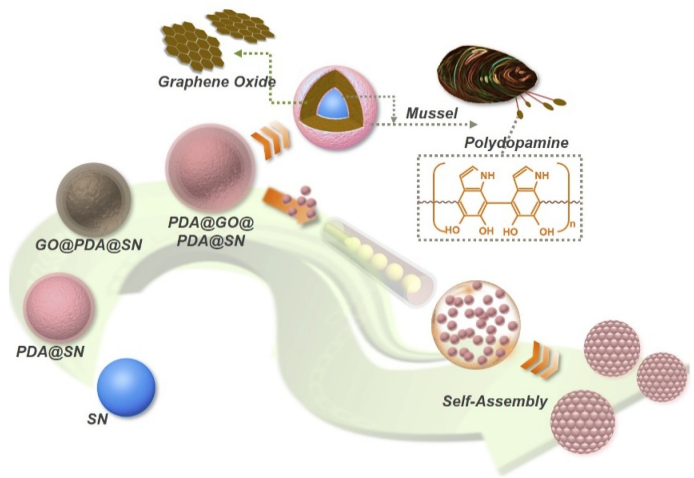
In this paper, inspired by melanosome-derived bright colors and super-adhesive mussels in nature, we present a new kind of SCPs assembled from polydopamine (PDA)-adhered multi-layer graphene oxide (GO) encapsulated silica nanoparticles (SNs), as illustrated in Scheme 1. GO, as a famous kind of graphene derivatives or related materials, has been widely utilized in various fields, such as electronics, biology, chemistry, medicine, and optics. GO has exhibited many remarkable properties, including extensive sources and accessibility, high surface area and refractive index, good dispersibility in water and organic solvents, unique photothermal effect, and abundant oxygen-containing functional groups (like epoxy and hydroxyl groups) on the surface [[26], [27], [28], [29]]. Besides, PDA is a main component of the self-produced adhesive protein of mussels that essentially contributes to their super-adhesivity, and it is often employed to optimize the materials’ adhesivity and combine different molecules or substances [[30], [31], [32], [33]].
Scheme 1.
The schematic of the modification process of the multi-layer PDA and GO encapsulated SNs, followed with the generation process of the modified-nanoparticle-derived SCPs.
Thus, it is conceivable that through the integration of PDA and GO, the multi-layer GO encapsulated SNs and their assembled novel SCPs will be readily achieved. As the elementary nanoparticles are encapsulated by GO layers, these particles are endowed with higher refractive index and broadband light absorption, therefore enhancing the color saturation and brightness of their derived SCPs. This phenomenon is similar with that the feathers of specific birds display extremely bright colors due to the melanosomes existed in their microstructures [34,35]. In addition, owing to the functional groups of GO, the generated SCPs can be easily functionalized without further complex modifications, which greatly simplifies the practical operation processes. Moreover, in our system, introducing the dopamine can not only implement the robust combination of GO and SNs, but also contribute to the enhancement of color saturation since the dark color of PDA. To summarize, the specific SCPs derived from the multi-layer GO encapsulated nanoparticles that we demonstrate here definitely possess brilliant structural color and multiple functions, thereby indicating versatile application prospects of the designed SCPs.
2. Experimental section
2.1. Materials
The silica nanoparticles (SNs) were bought from Nanjing Dongjian Biological Technology Co., Ltd. Graphene oxide (GO) dispersion solution (XF224-1, 0.5 mg/mL) was purchased from Nanjing XFNANO Materials Tech. Co., Ltd. Dopamine (DA) hydrochloride and tris base were purchased from Sigma Aldrich Co. Hydrochloric acid (HCl), ethanol absolute, and n-hexane were purchased from Sinopharm Chemical Reagent Co., Ltd. Dimethyl silicone oil was bought from Nanjing Wanqing Co. Ltd. The oligonucleotide sequences purified by high-performance liquid chromatography were obtained from Thermo Fisher Scientific Co., Ltd (Shanghai, China). The phosphate buffered saline (PBS) buffer was prepared in our lab. Deionized water with a resistivity of 18.2 MΩ cm−1 was obtained from a Millipore Milli-Q system and it was used during the whole work. All the chemical reagents were of the best grade available and used as received. The purity of the employed chemical reagents was all analytically pure.
