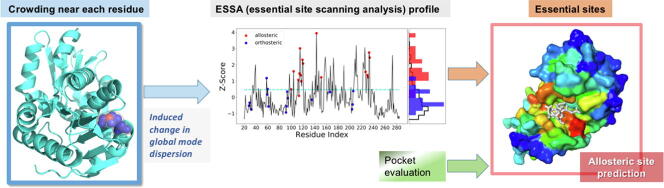
Graphical abstract
Keywords: Elastic network model, Functional dynamics, Essential sites, Allostery, Druggability, Protein–ligand interaction, Global hinge, Allosteric site prediction
Abstract
Despite the wealth of methods developed for exploring the molecular basis of allostery in biomolecular systems, there is still a need for structure-based predictive tools that can efficiently detect susceptible sites for triggering allosteric responses. Toward this goal, we introduce here an elastic network model (ENM)-based method, Essential Site Scanning Analysis (ESSA). Essential sites are here defined as residues that would significantly alter the protein’s global dynamics if bound to a ligand. To mimic the crowding induced upon substrate binding, the heavy atoms of each residue are incorporated as additional network nodes into the α-carbon-based ENM, and the resulting shifts in soft mode frequencies are used as a metric for evaluating the essentiality of each residue. Results on a dataset of monomeric proteins indicate the enrichment of allosteric and orthosteric binding sites, as well as global hinge regions among essential residues, highlighting the significant role of these sites in controlling the overall structural dynamics. Further integration of ESSA with information on predicted pockets and their local hydrophobicity density enables successful predictions of allosteric pockets for both ligand-bound and -unbound structures. ESSA can be efficiently applied to large multimeric systems. Three case studies, namely (i) G-protein binding to a GPCR, (ii) heterotrimeric assembly of the Ser/Thr protein phosphatase PP2A, and (iii) allo-targeting of AMPA receptor, demonstrate the utility of ESSA for identifying essential sites and narrowing down target allosteric sites identified by druggability simulations.
1. Introduction
Experimental studies including NMR [1], inelastic neutron scattering [2], hydrogen exchange mass spectroscopy [3], and femtosecond optical Kerr-effect spectroscopy [4] provide insights into the effects of ligand binding on protein structure and dynamics, including the conformational flexibility and fluctuations of the protein. Features of fluctuation dynamics, akin to vibrational modes of motions in molecular systems, can be computationally characterized by normal mode analyses (NMAs) [5] and experimentally observed by inelastic neutron scattering [2], optical microscopy [6] and extraordinary acoustic Raman spectroscopy [7], [8]. Of interest is the lowest frequency end of the vibrational spectrum, which is usually populated by modes that collectively engage large substructures, if not the entire structure, at a relatively low energetic cost, hence named as global modes or soft modes. NMAs using elastic network models (ENMs) [9], [10], [11] have shown that soft modes usually guide/facilitate the cooperative transitions between the apo and holo states of allosteric proteins [12], [13], [14]. This observation led to increased interest in exploring the soft modes and their alterations, toward rational design of allosteric modulators.
The X-ray crystallographic data deposited in the Protein Data Bank [15] contain information, not only on mean coordinates, but also on the fluctuations in the atomic positions of individual residues as well as the anisotropy of these fluctuations [16], [17], [18]. These fluctuations, known as Debye-Waller/B-factors, generally depend on the resolution of the structure [19], the crystallization temperature [20], [21], and crystal contacts that may bias the measurements [22], [23]. In addition to fluctuation dynamics, crystallographic data can indirectly provide information on large-scale conformational variability, reflected by the structures resolved for the same protein (or its structural homologues) under different conditions [14], [20], [21], [24], e.g. in ligand-bound or -unbound states, or at different stages of an allosteric cycle. Previous studies have shown that such conformational variabilities can be satisfactorily described by the global modes via ENM analysis of a representative conformer from the ensemble [13], [14], [25]. The time scale of these global motions is of the order of 100 s of nanoseconds or longer, depending on the size of the protein, as comparisons with microseconds molecular dynamics (MD) simulations have shown [26], [27].
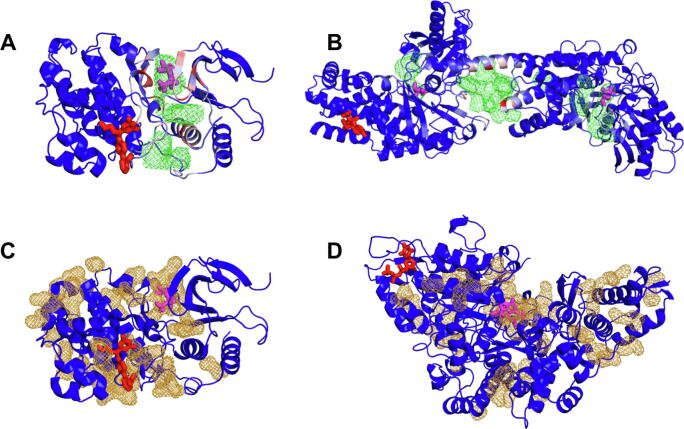
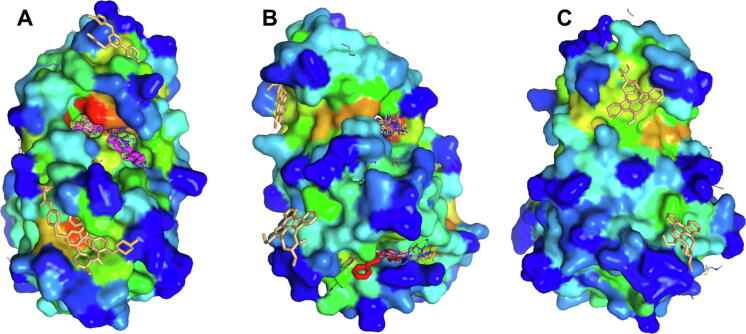
Structure-based computational studies for uncovering allosteric communication mechanisms and predicting allosteric sites utilize a plethora of models and methods [28], [29], [30]. Among them, several stand out using ENMs [31], [32], [33], [34], [35], MD simulations [36], [37] and graph theory [38]. Studies that combine ENM-based features with machine learning methods have shown significant success in identifying allosteric sites [39], [40], [41]. Most approaches for detecting allosteric binding sites require a priori information on cavities and/or the orthosteric site in assigning a quantitative measure, or score, to possible sites. Cavity detection methods have shown considerable success for identifying small molecule binding sites. Yet, not all allosteric sites observed in experiments are captured by these methods. Fig. 1 illustrates a few such cases. Therein, the pockets predicted by ConCavity [42] and Fpocket [43], two useful and widely used software packages for detecting cavities/pockets that potentially bind small ligands, are shown. Panels A and B, generated for two kinases, Jnk-1 (PDB id: 3o2m [44]) from the mitogen-activated protein kinase (MAPK) family and hexokinase I (PDB id: 1cza [45]), respectively, show that the cavities predicted by ConCavity (in green meshes) do accurately include the binding site for the orthosteric ligand (magenta sticks) but do not account for the binding site of the allosteric ligand (red sticks in both panels). Panels C and D show that the pockets predicted by Fpocket (gold meshes) account for the binding sites of the orthosteric and allosteric ligands of Jnk-1 (panel C), but not the allosteric ligand binding site of ribonucleotide reductase R1 (PDB id: 4r1r [46]) (panel D). Protein-protein interactions (PPI) interfaces are even more challenging to accurately detect due to their relatively large size and flat surface properties.
Fig. 1.
