Abstract
Since cancer is the leading cause of death worldwide, there is an urgent need to understand the mechanisms underlying cancer progression and the development of cancer inhibitors. Signal transducer and activator of transcription 3 (STAT3) is a major transcription factor that regulates the proliferation and survival of various cancer cells. Here, dual-specificity phosphatase 3 (DUSP3) was identified as a regulator of STAT3 based on an interaction screening performed using the protein tyrosine phosphatase library. DUSP3 interacted with the C-terminal domain of STAT3 and dephosphorylated p-Y705 of STAT3. In vitro dephosphorylation assay revealed that DUSP3 directly dephosphorylated p-STAT3. The suppressive effects of DUSP3 on STAT3 were evaluated by a decreased STAT3-specific promoter activity, which in turn reduced the expression of the downstream target genes of STAT3. In summary, DUSP3 downregulated the transcriptional activity of STAT3 via dephosphorylation at Y705 and also suppressed the migratory activity of cancer cells. This study demonstrated that DUSP3 inhibits interleukin 6 (IL-6)/STAT3 signaling and is expected to regulate cancer development. Novel functions of DUSP3 discovered in IL-6/STAT3 signaling regulation would help expand the understanding of cancer development mechanisms.
Keywords: Dephosphorylation, Dual-specificity phosphatase 3, Protein tyrosine phosphatase, Signal transducer and activator of transcription 3, Signal transduction
INTRODUCTION
During cancer development, chronic inflammation reportedly favors carcinogenesis, malignant transformation of cancer cells, and cancer metastasis (1). Typically, pro-inflammatory cytokines produced owing to cancer-associated inflammation not only help cancer cell growth but also cause epithelial–mesenchymal transition that increases the metastatic potential of cancer cells (2). Cancer development is associated with several signaling pathways. Among the signaling pathways, the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling is frequently dysregulated in various types of cancer (3). STAT3 is one of seven STAT transcription factor family members activated by interleukin 6 (IL-6), epidermal growth factor (EGF), platelet-derived growth factor (PDGF), growth hormone, or erythropoietin, and it regulates biological responses related to inflammation, metabolism, and carcinogenesis (4-6). Phosphorylation of STAT3 at Tyr-705 (Y705) promotes homo/heterodimer formation with other STATs and induces nuclear translocation (4). Moreover, the phosphorylation of STAT3 increases its transcriptional activity (4). Upregulated STAT3 activity suppresses cancer cell apoptosis by increasing the expression of STAT3 downstream genes, such as Bcl-2, Bcl-XL, survivin, and Mcl-1, which function in cell survival (7). Recently, it was reported that STAT3 is correlated with the Warburg effect, which is reprogramming of glucose metabolism for inducing aerobic glycolysis in cancer cells during cancer development (8). IL-6-induced STAT3 activation increases the expression of hexokinase 2 (HK2) and pyruvate dehydrogenase kinase 1, which promotes aerobic glycolysis and suppresses TCA cycle (8, 9). In normal liver, STAT3 is activated mainly by IL-6, which further regulates glucogenesis or liver regeneration, whereas oncogenic STAT3 induces high proliferation and migration, and contributes to the development of hepatocellular carcinoma (HCC) (10). Furthermore, hyperactivation of STAT3 often leads to poor prognosis and even chemoresistance in HCC (11). Therefore, inhibition of constitutively activated STAT3 in cancer cells is an attractive anti-tumor strategy.
