Abstract
Introduction
We report the results of a molecular surveillance study carried out on methicillin-susceptible Staphylococcus aureus (MSSA) isolated in a one-year duration from Hospital Canselor Tuanku Muhriz (HCTM), a tertiary hospital located in Kuala Lumpur, Malaysia.
Methods
The first strain isolated from each MSSA infection in HCTM during the year 2009 was included into the study. Antimicrobial susceptibility testing and agr group typing were carried out for all strains; virulence gene (cna, seh, TSST-1 and PVL) typing results of the strains were obtained from a previous study. Pulsed-field gel electrophoresis (PFGE) was done on selected strains from the orthopedic ward. Relationship(s) between different typing methods used in the study was investigated, where a p value of <0.05 indicated significant association between typing methods.
Results
A total of 880 MSSA strains were included into the study. The strains were generally susceptible to most antibiotics, with most of them carrying cna and agr-I (51.6%, n=454; 39.8%, n=350, respectively). A total of 17 PFGE pulsotypes were identified using an 80% similarity cut-off value, where the main pulsotype (pulsotype E) consisted of 24 isolates (23.5%). agr-III strains were found to be usually positive for both cna and seh (p<0.05). In addition, some PFGE pulsotypes were also characteristic of certain virulence genes or agr groups.
Conclusions
We did not identify a dominant MSSA clone circulating in HCTM in 2009. Nevertheless, results from this molecular surveillance will provide good baseline data for the hospital’s second S. aureus surveillance planned for the year 2020.
Keywords: Molecular surveillance, methicillin-susceptible S. aureus (MSSA), agr typing, virulence gene typing, PFGE
Introduction
Staphylococcus aureus is a major nosocomial pathogen whose pathological importance makes it a longstanding subject of epidemiological investigation. Epidemiological studies on S. aureus are usually performed for methicillin-resistant S. aureus (MRSA) as they are mostly multidrug-resistant with limited therapeutic options.1 On the other hand, less emphasis has been placed on methicillin-susceptible S. aureus (MSSA), even though they have a higher hospital prevalence than MRSA.2 MSSA has also been reported to harbor various virulence genes, where it has been observed that these bacteria might carry more virulence genes compared to MRSA.3
Molecular surveillance of Malaysian MRSA strains have been initiated in the past decade, however, studies that report on Malaysian MSSA genotyping remain few.4-6 Hospital Canselor Tuanku Muhriz (HCTM) is a 1000-bed teaching hospital located in Kuala Lumpur, Malaysia. In 2009, the hospital initiated a molecular surveillance study on S. aureus isolated from the hospital in a one-year duration. Results from the MRSA surveillance have been described and published.7 In this report, we report results of the molecular surveillance carried out on MSSA isolated from HCTM in the same year.
Methods
Study setting and bacterial strains
In 2009, MSSA infections from all wards of HCTM were identified and recorded by the hospital’s Department of Laboratory and Diagnostic Services. The first isolate from each infection was then segregated, colony-purified and established as a strain to be used in this study. The strains were stocked from time of isolation in Brain Heart Infusion broth with 40% glycerol at -70°C. Experiments on the strains were performed in 2010, after all MSSA strains had been collected for the surveillance.
Antimicrobial resistance testing
Antibiotic resistance profiles of the collected strains were determined by disk diffusion method on Mueller-Hinton agar according to the recommendations of Clinical Laboratory Standards Institute (CLSI).8 Antimicrobial agents tested were ciprofloxacin (5 µg), erythromycin (15 µg), fusidic acid (10 µg), gentamicin (10 µg), cefoxitin (10 µg) and penicillin (10 units) (Thermo Fisher Scientific Inc., Massachusetts, USA).
Chromosomal DNA isolation for virulence gene and agr typing
Chromosomal DNA of each MSSA strain was extracted using DNeasy Blood & Tissue Kit (Qiagen Inc., California, USA) according to the manufacturer’s instruction. Extracted DNA samples were stored at -20°C.
Virulence gene typing
A modified multiplex-PCR protocol was used to determine the presence of four virulence genes (cna, seh, PVL and TSST-1) in our collection of MSSA strains. Primer sequences for the tested genes are shown in Table 1; cycling conditions were carried out as described previously.9 Results for the typing have been published,9 in this current surveillance study, results from the typing were used for association analysis between typing methods.