2.2. Synthesis of the GO and PDA encapsulated multi-layer SNs
The SNs with the sizes of 263, 230, and 193 nm were chosen in this experiment. The original SNs were thoroughly washed by deionized water through 6800 rpm centrifugation. After being washed, the original SNs were prepared into aqueous dispersion at the concentration of 0.1 g/mL (1) Then the SNs were introduced into an alkaline reaction environment (pH = 8.5) for PDA encapsulating. The dopamine hydrochloride aqueous solution was prepared at the concentration of 13.3 mg/mL. The total volume of the reaction system was 10 mL, which consisted of 1.5 mL prepared dopamine hydrochloride aqueous solution (13.3 mg/mL, the total mass of dopamine in the reaction system was 20 mg to ensure the thickness of PDA layer), 2.03 mL original SNs aqueous dispersion (0.1 g/mL), 5 mL tris base (0.1 mol/L), and 1.47 mL HCl (0.1 mol/L). The encapsulating reaction was carried out in a glass container with continuously stirring for 6 h. Then the system was centrifuged at 6800 rpm for 10 min to remove the reaction residuals. After removing the supernatant, the remaining SNs were coated with PDA layers. (2) The remaining PDA-encapsulated SNs were resuspended by GO dispersion. The resuspension was poured into a glass container and stirred for 6 h to accomplish the GO encapsulating reaction. Due to the adhesivity of PDA layers, the GO layers were able to encapsulate the SNs. Then the system was also centrifuged with 6800 rpm for 10 min to remove the reaction residuals. Through repeating step (1) and step (2) sequentially, the SNs with multiple PDA and GO layers were obtained.
2.3. Generation of SCPs from the designed multi-layer nanoparticles
We used microfluidic technology to generate the SCPs. The aqueous dispersion of the designed PDA and GO encapsulated multi-layer SNs was employed as the dispersed phase, and the silicone oil (50 cs) was employed as the continuous phase. The concentration of the dispersed phase was 0.2 g/mL. The flow rates of the dispersed phase and continuous phase were 0.5 and 9 mL/h respectively. The dispersed phase would be sheared into droplets by the continuous phase at the orifice of the microfluidic device owing to the existence of fluid shear force. The formed droplets were collected in a plastic container filled with silicone oil (500 cs). Then the container was placed in an oven (75 °C) overnight to realize the self-assembly of the nanoparticles in the droplets. After finishing the solvent evaporation and self-assembly processes, the generated SCPs should be thoroughly washed by n-hexane in order to remove the residual silicone oil, since the silicone oil could be readily soluble in the n-hexane, and the n-hexane would not remain in the SCPs after its thorough evaporation.
2.4. Classical sandwich method for nucleic acid detection through generated SCPs
The generated SCPs were thoroughly washed by PBS buffer. The original concentration of the oligonucleotide sequences was all 100 μM. The sequences used in this part was demonstrated in Table S1. The probes were tenfold diluted, and the targets and labels were hundredfold diluted. First, the SCPs were incubated with the probe solution for 6 h with continuously stirring. Then, after washing by PBS buffer, the probe-linked SCPs were incubated with target and label solution for 1 h with continuously stirring. Finally, the SCPs were washed by PBS buffer.
2.5. Hybridization chain reaction (HCR) amplification method for nucleic acid detection through generated SCPs
The generated SCPs were thoroughly washed by PBS buffer. The original concentration of the oligonucleotide sequences was all 100 μM. The sequences used in this part was demonstrated in Table S2. The probe 1 and H1 & H2 were tenfold diluted, the target 1 and initiator 1 were hundredfold diluted. First, the SCPs were incubated with the probe 1 solution for 6 h with continuously stirring. Then, after washing by PBS buffer, the probe-linked SCPs were incubated with target 1 and initiator 1 solution for 1 h with continuously stirring. After washing by PBS buffer, the SCPs were then incubated with H1 & H2 for 1 h with continuously stirring. Finally, the SCPs were washed by PBS buffer.
2.6. Multiplexed nucleic acid detection through generated SCPs
The generated SCPs with three different colors (red, green, and blue) were used in this section and the detailed information of the employed oligonucleotide sequences were demonstrated in Table S2. The SCPs with red, green, and blue structural colors were coupled with probe 1, probe 2, and probe 3, respectively. The coupling method was the same as the last part (HCR amplification method). Then the specific experiments were divided into three groups. In the reaction system of group 1, three kinds of probe-coupled SCPs were incubated with target 1 and initiator 1. In the reaction system of group 2, three kinds of probe-coupled SCPs were incubated with target 2 and initiator 2. In the reaction system of group 3, three kinds of probe-coupled SCPs were incubated with target 3 and initiator 3. After incubating with targets and initiators, FAM labeled H1 and H2 were used to amplify the fluorescent signals. The specific incubation methods were the same as the last experimental section.
