Abstract
Ferroptosis is a recently discovered form of programmed cell death, but its regulatory mechanisms are not fully understood. In the current study, we reported that the BRD7-P53-SLC25A28 axis played a crucial role in regulating ferroptosis in hepatic stellate cells (HSCs). Upon exposure to ferroptosis inducers, bromodomain-containing protein 7 (BRD7) protein expression was remarkably increased through the inhibition of the ubiquitin-proteasome pathway. CRISPR/Cas9-mediated BRD7 knockout conferred resistance to HSC ferroptosis, whereas specific BRD7 plasmid-mediated BRD7 overexpression facilitated HSC ferroptosis. Interestingly, the elevated BRD7 expression exhibited to promote p53 mitochondrial translocation via direct binding with p53 N-terminal transactivation domain (TAD), which may be the underlying mechanisms for BRD7-enhanced HSC ferroptosis. Site-directed mutations of serine 392 completely blocked the binding of BRD7 to p53, and, in turn, prevented p53 mitochondrial translocation and HSC ferroptosis. Importantly, mitochondrial p53 interacted with solute carrier family 25 member 28 (SLC25A28) to form complex and enhanced the activity of SLC25A28, which could lead to the abnormal accumulation of redox-active iron and hyperfunction of electron transfer chain (ETC). SLC25A28 knockdown impaired BRD7-or p53-mediated ferroptotic events. In mice, erastin treatment ameliorated pathological damage of liver fibrosis through inducing HSC ferroptosis. HSC-specific blockade of BRD7-P53-SLC25A28 axis could abrogate erastin-induced HSC ferroptosis. Of note, we analyzed the effect of sorafenib on HSC ferroptosis in advanced fibrotic patients with hepatocellular carcinoma receiving sorafenib monotherapy. Attractively, BRD7 upregulation, p53 mitochondrial translocation, combination of SLC25A28 and p53, and ferroptosis induction occurred in primary human HSCs. Overall, these findings reveal novel signal transduction and regulatory mechanism of ferroptosis, and also suggest BRD7-P53-SLC25A28 axis as potential targets for liver fibrosis.
Keywords: Hepatic stellate cell, BRD7, P53, SLC25A28, Ferroptosis
1. Introduction
Liver fibrosis serves as a complex pathological event associated with excessive synthesis of extracellular matrix (ECM), and its final stage of cirrhosis has high recurrence rates and death rates without effective intervention measures [1]. The transdifferentiation of HSCs into matrix-producing myofibroblasts is a central event in liver fibrosis [2,3]. Therefore, targeting to scavenge HSCs is considered therapeutic approaches to reverse liver fibrosis. We previously showed that targeting of HSCs, including induction of apoptosis [4], necroptosis [5], senescence [6] and lipocyte phenotype [7], and inhibition of the pericyte function [8], proinflammatory function [9], glycolytic function [10] and contractile function [11], could relieve the pathological changes of liver fibrosis. Recently, we reported that ferroptosis can be considered as a new strategy to eliminate HSCs [12,13]. Activation of autophagy may be required for the RNA-binding protein ELAVL1/HuR and ZFP36/TTP to regulate ferroptosis in HSCs [12,13]. Moreover, we demonstrated that artemether treatment may ameliorate liver fibrosis and inhibit HSC activation by inducing HSC ferroptosis in a p53-dependent manner [14]. In the current study, we aimed to determine the function of BRD7 in the induction of ferroptosis, and to further identify BRD7-dependent ferroptosis as potential targets for liver fibrosis.
Ferroptosis is a genetically encoded program of cell death that results from iron-dependent lipid peroxidation, but it is different from classical programmed cell death including apoptosis, senescence, necrosis and pyroptosis [15]. It is characterized morphologically by the presence of smaller than normal mitochondria with condensed mitochondrial membrane densities, reduction or vanishing of mitochondria crista, and outer mitochondrial membrane rupture [15]. Importantly, glutathione peroxidase 4 (GPX4) inhibition (e.g., RSL3, FIN56, FINO2, and altretamine), system inhibition (e.g., sorafenib, erastin, and sulfasalazine), glutathione (GSH) depletion (e.g., BSO), and physiological conditions (e.g., high extracellular glutamate, amino acid starvation, and cystine deprivation) are all reported to trigger ferroptosis [16]. By contrast, lipophilic antioxidants (e.g., siderostatin-1, liproxstatin-1, and vitamin E), iron chelating agents (e.g. deferrioxamine), lipid peroxidation inhibitors (e.g. eugenol), and consumption of polyunsaturated fatty acyl phospholipids (PUFA-PLS) (e.g. arachidonic acid) could inhibit ferroptosis [16]. Mechanistically, p53 signaling [17], autophagy signaling [18], NF2-YAP signaling [19], p62-Keap1-NRF2 signaling [20], and glutaminolysis metabolism signaling [21] are mainly involved in the regulation of ferroptosis. Mitochondria take the center role in iron metabolism, as well as substance and energy metabolism as its the major organelle in iron utilization, catabolic and anabolic pathways [22]. Interestingly, whether mitochondrial iron metabolism plays a crucial role in ferroptosis is worth further study.
BRD7 is a crucial subunit of the PBAF (polybromo-associated BRG1-associated factor) chromatin remodeling complex, which is involved in transcriptional regulation through interactions with acetylated histones in chromatin [23]. BRD7 is originally identified as a tumor suppressor that inhibits tumor cell growth by negatively regulating the β-catenin and ERK signaling [23]. Moreover, ectopic expression of BRD7 inhibits cell cycle progression from G1 to S phase by transcriptionally regulating some cell cycle related genes including E2F3 gene [24]. Recently, BRD7 is found to be a transcriptional cofactor for the tumor suppressor protein p53 [25]. BRD7 is required for efficient p53-mediated transcription of a subset of target genes [25]. BRD7 interacts with p53 and p300, and is recruited to target gene promoters, affecting histone acetylation, p53 acetylation and promoter activity [25]. Additionally, BRD7 has been shown to be involved in the regulation of multiple cell fate and disease progression [[26], [27], [28]]. BRD7 may mediate hyperglycaemia-induced myocardial apoptosis via endoplasmic reticulum stress signaling [26]. Furthermore, BRD7 affects PI3K-mediated chromatin remodeling, and regulates p53-dependent replicative senescence [27]. Besides, BRD7 could inhibit the Warburg effect and tumor progression through inactivation of HIF1α/LDHA axis in breast cancer [28]. However, the regulatory mechanism of BRD7 on ferroptosis is still unclear.
In the present study and for the first time, we investigated novel molecular mechanism and signaling of ferroptosis in HSCs. We found that inhibition-, GPX4 inhibition-, and GSH depletion-mediated BRD7 upregulation could trigger p53 mitochondrial translocation via direct binding with N-terminal transactivation domain, thus promoting the accumulation of mitochondrial iron and the hyperfunction of electron transfer chain, and finally resulting in HSC ferroptosis. Our results indicated that BRD7 may be a critical and novel regulator of ferroptosis in liver fibrosis.
2. Materials and methods
2.1. Human liver specimens
According to our previous reports [12,13], We retrospectively analyzed 37 liver biopsy samples from cirrhotic patients without any treatment, and 24 partial hepatectomy samples from cirrhotic patients complicated with hepatocellular carcinoma (HCC) receiving sorafenib monotherapy (Nexavar; Bayer Healthcare Pharmaceuticals, Leverkusen, Germany) in Nanjing Hospital Affiliated to Nanjing University of Chinese Medicine from September 2014 to July 2019. The diagnosis of liver fibrosis and HCC was based on the criteria of the American Association for the Study of Liver Diseases (AASLD) [29]. An initial sorafenib dosage of 400 mg was administered orally twice daily, after breakfast and dinner [30]. Subsequently, discontinuations and dosage reductions of sorafenib were based on tolerance. Treatment was continued until clinical disease progression or unacceptable drug-related toxicity occurred, or upon withdrawal of consent. Informed consent in writing was obtained from patients. This study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki Principles, and was approved by the review committee of the Nanjing Hospital Affiliated to Nanjing University of Chinese Medicine. For detailed ethical proof, please refer to Figs. S1 and S2.
2.2. Primary human HSC isolation and characterization
Primary human HSCs were isolated from the collected human liver tissue by laser capture microdissection (LCM) as described previously [12,13]. In brief, frozen human liver tissue was sectioned at 10 μm with a cryostat and stained with immunofluorescence for desmin (Abcam, ab15200). Once the tissue was stained, the desmin-positive cells were located using specialized microdissection software. Then, a specific CapSure® LCM Cap (Thermo Fisher Scientific, LCM0211) was placed on the section. Pulsing the laser through the cap caused the thermoplastic film to form a thin protrusion that bridges the gap between the cap and tissue and adheres to the desmin-positive cells. Lifting of the cap could remove the target cells attached to the cap. Purification and characterization of the obtained human HSCs were confirmed by detection of ACTA2 (actin alpha 2, smooth muscle) and PDGFRβ (platelet derived growth factor receptor beta).
2.3. Animals and experimental design
Eight-week-old male C57BL/6 mice were purchased from Nanjing Medical University (Nanjing, China). Sixty mice were randomly divided into six groups of ten animals each with comparable mean body weight. Mice of six groups were treated with Sham, BDL + VA-Lip-Control-shRNA, BDL + VA-Lip-Control-shRNA + erastin, BDL + VA-Lip-BRD7-shRNA + erastin, BDL + VA-Lip-P53-shRNA + erastin or BDL + VA-Lip-SLC25A28-shRNA + erastin, respectively. Mice were anesthetized with isoflurane. A midline laparotomy was performed, and the common bile duct was ligated close to the liver hilus immediately below the bifurcation with 3-0 surgical silk and cut between the ligatures as described previously [12,13]. Controls underwent a sham operation that consisted of exposure, but not ligation, of the common bile duct. Erastin (30 mg/kg, once every other day) (Selleck Chemicals, S7242) was suspended in sterile phosphate-buffered saline (PBS; Sigma, P5368) and given by intraperitoneal injection for two weeks after the BDL operation. VA-Lip-Control-shRNA, VA-Lip-BRD7-shRNA, VA-Lip-P53-shRNA, and VA-Lip-SLC25A28-shRNA (0.75 mg/kg) were administered intravenously 3 times a week for two weeks after the BDL operation. The livers were collected two weeks after surgery under general anesthesia. A small portion of the liver was removed for histopathological studies by fixation with 10% formalin and subsequent embedment with paraffin. The remaining liver was cut in pieces and rapidly frozen with liquid nitrogen for extraction of total RNA and proteins. All experimental procedures were approved by the institutional and local committee on the care and use of animals of Nanjing University of Chinese Medicine (Nanjing, China), and all animals received humane care according to the National Institutes of Health (USA) guidelines.
2.4. Detection of serum liver fibrosis markers
Serum levels of liver fibrosis markers including glutamic-pyruvic transaminase (GPT/ALT), glutamic-oxaloacetic transaminase 1 (GOT1/AST), hyaluronic acid (HA), laminin (LN), type IV collagen (IV–C), and procollagen III (PC-III) were detected by enzyme-linked immunosorbent assay (ELISA) using commercially available kits (YIFEIXUE BIO TECH, Nanjing, China) according to the manufacturer's instructions. In brief, blood samples were collected from the abdominal aorta and immediately centrifuged at 3500×g for 15 min at 4 °C. The serum was obtained and frozen at −20 °C until analysis. Each serum sample was analyzed in duplicate and absorbance values were measured at 450 nm using a microplate reader (Thermo Fisher Scientific, 1410101). Based on standard curves run in duplicate on each plate, serum levels of liver fibrosis markers were determined with Graph Pad software.
2.5. Histological analysis
The liver tissue samples were immobilized in fixation buffer (Thermo Fisher Scientific, 28906) for 2 days, and then were transferred to ethanol of different concentration and embedded in paraffin in preparation for histopathological analysis. 4-μm thin sections were stained with H&E, Sirius red, and Masson for histological study. H&E, Sirius red, and Masson-stained areas from 10 random fields were quantified with ImageJ software (NIH, Bethesda, MD, USA). The liver fibrosis stage was assessed by Ishak scale [31].
2.6. Primary mouse hepatocyte isolation and characterization
Primary mouse hepatocytes were isolated from mouse liver according to our previous reports [12,13]. In brief, the livers of the mice were first perfused in situ via the portal vein with Ca2+- and Mg2+-free Hanks' balanced salt solution (HBSS; sigma; H1641) supplemented with 0.5 mM EGTA (Sigma, E3889) and 25 mM HEPES (Sigma, H3375) at 37 °C. Then, the buffer was replaced with 0.1% collagenase I solution (Sigma, C0130) in HBSS (containing 4 mM CaCl2, 0.8 mM MgSO4). After a few minutes of perfusion, the liver was excised rapidly from the body cavity and dispersed into cold HBSS. The cell suspension generated was filtered through a sterile 70-μm pore size nylon cell strainer (Sigma, CLS431751) and spun three times at 30×g for 4 min. The pellets were suspended in Dulbecco's modified essential medium (DMEM; Gibco, 12491-015) containing 10% fetal bovine serum (FBS; Gibco, 10099141) for primary hepatocyte culture. Purification and characterization of the obtained hepatocytes were confirmed by detection of albumin [32].
2.7. Primary mouse HSC isolation and characterization
According to our previous reports [12,13], primary mouse HSCs were separated from mouse liver. In brief, the livers of the mice were first perfused in situ with DMEM-free containing 1 mg/ml collagenase IV (Vetec, V900893) and 2 mg/ml pronase (Roche, PRON-RO) following HBSS including 0.5 mM EDTA (Sigma, E6758). After a few minutes of perfusion, the livers were removed and the digested hepatic cells were dispersed in DMEM-free. Next, DNase enzymes (Sigma, D4263) were added to prevent filamentous gelatinous material, and the undigested debris was removed through a filter. The filtrates were centrifuged at 50×g in a centrifuge tube for 5 min at 4 °C. The supernatant was collected following gradient centrifugation with 25% Nycodenz (Sigma, D2158) to isolate primary HSCs. Cells were washed and plated on 60 mm diameter tissue culture dishes (Sigma, CLS430599). Purification and characterization of the obtained HSCs were confirmed by detection of ACTA2 and PDGFRβ.