Table 1. List of primer sequences used for virulence genes and agr typing in this study.
| Name | Primer sequence | Product size (bp) | Reference |
|---|---|---|---|
| cna-F | 5’ ACACCAGACGGTGCAACAATTA 3’ | 815 | (9) |
| cna-R | 5’ AGCAATACCGTTTGCATCTGTTA 3’ | ||
| seh-F | 5’ ATTCACATCATATGCGAAAGCAG 3’ | 555 | (9) |
| seh-R | 5’ ATGTCGAATGAGTAATCTCTAG 3’ | ||
| pvl-F | 5’ ATGTCTGGACATGATCCAA 3’ | 970 | (9) |
| pvl-R | 5’ AACTATCTCTGCCATATGGT 3’ | ||
| tsst1-F | 5’ TGATATGTGGATCCGTCAT 3’ | 387 | (9) |
| tsst1-R | 5’ AAACACAGATGGCAGCAT 3’ | ||
| agr1-F | 5’ ATGCACATGGTGCACATGC 3’ | 441 | (10) |
| agr1-R | 5’ GTCACAAGTACTATAAGCTGCGAT 3’ | ||
| agr2-F | 5’ ATGCACATGGTGCACATGC 3’ | 575 | (10) |
| agr2-R | 5’ TATTACTAATTGAAAAGTGGCCATAGC 3’ | ||
| agr3-F | 5’ ATGCACATGGTGCACATGC 3’ | 323 | (10) |
| agr3-R | 5’ GTAATGTAATAGCTTGTATAATAATACCCA G 3’ | ||
| agr4-F | 5’ ATGCACATGGTGCACATGC 3’ | 659 | (10) |
| agr4-R | 5’ CGATAATGCCGTAATACCCG 3’ |
Characterization of agr
The agr genotype of each MSSA strain was determined using a multiplex PCR protocol, using primers listed in Table 1.10 Clinical strains MSSA 180, MSSA 188, MSSA 130 and MSSA 25 were used as controls for agr-I, agr-II, agr-III and agr-IV respectively; all agr sequences of these strains have been confirmed by sequencing. Cycling conditions were as follows: one cycle of pre-denaturation for 4 min at 94°C; 30 cycles of 94°C for 2 min (denaturation), 58°C for 1 min (annealing) and 72°C for 2 min (extension); and a final extension at 72°C for 5 min. PCR products were analyzed by gel electrophoresis with a 1.5% agarose gel (Sigma-Aldrich, St. Louis, USA).
Pulsed-field gel electrophoresis (PFGE) typing
PFGE typing by SmaI macrorestriction for a subset of 120 strains isolated from the orthopedic ward was performed according to methods described previously.11 Samples were chosen from the orthopedic ward, as the ward had the highest number of MSSA infections in 2009. Briefly, agarose plugs containing chromosomal DNA of tested strains were prepared and restricted by SmaI prior separation by PFGE with a CHEF-DR III system (Bio-Rad Laboratories, Inc., California, USA). Settings for the PFGE were as follows: initial switch time, 5 seconds; final switch time, 40 seconds; included angle, 120°; voltage, 6 V/cm or 200 V for a total running time of 22 hours. Fingerprinting II software (version 1.0; Bio-Rad Laboratories, Inc.) was used to analyze the banding patterns. Similarity index was determined for the strains using the Dice coefficient with a 0.5% band tolerance. A cut-off value of 80% genetic similarity was chosen for discrimination between distinct clusters of strains. Genetic relatedness of strains was determined according to the criteria established.12
Association between molecular typing
Association between virulence gene, agr and PFGE typing for the tested strains were determined using Statistical Package for the Social Sciences (SPSS) (version 15.0; IBM, Inc., Chicago, Illinois, USA). Variables were analyzed using Fisher’s exact test, where a p value of <0.05 was considered statistically significant.
Results
From January to December 2009, a total of 880 MSSA-related infections from various wards of HCTM were reported. Antimicrobial susceptibility testing showed that most strains retained their susceptibilities to the antibiotics tested (ciprofloxacin, 91.8%; erythromycin, 88.9%; fusidic acid, 85.8%; gentamicin, 97.1%; cefoxitin, 100%). The only antibiotic to which they were usually resistant was penicillin (79.1% of the isolates).