Several protein tyrosine phosphatases (PTPs) reportedly regulate the JAK/STAT signaling pathway: dual-specificity phosphatase 22 (DUSP22), PTP receptor-type D (PTPRD), PTPRT, PTPRK, PTPRO, Src homology region 2 (SH2) domain-containing phosphatase 1 (SHP1), SHP2, PTP non-receptor type 9 (PTPN9), and PTPN2 (12-14). Despite the importance of PTPs in regulating STAT3 signaling for cancer development, the relationship between STAT3 and PTPs still remains poorly understood. DUSP3, also known as vaccinia H1-related phosphatase (VHR), is one of the atypical PTP members. DUSP3 was initially discovered to dephosphorylate receptor-type kinases such as EGFR and PDGFR in vitro (15). Recently, it was reported that DUSP3 inhibits non-small cell lung cancer (NSCLC) via dephosphorylation of EGFR and ErbB2 (16). Additionally, DUSP3 was reported to dephosphorylate ERK and JNK, although its effects on MAPKs were weaker than those of other DUSPs (17, 18). Although various biological roles of DUSP3 have been identified, the functions of DUSP3 in JAK/STAT signaling are relatively unknown. In this study, the regulatory roles of DUSP3 in the IL-6/STAT3 signaling pathway as well as the changes caused by DUSP3 in IL-6-induced STAT3 transcriptional activity were examined.
RESULTS
Identification of DUSP3 as a phosphatase targeting STAT3
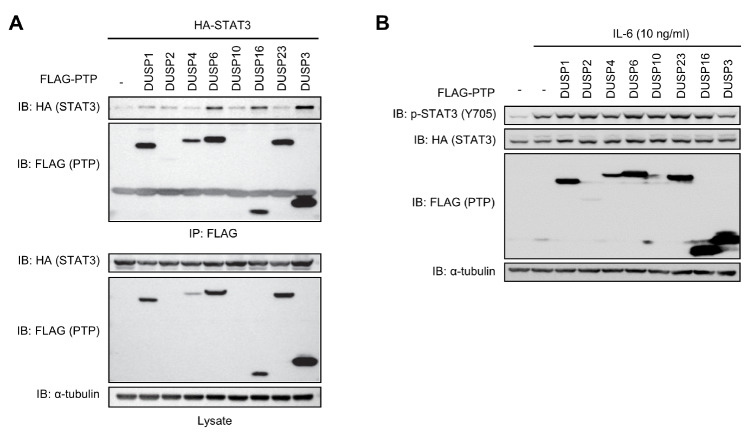
When STAT3 is phosphorylated at Y705 by kinases, such as JAK2, Src, and EGFR, the dimerization and transcriptional activity of STAT3 are enhanced (4). To identify the PTPs capable of regulating Y705 phosphorylation of STAT3, the following eight PTPs expected to interact with STAT3 were chosen using a protein–protein interaction prediction tool: DUSP1, DUSP2, DUSP3, DUSP4, DUSP6, DUSP10, DUSP16, and DUSP23. When these FLAG-tagged DUSP expression plasmids were transfected into HEK 293 cells with a HA-STAT3 expression plasmid, all of the abovementioned DUSPs, except DUSP2 and DUSP10, were expressed. The interactions between DUSPs and STAT3 were assayed by co-immunoprecipitating with anti-FLAG-conjugated beads followed by conducting immunoblotting analysis using an anti-HA specific antibody (Fig. 1A). Among all DUSP candidates, only DUSP6, DUSP16, and DUSP3 were found to interact with STAT3. However, of those STAT3-interacting candidates, only DUSP3 significantly suppressed IL-6-induced Y705 phosphorylation of STAT3 (Fig. 1B). These data suggest that DUSP3 is capable of binding to STAT3 and is likely to suppress p-Y705 of STAT3. Therefore, DUSP3 was chosen for further investigation.
Fig. 1.
Screening of STAT3-targeting phosphatases. HEK 293 cells were co-transfected with FLAG-PTPs and HA-STAT3. (A) The interactions between PTPs and STAT3 were assayed by co-immunoprecipitation with anti-FLAG conjugated beads, and STAT3 that interacted with immunoprecipitated FLAG-PTPs was subjected to immunoblotting analysis using an anti-HA specific antibody. (B) After transfection, cells were starved with serum-free medium for 12 h and stimulated with IL-6 (10 ng/mL) for 30 min. Phosphorylation and expression levels of STAT3 and FLAG-PTPs were analyzed by immunoblotting performed using specific antibodies.