2.8. Primary mouse liver macrophage isolation and characterization
Primary mouse macrophages were isolated from mouse liver according to our previous reports [12,13]. In brief, the livers of the mice were first perfused in situ with Ca2+- and Mg2+-free HBSS containing 2.5 mM EGTA via portal vein and then they were perfused again with 0.05% collagenase I HBSS solution. The digested livers were dissected and then gently teased with forceps until they were in solution. The cell suspensions were filtered through a 70-μm nylon cell strainer. Nonparenchymal cells were separated from the hepatocytes by 1 cycle of differential centrifugation (300×g for 5 min). The supernatant was centrifuged further (300×g for 5 min and 2 cycles of 1200×g for 5 min) to obtain nonparenchymal cells. The obtained nonparenchymal cells were resuspended in DMEM with 2% FBS, and separated by centrifugation on a 25 %–50% Percoll (Sigma, P4937) gradient. The macrophage fraction located at the interface of the 25 %–50% Percoll layer was seeded in DMEM containing 10% FBS and 10 mM HEPES. Purification and characterization of the obtained macrophages were confirmed by detection of ADGRE1 (adhesion G protein-coupled receptor E1).
2.9. Primary mouse liver sinusoidal endothelial cell isolation and characterization
According to our previous reports [13], primary liver sinusoidal endothelial cells (LSECs) were isolated from mouse liver. In brief, the livers of the mice were first washed with serum-free DMEM, minced into small pieces with a sterilized razor blade in a 60 mm tissue culture dish, and digested with 2 ml of 0.05% collagenase I (Sigma, C0130) at 37 °C. After digestion, cells were rinsed with 5 ml of DMEM containing 10% FBS, centrifuged for 5 min at 400×g, and resuspended in 5 ml of DMEM with 10% FBS. After filtration through a double layer of 70 μm pore size nylon cell strainer (Sigma, CLS431751), cells were centrifuged at 400×g for 10 min and rinsed twice with DMEM containing 10% FBS. Cells were resuspended in 1 ml of DMEM with 10% FBS and incubated with magnetic-beads coated with anti-PECAM1/CD31 (platelet and endothelial cell adhesion molecule 1) antibody (Abcam, ab28364) for 1 h at 4 °C on a rocker. After incubation, cells bound to the magnetic beads were collected with a magnetic tube holder and washed 6 times with 1 ml of DMEM containing 10% FBS. Purification and characterization of the obtained LSECs were confirmed by detection of PECAM1 (platelet and endothelial cell adhesion molecule 1).
2.10. Reagents and antibodies
Necrostatin-1 (S8037), ferrostatin-1 (S7243), Z-VAD-FMK (S7023), RSL3 (S8155), erastin (S7242), deferoxamine (S5742), eugenol (S4706), necrostatin-1 (S8037), necrosulfonamide (S8251), and rotenone (S2348) were bought from Selleck Chemicals. Buthionine sulfoximine (BSO, B2515), diethyl butylmalonate (DBM, 112038), NaN3 (S8032), MG-132 (M7449), CHX (C7698), and DMSO (156914) were obtained from Sigma-Aldrich. Antimycin (1397-94-0) was purchased from Santa Cruz Biotechnology. Phosphate-buffered saline (10010023) and Opti MEM medium (51985034) were obtained from Gibco BRL. Primary antibodies against BRD7 (ab56036), phospho-p53 (ab33889), p53 (ab131442), SLC25A28 (ab80467), VDAC (ab14734), Lamin B (ab16048), and β-actin (ab179467) were obtained from Abcam Technology. Anti-mouse IgG (7076) and anti-rabbit IgG (7054) were bought from Cell Signaling Technology.
2.11. Cell culture and drug treatment
HSC-LX2 (BNCC337957) and HSC-T6 (BNCC337976) cells were purchased from BeNa culture collection (Beijing, China). Cells were cultured in DMEM with 10% FBS, 1% antibiotics, and maintained at 37 °C in a humidified incubator of 5% CO2 and 95% air. Cell morphology was assessed using an inverted microscope with a Leica Qwin System (Leica, Germany). Erastin, RSL3, and BSO were dissolved in DMSO at a concentration of 10 mM and stored in a dark-colored bottle at −20 °C. The stock was diluted to the required concentration with DMSO when needed. Prior to the drug treatment, cells were grown to 70% confluence, and then exposed to drug at the indicated concentrations for different periods of time (0–24 h). Cells grown in a medium containing an equivalent amount of DMSO without drugs served as a control.
2.12. Plasmid construction and transfection
BRD7 shRNA (sc-92998, sc-141741), p53 shRNA (sc-44218, sc-45917), SLC25A28 shRNA (sc-90800, sc-149448), and control shRNA were obtained from Santa Cruz Biotechnology. The pcDNA3.1-BRD7 plasmid (NM_001108440.1, NM_ 013263.5), pcDNA3.1-P53 plasmid (NM_030989.3, NM_000546.5), pcDNA3.1-SLC25A28 plasmid (NM_00110 9515.1, NM_031212.4) and control pcDNA3.1 vector were purchased from Hanbio (Shanghai, China). The different p53 domains were constructed by PCR and cloned into pcDNA3.1-myc vector. The resulting plasmid was verified by sequencing. Transfections were performed with Lipofectamine™ 3000 (Invitrogen, L3000008) according to the manufacturer's instructions. VA-Lip-BRD7-shRNA, VA-Lip-P53-shRNA, VA-Lip-SLC25A28-shRNA and VA-Lip-Control-shRNA were prepared according to our previous reports [12,13]. In brief, 5 mg VA was added into 50 μl DMSO to form VA solution. 280 nmol VA solution and 0.14 μmol lipotrust solution (Hokkaido System Science, LEO-01) were mixed by vortexing in a 1.5-ml tube at 25 °C. 12.24 nmol BRD7 shRNA, p53 shRNA, SLC25A28 shRNA or control shRNA was added into VA-Lip solution with stirring at 25 °C. The VA-Lip solution was filtered. Fractions were collected and the material trapped in the filter was reconstituted with PBS to achieve the desired dose for in vivo use.
2.13. CRISPR/Cas9-mediated BRD7 knockout and S392A mutant
According to a reported protocol [33,34], CRISPR/Cas9 system was used to knockout BRD7 and construct S392A mutant. In brief, to knockout BRD7 in HSC cells, BRD7 CRISPR/Cas9 KO plasmid (sc-416299) and control plasmid were purchased from Santa Cruz Biotechnology. BRD7 CRISPR/Cas9 KO plasmid consists of a pool of 3 plasmids, each encoding the Cas9 nuclease and a target-specific 20 nt guide RNA (gRNA) designed for maximum knockout efficiency. To construct S392A mutant, regions of interest of the p53 gene from HSC cells were first sequenced. Sequences were analyzed with the online analysis tools available on benchling.com to identify and score CRISPR sgRNAs according to their specificity and efficiency. Each selected gRNA was cloned independently into the BbsI restriction sites of the pSpCas9 (BB)-2A-Puro (PX459) V2.0 vector (Addgene, 62988). To introduce the desired mutation into the genome by homologous recombination, we used a synthetic single-stranded DNA oligonucleotide (ssODN) of 200 bases as repair template. The ssODN was purchased from Integrated DNA Technologies (Liège, Belgium). Transfections were performed with Lipofectamine™ 3000 (Invitrogen, L3000008) according to the manufacturer's instructions. Cells were selected using 0.5 μg/ml puromycin for three days after transfection. Transfection efficiency was verified by real-time PCR.
2.14. Cell viability assay
Cell viability was examined with a CCK8 Cell Counting Kit (Beyotime Institute of Biotechnology, C0042) according to the manufacturer's instructions. In brief, HSC cells were plated in a 96-well plate (Sigma, CLS9898) and exposed to various concentrations of the cytotoxic compounds for the indicated times. The 10 μl CCK8 reagents were added to each well and incubated at 37 °C in 5% CO2 for 4 h, and then the plates were measured at 450 nm using a microplate reader (Thermo Fisher Scientific, 1410101).
2.15. Lipid peroxidation assay
The lipid peroxidation product malondialdehyde (MDA) concentration in cell lysates was assessed using a Lipid Peroxidation Assay Kit (Abcam, ab118970) according to the manufacturer's instructions. In brief, HSC cells were plated in a 6-well plate (NEST Biotechnology, 723001) and exposed to various concentrations of the cytotoxic compounds for the indicated times. Then, HSC cells were homogenized on ice in 300 μl MDA Lysis Buffer with 3 μl 100 × butylated hydroxytoluene and centrifuged at 13,000×g and 4 °C for 10 min to remove insoluble material. Subsequently, 200 μl supernatant from each homogenized sample was transferred to a microcentrifuge tube, and 600 μl thiobarbituric acid solution (Sigma-Aldrich, T5500) was added. Following incubation at 95 °C for 60 min and cooling to room temperature using an ice bath for 10 min, the 200 μl reaction mixture was transferred into a 96-well microplate for colorimetric analysis. The absorbance was measured at a wavelength of 532 nm and MDA levels were calculated using standard curve analysis.
2.16. Lipid ROS detection
Intracellular lipid ROS levels were measured using a peroxide-sensitive fluorescent probe C11-BODIPY (Thermo Fisher Scientific, D3861) according to the manufacturer's instructions. In brief, HSC cells were plated in a 6-well plate (NEST Biotechnology, 723001) and exposed to various concentrations of the cytotoxic compounds for the indicated times. Then, cells were incubated with C11-BODIPY at a final concentration of 10 μM in medium without FBS at 37 °C for 30 min and washed 3 times with medium. The levels of lipid ROS were determined by flow cytometer (Beckman, CytoFLEX).
2.17. Iron assay
The relative iron concentration in cell lysates was assessed using an Iron Assay Kit (Abcam, ab83366) according to the manufacturer's instructions. In brief, HSC cells were plated in a 6-well plate (NEST Biotechnology, 723001) and exposed to various concentrations of the cytotoxic compounds for the indicated times. Then, cells were immediately homogenized with 5 vol of Iron Assay buffer. The insoluble material was wiped off via the centrifugation at 13,000×g for 15 min at 4 °C. Sample wells were added iron reducer to reduce the switch from Fe3+ to Fe2+. The mixture was gently mixed via pipetting, and then reacted for 30 min in the dark. Subsequently, 100 μl iron probe was added into the standard and test samples, and the thoroughly mixed sample was incubated for 1 h at room temperature darkly. In the end point, the absorbance was determined at 593 nm using a microplate reader (Thermo Fisher Scientific, 1410101).
2.18. Glutathione assay
The relative GSH concentration in cell lysates was assessed using a Glutathione Assay Kit (Sigma, CS0260) according to the manufacturer's instructions. In brief, HSC cells were plated in a 6-well plate (NEST Biotechnology, 723001) and exposed to various concentrations of the cytotoxic compounds for the indicated times. Then, cells were homogenized in 50 mM MES buffer (Sigma-Aldrich, M8250) containing 1 mM EDTA (Sigma-Aldrich, 03620) and centrifuged at 10,000×g for 15 min at 4 °C. The supernatants were mixed with GSH detection working solution along with standards in 96-well plates and incubated for 25 min at room temperature. The absorbance was measured at 412 nm using a microplate reader (Thermo Fisher Scientific, 1410101).
2.19. Separation of nucleus, cytoplasm and mitochondria
Mitochondria were isolated from HSC cells using a Cell Mitochondria Isolation Kit (Abcam, ab110170), and nucleus were isolated from HSC cells using a Nuclei Isolation Kit (Sigma-Aldrich, NUC201) according to the manufacturer's instructions. In brief, HSC cells were plated in a 6-well plate (NEST Biotechnology, 723001) and exposed to various concentrations of the cytotoxic compounds for the indicated times. Then, cells were washed with pre-cooled PBS and lysed with a Cell Mitochondria Isolation buffer on ice. Mitochondria and cytoplasm were separated by grinding followed by centrifugation at 600×g for 10 min at 4 °C. Then the supernatant was further centrifuged at 11,000×g for 10 min at 4 °C. The pellet was collected as the mitochondria-enriched fraction, and it was then further resuspended in mitochondrial lysis buffer. The remaining supernatant was centrifuged 12,000×g for 10 min at 4 °C as cytosolic proteins. For nuclear separation, the cells were washed twice with cold PBS and centrifuged at 500×g for 3 min. The cell pellet was suspended in 200 ml of cytoplasmic extraction reagent I by vortexing. The suspension was incubated on ice for 10 min followed by the addition of 11 ml of a second cytoplasmic extraction reagent II, vortexed for 5 s, incubated on ice for 1 min and centrifuged for 5 min at 16,000×g. The supernatant fraction (cytoplasmic extract) was transferred to a pre-chilled tube. The insoluble pellet fraction containing crude nuclei was resuspended in 100 ml of nuclear extraction reagent by vortexing during 15 s and incubated on ice for 10 min, then centrifuged for 10 min at 16,000×g. The resulting supernatant constituting the nuclear extract was used for the subsequent experiments.
2.20. Detection of mitochondrial complexes enzymatic activity
Enzymatic activities of complexes I, II, III and IV were measured in mitochondria from HSC cells according to a reported protocol [35]. In brief, mitochondria were isolated from 5 × 106 HSC cells using the Mitochondria Isolation Kit (Abcam, ab110170). Then, complex I activity was determined using Complex I Enzyme Activity Microplate Assay Kit (Abcam, ab109721). Isolated mitochondria were added to the pre-coated with anti-complex I antibody microplate, and were incubated for 3 h at room temperature. Next, the assay solution containing NADH and dye was added, and absorbance was measured at 450 nm in kinetic mode for 5 min. Complex II activity was detected using Complex II Enzyme Activity Microplate Assay Kit (Abcam, ab109908). Isolated mitochondria were added to the pre-coated with anti-complex II antibody microplate, and were incubated for 3 h at room temperature. The assay solution containing succinate, ubiquinone 2 and DCIP dye was added, and absorbance was measured at 600 nm in kinetic mode for 40 min. Furthermore, complex III activity was determined using MitoTox™ Complex II + III OXPHOS Activity Assay Kit (Abcam, ab109905). Isolated mitochondria were incubated with oxidized cytochrome c on pre-coated 96-well microplate, and absorbance was measured at 550 nm in kinetic mode for 5 min at room temperature. Besides, complex IV activity was examined using Complex IV Rodent Enzyme Activity Microplate Assay Kit (Abcam, ab109911). Isolated mitochondria were incubated for 3 h at room temperature in microplate pre-coated with anti-Complex IV antibody after adding reduced cytochrome c, and absorbance was measured at 550 nm in kinetic mode for 5 min.