Comparison of computationally predicted ligand-binding pockets with known ligand-binding sites. Pockets identified by ConCavity(A-B) and Fpocket (C-D) are displayed by green and gold meshes, respectively, and the allosteric (red) and orthosteric (magenta) ligands are shown in sticks. (A) The orthosteric ligand of Jnk-1 (PDB id: 3o2m) coincides with the meshes; whereas the allosteric ligand does not. (B) Similarly, the allosteric ligand in hexokinase I (PDB id: 1cza) cannot be captured by the meshes, in contrast to the two orthosteric ligands (magenta sticks) that fall into two distant pockets. (C) Pockets detected by Fpocket for Jnk-1 (as in panel A) contact the allosteric ligand on both sides but do not fully enclose it. (D) No pocket is detected around the allosteric ligand of ribonucleotide reductase R1 (PDB id: 4r1r). The ligand binds to a mobile loop that is stabilized it in the complex. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We recently developed a residue-specific ENM [47] to assess the specific effect of ligand binding on the vibrational spectrum uniquely accessible to each protein [48]. Application of this method to a dataset of 315 proteins resolved in ligand-bound form revealed an increase in global modes’ frequencies upon ligand binding, for 87% of the examined proteins. This increase was observed by evaluating the mode spectrum of the liganded protein in comparison to that of its holo form, the latter being generated by removing the ligand(s) in silico. These positive shifts in frequencies were attributed to a narrowing of the harmonic well along soft modes upon ligand binding. In addition to allosteric site residues, several other residues exhibited a high sensitivity to substrate/drug-binding. Our ENM analysis of molecular dynamics (MD) trajectories generated for triosephosphate isomerase [49] also pointed to constraints on the functional modes of the enzyme experienced in the presence of an allosteric inhibitor. There is a need to systematically assess such sites that could be selectively targeted to effectively alter the global dynamics of the target protein.
Motivated by these studies that exposed the structure-encoded sensitivity of global dynamics to ligand binding at functional sites, we present here the new ENM-based methodology, the so-called Essential Site Scanning Analysis (ESSA). Global modes of motion have been traditionally defined [50] as ‘essential dynamics’. Here, essential residues are defined as those that alter the essential/global dynamics if bound to a ligand. ESSA serially probes the potential of all residues for altering the frequency dispersion of global modes in response to possible ligand binding. This is mimicked by increasing the local density of the network in the vicinity of a scanned residue, which is achieved by addition of extra nodes at its heavy atoms’ loci, on top of the original coarse-grained network. We aim to answer the following questions in this study: Can we detect, using ESSA, a set of residues that can play key roles in altering the global dynamics of proteins? Do these key residues occupy structural loci that correspond to ligand binding sites, including PPI sites? Can we use the outputs from ESSA to narrow down the multiple sites identified by druggability simulations to filter those with the strongest impact on structural dynamics, if targeted? A critical assessment of the functional significance of ESSA outputs will indeed disclose the essential role of ESSA-predicted residues in binding allosteric or orthosteric ligands, serving as key mechanical sites, or mediating PPIs. Furthermore, a new protocol that integrates Fpocket outputs with ESSA is shown to be successful in allosteric site prediction.
2. Materials and methods
2.1. Datasets
We first formed a dataset of 25 monomeric proteins by merging two curated datasets [31], [32], which are composed of crystal structures resolved in the presence of allosteric and orthosteric ligands. Supplementary Table S1 describes this so-called Dataset I. The table lists the PDB ids of the complexes (1st column), the name of the protein (2nd column), and the corresponding allosteric and orthosteric ligands (columns 3 and 4). In 17 of the examined proteins, both types of ligands were bound to the protein. The remaining eight had a ligand bound to either the orthosteric or the allosteric site. In those cases, we used the structural data resolved for the same protein in the presence of the 2nd type of ligand (see the corresponding PDB id and root-mean-square deviation (RMSD) from the original complex) to reconstruct a ternary complex with both ligands. Four of the proteins contained multiple ligands, summing up to a total of 28 allosteric and 26 orthosteric ligand-binding sites in the dataset.
In addition, we considered a second dataset, Dataset II, composed of 24 structures, mainly the bound and unbound forms of 12 proteins from the Dataset I, for which there was an apo structure resolved for the same protein with at least 90% sequence identity. Supplementary Table S2 lists the PDB ids of the ligand-bound proteins (top portion) and their apo counterparts (bottom portion). We used this dataset for further assessing the performance of ESSA for prediction.
2.2. Generation of ESSA profiles
We conducted the ESSA using two widely used ENMs, namely the Gaussian network model (GNM) [51], [52], [53] and the anisotropic network model (ANM) [54], [55], [56] with the ProDy [57] default parameters for inter-residue contact threshold distances (respective cutoff radii of 10 and 15 Å) between node pairs connected by elastic springs. Both network models, GNM and ANM, identify the positions of network nodes with those of the Cα- atoms, and use uniform force constants of 1 kcal/(mol.Å2) for all the springs of the network. These models yield a spectrum of N-1 (GNM) or 3 N-6 (ANM) nonzero modes of harmonic motions for a protein of N residues. Each mode is characterized by a frequency and mode shape described by the respective eigenvalue, and eigenvector, , of the Kirchhoff (GNM) or Hessian (ANM) matrices describing the network topology. We refer to these quantities as the reference frequencies and mode shapes (in the unbound state).
To mimic the local ‘crowding’ induced by ligand binding near residue i, we populated the vicinity of residue i by adding extra nodes at the positions of all its heavy atoms (other than the α-carbon that already exists in the reference network). The extra nodes were treated as ‘environment’ and ESSA was performed for the system (ENM composed of N nodes in the presence of this environment/perturbation) using the reduced model approach [58] implemented in ProDy [57]; and repeated for all residues 1 ≤ i ≤ N. Extra nodes and their interactions were treated using the same parameters as those of the original/reference network. We evaluated the percent shift in the eigenvalue of the global mode in response to the crowding near residue i using
| (2.2.1) |
Note that the order of the modes may differ in the reference and perturbed network, and the difference in Eq. (2.2.1) is evaluated after reordering the modes to select the best-matching modes between the two networks (using the ProDy function ‘matchModes’). In order to increase the robustness of the results, an effective shift in global mode frequencies is defined for each residue as the mean over the softest ten modes (1 ≤ ≤ 10), and a z-score (is assigned to quantify the effect of ligand binding near residue on global dynamics
| (2.2.2) |
Here and denote the respective mean and the standard deviation of over all residues. The plot of zi - scores as a function of residue index i is referred to as the ESSA profile. We then analyze whether this profile obtained in the absence of ligands can distinguish ligand-binding residues (within 4.5 Å atom–atom distance from the ligand) for the complexes in our dataset (see 3.1, 3.2).
2.3. Protocol for ESSA-based prediction of allosteric sites
We developed a new protocol for predicting allosteric sites in apo structures, as well as complexes after removing bound ligands in silico. Here, we first determine the pockets of the specific structure using Fpocket [43] with its default parameters. Secondly, an ESSA score is assigned to each pocket, which corresponds to the median (or the maximum) of the z-scores of the residues lining that pocket. Then, we rank-order the pockets based on these scores. As allosteric sites usually have relatively higher hydrophobic density [40], we use this feature, provided by Fpocket, for further screening. For this step, we determine a local hydrophobic density (LHD) z-score for each pocket based on all the pockets detected in the structure and then retain only those with positive LHD z-scores. In the case of very close ESSA values, we use LHD to refine the ranking further. As a result of these steps, all pockets are rank ordered in terms of their allosteric potential.
3. Results and discussion
3.1. Essential residues identified by ESSA to influence global dynamics are enriched in ligand-binding sites
We performed an ESSA for each protein in our Dataset I. Mainly, we evaluated the change in the eigenvalue dispersion of global modes as a metric for probing essential sites that may elicit considerable alterations in dynamics if bound to a ligand.
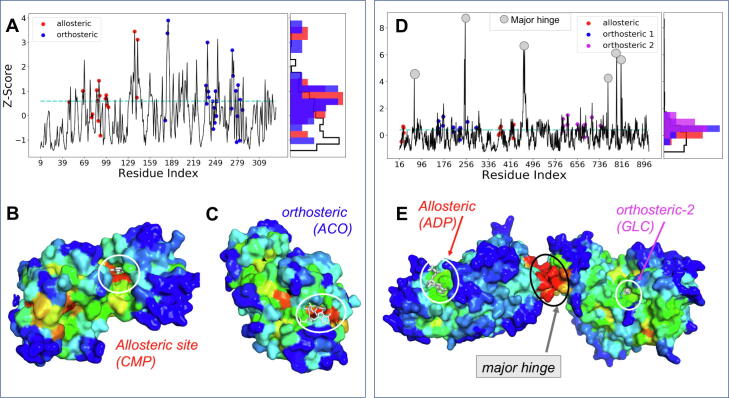
Fig. 2A-C presents the results for lysine acetyltransferase, an allosteric enzyme (PDB id: 4avb [59]). Panel A displays the corresponding ESSA profile, and panels B and C show its 3D structure viewed from two different perspectives to view its allosteric and orthosteric sites. The surface of the protein is color-coded by ‘essentiality’ (zi -score) predicted by ESSA for each residue i from blue (lowest) to red (highest). Both allosteric and orthosteric sites are detected as essential sites (colored red) in panels B and C. The histogram on the right ordinate (panel A) shows the individually normalized distributions (densities) for allosteric, orthosteric and the remaining residues.
