Abstract
Despite multiple recent advances, the diagnosis and management of lung cancer remain challenging and it continues to be the deadliest malignancy. In 2011, the National Lung Screening Trial (NLST) reported 20% reduction in lung cancer related mortality using annual low dose chest computed tomography (CT). These results led to the approval and nationwide establishment of lung cancer CT-based lung cancer screening programs. These findings have been further validated by the recently published Nederlands-Leuvens Longkanker Screenings Onderzoek (NELSON) and Multicentric Italian Lung Detection (MILD) trials, the latter showing benefit of screening even beyond the 5 years. However, the implementation of lung cancer screening has been impeded by several challenges, including the differentiation between benign and malignant nodules, the large number of false positive studies and the detection of indolent, potentially clinically insignificant lung cancers (overdiagnosis). Hence, the development of non-invasive strategies to accurately classify and risk stratify screen-detected pulmonary nodules in order to individualize clinical management remains a high priority area of research. Radiomics is a recently coined term which refers to the process of imaging feature extraction and quantitative analysis of clinical diagnostic images to characterize the nodule phenotype beyond what is possible with conventional radiologist assessment. Even though it is still in early phase, several studies have already demonstrated that radiomics approaches are potentially useful for lung nodule classification, risk stratification, individualized management and prediction of overall prognosis. The goal of this review is to summarize the current literature regarding the radiomics of screen-detected lung nodules, highlight potential challenges and discuss its clinical application along with future goals and challenges.
Keywords: Lung cancer, radiomics, imaging biomarker, risk stratification, pulmonary nodule
Introduction
Lung cancer remains the deadliest malignancy in the United States (US) and globally. Non-screen detected cases continue to be most commonly diagnosed at advanced, inherently difficult to treat disease stages. In contrast, the increased utilization of advanced cross-sectional high-resolution CT (HRCT) imaging has resulted in the increased incidental detection of indolent lesions of the lung adenocarcinoma spectrum and the more frequent diagnosis of multifocal lung cancer (1,2).
Over the last decades, there has been considerable emphasis on the development of effective lung cancer screening to facilitate the early diagnosis and treatment of lung cancer. In 2011, the National Lung Cancer Screening Trial (NLST) demonstrated a 20% reduction in lung cancer related mortality, resulting in the implementation of low-dose HRCT based lung cancer screening throughout the US (1). The NLST results were confirmed by the more recently reported Nederlands-Leuvens Longkanker Screenings Onderzoek (NELSON) study, and the long-term follow-up of the Multicentric Italian Lung Detection (MILD) study (3,4).
However, there are several remaining challenges. The NLST demonstrated a very high false positive rate, with most (>95%) of the detected pulmonary nodules being benign (1). While false positive rates are lower for larger nodules, increasing the nodule diameter also decreases the sensitivity of lung cancer screening (5). In addition, a significant subgroup (10–60%) of the screen or incidentally detected lung cancers represent indolent lesions and overdiagnosed lung cancers, of unknown clinical significance (3,6,7). Invasive diagnosis and treatment of overdiagnosed lung cancers are associated with preventable mortality, morbidity and substantial health care costs. Similar challenges also apply to the rapidly increasing numbers of incidentally detected lung nodules diagnosed by the ever-increasing number of cross-sectional imaging studies obtained for different indications (8). Ideally these challenges would be addressed using non-invasive biomarkers, including imaging biomarkers, to facilitate the accurate diagnosis, classification and risk-stratification of screen and incidentally detected lung cancers. In this context, most recent imaging biomarker research has focused on various “radiomics” approaches.
Radiomics refers to the process of identification, extraction, quantification and analysis of imaging features from radiologic images with the goal of better characterizing the phenotype of a given lung nodule in a way not otherwise possible with the naked eye. This approach is particularly attractive because rather than requiring the acquisition of additional diagnostic imaging, it can be applied to standard or already existing HRCT and positron emission tomography (PET) images, the two of the most commonly used imaging modalities in the diagnosis of lung cancer.
The goal of this review is to discuss the concept of radiomics, provide an update on its advancement and its application in the clinical setting to answer the most common questions associated with diagnosis, risk stratification, treatment response and prognosis of patients with screen and incidentally detected pulmonary nodules and lung cancer.
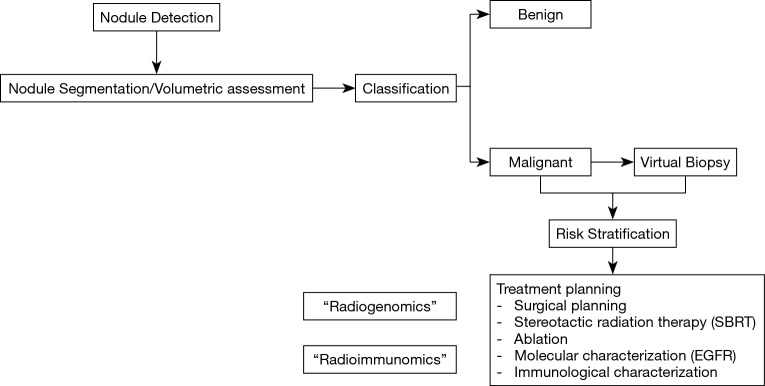
Workflow of radiomics for the analysis of pulmonary nodules and lung cancer (Figure 1)
Figure 1.
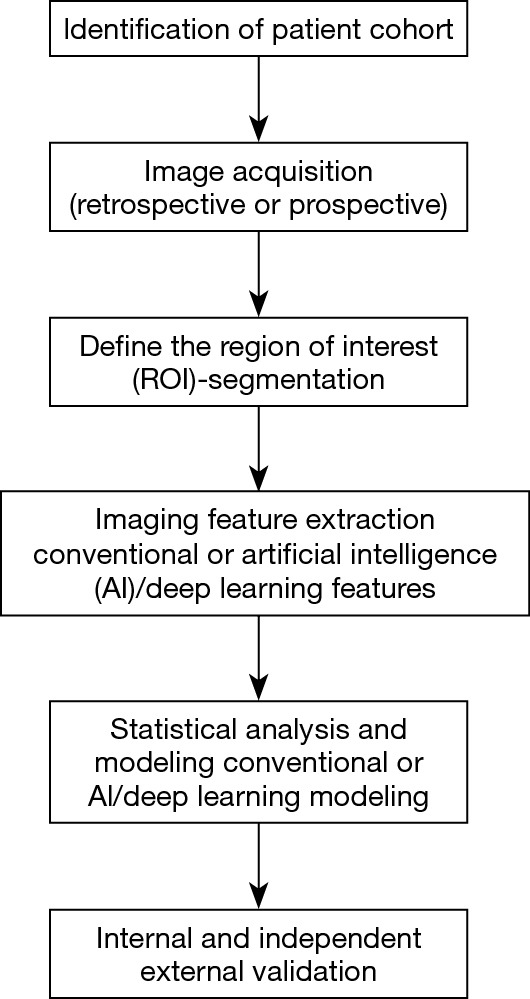
Workflow of radiomics for the analysis of pulmonary nodules and lung cancer.
