Abstract
G protein–coupled receptors (GPCRs) are biologic switches that transduce extracellular stimuli into intracellular responses in the cell. Temporally resolving GPCR transduction pathways is key to understanding how cell signaling occurs. Here, we investigate the kinetics and dynamics of the activation and early signaling steps of the CXC chemokine receptor (CXCR) 4 in response to its natural ligands CXC chemokine ligand (CXCL) 12 and macrophage migration inhibitory factor (MIF), using Förster resonance energy transfer–based approaches. We show that CXCR4 presents a multifaceted response to CXCL12, with receptor activation (≈0.6 seconds) followed by a rearrangement in the receptor/G protein complex (≈1 seconds), a slower dimer rearrangement (≈1.7 seconds), and prolonged G protein activation (≈4 seconds). In comparison, MIF distinctly modulates every step of the transduction pathway, indicating distinct activation mechanisms and reflecting the different pharmacological properties of these two ligands. Our study also indicates that CXCR4 exhibits some degree of ligand-independent activity, a relevant feature for drug development.
SIGNIFICANCE STATEMENT
The CXC chemokine ligand (CXCL) 12/CXC chemokine receptor (CXCR) 4 axis represents a well-established therapeutic target for cancer treatment. We demonstrate that CXCR4 exhibits a multifaceted response that involves dynamic receptor dimer rearrangements and that is kinetically embedded between receptor–G protein complex rearrangements and G protein activation. The alternative endogenous ligand macrophage migration inhibitory factor behaves opposite to CXCL12 in each assay studied and does not lead to G protein activation. This detailed understanding of the receptor activation may aid in the development of more specific drugs against this target.
Introduction
G protein–coupled receptors (GPCRs) transduce signals of diverse nature from the extracellular side into specific responses within the cell through a succession of biochemical events. Generally, binding of an agonist to a receptor causes structural changes in the transmembrane (TM) helices that stabilize the receptor in an active conformation. This is followed by interaction with and subsequent activation of heterotrimeric G proteins, which modulate the activity of different downstream effectors. Receptors can then be phosphorylated by kinases and internalized, resulting in degradation or recycling to the plasma membrane (Hilger et al., 2018).
Crystal structure analysis has provided enormous insights into the molecular mechanisms involved in GPCR activation. However, the detailed temporal dynamics of these changes cannot be resolved in these studies. In this aspect, the use of Förster resonance energy transfer (FRET)-based approaches represents a tool to investigate the dynamics and kinetics of GPCR activation and their downstream signaling events in real time and in intact cells (Lohse et al., 2012). The most common structural characteristic of receptor activation is a large outward shift of the intracellular part of TM domain VI (Altenbach et al., 2008). This unique feature has been the basis for the development of FRET sensors for many receptors, which can report ligand-induced structural rearrangements in a temporal manner (Lohse et al., 2014; Stumpf and Hoffmann, 2016; Wright et al., 2018). These sensors together with other FRET-based approaches have helped to understand the distinct mechanisms of activation between different ligand types (Vilardaga et al., 2005), allosterism (Messerer et al., 2017), and receptor classes (Vilardaga et al., 2003). Most studies have found activation time constants of monomeric GPCRs on the order of 30–50 milliseconds (Hoffmann et al., 2005; Rochais et al., 2007; Reiner et al., 2010; Ziegler et al., 2011). However, there are apparent differences between various receptor types. Thus, activation of class B parathyroid hormone receptor (PTHR) 1 by its large agonist PTH(1–34) is about 20-fold slower (Vilardaga et al., 2003). Another specific case is the activation in dimeric receptors. In a recent study aiming at resolving rapid activation steps of metabotropic glutamate receptors (mGluRs), it was shown that an initial rearrangement of the dimer structure occurs within 1 to 2 milliseconds, whereas conformational changes in the 7-helix TM structure occur within 20 milliseconds (Grushevskyi et al., 2019). Another open question regarding activation in receptor dimers is how the two protomers influence each other. An early study of α2-adrenergic/μ-opioid heterodimers suggested that in such a dimer only one of the two protomers is active (Vilardaga et al., 2008). Likewise, initial studies of mGluRs indicated that only one of the two protomers becomes activated (Hlavackova et al., 2005). However, it now appears clear that activation of both protomers is required for full activation (Kniazeff et al., 2004; Grushevskyi et al., 2019).
The CXC chemokine receptor (CXCR) 4 appears to be a facultative homodimer, and dimer formation may contribute to its signaling function. CXCR4 and its cognate ligand CXCL12, formerly known as stromal cell-derived factor-1, are key players in cell migration in different contexts (Alsayed et al., 2007; Yang et al., 2013). A two-step binding mechanism of chemokines to their chemokine receptors has been proposed (Qin et al., 2015). Binding of CXCL12 preferentially results in activation of Gi proteins as well as β-arrestin recruitment (Heuninck et al., 2019). The CXCR4/CXCL12 axis is an important therapeutic target for the development of new drugs because of its involvement in several types of cancer (Guo et al., 2016; Adlere et al., 2019; Bobkov et al., 2019) in addition to the well-proven role of CXCR4 as a human immunodeficiency virus coreceptor (Tsibris and Kuritzkes, 2007). Recently, macrophage migration inhibitory factor (MIF) has also been demonstrated to bind to CXCR4, but the binding mode and downstream consequences observed greatly differ from those of CXCL12. For instance, MIF binding to the receptor does not result in β-arrestin recruitment (Bernhagen et al., 2007; Rajasekaran et al., 2016; Lacy et al., 2018). The biomedical importance of CXCR4 and its ligands, their dynamic monomer/dimer equilibrium, the suggested two-step activation mechanism, and the existence of several endogenous agonists with diverse effects all make the spatio-temporal analysis of CXCR4 signaling particularly interesting.
Therefore, we set out to analyze the early steps of CXCR4 activation in response to the two distinct ligands CXCL12 and MIF. We have combined several FRET approaches to elucidate the temporal aspects of the receptor-mediated early signaling events with subsecond resolution. Our data show that these two ligands modulate the receptor’s activity oppositely and that, in the absence of agonist, CXCR4 exhibits some degree of constitutive activity.
Materials and Methods
Plasmid DNA Constructs
Human CXCR4 with YFP fused to the C terminus was a kind gift of F. Bachelerie (INSERM, Université Paris-Sud, France; Levoye et al., 2009). The α2A-adrenergic receptor (AR) has been previously described (Bünemann et al., 2003). The G protein subunits Gαi1, Gαi1–cyan fluorescent protein (CFP), Gβ1, Gγ2, and Gγ2-CFP and the α2A-AR-YFP (labeled at the C terminus) have been previously described (Hein et al., 2005). The G protein sensors for Gi1, Gi2, Gi3, and Gq have been previously described (Adjobo-Hermans et al., 2011; Van Unen et al., 2016). The cAMP-responsive element (CRE)-Luc construct has been previously described (Watts et al., 2013).
Human CXCR4 and human CXCR4 with three hemagglutinin (HA) tags fused to the N terminus (3HA-CXCR4) are in pcDEF3 (Vrije Universiteit, Amsterdam, The Netherlands). The 3HA-CXCR4 construct was employed to label the receptor with enhanced CFP (BD Biosciences). For that, the fluorescent protein was fused in-frame to the C terminus of CXCR4 by standard PCR extension overlap technique. The C-terminal stop codon of the receptor and the initial methionine codon of the fluorescent protein were deleted. The resulting construct was named CXCR4-CFP and contained the fluorescent protein after S352. To create the different receptor sensors, the fluorescein arsenical hairpin binder (FlAsH)-binding motif CCPGCC was inserted at different positions within the intracellular loop-3 (ICL-3) of the receptor in the CXCR4-CFP construct: between amino acids L226 and S227 (CXCR4-FlAsH226-CFP), between amino acids H228 and S229 (CXCR4-FlAsH228-CFP), or between amino acids S229 and K230 (CXCR4-FlAsH229-CFP). Cloning was performed using site-directed mutagenesis by overlap extension PCR. To create analogous constructs only containing the FlAsH-binding motif in different positions of ICL-3 (CXCR4-FlAsH226, CXCR4-FlAsH228, and CXCR4-FlAsH229), the CFP was removed from the C terminus of the corresponding receptor sensors via standard PCR.
The Pfu polymerase was used for amplifying DNA in the PCRs, and the T4 ligase enzyme was used for ligation. Escherichia coli DH5α (Invitrogen) was used as a host to clone all the genes described. All constructs were verified by sequencing (Eurofins Genomix GmbH, Germany).
Ligands
Recombinant human CXCL12 was purchased from Peprotech (300-28A); recombinant human MIF was purchased from Peprotech (300-69); norepinephrine was purchased from Sigma Aldrich (A9512); and IT1t was purchased from Tocris (4596). AMD3100 was purchased from Sigma Aldrich (A5602), and CXCL12-AlexaFluor647 was from Almac (CAF-11).
Cell Lines and Cell Culture
Human embryonic kidney cell 293 (HEK293) and HEK293T cell lines (American Type Culture Collection) (CRL-1573 and CRL-3216) were cultured using Dulbecco’s modified Eagle’s medium supplemented with 4.5 g/l glucose (Gibco), 10% (v/v) FBS (Biochrom), 1% penicillin/streptomycin (Gibco), and 1% L-glutamine (PanBiotech). Cells were kept in a humidified 7% CO2 atmosphere at 37°C. For routine maintenance, cells were split every 2 or 3 days by rinsing them with Dulbecco’s phosphate-buffered saline (DPBS) (Gibco) and then trypsin-EDTA (PanBiotech). Cell lines were routinely tested for mycoplasma contamination by PCR using a primer set specific for the highly conserved 16S ribosomal RNA–coding region in the mycoplasma genome. Cells have not been authenticated.
The N-terminal NanoLuc (NLuc)-labeled CXCR4 receptor construct was created from a previously described construct (Stoddart et al., 2015) and was used for the binding competition assay. Briefly, the adenosine A1 receptor cDNA was replaced with that encoding the human CXCR4 (NM_003467.2). The resultant construct fused NLuc in-frame with the membrane signal sequence from the 5-HT3A receptor within pcDNA3.1 to generate signal-NLuc. The human CXCR4 sequence (methionine start signal removed) was then fused to the 3′ end of signal-NLuc via a short Gly-Ser linker. Stable mixed-expression populations of HEK293G cell lines [HEK293 expressing the GloSensor (20F) cAMP biosensor; Promega Corporation] expressing the NLuc-CXCR4 cDNA construct were created by transfecting cells with FuGENE HD (Promega Corporation) following the manufacturer’s instructions. Cells were then subject to antibiotic selection using G418 disulfate salt (1 mg/ml; Sigma Aldrich) for 2 to 3 weeks, screened for luminescence expression, and expanded. The HEK293G-NLuc-CXCR4 cell line was cultured using Dulbecco’s modified Eagle’s medium (Sigma Aldrich) supplemented with 10% FBS (Sigma Aldrich). Cells were kept in a humidified 5% CO2 atmosphere at 37°C. For routine maintenance, cells were split every 2 or 3 days by using DPBS (Lonza) and trypsin-EDTA (0.25% w/v versene; Lonza).
FlAsH Labeling
FlAsH labeling was done as previously described (Hoffmann et al., 2010) right before the measurements. In brief, transfected cells grown on 24-mm glass coverslips (Hartenstein, Würzburg, Germany) or 40-mm WillCo dishes (WillCo Wells) were washed twice with labeling buffer (150 mM NaCl, 10 mM HEPES, 2.5 mM KCl, 4 mM CaCl2, 2 mM MgCl2 freshly supplemented with 1.8 g/l glucose; pH 7.3). Subsequently, cells were incubated for 1 hour at 37°C with labeling buffer containing 1 µM FlAsH (commercially available from Thermo Fisher Scientific, Darmstadt, Germany as TC-FlAsH labeling kit) and 12.5 μM 1,2-ethanedithiol (Sigma Aldrich). Cells were rinsed twice and incubated for 10 minutes at 37°C in labeling buffer containing 250 μM 1,2-ethanedithiol to reduce nonspecific FlAsH labeling. Finally, cells were washed twice and kept in media at 37°C until measurement.