Fig. 2.
ESSA highlights the essential sites of lysine acetyltransferase (A-C) and hexokinase I (D-E). (A) z-scores calculated for lysine acetyltransferase (PDB id: 4avb) give a measure of the extent of frequency shift in the global modes induced by each residue (abscissa). Highest values refer to those sites inducing the highest shift, if targeted by a ligand. Allosteric sites/residues (within 4.5 Å from any ligand atom) are indicated by red circles, and orthosteric sites by blue circles. The horizontal line (dashed cyan) indicates the top quartile. The individually normalized distributions of z-scores for allosteric (red bars), orthosteric (blue bars) and other (black contour, no color) residues are shown along the right ordinate. (B-C). Lysine acetyltransferase viewed from two different perspectives, color-coded by z-scores from red (highest) to blue (lowest). The diagrams display (B) the allosteric ligand and (C) the orthosteric ligand, both in white sticks (enclosed in white circles). (D-E) Similar results for hexokinase I (PDB id: 1cza), bound to one allosteric (red circles) and two orthosteric ligands (blue and magenta circles). We note in this case sharp peaks in the ESSA profile corresponding to the global hinge center (gray circles in D, central red region in E). Results are obtained using the GNM in ESSA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As a quantitative analysis, we rank-ordered the residues based on their z-scores and examined those lying in the top quartile (top 25%, zi > 0.59), which is separated by the cyan dashed line in panel A. Residues known to coordinate one or more allosteric ligands (indicated by red circles in panel A) stand out in terms of their ability to shape the global dynamics. 59% of those residues lies in this upper quartile. Likewise, 48% of those binding the orthosteric ligand ACO lie in the top quartile of the zi -scores, in support of the enrichment of both allosteric and orthosteric sites among essential residues detected by ESSA.
The examination of the entire dataset will show below that allosteric and orthosteric sites generally exhibit higher zi-scores than the rest of the protein, in line with their dominant role in defining functional dynamics. However, we note that the zi -scores vary over a broad range (as could be seen in the histograms on the right ordinate of Fig. 2A) and not all proteins’ allosteric or orthosteric sites are distinguished by high scores. For example, the orthosteric site residues of glutamate racemase (PDB id: 2jfn [60]) are observed to undergo minimal changes in their global dynamics upon ligand binding, as illustrated in Supplementary Fig. S1 panels A-C, while. In contrast, 94% of residues that bind the allosteric ligand of glutamate racemase lie in the top quartile (zi > 0.47) in this case, in support of the evolutionary selection of allosteric sites from amongst the residues that possess a strong structure-encoded ability to modulate the global dynamics. Likewise, Fig. S1 shows that the allosteric site of Jnk-1 (not detected by ConCavity in Fig. 1A) is clearly distinguished by the peaks in the ESSA profile (panel D) as well as the color-coded diagram (panel E), while the zi-scores for the orthosteric site indicate a moderate effect (panel F).
3.2. Global hinges of multidomain proteins are distinguished as essential sites
In addition to allosteric and orthosteric sites, previous studies have shown that the global hinge centers play a critical role in mediating the global dynamics and are often evolutionarily exploited due to their essential mechanical role [61], [62], [63]. We examined whether such centers were detected by ESSA.
Fig. 2D-E presents the results for hexokinase I, a two-domain enzyme also illustrated in Fig. 1B. This enzyme contains a central hinge at the interface between two domains, and three sites of interest, one allosteric and two orthosteric (panel E). Hinge site residues are distinguished by high peaks in panel A, consistent with their key mechanical roles mediating the relative movements of the two domains. Furthermore, careful examination shows that 42% of allosteric- and 73 and 80% of the two orthosteric ligand-binding residues lie in the top quartile of rank-ordered list of essential residues (z-score > 0.39).
The case studies in subsections 3.1 and 3.2 show that allosteric and orthosteric sites, as well as key mechanical sites, tend to lie among those identified by ESSA to be essential to modulating global modes. Yet, individual cases show differences, e.g. allosteric residues are distinguished in glutamate racemase (Fig. S1A); whereas key mechanical residues overshadow others in hexokinase I (Fig. 2D). Toward understanding the general behavior, we undertake a systematic study of all proteins in Dataset I, next.
3.3. Dataset analysis points to the propensity of ligand-binding sites to alter the dispersion of global modes
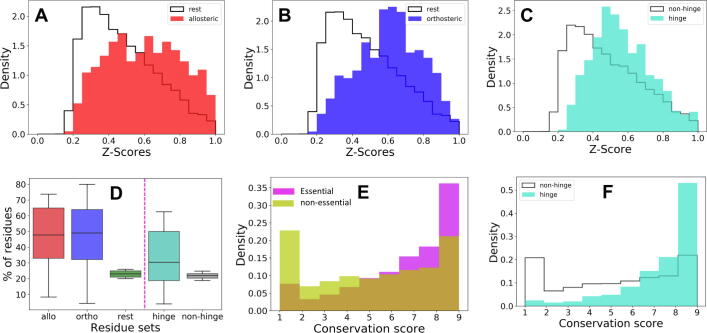
We performed a systematic ESSA for all proteins in the Dataset I. As described in Methods this dataset includes monomeric proteins with known allosteric- and orthosteric-ligand binding loci compiled in earlier datasets [31], [32], summarized in Table S1. Our analysis led to the results presented in Fig. 3. The z-scores are convoluted therein with a sigmoid function,, to map them into the interval [0,1] and obtain the cumulative distributions for the dataset.
Fig. 3.
Statistical analyses point to the enrichment of ligand-binding and hinge residues among the sites predicted by ESSA to be essential. Comparison of the z-score histograms obtained for (A) allosteric-ligand binding sites (red bars) and (B) orthosteric-ligand binding sites (blue bars), compared to that of the rest of the residues that do not interact with the ligands (black contour). The z-scores have been normalized to lie in the interval [0,1] for all 25 proteins in the dataset. (C) Z-score histograms comparing hinge and non-hinge sites/residues. (D) Box plots showing the percentage of ligand-binding residues and hinge residues in the upper quartile of z-scores for each protein. Allo and ortho-ligand binding residue sets are more populated in the upper quartile compared to the rest of the residues, i.e. they induce more pronounced effects on protein dynamics. (E) Distributions of conservation scores for the essential residues (in the top quartile of z-scores) obtained with ESSA (magenta) and the remaining (yellow). (F) Conservation scores of hinge residues predicted by the GNM based on softest 10 modes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In Fig. 3 panels A and B, the normalized distributions of z-scores are shown for three non-overlapping residue sets, namely those occupying allosteric sites (panel A) and those at the orthosteric sites (panel B) compared to the rest of the residues that do not interact with these specific ligands. We clearly observe a relatively high population of ligand-binding residues in the high z-score region for both allosteric and orthosteric sites. The respective mean values (and standard deviations) of the z-scores for the sets of allosteric, orthosteric, and rest are 0.60 ± 0.21, 0.61 ± 0.18, and 0.48 ± 0.19.