2.7. Characterization
The microstructures of the different kinds of nanoparticles were observed by a transmission electron microscope (JEOL, JEM-2100). The microstructures of the derived SCPs were observed by a scanning electron microscope (Hitachi S–300 N). The optical images of the generated SCPs were captured by a stereomicroscope (NOVEL NTB-3A, Ningbo Yongxin Optics Co., Ltd., China) equipped with a color CCD camera (Media Cybernetics Evolution MP 5.0 RTV). The reflection spectra of the generated SCPs were obtained through an optical microscope (Olympus BX51) equipped with a fiberoptic spectrometer (Ocean Optics QE65000). The fluorescent intensity curves were obtained through a fluorescent microscope (Olympus, CKX41) equipped with a fiberoptic spectrometer (Ocean Optics QE65000). The fluorescent images were captured by a stereoscopic microscope (Olympus, SZX16) equipped with the camera under a mercury lamp (Olympus, U-RFL-T) and a halogen lamp (Olympus, LG-PS2).
3. Results and discussion
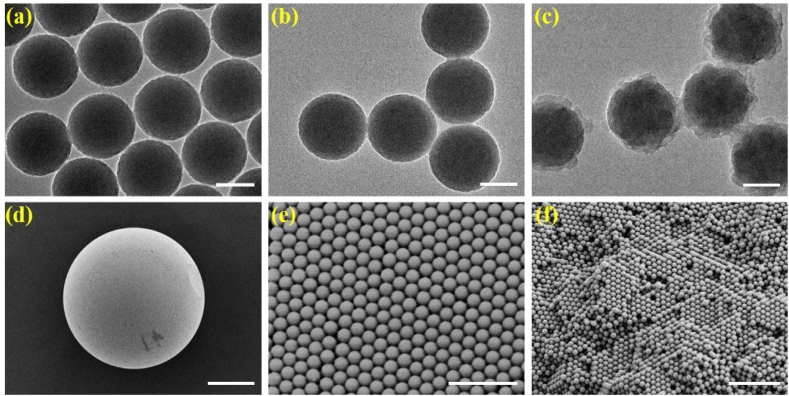
In a typical experiment, the multi-layer SNs were first prepared. The unmodified SNs were originally coated with a PDA layer through the polymerization process of dopamine monomers. Then, due to the super adhesivity of PDA, the GO layer could be easily adhered onto the nanoparticles, as shown in Fig. 1a–c and S1. The transmission electron microscope (TEM) images demonstrated that after the modification process, the SNs with smooth surface were successfully encapsulated by the rough GO layer. In addition, through repeating the preceding steps, SNs coated by a larger amount of GO were obtained, and Fig. S1 showed the comparison between the multi-layer SNs with different amount of GO encapsulated. After the generation of the elementary multi-layer SNs, these nanoparticles were employed to fabricate specific SCPs through microfluidic technology. We used the aqueous dispersion of the prepared multi-layer SNs as the dispersed phase, and silicone oil (50 cs) for the continuous phase. The droplets containing abundant nanoparticles were generated through the fluid shear force, and these multi-layer nanoparticles could be finally self-assembled into the SCPs after solvent evaporation process. It was found that these SCPs had excellent sphericity and monodispersity, since the coefficient of variation (CV) value, i.e. the ratio of the standard deviation to the mean, was only calculated to be 1.54%, which was much lower than 5% (the common CV value for representing good monodispersity) and thus confirmed the favorable uniformity of the particles (Fig. S2). The microstructures of the resultant SCPs were also characterized by a scanning electron microscope (SEM), and the SEM images showed that both of the surface and interior of the SCPs had ordered hexagonal-close-packing structure, as shown in Fig. 1d–f and S1, which indicated the exquisite assembly of the nanoparticles.
Fig. 1.
(a–c) TEM images of unmodified SNs (a), PDA encapsulated SNs (b), and GO and PDA encapsulated SNs (c); (d–f) SEM images of the single resultant SCP (d), surface microstructure of the resultant SCP (e), and interior microstructure of the resultant SCP. The scale bars are 100 nm in (a)–(c), 100 μm in (d), 1 μm in (e), and 2 μm in (f).