Confocal Microscopy
HEK293 cells were seeded on 24-mm glass coverslips, which had been previously coated with poly-D-lysine (1 mg/ml; MP Biomedicals) for 30 minutes and washed once with DPBS, and placed in wells of a six-well plate. After 6 hours, and for each well, cells were transfected with 0.5 µg of the different CXCR4 sensors (CXCR4-FlAsH226-CFP, CXCR4-FlAsH228-CFP, or CXCR4-FlAsH229-CFP) using Effectene (Qiagen), following the manufacturer’s instructions. Prior to transfection, the media of the cells was exchanged for fresh media. Cells were analyzed 48 hours after transfection. When indicated, cells were FlAsH-labeled right before the measurement. During the experiment, cells were kept in imaging buffer (140 mM NaCl, 5.4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES; pH 7.3).
Confocal microscopy was performed using a Leica TCS SP8 system (Leica Microsystems, Germany) with an Attofluor holder (Molecular Probes), and images were taken with a 63× water objective (numerical aperture 1.4). CFP was excited with a diode laser line at 442 nm, and fluorescence intensities were recorded from 470 to 550 nm. FlAsH was excited with an argon laser line at 514 nm, and fluorescence intensities were recorded from 525 to 600 nm. Images were acquired at 1024 × 1024–pixel format, line average 3 and 400 Hz. To avoid bleed through, parallel images of CFP and FlAsH were taken in sequential scan. Images were analyzed using the Leica Application Suite Advanced Fluorescence Software (Leica Microsystems).
Single-Cell FRET Experiments
Microscopic FRET Set-Up.
FRET measurements were performed on an inverted microscope (Zeiss Axiovert 200; Zeiss) equipped with an oil immersion 63× objective lens and a dual-emission photometric system (Till Photonics). The transfected cells were excited with light from a polychrome IV (Till Photonics) at a frequency of 10 Hz with 40 milliseconds illumination out of a total time of 100 milliseconds. Emission of the donor (CFP or mTurquoise2; 480 ± 20 nm) and acceptor (YFP, FlAsH, or Venus; 535 ± 15 nm) and the FRET ratio (Facceptor/Fdonor) were monitored simultaneously (beam splitter DCLP 505 nm) upon excitation at 436 ± 10 nm (beam splitter DCLP 460 nm). Fluorescence signals were detected by photodiodes, digitalized using an analog-digital converter (Digidata 1440A; Axon Instruments), and stored with Clampex 9.0 software (Molecular Devices, Sunnyvale, CA).
Determination of FRET Efficiency.
Determination of the FRET efficiency was performed as previously described (Jost et al., 2008). HEK293 cells were prepared in 24-mm glass coverslips, as described for confocal microscopy. To study the basal intramolecular FRET, cells were transfected with 0.5 µg of CXCR4-FlAsH226-CFP, CXCR4-FlAsH228-CFP, or CXCR4-FlAsH229-CFP sensor per well. To study the basal intermolecular FRET, cells were transfected with 0.3 µg of CXCR4-CFP and 0.3 µg of CXCR4-FlAsH228 per well. Transfection was performed using Effectene. Prior to the measurement, cells were FlAsH-labeled. Then, the coverslips were mounted on an Attofluor holder and kept in 999 µl of imaging buffer. During the measurement (after approximately 30 seconds of recording), 1 µl of British anti-Lewisite (BAL) (Fluka) was added to yield a final BAL concentration of 5 mM, which displaced FlAsH from its binding site in the receptors’ ICL-3. The FRET efficiency was calculated by analyzing the increase in the CFP emission [CFP fluorescence (FCFP)] due to dequenching using the formula (FCFP,max − FCFP,min)/FCFP,max and the Clampfit software (Molecular Devices). Data are shown as a scatter plot with mean and S.D., in which each dot represents an individual cell. At least three independent repeats were performed for each condition. Measurements were performed using the microscopic FRET set-up described above.
Kinetic Measurements.
For kinetic experiments, HEK293 cells were seeded on 40-mm WillCo dishes previously coated with poly-D-lysine for 30 minutes and washed once with DPBS. Six hours later, cells were transfected with the appropriate construct(s) using Effectene. For measuring receptor activation, 0.7 µg of each sensor (CXCR4-FlAsH226-CFP, CXCR4-FlAsH228-CFP, or CXCR4-FlAsH229-CFP) were transfected per dish, and FlAsH labeling was performed before the measurement. For receptor/G protein interaction studies, 0.6 µg of CXCR4-YFP, 0.7 µg of Gαi1, 0.3 µg of Gβ1, and 0.2 µg of Gγ2-CFP were transfected per dish. When indicated, 0.6 µg of CXCR4-YFP, 0.7 µg of Gαi1-CFP, 0.3 µg of Gβ1, and 0.2 µg of Gγ2 were transfected instead. For measuring G protein activation, 0.4 µg of receptor (CXCR4 or α2A-AR) or empty plasmid and 0.75 µg of G protein sensor (Gi1, Gi2 or Gi3) were transfected per dish. For measuring rearrangements within dimers, 0.5 µg of CXCR4-CFP and 0.5 µg of CXCR4-YFP were transfected per dish. Prior to transfection, the media of the cells was exchanged for fresh media. FRET measurements were performed 48 hours after transfection using the microscopic FRET set-up.
During the experiments, cells were kept in imaging buffer. Then, using the BioPen microfluidic system (Fluicell, Gothenburg, Sweden; Ainla et al., 2010, 2012; Wright et al., 2018), single cells were exposed to the different solutions, as follows: cells were initially washed (approximately for 10–20 seconds) with imaging buffer to establish a baseline. Then, cells were stimulated with buffer supplemented with the appropriate ligand (CXCL12, MIF, or norepinephrine) at the indicated concentrations in the figures, until the response reached a plateau. Then, cells were washed again with imaging buffer, ideally, until reversal of the ligand-induced signal was observed (return to baseline). When indicated, a second stimulation and wash-out were performed. The duration of the stimuli is indicated in the figures as horizontal black lines. All the solutions contained 0.1% bovine serum albumin (BSA) (AppliChem).
For determining the kinetics of each process (τ values; apparent on- or off-rate), the FRET ratio of individual experiments was fitted to a one-component exponential decay function using the Clampfit software. For each case, τ values are shown as a scatter plot with median and interquartile range (IQR), in which N is the total number of individual cells measured in several independent experiments, as indicated in the figure legends. When required, statistical significance was tested using Mann-Whitney test or Kruskal-Wallis test. For determining the amplitude of the signals, maximum and minimum values were considered, and data are reported as mean ± S.E.M.
The emission of the acceptor fluorophores was corrected for that attributable to bleed through (spillover of donor into the 535-nm channel) and direct excitation (acceptor emission at 436 nm excitation). FRET was normalized and corrected for photobleaching. Processing of the data were done with OriginPro 2016 (OriginLab Corporation, Northampton, MA).
Acceptor Photobleaching Experiments.
HEK293 cells were prepared in 24-mm glass coverslips as previously described for confocal microscopy. Six hours later, cells were transfected with 0.6 µg of CXCR4-YFP or α2A-AR-YFP, 0.7 µg of Gαi1, 0.3 µg of Gβ1, and 0.2 µg of Gγ2-CFP, per dish, using Effectene. Prior to transfection, the media of the cells was exchanged for fresh media. FRET measurements were performed 48 hours after transfection using the microscopic FRET set-up. For the experiment, CFP and YFP fluorescence intensities were measured at 10 Hz for 40 seconds with 20 milliseconds illumination out of a total time of 100 milliseconds. Then, the donor was photobleached by illuminating for 9 minutes at 490 nm with 90 milliseconds illumination out of a total time of 100 milliseconds. Subsequently, the CFP and YFP fluorescence intensities were measured again for 40 seconds with 20 milliseconds illumination out of a total time of 100 milliseconds. During the measurements, cells were kept in 600 µl of imaging buffer. When indicated, cells expressing CXCR4 and the G protein subunits were pretreated with media containing 10 µM IT1t for 4 hours and kept in 600 µl imaging buffer containing 10 µM IT1t during the measurement. The change in the FCFP was calculated by considering the values of CFP fluorescence right before and after the bleaching process, using the formula [(FCFP,after − FCFP,before)/FCFP,after] and the Clampfit software. For each condition, three independent repeats were performed. Data are shown as a box plot in which the whiskers represent maximum and minimum values. Statistical significance between groups was tested using unpaired t test.
CRE-Luc Reporter-Gene Assay
The signaling properties of the different CXCR4 constructs were characterized using a CRE-driven reporter-gene assay. HEK293T cells were transfected with the CXCR4 constructs (CXCR4, CXCR4-CFP, or CXCR4-FlAsH228-CFP) and the CRE-Luc construct using 25-kDa linear polyethylenimine (PEI) (Polysciences) as DNA carrier agent in a 1:10 ratio (DNA/PEI). For transfection, 2 µg of DNA were combined with 12 µg of PEI in a total volume of 250 µl 150 mM NaCl and incubated for 20 minutes at room temperature. Then, 1 million resuspended cells were added to the DNA/PEI mix, and 80 µl were seeded at a density of 32,000 cells/well in a white, flat-bottom 96-well plate (Greiner Bio One). Analysis was done 24 hours after seeding the cells using Mithras LB940 (Berthold Technologies).
The day of the measurement, cells were incubated in media or media supplemented with 1 µM forskolin (FSK) (LC Laboratories) and increasing concentrations of CXCL12 (as indicated in the figure) and incubated for 6 hours at 37°C. Subsequently, the media was substituted for 25 µl of luciferase assay reagent [45 mM Tris H3PO4 pH 7.8, 45% glycerol, 3% Triton X-100, 1 mM DTT, 20 mM MgCl2, 800 µM D-Luciferin (Duchefa), 80 µM ATP] and incubated 20 minutes in the dark. Finally, the plate luminescence was measured and quantified.
The data from concentration-response curves were fitted to a three-parameter sigmoidal model using GraphPad. For each construct tested, four independent experiments were performed in quadruplicate. Means of EC50 values and asymmetric 95% confidence interval (CI) are reported in the text and have been calculated based on the logarithmic values (logEC50).
Binding Competition Assay
Twenty-four hours prior to assay, HEK293G cells stably expressing NLuc-CXCR4 were seeded on poly-D-lysine–coated white flat-bottom 96-well plates at a density of 40,000 cells/well. On the day of the assay, plating medium was removed and replaced with Hanks’ balanced salt solution/0.1% protease-free BSA (Hanks’ balanced salt solution composition 10 mM HEPES, 146 mM NaCl, 5 mM KCl, 1 mM MgS04, 2 mM sodium pyruvate, 1.3 mM CaCl2, and 1.5 mM NaHCO3; pH 7.4 with the addition of 10 mM glucose after autoclaving). A fixed concentration of the fluorescent ligand CXCL12-AlexaFluor647 (10 nM) was added simultaneously in triplicate wells, alongside increasing concentrations of unlabeled CXCL12, AMD3100, or MIF for 120 minutes at 37°C. The NLuc substrate was then added (50× dilution), and plates were left to equilibrate in the dark for 5 minutes.
Sequential dual luminescence and fluorescence readings were recorded using a PHERAstar FS plate reader (BMG Labtech, Germany) with 460 nm (80-nm bandpass; donor NLuc emission) and >610 nm longpass filters (acceptor CXCL12-AF647 emission). Raw bioluminescence resonance energy transfer ratios were calculated from the ratio of acceptor to donor emission values. Data were fitted to a three-parameter sigmoidal model using GraphPad. Data were pooled from five-sixths independent experiments (six independent experiments with CXCL12 and MIF ligands, of which five experiments also used AMD3100) and are expressed as mean ± S.E.M.
G Protein Activation in 96-Well Plates
HEK293T cells were seeded in 100-mm plates and grown until they reached 60%–65% confluency. Then, cells were transfected with 1.5 μg of receptor (CXCR4, CXCR4-FlAsH226, CXCR4-FlAsH228, or CXCR4-FlAsH229) and 3 μg of FRET-based G protein sensor (Gi1, Gi2, Gi3, or Gq) plasmid using Effectene. When indicated, empty plasmid was transfected instead of receptor. Prior to transfection, the media of the cells were replaced by fresh media. Twenty-four hours after transfection, cells were harvested in culture media and seeded at a density of 30,000 cells/well in black flat-bottom 96-well plates (Brand). The plates had been preincubated with poly-D-lysine (1 mg/ml) for 30 minutes and washed once with DPBS. Analysis was done 24 hours after seeding the cells.