We also examined the z-score histogram for residues participating in hinge centers (shortly called hinge residues) evaluated for all dataset proteins using ProDy. We determined the hinge residues by calculating the residue mean-square fluctuations (MSFs) driven by the softest ten GNM modes and selecting those residues (N/10 of them for each protein) exhibiting the lowest MSFs. Fig. 3C compares the histograms for hinge residues and non-hinge residues. Even though some hinge residues are located at highly flexible linkers such that the effect of increased crowding in their vicinity would be small, we still observe an overrepresentation of hinge sites among essential (high z-scoring) residues consistent with their overall mechanical role: the mean z-scores and standard deviations are 0.56 ± 0.17 and 0.48 ± 0.20 for the respective hinge and non-hinge subsets.
As an additional assessment of the identification of essential residues by ESSA, we calculated the percentage of residues in each set that have z-scores lying in the top quartile. Fig. 3D shows the results in box plots for all residues belonging to three sets (a total of 421 and 444 residues in allosteric and orthosteric sites, and 9,017 residues in the rest) on the left of the dashed pink line. Red and blue boxes clearly demonstrate that the respective allosteric- and orthosteric-ligand binding sites populate this top quartile, and therefore have an impact on global modes. In contrast, the remaining residues, shown in green box, have percentages sharply located around 24%. We also display the results for hinge residues (a total of 976 residues) on the right side, which also reveals the enrichment of essential residues at hinge centers, although this effect is not as strong as that of allosteric and orthosteric sites.
Finally, we examined whether the essential residues identified by ESSA tend to be evolutionarily conserved. We evaluated the conservation scores of the two mutually exclusive subsets of residues: those with z-scores in the top quartile (2,472 of them) and other residues (7,386 of them), using ConSurf [64]. As can be seen in the normalized distributions of conservation scores (where the upper limit refers to fully conserved sites) displayed in Fig. 3E, essential residues (magenta bars) exhibit a higher propensity to be evolutionary conserved than others (yellow bars). Similarly, Fig. 3F shows that the hinges are significantly more conserved than the non-hinges.
Overall, this analysis demonstrates that ligand-binding residues, orthosteric or allosteric, are selected amongst those essential sites that possess a relatively higher propensity to elicit global changes in protein dynamics. Likewise, residues participating in hinge centers generally colocalize with essential sites. ESSA emerges as a simple, efficient tool for a quick assessment of the potential functional role of residues as ligand-binding or hinge sites in any protein, which could be readily exploited in more extensive, structural-proteome scale analyses, including multimeric complexes.
3.4. ESSA integrated with Fpocket successfully predicts allosteric sites
To assess whether ESSA scores could be utilized for detecting allosteric pockets, we developed the new protocol described in Section 2.3 and applied it to the Dataset II of 12 proteins that have been resolved in both apo and holo forms (Table S2). Below we present the results from our comparative analysis and discuss the performance of this protocol.
3.4.1. Comparison with other methods
Results from ESSA-based evaluation are summarized in Table S2 along with those from two other ENM-based tools available online, PARS [33] and AllositePro [40]. These servers do not require prior input on orthosteric site(s), which is also the case in our algorithm. We chose a stringent criterion for ESSA-based predictions, such that only the allosteric sites detected in the top 3 predictions among all pockets are labelled as successful. Note that the total number of pockets may be up to 50 in some proteins. Calculations performed using the holo structure (in which the ligand has been removed in silico) as input showed that 10 out of 14 allosteric sites present in Dataset II are successfully predicted by ESSA, as opposed to the 2/14 and 8/14 success rates of PARS and AllositePro, respectively. More importantly, in the more challenging test of using apo structures as input, ESSA shows 7/14 success rate, which is well above those (both 2/14) of PARS and AllositePro.
3.4.2. Cryptic sites
Conformational rearrangements in apo structures may result in cryptic pockets, i.e. those unavailable in the apo form, but becoming accessible upon conformational changes. In our dataset, there are four cryptic allosteric sites (see Table S2), which could not be detected by Fpocket and thus led to unsuccessful predictions. Not surprisingly, these sites could not be detected by the other two tools either. We examined the possibility of capturing such sites upon repeating the analysis for alternative conformers sampled along the first three global modes, using our ClustENM algorithm [65]. The cryptic pockets of beta-lactamase (PDB id: 1zg4 [66]) and MAP kinase 14 (PDB id: 5uoj [67]) became exposed, leading to successful ESSA predictions. Fig. S2 displays the results for beta-lactamase.
3.4.3. Ligand bound to hinge site
Another unsuccessful case in our dataset was ribonuclease reductase R1 protein (PDB id: 4r1r [46]) in Fig. 1D, where the ligand binds to a hinge site at the base of a flexible loop, thereby stabilizing the loop in the complex. This site could not be predicted by any of the tools, as no pockets could be detected at this allosteric hinge site in either the apo or the holo structure resolved for this enzyme. We note, however, that the essentiality of this site is captured by the ESSA profile (Fig. S3). Interestingly, the use of ESSA alone, rather than combined ESSA/Fpocket protocol, would have been sufficient to unambiguously capture this allosteric site.
3.5. ESSA can detect multiple binding sites resolved in alternative structures
We would like to point to an inherent problem in the assessment of prediction algorithms based on current benchmarks or datasets, including the one in this study. In general, a single complex structure is used for each protein among those available in these datasets. However, there may be different binding sites that are resolved in alternative structures for the same protein or in homologous structures. Therefore, some pockets that appear to be ‘false positives’ may in fact be pointing to true positives, either observed in another structure or not resolved yet. In the following, we present two such examples from our dataset.
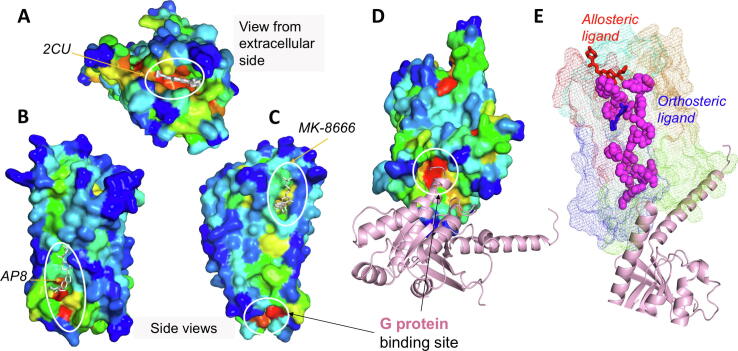
3.5.1 G-protein coupled receptors (GPCR) present multiple ligand-binding sites accounted for by ESSA. Alternative binding sites exist at different regions of GPCRs, including intracellular, extracellular and lipid-exposed (transmembrane) allosteric regions, as well as the buried orthosteric site [68]. The only GPCR in our dataset was the muscarinic acetylcholine receptor (PDB id: 4mqt [69]) resolved in the presence of an allosteric (2CU, LY2119620) and an orthosteric (iperoxo) ligand. Here, the allosteric site exposed to the extracellular medium (Fig. 4A), and the buried orthosteric site (not shown) stood out in terms of their ESSA scores. Furthermore, ESSA performed for another GPCR, the free fatty acid receptor 1 GPR40 (PDB id: 5tzy [70]), detected the lipid-facing allosteric sites at two different locations. Fig. 4B and C show the corresponding binding poses of agoPAM (AP8) and the partial agonist (MK-8666) in excellent agreement with ESSA-predicted hot spots. We also note the G protein binding site as an essential site, which will be revisited below.
Fig. 4.
ESSA correctly identifies the multiple ligand-binding sites on GPCRs. (A) ESSA results for muscarinic acetylcholine receptor (PDB id: 4mqt). The GPCR is color-coded by the ESSA profile. The extracellularly-exposed site that binds the allosteric ligand 2CU (white sticks) is correctly detected as an essential site (orange-to-red). (B-C) ESSA results for the free fatty acid receptor 1 GPR40. Two different views of the GPR40 complex (PDB id: 5tzy) show allosteric ligands in the transmembrane region, namely (B) the full allosteric agonist AP8 and (C) the partial agonist MK-8666. The residues interacting with either ligand are predicted to significantly affect the global motions dispersion, if bound to those ligands. We note at the bottom another essential site strongly captured by ESSA, which is the G protein binding site. The latter is more clearly shown for muscarinic acetylcholine receptor in (D), where the GPCR is tilted to display its intracellular vestibule and its bound G protein (aligned from PDB id: 6oik). (E) The residues with top 5% BC score (magenta spheres) form a direct path between the extracellular and intracellular regions in muscarinic acetylcholine receptor, passing through the allosteric (red sticks) and orthosteric (blue sticks) ligand-binding sites, all the way to the G protein binding site. The mesh surface is color-coded to show the domains subject to coupled dynamics. 93% of top-scoring BC residues are highly conserved (ConSurf score > 7). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5.2c-Abl kinase, another highly promiscuous enzyme with experimentally proven ligand-binding sites captured by ESSA. Fig. 5 illustrates the results for c-Abl kinase (PDB id: 3pyy [71]). As can be seen, the essential sites include both the orthosteric (magenta) and allosteric (red, myristoylation) sites that proved to be effective in cancer treatment when administered in combination. Additionally, several other ligands bound to homologous structures retrieved from the PDB reside close to other high-scoring regions.