DUSP3 interacts with the C-terminal domain of STAT3
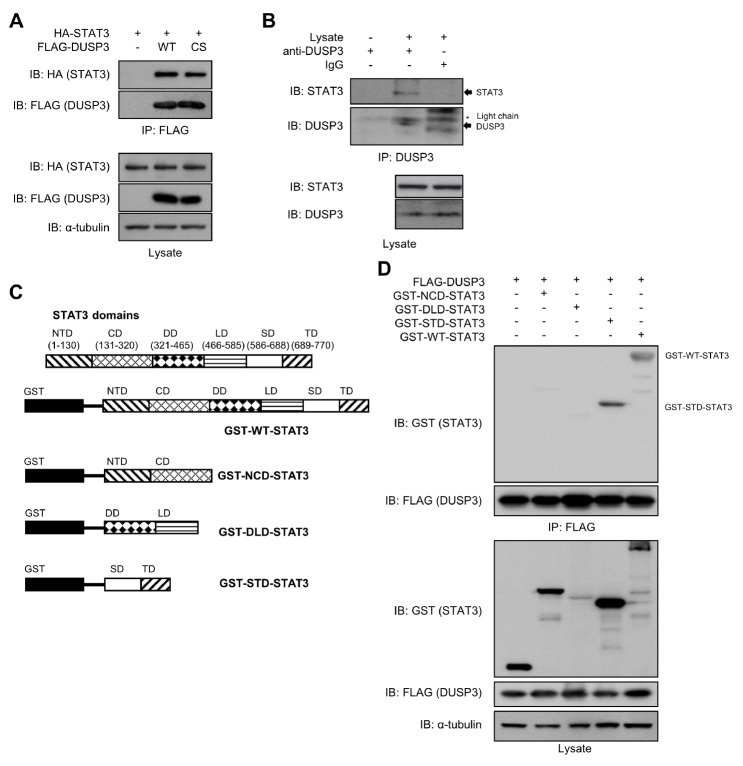
To further investigate the interaction between DUSP3 and STAT3, FLAG-DUSP3 wild-type (WT) or the catalytically inactive C124S mutant (CS) was co-expressed with HA-STAT3 in HEK 293 cells. Both WT and CS DUSP3 proteins interacted with STAT3 in these cells, regardless of the catalytic activity of DUSP3 (Fig. 2A). Additionally, endogenous DUSP3 and STAT3 interacted with each other (Fig. 2B). To systematically analyze the interaction between DUSP3 and STAT3, the truncated constructs of STAT3 were designed based on the STAT3 domains (Fig. 2C). The interaction between DUSP3 and each GST-tagged truncated form of STAT3 was analyzed via co-immunoprecipitation. Among the truncated forms of STAT3, the SH2-transactivation domain (STD) of STAT3 interacted with DUSP3 as strong as WT-STAT3 interacted with DUSP3 (Fig. 2D). These results indicate that DUSP3 interacts with STAT3 in cells, and this interaction is dependent on the STD of STAT3.
Fig. 2.
Interaction between DUSP3 and STAT3. (A) HEK 293 cells were co-transfected with FLAG-DUSP3 WT or CS and HA-STAT3. The interaction between FLAG-DUSP3 and STAT3 was analyzed by co-immunoprecipitation with anti-FLAG conjugated beads, and the bound STAT3 was subjected to immunoblotting analysis using an anti-HA specific antibody. (B) Hep3B cells were stimulated with IL-6 (100 ng/mL) for 1 h. The interaction between DUSP3 and STAT3 was analyzed by co-immunoprecipitation with anti-DUSP3-specific antibody and protein agarose A/G beads, and the bound STAT3 was subjected to immunoblotting analysis using an anti-STAT3 specific antibody. (C) Human STAT3 consists of 6 domains: N-terminal domain, a coiled-coil domain, DNA-binding domain, linker domain, SH2 domain, and transactivation domain. The constructs of the STAT3 truncated forms were designed to include two domains in each construct. (D) HEK 293 cells were co-transfected with FLAG-DUSP3 and GST-tagged truncated or WT-STAT3. The interaction between DUSP3 and GST-STAT3 constructs was observed by co-immunoprecipitation and immunoblotting analysis. WT, wild type; CS, C124S; ND, N-terminal domain; CD, coiled-coil domain; DD, DNA-binding domain; LD, linker domain; SD, SH2 domain; TD, transactivation domain; NCD, N-terminal-coiled-coil domain; DLD, DNA-binding-linker domain; STD, SH2-transactivation domain.