To generate concentration-response curves in a microplate reader, cells were incubated at 37°C over 30 minutes with 90 µl of imaging buffer containing 0.1% BSA. Afterward, the fluorescence was read for 5 minutes to determine the preread signal. After the reading, 10 µl of buffer or increasing concentrations of CXCL12 (indicated in the figure) were added to the wells for a total assay volume of 100 µl. Fluorescence was read again for 20 minutes to determine the postread signal. During the measurement, cells were kept at 37°C. CXCL12 was prepared in imaging buffer containing 0.1% BSA. The FRET change produced by each concentration of ligand tested was corrected for the signal obtained in vehicle (nonligand; buffer)-treated cells. To determine the EC50 values for G protein activation, data were fitted to a three-parameter sigmoidal model using GraphPad. N = 5 independent experiments performed in quadruplicate. Means of EC50 values and asymmetric 95% CI are reported in the text and have been calculated based on the logarithmic values (logEC50). Statistical significance was tested using one-way ANOVA followed by Tukey’s test on the logEC50 values.
To investigate basal G protein activity, HEK293T cells transfected with CXCR4 and the Gi2 sensor were seeded at a density of 15,000 cells/well after the same protocol as described above for concentration-response curves. Three different conditions were analyzed in parallel: 1) buffer, 2) IT1t treatment, and 3) CXCL12 treatment. For 1), cells were incubated with 100 µl of imaging buffer for 4 hours. For 2), cells were incubated with 100 µl of imaging buffer supplemented with 100 µM IT1t for 4 hours. For 3), cells were incubated with 90 µl of imaging buffer for 4 hours, and 2 minutes before the measurement, 10 µl of 1 µM CXCL12 were added (final CXCL12 concentration: 100 nM). All the solutions contained 0.1% BSA. After the treatments, fluorescence was read for 8 minutes. The FRET detected under the three different conditions was normalized to “buffer treatment,” which was set to 100. Three independent repeats were performed in quadruplicate. Data are shown as a bar graph with S.D. Statistical significance between groups was tested using unpaired t test.
Experiments were performed using Synergy Neo2 Multi-Mode Microplate Reader (Biotek) with Gen5 Data Analysis Software. During the measurements, cells were excited at 420/50 nm (Biotek CFP-YFP Filter; 1035013), and emission was monitored at 485/20 and 540/25 nm (Biotek CFP-YFP Filter; 1035043).
Quantification and Statistical Analysis
Statistical analyses were performed using GraphPad Prism software (version 7). The statistical tests applied can be found in the figure legends and Materials and Methods section. When normal distribution could be assumed, t test or ANOVA followed by Tukey’s test was performed. When normal distribution could not be assumed, Mann-Whitney test or Kruskal-Wallis test was performed. The minimum criterion for statistical significance was P * 0.05. It should be noted that because of the exploratory nature of our research, the P values reported are descriptive and should not be interpreted as hypothesis testing.
Results
Development and Functional Characterization of FRET-Based CXCR4 Sensors.
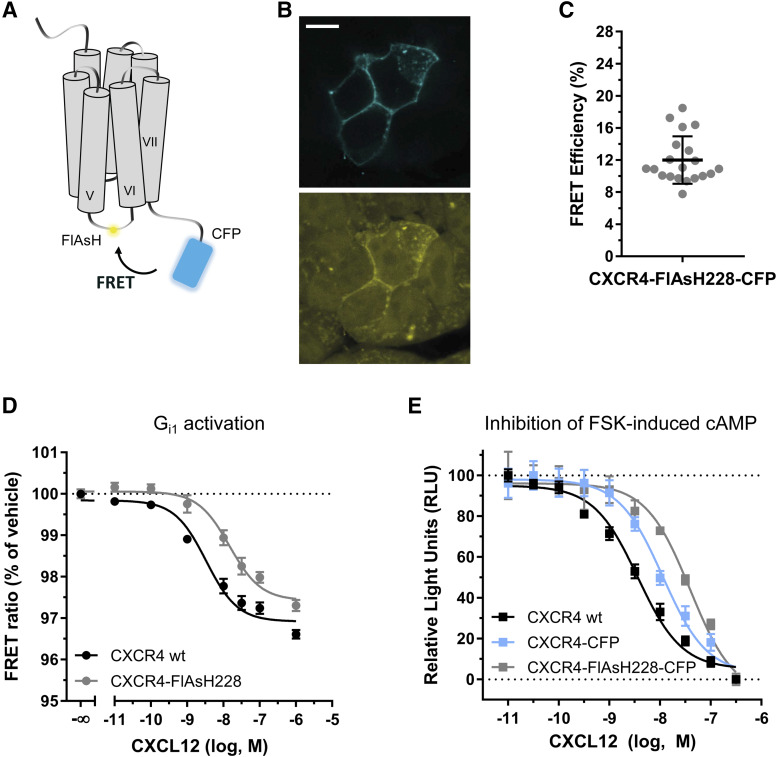
To study the conformational movements that CXCR4 undergoes during ligand-induced activation, we generated three different FRET sensors for CXCR4 with modifications in different positions. Addition of FRET-compatible fluorescent proteins or tags into the C terminus and intracellular loops has been established before in several GPCRs to investigate their activation (Stumpf and Hoffmann, 2016). Conformational changes induced upon ligand binding alter the distance and/or orientation between the fluorophores, resulting in changes in FRET that can be monitored in real time (Fig. 1A). Therefore, a CFP was fused to the C-terminal end of CXCR4, and the short FlAsH-binding motif CCPGCC was inserted in three different positions within the ICL-3: between L226 and S227 to generate the CXCR4-FlAsH226-CFP sensor, between H228 and S229 to generate the CXCR4-FlAsH228-CFP sensor, or between S229 and K230 to generate the CXCR4-FlAsH229-CFP sensor. The receptor constructs were characterized for correct functional properties. In particular, the CXCR4-FlAsH228-CFP sensor was chosen for full characterization, since it produced the highest FRET change upon agonist binding, as shown in the following section. Analysis by confocal microscopy showed that all three CXCR4 sensors localized to the plasma membrane (Fig. 1B upper panel and Supplemental Fig. 1A). Along with this, we observed that FlAsH specifically binds to the FlAsH-binding motif in ICL-3 (Fig. 1B lower panel), as previously reported for analogous receptor sensors (Hoffmann et al., 2005, 2010).
Fig. 1.
Principle and functional characterization of the CXCR4-FlAsH228-CFP sensor. (A) Schematic depicting the intramolecular FRET-based sensor. Ligand-induced conformational changes in CXCR4 are monitored as changes in FRET. (B) Representative confocal images of HEK293 cells transiently expressing CXCR4-FlAsH228-CFP and FlAsH-labeled prior to the measurement. Upper panel shows CFP emission. Lower panel shows FlAsH emission. Scale bar, 10 µm. (C) Intramolecular FRET efficiency of CXCR4-FlAsH228-CFP as determined by BAL treatment. Values were calculated from the increase in the CFP fluorescence upon BAL addition. Data shows mean ± S.D. of 20 cells measured on 4 independent experimental days. A representative individual experiment is shown in Supplemental Fig. 2. (D) Gi1 activation via CXCR4 or CXCR4-FlAsH228 in response to increasing concentrations of CXCL12. Data show mean ± S.E.M. and are representative of n = 3 independent experiments conducted in quadruplicate. In this particular experiment, EC50 = 3.3 and 14.7 nM for CXCR4 and CXCR4-FlAsH228, respectively. (E) Inhibition of FSK-induced cAMP accumulation in response to increasing concentrations of CXCL12 by HEK293T cells expressing CXCR4, CXCR4-CFP, or CXCR4-FlAsH228-CFP. Data show mean ± S.E.M. and are representative of n = 4 independent experiments conducted in triplicate. In this particular experiment, EC50 = 7.3, 11.3, and 39.0 nM for CXCR4, CXCR4-CFP, and CXCR4-FlAsH228-CFP, respectively. Characterization of the CXCR4-FlAsH226-CFP and CXCR4-FlAsH229-CFP sensors is presented in Supplemental Fig. 1. RLU, relative light unit.
To investigate intramolecular FRET in the sensors, we performed acceptor-bleaching experiments using BAL (2,3-dimercapto-1-propanol) in cells transfected with the sensors and FlAsH-labeled prior to the measurement. BAL displays very high affinity for arsenicals and, at high concentrations, displaces FlAsH from its binding site. Addition of 5 mM BAL led to a decrease in the FlAsH signal and dequenched CFP, proving the existence of basal intramolecular FRET (Supplemental Fig. 2A). FRET efficiency was calculated based on the increase in the CFP emission and was determined to be 12.0% ± 3.0% (mean ± S.D.) for the CXCR4-FlAsH228-CFP sensor (Fig. 1C). The constructs CXCR4-FlAsH226-CFP and CXCR4-FlAsH229-CFP showed FRET efficiencies of 5.0% ± 2.7% and 16.5% ± 4.3%, respectively (Supplemental Fig. 1B). No basal intermolecular FRET was detected in cells coexpressing CXCR4-CFP and CXCR4-FlAsH228 (Supplemental Fig. 2B), which indicates that the observed signal results from intramolecular FRET and not from crosstalk between adjacent receptors or between protomers of a dimer.
We then characterized the constructs for their ability to induce G protein signaling in response to CXCL12, since they contain the FlAsH-binding sequence in ICL-3, a region generally known to be important for G protein interaction (Hu et al., 2010; Chung, 2013). For this purpose, we used a FRET-based Gi1 sensor that measures G protein activation as a change in FRET between the labeled Gα and Gγ subunits (van Unen et al., 2016). HEK293 cells cotransfected with the Gi1 sensor and either CXCR4 or CXCR4-FlAsH228 were stimulated with increasing concentrations of CXCL12 in a 96-well plate assay format. Results showed that CXCR4-FlAsH228 preserves the ability to activate Gi1 (EC50 = 15.5 [9.3–25.9] nM; mean and asymmetric 95% CI) although with lower potency than the wild-type (wt) receptor (EC50 = 3.4 [2.3–4.9] nM; Fig. 1D). The CXCR4-FlAsH226 and CXCR4-FlAsH229 constructs also activated Gi1 with very similar potencies to CXCR4-FlAsH228 (EC50 = 18.1 [5.3–62.2] and 17.1 [4.9–59.3] nM, respectively; Supplemental Fig. 1C). Subsequently, and to examine the functionality of the complete sensor, we assessed the capacity of CXCL12 to inhibit FSK-induced cAMP accumulation in cells expressing CXCR4-FlAsH228-CFP in comparison with cells expressing CXCR4-CFP or the wt receptor. The results showed that cells expressing the sensor exhibited lower potency for the inhibition of FSK-induced cAMP production (EC50 = 19.0 [3.5–102.7] nM) in comparison with the wt (EC50 = 2.2 [0.6–8.0] nM). We also noted that the presence of CFP at the end of the C terminus contributes to an intermediate potency of the sensor with regards to Gi signaling (EC50 = 10.6 [1.9–57.9] nM) (Fig. 1E). Most importantly, the maximal activity of the sensor was comparable to the wt receptor. On the basis of this validation, these three constructs can serve as sensors to monitor ligand-induced intramolecular conformational changes.
The FRET-Based CXCR4 Sensors Report the Dynamics and Kinetics of Receptor Activation and Deactivation.