Fig. 5.
ESSA of c-Abl kinase points to sites whose simultaneous targeting proved effective. c-Abl kinase surface color-coded by z-scores (shown from three different perspectives using the PDB id: 3pyy. Panel (A-C) show the allosteric (red sticks in B; 3YY), orthosteric (magenta sticks in A, STI) and other ligands (light orange sticks) retrieved from homologous structures (all three panels) located at essential sites predicted by ESSA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Overall, this analysis suggests that some of the high-scoring regions which do not necessarily overlap with known ligand/drug-binding sites, could actually point to hot spots that have not yet been resolved in the presence of ligands by experiments, rather than being false positives, and those sites could provide valuable hypotheses for polypharmacological strategies.
3.6. Essential sites preferentially locate at PPI interfaces
We also examined whether the essential sites identified by ESSA for modulating protein dynamics could also play a role in mediating the interfacial sites between protein pairs.
3.6.1. GPCR
As a first example, we considered the interaction of the muscarinic acetylcholine receptor, with its cognate G protein. Interestingly, the G protein binding site is identified as an essential site (Fig. 4D), shown with the G-protein aligned from a more recent structure (PDB id: 6oik [72]). This study suggests that the sites susceptible to elicit cooperative changes in global modes upon binding substrates, including PPI sites, could be located by ESSA.
As a further analysis, inspired by a recent study [41], we investigated whether the loci of essential sites identified above could be further consolidated by performing a graph theoretical analysis of signal propagation across the GPCR. Essentially, we asked whether the significance of the identified sites could be further assessed by evaluating the betweenness centrality [73] (BC) of the GNM nodes that represent the GPCR. BC measures the importance of a node as a bridge in signal communication by calculating the fraction of shortest paths passing through that node. Strikingly, the residues with top 5% centrality scores (evaluated by NetworkX [74]) were found to form a path connecting the allosteric site to the G-protein binding site, also passing through the orthosteric site (Fig. 4E).
3.6.2. Protein phosphatase 2A (PP2A)
The Ser/Thr phosphatase PP2A is composed of a PR65 HEAT repeat scaffold (gray), a regulatory subunit (salmon) and a catalytic subunit (magenta), shown in Fig. 6A. In this hetero-trimeric PP2A holoenzyme (PDB id: 2iae [75]), the horseshoe-shaped scaffold interacts simultaneously with the catalytic and regulatory subunits, holding them in close proximity. The regulatory subunit also forms pseudo-HEAT repeats and interacts with the catalytic subunit near its active site. We performed a two-fold ESSA. First, we scanned the scaffold, and we found that the essential sites are populated in the interior surface of the horseshoe, with one of the prominent regions lying next to the catalytic subunit binding site (see Fig. 6B). Next, we scanned the catalytic subunit, which yielded as essential sites to three hot spots corresponding to the binding interfaces with the other subunits, shown by white arrows (Fig. 6C). The opposite view in panel D further shows that the peptide-binding region on the catalytic subunit is correctly identified.
Fig. 6.
Essential sites predicted for heterotrimeric PP2A holoenzyme. (A) The complex (PDB id: 2iae) is composed of the PR65 heat-repeat scaffold (gray), the catalytic subunit (magenta) and the regulatory subunit (salmon) with a peptide (orange sticks, within black circle) bound to catalytic subunit. (B) Scanning of the PR65 scaffold shows a hot spot at the catalytic unit binding site (within white circle). (C) Scanning of the catalytic subunit indicates hot spots for binding (white arrows) to the scaffold PR65 (gray surface) and regulatory (salmon surface) subunits, and (D) and another region (cyan) shown from a different perspective that coincides with the peptide-binding site (peptide in orange sticks). ESSA results were obtained using the ANM. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As protein–protein interfaces may cover large portions of the protein surface, it may be of interest to determine how discriminative the ESSA scores are in comparison to a randomized signal. Fig. S4 presents such a comparison of ESSA profiles (original vs. shuffled). The interfacial residues are populated on the peaks of the original ESSA profile and the color-coded diagram highlights the hot spots formed by spatially contiguous residues consistent with the known PPIs (panels A-C). No such properties can be observed upon shuffling the residues (panels D-F). Further investigation on a larger dataset will be pursued in order to determine the utility of ESSA in terms of PPIs.
3.7. ESSA assists in selecting target sites in combination with druggability simulations
In recent years significant efforts have focused on conducting druggability simulations which proved to be particularly useful for identifying potentially allosteric sites and estimating binding free energies and entropies [76], [77], [78]. They are MD simulations carried out in the presence of an ensemble of solute (or co-solvent) molecules which serve as chemical probes in an aqueous environment and help elucidate ‘druggable’ sites on the target protein surface while the protein is able to sample its conformational space. Prior studies showed that druggability trajectories point to numerous sites which could benefit from narrowing down with the help of additional tools that identify essential sites, especially for large complexes. We investigated the potential utility of ESSA for this aim. As will be presented next ESSA can be applied to multimeric systems (no modification required) with high computationally efficiency.
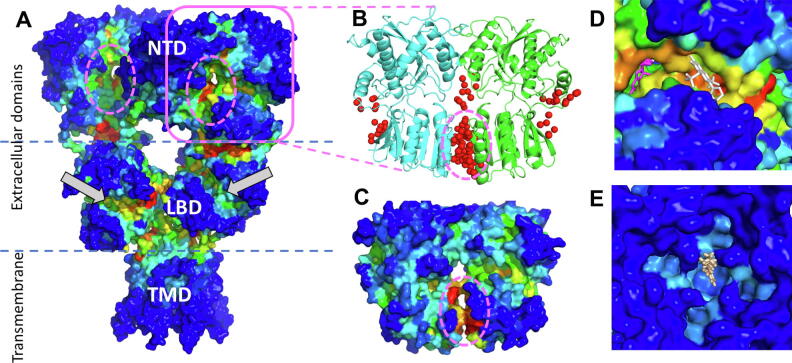
Application of ESSA to AMPA receptor (PDB id: 3kg2 [79]), a tetrameric complex composed of multiple domains (Fig. 7A), pointed to several essential sites located at domain interfaces (delimited by the dashed lines), as well as intra-domain regions. Notably, essential sites coincided with the ligand (glutamate) binding clefts of the dimers in the ligand-binding domain (LBD) (indicated by the gray arrows; see also panel D). It is interesting to note that the only region within the transmembrane domain (TMD) that showed a moderate signal coincided with the binding site for a channel blocker (panel E).
Fig. 7.