2.21. Detection of SLC25A28 activity
SLC25A28 activity was measured as the rate of mitochondrial iron uptake according to a reported protocol [36]. In Brief, 1 μCi 55FelC3 was incubated with 2 μg iron-free enterobactin (1 mg/ml in DMSO) at room temperature for 3 h, followed by the addition of purified mitochondria from HSC cells. The samples were incubated for 4 h at room temperature, and the amount of 55Fe in lysed mitochondria was determined by liquid scintillation.
2.22. Western blot analysis
For Western blot analysis, HSC cells were washed 3 times with PBS and then used for extraction of total proteins. Protein extracts were prepared by mammalian lysis buffer (Sigma-Aldrich, MCL1). Protein concentrations were measured by the Pierce™ BCA Protein Assay Kit (Thermo Scientific, 23250). Protein extracts were separated by SDSPAGE (Sigma-Aldrich, PCG2001) and transferred onto polyvinylidene fluoride (PVDF) membranes (Sigma-Aldrich, P2938). Then the PVDF membranes were blotted individually with appropriate primary antibodies and appropriate secondary antibodies. Protein bands were visualized using the chemiluminescence system (Merck Millipore, Darmstadt, Germany). Densitometry analyses were performed using ImageJ software.
2.23. Immunoprecipitation assay
For immunoprecipitation assay, HSC cells were lysed and sonicated in RIPA buffer (Sigma-Aldrich, R0278) for 30 min at 4 °C. The homogenates were centrifuged at 13,000×g for 30 min at 4 °C. About 10% of the supernatant was harvested for Western blot analysis as inputs, while the remaining cell lysate was incubated with indicated antibodies overnight at 4 °C. Protein A/G plus agarose beads (Santa Cruz Biotechnology, sc-2003) were added at 4 °C for another 2 h. The immunoprecipitation beads were washed with lysis buffer for 5 times, followed by Western blot analysis. Every experiment was repeated at least 3 times independently, and all the band detection is within the linear range.
2.24. RNA isolation and real-time PCR
Total RNA was isolated using TRIzol™ Reagent (Invitrogen, 15596018) and reverse transcribed using SuperScript™ III Platinum™ One-Step qRT-PCR Kit (Invitrogen, 11732088). Quantitative real-time PCR was performed using Fast SYBR™ Green Master Mix (Thermo Fisher Scientific, 4385610). The amounts of transcript were normalized to those for glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Melting curves were run to ensure amplification of a single product. Primer sequences were available on request.
2.25. Transmission electron microscopy
Transmission electron microscopy assay was performed according to our previous reports [12,13]. In brief, HSC cells were seeded onto 4-well Chambered Coverglass (Thermo Scientific, 155382) at a density of 2 × 104 cells/mL (14,000 cells/well). Images were acquired using the Thermo Scientific™ Talos™ F200C transmission electron microscope.
2.26. Immunofluorescence analysis
Immunofluorescence analysis was performed according to our previous reports [7]. In brief, HSC cells were fixed by addition of 15 μl of 16% paraformaldehyde (Sigma, P6148) directly to the 50 μl cell culture medium in each well (final concentration of formaldehyde was 3.7%). Samples were incubated for 30 min at room temperature. Well contents were aspirated and 50 μl/well of PBS containing 0.5% bovine serum albumin, 0.5% Triton X-100 (Sigma-Aldrich, T8787) was added to each well. Samples were incubated for 30 min at room temperature. Well contents were aspirated and washed 3 times with 100 μl/well of PBS. Supernatant was aspirated from the wells, and 25 μl/well of primary antibody (p53 antibody, Abcam, ab131442; SLC25A28 antibody, Abcam, ab80467) diluted 1:500 in Antibody Dilution Buffer (Sigma-Aldrich, U3510) was dispensed. Samples were incubated for 2 h at room temperature. Well contents were aspirated and washed 3 times with 100 μl/well of PBS. 25 μl/well of secondary antibody solution (Sigma-Aldrich, F4890) diluted 1:1000 and 1 μg/ml Hoechst 33342 (Sigma-Aldrich, B2261) diluted in Antibody Dilution Buffer were dispensed into each well. Samples were incubated for 2 h at room temperature. All the images were captured with the fluorescence microscope and representative images were shown. The software ImageJ was used to quantitate the fluorescent intensity on the micrographs.
2.27. Mitotracker green staining
Mitotracker green staining was performed according to a reported protocol [37]. In brief, HSC cells were plated in a 6-well plate (NEST Biotechnology, 723001) and exposed to various concentrations of the cytotoxic compounds for the indicated times. Then, cells were incubated with 50 nM MitoTracker Green (Beyotime, C1048) and 10 μM Hoechst 33342 (Sigma-Aldrich, B2261) to visualize mitochondria and nuclei, respectively. Then, the loading solution was removed, and the cell monolayers were washed 3 times with PBS and examined by confocal microscopy (Zeiss, LSM-800).
2.28. Trypan blue staining
HSC-LX2 cells were cultured in 6-well plate (NEST Biotechnology, 723001) for 24 h, and then treated with vehicle, erastin, erastin + ZVAD-FMK, erastin + necrostatin-1, and erastin + ferrostatin-1. After the indicated treatment times, cell suspension and 0.4% trypan blue solution (Sigma, T8154) were mixed in 9:1 ratio. After 3 min, the counting plate containing the live cells (no cytoplasmic fluorescence) and dead cells (blue cytoplasmic fluorescence) were counted. The trypan blue-positive ratio from 10 random fields was quantified with ImageJ software.
2.29. Statistical analyses
Statistical analysis was conducted by Student's t-test (two-group comparison) or one-way analysis of variance followed by Student-Newman-Keuls test (more than two groups). Matched controls were used in individual cell experiments and animal experiments and repeated 3 times. Data were expressed as mean ± standard error of the mean (SEM). In the retrospective cohort study, quantitative values were expressed as means with standard deviations or medians was expressed as range, while categorical values were expressed as percentage numbers. Categorical variables were analyzed using the chi-squared test or Fisher's exact test as appropriate. Statistical significance was indicated at P < 0.05. All statistical analyses were performed using the SPSS program (version 20.0; IBM, Somers, NY, USA).
3. Results
3.1. BRD7 protein levels were upregulated during HSC ferroptosis
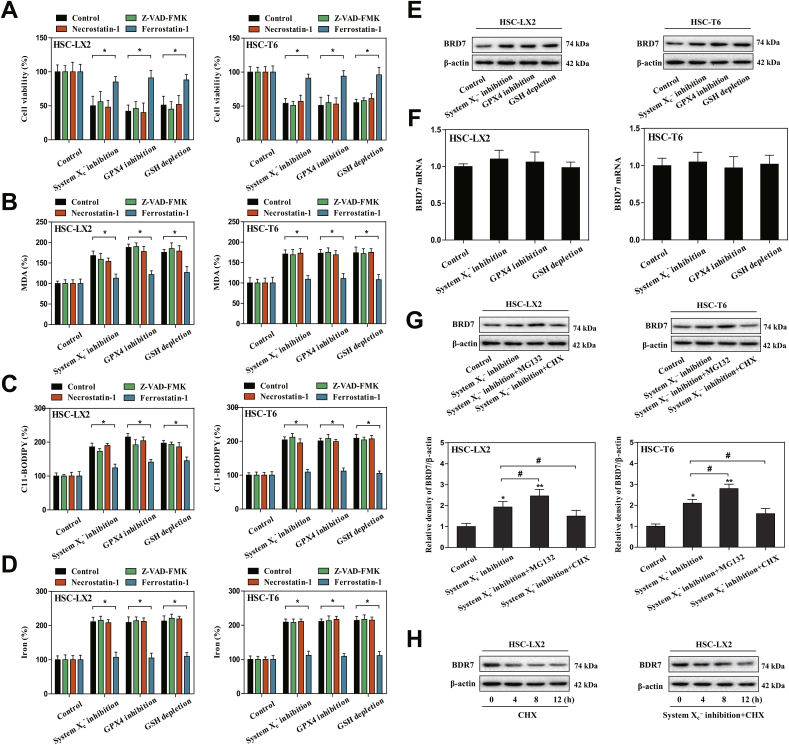
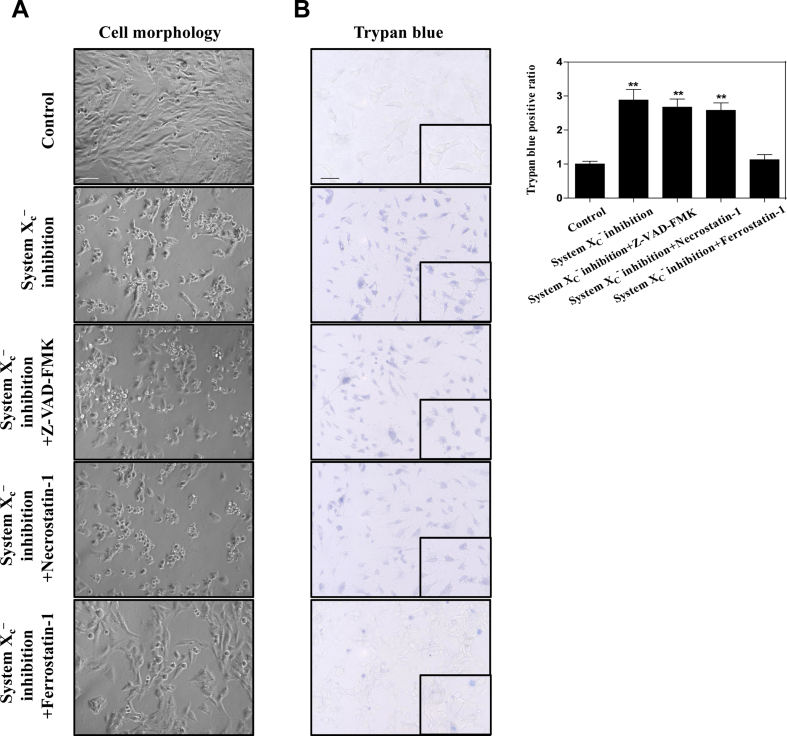
We previously reported that system inhibition by preclinical drug erastin or clinical drug sorafenib, GPX4 inhibition by RSL3, and GSH depletion by BSO can trigger ferroptosis in rat and human HSC lines (HSC-T6 and HSC-LX2) [12,13]. Consistent with previous results, we found that system inhibition-, GPX4 inhibition-, and GSH depletion-induced cell death in rat and human HSC lines were completely impaired by ferrostatin-1 (a potent ferroptosis inhibitor) but not necrostatin-1 (a potent necroptosis inhibitor) and ZVAD-FMK (a potent apoptosis inhibitor) (Fig. 1A). Moreover, morphological observation and cellular reactive trypan blue staining confirmed that system inhibition-induced cell death was rescued by ferroptosis inhibitor but not apoptosis inhibitor and necroptosis inhibitor (Figs. S3A and B). It is well known that lipid peroxidation, lipid ROS generation, and redox-active iron overload occurred simultaneously and were mutually amplifying events during ferroptosis [38]. Interestingly, lipid peroxidation product MDA (Fig. 1B), lipid ROS generation (Fig. 1C), and iron accumulation (Fig. 1D) were all induced during HSC ferroptosis by GSH depletion, GPX4 inhibition, and system inhibition. Noteworthy, it is ferrostatin-1 rather than ZVAD-FMK and necrostatin-1 that reversed the classical ferroptotic events (Fig. 1B–D). Collectively, these findings fully confirmed that system inhibition by erastin, GPX4 inhibition by RSL3, and GSH depletion by BSO can trigger HSC ferroptosis in vitro.
Fig. 1.
BRD7 protein levels were upregulated during HSC ferroptosis. HSC-LX2 and HSC-T6 cells were exposed to erastin (10 μM), RSL3 (2.5 μM), and BSO (200 μM) with or without the indicated inhibitors (ZVAD-FMK, 10 μM; necrostatin-1, 10 μM; ferrostatin-1, 1 μM) for 24 h. (A) Cell viability were determined by CCK8 kit (n = 3 in every group, *P < 0.05). (B-D) The levels of MDA, lipid ROS, and iron were determined by commercial kits (n = 3 in every group, *P < 0.05). (E, F) HSC-T6 and HSC-LX2 cells were exposed to erastin (10 μM), RSL3 (2.5 μM), and BSO (200 μM) for 24 h. BRD7 protein and mRNA levels were examined by Western blot and real-time PCR (n = 3 in every group). (G) HSC-LX2 and HSC-T6 cells were exposed to erastin (10 μM) with or without cycloheximide (CHX, 20 μg/ml) or MG-132 (5 μM) for 24 h, and BRD7 protein level was determined by Western blot (n = 3 in every group, *P < 0.05, **P < 0.01, #P < 0.05). (H) HSC-LX2 cells were treated with CHX (20 μg/ml) for 12 h or treated with erastin (10 μM) and CHX (20 μg/ml) for 12 h. BRD7 protein level was examined by Western blot 0, 4, 8, and 12 h after the end of cell exposure to erastin and CHX (n = 3 in every group).