Patient care in oncology is becoming increasingly personalized. Even though radiomics is still in its infancy, it has already demonstrated great promise by extracting additional important information from conventional radiological images. Moreover, by being reproducible and quantifiable, radiomics eliminates intra and inter-observer variability and facilitates the consistent, quantitative comparison between different patients. It not only focuses on the location, shape, size and density of the lesion, which represent the most readily evaluated lesion characteristics by a radiologist, but also extracts features such as volume, textural, surface characteristics and subtle changes in the surrounding lung parenchyma as well as other features that are beyond the capability of the eye of the human reader (9). The development of all radiomics approaches for the analysis of lung nodules requires a series of coordinated steps with input from a multidisciplinary team including the clinician, radiologist and the bioimaging developer/scientist.
Image acquisition
Image acquisition refers to obtaining the image of interest, most commonly from CT or PET images for patients with pulmonary nodules and lung cancer. However, radiomics approaches can be applied to other imaging modalities as well. The biggest challenge is the heterogeneity of image acquisition across different institutions. Chest CT scans are characterized by a wide variety of variables including scanner manufacturer, radiation dose, acquisition protocol, pixel size, slice thickness, intravenous contrast and the depth of the breath hold among others. In addition, post image acquisition processing requires data reconstruction which also has varying parameters (9,10). Currently, there are no accepted standards for chest CT acquisition and post-acquisition processing across different institutions which results in significant variability among clinical chest CT scans acquired during routine clinical practice. While this phenomenon represents a challenge for human readers, its impact is particularly challenging in radiomics which has resulted in difficulties in interpretation, validation and generalization of the results of radiomics studies using otherwise homogeneous datasets. The clinical applicability of radiomics models increases significantly if the findings can be replicated in heterogeneous “real life” datasets.
Image segmentation
Once the image has been acquired, a region of interest containing the lung nodule or lung cancer is isolated from the surrounding normal parenchyma with a process called segmentation. This is performed by outlining the margins of the lesion which is relatively simple in solid nodules but becomes more complex in sub-solid and ground glass opacities (9,10). Segmentation can be performed using three different approaches: manual, semi-automated or automated. Manual segmentation is labor intensive, subject to intra- and inter-reader variability and time consuming. Semi-automated and automated segmentations can be extremely accurate; however, it can be complicated in nodules in close proximity to adjacent structures such as hilum, pleura or adjacent vessel where a lesion may need to be outlined manually for accurate segmentation. Despite this, due to lack of standardization in semi-automated and automated segmentation, manual segmentation is still heavily relied upon across different studies. Machine and deep-learning approaches have also been used to automate manual lesion segmentation (11). While many radiomics approaches rely on segmentation of the lesion, others have focused on analyzing larger lung volumes, regions of interest, including the lesion circumventing some of the challenges of segmentation.
Feature extraction, selection and modeling
Following image acquisition, lesion segmentation or selection of a region of interest, a vast amount of phenotypic features are extracted. These can be broadly divided into semantics; those that are visible to the eye such as location, shape, size, presence of pleural effusion or lymph node involvement, and agnostics; those that are not obviously visible to the eye such as tumor heterogeneity, volume, skewness, degree of asymmetry and other features (12). Radiomics feature extraction is most commonly achieved using a variety of commercial or open source software packages. Semantics and agnostic features are usually unrelated and provide complementary information regarding the region of interest. Among these features, through data analysis, the features that are most clinically relevant to the set outcome are selected and integrated into a model to evaluate the desired clinical outcome. Artificial intelligence and deep learning are frequently used to generate, process and analyze the massive amounts of data related to radiomics projects. Deep learning approaches such as convolutional neuronal networks represent a subset of machine learning algorithms. The purpose of machine learning is to identify and classify radiomics features to facilitate lung nodule/mass segmentation or diagnostic/prognostic classification. Machine learning algorithms can be either supervised or unsupervised and linear or non-linear in nature. In contrast, deep learning algorithms do not require a separate feature extraction or engineering step in order to learn the relationship between the radiomics input and the corresponding clinical variable (13,14).
To ensure clinical applicability, it is crucial that the model is trained using cases with verified clinical outcomes and tested and validated in order to assess clinical application. Again, the differences in the radiomics programs, extracted features and analysis approaches and uncertainties about the clinical gold standard continue to be potential barriers to the clinical implementation of radiomics models.
Clinical application of radiomics for pulmonary nodules and lung cancer
The successful clinical application of radiomics in the clinical setting provides clinician with additional information regarding patient’s disease process which can assist in decision making along with reducing cost and overall morbidity of the patient. This information is typically extracted from already existing imaging studies which is non-invasive and cost effective. Radiomics can be applied during several stages of the evaluation of patients with pulmonary nodules and lung cancer (Figure 2).
Figure 2.
Utility of radiomics at different stages of patient evaluation for pulmonary nodules and lung cancer.
Radiomics classification of pulmonary nodules
Once an indeterminate pulmonary nodule has been identified, the next step is to determine its probability of malignancy. Several clinical calculators are currently available (15-17). These models use clinical variables and imaging features to classify pulmonary nodules as low (<5%), intermediate (10–65%) and high (>65%) probability of lung cancer. However, these models remain suboptimal and do not correlate well with each other. Consequently, there continues to be an urgent need for better biomarkers. Several recent studies have explored the potential role of radiomics in the classification and risk stratification of indeterminate pulmonary nodules (Table 1). While many of these studies have reported very promising results, the extracted radiomics features included in these models vary significantly between the different studies. Many of these studies are also limited by variability in image acquisition, lack of stability of the imaging features, small numbers of scans in relationship to the extracted imaging features (type I error) and a lack of external validation. In models using larger heterogenous datasets such as the NLST dataset, the inclusion of temporal changes (delta-radiomics) and model validation in true external datasets will likely be needed to successfully translate this approach into the clinic. We have recently identified a model using 8 among 57 pre-defined semantic imaging features, representative of nodule shape, surface characteristics, texture and location, to successfully differentiate benign from malignant lung nodules in a subset of the 726 NLST screen-detected malignant and benign pulmonary nodules. Interestingly, our model was independent of nodule size, which frequently represents one of the major features in other models. The AUC of our model was excellent at 0.94. We have applied our model in a blinded fashion to an independent external dataset of 170 consecutive incidentally detected benign (n=78) and malignant (n=92) lung nodules at Vanderbilt University. The AUC was 0.90 and the model outperformed the Brock University model in this dataset (AUC 0.87). The clinical use of our Mayo Clinic Radiomics model would result in 11% and 16% benign resections rates if applied to nodules with an intermediate probability of lung cancer (10–60%) by the Brock University model in the NLST and Vanderbilt datasets, respectively (28).