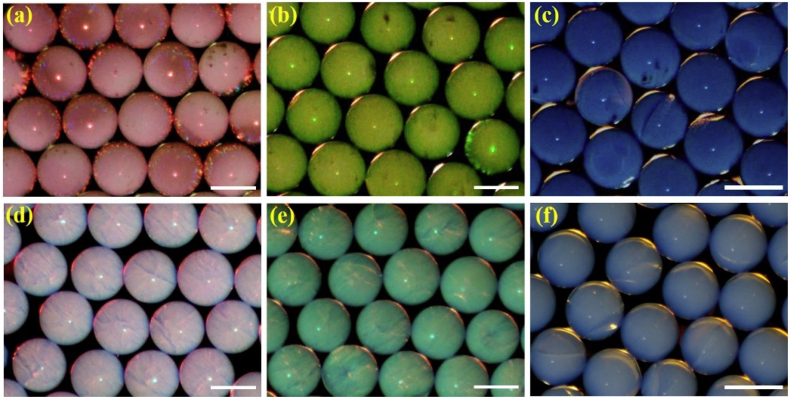
Because of this special hexagonal-close-packing structure, the generated SCPs were endowed with typical photonic band gaps (PBGs). The existence of these PBGs can result in the location and reflection of lights with certain wavelengths rather than the light propagation across the materials. Thus, the generated SCPs derived from the multi-layer nanoparticles displayed vivid structural colors, as shown in Fig. 2a–c. We also compared these SCPs derived from modified nanoparticles with the ones from primal nanoparticles, and the microscope images depicted that the former kind of SCPs showed brighter and more vivid structural colors, while the latter had paler and dim colors (Fig. 2d–f). This phenomenon could be ascribed to the PDA and GO layers of the modified SNs. Inspired by that some kind of birds' feathers possess bright colors, the GO and PDA layers could serve as the dark elements resembling the melanosomes in feathers, which greatly increased the refractive index and broadband light absorption of the nanoparticles, thus ultimately improving the color saturation of the SCPs. We demonstrated the SCPs with three different colors (red, green, and blue), which were assembled from modified nanoparticles with 263, 230, and 193 nm silica cores respectively. According to the Bragg's equation (λ = 1.633dnaverage, in which d is the center-to-center distance between nearest nanoparticles/nanovoids and naverage is the average refractive index of the materials), with the increasing diameter of the elementary nanoparticles, the reflection peak position of their derived microparticles will be red-shifted correspondingly. Therefore, it could be inferred that the reflection peak position of the SCPs generated from the multi-layer nanoparticles would red shift compared with the ones from the unmodified nanoparticles with the same silica cores, since the existence of the PDA and GO layers would enlarge the nanoparticles diameter. The reflection peak images showed in Fig. S3 just confirmed this inference. In addition, the reflection intensity of the SCPs derived from the multi-layer nanoparticles was relatively lower than the ones from the unmodified nanoparticles, due to the lights absorption of the PDA and GO layers.
Fig. 2.
(a–c) SCPs derived from the designed multi-layer PDA and GO encapsulated SNs with red (a), green (b), and blue (c) structural colors; (d–f) SCPs derived from the unmodified SNs with red (d), green (e), and blue (f) structural colors. The scale bars are 200 μm in (a)–(b) and (d)–(e), and 300 μm in (c) and (f). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
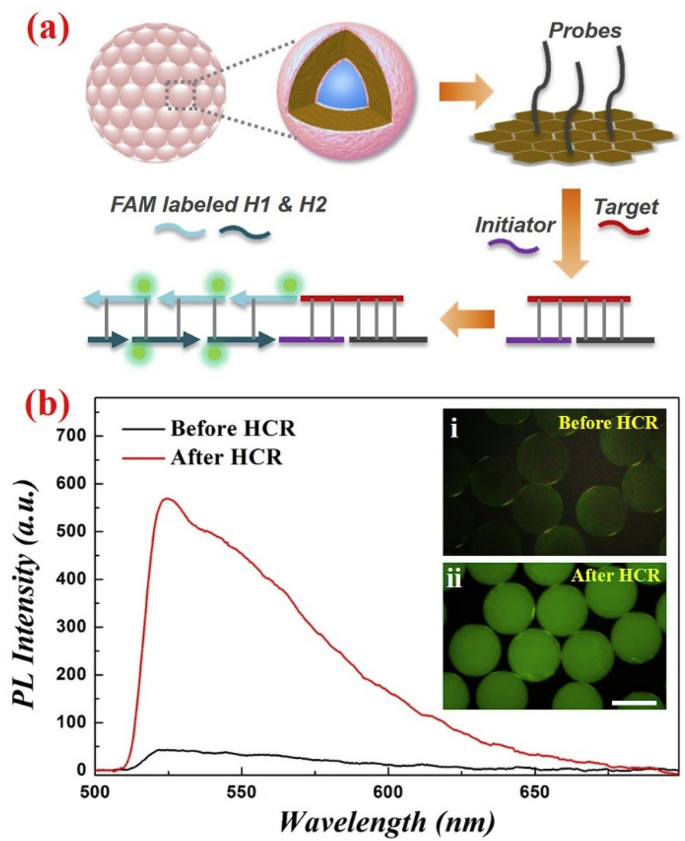
As mentioned above, GO has abundant epoxy groups which can be coupled with the amino groups under room temperature. Also, owing to the typical π−π stacking interaction of GO, it can easily bind with biomolecules [36,37]. Hence, in this work, GO could not only serve as the dark element to improve the color saturation, but also could be employed as a connecting bridge between the SCPs and other biomolecules, thereby providing great convenience for the biomolecular detections. Therefore, we used the designed SCPs for conducting nucleic acid detection. Two different detection strategies were compared in this part, the classical sandwich method and the hybridization chain reaction (HCR) amplification method. In both strategies, the amino-modified detection probes were first bound with the SCPs because of the GO in the particles. Due to the existing epoxy groups and π−π stacking interaction of GO, the probes could be successfully combined to the particles. Afterwards, applying the classical sandwich approach, the detection targets and FAM-decorated labels were introduced into the reaction system, and these three kinds of strands (probes, targets, and labels) could be bonded together based on the complementary base pairing rules (The specific nucleic acid sequence information was