DUSP3 directly dephosphorylates STAT3
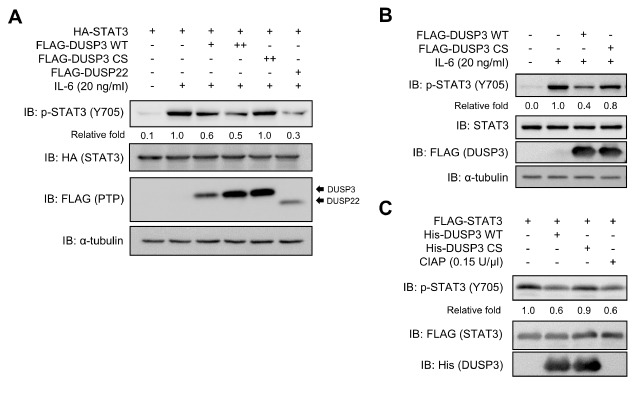
To investigate the regulatory effects of DUSP3 on IL-6-induced STAT3 phosphorylation, the phosphorylation levels of STAT3 along with the expression of either FLAG-DUSP3 WT or CS were analyzed. The expression of DUSP3 WT dose-dependently suppressed IL-6-induced STAT3 phosphorylation, whereas that of DUSP3 CS did not (Fig. 3A). The expression of DUSP3 WT reduced STAT3 Y705 phosphorylation as much as DUSP22, a specific phosphatase for STAT3. Furthermore, STAT3 phosphorylation was inhibited by DUSP3 in SK-Hep1 hepatoma cells (Fig. 3B). To evaluate whether DUSP3 directly dephosphorylates p-Y705 of STAT3, the purified recombinant DUSP3 WT or CS was mixed with the FLAG-STAT3 protein purified from IL-6-stimulated HEK 293 cells. As shown in Fig. 3C, the recombinant DUSP3 WT directly dephosphorylated p-STAT3 in vitro, whereas the CS mutant of DUSP3 failed to dephosphorylate p-STAT3. The decrease in the extent of STAT3 phosphorylation caused by DUSP3 WT was similar to that of STAT3 caused by non-specific calf intestinal alkaline phosphatase (CIAP), which was used as control (Fig. 3C). These results suggest that STAT3 is an intracellular substrate of DUSP3, and STAT3 dephosphorylation by DUSP3 is dependent on the catalytic activity of DUSP3.
Fig. 3.
Direct dephosphorylation of p-STAT3 by DUSP3 both in cell and in vitro. (A) HEK 293 cells were co-transfected with FLAG-PTPs and HA-STAT3. DUSP22 was used as positive control. (B) SK-Hep1 cells were transfected with FLAG-DUSP3 WT or CS. (A, B) After transfection, cells were starved with serum-free medium for 12 (HEK 293) or 24 h (SK-Hep1) and stimulated with IL-6 for 30 min. Phosphorylation and expression levels of STAT3 and FLAG-PTPs were subjected to immunoblotting analysis using specific antibodies. (C) Recombinant DUSP3 WT or CS was mixed with the p-STAT3 purified from HEK 293 cells and incubated for 1 h at 30°C. Phosphorylation and expression levels of each protein were subjected to immunoblotting analysis using specific antibodies. Non-specific phosphatase CIAP was used as positive control. CS, C124S; CIAP, calf intestinal alkaline phosphatase.