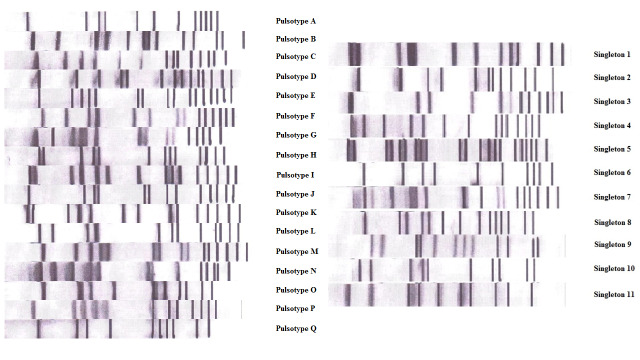
For molecular typing, 51.6% (n=454) of the MSSA strains in our study harbored cna, 21.8% (n=192) carried seh, while the prevalence for PVL and TSST-1 was 10.23% (n=90) and 6.82% (n=60), respectively. Almost half of the strains (41.1%, n=362) were not carrying any of the tested virulence genes. Table 2 shows virulence gene profile of the tested strains. Meanwhile, for our agr typing experiments, 39.8% (n=350), 12.7% (n=112), 28.0% (n=246) and 4.3% (n=39) of our strains were detected as agr-I, -II, -III or -IV, respectively. The remaining 133 (15.1%) strains were not agr-typeable. Meanwhile, PFGE pulsotype analysis of selected strains identified 17 pulsotypes (A-Q) and 11 singletons, when an 80% of cut-off value in similarity was employed (Figure 1). The majority (23.5%, n=24) of these strains were designated as pulsotype E.
Table 2. Virulence gene profile of MSSA strains in this study.
| Virulence gene profile | Number of strains (n) | Percentage (%) |
|---|---|---|
| cna | 202 | 23.0 |
| cna + seh | 167 | 19.0 |
| cna + PVL | 38 | 4.3 |
| cna + PVL + TSST-1 | 1 | 0.1 |
| cna + seh + PVL | 23 | 2.6 |
| cna + seh + TSST-1 | 2 | 0.2 |
| cna + TSST-1 | 21 | 2.4 |
| PVL | 28 | 3.2 |
| TSST-1 | 36 | 4.1 |
| No virulence gene | 362 | 41.1 |
| Total | 880 | 100.0 |
Figure 1. Pulsotypes of MSSA strains isolated from the orthopedic ward.
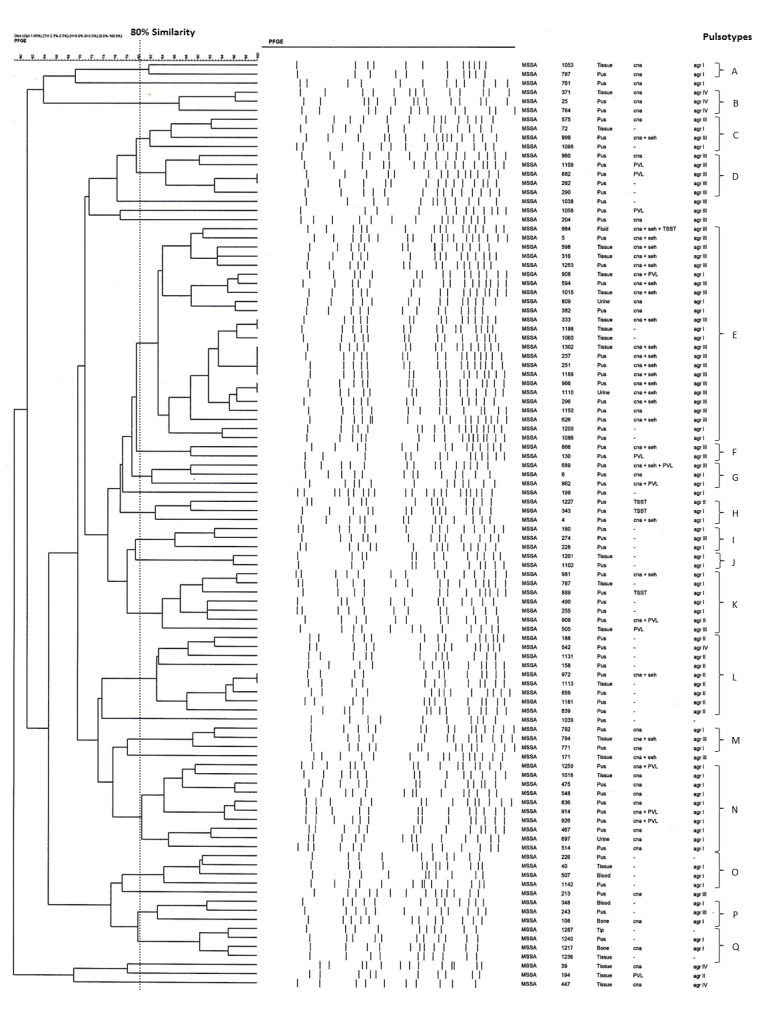
We proceeded to investigate if any association could be found between the 3 molecular typing methods (virulence gene, agr and PFGE). Interestingly, we noticed that most of the agr-III strains were also positive for both cna and seh (p<0.05). In addition, certain PFGE pulsotypes were strongly associated with a particular agr group or virulence gene cluster: pulsotype E with the carriage of cna, seh and agr-III; pulsotype N with cna and agr-I; and pulsotype L which harbored none of the tested virulence genes, but was determined as agr-II (Figure 2).