demonstrated in Table S1). Although the fluorescent detection signal could be observed by a microscope, the signal intensity obtained by this method was extremely weak, which was not desirable for the nucleic acid detection. As a replacement, the second strategy, i.e. the HCR amplification method, was employed in the following nucleic acid detection. The schematic of the HCR amplification process was illustrated in Fig. 3a. After the combination of detection probes with the SCPs, the targets and initiators were introduced into the reaction system, in which the initiator was utilized for triggering the HCR process. Then the addition of two kinds of FAM-labeled hairpins (H1 and H2) would propagate this chain reaction through the hybridization events and finally form a long DNA strands with strong fluorescent signals. Fig. 3b showed a comparison of the detection results between these two strategies, and it was obvious that the HCR amplification method had a much stronger fluorescent signal, which was proved by both fluorescent images and the photoluminescence intensity curves. In addition, it is worth noting that since the nucleic acid coupling would not affect the structure and the average refractive index of the designed SCPs, the corresponding reflection spectra of these particles before and after the detection process would not obviously change, as shown in Fig. S4.
Fig. 3.
(a) The principle illustration of the HCR amplification method; (b) the photoluminescence (PL) intensity curves and fluorescent images of the SCPs before and after HCR amplification method. The scale bar is 200 μm.
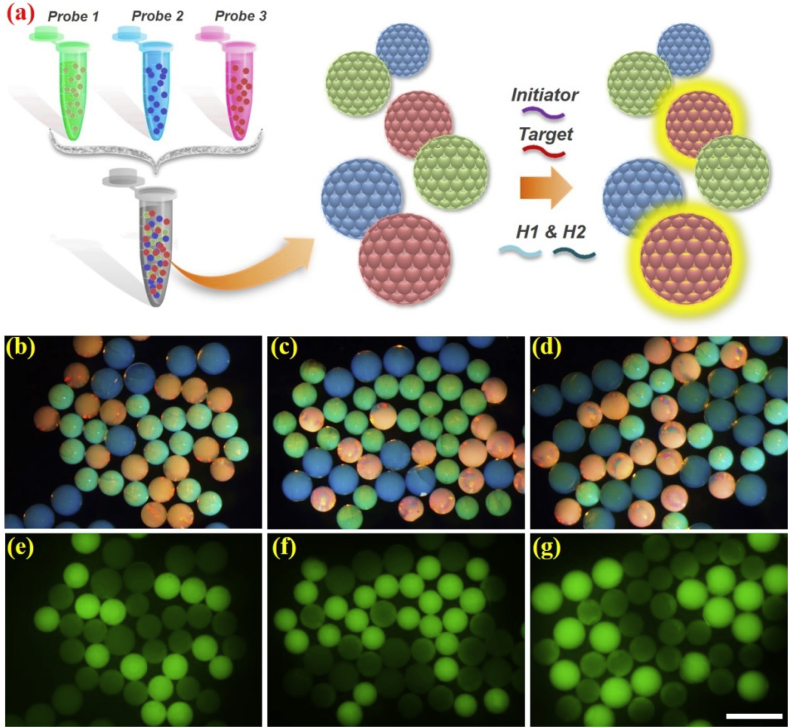
After confirming the feasibility of the designed SCPs as the platform for nucleic acid detection, we also devoted to broadening their applications in multiplexed bioassays. The principle of the multiplexed nucleic acid analysis was illustrated in Fig. 4a. We selected SCPs with different structural colors as detection barcodes, and each kind of them was coupled with corresponding independent nucleic acid probes. These different kinds of barcodes would be placed in one reactor, and then the target sequences and initiators would be introduced into the reaction system. With the assistance of H1 and H2 for signal amplification, the target sequences could be recognized through comparing the optical and fluorescent images. Specifically, SCPs with red, green, and blue colors were chosen in the following experiment, and they were combined with nucleic acid probe 1, probe 2, and probe 3, respectively (The specific nucleic acid sequence information was demonstrated in Table S2). These formed barcodes were divided into three groups (group 1, group 2, and group 3) and each group was responsible for detecting one kind of nucleic acid targets (target 1, target 2, and target 3). As shown in Fig. 4b–g, because only one kind of probes could be successfully paired with the introducing target sequences in each group (probe 1 loaded red barcodes paired with target 1, probe 2 loaded green barcodes paired with target 2, and probe 3 loaded blue barcodes paired with target 3), it was observed that only barcodes with single specific color exhibited strong fluorescent signals. The corresponding optical density (OD) value histogram was also consistent with these results (Fig. S5). Besides, it is worth mentioning that owing to the formation mechanism and special characteristics of structural colors, the intrinsic colors of the barcodes could remain stable and constant during the whole detection process without any interference of the fluorescent signals or reaction environment, which demonstrated the effectivity and robustness of these SCPs as the nucleic acid detection barcodes. These results indicated the capacity of the designed SCPs for multiplexed bioassays, and these particles were confirmed to be an ideal platform due to their characteristic optical properties, stability, and feasible signal amplification through HCR.