STAT3 transcriptional activity is regulated by DUSP3
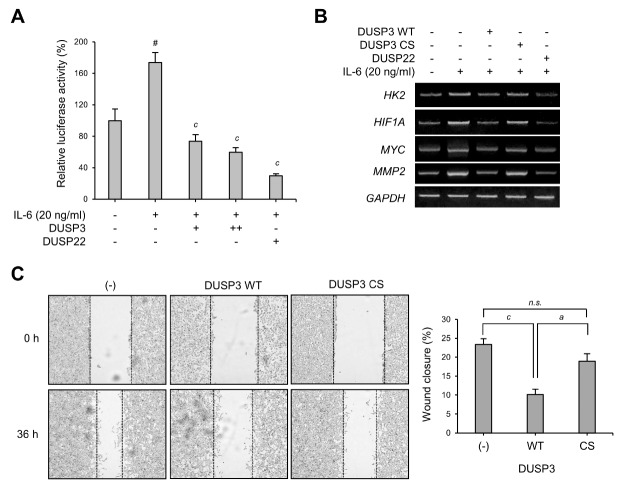
The phosphorylation of STAT3 at Y705 regulates STAT3 transcriptional activity and nuclear translocation (19). To investigate whether STAT3 dephosphorylation by DUSP3 alters the transcriptional activity of STAT3, a STAT3-specific reporter assay was performed. DUSP3 overexpression significantly decreased STAT3-specific reporter activity in IL-6-induced HEK 293 cells (Fig. 4A). DUSP22, known as a regulator of STAT3, also inhibited STAT3-specific reporter activity as expected. DUSP3-mediated STAT3 transcriptional activity suppression was observed along with a reduced expression of HK2, HIF1A, MYC, and MMP2, which are downstream target genes of STAT3 (Fig. 4B). Furthermore, the stable expression of DUSP3 WT reduced wound healing ability of IL-6-treated SK-Hep1 cells compared with the control (Fig. 4C), which suggests that the suppression of STAT3 by DUSP3 even affects the migration activities of cancer cells. Altogether, these results show that STAT3 dephosphorylation at Y705 by DUSP3 downregulates STAT3 transcriptional activity and its associated cellular physiology.
Fig. 4.
Regulation of STAT3 transcriptional activity by DUSP3. (A) Luciferase reporter assay was performed using HEK 293 cells transfected with STAT3-specific reporter, gWIZ-GFP, and FLAG-PTP as indicated. The transfected cells were stimulated with IL-6 (20 ng/mL) for 24 h before lysis, and luciferase activity was subsequently measured using a luciferase assay system (Promega, Madison, WI, USA). DUSP22, a known STAT3 activity inhibitor, was used as control. STAT3-specific reporter activity was normalized by GFP expression levels and is shown as a bar graph. The bar graph represents the mean ± SEM and was analyzed using one-way ANOVA for the significance between the three independent experiments; #P < 0.0001 vs. IL-6-untreated control groups; cP < 0.001 vs. IL-6-treated groups. (B) Total RNAs were isolated and reverse-transcribed into cDNA. The mRNA expression levels of HK2, HIF1A, MYC, MMP2, and GAPDH were examined by RT-PCR. (C) SK-Hep1 cells that stably express DUSP3 WT or CS were seeded into 6-well plates and grown until confluent. A wound was created with SPL Scratcher, and then cells were incubated in IL-6 (20 ng/mL)-containing 1% FBS media. Images were taken at 0 and 36 h. The wound closure values were quantified by measuring the percentage of wound size compared with those at 0-h point for each sample (0%). Data are analyzed using one-way ANOVA with Dunnett’s multiple comparison test; cP < 0.001 vs. control and aP < 0.05 vs. DUSP3 WT group. n.s., not significant; CS, C124S; HK2, hexokinase 2.