Figure 2. Association between PFGE pulsotypes with toxin and agr genotype in MSSA strains used in this study. Pulsotype E was strongly associated with the carriage of cna, seh and agr-III; pulsotype N with cna and agr-I; while pulsotype L harbored none of the tested toxin genes but was typed as agr-II.
Discussion
Molecular surveillance is not new in public health; it has been performed for pathogens such as Mycobacterium tuberculosis and the human immunodeficiency viruses.13,14 In the recent decade, many hospitals have also initiated molecular surveillance for important nosocomial pathogens.15,16 This study was part of a Staphylococcus aureus molecular surveillance initiated by HCTM in 2009. The total number of S. aureus infections in 2009 for the hospital was 1,198, where 73.5% (n=880) were MSSA infections, and 13.5% (n=318) were MRSA. Although most MRSAs (86.8%) isolated during this surveillance were multidrug-resistant, with resistance towards ciprofloxacin, erythromycin and gentamicin, MSSAs isolated from our hospital in 2009 were mostly (more than 80%) susceptible to tested antibiotics; cefoxitin remains the treatment of choice for MSSA infections in HCTM.7
Various methods have been established for S. aureus molecular typing and surveillance. These methods include SCCmec typing (for MRSA only), virulence gene characterization, agr typing, pulsed-field gel electrophoresis typing (PFGE) and multilocus sequence typing (MLST).17 In our study, virulence genes (cna, seh, TSST-1 and PVL), agr and PFGE genotyping were carried out for the strains. We found cna to be the most prevalent virulence gene carried by HCTM MSSA in 2009; interestingly, the same observation was also found in our MRSAs isolated in the same study duration (MRSA, 94.0%, n=299; MSSA, 51.6%, n=454).7 The gene is a virulence factor which codes for collagen adhesion; this protein is important for the attachment of S. aureus to cells or extracellular matrices.18 MSSA strains isolated from Hospital Kuala Lumpur (HKL), the largest hospital in the capital, also appeared to harbor a similar prevalence of cna (52.3%, n=140).4 Nonetheless, the gene was not detected in a study on both clinical and community MSSA strains isolated from another university hospital (University Malaya Medical Centre, UMMC) located at the border of Kuala Lumpur, in the same year.5 Remarkably, the prevalence of seh, TSST-1 and PVL in MSSAs isolated from HCTM, HKL and UMMC were only below or approximately 10% difference from one hospital to another. Similarly, agr distribution for HCTM MSSAs was similar to HKL and UMMC, with a majority of the strains being typed as agr-I, followed by agr-III. Comparatively, HCTM MRSAs were dominantly agr-I (94.4%, n = 237).7
Even though there seemed to be little differences between MSSA strains isolated from hospitals in Kuala Lumpur based on their virulence and agr genotypes, genetic diversity of our MSSA strains was revealed via PFGE typing. PFGE has been used as the gold standard for bacteria typing; the platform was and is still widely being used in many reference laboratories for pathogen characterization and epidemiological surveillance.19 In contrast to nosocomial MRSA where the dissemination of a single major PFGE clone in many hospital settings has been commonly observed,7,20-23 PFGE typing of nosocomial MSSA has mostly revealed very diverse results, with the occurrence of many sporadic clones in the same setting within fixed duration of the studies.4,24 For our study, even though PFGE typing was only conducted for strains isolated from the orthopedic ward, as many as 17 pulsotypes were identified, compared to only 5 common MRSA pulsotypes identified in all HCTM medical, surgical and intensive care units combined.7,25 Incidentally, these 5 MRSA pulsotypes were also identified to be circulating in the orthopedic ward in 2009 (unpublished data).