Table 1. Summary of recent studies with radiomic models to identify benign vs. malignant pulmonary nodules.
| Study | Number of scans (benign vs. malignant) | Conventional radiomics vs. deep learning | Number of features/model description | Internal vs. external validation | Model’s performance |
|---|---|---|---|---|---|
| Chen et al. (18) | 33 benign 42 malignant |
Conventional radiomics | - Support vector machine (SVM) was used as the classifier - 76 out of 750 features were significantly different between benign and malignant nodules - Accuracy for the selected 4-feature signature (SFS) was the highest |
Internal | For SFS: Accuracy: 84% Sensitivity: 92.85% Specificity: 72.73% |
| Ardila et al. (19) | Training dataset from NLST: 6,630 benign 86 malignant Independent validation set: 1,112 benign 27 malignant |
Deep convolutional neural network | -1,024 radiomics features - compared to expert radiologists |
External | AUC of training dataset: 0.944 AUC of validation dataset: 0.955 |
| Delzell et al. (20) | 90 benign 110 malignant |
Conventional radiomics | - 416 radiomic features - Combinations of the six feature selection methods and twelve classifiers were investigated by implementing a 10-fold repeated cross-validation framework with five repeats |
Internal | Values for the best selection method and classifier combination: AUC: 0.747 Sensitivity: 61.6% Specificity: 72.9% |
| Hawkins et al. (21) | NLST dataset: 328 benign 170 malignant |
Conventional radiomics | - 219 radiomic features with best model identifying 23 stable features - J48, JRIP (RIPPER), Naïve Bayes, support vector machines (SVMs), and random forest(s) classifiers tested |
Internal | Best models used random forests classifier with accuracy of predicting nodules becoming cancerous in 1 and 2 years: 80% (AUC 0.83) and 79% (AUC 0.75), respectively. |
| He et al. (22) | 60 benign 180 malignant Total: 240 (120 in primary cohort, 120 in validation cohort) |
Conventional radiomics | - 150 radiomic features - Least Absolute Shrinkage and Selection Operator Method (LASSO) logistic regression model used - Divided into four groups: Group 1 = non-contrast + 1.25 mm + standard convolution kernel; Group 2 = contrast enhancement + 1.25 mm + standard convolution kernel; Group 3 = non-contrast + 5 mm + standard convolution kernel; Group 4 = non-contrast + 5 mm + lung convolution kernel |
Internal | Group 1 had best performance: AUC: 0.862 Primary cohort: Sensitivity: 94.4% Specificity: 63.3% Accuracy: 85.8% Validation cohort: Sensitivity: 92.2% Specificity: 56.7% Accuracy: 83.3% |
| Peikert et al. (23) | NLST dataset 318 benign 408 malignant |
Conventional radiomics | - LASSO logistic regression model used - 8 out of 57 features selected |
Internal | AUC: 0.939 |
| Uthoff et al. (24) | Training cohort: 289 benign 74 malignant Validation cohort: 50 benign 50 malignant |
Machine learning/Artificial neural network | - Features of parenchyma surrounding the nodule were included | Internal and External | Best performing tool’s performance on validation cohort: AUC: 0.965 Accuracy: 98% Sensitivity: 100% Specificity: 96% |
| Xu et al. (25) | 192 benign 181 malignant |
Conventional radiomics | - 1160 radiomic features - Lesions classified in 3 groups based on size: T1a, T1b and T1c - Developed 3 radiomic models to predict malignancy in each group - Fivefold cross-validation was used |
Internal | Model 1 for T1a: AUC: 0.84 Accuracy: 77% Sensitivity: 89% Specificity: 74% Model 2 for T1b: AUC: 0.78 Accuracy: 73% Sensitivity: 74% Specificity: 71% Model 3 for T1c: AUC: 0.79 Accuracy: 76% Sensitivity: 77% Specificity: 73% |
| Mao et al. (26) | Training cohort: 156 benign 40 malignant Validation cohort: 75 benign 23 malignant |
Conventional radiomics | - 11 out of 385 radiomic features identified - LASSO logistic regression model used |
Internal | Training cohort: AUC: 0.953 Validation cohort: AUC: 0.97 Accuracy: 89.8% Sensitivity: 81% Specificity: 92.2% |
| Choi et al. (27) | 31 benign 41 malignant |
Conventional radiomics | - 103 radiomic features - SVM-LASSO model with ten-fold cross validation - Best model had 2 radiomic features |
Internal | AUC: 0.89 Accuracy: 84.6% |
A Google group in collaboration with clinical investigators also recently proposed a deep learning algorithm using patients’ current and prior CT images to calculate volumes and assess the malignancy risk. They created this model on a large cohort of 6,716 patients from NLST achieving a similar impressive AUC of 0.94. This was validated on 1,139 other cases with similar results. This model also outperformed the radiologists when prior CT images were not available and reduced the false positive and false negative risk to 11% and 5%, respectively (19).
Virtual biopsy and non-invasive risk stratification of malignant pulmonary nodules
In malignant pulmonary nodules of the lung adenocarcinoma spectrum, the degree of histopathological invasion is closely correlated with patient outcomes and facilitates patient management and prognostication. Adenocarcinomas present a clinical and histological spectrum ranging from indolent (non-invasive) to aggressive (invasive) lesions. Histologically, these lesions can be divided into three groups based on invasiveness: adenocarcinoma in situ (AIS) with no invasion, minimally invasive adenocarcinoma (MIA, ≤5 mm invasive focus) and invasive adenocarcinoma (IA, ≥5 mm invasive focus). Unfortunately, this histological classification entails surgical resection of the lesion and is not feasible based on small needle and/or core biopsies which limits its use for patient management. While indolent lesions would lend themselves to continued surveillance, limited resection, ablation or stereotactic body radiation therapy (SBRT), aggressive lesions should be treated with a standard surgical approach (lobectomy) if feasible. Consequently, non-invasive virtual biopsy tools and risk-stratification approaches are urgently needed. Several investigators have used radiomics approaches to address these issues (Table 2).