BRD7, a well-established bromodomain-containing factor, plays pivotal roles in the regulation of cell fate, involving in cellular growth, apoptosis, senescence, pyroptosis, and necrosis [[26], [27], [28]]. Whether BRD7 was involved in controlling HSC ferroptosis? To verify this possibility, we first determined BRD7 mRNA and protein levels during HSC ferroptosis. Remarkably, the protein expression but not mRNA expression of BRD7 was significantly upregulated in the induction of ferroptosis (Fig. 1E and F), suggesting that induction of BRD7 protein in ferroptosis may occur in a transcription-independent manner. To further confirm this potential mechanism in BRD7 protein induction during HSC ferroptosis, the chemical protein synthesis inhibitor cycloheximide (CHX) was used to limit BRD7 protein synthesis, and a selective 26S proteosomal inhibitor MG-132 was utilized to prevent BRD7 protein degradation. As a result, CHX treatment completely abrogated the increased BRD7 protein level, whereas MG-132 treatment markedly augmented the protein expression of BRD7 in the system inhibition-mediated ferroptosis (Fig. 1G). Additionally, the stability of BRD7 protein in ferroptosis was also analyzed. As expected, ferroptotic stimulator erastin dramatically prolonged the half-life of BRD7 protein (Fig. 1H). Overall, these data thoroughly showed that BRD7 protein expression was upregulated via inhibition of ubiquitination-peroxisome pathway during HSC ferroptosis.
3.2. Upregulated BRD7 expression contributes to HSC ferroptosis
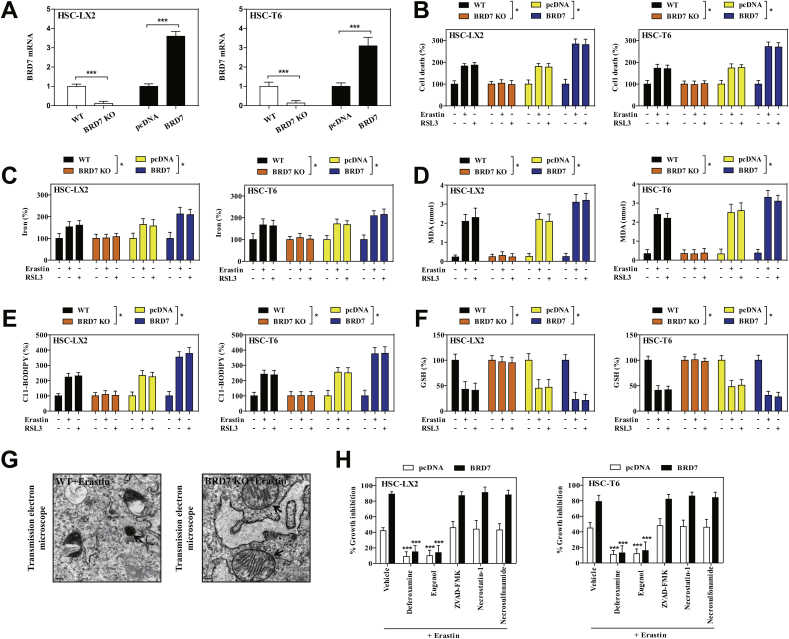
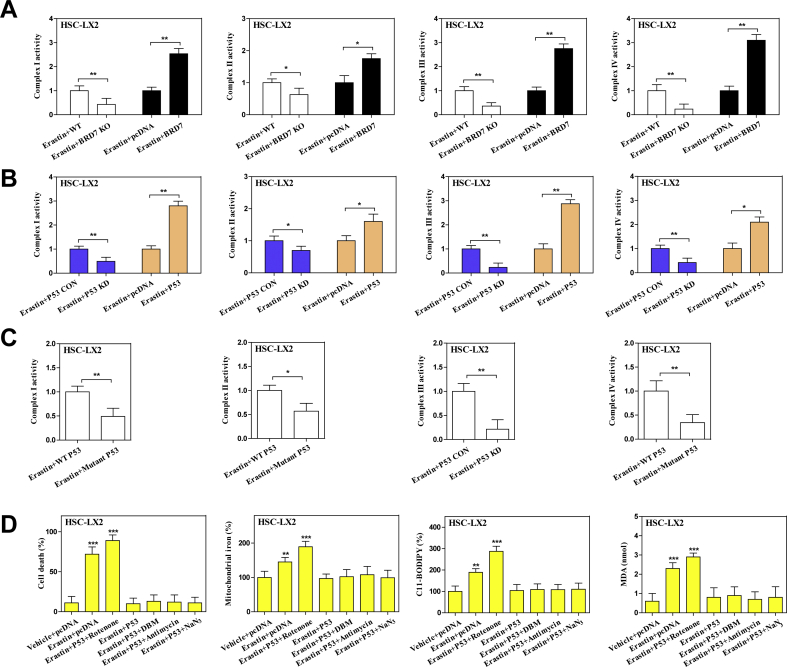
To determine unambiguously whether upregulated BRD7 expression contributes to HSC ferroptosis, the modified inducible CRISPR/Cas9 system was used to establish BRD7 knockout HSC lines, and the specific BRD7 plasmid was utilized to construct BRD7 knockin HSC lines (Fig. 2A). It is worth noting that CRISPR/Cas9-mediated BRD7 knockout greatly reduced the sensitivity of HSCs to ferroptotic death induced by system inhibition or GPX4 inhibition, whereas BRD7 plasmid mediated BRD7 knockin contributed to ferroptotic death (Fig. 2B). Furthermore, the knockout of BRD7 dramatically abolished system inhibition- or GPX4 inhibition-mediated iron accumulation (Fig. 2C), lipid peroxidation (Fig. 2D), lipid ROS production (Fig. 2E), and GSH depletion (Fig. 2F). In striking contrast, the knockin of BRD7 contributed to classical ferroptotic events (Fig. 2C–F). It is well known that mitochondrial shrinkage and loss of mitochondrial ridge are typical features of ferroptotic cells [15]. As expected, system inhibition by erastin evidently shanked mitochondria and decreased the number of ridges in wide type HSC-LX2, whereas CRISPR/Cas9-mediated BRD7 knockout completely abolished the typical changes of ferroptotic cells (Fig. 2G). Besides, it should be noted that several ferroptosis inhibitors (iron chelator deferoxamine and lipid peroxidation inhibitor eugenol) significantly reversed system inhibition-mediated growth inhibition in BRD7-overexpressed HSCs (Fig. 2H). However, ZVAD-FMK (a pan-caspase inhibitor), necrostatin-1 (a potent necroptosis inhibitor that targets RIPK1), and necrosulfonamide (a potent necroptosis inhibitor that targets MLKL) did not remarkably reverse this process (Fig. 2H). Altogether, these results supported that upregulated BRD7 protein may contribute to ferroptosis, but not necroptosis and apoptosis.
Fig. 2.
Upregulated BRD7 protein level contributed to HSC ferroptosis. (A) The modified inducible CRISPR/Cas9 system was used to establish BRD7 knockout HSC lines, and the specific BRD7 plasmid was utilized to construct BRD7 knockin HSC lines. Transfection efficiency was verified by real-time PCR (n = 3 in every group, ***P < 0.001) (B–F) BRD7 knockout and knockin HSC lines were exposed to erastin (10 μM) or RSL3 (2.5 μM) for 24 h. Cell death was determined by CCK8 kit (n = 3 in every group, *P < 0.05). The levels of iron, MDA, lipid ROS, and GSH were determined by commercialized kits (n = 3 in every group, *P < 0.05). (G) BRD7 wild type (WT) and knockout (KO) HSC-LX2 cells were exposed to erastin (10 μM) for 24 h. The typical changes of ferroptotic cells were examined by transmission electron microscope. (H) BRD7 knockin HSC lines were exposed to erastin (10 μM) with or without the indicated inhibitors (Deferoxamine, 100 μM; eugenol, 2 μM; ZVAD-FMK, 10 μM; necrostatin-1, 10 μM; necrosulfonamide, 5 μM) for 24 h. Cell viability were determined by CCK8 kit (n = 3 in every group, ***P < 0.001).
3.3. Mitochondrial translocation of p53 mediated BRD7-enhanced HSC ferroptosis
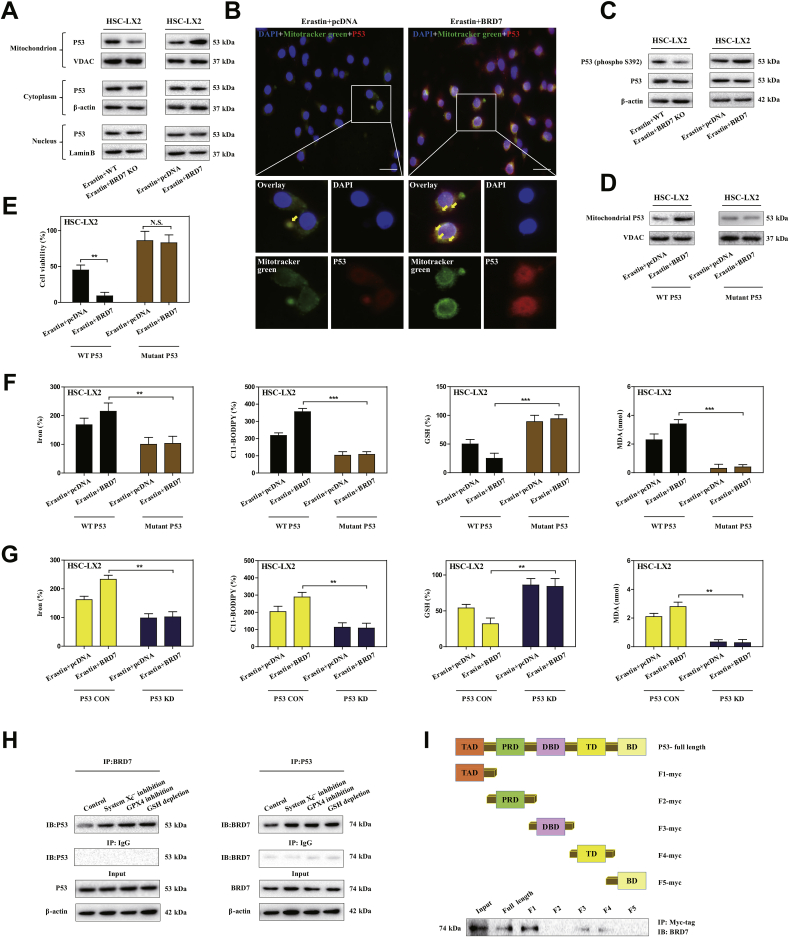
BRD7 has been demonstrated to be required for p53 to play its biological function [25]. Does upregulated BRD7 expression contribute to HSC ferroptosis in a p53-dependent mechanism? To verify the possibility, we analyzed the expression level of p53 and subcellular localization. Interestingly, BRD7 knockout significantly reduced the expression of p53 in mitochondria rather than in cytoplasm and nucleus, whereas BRD7 knockin markedly increased the expression of p53 in mitochondria but not in cytoplasm and nucleus (Fig. 3A). Moreover, confocal imaging confirmed that BRD7 knockin promoted co-localization of p53 (red fluorescence) and mitochondria (mitotracker green) during HSC ferroptosis (Fig. 3B). All these results suggest that mitochondrial translocation of p53 is essential for BRD7-enhanced HSC ferroptosis. It has been reported that the phosphorylation of serine 392 is necessary for p53 mitochondrial translocation [34]. Therefore, we next investigated the impact of BRD7 knockout and knockin on serine 392 phosphorylation. As expected, BRD7 knockin noticeably triggered serine 392 phosphorylation, but BRD7 knockout completely counteracted this effect in ferroptosis (Fig. 3C). According to a reported protocol [34], we also used CRISPR/Cas9 system to substitute serine 392 by a non-phosphorylatable alanine (S392A mutant) in HSC-LX2 cells. Attractively, the S392A mutant completely impaired the BRD7 knockin-mediated p53 mitochondrial translocation (Fig. 3D), and, in turn, reduced the ferroptotic cell death during HSC ferroptosis (Fig. 3E). Additionally, p53 knockdown and S392A mutant both can reduced the classical ferroptotic events by BRD7 knockin, including redox-active iron overload, lipid ROS accumulation, GSH depletion, and lipid peroxidation (Fig. 3F and G). Subsequently, we tested whether BRD7 knockin induced serine 392 phosphorylation and p53 mitochondrial translocation through direct binding to p53. Immunoprecipitation (IP) analysis clearly indicated that system inhibition, GPX4 inhibition, and GSH depletion could promoted the direct binding of BRD7 and p53 (Fig. 3H). It is well known that the p53 protein consists of five main domains, namely the N-terminal transactivation domain (TAD), the Pro-rich domain (PRD), the central DNA binding domain (DBD), the tetramerization domain (TD) and the C-terminal basic domain (BD) [39]. Which domain of p53 protein can bind directly to BRD7? To address this question, myc-tagged plasmids which contained the full length p53, as well as five separated domains (TAD, PRD, DBD, TD, and BD) were constructed, respectively. After co-transfecting HSC-LX2 cells with each p53 constructs and BRD7, antibodies against myc-tag and BRD7 were used for IP and Western blot, respectively. More importantly, BRD7 could directly bind to p53 through its TAD domain, but not PRD, DBD, TD, and BD domain (Fig. 3I). Taken together, these findings fully revealed that mitochondrial translocation of p53 may mediate BRD7-enhanced HSC ferroptosis.
Fig. 3.