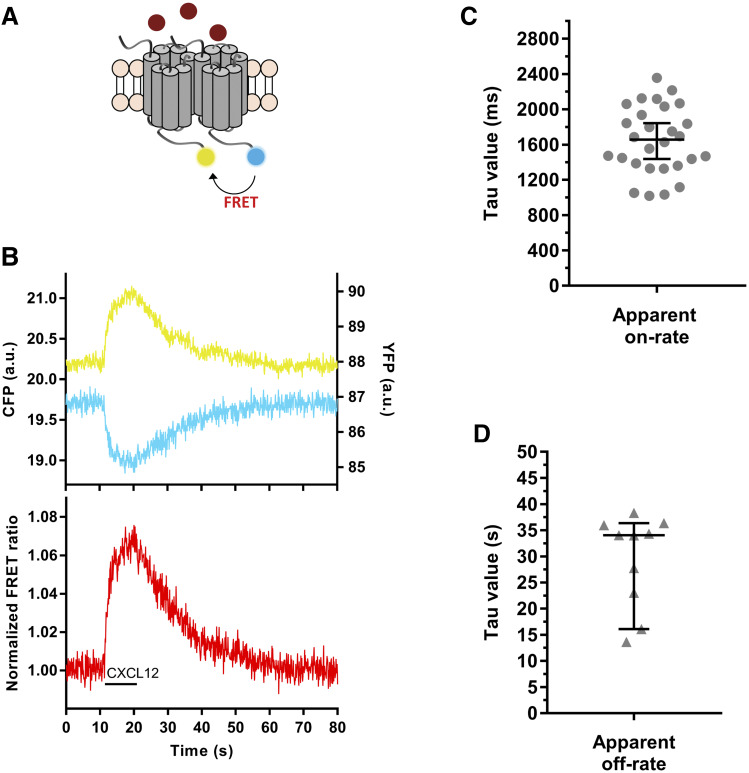
The CXCR4 sensors were used for further FRET experiments in single cells. To investigate the conformational changes upon receptor activation, HEK293 cells transiently expressing the receptor sensors and FlAsH-labeled prior to the measurements were stimulated with 30 µM CXCL12. A saturating concentration of ligand is required to determine the maximal and fastest kinetics of the process as well as to ensure that ligand diffusion to receptors is not time-limiting. A BioPen microfluidic device (Fluicell) was employed for delivering the solutions. This perfusion system allows the precise targeting of one or a few cells without affecting the surrounding environment (Ainla et al., 2010, 2012; Wright et al., 2018). A more detailed description of the stimulation and wash-out steps is described in the Materials and Methods section.
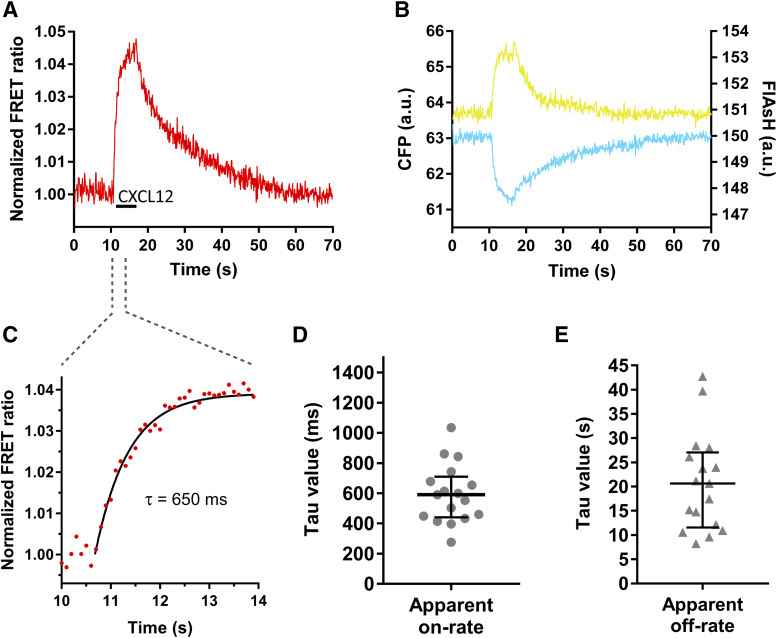
Stimulation of cells expressing the CXCR4-FlAsH228-CFP sensor with CXCL12 induced a rapid increase in the FlAsH signal and a simultaneous decrease in the CFP signal, which led to an increase in the FRET ratio by 2%–5% (2.8% ± 0.7%; mean ± S.D.) (Fig. 2, A and B; Supplemental Fig. 1G). Using this sensor, activation of CXCR4 occurred with a time constant τ = 590 (440–710) milliseconds (median and IQR) in response to CXCL12 (Fig. 2, C and D). Similar responses were observed when the CXCR4-FlAsH226-CFP and CXCR4-FlAsH229-CFP sensors were used to resolve the kinetics of receptor activation [τ = 600 (360–820) and τ = 530 (400–660) milliseconds, respectively; Supplemental Fig. 1D]. However, the amplitudes of the ligand-induced FRET signals with these two sensors were smaller than with CXCR4-FlAsH228-CFP (2.1% ± 0.6% for CXCR4-FlAsH226-CFP and 1.7% ± 0.6% for CXCR4-FlAsH229-CFP; Supplemental Fig. 1, E–G). After stimulation, rapid superfusion of the cells with buffer removed the ligand, and the FRET signal returned to baseline, allowing the off-kinetics of the receptor to be determined. Deactivation of CXCR4 after CXCL12 stimulation occurred with τ = 20.7 (11.6–27.1) seconds (Fig. 2E). In summary, these sensors faithfully monitor the ligand-induced structural rearrangements within the TM domain that reflect the activation of the receptor and have shown that the CXCL12-induced activation of CXCR4 is slower in comparison with other class A GPCRs and more similar to the activation of the class B PTHR.
Fig. 2.
The CXCR4-FlAsH228-CFP sensor reports the dynamics and kinetics of receptor activation and deactivation. (A and B) Representative traces of the FRET response from a single HEK293 cell expressing the CXCR4-FlAsH228-CFP sensor and stimulated with 30 µM CXCL12 for the indicated period of time (black line). Left panel shows corrected and normalized FRET ratio. Right panel shows corrected FlAsH (yellow) and CFP (cyan) emissions. (C) Kinetic analysis of receptor activation. The FRET change was fitted to a one-component exponential decay function to obtain the time constant τ. (D and E) On-kinetics of CXCR4 in response to CXCL12 (D) and off-kinetics of CXCR4 upon wash-out of the ligand with buffer (E). τ values from individual experiments are represented in a scatter plot. Data show median and IQR of 17 cells measured on 4 independent experimental days. a.u., arbitrary units.
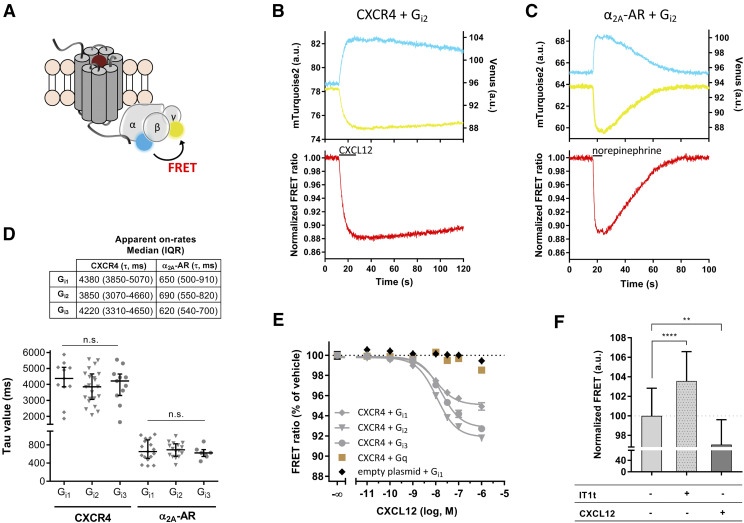
CXCR4 and Gi Proteins Remain within FRET Distance in the Absence of Agonist.
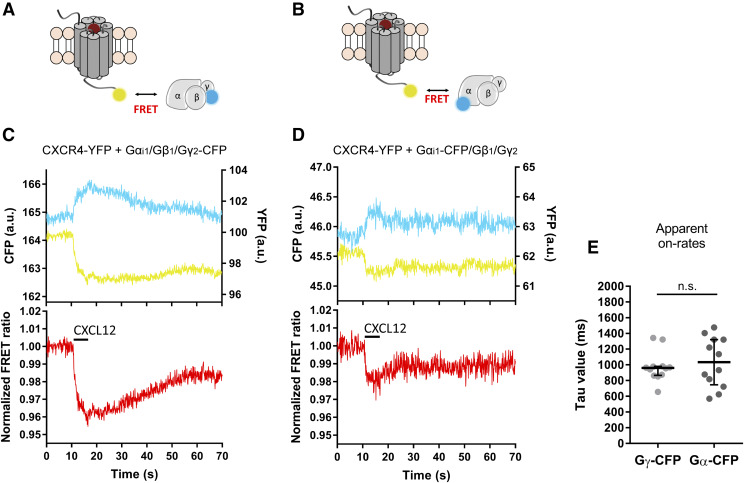
Generally, activation of a GPCR results in the recruitment of G proteins to the agonist-bound receptor. To determine the speed of coupling, we employed the constructs CXCR4-YFP and Gγ2-CFP, which are coexpressed with the Gαi1 and Gβ1 subunits for proper stoichiometry and localization of the G proteins to the plasma membrane (Fig. 3A). Surprisingly, and in contrast to what has been described for most GPCRs (Hein et al., 2005, 2006; Ferrandon et al., 2009; Hoffmann et al., 2012), stimulation of CXCR4 with CXCL12 (30 µM) resulted in a decrease in the FRET signal with an amplitude of 3.3 ± 1.0% (Fig. 3C). This rearrangement occurred at a speed of τ = 960 (870–980) milliseconds (Fig. 3E). To further investigate this effect, we examined the interaction of CXCR4-YFP with the G protein CFP-labeled at Gαi1 (Gαi1-CFP) (Fig. 3B). Again, a decrease in the agonist-dependent FRET signal was detected upon receptor activation with an amplitude of 2.7% ± 0.9% (Fig. 3D) and at a speed of τ = 1030 (750–1320) milliseconds (Fig. 3E).
Fig. 3.
CXCL12 induces rearrangements between CXCR4 and the Gi1 protein. (A and B) Schematic depicting the settings employed to investigate the interaction between the receptor and the Gi1 protein. HEK293 cells were transfected with CXCR4-YFP and Gαi1/Gβ1/Gγ2-CFP (A) or Gαi1-CFP/Gβ1/Gγ2 (B). (C and D) Representative traces of the FRET response from a single HEK293 cell expressing CXCR4-YFP and the G protein CFP-labeled at the Gγ2 (C) or Gαi1 subunit (D) and stimulated with 30 µM CXCL12 for the indicated period of time (black line). Upper panels show corrected YFP (yellow) and CFP (cyan) emissions. Lower panels show corrected and normalized FRET ratios. (E) On-kinetics of the interaction of CXCR4 with the Gi1 protein upon CXCL12 stimulation as measured in the two settings. τ values from individual experiments are represented in a scatter plot. Data show median and IQR of 12 cells for each setting, measured on at least 3 independent experimental days. Statistical significance was tested using Mann-Whitney test. a.u.,arbitrary units; n.s., nonsignificant.
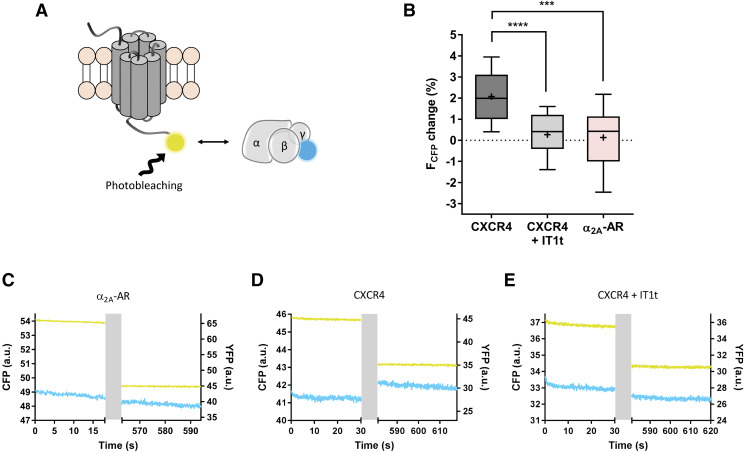
Whether receptors are preassembled with G proteins or only associate upon agonist exposure is still a matter of debate and, so far, it seems to be dependent on the specific receptor/G protein pair (Hein and Bünemann, 2009; Andressen et al., 2018). To further study a potential interaction of CXCR4 with the Gi protein in the absence of agonist, we performed acceptor photobleaching experiments in cells expressing CXCR4-YFP, Gγ2-CFP, Gβ1, and Gαi1 (Fig. 4A). In parallel, the YFP-labeled α2A-AR was tested under the same conditions. CFP and YFP emission intensities were measured prior to and after photobleaching the YFP, and the changes detected in the FCFP were quantified. After photobleaching, no significant increase in FCFP was detected with the α2A-AR-YFP [ΔFCFP = 0.4 (−1.0 to 1.1) %; median and IQR; Fig. 4, B and C], in agreement with published data (Hein et al., 2005). In contrast, a significant increase in FCFP was detected with CXCR4-YFP [ΔFCFP = 2.0 (1.0–3.1) %; Fig. 4, B and D], indicating the existence of basal energy transfer prior to the photobleaching process. When such cells were treated with 10 µM of the small molecule IT1t (antagonist), no significant change in FCFP upon YFP photobleaching was detected [ΔFCFP = 0.4 (−0.4 to 1.2) %; Fig. 4, B and E]. These findings suggest that although there is no specific interaction between α2A-AR and the Gi proteins in the absence of agonist under these experimental conditions, CXCR4 resides within FRET distance of the Gi protein, which might reflect a potential interaction between these two partners in the absence of agonist. Such behavior is consistent with other studies that suggest that CXCR4 exhibits some degree of basal receptor activity (Mishra et al., 2016; Mona et al., 2016).