DISCUSSION
STAT3 is an oncogene that is constitutively activated in most cancer cells (3). Several factors alter STAT3 activity. An enhanced IL-6 production by cancer cells induces STAT3 phosphorylation (20). Increased expression of growth factor receptors also causes JAK/STAT activation (21). Moreover, the gain-of-function mutations or JAK amplification lead to the constitutive activation of STAT3 in cancer cells (22). Because STAT3 plays an essential role in cancer development, many studies have aimed to discover molecules that can inhibit the aberrant activation of STAT3 (8, 20, 23). In particular, dephosphorylation of STAT3 by phosphatases exerts an essential negative effect on the JAK/STAT signaling pathway (12). In this study, STAT3 dephosphorylation and the subsequent reduction in STAT3 transcriptional activity were observed by DUSP3 expression (Fig. 1B, 3, and 4). Comparing the expression levels of phosphatases with the levels of STAT3 bound to each phosphatase, DUSP16 showed a higher binding affinity for STAT3 than DUSP3 or DUSP6. However, only DUSP3, not DUSP6 or DUSP16, induced dephosphorylation of STAT3 among phosphatases that interacted with STAT3. A previous study has shown that DUSP3 inhibits STAT5a transcriptional activity via STAT5a dephosphorylation; however, the inhibitory effect of DUSP3 on STAT5a was selectively observed only when Y138 of DUSP3 was phosphorylated by IFNβ-induced Tyk2 (24). Based on the results of previous and present studies, the suppression of IL-6/JAK/STAT3 signaling by DUSP3 may be associated with the upregulation of IL-6/Tyk2/DUSP3 activation (25). Consequently, it might be assumed that DUSP3 functions as a negative feedback regulator of JAK/STAT signaling via ligand/Tyk2/DUSP3 signal transduction.
STAT3-regulating PTPs suppress STAT3 activation via different processes. For example, DUSP22, SHP1, and SHP2 are present in the cytoplasm of cells, and they dephosphorylate STAT3 as STAT3 expression is increased by stimulators such as IL-6 and leukemia inhibitory factor (13, 26, 27). Similarly, PTPN9 is also present in the cytoplasm and induces STAT3 dephosphorylation (28). Because the receptor-type PTPs, such as PTPRD, PTPRT, PTPRK, and PTPRO, are attached to the plasma membrane, they dephosphorylate STAT3 close to the membrane (29-31). Given that PTPN2 harbors a nuclear localization sequence, it shuttles between the cytoplasm and nucleus in response to various growth factors or cytokines (32, 33). Therefore, PTPN2 has the potential to translocate to the nucleus and dephosphorylate STAT3 in the nucleus (34). Unlike these PTPs, DUSP3, which has been identified to interact with STAT3 in this study (Fig. 1A and 2), is ubiquitously expressed and present mainly in the nucleus (35). Furthermore, based on a previous report regarding DUSP3-mediated regulation of IFNβ-induced STAT5a activation, DUSP3 is also likely to selectively regulate STAT3 (24). Altogether, it can be assumed that the mechanism underlying the regulation of IL-6-activated STAT3 by DUSP3 is different from the mechanism previously reported for STAT3-targeting PTPs.
In this study, the regulatory role of DUSP3 was identified in IL-6/JAK/STAT3 signaling. According to recent studies, DUSP3 deficiency can promote metastatic growth through Lewis lung carcinoma (LLC)-bearing lungs toward the recruitment of macrophages in an LLC experimental model (36). In addition, DUSP3 deficiency confers tolerance to LPS-induced septic shock by increasing M2 macrophages in DUSP3−/− mice (37). On the other hand, constitutive activation of STAT3 has been known to induce metastatic growth in various cancer cells (23). Furthermore, STAT3 activation is reportedly associated with M2 polarization of macrophages (38, 39). Taken together, DUSP3 may be an essential regulator of STAT3-mediated metastatic growth and cancer cell survival, which is also shown in Fig. 4C with reduced migration activities of cancer cells caused by DUSP3 WT expression.
In conclusion, our study demonstrated that DUSP3 directly dephosphorylated p-STAT3 at Y705 and downregulated STAT3 activity. Moreover, the function of DUSP3 in STAT3 activity regulation is expected to contribute to the understanding of STAT3-mediated cancer development.
MATERIALS AND METHODS
Detailed information is provided in the Supplementary Information.