Mitochondrial translocation of p53 mediated BRD7-enhanced HSC ferroptosis. (A) HSC-LX2 cells with BRD7 knockout and knockin were exposed to erastin (10 μM) for 24 h. Then, the mitochondria, cytoplasm and nucleus were extracted by commercialized kits, respectively. The levels of p53 in cytoplasm, mitochondria, and nucleus were determined by Western blot (n = 3 in every group). (B) HSC-LX2 cells with BRD7 knockin were exposed to erastin (10 μM) for 24 h. The co-localization of p53 (red fluorescence) and mitochondria (mitotracker green) was determined by confocal imaging. Scale bars are 100 μm. Representative photographs were shown (n = 3 in every group). (C) HSC-LX2 cells with BRD7 knockout and knockin were exposed to erastin (10 μM) for 24 h. The expression of total p53 and phosphorylated p53 was detected by Western blot (n = 3 in every group). Serine 392 was substituted by a non-phosphorylatable alanine (S392A mutant) with CRISPR/Cas9 system in HSC-LX2 cells with BRD7 knockin. Then, HSC-LX2 cells were exposed to erastin (10 μM) for 24 h. (D) The levels of p53 in mitochondria were examined by Western blot (n = 3 in every group). (E) Cell viability were determined by CCK8 kit (n = 3 in every group, **P < 0.01, N·S., not significant). (F) The levels of iron, lipid ROS, GSH, and MDA were determined by commercial kits (n = 3 in every group, **P < 0.01, ***P < 0.001). (G) HSC-LX2 cells were transfected with p53 shRNA and BRD7 plasmid, and were exposed to erastin (10 μM) for 24 h. The levels of iron, lipid ROS, GSH, and MDA were determined by commercial kits (n = 3 in every group, **P < 0.01). (H) HSC-LX2 cells were exposed to erastin (10 μM), RSL3 (2.5 μM), and BSO (200 μM) for 24 h. The combination of BRD7 and p53 was examined by immunocoprecipitation (n = 3 in every group). (I) The full-length and different domains of p53 with myc tag were constructed, and then were cotransfected with pcDNA3.1-BRD7 into HSC-LX2 cells. Cell lysates were immunoprecipitated with anti-myc antibody and blotted with anti-BRD7 antibody (n = 3 in every group). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Mitochondrial p53 interacted with SLC25A28 to disorganize iron homeostasis in BRD7-enhanced HSC ferroptosis
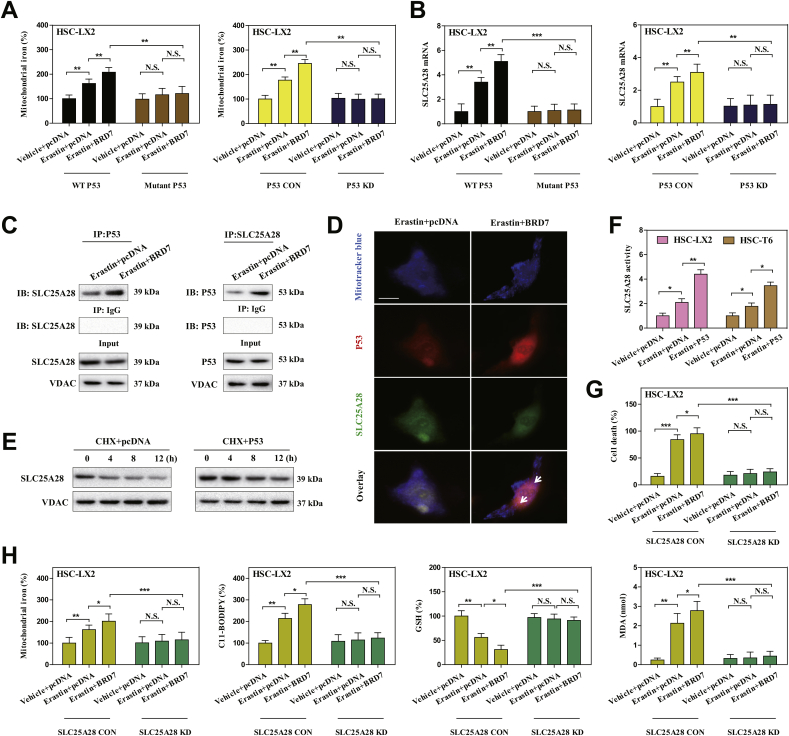
As the mitochondria serves as main hubs of iron storage and utilization [40], is the p53 mitochondrial translocation involved in the regulation of mitochondrial iron homeostasis? To test this possibility, we isolated mitochondria of ferroptotic cells and further analyzed the effect of S392A mutant and p53 knockdown on mitochondrial iron metabolism. As a result, compared to control cells, erastin treatment apparently increased mitochondrial iron content, whereas S392A mutant and p53 knockdown completely reduced mitochondrial iron accumulation in ferroptosis (Fig. 4A). These data suggest that mitochondrial p53 may regulate mitochondrial iron homeostasis during HSC ferroptosis. To further define the direct target of mitochondrial p53 in ferroptosis, we performed an unbiased screen in mitochondrial extract of S392A mutant HSC-LX2 cells, involving in iron trafficking to mitochondria (such as SLC25A28, SLC25A39, LC22A4, TMEM14C, ABCB10, ferrochelatase, and sideroflexin-1), iron transfer out of mitochondria (such as ABCB7, HEBP1, and FLVCR1), mitochondrial iron stockpile (such as mitochondrial ferritin), and mitochondrial iron usage (such as iron-sulfur cluster biogenesis and heme synthesis) [41]. Of note, we identified a candidate target SLC25A28 in iron trafficking, but not mitochondrial iron utilization, mitochondrial iron storage, and iron transfer, was evidently inhibited upon S392A mutant or p53 knockdown according to the fold change (Fig. 4B). Moreover, IP analysis clearly supported that BRD7 knockin promoted direct binding of p53 to SLC25A28 in mitochondria (Fig. 4C). Further, confocal imaging confirmed that p53 (red fluorescence) and SLC25A28 (green fluorescence) overlapped well in mitochondria (mitotracker blue) in BRD7-enhanced HSC ferroptosis (Fig. 4D). Dose the binding of p53 enhance the stability and activity of SLC25A28? Indeed, Western blot analysis showed that the half-life of SLC25A28 was evidently prolonged in HSC-T6 cells transfected with p53-overexpressing constructs (Fig. 4E). Besides, p53 knockin also significantly enhanced the activity of SLC25A28 in ferroptosis (Fig. 4F). Noteworthy, we constructed specific small interfering RNA against SLC25A28 for reverse validation. As expected, shRNA-mediated knockdown of SLC25A28 completely abolished the mitochondrial iron accumulation, and, in turn, impaired BRD7 knockin-enhanced ferroptotic cell death (Fig. 4G) and classical ferroptotic events (Fig. 4H). Collectively, these results indicated that mitochondrial p53 interacted with SLC25A28 to trigger mitochondrial iron overload in BRD7-enhanced HSC ferroptosis.
Fig. 4.
Mitochondrial p53 interacted with SLC25A28 to trigger iron accumulation in BRD7-enhanced HSC ferroptosis. The mutant p53 or p53 KD HSC-LX2 cells were transfected with BRD7 plasmid, and were exposed to erastin (10 μM) for 24 h. (A) Mitochondrial iron levels were determined by iron assay kit (n = 3 in every group, **P < 0.01, N·S., not significant). (B) SLC25A28 expression was determined by real-time PCR (n = 3 in every group, **P < 0.01, ***P < 0.001, N·S., not significant). HSC-LX2 cells were transfected with BRD7 plasmid, and then were exposed to erastin (10 μM) for 24 h. (C) The combination of p53 and SLC25A28 was examined by immunocoprecipitation (n = 3 in every group). (D) The co-localization of p53 (red fluorescence), SLC25A28 (green fluorescence), and mitochondria (mitotracker blue) was determined by confocal imaging. Scale bars are 100 μm. Representative photographs were shown (n = 3 in every group). (E) HSC-LX2 cells were transfected with p53 plasmid, and were exposed to erastin (10 μM) with or without cycloheximide (CHX, 20 μg/ml) for 24 h. The levels of SLC25A28 were evaluated 0, 4, 8, and 12 h after the end of cell exposure to erastin and CHX (n = 3 in every group). (F) The indicated cells were transfected with p53 plasmid, and were exposed to erastin (10 μM) for 24 h. The SLC25A28 activity was examined as the rate of mitochondrial iron uptake (n = 3 in every group, *P < 0.05, **P < 0.01). (G, H) HSC-LX2 cells were transfected with BRD7 plasmid and SLC25A28 shRNA, and were exposed to erastin (10 μM) for 24 h. Cell death and the levels of mitochondrial iron, lipid ROS, GSH, and MDA were determined by commercial kits (n = 3 in every group, *P < 0.05, **P < 0.01,***P < 0.001, N·S., not significant). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. BRD7-P53-SLC25A28 axis disturbed electron transport chain to aggravate lipid peroxidation in BRD7-enhanced HSC ferroptosis
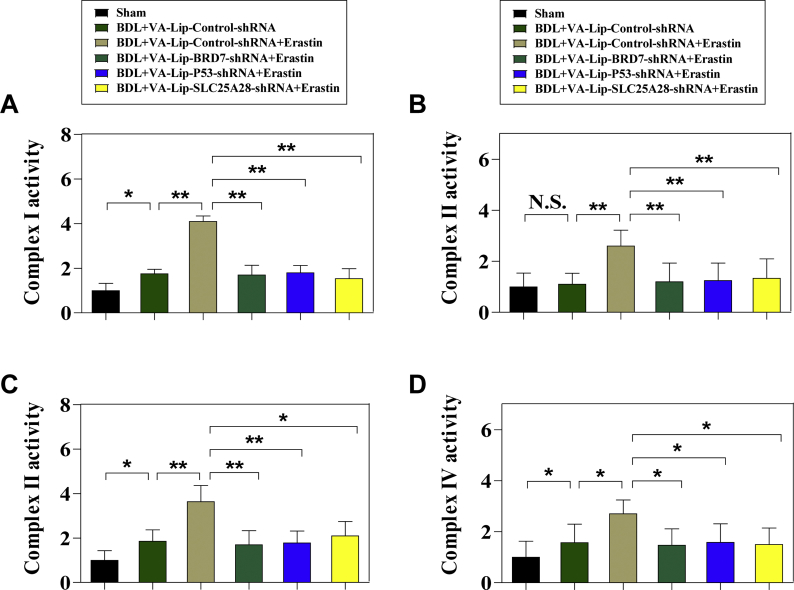
It should be noted that iron is a crucial component of iron-sulfur clusters and heme, and is essential for enzymes participating in mitochondria electron transport chain (ETC) (Fig. 5A). Dose mitochondrial iron accumulation by BRD7-P53-SLC25A28 axis disturb ETC in BRD7-enhanced HSC ferroptosis? To address this question, we determined the impact of SLC25A28 knockdown and knockin on mitochondria biogenesis during HSC ferroptosis. Interestingly, neither SLC25A28 knockdown nor SLC25A28 knockin affected mitochondrial mass (Fig. 5B). Furthermore, SLC25A28 knockdown and SLC25A28 knockin did not affect mitochondrial DNA (mtDNA) level compared to controls (Fig. 5C). Notably, we further investigated the impact of SLC25A28 knockdown or SLC25A28 knockin on electron transport activity of protein complexes embedded in the inner-membrane of mitochondria. As expected, SLC25A28 knockdown significantly reduced activities of complexes I, II, III, and IV, whereas SLC25A28 knockin dramatically increased electron transport activities of these complexes in ferroptosis (Fig. 5D). Similarly, BRD7 knockin (Fig. S4A) and p53 knockin (Fig. S4B) also promoted electron transport activities of complexes, whereas BRD7 knockout (Fig. S4A), p53 knockdown (Fig. S4B), and S392A mutant (Fig. S4C) blocked the function of ETC. Besides, we also detected the expression of several subunits of mitochondrial complexes in BRD7-enhanced HSC ferroptosis. Our results showed that the expression of NDUFB8 (complex I), SDHB (complex II), MTCO1 (complex III), and UQCRC2 (complex IV) was significantly increased in BRD7-overexpressing HSC-T6 cells upon treatment with erastin (Fig. 5E). In contrast, BRD7 knockout dramatically inhibited the expression of these subunits in ferroptosis (Fig. 5E). These data suggested that BRD7-P53-SLC25A28 axis disturbed the function of mitochondrial complexes via inhibiting subunit levels rather than mtDNA replication during HSC ferroptosis. Importantly, we utilized an array of ETC inhibitors to further determine whether ETC is required for BRD7-enhanced HSC ferroptosis. As a result, inhibitors of mitochondrial complex I (rotenone), complex II (DBM), complex III (antimycin), and complex IV (NaN3) all impaired BRD7 knockin (Fig. 5F), p53 knockin (Fig. S4D), or SLC25A28 knockin (Fig. 5G) enhanced ferroptotic cell death and classical ferroptotic events. Overall, these data showed that mitochondrial iron accumulation by BRD7-P53-SLC25A28 axis resulted in the hyperfunction of ETC in BRD7-enhanced HSC ferroptosis, which could further lead to lipid peroxidation and HSC ferroptosis.
Fig. 5.
BRD7-P53-SLC25A28 axis disturbed electron transport chain to aggravate lipid peroxidation in BRD7-enhanced HSC ferroptosis. (A) Scheme indicated mitochondrial electron transport chain (ETC) complexes and their inhibitors. HSC-LX2 cells were transfected with SLC25A28 shRNA or SLC25A28 plasmid, and were exposed to erastin (10 μM) for 24 h. (B) Mitochondria mass was determined by plate reader-based quantitation of Mito Tracker (n = 3 in every group, N·S., not significant). (C) mtDNA copy number was examined by real-time PCR (n = 3 in every group, N·S., not significant). (D) The enzymatic activities of mitochondrial complexes were analyzed by commercial kits (n = 3 in every group, *P < 0.05, **P < 0.01, ***P < 0.001). (E) BRD7 knockout or knockin HSC-LX2 cells were exposed to erastin (10 μM) for 24 h. Subunits expression of mitochondrial complexes was detected by real-time PCR (n = 3 in every group, *P < 0.05, **P < 0.01, ***P < 0.001). (F) HSC-LX2 cells were transfected with BRD7 plasmid, and were exposed to erastin (10 μM) with or without ETC inhibitors (rotenone, 10 mM; diethyl butylmalonate (DBM), 2 mM; Antimycin A (Anti A), 50 mM; and NaN3, 15 mM) for 24 h. Cell death and the levels of mitochondrial iron, lipid ROS, and MDA were determined by commercial kits (n = 3 in every group, **P < 0.01, ***P < 0.001). (G) HSC-LX2 cells were transfected with SLC25A28 plasmid, and were exposed to erastin (10 μM) with or without ETC inhibitors (rotenone, 10 mM; DBM, 2 mM; Antimycin A (Anti A), 50 mM; and NaN3, 15 mM) for 24 h. Cell death and the levels of mitochondrial iron, lipid ROS, and MDA were determined by commercial kits (n = 3 in every group, **P < 0.01, ***P < 0.001).