Fig. 4.
CXCR4 resides within FRET distance from Gi proteins in the absence of agonist. (A) Schematic of acceptor photobleaching experiments. Donor and acceptor emissions were measured prior to and after photobleaching the YFP in HEK293 cells expressing the constructs Gαi1/Gβ1/Gγ2-CFP and α2A-AR-YFP or CXCR4-YFP. The latter was also measured in the presence of IT1t. (B) The change in the FCFP after YFP photobleaching from individual experiments in each condition is shown as a box plot. N = 19, 13, and 12 cells for CXCR4, CXCR4 + IT1t, and α2A-AR, respectively, measured on 3 independent experimental days. Statistical significance was tested using unpaired t test (***P ≤ 0.001; ****P ≤ 0.0001). (C–E) Representative CFP (cyan) and YFP (yellow) traces from individual experiments. The YFP photobleaching period is indicated in gray. a.u., arbitrary units.
CXCR4 Induces Prolonged Gi Protein Activation in Response to CXCL12.
To investigate the kinetics of Gi protein activation, we used FRET-based sensors for Gi1, Gi2, and Gi3 (van Unen et al., 2016). These sensors provide a real-time read-out for G protein activation by the loss of FRET between the Gγ- and Gα-labeled subunits in response to agonist stimulation of the receptor (Fig. 5A). HEK293 cells were cotransfected with CXCR4 or α2A-AR and one of the three G protein sensors. Then, single cells were stimulated with the appropriate ligand, either CXCL12 (30 µM) or norepinephrine (100 µM), using the BioPen. We detected activation of all three G protein subtypes via both receptors in response to their respective agonists, with no significant differences among the G protein subtypes. Upon stimulation, an immediate decrease in the FRET ratio was observed in every case (Fig. 5, B and C; Supplemental Fig. 3, A–D). Interestingly, the kinetic profiles of G protein activation via the two receptors were remarkably different. For CXCR4, G protein activation occurred with τ ≈ 4 seconds, whereas for α2A-AR, G protein activation occurred with τ ≈ 650 milliseconds (Fig. 5D), which is almost an order of magnitude faster and in agreement with published data (Hein et al., 2005; Van Unen et al., 2016). After ligand exposure, cells were continuously washed with buffer to remove the ligands. In the case of the α2A-AR, washing resulted in the FRET signal returning rapidly to baseline, which suggests that the G proteins rapidly adopt once again the inactive conformation upon removal of the ligand. However, CXCL12 induced a prolonged Gi activation via CXCR4, reflected by slower G protein off-kinetics that could not be resolved in the time frame of the experiment (Fig. 5, B and C, lower panels; Supplemental Fig. 3, A–D). These observations suggest different G protein activation by these two receptors, even though they both belong to class A GPCRs.
Fig. 5.
CXCR4 activates Gi proteins in response to CXCL12. (A) FRET-based sensors for Gi1, Gi2, Gi3, and Gq were employed to study G protein activation. A loss of FRET between the Gγ-Venus and Gα-mTurquoise2 subunits is detected upon activation. (B and C) Representative traces of the FRET response from a single HEK293 cell expressing the Gi2 sensor and CXCR4 (B) or α2A-AR (C) and stimulated with 30 μM CXCL12 or 100 µM norepinephrine, respectively (black line). Upper panels show corrected Venus (yellow) and mTurquoise2 (cyan) emissions. Lower panels show corrected and normalized FRET ratios. Supplemental Fig. 3 shows activation of Gi1 and Gi3. (D) Kinetics of Gi1, Gi2, and Gi3 protein activation via CXCR4 or α2A-AR in response to CXCL12 and norepinephrine, respectively. Table shows median and IQR. τ values from individual experiments are represented in a scatter plot with median and IQR. For CXCR4, n = 11, 22, and 11 cells for Gi1, Gi2, and Gi3 activation, respectively, measured on at least 3 independent experimental days. For α2A-AR, n = 17, 16, and 7 cells for Gi1, Gi2, and Gi3 activation, respectively, measured on at least 2 independent experimental days. Statistical significance was tested using Kruskal-Wallis test. n.s., nonsignificant. (E) CXCR4-mediated activation of Gi1, Gi2, Gi3, and Gq activation in response to increasing concentrations of CXCL12. As a control, empty plasmid was transfected instead of receptor. Data show mean ± S.E.M. and are representative of n = 5 independent experiments conducted in quadruplicate. In this particular experiment, EC50 values were 9.7, 10.8, and 17.1 nM for Gi1, Gi2, and Gi3, respectively. (F) FRET of cells expressing the CXCR4 and Gi2 sensor upon treatment with buffer, buffer supplemented with 100 µM IT1t or 100 nM CXCL12. Data are representative of three independent experiments. Data show mean ± S.D. and are normalized to buffer treatment. N = 30, 30, and 15 wells (containing 15,000 cells each) for the treatment with buffer, IT1t, and CXCL12, respectively. Statistical significance was tested using unpaired t test (**P ≤ 0.01; ****P ≤ 0.0001). a.u., arbitrary units.
Further experiments were performed in a 96-well plate FRET reader to test the effects of CXCL12-induced CXCR4 activation on G proteins. We observed a concentration-dependent activation of Gi1, Gi2, and Gi3 in cells expressing CXCR4 (Fig. 5E, gray lines). The EC50 values obtained for CXCL12 were 6.7 [2.9–15.4] nM for Gi1 activation, 9.9 [4.8–20.2] nM for Gi2 activation, and 18.6 [8.4–40.9] nM for Gi3 activation. Therefore, the isoforms Gi1 and Gi2 were activated with significantly higher potency than Gi3, which is in accordance with published data (Kleemann et al., 2008). The EC50 values obtained are also in good agreement with the potency of CXCL12 at CXCR4 described in other systems (Gupta et al., 2001; Rosenkilde et al., 2004; Kleemann et al., 2008; Levoye et al., 2009). No G protein activation was detected in cells expressing the G protein sensors but not CXCR4 (Fig. 5E, black data points; controls for Gi2 and Gi3 not shown). Although the CXCL12/CXCR4 axis has also been shown to activate Gq proteins in some contexts (Soede et al., 2001; Shi et al., 2007), we did not detect activation of Gq using a FRET-based Gq sensor (Fig. 5E, brown data points; Adjobo-Hermans et al., 2011).
Since our data suggest a potential interaction between CXCR4 and G proteins in the absence of agonist, we next analyzed the effects of IT1t versus those of CXCL12 on the Gi2 FRET sensor in CXCR4-expressing cells. FRET was analyzed under different conditions: buffer treatment, 100 µM IT1t treatment, or 100 nM CXCL12 treatment. We observed that IT1t caused a significantly higher FRET, and CXCL12 caused a significantly lower FRET compared with buffer (Fig. 5F). These data support the notion that CXCR4 activates G proteins in the absence of agonist or, in other words, that it exhibits some degree of basal receptor activity that can be blocked by IT1t, thus suggesting that this molecule would act as inverse agonist on CXCR4.
CXCR4 Homodimer Rearrangement Kinetically Precedes G Protein Activation.
Despite the fact that GPCR monomers have been demonstrated to be functional units (Whorton et al., 2007), increasing evidence has emerged for a functional role of dimers and higher oligomeric structures (Milligan et al., 2019). Therefore, we next investigated the kinetics of the rearrangement that occurs between CXCR4 protomers upon binding of CXCL12. To do so, HEK293 cells were transfected with CXCR4-CFP and CXCR4-YFP and stimulated with the agonist CXCL12, again using the BioPen (Fig. 6A).
Fig. 6.
CXCR4 homodimers undergo conformational changes in response to CXCL12. (A) The rearrangement between CXCR4 protomers was investigated in HEK293 cells cotransfected with CXCR4-CFP and CXCR4-YFP, with the fluorophores fused to the C termini. (B) Representative traces of the FRET response from a single HEK293 cell expressing CXCR4-YFP and CXCR4-CFP and stimulated with 30 μM CXCL12 (black line). Upper panel shows corrected YFP (yellow) and CFP (cyan) emissions. Lower panel shows corrected and normalized FRET ratio. (C and D) On-kinetics of the rearrangement between CXCR4 protomers in response to CXCL12 (C) and off-kinetics upon wash-out of the ligand with buffer (D). τ alues from individual experiments are represented in a scatter plot with median and IQR. N = 28 and 10 cells, respectively, measured on 4 independent experimental days. a.u., arbitrary units.
Upon stimulation of single cells with CXCL12 (30 µM), a large increase in the FRET signal was detected with an amplitude of 7.5% ± 2.8%, which suggests an approximation of the two fluorophores in the active state of the receptor (Fig. 6B). Interestingly, this rearrangement occurred at a speed of τ = 1660 (1370–2010) milliseconds (Fig. 6C), which indicates that this movement is achieved faster than the activation of the G proteins. Upon wash-out of the ligand with buffer, the signal returned to baseline, which allowed the determination of the off-kinetics of this process to be τ = 34.1 (21.3–36.1) seconds (Fig. 6D). Altogether, these data suggest that changes in CXCR4 homodimer occur slower than activation of a CXCR4 protomer. In contrast to our observations for CXCR4, several reports demonstrated that for the class C mGluR1, intersubunit rearrangements between protomers precedes intrasubunit rearrangements (Hlavackova et al., 2012; Grushevskyi et al., 2019). This distinct behavior might be related to the different nature of the receptor dimers and might be a characteristic activation feature of class C GPCRs, for which dimerization is obligatory for function (Kniazeff et al., 2011; Zhang et al., 2014).
MIF Induces Distinct Structural Rearrangements in CXCR4 and Does Not Activate Gi Proteins.
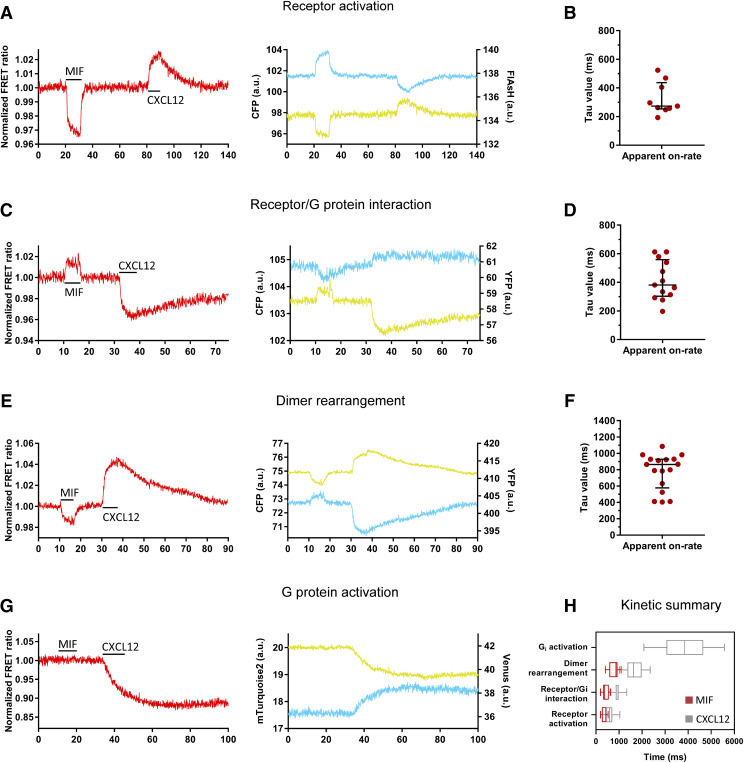
MIF has been recently described to be able to bind and signal via CXCR4 as a partial agonist (Rajasekaran et al., 2016). Therefore, the settings that have been earlier established in this work to investigate the CXCL12-induced activation of CXCR4 were employed to investigate each step of the transduction pathway of this receptor in response to MIF. During the measurements, single cells expressing the construct(s) of interest were stimulated with 100 µM MIF, followed by a short wash-out with buffer and subsequent stimulation with 30 µM CXCL12.