Noting the difference in MSSA genomic diversity presented via different genotyping methods, we attempted to investigate if associations between the typing methods of virulence genes, agr and PFGE for MSSA could be found. While we identified some significant associations between certain PFGE pulsotypes with a particular agr group or virulence gene cluster, as this is the first MSSA surveillance study in HCTM, it remains to be observed if these pulsotypes will still be prevalent in our follow up surveillance in 2020, and if there has been successful dissemination of these pulsotypes in our hospital.
Conclusions
In conclusion, we presented the results of the first MSSA molecular surveillance for our hospital which was carried out in 2009. No dominant MSSA clone was identified. The strains were found to be genetically diverse via PFGE typing; a majority of them harbored the cna gene and were agr-I. Associations between specific groups of agr, virulence genes and PFGE pulsotypes could be made for these MSSAs. The next molecular surveillance for HCTM MSSA is being planned for 2020; comparison of results from these two surveillance studies will reveal, if any, changes in molecular genotypes of MSSA circulating in our hospital.
Acknowledgements
The authors would like to acknowledge the staff from the Department of Laboratory Diagnostic Services, Hospital Canselor Tuanku Muhriz, for their help in retrieval of study samples and antibiotic susceptibility testing results.
Footnotes
Authors’ contributions statement: HFS carried out the experiments and drafted the manuscript. HFS, MAHI and TLT analyzed the results and edited the manuscript. NAMS and AN helped in carrying out the experiments. Both HN and SH conceived the study, participated in its design and coordination, and edited the manuscript. All authors read and approved the final version of the manuscript.
Conflicts of interest: All authors – none to declare.
Funding: This research was supported by the UKM Research University Grant UKM-GUP-TKP-08-19-067 and UKM Medical Centre Fundamental Research Fund FF-001-2012.
References
- 1.Choo EJ, Chambers HF. Treatment of methicillin-resistant Staphylococcus aureus bacteremia. Infect Chemother. 2016;48:267–73. doi: 10.3947/ic.2016.48.4.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–61. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu C, Chen ZJ, Sun Z, et al. Molecular characteristics and virulence factors in methicillin-susceptible, resistant, and heterogeneous vancomycin-intermediate Staphylococcus aureus from central-southern China. J Microbiol Immunol Infect. 2015;48:490–6. doi: 10.1016/j.jmii.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Ghasemzadeh-Moghaddam H, Ghaznavi-Rad E, Sekawi Z, et al. Methicillin-susceptible Staphylococcus aureus from clinical and community sources are genetically diverse. Int J Med Microbiol. 2011;301:347–53. doi: 10.1016/j.ijmm.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Lim KT, Hanifah YA, Yusof MYM, Thong KL. Characterisation of the virulence factors and genetic types of methicillin susceptible Staphylococcus aureus from patients and healthy individuals. Indian J Microbiol. 2012;52:593–600. doi: 10.1007/s12088-012-0286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghasemzadeh-Moghaddam H, Neela V, Goering R, Mariana NS. Methicillin sensitive Staphylococcus aureus (MSSA) isolates as a potential source for the emergence of USA 300 methicillin resistant Staphylococcus aureus (MRSA) in Malaysia. Trop Biomed. 2012;29:429–33. [PubMed] [Google Scholar]
- 7.Noordin A, Sapri HF, Mohamad Sani NA, et al. Antimicrobial resistance profiling and molecular typing of methicillin-resistant Staphylococcus aureus isolated from a Malaysian teaching hospital. J Med Microbiol. 2016;65:1476–81. doi: 10.1099/jmm.0.000387. [DOI] [PubMed] [Google Scholar]
- 8.Clinical Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: Nineteenth informational (supp 2009:M100-S19). Wayne, PA, USA: CLSI; 2009. [Google Scholar]