3.6. HSC-specific blockade of BRD7-P53-SLC25A28 axis impaired erastin-induced HSC ferroptosis in murine liver fibrosis
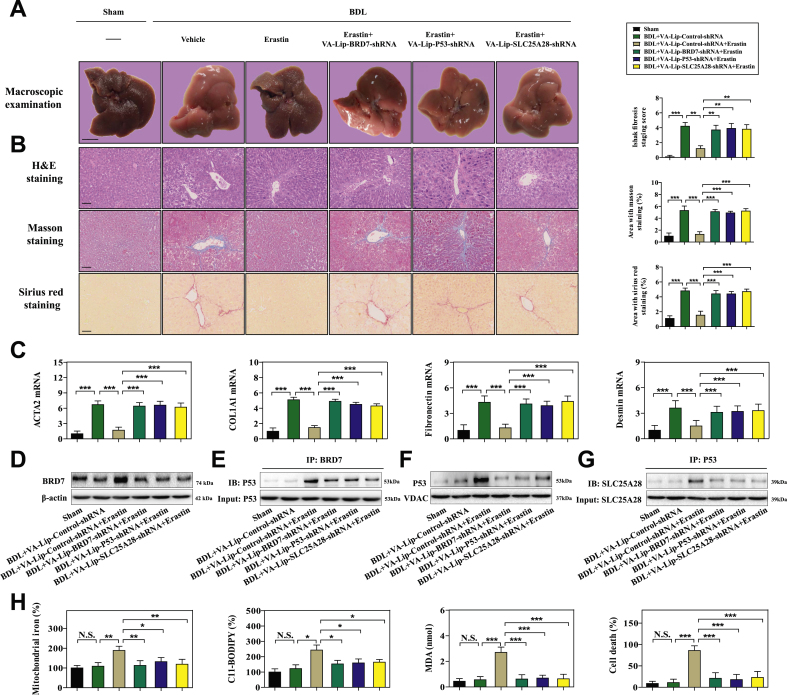
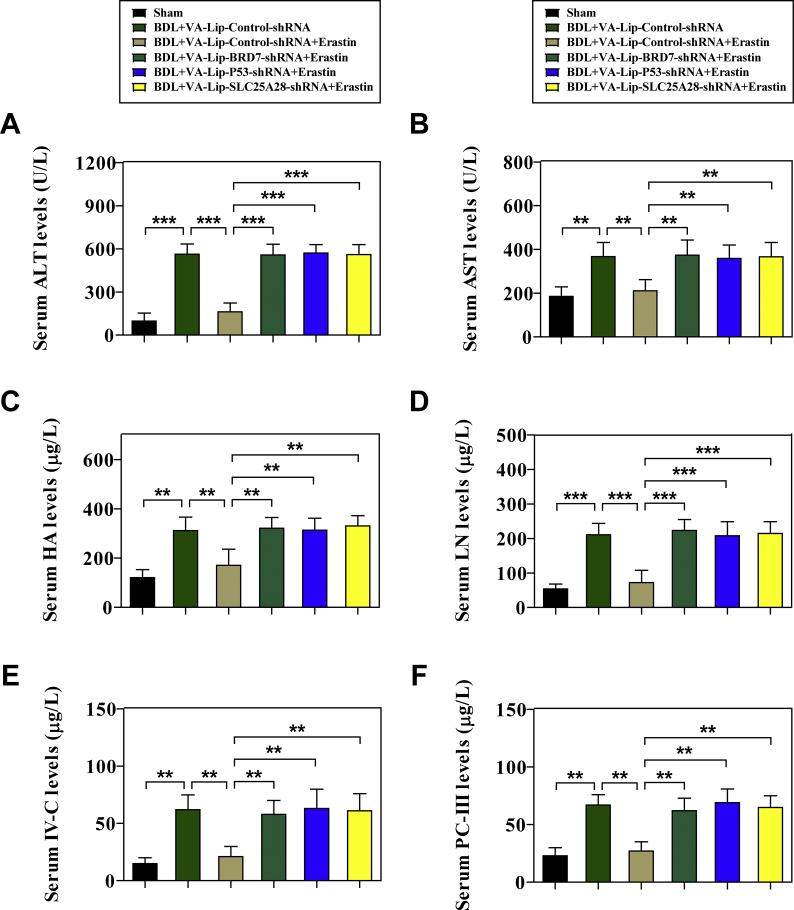
To evaluate whether BRD7-P53-SLC25A28 axis regulates ferroptosis in vivo, a classical murine liver fibrosis model induced by bile duct ligation (BDL) was established [12,13]. Noteworthy, vitamin A-coupled liposomes containing BRD7 shRNA (VA-Lip-BRD7-shRNA), p53 shRNA (VA-Lip-P53-shRNA), and SLC25A28 shRNA (VA-Lip-SLC25A28-shRNA) were used to block the BRD7-P53-SLC25A28 axis in HSCs [12,13]. First, we determined the effect of BRD7-P53-SLC25A28 axis on the pathological changes of liver fibrosis in vivo. As a result, severe fibrotic liver injury appeared in the liver of model group mice, but erastin treatment ameliorated BDL-mediated fibrotic pathological changes (Fig. 6A). Intriguingly, the improving effect of erastin on liver fibrosis was evidently damaged by treatment with VA-Lip-BRD7-shRNA or VA-Lip-P53-shRNA or VA-Lip-SLC25A28-shRNA (Fig. 6A). Furthermore, histopathologic analysis including hematoxylin and eosin (H&E), masson, and sirius red staining clearly indicated that collagen overdeposited in fibrotic livers of model group, but erastin treatment markedly reduced collagen deposition (Fig. 6B). Of note, VA-Lip-BRD7-shRNA or VA-Lip-P53-shRNA or VA-Lip-SLC25A28-shRNA almost reversed the effect of erastin on collagen deposition (Fig. 6B). Moreover, serum levels of liver fibrosis markers including glutamic-pyruvic transaminase (GPT/ALT), glutamic-oxaloacetic transaminase 1 (GOT1/AST), hyaluronic acid (HA), laminin (LN), type IV collagen (IV–C), and procollagen III (PC-III) were increased in model group compared with sham group, but erastin treatment significantly reduced the elevated serum levels of liver fibrosis markers (Figs. S5A–F). Interestingly, treatment with VA-Lip-BRD7-shRNA or VA-Lip-P53-shRNA or VA-Lip-SLC25A28-shRNA completely impaired the inhibitory effects of erastin (Figs. S5A–F). Furthermore, real-time PCR analysis confirmed that intraperitoneal injection of erastin for 2 weeks prominently decreased the mRNA expression of ACTA2, COL1A1 (collagen type I alpha 1 chain), fibronectin, and desmin (Fig. 6C), but VA-Lip-BRD7-shRNA or VA-Lip-P53-shRNA or VA-Lip-SLC25A28-shRNA impaired the inhibitory effect of erastin (Fig. 6C).
Fig. 6.
HSC-specific blockade of BRD7-P53-SLC25A28 axis impaired erastin-mediated HSC ferroptosis in murine liver fibrosis. Mice of 6 groups were treated with Sham, BDL + VA-Lip-Control-shRNA, BDL + VA-Lip-Control-shRNA + erastin, BDL + VA-Lip-BRD7-shRNA + erastin, BDL + VA-Lip-P53-shRNA + erastin, or BDL + VA-Lip-SLC25A28-shRNA + erastin. (A, B) The fibrotic injury was observed by macroscopic examination and histopathological examination (n = 6 in every group, **P < 0.01, ***P < 0.001). (C) The levels of ACTA2, COL1A1, fibronectin, and desmin were examined by real-time PCR in fibrotic liver tissues (n = 6 in every group, ***P < 0.001). Primary HSCs were also isolated from fibrotic livers. (D) The levels of BRD7 were detected by Western blot (n = 6 in every group). (E) The combination of BRD7 and p53 was examined by immunocoprecipitation (n = 6 in every group). (F) The levels of p53 in mitochondria were detected by Western blot (n = 6 in every group). (G) The combination of SLC25A28 and p53 was examined by immunocoprecipitation (n = 6 in every group). (H) The levels of mitochondrial iron, lipid ROS MDA, and cell death were determined by commercial kits (n = 6 in every group, *P < 0.05, **P < 0.01, ***P < 0.001, N·S., not significant).
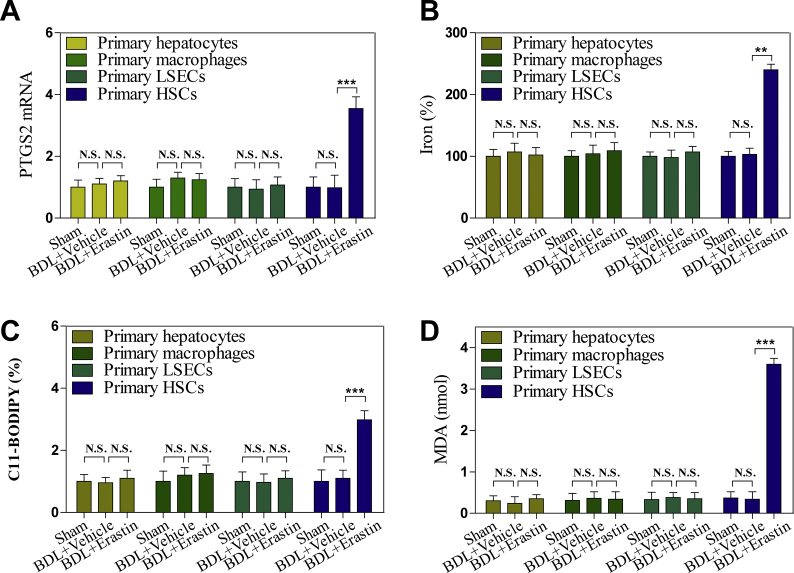
Which specific cells have undergone ferroptosis following erastin treatment? To clarify this issue, we further isolated the primary hepatocytes, macrophages, liver sinusoidal endothelial cells (LSECs), and HSCs from fibrotic livers. Importantly, erastin treatment evidently increased the expression of the ferroptosis marker PTGS2 (prostaglandin-endoperoxide synthase 2) in primary HSCs but not in primary hepatocytes, LSECs, and macrophages (Fig. S6A). Further, erastin treatment could trigger redox-active iron overload (Fig. S6B), lipid ROS accumulation (Fig. S6C), and MDA production (Fig. S6D) in primary HSCs, but not in primary hepatocytes, LSECs, and macrophages. These data fully suggest that ferroptosis inducer erastin may improve liver fibrosis by inducing ferroptosis in HSCs, but not in hepatocytes, LSECs, and macrophages.
To further evaluate the regulatory function of BRD7-P53-SLC25A28 axis in erastin-mediated HSC ferroptosis, we isolated primary HSCs from fibrotic livers of mice treated with sham, BDL + VA-Lip-Control-shRNA, BDL + VA-Lip-Control-shRNA + erastin, BDL + VA-Lip-BRD7-shRNA + erastin, BDL + VA-Lip-P53-shRNA + erastin, and BDL + VA-Lip-SLC25A28-shRNA + erastin, respectively. Consistent with in vitro results, system inhibition by erastin could promote BRD7 protein expression (Fig. 6D), the binding of BRD7 and p53 (Fig. 6E), p53 mitochondrial translocation (Fig. 6F), the interaction between p53 and SLC25A28 (Fig. 6G), mitochondrial iron accumulation (Fig. 6H), ETC hyperfunction (Figs. S7A–D), the increased lipid peroxidation (Fig. 6H), and in turn, ferroptotic cell death (Fig. 6H). More importantly, HSC-specific blockade of BRD7-P53-SLC25A28 axis by VA-Lip-BRD7-shRNA or VA-Lip-P53-shRNA or VA-Lip-SLC25A28-shRNA completely abrogated erastin-induced BRD7 upregulation, p53 mitochondrial translocation, mitochondrial iron accumulation, ETC hyperfunction, lipid peroxidation, and ferroptotic cell death (Fig. 6D–H and Figs. S7A–D). Altogether, these data support the hypothesis that HSC-specific blockade of BRD7-P53-SLC25A28 axis impairs erastin-induced HSC ferroptosis in murine liver fibrosis.
3.7. BRD7-P53-SLC25A28 axis regulates ferroptosis in human HSCs from fibrotic patients with sorafenib monotherapy
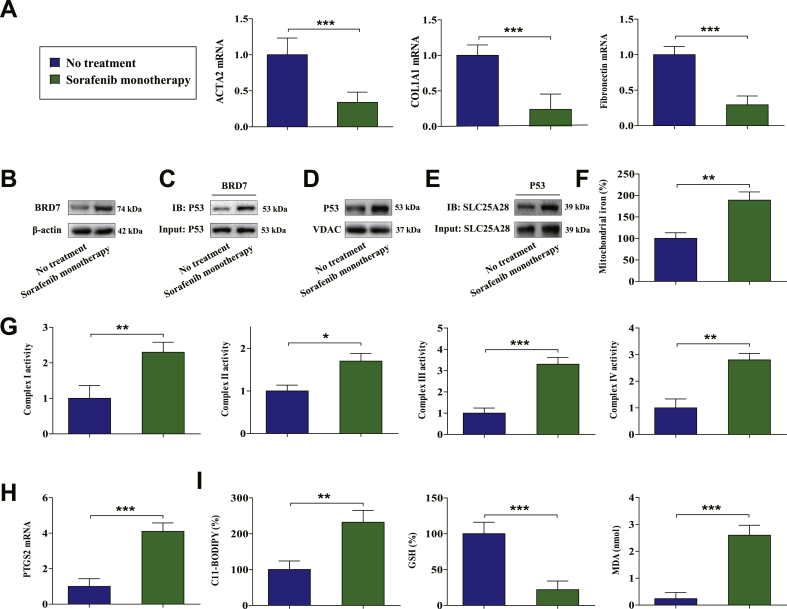
To verify this underlying mechanism in clinical, 24 partial hepatectomy samples from cirrhotic patients complicated with HCC who received sorafenib monotherapy and 37 liver biopsy samples from cirrhotic patients who did not receive any treatment were analyzed. Interestingly, sorafenib monotherapy dramatically reduced the levels of fibrosis markers ACTA2, COL1A1, and fibronectin (Fig. 7A). Furthermore, Western blot analysis indicated that the levels of BRD7 were substantially upregulated in human HSCs following sorafenib monotherapy (Fig. 7B). Moreover, sorafenib monotherapy promoted the binding of BRD7 and p53 in primary human HSCs (Fig. 7C). More importantly, we further isolated mitochondria from sorafenib-treated and untreated human HSCs. As expected, sorafenib monotherapy triggered p53 mitochondrial translocation (Fig. 7D), the interaction between p53 and SLC25A28 (Fig. 7E), mitochondrial iron accumulation (Fig. 7F), and ETC hyperfunction (Fig. 7G) in primary human HSCs. Besides, sorafenib monotherapy may lead to the upregulation of PTGS2 (Fig. 7H) and ferroptotic events including lipid ROS generation (Fig. 7I), GSH depletion (Fig. 7I), and lipid peroxidation (Fig. 7I) in human HSCs. Overall, these results indicated that the BRD7-P53-SLC25A28 axis regulates ferroptosis in human HSCs from fibrotic patients with sorafenib monotherapy.