First, receptor activation was investigated in cells transfected with the CXCR4-FlAsH228-CFP sensor and FlAsH-labeled prior to the measurement. In contrast to CXCL12, which produced an increase in the FRET signal, MIF binding to the sensor led to a decrease in the FRET ratio at a speed of τ = 270 (250–440) milliseconds (Fig. 7, A and B). This suggests different conformational changes in CXCR4 by these two ligands. Washing of the cells with buffer after MIF stimulation rapidly returned the FRET signal to baseline, indicating that MIF can be easily removed from the receptor and suggesting a low ligand affinity. Subsequent stimulation with CXCL12 produced a response that was indistinguishable from the response observed when cells were directly stimulated with CXCL12, indicating that this response is not affected by the prior stimulation of the cells with MIF (Supplemental Fig. 4A). Second, the interaction of CXCR4 with the Gi protein was examined in cells transfected with CXCR4-YFP and the G protein CFP-labeled at the Gγ2 subunit. Upon stimulation with MIF, an increase in the FRET signal was observed with τ = 380 (300–560) milliseconds (Fig. 7, C and D). Thus, again, MIF produced a response that is opposite to that of CXCL12, suggesting that the rearrangement of the Gi protein with CXCR4 is different when the receptor is bound to MIF and CXCL12 and further supports the idea that the receptor adopts distinct conformations when bound to these two ligands. Upon washing of the cells with buffer, the FRET signal rapidly returned to baseline, and the cells could be subsequently stimulated with CXCL12, inducing a similar response to when cells are directly stimulated with this ligand (Supplemental Fig. 4B). Third, we investigated the movement between CXCR4 protomers upon MIF binding in cells transfected with CXCR4-CFP and CXCR4-YFP. We observed an MIF-induced decrease in the FRET signal that occurred at a speed of τ = 870 (580–930) milliseconds (Fig. 7, E and F). Again, the signal induced by MIF moved in the opposite direction, indicating once again different conformational changes in the receptor induced by these two ligands. The response induced by CXCL12 was also not affected by prior stimulation of the cells with MIF (Supplemental Fig. 4C). Fourth, cells transfected with CXCR4 and the Gi2 sensor were employed to investigate receptor-mediated G protein activation. Surprisingly, stimulation of the cells with MIF did not result in a change in FRET in the G protein sensor (Fig. 7G), indicating a lack of CXCR4-mediated G protein activation in response to this ligand. As a positive control, subsequent stimulation of the same cells with CXCL12 led to the activation of the G protein. A summary of the kinetics of CXCR4 activation and signaling in response to MIF in comparison with CXCL12 is shown in Fig. 7H.
Fig. 7.
MIF induces structural rearrangements in CXCR4 but does not lead to Gi protein activation. (A, C, E, and G) Representative traces of the FRET response from a single HEK293 cell transiently expressing: CXCR4-FlAsH228-CFP sensor (A), CXCR4-YFP and Gαi1/Gβ1/Gγ2-CFP (C), CXCR4-CFP and CXCR4-YFP (E) or CXCR4 and Gi2 sensor (G), which were stimulated with 100 µM MIF and then followed by wash-out and then stimulation with 30 μM CXCL12. Upper panels show corrected acceptor (yellow) and donor (cyan) emissions. Lower panels show corrected and normalized FRET ratios. (B, D, and F) On-kinetics of receptor activation (n = 9 cells) (B), receptor/G protein interaction (n = 13 cells) (D) and rearrangement between CXCR4 protomers (n = 17 cells) (F) in response to 100 µM MIF. τ values from individual experiments are represented in a scatter plot with median and IQR. Measurements were performed on at least 2 independent experimental days. (H) Comparison of CXCR4 on-kinetics in response to CXCL12 and MIF. Data from receptor activation belong to Figs. 2D and 7B. Data from receptor/G protein interaction belong to Figs. 3E (Gγ-labeled) and 7D. Data from protomers rearrangement belong to Figs. 6C and 7F. Data from G protein activation belong to Fig. 5D (Gi2 sensor). Data are shown as a box plot in which the whiskers represent maximum and minimum values. a.u., arbitrary units.
Discussion
The temporal resolution of individual biochemical steps that comprise a signaling cascade is important to understand cellular signaling. In this study, we report the kinetic and dynamic properties of early activation events in the CXCR4 transduction pathway in response to CXCL12 and MIF. Based on known movements of the TM domains that occur in GPCRs upon activation (Latorraca et al., 2017), we developed FRET-based sensors that are able to faithfully report the ligand-induced conformational changes in CXCR4. These constructs localize to the plasma membrane and preserve the ability to activate G proteins (Fig. 1), representing the first FRET sensors developed in the field of chemokine receptors.
The CXCR4 FRET sensors exhibited activation kinetics of τ ≈ 600 milliseconds in response to binding of the peptide agonist CXCL12 (Fig. 2). This is considerably slower than other class A receptors in response to small molecules, which exhibit activation time constants in the range of 2–50 milliseconds (Vilardaga et al., 2003; Hoffmann et al., 2005; Rochais et al., 2007; Reiner et al., 2010; Ziegler et al., 2011; Grushevskyi et al., 2019). Interestingly, CXCL12-induced activation of CXCR4 occurred in the same time range as the activation kinetics determined for the class B PTHR in response to the peptide PTH(1–34) (τ ≈ 1 second; Vilardaga et al., 2003). These differences in the activation kinetics might depend on intrinsic properties of the receptors, the type of ligand, and its binding mode. In this case, the binding of chemokines to their chemokine receptors proceeds via a complex mechanism that involves the formation of an extensive protein-protein interface with multiple recognition sites throughout the receptor (Wu et al., 2010; Kleist et al., 2016; Gustavsson et al., 2017). Likewise, PTH(1–34) was described to bind in two steps to PTHR (Castro et al., 2005).
The next signaling step we investigated led to the interesting observation of a decrease in the FRET signal between CXCR4-YFP and the CFP-labeled G protein upon CXCL12 stimulation (Fig. 3). Most receptors tested using this setting reported an increase in the FRET signal upon agonist stimulation, which is interpreted as G protein recruitment by the receptor (Hein et al., 2005, 2006). A recent study comparing the propensity of the two serotonin receptors 5-HT4 and 5-HT7 to associate with G proteins showed that 5-HT7 preassociates with Gs, whereas 5-HT4 interacts with Gs in an agonist-dependent manner. Respectively, a decrease and increase in the FRET signals were detected (Andressen et al., 2018). Accordingly, our data might suggest that CXCL12 stimulation of CXCR4 does not lead to recruitment of the G protein but rather to a rearrangement or dissociation of a preformed complex.
The spontaneous ligand-independent transition of GPCRs between inactive and active conformations is called basal or constitutive activity and is a common property of wt GPCRs but can also be a result of receptor mutations, which can cause a variety of diseases (Seifert and Wenzel-Seifert, 2002). Our results suggest a significant degree of basal activity for CXCR4. Photobleaching experiments in cells expressing CXCR4-YFP and CFP-labeled Gi1 protein showed a ligand-independent energy transfer between these two proteins (Fig. 4). Although the nature of their interaction is unknown, these data suggest close proximity or potential interaction between CXCR4 and the Gi protein prior to agonist stimulation. This hypothesis is further supported by the basal activation of Gi proteins observed in the presence of CXCR4, which was abolished by addition of IT1t, hence indicating inverse agonist properties for this compound (Fig. 4; Fig. 5F). Considering that an estimated 85% of antagonists turn out to be inverse agonists when tested in contexts in which receptors exhibit constitutive activity (Kenakin, 2004), it is not surprising that IT1t, which has been considered a CXCR4 antagonist until now, exhibits inverse agonist properties in our assays. Although our findings indicate that CXCR4 exhibits constitutive activity (at least in regard to G protein signaling), stimulation of the cells with CXCL12 further activated G proteins, thus indicating that this activity was not saturated. The existence of basal CXCR4 activity is in agreement with observations in previous studies (Mishra et al., 2016; Mona et al., 2016). Since overexpression of CXCR4 has been widely reported for a large number of cancers and other pathologies (Müller et al., 2001; Balkwill, 2004; Darash-Yahana et al., 2004; Chatterjee et al., 2014b; Zhao et al., 2015), the degree of spontaneous activity might reach a significant level in these contexts and become therapeutically relevant. On the basis of our observations, the degree of basal activity of CXCR4 should be validated in such pathophysiological systems and in vivo. An in-depth understanding of the biology of CXCR4 in this respect should assist in the design of improved CXCR4-targeting drugs.
When measuring G protein activation, we made an intriging observation related to the G protein off-kinetics. Even after persistent wash-out of CXCL12 from the receptor, CXCR4-mediated G protein activation remained for a long time (Fig. 5B), whereas this was not the case for the norepinephrine/α2A-AR axis (Fig. 5C) or other ligand/receptor pairs tested using these same G protein sensors (van Unen et al., 2016; Oehler et al., 2017; Grundmann et al., 2018). The nature of this observation is currently unknown to us, but several factors might play a role in this process. A possible explanation might be related to ligand residence time that might contribute to prolonged signaling (Hoffmann et al., 2015; Hothersall et al., 2016). Compared with norepinephrine and α2A-AR, CXCL12 remains bound to CXCR4 for a significantly longer time period. This assumption is based on our observed slower CXCR4 off-kinetics upon CXCL12 stimulation and wash-out with buffer, which was roughly ≈21 seconds (Fig. 2E), hence approximately 10 times slower than the α2A-AR sensor (Hein et al., 2005). However, this deactivation of the CXCR4 receptor was still much faster than the G protein deactivation. Alternatively, the different receptors might induce distinct active conformations of the G protein, which would display different lifetimes and GTPase activities (Furness et al., 2016). To mechanistically explain these differences in G protein off-kinetics, further research is required.
Our study provides insights into the distinct mechanisms by which MIF and CXCL12 regulate the activity of CXCR4 and presents evidence for their different pharmacological properties. Our data indicate that binding of CXCL12 and MIF induces distinct structural rearrangements in CXCR4, which can be distinguished dynamically and kinetically at each step of the signaling cascade (Fig. 7). This suggests different binding modes of these two ligands to the same receptor, as proposed by other studies (Rajasekaran et al., 2016; Lacy et al., 2018). The extensive interaction interface between CXCL12 and CXCR4 involves interactions between the core of the ligand and the receptor N terminus as well as the ligand N terminus with the receptor TM cavity (Qin et al., 2015). In contrast, MIF binds to the N-terminal region of CXCR4 at different residues than CXCL12 and not within the TM cavity (Rajasekaran et al., 2016). Their different binding modes are also supported by the incomplete displacement of CXCL12 from CXCR4 by MIF (Supplemental Fig. 5; Bernhagen et al., 2007). Furthermore, and in line with our distinct FRET signals (MIF opposite to CXCL12) but in contrast to other studies, we did not detect Gi protein activation via CXCR4 in response to MIF (Fig. 7G). Although our observations would suggest inverse agonist properties for MIF, functional data in other studies show that MIF acts as a CXCR4 allosteric partial agonist and mediates at least some of its functions via Gi proteins (Bernhagen et al., 2007; Klasen et al., 2014; Rajasekaran et al., 2016). A possible explanation for these distinct observations could be the cellular context. Although MIF can bind to CXCR4 alone, its binding and function appear to be facilitated by the presence of the single-pass membrane-receptor CD74 (cluster of differentiation 74), which together with CXCR4 forms a functional high-affinity heteromeric complex for MIF (Bernhagen et al., 2007; Schwartz et al., 2009; Klasen et al., 2014). HEK293 cells lack endogenous expression of CD74 (Schwartz et al., 2009), and, in our experiments, MIF was rapidly washed away from the receptor upon addition of buffer (Fig. 7, A, C, and E). The presence of CXCR4/CD74 complexes might be crucial in determining MIF signaling. For example, in platelets, which lack CD74, MIF could bind to CXCR4 but did not lead to extracellular signal-regulated kinase phosphorylation, whereas MIF-induced extracellular signal-regulated kinase phosphorylation was detected in monocytes, which express both receptors (Chatterjee et al., 2014a).