Table 2. Summary of recent studies with radiomics model that can be used as ‘virtual biopsy’ tools. MIA: minimally invasive adenocarcinoma; IA: invasive adenocarcinoma, AAH: atypical adenomatous hyperplasia, AIS: adenocarcinoma insitu.
| Study | Dataset | Model description | Model performance |
|---|---|---|---|
| Wu et al. (29) | Training set: 152 adenocarcinoma 51 squamous cell carcinoma Validation set: 62 adenocarcinoma 90 squamous cell carcinoma |
- Three classifiers: random Forests, Naive Baye’s, and K-nearest neighbors were evaluated. - 67 out of 440 features selected in multivariate analysis |
Naive Baye’s classifier performed the best with AUC 0.72 in identifying adenocarcinoma and squamous cell carcinoma |
| Digumarthy et al. (30) | 69 adenocarcinoma 25 squamous cell carcinoma |
- 11 radiomic features - Radiomic analysis comprised an initial image filtration step followed by quantification of texture within the lesion |
- 3/11 radiomic features were significantly different between adenocarcinoma and squamous cell carcinoma (AUC 0.686–0.744) - For probability variables, ROC analysis showed higher AUC value for radiomics (AUC 0.800) than clinical (AUC 0.780) and imaging (AUC 0.694) for differentiating adenocarcinomas and squamous cell carcinomas |
| Chae et al. (31) | 58 invasive pulmonary adenocarcinoma (7 MIA and 51 IA) 28 pre-invasive pulmonary adenocarcinoma (4 AAH and 24 AIS) |
- Investigate the value of computerized three-dimensional texture analysis for differentiation of preinvasive lesions from invasive pulmonary adenocarcinomas - Three-layered artificial neural networks (ANNs) with a back-propagation algorithm used |
- Smaller mass (adjusted OR: 0.092) and higher kurtosis (adjusted OR: 3.319) were significant differentiators of preinvasive lesions from invasive lesions (P<0.05). - ANNs model showed excellent accuracy in differentiation of preinvasive lesions from invasive lesions (AUC 0.981). |
| Li et al. (32) | 77 invasive pulmonary adenocarcinoma (37 MIA and 40 IA) 32 pre-invasive pulmonary adenocarcinoma (22 AAH and 10 AIS) |
- Stepwise model selection that mixed both forward and backward methods of variable selection using Akaike’s information criterion (AIC) was used to select the final predictive model | - Voxel count feature was significantly different between the invasive and preinvasive Lesions (82.5% sensitivity and 62.5% specificity) - Correlation feature predicted preinvasive lesions and MIAs better (sensitivity 81.1% and specificity 53.1%) |
| Son et al. (33) | 26 IA 9 MIA 4 AIS |
- Looked into utility of iodine enhanced imaging and virtual non-contrast (VNC) imaging in differentiating histologic subtypes of adenocarcinoma | The power of diagnosing IA improved after adding the iodine-enhanced imaging parameters compared to VNC imaging alone (AUC 0.959 vs. 0.888) |
| Maldonado et al. (34) | Training set: 2 AIS 20 MIA 32 IA Validation set: 1 AIS 10 MIA 75 IA |
- Development of computer-aided nodule assessment and risk yield (CANARY) software - Nonparametric Spearman correlation was used to analyze the relationship between histopathologic and radiologic invasion as determined by CANARY |
- Identified nine unique exemplars - Spearman R =0.87, P<0.0001 and 0.89 and P<0.0001 for the training and the validation set, respectively |
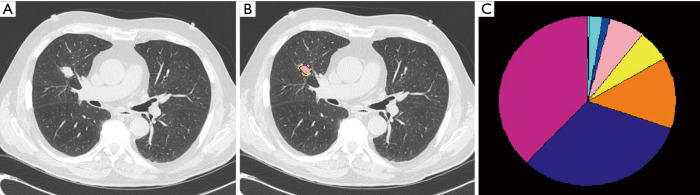
To address this problem, we developed a machine learning tool: Computer Aided Nodule Analysis and Risk Yield (CANARY), at Mayo Clinic, Rochester, MN, USA. CANARY identified 9 unique exemplars (radiomic fingerprints) that characterize the lung adenocarcinoma spectrum. As a virtual biopsy tool, CANARY has shown to correlate directly with invasion of adenocarcinoma (34). Adenocarcinoma lesions naturally cluster into three separate CANARY groups. These three clusters directly correlated with the disease-free survival with cluster 1 (good) having a 5-year survival of 100%, cluster 2 (intermediate) of 72.7% and cluster 3 (poor) of 51.4%. Moreover, this outcome prediction was shown to be better as compared to the pathologic TNM staging system (P<0.0001 vs. 0.55, respectively) (35). This approach was validated in the real-life clinical NLST dataset (36) (Figure 3).
Figure 3.
Computer Aided Nodule Analysis and Risk Yield (CANARY) of lung adenocarcinomas. (A) Representative axial CT scan showing the nodule of interest; (B) through CANARY, 9 natural clusters have been identified using automated clustering representing the basic radiologic building blocks of these lesions. The most central Region of Interest (ROI) of each cluster was selected as the cluster’s texture exemplar and the exemplars were color coded as Indigo, Green, Red, Pink, Yellow, Cyan, Blue, Orange and Violet; (C) when processing a new nodule, each voxel and its surrounding ROI is compared with the 9 exemplars and the voxel is color coded to the nearest exemplar. The relative distribution of these exemplars is displayed in a glyph.
To further improve CANARY as a virtual biopsy tool, we developed a computerized scoring system named Score Indicative of Lung Cancer Aggression (SILA), a cumulative aggregate of normalized distributions of the CANARY exemplars. SILA was able to differentiate between indolent and invasive adenocarcinoma (P<0.0001). But even beyond that SILA achieved a greater level of granularity, discriminating between different histopathological invasion depths in invasive adenocarcinoma (37). Such a discriminating ability also proved to be beneficial in predicting long-term patient outcomes.