Fig. 7.
BRD7-P53-SLC25A28 axis regulated ferroptosis in human HSCs from fibrotic patients with sorafenib monotherapy. (A) The levels of fibrosis markers ACTA2, COL1A1, and fibronectin were examined in sorafenib-treated hepatectomy samples and untreated liver biopsy samples (No treatment, n = 37; sorafenib monotherapy, n = 24; ***P < 0.001). Primary human HSCs were isolated from the collected liver tissue by laser capture microdissection. (B) The levels of BRD7 were examined by Western blot (No treatment, n = 37; sorafenib monotherapy, n = 24). (C) The combination of BRD7 and p53 was determined by immunocoprecipitation (No treatment, n = 37; sorafenib monotherapy, n = 24). (D) The levels of p53 in mitochondria were examined by Western blot (No treatment, n = 37; sorafenib monotherapy, n = 24). (E) The combination of SLC25A28 and p53 was determined by immunocoprecipitation (No treatment, n = 37; sorafenib monotherapy, n = 24). (F) Mitochondrial iron levels were determined by iron assay kit (No treatment, n = 37; sorafenib monotherapy, n = 24; **P < 0.01). (G) The enzymatic activities of mitochondrial complexes were analyzed by commercial kits (No treatment, n = 37; sorafenib monotherapy, n = 24; *P < 0.05, **P < 0.01, ***P < 0.001). (H) The expression of PTGS2 was examined by real-time PCR (No treatment, n = 37; sorafenib monotherapy, n = 24; ***P < 0.001). (I) The levels of lipid ROS, GSH, and MDA were determined by commercial kits (No treatment, n = 37; sorafenib monotherapy, n = 24; **P < 0.01, ***P < 0.001).
4. Discussion
Ferroptosis is a newly defined programmed cell death process with the hallmark of the accumulation of iron-dependent lipid peroxides [15]. Ferroptosis is distinct from other already established programmed cell death and has unique morphological and bioenergetic features [15]. The physiological and pathological role of ferroptosis during development has not been well characterized. Interestingly, more and more studies showed that ferroptosis may be an attractive strategy to prevent and treat liver fibrosis [42,43]. Kong Z et al. reported that artesunate alleviates liver fibrosis by regulating ferroptosis signaling pathway [42]. Moreover, Sui M et al. demonstrated that magnesium isoglycyrrhizinate ameliorates liver fibrosis and HSC activation by regulating ferroptosis signaling pathway [43]. Furthermore, Wang L et al. showed that p53-dependent induction of ferroptosis is required for artemether to alleviate carbon tetrachloride-induced liver fibrosis and HSC activation [14]. Noteworthy, we recently discovered that activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in HSCs [13]. Consistent with previous studies, we currently observed that induction of HSC ferroptosis can effectively improve the pathological changes of liver fibrosis. Although ferroptosis offers new hope for the prevention and treatment of liver fibrosis, there are still many drawbacks that need to be monitored. Wang H et al. reported that the ferroptosis stimulant ferric citrate potently induces ferroptosis in murine primary hepatocytes and bone marrow-derived macrophages, which may be a disadvantage for the improvement of liver injury [44]. Therefore, drug design for targeting induction of ferroptosis in HSCs but not in hepatocytes, LSECs, and macrophages is a new trend in the treatment of liver fibrosis.
Bromodomain-containing proteins (BRDs) are evolutionarily conserved protein modules that recognize acetyl-lysine motifs [45]. BRD7 is a vital member of the BRD family and is frequently involved in transcriptional regulation through protein-protein interactions with other transcription factors, such as p53, BRCA1 and Smads [23]. It is worth noting that BRD7 plays an indispensable role in the regulation of cell fate. Wang XM et al. reported that BRD7 mediates hyperglycaemia-induced myocardial apoptosis via endoplasmic reticulum stress signaling pathway [46]. Moreover, Drost J et al. showed that BRD7 suppresses tumorigenicity by serving as a p53 cofactor required for the efficient induction of p53-dependent oncogene-induced senescence [25]. Furthermore, Liu Y et al. demonstrated that BRD7 expression and c-myc activation form a double-negative feedback loop that controls the cell proliferation and tumor growth of nasopharyngeal carcinoma by targeting oncogenic miR-141 [47]. Besides, Peng C et al. revealed that BRD7 suppresses the growth of nasopharyngeal carcinoma cells through negatively regulating beta-catenin and ERK pathways [48]. In the present study, we first studied the regulatory role of BRD7 in ferroptosis. Upon induction of ferroptosis, BRD7 protein expression was evidently elevated through the inhibition of the ubiquitin-proteasome pathway. CRISPR/Cas9-mediated BRD7 knockout confers resistance to ferroptosis, whereas specific BRD7 plasmid-mediated BRD7 overexpression contributed to classical ferroptotic events characterized by redox-active iron accumulation, GSH depletion, and lipid peroxidation. Similarly, HSC-specific knockdown of BRD7 by VA-Lip-BRD7-shRNA also impaired HSC ferroptosis in murine liver fibrosis. More importantly, upregulated BRD7 appeared to promote p53 mitochondrial translocation via direct binding with p53 N-terminal transactivation domain, which was the underlying mechanism for BRD7-enhanced ferroptosis. Although more experiments are needed to determine the exact role of BRD7 in ferroptosis, our results indicate a novel function of BRD7 in addition to controlling apoptosis, proliferation, and senescence.
Several lines of evidence showed an important relationship between p53 and ferroptosis [17,49]. P53 can regulate the expression of ferroptosis-related genes in the nucleus, and also directly control the activity of ferroptosis-related molecules in cytoplasm. It should be noted that p53 can enhance ferroptosis by inhibiting the expression of SLC7A11 or by enhancing that of SAT1, whereas p53 can also suppress ferroptosis through the direct inhibition of DPP4 activity or by the induction of CDKN1A/p21 expression [17,49]. Jiang L et al. reported that ferroptosis is identified as a p53-mediated activity during tumor suppression [17]. Mechanistically, p53 inhibits cystine uptake and sensitizes cells to ferroptosis by transcriptional repressing SLC7A11 expression, a key component of the cystine/glutamate antiporter [17]. Moreover, Ou Y et al. demonstrated that activation of SAT1 may engage polyamine metabolism with p53-mediated ferroptotic responses [49]. SAT1 is identified as a transcription target of p53, and CRISPR/Cas9-mediated knockout of SAT1 expression partially abrogates p53-mediated ferroptosis [49]. By contrary, Xie Y et al. revealed that the tumor suppressor p53 limits ferroptosis by blocking DPP4 activity [50]. Furthermore, Tarangelo A et al. reported that p53-mediated CDKN1A expression delays the onset of ferroptosis in response to subsequent cystine deprivation in cancer cells [51]. Although the mechanism of p53 regulating ferroptosis in nucleus and cytoplasm has been addressed, little is known about the mechanism of p53 regulating ferroptosis in mitochondria. In the present study, we found that mitochondrial translocation of p53 mediated BRD7-enhanced HSC ferroptosis. Interestingly, BRD7 knockout significantly reduced the expression of p53 in mitochondria rather than in the cytoplasm and nucleus, whereas BRD7 knockin markedly increased the expression of p53 in mitochondria but not in the cytoplasm and nucleus. It has been reported that serine 392 phosphorylation modulates p53 mitochondrial translocation [34]. Indeed, BRD7 knockin noticeably triggered serine 392 phosphorylation, but BRD7 knockout completely counteracted this effect in ferroptosis. Attractively, the S392A mutant completely impaired the BRD7 knockin mediated p53 mitochondrial translocation and ferroptotic cell death and classical ferroptotic events during HSC ferroptosis. Our results reveal the potential mechanism of p53 regulating ferroptosis from a new perspective.
Mitoferrin, a mitochondrial protein, has been reported to mediate ferrous iron transport across the mitochondrial inner membrane [52]. Mitoferrin has two isoforms: SLC25A37 (mitoferrin-1) and SLC25A28 (mitoferrin-2), which are localized on the inner mitochondrial membrane and function as a requisite importer of iron for mitochondrial heme and iron-sulfur cluster [52]. SLC25A37 is mainly distributed in erythroid cells with low levels in other tissues, whereas SLC25A28 is ubiquitously distributed [52]. The role of SLC25A28 in ferroptosis has not been elucidated, but its function in other diseases has been studied. Hung HI et al. reported that SLC25A28-dependent mitochondrial iron uptake sensitizes human head and neck squamous carcinoma cells to photodynamic therapy [53]. Moreover, Wang C et al. showed that SLC25A28 participates in mitochondrial ROS-dependent mechanisms underlying As2O3-mediated damage in glioma cells [54]. In the current study, we first explored the role of SLC25A28 in ferroptosis. Of note, we identified a crucial target SLC25A28 in iron trafficking, but not mitochondrial iron utilization, mitochondrial iron storage, and iron transfer, was evidently inhibited upon S392A mutant or p53 knockdown according to the fold change. Moreover, BRD7 knockin promoted direct binding of p53 to SLC25A28 in mitochondria. Further, shRNA-mediated knockdown of SLC25A28 completely abolished the mitochondrial iron accumulation, and, in turn, impaired BRD7 knockin-enhanced ferroptotic cell death and classical ferroptotic events. With a better understanding of SLC25A28-dependent ferroptosis, SLC25A28 may have potential as a target of antifibrotic therapy.
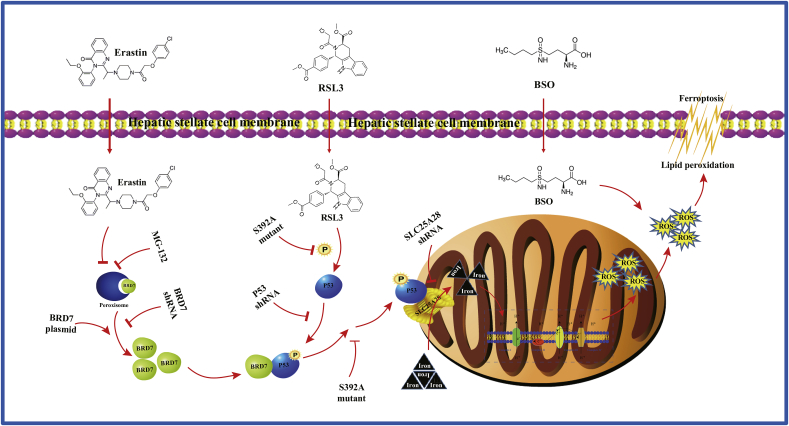
In conclusion, our findings confirm that BRD7-P53-SLC25A28 axis involves in mediating ferroptosis via mitochondrial iron metabolism pathway (Fig. 8). Further exploring the molecular mechanism of apoptosis will provide an alternative and effective strategy for eliminating HSCs.
Fig. 8.
The BRD7-P53-SLC25A28 axis may regulate ferroptosis in hepatic stellate cells. inhibition-, GPX4 inhibition-, and GSH depletion-mediated BRD7 upregulation triggers p53 mitochondrial translocation via direct binding with N-terminal transactivation domain, thus aggravating the accumulation of mitochondrial iron, the hyperfunction of electron transfer chain, lipid peroxidation, and eventually leading to iron-dependent ferroptosis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31401210, 81870423), the Joint Project of Jiangsu Key Laboratory for Pharmacology and Safety Evaluation of Chinese Materia Medica and Yangtze River Pharmaceutical (JKLPSE202005), the Major Project of the Natural Science Research of Jiangsu Higher Education Institutions (19KJA310005), the Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Natural Science Foundation of Jiangsu Province (BK20200056) and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_0165).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101619.
Contributor Information
Peng Cao, Email: cao_peng@njucm.edu.cn.