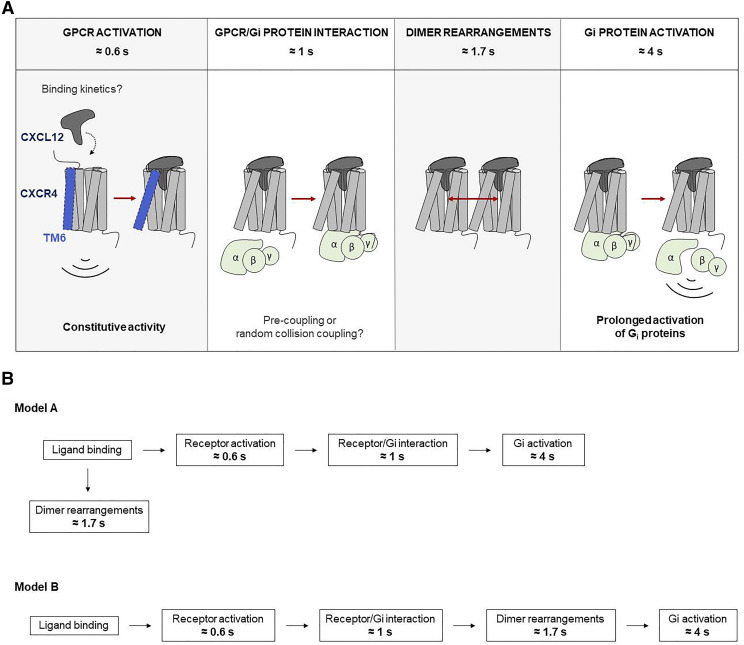
In summary, we provide kinetic data from ensemble measurements for the early signaling steps of the human chemokine receptor CXCR4. Upon CXCL12 engagement, structural rearrangements within the TM domain (τ ≈ 600 milliseconds) are kinetically followed by rearrangements between CXCR4 and the G protein in the intracellular side that occur at a speed of τ ≈ 1 seconds. Furthermore, a structural rearrangement between CXCR4 protomers occurs with τ ≈ 1.7 seconds, whereas G protein activation by CXCR4 finally occurs with τ ≈ 4 seconds. These kinetic events are summarized in Fig. 8. It is tempting to speculate that the rearrangement between protomers precedes G protein activation and to suggest that conformational changes in CXCR4 homodimers, when present, play a possible role in the signaling activation course of this receptor as depicted in model B. However, we need to emphasize that mechanistic interpretation needs to be based on measuring microscopic rate constants, and hence, model A, in which dimer rearrangement offers an alternative pathway, is also compatible with our dataset.
Fig. 8.
Summary of the findings in this manuscript regarding the CXCL12/CXCR4 axis. The upper part of the figure shows the kinetics of each step of the signaling cascade investigated using FRET. The main findings regarding the CXCL12/CXCR4 axis are: 1) Activation kinetics of CXCR4 upon CXCL12 binding are slower than other class A GPCRs; 2) Rearrangements within dimers occur faster than activation of Gi proteins; 3) This axis leads to a prolonged activation of Gi proteins; and 4) CXCR4 exhibits some degree of constitutive activity. It is tempting to speculate that the rearrangement between protomers precedes G protein activation, which might suggest that conformational changes in CXCR4 homodimers, when present, play a possible role in the signaling activation course of this receptor as depicted in model B. However, we need to emphasize that mechanistic interpretation needs to be based on measuring microscopic rate constants, and hence, model A, in which the dimer rearrangement offers an alternative pathway, is also compatible with our dataset. a.u., arbitrary units.
Acknowledgments
We thank all colleagues from ONCORNET for helpful scientific discussions.
Abbreviations
- AR
adrenergic receptor
- BAL
British anti-Lewisite
- BSA
bovine serum albumin
- CD74
cluster of differentiation 74
- CFP
cyan fluorescent protein
- CI
confidence interval
- CRE
cAMP-responsive element
- CXCL
CXC chemokine ligand
- CXCR
CXC chemokine receptor
- DPBS
Dulbecco’s phosphate-buffered saline
- FCFP
CFP fluorescence
- FlAsH
fluorescein arsenical hairpin binder
- FRET
Förster resonance energy transfer
- FSK
forskolin
- GPCR
G protein–coupled receptor
- HA
hemagglutinin
- HEK293
human embryonic kidney cell 293
- 5-HT
5-hydroxytryptamine
- ICL-3
intracellular loop-3
- IQR
interquartile range
- Luc
luciferase
- mGluR
metabotropic glutamate receptor
- MIF
macrophage migration inhibitory factor
- NLuc
NanoLuc
- PCR
polymerase chain reaction
- PEI
polyethylenimine
- PTH
parathyroid hormone
- PTHR
PTH receptor
- TM
transmembrane
- wt
wild type
- YFP
yellow fluorescent protein
Authorship Contributions
Participated in research design: Perpiñá-Viciano, Zarca, Kilpatrick, Hoffmann.
Conducted experiments: Perpiñá-Viciano, Zarca, Caspar, Kilpatrick.
Performed data analysis: Perpiñá-Viciano, Zarca, Caspar, Kilpatrick.
Wrote or contributed to the writing of the manuscript: Perpiñá-Viciano, Işbilir, Kilpatrick, Hill, Smit, Lohse, Hoffmann.
Footnotes
This work was supported by the European Union’s Horizon 2020 Program under grant agreement 641833 (Oncornet); the Deutsche Forschungsgemeinschaft [Grant TR166 ReceptorLight project C02]; and the Medical Research Council [MR/N020081/1].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Adjobo-Hermans MJ, Goedhart J, van Weeren L, Nijmeijer S, Manders EM, Offermanns S, Gadella TWJ., Jr. (2011) Real-time visualization of heterotrimeric G protein Gq activation in living cells. BMC Biol 9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlere I, Caspar B, Arimont M, Dekkers S, Visser K, Stuijt J, de Graaf C, Stocks M, Kellam B, Briddon S, et al. (2019) Modulators of CXCR4 and CXCR7/ACKR3 function. Mol Pharmacol 96:737–752. [DOI] [PubMed] [Google Scholar]
- Ainla A, Jansson ET, Stepanyants N, Orwar O, Jesorka A. (2010) A microfluidic pipette for single-cell pharmacology. Anal Chem 82:4529–4536. [DOI] [PubMed] [Google Scholar]
- Ainla A, Jeffries GD, Brune R, Orwar O, Jesorka A. (2012) A multifunctional pipette. Lab Chip 12:1255–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsayed Y, Ngo H, Runnels J, Leleu X, Singha UK, Pitsillides CM, Spencer JA, Kimlinger T, Ghobrial JM, Jia X, et al. (2007) Mechanisms of regulation of CXCR4/SDF-1 (CXCL12)-dependent migration and homing in multiple myeloma. Blood 109:2708–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. (2008) High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA 105:7439–7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andressen KW, Ulsund AH, Krobert KA, Lohse MJ, Bünemann M, Levy FO. (2018) Related GPCRs couple differently to Gs: preassociation between G protein and 5-HT7 serotonin receptor reveals movement of Gαs upon receptor activation. FASEB J 32:1059–1069. [DOI] [PubMed] [Google Scholar]
- Balkwill F. (2004) The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol 14:171–179. [DOI] [PubMed] [Google Scholar]
- Bernhagen J, Krohn R, Lue H, Gregory JL, Zernecke A, Koenen RR, Dewor M, Georgiev I, Schober A, Leng L, et al. (2007) MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med 13:587–596. [DOI] [PubMed] [Google Scholar]
- Bobkov V, Arimont M, Zarca A, De Groof TWM, van der Woning B, de Haard H, Smit MJ. (2019) Antibodies targeting chemokine receptors CXCR4 and ACKR3. Mol Pharmacol 96:753–764. [DOI] [PubMed] [Google Scholar]
- Bünemann M, Frank M, Lohse MJ. (2003) Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc Natl Acad Sci USA 100:16077–16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro M, Nikolaev VO, Palm D, Lohse MJ, Vilardaga JP. (2005) Turn-on switch in parathyroid hormone receptor by a two-step parathyroid hormone binding mechanism. Proc Natl Acad Sci USA 102:16084–16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Borst O, Walker B, Fotinos A, Vogel S, Seizer P, Mack A, Alampour-Rajabi S, Rath D, Geisler T, et al. (2014a) Macrophage migration inhibitory factor limits activation-induced apoptosis of platelets via CXCR7-dependent Akt signaling. Circ Res 115:939–949. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Behnam Azad B, Nimmagadda S. (2014b) The intricate role of CXCR4 in cancer. Adv Cancer Res 124:31–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KY. (2013) Structural aspects of GPCR-G protein coupling. Toxicol Res 29:149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darash-Yahana M, Pikarsky E, Abramovitch R, Zeira E, Pal B, Karplus R, Beider K, Avniel S, Kasem S, Galun E, et al. (2004) Role of high expression levels of CXCR4 in tumor growth, vascularization, and metastasis. FASEB J 18:1240–1242. [DOI] [PubMed] [Google Scholar]
- Ferrandon S, Feinstein TN, Castro M, Wang B, Bouley R, Potts JT, Gardella TJ, Vilardaga JP. (2009) Sustained cyclic AMP production by parathyroid hormone receptor endocytosis. Nat Chem Biol 5:734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness SGB, Liang YL, Nowell CJ, Halls ML, Wookey PJ, Dal Maso E, Inoue A, Christopoulos A, Wootten D, Sexton PM. (2016) Ligand-dependent modulation of G protein conformation alters drug efficacy. Cell 167:739–749.e11. [DOI] [PubMed] [Google Scholar]
- Grundmann M, Merten N, Malfacini D, Inoue A, Preis P, Simon K, Rüttiger N, Ziegler N, Benkel T, Schmitt NK, et al. (2018) Lack of beta-arrestin signaling in the absence of active G proteins. Nat Commun 9:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grushevskyi EO, Kukaj T, Schmauder R, Bock A, Zabel U, Schwabe T, Benndorf K, Lohse MJ. (2019) Stepwise activation of a class C GPCR begins with millisecond dimer rearrangement. Proc Natl Acad Sci USA 116:10150–10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Wang Y, Liu J, Mok SC, Xue F, Zhang W. (2016) CXCL12/CXCR4: a symbiotic bridge linking cancer cells and their stromal neighbors in oncogenic communication networks. Oncogene 35:816–826. [DOI] [PubMed] [Google Scholar]
- Gupta SK, Pillarisetti K, Thomas RA, Aiyar N. (2001) Pharmacological evidence for complex and multiple site interaction of CXCR4 with SDF-1alpha: implications for development of selective CXCR4 antagonists. Immunol Lett 78:29–34. [DOI] [PubMed] [Google Scholar]
- Gustavsson M, Wang L, van Gils N, Stephens BS, Zhang P, Schall TJ, Yang S, Abagyan R, Chance MR, Kufareva I, et al. (2017) Structural basis of ligand interaction with atypical chemokine receptor 3. Nat Commun 8:14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein P, Bünemann M. (2009) Coupling mode of receptors and G proteins. Naunyn Schmiedebergs Arch Pharmacol 379:435–443. [DOI] [PubMed] [Google Scholar]
- Hein P, Frank M, Hoffmann C, Lohse MJ, Bünemann M. (2005) Dynamics of receptor/G protein coupling in living cells. EMBO J 24:4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein P, Rochais F, Hoffmann C, Dorsch S, Nikolaev VO, Engelhardt S, Berlot CH, Lohse MJ, Bünemann M. (2006) Gs activation is time-limiting in initiating receptor-mediated signaling. J Biol Chem 281:33345–33351. [DOI] [PubMed] [Google Scholar]
- Heuninck J, Perpiñá Viciano C, Işbilir A, Caspar B, Capoferri D, Briddon SJ, Durroux T, Hill SJ, Lohse MJ, Milligan G, et al. (2019) Context-dependent signaling of CXC chemokine receptor 4 and atypical chemokine receptor 3. Mol Pharmacol 96:778–793. [DOI] [PubMed] [Google Scholar]