Cell culture
HEK 293, HEK 293T, Hep3B, and SK-Hep1 cells were obtained from ATCC (Manassas, VA, USA). Cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Welgene, Seoul, Korea) supplemented with 10% fetal bovine serum (FBS; Welgene), 50 units/mL penicillin, and 50 μg/mL streptomycin (Gibco-BRL, Grand Island, NY, USA) at 37°C under a 5% CO2 atmosphere.
Prediction of the interaction between STAT3 and PTPs
The online transcriptional regulation interactions (TRI) tool was used to predict the interaction between STAT3 and PTPs based on protein sequences (https://www.vin.bg.ac.rs/180/tools/tfpred.php) (40). Among the 107 human PTPs, 99 PTPs except for the PTPs already reported to regulate STAT3 signaling (namely DUSP22, PTPRD, PTPRT, PTPRK, PTPRO, SHP1, SHP2, PTPN9, and PTPN2) were used to analyze whether PTPs could interact with STAT3 (12, 13).
Statistical analysis
All data were obtained from three independent experiments and were expressed as mean ± standard error of the mean (SEM). Statistical analyses were conducted using one-way ANOVA with Dunnett’s multiple comparison test by employing Prism 3.0 (GraphPad Software, San Diego, CA, USA).
Supplemental Materials
ACKNOWLEDGEMENTS
This study was supported by the National Research Foundation of Korea (NRF) grant funded by Ministry of Science and ICT (NRF-2018R1A2B6005084 and 2015R1A5A1008958) and the Chung-Ang University Graduate Research Scholarship in 2018.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Multhoff G, Molls M, Radons J. Chronic Inflammation in Cancer Development. Front Immunol. 2011;2:Article 98. doi: 10.3389/fimmu.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeNardo DG, Johansson M, Coussens LM. Immune Cells as Mediators of Solid Tumor Metastasis. Cancer Metastasis Rev. 2008;27:11–18. doi: 10.1007/s10555-007-9100-0. [DOI] [PubMed] [Google Scholar]
- 3.Lee H, Jeong AJ, Ye SK. Highlighted STAT3 as a potential drug target for cancer therapy. BMB Rep. 2019;52:415–423. doi: 10.5483/BMBRep.2019.52.7.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darnell JE. STATs and Gene Regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 5.Park Y, Kim J. Regulation of IL-6 signaling by miR-125a and let-7e in endothelial cells controls vasculogenic mimicry formation of breast cancer cells. BMB Rep. 2019;52:214–219. doi: 10.5483/BMBRep.2019.52.3.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoue H, Ogawa W, Ozaki M, et al. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nat Med. 2004;10:168–174. doi: 10.1038/nm980. [DOI] [PubMed] [Google Scholar]
- 7.Yu H, Jove R. The STATs of Cancer-New Molecular Targets Come of Age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 8.Demaria M, Giorgi C, Lebiedzinska M, et al. A STAT3-Mediated Metabolic Switch is Involved in Tumour Transformation and STAT3 Addiction. Aging (Albany NY) 2010;2:823–842. doi: 10.18632/aging.100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Liu T, Zhao L, et al. Ginsenoside 20 (S)-Rg3 Inhibits the Warburg Effect through STAT3 Pathways in Ovarian Cancer Cells. Int J Oncol. 2015;46:775–781. doi: 10.3892/ijo.2014.2767. [DOI] [PubMed] [Google Scholar]
- 10.Svinka J, Mikulits W, Eferl R. STAT3 in hepatocellular carcinoma: new perspectives. Hepat Oncol. 2014;1:107–120. doi: 10.2217/hep.13.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, Lee M, Kim JB, et al. 17beta-estradiol exerts anticancer effects in anoikis-resistant hepatocellular carcinoma cell lines by targeting IL-6/STAT3 signaling. Biochem Biophys Res Commun. 2016;473:1247–1254. doi: 10.1016/j.bbrc.2016.04.049. [DOI] [PubMed] [Google Scholar]