Fig. 4.
(a) The principle illustration of the multiplexed nucleic acid detection; (b–g) the optical and fluorescent images of the multiplexed detection results. (b) and (e) demonstrated the detection result of group 1; (c) and (f) demonstrated the detection result of group 2; (d) and (g) demonstrated the detection result of group 3. The scale bar is 500 μm.
4. Conclusion
In conclusion, we have reported a kind of bioinspired SCPs generated from the multi-layer PDA and GO encapsulated SNs, which were capable for complicated multiplexed bioassays. The elementary multi-layer nanoparticles were formed through coating the silica nanocores with GO layers supporting by PDA's super-adhesivity. Then the SCPs were fabricated through microfluidics, followed with the self-assembly of the multi-layer nanoparticles during the solvent evaporation process. Compared with SCPs generated from conventional unmodified SNs, the resultant SCPs displayed higher saturation and brighter structural colors. In addition, due to the excellent properties of GO additives, i.e. the inherent functional groups and the typical π−π stacking interaction of GO which were beneficial for the binding of target biomolecules, the designed SCPs were imparted with more advanced functions, such as multiplexed nucleic acid detections. Moreover, owing to the robustness of intrinsic structural colors of the SCPs that cannot be affected by fluorescence or environmental factors, the detection process was stable and constant. These results indicated the promising prospects of the designed SCPs.
CRediT authorship contribution statement
Yuxiao Liu: Methodology, Validation, Formal analysis, Investigation, Data curation, Writing - original draft. Yuetong Wang: Investigation, Writing - review & editing. Yu Wang: Investigation, Writing - review & editing. Yilai Shu: Investigation, Writing - review & editing. Hui Tan: Investigation, Writing - review & editing. Yuanjin Zhao: Conceptualization, Methodology, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (grants 61927805, 81822011 and 51773119), the Natural Science Foundation of Jiangsu (Grant no. BE2018707), the Science Foundation of Guangdong Province (2019A1515011750), and the Scientific Research Foundation of the Graduate School of Southeast University (Grant No. YBPY1873).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2020.06.013.
Contributor Information
Yilai Shu, Email: yilai_shu@fudan.edu.cn.
Hui Tan, Email: huitan@email.szu.edu.cn.