ESSA and druggability analysis of AMPA receptor. (A) AMPA receptor (PDB id: 3 kg2) color-coded by ESSA results. The tetrameric receptor is composed of two extracellular domains (N-terminal (NTD), ligand-binding (LBD)), a transmembrane domain (TMD) and a C-terminal domain (not resolved). Essential sites are at the interdomain boundaries and a few intradomain regions, such as the ligand-binding sites of the LBD (shown by gray arrows) and the interface of NTD forming dimers (enclosed in ellipses) (B) Druggable sites of NTD dimer (PDB id: 3o21, monomers colored green and cyan) from druggability simulations with various probes (red spheres). The dimer in (A) is shown within the magenta square. (C) ESSA results for the NTD dimer. Dashed ellipse encloses the essential residues predicted by ESSA. (D) Essential site predicted for the LBD domain has been observed to bind multiple ligands such as the competitive agonist ZK 200,775 (white sticks, PDB id: 3 kg2) and positive allosteric modulator cyclothiazide (magentas sticks, aligned from PDB id: 6dlz). (E) Channel blocker spider toxin analog NASPM (aligned from PDB id: 6 dm1) shown from the intracellular side. ESSA results were obtained using ANM. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The interfacial region between the dimers in the N-terminal domain (NTD; one of which is enclosed by magenta ellipse in Fig. 7A) is in excellent agreement with known ligand binding sites of the tetramer, and previous druggability results [80] for the GluA3 NTD dimer (PDB id: 3o21 [81]). Fig. 7B illustrates the results from these simulations, with the probes (shown in red spheres) preferentially binding at a few sites. Among them, those between the lower lobes of the monomers forming the NTD dimers has been experimentally shown to bind allosteric ligands. The latter stands out in ESSA (applied to both the dimer and the whole receptor in panels C and A, respectively) as an essential site for ligand binding.
This analysis demonstrates the utility of ESSA for selecting amongst multiple sites pointed by druggability simulations to be druggable. ESSA identifies those essential sites that impact the dynamics. Thus, combination of ESSA with druggability simulations would help identify druggable sites that also impact the dynamics. Alternatively, ESSA can be efficiently used for scanning large complexes/assemblies and identifying essential sites, whose druggability simulations may then be explored by more focused simulations using only the specific subunits. Such an approach seems valuable for narrowing down the alternatives for allo-targeting purposes.
4. Conclusion
In the current study, we present an efficient ENM-based scanning methodology, ESSA, for identifying essential sites of proteins, i.e. those playing a dominant role in defining the dispersion of global modes. This modification of the soft modes can originate from small molecule binding or PPIs. First, we illustrated the functional significance of such sites via a statistical analysis on a dataset of proteins. Allosteric and orthosteric ligand-binding residues as well as hinge residues were found to be populated in such key regions that induce cooperative effect on the protein dynamics.
Second, we designed an integrated protocol, which combined ESSA with Fpocket so as to make predictions on potential allosteric sites. Tests performed on a dataset including not only holo but also apo structures showed that the new approach outperformed two other ENM-based allosteric-site prediction algorithms, PARS and AllositePro. In the case of cryptic pockets observed in some apo structures, generation of new conformers using ClustENM [65] allowed us to identify transient pockets and detect associated allosteric sites using ESSA. The integration of ESSA and ClustENM has potential to study cryptic pockets at the proteome level.
Another utility of ESSA is the identification of multiple sites, which may provide insight for new ligand- or drug-binding sites for target proteins. Different allosteric binding sites of GPCRs such as those at the extracellular or lipid-facing regions as well as the orthosteric binding site simultaneously stand out in the ESSA profile. Likewise, different ligands bound to homologous structures of c-Abl kinase retrieved from the PDB were shown to reside close to high-scoring regions as well. This suggests that some of the high-scoring regions predicted by ESSA, which have not (yet) been experimentally confirmed, are not necessarily false positives. Simultaneous targeting of multiple essential sites could be exploited for polypharmacological purposes. Moreover, it is shown for AMPA receptor that ESSA can narrow down the set of druggable sites indicated by druggability simulations to those that impact the dynamics.
Furthermore, ESSA can help identifying epitopes for binding substrate proteins or assembling subunits, as presented for GPCR-G protein and trimeric PP2A subunits. This approach provides a unifying picture for identifying ligand-binding sites and effect on protein dynamics which needs to be further assessed on a dataset of protein–protein complexes.
Finally, ESSA lends itself to high-throughput analysis and could be applied at the structural proteome level due to its computational efficiency. A recent study demonstrated the utility of considering ENM-based structural dynamics in a machine learning algorithm for an improved assessment of the effect of point mutations (missense variants), neutral or deleterious, on protein function [82], [83]. It remains to be seen whether the ESSA profiles, possibly equipped with pocket information, could serve as additional ENM-based criteria for further improving the accuracy of such pathogenicity predictors.
CRediT authorship contribution statement
Burak T. Kaynak: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Writing - original draft, Visualization. Ivet Bahar: Conceptualization, Formal analysis, Writing - review & editing, Resources, Visualization, Supervision, Funding acquisition. Pemra Doruker: Conceptualization, Methodology, Validation, Investigation, Formal analysis, Investigation, Writing - original draft, Visualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
NIH grant P41 GM103712 and HFSP award RGP0027/2020 are gratefully acknowledged by IB.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.06.020.
Contributor Information
Burak T. Kaynak, Email: burak.kaynak@pitt.edu.
Ivet Bahar, Email: bahar@pitt.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Stone M.J. NMR relaxation studies of the role of conformational entropy in protein stability and ligand binding. Acc Chem Res. 2001;34(5):379–388. doi: 10.1021/ar000079c. [DOI] [PubMed] [Google Scholar]
- 2.Balog E., Becker T., Oettl M., Lechner R., Daniel R., Finney J. Direct determination of vibrational density of states change on ligand binding to a protein. Phys Rev Lett. 2004;93(2) doi: 10.1103/PhysRevLett.93.028103. [DOI] [PubMed] [Google Scholar]
- 3.Konermann L., Pan J., Liu Y.H. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev. 2011;40(3):1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 4.Turton D.A., Senn H.M., Harwood T., Lapthorn A.J., Ellis E.M., Wynne K. Terahertz underdamped vibrational motion governs protein-ligand binding in solution. Nat Commun. 2014;5:3999. doi: 10.1038/ncomms4999. [DOI] [PubMed] [Google Scholar]
- 5.Cui Q., Bahar I. Chapman & Hall; Boca Raton: 2006. Normal mode analysis : theory and applications to biological and chemical system. [Google Scholar]
- 6.Acbas G., Niessen K.A., Snell E.H., Markelz A.G. Optical measurements of long-range protein vibrations. Nat Commun. 2014;5:3076. doi: 10.1038/ncomms4076. [DOI] [PubMed] [Google Scholar]
- 7.DeWolf T., Gordon R. Theory of Acoustic Raman Modes in Proteins. Phys Rev Lett. 2016;117(13) doi: 10.1103/PhysRevLett.117.138101. [DOI] [PubMed] [Google Scholar]
- 8.Wheaton S., Gelfand R.M., Gordon R. Probing the Raman-active acoustic vibrations of nanoparticles with extraordinary spectral resolution. Nat Photon. 2015;9(1):68. [Google Scholar]