- 9.Hassriana Fazilla S, Nurul Azirah MS, Ainihayati N, Hui-min N, Salasawati H. Virulence gene typing of methicillin-susceptible Staphylococcus aureus (MSSA) isolates in Universiti Kebangsaan Malaysia Medical Centre (UKMMC). Asia-Pacific J Mol Med. 2011;1:1–4. [Google Scholar]
- 10.Gilot P, Lina G, Cochard T, Poutrel B. Analysis of the genetic variability of genes encoding the RNA III-activating components agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J Clin Microbiol. 2002;40:4060–7. doi: 10.1128/JCM.40.11.4060-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jamaluddin TZ, Kuwahara-Arai K, Hisata K, et al. Extreme genetic diversity of methicillin-resistant Staphylococcus epidermidis strains disseminated among healthy Japanese children. J Clin Microbiol. 2008;46:3778–83. doi: 10.1128/JCM.02262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchini A, Andrés M, Fiebig L, Albrecht S, Hauer B, Haas W. Assessment of the use and need for an integrated molecular surveillance of tuberculosis: an online survey in Germany. BMC Public Health. 2019;19:321. doi: 10.1186/s12889-019-6631-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ragonnet-Cronin M, Hu YW, Morris SR, Sheng Z, Poortinga K, Wertheim JO. HIV transmission networks among transgender women in Los Angeles County: network analysis of surveillance data. Lancet HIV. 2019;6:e164–72. doi: 10.1016/S2352-3018(18)30359-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wendel AF, Malecki M, Otchwemah R, Tellez-Castillo CJ, Sakka SG, Mattner F. One-year molecular surveillance of carbapenem-susceptible A. baumannii on a German intensive care unit: diversity or clonality. Antimicrob Resist Infect Control. 2018;7:145. doi: 10.1186/s13756-018-0436-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crago B, Ferrato C, Drews SJ, et al. Surveillance and molecular characterization of non-tuberculous mycobacteria in a hospital water distribution system over a three-year period. J Hosp Infect. 2014;87:59–62. doi: 10.1016/j.jhin.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31:e00020–18. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Rivas JM, Brown EL, Liang X, Höök M. Virulence potential of the staphylococcal adhesin CNA in experimental arthritis is determined by its affinity for collagen. J Infect Dis. 2004;189:2323–33. doi: 10.1086/420851. [DOI] [PubMed] [Google Scholar]
- 19.Neoh HM, Tan XE, Sapri HF, Tan TL. Pulsed-field gel electrophoresis (PFGE): A review of the "gold standard" for bacteria typing and current alternatives. Infect Genet Evol. 2019;74:103935. doi: 10.1016/j.meegid.2019.103935. [DOI] [PubMed] [Google Scholar]
- 20.Aires de Sousa M, Conceição T, Simas C, de Lencastre H. Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J Clin Microbiol. 2005;43:5150–7. doi: 10.1128/JCM.43.10.5150-5157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aires de Sousa M, Crisóstomo MI, Sanches IS, et al. Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J Clin Microbiol. 2003;41:159–63. doi: 10.1128/JCM.41.1.159-163.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghaznavi-Rad E, Nor Shamsudin M, Sekawi Z, et al. Predominance and emergence of clones of hospital-acquired methicillin-resistant Staphylococcus aureus in Malaysia. J Clin Microbiol. 2010;48:867–72. doi: 10.1128/JCM.01112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norazah A, Lim VK, Rohani MY, Alfizah H, Koh YT, Kamel AG. A major methicillin-resistant Staphylococcus aureus clone predominates in Malaysian hospitals. Epidemiol Infect. 2003;130:407–11. doi: 10.1017/s095026880300832x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Dijk Y, Wielders CL, Fluit AC, Paauw A, Diepersloot RJ, Mascini EM. Genotyping of clinical methicillin-susceptible Staphylococcus aureus isolates in a Dutch teaching hospital. J Clin Microbiol. 2002;40:663–5. doi: 10.1128/JCM.40.2.663-665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan XE, Neoh H, Salasawati H, Noraziah MZ. Pulsed-field gel electrophoresis typing reveals clonal spreading and possible microevolution of methicillin-resistant Staphylococcus aureus strains isolated from a university teaching hospital [Google Scholar]