- Hilger D, Masureel M, Kobilka BK. (2018) Structure and dynamics of GPCR signaling complexes. Nat Struct Mol Biol 25:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavackova V, Goudet C, Kniazeff J, Zikova A, Maurel D, Vol C, Trojanova J, Prézeau L, Pin JP, Blahos J. (2005) Evidence for a single heptahelical domain being turned on upon activation of a dimeric GPCR. EMBO J 24:499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavackova V, Zabel U, Frankova D, Bätz J, Hoffmann C, Prezeau L, Pin JP, Blahos J, Lohse MJ. (2012) Sequential inter- and intrasubunit rearrangements during activation of dimeric metabotropic glutamate receptor 1. Sci Signal 5:ra59. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Castro M, Rinken A, Leurs R, Hill SJ, Vischer HF. (2015) Ligand residence time at G-protein-coupled receptors-why we should take our time to study it. Mol Pharmacol 88:552–560. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Gaietta G, Bünemann M, Adams SR, Oberdorff-Maass S, Behr B, Vilardaga JP, Tsien RY, Ellisman MH, Lohse MJ. (2005) A FlAsH-based FRET approach to determine G protein-coupled receptor activation in living cells. Nat Methods 2:171–176. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Gaietta G, Zürn A, Adams SR, Terrillon S, Ellisman MH, Tsien RY, Lohse MJ. (2010) Fluorescent labeling of tetracysteine-tagged proteins in intact cells. Nat Protoc 5:1666–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Nuber S, Zabel U, Ziegler N, Winkler C, Hein P, Berlot CH, Bünemann M, Lohse MJ. (2012) Comparison of the activation kinetics of the M3 acetylcholine receptor and a constitutively active mutant receptor in living cells. Mol Pharmacol 82:236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothersall JD, Brown AJ, Dale I, Rawlins P. (2016) Can residence time offer a useful strategy to target agonist drugs for sustained GPCR responses? Drug Discov Today 21:90–96. [DOI] [PubMed] [Google Scholar]
- Hu J, Wang Y, Zhang X, Lloyd JR, Li JH, Karpiak J, Costanzi S, Wess J. (2010) Structural basis of G protein-coupled receptor-G protein interactions. Nat Chem Biol 6:541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost CA, Reither G, Hoffmann C, Schultz C. (2008) Contribution of fluorophores to protein kinase C FRET probe performance. ChemBioChem 9:1379–1384. [DOI] [PubMed] [Google Scholar]
- Kenakin T. (2004) Efficacy as a vector: the relative prevalence and paucity of inverse agonism. Mol Pharmacol 65:2–11. [DOI] [PubMed] [Google Scholar]
- Klasen C, Ohl K, Sternkopf M, Shachar I, Schmitz C, Heussen N, Hobeika E, Levit-Zerdoun E, Tenbrock K, Reth M, et al. (2014) MIF promotes B cell chemotaxis through the receptors CXCR4 and CD74 and ZAP-70 signaling. J Immunol 192:5273–5284. [DOI] [PubMed] [Google Scholar]
- Kleemann P, Papa D, Vigil-Cruz S, Seifert R. (2008) Functional reconstitution of the human chemokine receptor CXCR4 with G(i)/G (o)-proteins in Sf9 insect cells. Naunyn Schmiedebergs Arch Pharmacol 378:261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleist AB, Getschman AE, Ziarek JJ, Nevins AM, Gauthier PA, Chevigné A, Szpakowska M, Volkman BF. (2016) New paradigms in chemokine receptor signal transduction: moving beyond the two-site model. Biochem Pharmacol 114:53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeff J, Prézeau L, Rondard P, Pin JP, Goudet C. (2011) Dimers and beyond: the functional puzzles of class C GPCRs. Pharmacol Ther 130:9–25. [DOI] [PubMed] [Google Scholar]
- Kniazeff J, Saintot PP, Goudet C, Liu J, Charnet A, Guillon G, Pin JP. (2004) Locking the dimeric GABA(B) G-protein-coupled receptor in its active state. J Neurosci 24:370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy M, Kontos C, Brandhofer M, Hille K, Gröning S, Sinitski D, Bourilhon P, Rosenberg E, Krammer C, Thavayogarajah T, et al. (2018) Identification of an Arg-Leu-Arg tripeptide that contributes to the binding interface between the cytokine MIF and the chemokine receptor CXCR4. Sci Rep 8:5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorraca NR, Venkatakrishnan AJ, Dror RO. (2017) GPCR dynamics: structures in motion. Chem Rev 117:139–155. [DOI] [PubMed] [Google Scholar]
- Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B. (2009) CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood 113:6085–6093. [DOI] [PubMed] [Google Scholar]
- Lohse MJ, Maiellaro I, Calebiro D. (2014) Kinetics and mechanism of G protein-coupled receptor activation. Curr Opin Cell Biol 27:87–93. [DOI] [PubMed] [Google Scholar]
- Lohse MJ, Nuber S, Hoffmann C. (2012) Fluorescence/bioluminescence resonance energy transfer techniques to study G-protein-coupled receptor activation and signaling. Pharmacol Rev 64:299–336. [DOI] [PubMed] [Google Scholar]
- Messerer R, Kauk M, Volpato D, Alonso Cañizal MC, Klöckner J, Zabel U, Nuber S, Hoffmann C, Holzgrabe U. (2017) FRET studies of quinolone-based bitopic ligands and their structural analogues at the muscarinic M1 receptor. ACS Chem Biol 12:833–843. [DOI] [PubMed] [Google Scholar]
- Milligan G, Ward RJ, Marsango S. (2019) GPCR homo-oligomerization. Curr Opin Cell Biol 57:40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RK, Shum AK, Platanias LC, Miller RJ, Schiltz GE. (2016) Discovery and characterization of novel small-molecule CXCR4 receptor agonists and antagonists. Sci Rep 6:30155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mona CE, Besserer-Offroy É, Cabana J, Leduc R, Lavigne P, Heveker N, Marsault É, Escher E. (2016) Design, synthesis, and biological evaluation of CXCR4 ligands. Org Biomol Chem 14:10298–10311. [DOI] [PubMed] [Google Scholar]
- Müller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. (2001) Involvement of chemokine receptors in breast cancer metastasis. Nature 410:50–56. [DOI] [PubMed] [Google Scholar]
- Oehler B, Mohammadi M, Perpiñá Viciano C, Hackel D, Hoffmann C, Brack A, Rittner HL. (2017) Peripheral interaction of resolvin D1 and E1 with opioid receptor antagonists for antinociception in inflammatory pain in rats. Front Mol Neurosci 10:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Kufareva I, Holden LG, Wang C, Zheng Y, Zhao C, Fenalti G, Wu H, Han GW, Cherezov V, et al. (2015) Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science 347:1117–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekaran D, Gröning S, Schmitz C, Zierow S, Drucker N, Bakou M, Kohl K, Mertens A, Lue H, Weber C, et al. (2016) Macrophage Migration Inhibitory Factor-CXCR4 Receptor Interactions: evidence for partial allosteric agonism in comparison with CXCL12 chemokine. J Biol Chem 291:15881–15895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner S, Ambrosio M, Hoffmann C, Lohse MJ. (2010) Differential signaling of the endogenous agonists at the beta2-adrenergic receptor. J Biol Chem 285:36188–36198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochais F, Vilardaga JP, Nikolaev VO, Bünemann M, Lohse MJ, Engelhardt S. (2007) Real-time optical recording of beta1-adrenergic receptor activation reveals supersensitivity of the Arg389 variant to carvedilol. J Clin Invest 117:229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkilde MM, Gerlach LO, Jakobsen JS, Skerlj RT, Bridger GJ, Schwartz TW. (2004) Molecular mechanism of AMD3100 antagonism in the CXCR4 receptor: transfer of binding site to the CXCR3 receptor. J Biol Chem 279:3033–3041. [DOI] [PubMed] [Google Scholar]
- Schwartz V, Lue H, Kraemer S, Korbiel J, Krohn R, Ohl K, Bucala R, Weber C, Bernhagen J. (2009) A functional heteromeric MIF receptor formed by CD74 and CXCR4. FEBS Lett 583:2749–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert R, Wenzel-Seifert K. (2002) Constitutive activity of G-protein-coupled receptors: cause of disease and common property of wild-type receptors. Naunyn Schmiedebergs Arch Pharmacol 366:381–416. [DOI] [PubMed] [Google Scholar]
- Shi G, Partida-Sánchez S, Misra RS, Tighe M, Borchers MT, Lee JJ, Simon MI, Lund FE. (2007) Identification of an alternative Galphaq-dependent chemokine receptor signal transduction pathway in dendritic cells and granulocytes. J Exp Med 204:2705–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soede RD, Zeelenberg IS, Wijnands YM, Kamp M, Roos E. (2001) Stromal cell-derived factor-1-induced LFA-1 activation during in vivo migration of T cell hybridoma cells requires Gq/11, RhoA, and myosin, as well as Gi and Cdc42. J Immunol 166:4293–4301. [DOI] [PubMed] [Google Scholar]
- Stoddart LA, Johnstone EKM, Wheal AJ, Goulding J, Robers MB, Machleidt T, Wood KV, Hill SJ, Pfleger KDG. (2015) Application of BRET to monitor ligand binding to GPCRs. Nat Methods 12:661–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf AD, Hoffmann C. (2016) Optical probes based on G protein-coupled receptors - added work or added value? Br J Pharmacol 173:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsibris AM, Kuritzkes DR. (2007) Chemokine antagonists as therapeutics: focus on HIV-1. Annu Rev Med 58:445–459. [DOI] [PubMed] [Google Scholar]
- van Unen J, Stumpf AD, Schmid B, Reinhard NR, Hordijk PL, Hoffmann C, Gadella TWJ, Jr., Goedhart J. (2016) A new generation of FRET sensors for robust measurement of Gαi1, Gαi2 and Gαi3 activation kinetics in single cells. PLoS One 11:e0146789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardaga JP, Bünemann M, Krasel C, Castro M, Lohse MJ. (2003) Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nat Biotechnol 21:807–812. [DOI] [PubMed] [Google Scholar]
- Vilardaga JP, Nikolaev VO, Lorenz K, Ferrandon S, Zhuang Z, Lohse MJ. (2008) Conformational cross-talk between α2A-adrenergic and μ-opioid receptors controls cell signaling. Nat Chem Biol 4:126–131. [DOI] [PubMed] [Google Scholar]
- Vilardaga JP, Steinmeyer R, Harms GS, Lohse MJ. (2005) Molecular basis of inverse agonism in a G protein-coupled receptor. Nat Chem Biol 1:25–28. [DOI] [PubMed] [Google Scholar]
- Watts AO, van Lipzig MM, Jaeger WC, Seeber RM, van Zwam M, Vinet J, van der Lee MM, Siderius M, Zaman GJ, Boddeke HW, et al. (2013) Identification and profiling of CXCR3-CXCR4 chemokine receptor heteromer complexes. Br J Pharmacol 168:1662–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton MR, Bokoch MP, Rasmussen SG, Huang B, Zare RN, Kobilka B, Sunahara RK. (2007) A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci USA 104:7682–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SC, Alonso Cañizal MC, Benkel T, Simon K, Le Gouill C, Matricon P, Namkung Y, Lukasheva V, König GM, Laporte SA, et al. (2018) FZD5 is a Gαq-coupled receptor that exhibits the functional hallmarks of prototypical GPCRs. Sci Signal 11:eaar5536. [DOI] [PubMed] [Google Scholar]
- Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. (2010) Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330:1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Edman LC, Sánchez-Alcañiz JA, Fritz N, Bonilla S, Hecht J, Uhlén P, Pleasure SJ, Villaescusa JC, Marín O, et al. (2013) Cxcl12/Cxcr4 signaling controls the migration and process orientation of A9-A10 dopaminergic neurons. Development 140:4554–4564. [DOI] [PubMed] [Google Scholar]
- Zhang XC, Liu J, Jiang D. (2014) Why is dimerization essential for class-C GPCR function? New insights from mGluR1 crystal structure analysis. Protein Cell 5:492–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Guo L, Zhao H, Zhao J, Weng H, Zhao B. (2015) CXCR4 over-expression and survival in cancer: a system review and meta-analysis. Oncotarget 6:5022–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler N, Bätz J, Zabel U, Lohse MJ, Hoffmann C. (2011) FRET-based sensors for the human M1-, M3-, and M5-acetylcholine receptors. Bioorg Med Chem 19:1048–1054. [DOI] [PubMed] [Google Scholar]