- 9.Tama F., Sanejouand Y.H. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001;14(1):1–6. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Bahar I., Lezon T.R., Yang L.W., Eyal E. Global dynamics of proteins: bridging between structure and function. Annu Rev Biophys. 2010;39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uyar A., Kantarci-Carsibasi N., Haliloglu T., Doruker P. Features of large hinge-bending conformational transitions. Prediction of closed structure from open state. Biophys J. 2014;106(12):2656–2666. doi: 10.1016/j.bpj.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinsen K. Analysis of domain motions by approximate normal mode calculations. Proteins. 1998;33(3):417–429. doi: 10.1002/(sici)1097-0134(19981115)33:3<417::aid-prot10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 13.Tobi D., Bahar I. Structural changes involved in protein binding correlate with intrinsic motions of proteins in the unbound state. Proc Natl Acad Sci USA. 2005;102(52):18908–18913. doi: 10.1073/pnas.0507603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakan A., Bahar I. The intrinsic dynamics of enzymes plays a dominant role in determining the structural changes induced upon inhibitor binding. Proc Natl Acad Sci USA. 2009;106(34):14349–14354. doi: 10.1073/pnas.0904214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein, F. C.; Koetzle, T. F.; Williams, G. J.; Meyer, E. F., Jr.; Brice, M. D.; Rodgers, J. R.; Kennard, O.; Shimanouchi, T.; Tasumi, M., The Protein Data Bank. A computer-based archival file for macromolecular structures. Eur J Biochem 1977, 80 (2), 319-24. [DOI] [PubMed]
- 16.Thorn A., Dittrich B., Sheldrick G.M. Enhanced rigid-bond restraints. Acta Cryst Section A. 2012;68(Pt 4):448–451. [Google Scholar]
- 17.Burden C.J., Oakley A.J. Anisotropic atomic motions in high-resolution protein crystallography molecular dynamics simulations. Phys Biol. 2007;4(2):79–90. doi: 10.1088/1478-3975/4/2/002. [DOI] [PubMed] [Google Scholar]
- 18.Eyal E., Chennubhotla C., Yang L.W., Bahar I. Anisotropic fluctuations of amino acids in protein structures: insights from X-ray crystallography and elastic network models. Bioinformatics. 2007;23(13):i175–i184. doi: 10.1093/bioinformatics/btm186. [DOI] [PubMed] [Google Scholar]
- 19.Schneider B., Gelly J.C., de Brevern A.G., Cerny J. Local dynamics of proteins and DNA evaluated from crystallographic B factors. Acta Cryst Section D. 2014;70(Pt 9):2413–2419. doi: 10.1107/S1399004714014631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tilton R.F., Jr., Dewan J.C., Petsko G.A. Effects of temperature on protein structure and dynamics: X-ray crystallographic studies of the protein ribonuclease-A at nine different temperatures from 98 to 320 K. Biochemistry. 1992;31(9):2469–2481. doi: 10.1021/bi00124a006. [DOI] [PubMed] [Google Scholar]
- 21.Fraser J.S., van den Bedem H., Samelson A.J., Lang P.T., Holton J.M., Echols N. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc Natl Acad Sci USA. 2011;108(39):16247–16252. doi: 10.1073/pnas.1111325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kundu, S.; Melton, J. S.; Sorensen, D. C.; Phillips, G. N., Jr., Dynamics of proteins in crystals: comparison of experiment with simple models. Biophys J 2002, 83 (2), 723-32. [DOI] [PMC free article] [PubMed]
- 23.Liu L., Koharudin L.M., Gronenborn A.M., Bahar I. A comparative analysis of the equilibrium dynamics of a designed protein inferred from NMR, X-ray, and computations. Proteins. 2009;77(4):927–939. doi: 10.1002/prot.22518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Townsend P.D., Jungwirth B., Pojer F., Bussmann M., Money V.A., Cole S.T. The crystal structures of apo and cAMP-bound GlxR from Corynebacterium glutamicum reveal structural and dynamic changes upon cAMP binding in CRP/FNR family transcription factors. PLoS ONE. 2014;9(12) doi: 10.1371/journal.pone.0113265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahar I., Lezon T.R., Bakan A., Shrivastava I.H. Normal mode analysis of biomolecular structures: functional mechanisms of membrane proteins. Chem Rev. 2010;110(3):1463–1497. doi: 10.1021/cr900095e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leioatts N., Romo T.D., Grossfield A. Elastic Network Models are Robust to Variations in Formalism. J Chem Theory Comput. 2012;8(7):2424–2434. doi: 10.1021/ct3000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gur M., Zomot E., Bahar I. Global motions exhibited by proteins in micro- to milliseconds simulations concur with anisotropic network model predictions. J Chem Phys. 2013;139(12) doi: 10.1063/1.4816375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greener J.G., Sternberg M.J. Structure-based prediction of protein allostery. Curr Opin Struct Biol. 2018;50:1–8. doi: 10.1016/j.sbi.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Wodak S.J., Paci E., Dokholyan N.V., Berezovsky I.N., Horovitz A., Li J. Allostery in Its Many Disguises: From Theory to Applications. Structure. 2019;27(4):566–578. doi: 10.1016/j.str.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Doruker P., Kaynak B., Zhang S., Krieger J., Li H. Intrinsic dynamics is evolutionarily optimized to enable allosteric behavior. Curr Opin Struct Biol. 2019;62:14–21. doi: 10.1016/j.sbi.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tee W.V., Guarnera E., Berezovsky I.N. Reversing allosteric communication: From detecting allosteric sites to inducing and tuning targeted allosteric response. Plos Comput Biol. 2018;14:(6). doi: 10.1371/journal.pcbi.1006228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X.M., Meng H., Lai L.H. Motions of Allosteric and Orthosteric Ligand-Binding Sites in Proteins are Highly Correlated. J Chem Inf Model. 2016;56(9):1725–1733. doi: 10.1021/acs.jcim.6b00039. [DOI] [PubMed] [Google Scholar]
- 33.Panjkovich A., Daura X. PARS: a web server for the prediction of Protein Allosteric and Regulatory Sites. Bioinformatics. 2014;30(9):1314–1315. doi: 10.1093/bioinformatics/btu002. [DOI] [PubMed] [Google Scholar]
- 34.Clarke D., Sethi A., Li S., Kumar S., Chang R.W.F., Chen J. Identifying Allosteric Hotspots with Dynamics: Application to Inter- and Intra-species Conservation. Structure. 2016;24(5):826–837. doi: 10.1016/j.str.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodgers T.L., Townsend P.D., Burnell D., Jones M.L., Richards S.A., McLeish T.C. Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors. PLoS Biol. 2013;11(9) doi: 10.1371/journal.pbio.1001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hacisuleyman A., Erman B. Entropy Transfer between Residue Pairs and Allostery in Proteins: Quantifying Allosteric Communication in Ubiquitin. Plos Comput Biol. 2017;13(1) doi: 10.1371/journal.pcbi.1005319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh S., Bowman G.R. Quantifying Allosteric Communication via Both Concerted Structural Changes and Conformational Disorder with CARDS. J Chem Theory Comput. 2017;13(4):1509–1517. doi: 10.1021/acs.jctc.6b01181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amor B.R., Schaub M.T., Yaliraki S.N., Barahona M. Prediction of allosteric sites and mediating interactions through bond-to-bond propensities. Nat Commun. 2016;7:12477. doi: 10.1038/ncomms12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greener J.G., Sternberg M.J. AlloPred: prediction of allosteric pockets on proteins using normal mode perturbation analysis. BMC Bioinf. 2015;16:335. doi: 10.1186/s12859-015-0771-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song K., Liu X., Huang W., Lu S., Shen Q., Zhang L. Improved Method for the Identification and Validation of Allosteric Sites. J Chem Inf Model. 2017;57(9):2358–2363. doi: 10.1021/acs.jcim.7b00014. [DOI] [PubMed] [Google Scholar]
- 41.Mishra S.K., Kandoi G., Jernigan R.L. Coupling dynamics and evolutionary information with structure to identify protein regulatory and functional binding sites. Proteins. 2019;87(10):850–868. doi: 10.1002/prot.25749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capra J.A., Laskowski R.A., Thornton J.M., Singh M., Funkhouser T.A. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. Plos Comput Biol. 2009;5(12) doi: 10.1371/journal.pcbi.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: An open source platform for ligand pocket detection. BMC Bioinf. 2009;10(1):168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Comess K.M., Sun C., Abad-Zapatero C., Goedken E.R., Gum R.J., Borhani D.W. Discovery and characterization of non-ATP site inhibitors of the mitogen activated protein (MAP) kinases. ACS Chem Biol. 2011;6(3):234–244. doi: 10.1021/cb1002619. [DOI] [PubMed] [Google Scholar]