In addition to CT, multiple groups have also looked at the utility of radiomics in PET scans for risk stratification. Arshad et al. identified a radiomics feature predictor FVX which was directly associated with overall survival in a multi-center study evaluating pre-treatment (radiation and chemotherapy) PET scans (38). Ahn et al., on the other hand evaluated pre-treatment PET scans of patients who underwent curative resection and identified contrast and busyness texture features to be the best two predictors of disease recurrence (39). We predict that virtual biopsy tools and non-invasive risk-stratification model supporting the personalized management of lung adenocarcinoma spectrum lesions will be implemented into clinical practice in the near future.
Role of radiomics in identifying mutations associated with lung cancer “Radiogenomics”
Once the patient suspected to have malignancy undergoes a biopsy, the treatment plan is largely dependent on two characteristics: histological subtype of malignancy and molecular analysis or mutations associated with the malignancy. There have been studies showing radiomics can predict both non-invasively.
Patients with lung adenocarcinoma frequently harbor driver mutations with epithelial growth factor receptor (EGFR) and Kristen rat sarcoma (KRAS) mutations being the most frequently identified abnormalities. Mutations are typically identified using tissue biopsies and hence, require an invasive procedure. The identification of these mutations is highly clinically significant as patients with EGFR mutations, ALK- and ROS1 translocations can be primarily treated with an EGFR inhibitor (TKI) and/or ALK-inhibitor, respectively. KRAS inhibitors are currently being evaluated in clinical trials. In contrast, tumors driven by these mutations appear to be less likely to respond to immunotherapy which has become an integral part of advanced stage lung cancer therapy.
Several studies have highlighted the utility of radiomics in trying to identify the presence of driver mutations non-invasively. We have demonstrated that the presence of the CANARY exemplars Yellow (Y) and green (G) were more likely in the presence of EGFR mutations (AUC of 0.77). These patients also tend to have significantly less fibrosis and low attenuation areas in the surrounding tumor free lung parenchyma. Combining these imaging features with the smoking status, we were able to achieve an AUC of 0.87 to identify EGFR mutations. Conversely, none of the CANARY exemplars or imaging features were found to be significantly associated with KRAS mutation (40). Digumarthy et al. showed that there were significant differences in 2 out of 11 radiomic features between EGFR mutant and wild type adenocarcinoma with AUC of 0.656–0.713 (30). Grossman et al. also reported several features in their radiomics model to be suggestive of EGFR and KRAS mutations along with one feature suggestive of TP53 mutation (41).
Li et al. evaluated a total of 51 patients (23 EGFR mutated, 28 wild type) with adenocarcinoma and classified them into four groups based on slice thickness (thin: 1 mm and thick: 5 mm) and two convolution kernels (smooth and sharp) yielding four groups: (I) Thin-Sharp, (II) Thin-Smooth, (III) Thick-Sharp, and (IV) Thick-Smooth. Prediction models were built using machine learning algorithms. Thin-Smooth model was the best predictive model for EGFR mutation with AUC of 0.83. The models using thick slices underperformed significantly while the effect of convolution kernel overall was insignificant. They concluded that high-resolution CT images could help to predict the EGFR mutational status (42). Rios Velazquez et al. also created a radiomics model based on a discovery cohort of 353 and training cohort of 352 patients with adenocarcinoma. They found sixteen radiomic features to be significantly associated with presence of EGFR mutation and ten features that were significantly associated with presence of KRAS mutation. They then developed radiomic signatures to identify patients with (I) EGFR+ and EGFR−, (II) KRAS+ and KRAS− and (III) EGFR+ and KRAS+ mutations. Clinical models comprising age, gender, smoking status, race, and clinical stage were also created to classify between these three groups. The radiomic signature was able to differentiate between EGFR+ and EGFR− cases with AUC of 0.69. This was similar to the performance of the clinical model of EGFR status which achieved AUC of 0.70. Combining the two signatures, an improved AUC of 0.75 was obtained which was significantly better than radiomic or clinical signatures alone. A KRAS+/KRAS− radiomic signature underperformed when compared to the clinical signature (AUC 0.63 vs. 0.75, respectively). The radiomic signature to discriminate between EGFR+ and KRAS+ tumors performed the best with AUC of 0.80 which further improved to 0.86 after combining it with the clinical signature (43). Weiss et al. showed that positive skewness and lower kurtosis was significantly associated with positive K-ras mutation (44).
A group from Shanghai Chest Hospital in China created two models to evaluate the ability of detecting EGFR mutation on CT chest images; (I) radiomics based model (MRadiomics) and (II) multi-level residual convolutional neural networks (MCNNs) based model (MMCNNS). They had 810 patients in the training set and 200 patients in the validation set with a total of 510 patients having EGFR mutation while the remaining 500 patients had wild type adenocarcinoma. The performance of MMCNNS was found to be better than MRadiomics with AUC of 0.810 as compared to 0.740, respectively (P=0.0255). This suggests that the performance of radiomics models can be heterogenous and further work to better understand the potential limitations of this technology is needed (45).
Other studies have similarly shown the usefulness of identifying genetic mutations in lung cancers via radiomics including from PET scans (46,47).
The role of radiomics in predicting treatment responses in lung cancer
Once a patient has been committed for treatment, it is challenging to predict how the tumor will respond to it. It is also difficult to predict the likelihood of recurrence or distant metastasis. The current standard of assessing tumor response is based on Response Evaluation on Criteria in Solid Tumors (RECIST) which is limited in its ability to monitor the treatment response accurately. There are several studies that support the notion of radiomics contributing to bridging this gap. These can be applied to patients undergoing various modalities of treatment including surgery, radiation, chemotherapy, targeted therapies and immunotherapy.