Shizhong Zheng, Email: nytws@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
References
- 1.De Magalhaes Filho C.D., Downes M., Evans R. Bile acid analog intercepts liver fibrosis. Cell. 2016;166:789. doi: 10.1016/j.cell.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Kostallari E., Hirsova P., Prasnicka A., Verma V.K., Yaqoob U., Wongjarupong N., Roberts L.R., Shah V.H. Hepatic stellate cell-derived platelet-derived growth factor receptor-alpha-enriched extracellular vesicles promote liver fibrosis in mice through SHP2. Hepatology. 2018;68:333–348. doi: 10.1002/hep.29803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lackner C., Tiniakos D. Fibrosis and alcohol-related liver disease. J. Hepatol. 2019;70:294–304. doi: 10.1016/j.jhep.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Bian M., Chen X., Zhang C., Jin H., Wang F., Shao J., Chen A., Zhang F., Zheng S. Magnesium isoglycyrrhizinate promotes the activated hepatic stellate cells apoptosis via endoplasmic reticulum stress and ameliorates fibrogenesis in vitro and in vivo. Biofactors. 2017;43:836–846. doi: 10.1002/biof.1390. [DOI] [PubMed] [Google Scholar]
- 5.Jia Y., Wang F., Guo Q., Li M., Wang L., Zhang Z., Jiang S., Jin H., Chen A., Tan S., Zhang F., Shao J., Zheng S. Curcumol Induces RIPK1/RIPK3 complex-dependent necroptosis via JNK1/2-ROS signaling in hepatic stellate cells. Redox Biol. 2018;19:375–387. doi: 10.1016/j.redox.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z., Yao Z., Zhao S., Shao J., Chen A., Zhang F., Zheng S. Interaction between autophagy and senescence is required for dihydroartemisinin to alleviate liver fibrosis. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z., Zhao S., Yao Z., Wang L., Shao J., Chen A., Zhang F., Zheng S. Autophagy regulates turnover of lipid droplets via ROS-dependent Rab25 activation in hepatic stellate cell. Redox Biol. 2017;11:322–334. doi: 10.1016/j.redox.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Zhang F., Lu S., He J., Jin H., Wang F., Wu L., Shao J., Chen A., Zheng S. Ligand activation of PPARγ by ligustrazine suppresses pericyte functions of hepatic stellate cells via SMRT-mediated transrepression of HIF-1α. Theranostics. 2018;8:610–626. doi: 10.7150/thno.22237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z., Guo M., Zhao S., Shao J., Zheng S. ROS-JNK1/2-dependent activation of autophagy is required for the induction of anti-inflammatory effect of dihydroartemisinin in liver fibrosis. Free Radic. Biol. Med. 2016;101:272–283. doi: 10.1016/j.freeradbiomed.2016.10.498. [DOI] [PubMed] [Google Scholar]
- 10.Wang F., Jia Y., Li M., Wang L., Shao J., Guo Q., Tan S., Ding H., Chen A., Zhang F., Zheng S. Blockade of glycolysis dependent contraction by oroxylin a via inhibition of lactate dehydrogenase-a in hepatic stellate cells. Cell Commun. Signal. 2019;17:11. doi: 10.1186/s12964-019-0324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu W., Lu C., Zhang F., Shao J., Yao S., Zheng S. Dihydroartemisinin counteracts fibrotic portal hypertension via farnesoid X receptor-dependent inhibition of hepatic stellate cell contraction. FEBS J. 2017;284:114–133. doi: 10.1111/febs.13956. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z., Guo M., Li Y., Shen M., Kong D., Shao J., Ding H., Tan S., Chen A., Zhang F., Zheng S. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. 2019;11:1–24. doi: 10.1080/15548627.2019.1687985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z., Yao Z., Wang L., Ding H., Shao J., Chen A., Zhang F., Zheng S. Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy. 2018;14:2083–2103. doi: 10.1080/15548627.2018.1503146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L., Zhang Z., Li M., Wang F., Jia Y., Zhang F., Shao J., Chen A., Zheng S. P53-dependent induction of ferroptosis is required for artemether to alleviate carbon tetrachloride-induced liver fibrosis and hepatic stellate cell activation. IUBMB Life. 2019;71:45–56. doi: 10.1002/iub.1895. [DOI] [PubMed] [Google Scholar]
- 15.Wang W., Green M., Choi J.E., Gijón M., Kennedy P.D., Johnson J.K., Liao P., Lang X., Kryczek I., Sell A., Xia H., Zhou J., Li G., Li J., Li W., Wei S., Vatan L., Zhang H., Szeliga W., Gu W., Liu R., Lawrence T.S., Lamb C., Tanno Y., Cieslik M., Stone E., Georgiou G., Chan T.A., Chinnaiyan A., Zou W. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alim I., Caulfield J.T., Chen Y., Swarup V., Geschwind D.H., Ivanova E., Seravalli J., Ai Y., Sansing L.H., Ste Marie E.J., Hondal R.J., Mukherjee S., Cave J.W., Sagdullaev B.T., Karuppagounder S.S., Ratan R.R. Selenium drives a transcriptional adaptive program to block ferroptosis and treat stroke. Cell. 2019;177:1262–1279. doi: 10.1016/j.cell.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 17.Jiang L., Kon N., Li T., Wang S.J., Su T., Hibshoosh H., Baer R., Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou W., Xie Y., Song X., Sun X., Lotze M.T., Zeh H.J., 3rd, Kang R., Tang D. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12:1425–1428. doi: 10.1080/15548627.2016.1187366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J., Minikes A.M., Gao M., Bian H., Li Y., Stockwell B.R., Chen Z.N., Jiang X. Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signaling. Nature. 2019;572:402–406. doi: 10.1038/s41586-019-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun X., Ou Z., Chen R., Niu X., Chen D., Kang R., Tang D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–184. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao M., Monian P., Quadri N., Ramasamy R., Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao M., Yi J., Zhu J., Minikes A.M., Monian P., Thompson C.B., Jiang X. Role of mitochondria in ferroptosis. Mol Cell. 2019;73:354–363. doi: 10.1016/j.molcel.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiu Y.H., Lee J.Y., Cantley L.C. BRD7, a tumor suppressor, interacts with p85α and regulates PI3K activity. Mol Cell. 2014;54:193–202. doi: 10.1016/j.molcel.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng C., Zhou J., Liu H.Y., Zhou M., Wang L.L., Zhang Q.H., Yang Y.X., Xiong W., Shen S.R., Li X.L., Li G.Y. The transcriptional regulation role of BRD7 by binding to acetylated histone through bromodomain. J. Cell. Biochem. 2006;97:882–892. doi: 10.1002/jcb.20645. [DOI] [PubMed] [Google Scholar]
- 25.Drost J., Mantovani F., Tocco F., Elkon R., Comel A., Holstege H., Kerkhoven R., Jonkers J., Voorhoeve P.M., Agami R., Del Sal G. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat. Cell Biol. 2010;12:380–389. doi: 10.1038/ncb2038. [DOI] [PubMed] [Google Scholar]
- 26.Wang X.M., Wang Y.C., Liu X.J., Wang Q., Zhang C.M., Zhang L.P., Liu H., Zhang X.Y., Mao Y., Ge Z.M. BRD7 mediates hyperglycaemia-induced myocardial apoptosis via endoplasmic reticulum stress signaling pathway. J. Cell Mol. Med. 2017;21:1094–1105. doi: 10.1111/jcmm.13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burrows A.E., Smogorzewska A., Elledge S.J. Polybromo-associated BRG1 associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14280–14285. doi: 10.1073/pnas.1009559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niu W., Luo Y., Wang X., Zhou Y., Li H., Wang H., Fu Y., Liu S., Yin S., Li J., Zhao R., Liu Y., Fan S., Li Z., Xiong W., Li X., Li G., Ren C., Tan M., Zhou M. BRD7 inhibits the Warburg effect and tumor progression through inactivation of HIF1α/LDHA axis in breast cancer. Cell Death Dis. 2018;9:519. doi: 10.1038/s41419-018-0536-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Muratori L., Muratori P., Lanzoni G., Ferri S., Lenzi M. Application of the 2010 American Association for the study of liver diseases criteria of remission to a cohort of Italian patients with autoimmune hepatitis. Hepatology. 2010;52:1857–1858. doi: 10.1002/hep.23924. [DOI] [PubMed] [Google Scholar]
- 30.Nagai H., Mukozu T., Kobayashi K., Amanuma M., Yoshimine N., Ogino Y.U., Matsui D., Daido Y., Matsukiyo Y., Matsui T., Wakui N., Momiyama K., Shinohara M., Higai K., Igarashi Y. Influence of sorafenib on host immunity in patients with liver cirrhosis with advanced hepatocellular carcinoma stratified by etiology. Anticancer Res. 2019;39:2183–2191. doi: 10.21873/anticanres.13333. [DOI] [PubMed] [Google Scholar]
- 31.Standish R.A., Cholongitas E., Dhillon A., Burroughs A.K., Dhillon A.P. An appraisal of the histopathological assessment of liver fibrosis. Gut. 2006;55:569–578. doi: 10.1136/gut.2005.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aparicio-Vergara M., Tencerova M., Morgantini C., Barreby E., Aouadi M. Isolation of Kupffer cells and hepatocytes from a single mouse liver. Methods Mol. Biol. 2017;1639:161–171. doi: 10.1007/978-1-4939-7163-3_16. [DOI] [PubMed] [Google Scholar]
- 33.Zhang F., Wen Y., Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum. Mol. Genet. 2014;23:R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 34.Castrogiovanni C., Waterschoot B., De Backer O., Dumont P. Serine 392 phosphorylation modulates p53 mitochondrial translocation and transcription independent apoptosis. Cell Death Differ. 2018;25:190–203. doi: 10.1038/cdd.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Báez A.L., Lo Presti M.S., Bazán P.C., Strauss M., Velázquez López D.A., Miler N., Rivarola H.W., Paglini-Oliva P.A. Analysis of mitochondrial enzymatic activity in blood lymphomonocyte fractions during infection with different Trypanosoma cruzi strains. Rev. Inst. Med. Trop. Sao Paulo. 2020;62:e15. doi: 10.1590/S1678-9946202062015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C., Zhang Y., Cheng X., Yuan H., Zhu S., Liu J., Wen Q., Xie Y., Liu J., Kroemer G., Klionsky D.J., Lotze M.T., Zeh H.J., Kang R., Tang D. PINK1 and PARK2 suppress pancreatic tumorigenesis through control of mitochondrial iron-mediated immunometabolism. Dev. Cell. 2018;46:441–455. doi: 10.1016/j.devcel.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keij J.F., Bell-Prince C., Steinkamp J.A. Staining of mitochondrial membranes with 10-nonyl acridine orange, MitoFluor Green, and MitoTracker Green is affected by mitochondrial membrane potential altering drugs. Cytometry. 2000;39:203–210. doi: 10.1002/(sici)1097-0320(20000301)39:3<203::aid-cyto5>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 38.Bertrand R.L. Iron accumulation, glutathione depletion, and lipid peroxidation must occur simultaneously during ferroptosis and are mutually amplifying events. Med. Hypotheses. 2017;101:69–74. doi: 10.1016/j.mehy.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 39.Kamada R., Toguchi Y., Nomura T., Imagawa T., Sakaguchi K. Tetramer formation of tumor suppressor protein p53: structure, function, and applications. Biopolymers. 2016;106:598–612. doi: 10.1002/bip.22772. [DOI] [PubMed] [Google Scholar]
- 40.Pandey A.K., Pain J., Dancis A., Pain D. Mitochondria export iron-sulfur and sulfur intermediates to the cytoplasm for iron-sulfur cluster assembly and tRNA thiolation in yeast. J. Biol. Chem. 2019;294:9489–9502. doi: 10.1074/jbc.RA119.008600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul B.T., Manz D.H., Torti F.M., Torti S.V. Mitochondria and iron: current questions. Expet Rev. Hematol. 2017;10:65–79. doi: 10.1080/17474086.2016.1268047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong Z., Liu R., Cheng Y. Artesunate alleviates liver fibrosis by regulating ferroptosis signaling pathway. Biomed. Pharmacother. 2019;109:2043–2053. doi: 10.1016/j.biopha.2018.11.030. [DOI] [PubMed] [Google Scholar]
- 43.Sui M., Jiang X., Chen J., Yang H., Zhu Y. Magnesium isoglycyrrhizinate ameliorates liver fibrosis and hepatic stellate cell activation by regulating ferroptosis signaling pathway. Biomed. Pharmacother. 2018;106:125–133. doi: 10.1016/j.biopha.2018.06.060. [DOI] [PubMed] [Google Scholar]
- 44.Wang H., An P., Xie E., Wu Q., Fang X., Gao H., Zhang Z., Li Y., Wang X., Zhang J., Li G., Yang L., Liu W., Min J., Wang F. Characterization of ferroptosis in murine models of hemochromatosis. Hepatology. 2017;66:449–465. doi: 10.1002/hep.29117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujisawa T., Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 2017;18:246–262. doi: 10.1038/nrm.2016.143. [DOI] [PubMed] [Google Scholar]
- 46.Wang X.M., Wang Y.C., Liu X.J., Wang Q., Zhang C.M., Zhang L.P., Liu H., Zhang X.Y., Mao Y., Ge Z.M. BRD7 mediates hyperglycaemia-induced myocardial apoptosis via endoplasmic reticulum stress signalling pathway. J. Cell Mol. Med. 2017;21:1094–1105. doi: 10.1111/jcmm.13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y., Zhao R., Wei Y., Li M., Wang H., Niu W., Zhou Y., Qiu Y., Fan S., Zhan Y., Xiong W., Zhou Y., Li X., Li Z., Li G., Zhou M. BRD7 expression and c-Myc activation forms a double-negative feedback loop that controls the cell proliferation and tumor growth of nasopharyngeal carcinoma by targeting oncogenic miR-141. J. Exp. Clin. Canc. Res. 2018;37:64. doi: 10.1186/s13046-018-0734-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng C., Liu H.Y., Zhou M., Zhang L.M., Li X.L., Shen S.R., Li G.Y. BRD7 suppresses the growth of nasopharyngeal carcinoma cells (HNE1) through negatively regulating beta-catenin and ERK pathways. Mol. Cell. Biochem. 2007;303:141–149. doi: 10.1007/s11010-007-9466-x. [DOI] [PubMed] [Google Scholar]
- 49.Ou Y., Wang S.J., Li D., Chu B., Gu W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E6806–E6812. doi: 10.1073/pnas.1607152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Y., Zhu S., Song X., Sun X., Fan Y., Liu J., Zhong M., Yuan H., Zhang L., Billiar T.R., Lotze M.T., Zeh H.J., 3rd, Kang R., Kroemer G., Tang D. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20:1692–1704. doi: 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- 51.Tarangelo A., Magtanong L., Bieging-Rolett K.T., Li Y., Ye J., Attardi L.D., Dixon S.J. p53 suppresses metabolic stress-induced ferroptosis in cancer cells. Cell Rep. 2018;22:569–575. doi: 10.1016/j.celrep.2017.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaw G.C., Cope J.J., Li L., Corson K., Hersey C., Ackermann G.E., Gwynn B., Lambert A.J., Wingert R.A., Traver D., Trede N.S., Barut B.A., Zhou Y., Minet E., Donovan A., Brownlie A., Balzan R., Weiss M.J., Peters L.L., Kaplan J., Zon L.I., Paw B.H. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;44:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 53.Hung H.I., Schwartz J.M., Maldonado E.N., Lemasters J.J., Nieminen A.L. Mitoferrin-2 dependent mitochondrial iron uptake sensitizes human head and neck squamous carcinoma cells to photodynamic therapy. J. Biol. Chem. 2013;288:677–686. doi: 10.1074/jbc.M112.422667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C., Chen X., Zou H., Chen X., Liu Y., Zhao S. The roles of mitoferrin-2 in the process of arsenic trioxide-induced cell damage in human gliomas. Eur. J. Med. Res. 2014;19:49. doi: 10.1186/s40001-014-0049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.