Yuanjin Zhao, Email: yjzhao@seu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Tadepalli S., Slocik J.M., Gupta M.K., Naik R.R., Singamaneni S. Bio-optics and bio-inspired optical materials. Chem. Rev. 2017;117(20):12705–12763. doi: 10.1021/acs.chemrev.7b00153. [DOI] [PubMed] [Google Scholar]
- 2.Ling S.J., Kaplan D.L., Buehler M.J. Nanofibrils in nature and materials engineering. Nat. Rev. Mater. 2018;3(4):18061. doi: 10.1038/natrevmats.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kristensen A., Yang J.K.W., Bozhevolnyi S.I., Link S., Nordlander P., Halas N.J., Mortensen N.A. Plasmonic colour generation. Nat. Rev. Mater. 2017;2(1):16088. [Google Scholar]
- 4.Li Z.W., Yin Y.D. Stimuli-responsive optical nanomaterials. Adv. Mater. 2019;31(15):1807061. doi: 10.1002/adma.201807061. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z.Y., Fu F.F., Yu Y.R., Shang Y.X., Wang H., Zhao Y.J. Cardiomyocytes actuated morpho butterfly wings. Adv. Mater. 2019;31(8):1805431. doi: 10.1002/adma.201805431. [DOI] [PubMed] [Google Scholar]
- 6.Wang S.Q., Dong H., Sun F.W., Zhang W.L., Liang Y., Tian L., Wang P., Yin X.P., Li G.T. Large-area fabrication of highly tunable hybrid plasmonic-photonic structures based on colloidal lithography and a photoreconfigurable polymer. Adv. Opt. Mater. 2019;7(19):1900483. [Google Scholar]
- 7.Goerlitzer E.S.A., Taylor R.N.K., Vogel N. Bioinspired photonic pigments from colloidal self-assembly. Adv. Mater. 2018;30(28):1706654. doi: 10.1002/adma.201706654. [DOI] [PubMed] [Google Scholar]
- 8.Fu Q.Q., Zhu H.M., Ge J.P. Electrically tunable liquid photonic crystals with large dielectric contrast and highly saturated structural colors. Adv. Funct. Mater. 2018;28(43):1804628. [Google Scholar]
- 9.Hou J., Li M.Z., Song Y.L. Patterned colloidal photonic crystals. Angew. Chem. Int. Ed. 2018;57(10):2544–2553. doi: 10.1002/anie.201704752. [DOI] [PubMed] [Google Scholar]
- 10.Fu F.F., Shang L.R., Chen Z.Y., Yu Y.R., Zhao Y.J. Bioinspired living structural color hydrogels. Sci. Robot. 2018;3(16) doi: 10.1126/scirobotics.aar8580. eaar8580. [DOI] [PubMed] [Google Scholar]
- 11.Arppe R., Sorensen T.J. Physical unclonable functions generated through chemical methods for anti-counterfeiting. Nat. Rev. Chem. 2017;1(4) UNSP 0031. [Google Scholar]
- 12.Martin-Palma R.J., Lakhtakia A. Progress on bioinspired, biomimetic, and bioreplication routes to harvest solar energy. Appl. Phys. Rev. 2017;4(2) [Google Scholar]
- 13.Zhang Y.Q., Fu Q.Q., Ge J.P. Photonic sensing of organic solvents through geometric study of dynamic reflection spectrum. Nat. Commun. 2015;6:7510. doi: 10.1038/ncomms8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu H.C., Zhao Y.J., Cheng Y., Xie Z.Y., Rong F., Li J.Q., Wang B.P., Fu D.G., Gu Z.Z. Tailoring colloidal photonic crystals with wide viewing angles. Small. 2013;9(13):2266–2271. doi: 10.1002/smll.201203179. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y., Kim H., Xu J.P., Hwang M.S., Tian D., Wang K., Zhang L.B., Liao Y.G., Park H.G., Yi G.R., Xie X.L., Zhu J.T. Responsive block copolymer photonic microspheres. Adv. Mater. 2018;30(21):1707344. doi: 10.1002/adma.201707344. [DOI] [PubMed] [Google Scholar]
- 16.Dong R.H., Liu Y., Mou L., Deng J.Q., Jiang X.Y. Microfluidics-based biomaterials and biodevices. Adv. Mater. 2019;31(45) doi: 10.1002/adma.201805033. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y.J., Chan Y.K., Lau L.H., Chao Y.C., Shih K.C., Lai S.M., Wong D., Shum H.C. Adhesion of silicone oil and emulsification: an in vitro assessment using a microfluidic device and 'eye-on-a-chip'. Acta Ophthalmol. 2019;97(3):313–318. doi: 10.1111/aos.13982. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X.X., Zhao Y.J. Wearable droplet microfluidics. Sci. Bull. 2019;64(20):1472–1473. doi: 10.1016/j.scib.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Ma X.D., Ozliseli E., Zhang Y.Z., Pan G.Q., Wang D.Q., Zhang H.B. Fabrication of redox-responsive doxorubicin and paclitaxel prodrug nanoparticles with microfluidics for selective cancer therapy. Biomater. Sci. 2019;7(2):634–644. doi: 10.1039/c8bm01333k. [DOI] [PubMed] [Google Scholar]