As mentioned earlier, CANARY has been shown to predict post-resection outcomes in early stage cancer by classifying the patients into Good, Intermediate and Poor categories (36). Aerts et al. evaluated 47 patients with early NSCLC before and 3 weeks post treatment with geftinib to analyze whether radiomics can identify a geftinib-responsive phenotype. On baseline scan, the radiomic feature Laws-Energy was significantly predictive of EGFR mutation status (AUC 0.67, P=0.03). Although no features were strongly predictive, particularly on post treatment scans, the change in features between the two scans (delta-radiomics) were strongly predictive particularly delta volume and delta maximum diameter (AUC range, 0.74–0.91) (48). Mattonen et al. looked into the effectiveness of radiomics in assessing the detection of local recurrence after stereotactic ablative radiation therapy (SABR) as compared to physicians. There were 15 patients with local recurrence matched with 30 patients without recurrence. The radiomic signature comprising 5 image-appearing features demonstrated high accuracy in discriminating the two groups with AUC of 0.85, classification error of 24%, false positive rate of 24% and false negative rate of 23%. This was in contrast to the physicians’ performance particularly when assessing likelihood of recurrence in <6 months post SABR where they called most of the changes as benign/no recurrence with mean error of 35% and high false negative rate of 99% (49). Coroller et al. looked to assess whether pre-treatment CT scan radiomics data was able to predict pathological response after neoadjuvant chemoradiation in patients with locally advanced NSCLC. There were seven features predictive of pathologic gross residual disease and one feature for complete pathological response. In contrast, no conventional imaging features were predictive (50). Similarly, Fave et al. also reported changes in radiomics textural and intensity features (delta radiomics) to be strongly predictive of tumor response to radiation therapy (51).
Dou et al. hypothesized that normal appearing peri-tumoral parenchyma may harbor microscopic tumor cells leading to distant metastases. They identified a peri-tumor 3 mm rim radiomic structure which correlated with risk of distant metastasis with moderate accuracy (CI =0.64, P value =2.4×10−5). This, however, was still better than a multivariable clinical model (CI =0.53), a visible tumor radiomics model (CI =0.59), or an exterior tissue model (CI =0.55) (52).
Immunotherapy has revolutionized the treatment of lung cancer, to the extent that it was recently approved for the management of small cell carcinoma as well (53). There is supporting evidence in the literature that radiomics can be used to predict and/or identify therapeutic response to immunotherapy. Khorrami et al. through machine learning, created a model comparing changes in the radiomic texture (DelRADx) of CT patterns both within the nodule and the parenchyma surrounding it before and after immune checkpoint inhibitor therapy. They divided 139 patients into a discovery set (D1, n=50) and two independent validation sets (D2, n=62; D3, n=27). A linear discriminant analysis (LDA) classifier achieved an AUC of 0.88, 0.85 and 0.81 in differentiating between responders and non-responders in D1, D2 and D3, respectively (54). A group from France developed a radiomic signature by combining contrast enhanced CT images with RNA-seq genomic data from tumor biopsies to assess CD8 cell tumor infiltration to identify the patients who are more likely to respond to anti-programmed cell death protein (PD)-1 or anti-programmed cell death ligand 1 (PD-L1) therapy. They reported higher baseline radiomics score for patients who responded to immunotherapy which was in turn associated with a higher survival rate (55). Along similar lines, Tang et al. created an immune pathology-informed model (IPIM) based on the quantitative parameters from pre-resection CT chest along with percent tumor PDL1 expression and density of tumor-infiltrating lymphocyte (via CD3 count) obtained through immunohistochemistry. Based on IPIM, 4 clusters (designated A-D) utilizing 4 radiomics features were identified. The IPIM designation was significantly associated with overall survival in both training (5-year OS: 61%, 41%, 50%, and 91%, for clusters A-D, respectively, P=0.04) and validation (5-year OS: 55%, 72%, 75%, and 86%, for clusters A-D, respectively, P=0.002) cohorts and immune pathology (all P<0.05) (56). Trebeschi et al. evaluated 1,055 primary and metastatic lesions from 203 patients with advanced melanoma and non-small cell lung cancer receiving anti PD-1 therapy. Based on their radiomics signature, immunotherapy response could be predicted with an AUC of up to 0.76 for both cancer types. They also reported significant associations with pathways involved in mitosis, suggesting that a favorable response to immunotherapy is directly related to the proliferative potential of the tumor (57).
Future directions and challenges for the clinical application of radiomics to pulmonary nodules and lung cancer
While radiomics approaches to the management of pulmonary nodules and lung cancer are extremely promising, it is very important to highlight some of the potential challenges related to the currently available data and the clinical implementation of radiomics. Many radiomics models are sensitive to variations in image acquisition protocols and efforts to standardize these protocols between different institutions remain very challenging limiting the clinical applicability of these radiomics models. In this context, radiomics approaches that are developed and independently validated in more heterogenous imaging datasets appear to be much more likely to be successfully implemented into clinical practice. Furthermore, many radiomics models use a large number of imaging features; however, are based on relatively small datasets raising the possibility of type I errors. Large, well curated imaging datasets of patients with well-defined clinical outcomes are urgently needed. This will most likely require national and international multi-institutional efforts. One way to accomplish this goal would be to develop shared imaging repositories of de-identified imaging datasets with associated clinical information using cloud based platforms. In addition, it would be very helpful to continue to try to understand the biological meaning of the identified radiomics features in different models. This approach may help to reconcile some of the differences between various radiomics models, lead to the development of more robust models and enhance our understanding of tumor biology. In summary, the initial data for radiomics in the evaluation and management of pulmonary nodules and lung cancer is extremely promising and continued efforts, in particular solving the challenges of image acquisition and the establishment of large imaging repositories for the development and validation of these models will likely result in the clinical implementation of these models for many aspects of the evaluation and management of pulmonary nodules and lung cancer in the near future.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editors (Fabien Maldonado and Robert Lentz) for the series “Novel Diagnostic Techniques for Lung Cancer” published in Journal of Thoracic Disease. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd.2020.03.105). The series “Novel Diagnostic Techniques for Lung Cancer” was commissioned by the editorial office without any funding or sponsorship. FM served as the unpaid Guest Editor of the series and serves as an unpaid editorial member of Journal of Thoracic Disease from Aug 2019 to Jul 2021. BJB reports other from CANARY (Imbio), outside the submitted work; In addition, BJB has a patent Mayo Clinic Radiomics Model, benign versus malignant nodules pending to none. RAK reports other from Imbio Inc., outside the submitted work. FM reports other from CANARY (Imbio), outside the submitted work; In addition, FM has a patent Mayo Clinic Radiomics Model, benign versus malignant nodules pending to none. TP reports other from CANARY (Imbio), outside the submitted work; In addition, TP has a patent Mayo Clinic Radiomics Model, benign versus malignant nodules pending to none. The other authors have no other conflicts of interest to declare.