- 45.Aleshin A.E., Kirby C., Liu X., Bourenkov G.P., Bartunik H.D., Fromm H.J. Crystal structures of mutant monomeric hexokinase I reveal multiple ADP binding sites and conformational changes relevant to allosteric regulation. J Mol Biol. 2000;296(4):1001–1015. doi: 10.1006/jmbi.1999.3494. [DOI] [PubMed] [Google Scholar]
- 46.Eriksson M., Uhlin U., Ramaswamy S., Ekberg M., Regnstrom K., Sjoberg B.M. Binding of allosteric effectors to ribonucleotide reductase protein R1: reduction of active-site cysteines promotes substrate binding. Structure. 1997;5(8):1077–1092. doi: 10.1016/s0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 47.Kaynak B.T., Findik D., Doruker P. RESPEC Incorporates Residue Specificity and the Ligand Effect into the Elastic Network Model. J Phys Chem B. 2018;122:5347–5355. doi: 10.1021/acs.jpcb.7b10325. [DOI] [PubMed] [Google Scholar]
- 48.Kaynak, B. T.; Doruker, P., Protein–Ligand Complexes as Constrained Dynamical Systems. J Chem Inf Model 2019, acs.jcim.8b00946. [DOI] [PubMed]
- 49.Kurkcuoglu Z., Findik D., Akten E.D., Doruker P. How an Inhibitor Bound to Subunit Interface Alters Triosephosphate Isomerase Dynamics. Biophys J. 2015;109(6):1169–1178. doi: 10.1016/j.bpj.2015.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amadei A., Linssen A.B., Berendsen H.J. Essential dynamics of proteins. Proteins. 1993;17(4):412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 51.Bahar I., Atilgan A.R., Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2:173–181. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 52.Haliloglu T., Bahar I., Erman B. Gaussian Dynamics of Folded Proteins. Phys Rev Lett. 1997;79:3090–3093. [Google Scholar]
- 53.Li H., Chang Y.Y., Yang L.W., Bahar I. iGNM 2.0: the Gaussian network model database for biomolecular structural dynamics. Nucleic Acids Res. 2016;44(D1):D415–D422. doi: 10.1093/nar/gkv1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doruker P., Atilgan A.R., Bahar I. Dynamics of proteins predicted by molecular dynamics simulations and analytical approaches: Application to alpha-amylase inhibitor. Proteins. 2000;40:512–524. [PubMed] [Google Scholar]
- 55.Atilgan A.R., Durell S.R., Jernigan R.L., Demirel M.C., Keskin O., Bahar I. Anisotropy of Fluctuation Dynamics of Proteins with an Elastic Network Model. Biophys J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eyal, E.; Lum, G.; Bahar, I., The anisotropic network model web server at 2015 (ANM 2.0). Bioinformatics 2015, 31 (9), 1487-9. [DOI] [PMC free article] [PubMed]
- 57.Bakan A., Meireles L.M., Bahar I. ProDy: Protein Dynamics Inferred from Theory and Experiments. Bioinformatics. 2011;27:1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hinsen K., Petrescu A.J., Dellerue S., Bellissent-Funel M.C., Kneller G.R. Harmonicity in slow protein dynamics. Chem Phys. 2000;261(1–2):25–37. [Google Scholar]
- 59.Lee H.J., Lang P.T., Fortune S.M., Sassetti C.M., Alber T. Cyclic AMP regulation of protein lysine acetylation in Mycobacterium tuberculosis. Nat Struct Mol Biol. 2012;19(8):811–818. doi: 10.1038/nsmb.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lundqvist T., Fisher S.L., Kern G., Folmer R.H., Xue Y., Newton D.T. Exploitation of structural and regulatory diversity in glutamate racemases. Nature. 2007;447(7146):817–822. doi: 10.1038/nature05689. [DOI] [PubMed] [Google Scholar]
- 61.Saldano T.E., Monzon A.M., Parisi G., Fernandez-Alberti S. Evolutionary Conserved Positions Define Protein Conformational Diversity. Plos Comput Biol. 2016;12(3) doi: 10.1371/journal.pcbi.1004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y., Bahar I. Sequence evolution correlates with structural dynamics. Mol Biol Evol. 2012;29(9):2253–2263. doi: 10.1093/molbev/mss097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Campitelli P., Modi T., Kumar S., Ozkan S.B. The Role of Conformational Dynamics and Allostery in Modulating Protein Evolution. Annu Rev Biophys. 2020;49:267–288. doi: 10.1146/annurev-biophys-052118-115517. [DOI] [PubMed] [Google Scholar]
- 64.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44(W1):W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kurkcuoglu Z., Bahar I., Doruker P. ClustENM: ENM-Based Sampling of Essential Conformational Space at Full Atomic Resolution. J Chem Theory Comput. 2016;12(9):4549–4562. doi: 10.1021/acs.jctc.6b00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stec B., Holtz K.M., Wojciechowski C.L., Kantrowitz E.R. Structure of the wild-type TEM-1 beta-lactamase at 1.55 A and the mutant enzyme Ser70Ala at 2.1 A suggest the mode of noncovalent catalysis for the mutant enzyme. Acta Cryst Section D. 2005;61(Pt 8):1072–1079. doi: 10.1107/S0907444905014356. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z., Harkins P.C., Ulevitch R.J., Han J., Cobb M.H., Goldsmith E.J. The structure of mitogen-activated protein kinase p38 at 2.1-A resolution. Proc Natl Acad Sci USA. 1997;94(6):2327–2332. doi: 10.1073/pnas.94.6.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wakefield A.E., Mason J.S., Vajda S., Keseru G.M. Analysis of tractable allosteric sites in G protein-coupled receptors. Sci Rep. 2019;9(1):6180. doi: 10.1038/s41598-019-42618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kruse A.C., Ring A.M., Manglik A., Hu J., Hu K., Eitel K. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504(7478):101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu J., Byrne N., Wang J., Bricogne G., Brown F.K., Chobanian H.R. Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat Struct Mol Biol. 2017;24(7):570–577. doi: 10.1038/nsmb.3417. [DOI] [PubMed] [Google Scholar]
- 71.Yang J., Campobasso N., Biju M.P., Fisher K., Pan X.Q., Cottom J. Discovery and characterization of a cell-permeable, small-molecule c-Abl kinase activator that binds to the myristoyl binding site. Chem Biol. 2011;18(2):177–186. doi: 10.1016/j.chembiol.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 72.Maeda S., Qu Q., Robertson M.J., Skiniotis G., Kobilka B.K. Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science. 2019;364(6440):552–557. doi: 10.1126/science.aaw5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Freeman L.C. Set of Measures of Centrality Based on Betweenness. Sociometry. 1977;40(1):35–41. [Google Scholar]
- 74.Hagberg Aric, A., Schult Daniel A., Swart Pieter J. Exploring network structure, dynamics, and function using NetworkX. In: Varoquaux Gäel, Vaught Travis., editors. Proceedings of the 7th Python in Science Conference (SciPy2008); Pasadena, CA USA; 2008. pp. 11–15. [Google Scholar]
- 75.Cho U.S., Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445(7123):53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- 76.Bakan A., Nevins N., Lakdawala A.S., Bahar I. Druggability Assessment of Allosteric Proteins by Dynamics Simulations in the Presence of Probe Molecules. J Chem Theory Comput. 2012;8(7):2435–2447. doi: 10.1021/ct300117j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghanakota P., Carlson H.A. Moving Beyond Active-Site Detection: MixMD Applied to Allosteric Systems. J Phys Chem B. 2016;120(33):8685–8695. doi: 10.1021/acs.jpcb.6b03515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ghanakota P., DasGupta D., Carlson H.A. Free Energies and Entropies of Binding Sites Identified by MixMD Cosolvent Simulations. J Chem Inf Model. 2019;59(5):2035–2045. doi: 10.1021/acs.jcim.8b00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sobolevsky A.I., Rosconi M.P., Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462(7274):745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee, J. Y.; Krieger, J.; Herguedas, B.; Garcia-Nafria, J.; Dutta, A.; Shaikh, S. A.; Greger, I. H.; Bahar, I., Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain. Structure 2019, 27 (2), 241-252 e3. [DOI] [PMC free article] [PubMed]
- 81.Sukumaran M., Rossmann M., Shrivastava I., Dutta A., Bahar I., Greger I.H. Dynamics and allosteric potential of the AMPA receptor N-terminal domain. EMBO J. 2011;30(5):972–982. doi: 10.1038/emboj.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ponzoni L., Bahar I. Structural dynamics is a determinant of the functional significance of missense variants. Proc Natl Acad Sci USA. 2018;115(16):4164–4169. doi: 10.1073/pnas.1715896115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ponzoni L., Penaherrera D.A., Oltvai Z.N., Bahar I. Rhapsody: Predicting the pathogenicity of human missense variants. Bioinformatics. 2020;36(10):3084–3092. doi: 10.1093/bioinformatics/btaa127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.