- 20.Choi T.M., Lee G.H., Kim Y.S., Park J.G., Hwang H., Kim S.H. Photonic microcapsules containing single-crystal colloidal arrays with optical anisotropy. Adv. Mater. 2019;31(18):1900693. doi: 10.1002/adma.201900693. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y.S., Wang H., Chen B.A., Liu H., Zhao Y.J. Emerging barcode particles for multiplex bioassays. Sci. China Mater. 2019;62(3):289–324. [Google Scholar]
- 22.Wang K., Li C., Li Z., Li H.Z., Li A., Li K.X., Lai X.T., Liao Q., Xie F., Li M.Z., Song Y.L. A facile fabrication strategy for anisotropic photonic crystals using deformable spherical nanoparticles. Nanoscale. 2019;11(30):14147–14154. doi: 10.1039/c9nr04735b. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X.X., Chen G.P., Bian F.K., Cai L.J., Zhao Y.J. Encoded microneedle arrays for detection of skin interstitial fluid biomarkers. Adv. Mater. 2019;31(37):1902825. doi: 10.1002/adma.201902825. [DOI] [PubMed] [Google Scholar]
- 24.Choi T.M., Je K., Park J.G., Lee G.H., Kim S.H. Photonic capsule sensors with built-in colloidal crystallites. Adv. Mater. 2018;30(43):1803387. doi: 10.1002/adma.201803387. [DOI] [PubMed] [Google Scholar]
- 25.Wang H., Zhao Z., Liu Y.X., Shao C.M., Bian F.K., Zhao Y.J. Biomimetic enzymes cascade reaction system in microfluidic electrospray microcapsules. Sci. Adv. 2018;4(6) doi: 10.1126/sciadv.aat2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi Y.H., Lee S.S., Lee D.M., Jeong H.S., Kim S.H. Composite microgels created by complexation between polyvinyl alcohol and graphene oxide in compressed double-emulsion drops. Small. 2020;16(9):1903812. doi: 10.1002/smll.201903812. [DOI] [PubMed] [Google Scholar]
- 27.Wang J., Chen G.P., Zhao Z., Sun L.Y., Zou M.H., Ren J.A., Zhao Y.J. Responsive graphene oxide microcarriers for controllable cell capture and release. Sci. China. Mater. 2018;61(10):1314–1324. [Google Scholar]
- 28.Cheng C.L., Cai Y.Q., Guan G.J., Yeo L., Wang D.Y. Hydrophobic-force-driven removal of organic compounds from water by reduced graphene oxides generated in agarose hydrogels. Angew. Chem. Int. Ed. 2018;57(35):11177–11181. doi: 10.1002/anie.201803834. [DOI] [PubMed] [Google Scholar]
- 29.Shao C.M., Liu Y.X., Chi J.J., Wang J., Zhao Z., Zhao Y.J. Photo-controllable inverse opal graphene hydrogel scaffolds with biomimetic enrichment capability for cell culture. Research. 2019;2019:9783793. doi: 10.34133/2019/9783793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee H., Dellatore S.M., Miller W.M., Messersmith P.B. Mussel-inspired surface chemistry for multifunctional coatings. Science. 2007;318(5849):426–430. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu P.M., Chen J.L., Zhang Z.X., Xie Z.Y., Du X., Gu Z.Z. Bio-inspired robust non-iridescent structural color with self-adhesive amorphous colloidal particle arrays. Nanoscale. 2018;10(8):3673–3679. doi: 10.1039/c7nr08056e. [DOI] [PubMed] [Google Scholar]
- 32.Li J., Sun C.L., An P.R., Liu X.Y., Don R.H., Sun J.H., Zhang X.Y., Xie Y.B., Qin C.G., Zheng W.F., Zhang H.L., Jiang X.Y. Construction of dopamine-releasing gold surfaces mimicking presynaptic membrane by on-chip electrochemistry. J. Am. Chem. Soc. 2019;141(22):8816–8824. doi: 10.1021/jacs.9b01003. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y.X., Shao C.M., Wang Y., Sun L.Y., Zhao Y.J. Bio-inspired self-adhesive bright non-iridescent graphene pigments. Matter. 2019;1(6):1581–1591. [Google Scholar]
- 34.Eliason C.M., Bitton P.P., Shawkey M.D. How hollow melanosomes affect iridescent colour production in birds. Proc. Biol. Sci. 2013;280(1767):20131505. doi: 10.1098/rspb.2013.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao M., Hu Z.Y., Wang Z., Li Y.W., Tormo A.D., Le Thomas N., Wang B.X., Gianneschi N.C., Shawkey M.D., Dhinojwala A. Bioinspired bright noniridescent photonic melanin supraballs. Sci. Adv. 2017;3(9) doi: 10.1126/sciadv.1701151. UNSP e1701151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen T., Li M.X., Liu J.Q. Pi-pi stacking interaction: a nondestructive and facile means in material engineering for bioapplications. Cryst. Growth Des. 2018;18(5):2765–2783. [Google Scholar]
- 37.Bian F.K., Wu J.D., Wang H., Sun L.Y., Shao C.M., Wang Y., Li Z.Y., Wang X.H., Zhao Y.J. Bio-inspired photonic barcodes with graphene oxide encapsulation for multiplex microrna quantification. Small. 2018;14(52):1803551. doi: 10.1002/smll.201803551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.