References
- 1.Aberle DR, Adams AM, Berg CD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. 10.1056/NEJMoa1102873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 3.de Koning HJ, van der Aalst CM, de Jong PA, et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N Engl J Med 2020;382:503-13. 10.1056/NEJMoa1911793 [DOI] [PubMed] [Google Scholar]
- 4.Pastorino U, Sverzellati N, Sestini S, et al. Ten-year results of the Multicentric Italian Lung Detection trial demonstrate the safety and efficacy of biennial lung cancer screening. Eur J Cancer 2019;118:142-8. 10.1016/j.ejca.2019.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yip R, Henschke CI, Yankelevitz DF, et al. CT screening for lung cancer: alternative definitions of positive test result based on the national lung screening trial and international early lung cancer action program databases. Radiology 2014;273:591-6. 10.1148/radiol.14132950 [DOI] [PubMed] [Google Scholar]
- 6.Heleno B, Siersma V, Brodersen J. Estimation of Overdiagnosis of Lung Cancer in Low-Dose Computed Tomography Screening: A Secondary Analysis of the Danish Lung Cancer Screening Trial. JAMA Intern Med 2018;178:1420-2. 10.1001/jamainternmed.2018.3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Lung Screening Trial Research Team . Lung Cancer Incidence and Mortality with Extended Follow-up in the National Lung Screening Trial. J Thorac Oncol 2019;14:1732-42. 10.1016/j.jtho.2019.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gould MK, Tang T, Liu IL, et al. Recent Trends in the Identification of Incidental Pulmonary Nodules. Am J Respir Crit Care Med 2015;192:1208-14. 10.1164/rccm.201505-0990OC [DOI] [PubMed] [Google Scholar]
- 9.Hassani C, Varghese BA, Nieva J, et al. Radiomics in Pulmonary Lesion Imaging. AJR Am J Roentgenol 2019;212:497-504. 10.2214/AJR.18.20623 [DOI] [PubMed] [Google Scholar]
- 10.Lee G, Bak SH, Lee HY. CT Radiomics in Thoracic Oncology: Technique and Clinical Applications. Nucl Med Mol Imaging 2018;52:91-8. 10.1007/s13139-017-0506-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aresta G, Jacobs C, Araújo T, et al. iW-Net: an automatic and minimalistic interactive lung nodule segmentation deep network. Sci Rep 2019;9:11591. 10.1038/s41598-019-48004-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563-77. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koçak B, Durmaz EŞ, Ateş E, et al. Radiomics with artificial intelligence: a practical guide for beginners. Diagn Interv Radiol 2019;25:485-95. 10.5152/dir.2019.19321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Court LE, Fave X, Mackin D, et al. Computational resources for radiomics. Transl Cancer Res 2016;5:340-8. 10.21037/tcr.2016.06.17 [DOI] [Google Scholar]
- 15.Swensen SJ, Silverstein MD, Ilstrup DM, et al. The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules. Arch Intern Med 1997;157:849-55. 10.1001/archinte.1997.00440290031002 [DOI] [PubMed] [Google Scholar]
- 16.McWilliams A, Tammemagi MC, Mayo JR, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med 2013;369:910. 10.1056/NEJMoa1214726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herder GJ, van Tinteren H, Golding RP, et al. Clinical prediction model to characterize pulmonary nodules: validation and added value of 18F-fluorodeoxyglucose positron emission tomography. Chest 2005;128:2490-6. 10.1378/chest.128.4.2490 [DOI] [PubMed] [Google Scholar]
- 18.Chen CH, Chang CK, Tu CY, et al. Radiomic features analysis in computed tomography images of lung nodule classification. PLoS One 2018;13:e0192002. 10.1371/journal.pone.0192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ardila D, Kiraly AP, Bharadwaj S, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 2019;25:954-61. 10.1038/s41591-019-0447-x [DOI] [PubMed] [Google Scholar]
- 20.Delzell DAP, Magnuson S, Peter T, et al. Machine Learning and Feature Selection Methods for Disease Classification With Application to Lung Cancer Screening Image Data. Front Oncol 2019;9:1393. 10.3389/fonc.2019.01393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawkins S, Wang H, Liu Y, et al. Predicting Malignant Nodules from Screening CT Scans. J Thorac Oncol 2016;11:2120-8. 10.1016/j.jtho.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, Huang Y, Ma Z, et al. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep 2016;6:34921. 10.1038/srep34921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peikert T, Duan F, Rajagopalan S, et al. Novel high-resolution computed tomography-based radiomic classifier for screen-identified pulmonary nodules in the National Lung Screening Trial. PLoS One 2018;13:e0196910. 10.1371/journal.pone.0196910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uthoff J, Stephens MJ, Newell JD, Jr, et al. Machine learning approach for distinguishing malignant and benign lung nodules utilizing standardized perinodular parenchymal features from CT. Med Phys 2019;46:3207-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, Lu L, E LN, et al. Application of Radiomics in Predicting the Malignancy of Pulmonary Nodules in Different Sizes. AJR Am J Roentgenol 2019;213:1213-20. 10.2214/AJR.19.21490 [DOI] [PubMed] [Google Scholar]
- 26.Mao L, Chen H, Liang M, et al. Quantitative radiomic model for predicting malignancy of small solid pulmonary nodules detected by low-dose CT screening. Quant Imaging Med Surg 2019;9:263-72. 10.21037/qims.2019.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi W, Oh JH, Riyahi S, et al. Radiomics analysis of pulmonary nodules in low-dose CT for early detection of lung cancer. Med Phys 2018;45:1537-49. 10.1002/mp.12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peikert T, Duan F, Rajagopalan S, et al. Independent validation of a novel high-resolution computed tomography-based radiomic classifier for indeterminate lung nodules. J Thorac Oncol 2019;14:S221-2. 10.1016/j.jtho.2019.08.439 [DOI] [Google Scholar]
- 29.Wu W, Parmar C, Grossmann P, et al. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front Oncol 2016;6:71. 10.3389/fonc.2016.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Digumarthy SR, Padole AM, Gullo RL, et al. Can CT radiomic analysis in NSCLC predict histology and EGFR mutation status? Medicine (Baltimore) 2019;98:e13963. 10.1097/MD.0000000000013963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chae HD, Park CM, Park SJ, et al. Computerized texture analysis of persistent part-solid ground-glass nodules: differentiation of preinvasive lesions from invasive pulmonary adenocarcinomas. Radiology 2014;273:285-93. 10.1148/radiol.14132187 [DOI] [PubMed] [Google Scholar]
- 32.Li W, Wang X, Zhang Y, et al. Radiomic analysis of pulmonary ground-glass opacity nodules for distinction of preinvasive lesions, invasive pulmonary adenocarcinoma and minimally invasive adenocarcinoma based on quantitative texture analysis of CT. Chin J Cancer Res 2018;30:415-24. 10.21147/j.issn.1000-9604.2018.04.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Son JY, Lee HY, Kim JH, et al. Quantitative CT analysis of pulmonary ground-glass opacity nodules for distinguishing invasive adenocarcinoma from non-invasive or minimally invasive adenocarcinoma: the added value of using iodine mapping. Eur Radiol 2016;26:43-54. 10.1007/s00330-015-3816-y [DOI] [PubMed] [Google Scholar]
- 34.Maldonado F, Boland JM, Raghunath S, et al. Noninvasive characterization of the histopathologic features of pulmonary nodules of the lung adenocarcinoma spectrum using computer-aided nodule assessment and risk yield (CANARY)--a pilot study. J Thorac Oncol 2013;8:452-60. 10.1097/JTO.0b013e3182843721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raghunath S, Maldonado F, Rajagopalan S, et al. Noninvasive risk stratification of lung adenocarcinoma using quantitative computed tomography. J Thorac Oncol 2014;9:1698-703. 10.1097/JTO.0000000000000319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maldonado F, Duan F, Raghunath SM, et al. Noninvasive Computed Tomography-based Risk Stratification of Lung Adenocarcinomas in the National Lung Screening Trial. Am J Respir Crit Care Med 2015;192:737-44. 10.1164/rccm.201503-0443OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varghese C, Rajagopalan S, Karwoski RA, et al. Computed Tomography-Based Score Indicative of Lung Cancer Aggression (SILA) Predicts the Degree of Histologic Tissue Invasion and Patient Survival in Lung Adenocarcinoma Spectrum. J Thorac Oncol 2019;14:1419-29. 10.1016/j.jtho.2019.04.022 [DOI] [PubMed] [Google Scholar]
- 38.Arshad MA, Thornton A, Lu H, et al. Discovery of pre-therapy 2-deoxy-2-18F-fluoro-D-glucose positron emission tomography-based radiomics classifiers of survival outcome in non-small-cell lung cancer patients. Eur J Nucl Med Mol Imaging 2019;46:455-66. 10.1007/s00259-018-4139-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahn HK, Lee H, Kim SG, et al. Pre-treatment 18F-FDG PET-based radiomics predict survival in resected non-small cell lung cancer. Clin Radiol 2019;74:467-73. 10.1016/j.crad.2019.02.008 [DOI] [PubMed] [Google Scholar]
- 40.Clay R, Kipp BR, Jenkins S, et al. Computer-Aided Nodule Assessment and Risk Yield (CANARY) may facilitate non-invasive prediction of EGFR mutation status in lung adenocarcinomas. Sci Rep 2017;7:17620. 10.1038/s41598-017-17659-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grossmann P, Stringfield O, El-Hachem N, et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife 2017. doi: . 10.7554/eLife.23421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Lu L, Xiao M, et al. CT Slice Thickness and Convolution Kernel Affect Performance of a Radiomic Model for Predicting EGFR Status in Non-Small Cell Lung Cancer: A Preliminary Study. Sci Rep 2018;8:17913. 10.1038/s41598-018-36421-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rios Velazquez E, Parmar C, Liu Y, et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res 2017;77:3922-30. 10.1158/0008-5472.CAN-17-0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss GJ, Ganeshan B, Miles KA, et al. Noninvasive image texture analysis differentiates K-ras mutation from pan-wildtype NSCLC and is prognostic. PLoS One 2014;9:e100244. 10.1371/journal.pone.0100244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li XY, Xiong JF, Jia TY, et al. Detection of epithelial growth factor receptor (EGFR) mutations on CT images of patients with lung adenocarcinoma using radiomics and/or multi-level residual convolutionary neural networks. J Thorac Dis 2018;10:6624-35. 10.21037/jtd.2018.11.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mei D, Luo Y, Wang Y, et al. CT texture analysis of lung adenocarcinoma: can Radiomic features be surrogate biomarkers for EGFR mutation statuses. Cancer Imaging 2018;18:52. 10.1186/s40644-018-0184-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Zhao X, Zhao Y, et al. Value of pre-therapy 18F-FDG PET/CT radiomics in predicting EGFR mutation status in patients with non-small cell lung cancer. Eur J Nucl Med Mol Imaging 2020;47:1137-46. 10.1007/s00259-019-04592-1 [DOI] [PubMed] [Google Scholar]
- 48.Aerts HJ, Grossmann P, Tan Y, et al. Defining a Radiomic Response Phenotype: A Pilot Study using targeted therapy in NSCLC. Sci Rep 2016;6:33860. 10.1038/srep33860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mattonen SA, Palma DA, Johnson C, et al. Detection of Local Cancer Recurrence After Stereotactic Ablative Radiation Therapy for Lung Cancer: Physician Performance Versus Radiomic Assessment. Int J Radiat Oncol Biol Phys 2016;94:1121-8. 10.1016/j.ijrobp.2015.12.369 [DOI] [PubMed] [Google Scholar]
- 50.Coroller TP, Agrawal V, Narayan V, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol 2016;119:480-6. 10.1016/j.radonc.2016.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fave X, Zhang L, Yang J, et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep 2017;7:588. 10.1038/s41598-017-00665-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dou TH, Coroller TP, van Griethuysen JJM, et al. Peritumoral radiomics features predict distant metastasis in locally advanced NSCLC. PLoS One 2018;13:e0206108. 10.1371/journal.pone.0206108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horn L, Mansfield AS, Szczęsna A, et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med 2018;379:2220-9. 10.1056/NEJMoa1809064 [DOI] [PubMed] [Google Scholar]
- 54.Khorrami M, Prasanna P, Gupta A, et al. Changes in CT Radiomic Features Associated with Lymphocyte Distribution Predict Overall Survival and Response to Immunotherapy in Non-Small Cell Lung Cancer. Cancer Immunol Res 2020;8:108-19. 10.1158/2326-6066.CIR-19-0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018;19:1180-91. 10.1016/S1470-2045(18)30413-3 [DOI] [PubMed] [Google Scholar]
- 56.Tang C, Hobbs B, Amer A, et al. Development of an Immune-Pathology Informed Radiomics Model for Non-Small Cell Lung Cancer. Sci Rep 2018;8:1922. 10.1038/s41598-018-20471-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trebeschi S, Drago SG, Birkbak NJ, et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol 2019;30:998-1004. 10.1093/annonc/mdz108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as