- 12.Kim M, Morales L, Jang IS, Cho YY, Kim D. Protein Tyrosine Phosphatases as Potential Regulators of STAT3 Signaling. Int J Mol Sci. 2018;19:E2708. doi: 10.3390/ijms19092708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekine Y, Tsuji S, Ikeda O, et al. Regulation of STAT3-Mediated Signaling by LMW-DSP2. Oncogene. 2006;25:5801–5806. doi: 10.1038/sj.onc.1209578. [DOI] [PubMed] [Google Scholar]
- 14.Hou J, Xu J, Jiang R, et al. Estrogen-sensitive PTPRO expression represses hepatocellular carcinoma progression by control of STAT3. Hepatology. 2013;57:678–688. doi: 10.1002/hep.25980. [DOI] [PubMed] [Google Scholar]
- 15.Ishibashi T, Bottaro DP, Chan A, Miki T, Aaronson SA. Expression Cloning of a Human Dual-Specificity Phosphatase. Proc Natl Acad Sci U S A. 1992;89:12170–12174. doi: 10.1073/pnas.89.24.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang JY, Yeh CL, Chou HC, et al. Vaccinia H1-Related Phosphatase (VHR) is a Phosphatase of ErbB Receptors and is Down-Regulated in Non-Small Cell lung Cancer. J Biol Chem. 2011;286:10177–10184. doi: 10.1074/jbc.M110.163295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Todd JL, Rigas JD, Rafty LA, Denu JM. Dual-Specificity Protein Tyrosine Phosphatase VHR Down-Regulates c-Jun N-terminal Kinase (JNK) Oncogene. 2002;21:2573–2583. doi: 10.1038/sj.onc.1205344. [DOI] [PubMed] [Google Scholar]
- 18.Todd JL, Tanner KG, Denu JM. Extracellular Regulated Kinases (ERK) 1 and ERK2 are Authentic Substrates for the Dual-Specificity Protein-Tyrosine Phosphatase VHR. A Aovel Role in Down-Regulating the ERK Pathway. J Biol Chem. 1999;274:13271–13280. doi: 10.1074/jbc.274.19.13271. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, McBride KM, Reich NC. STAT3 Nuclear Import is Independent of Tyrosine Phosphorylation and Mediated by Importin-α3. Proc Natl Acad Sci U S A. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bromberg J, Wang TC. Inflammation and Cancer: IL-6 and STAT3 Complete the Link. Cancer Cell. 2009;15:79–80. doi: 10.1016/j.ccr.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar J, Fraser F, Riley C, Ahmed N, McCulloch D, Ward A. Granulocyte Colony-Stimulating Factor Receptor Signalling via Janus Kinase 2/Signal Transducer and Activator of Transcription 3 in Ovarian Cancer. Br J Cancer. 2015;113:1642–1643. doi: 10.1038/bjc.2015.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puente XS, Pinyol M, Quesada V, et al. Whole-Genome Sequencing Identifies Recurrent Mutations in Chronic Lymphocytic Leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devarajan E, Huang S. STAT3 as a Central Regulator of Tumor Metastases. Curr Mol Med. 2009;9:626–633. doi: 10.2174/156652409788488720. [DOI] [PubMed] [Google Scholar]
- 24.Hoyt R, Zhu W, Cerignoli F, Alonso A, Mustelin T, David M. Cutting Edge: Selective Tyrosine Dephosphorylation of Interferon-Activated Nuclear STAT5 by the VHR Phosphatase. J Immunol. 2007;179:3402–3406. doi: 10.4049/jimmunol.179.6.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stahl N, Boulton TG, Farruggella T, et al. Association and Activation of Jak-Tyk Kinases by CNTF-LIF-OSM-IL-6 Beta Receptor Components. Science. 1994;263:92–95. doi: 10.1126/science.8272873. [DOI] [PubMed] [Google Scholar]
- 26.Han Y, Amin HM, Franko B, Frantz C, Shi X, Lai R. Loss of SHP1 Enhances JAK3/STAT3 Signaling and Decreases Proteosome Degradation of JAK3 and NPM-ALK in ALK+ Anaplastic Large-Cell Lymphoma. Blood. 2006;108:2796–2803. doi: 10.1182/blood-2006-04-017434. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Friedman JM. Leptin Receptor Activation of SH2 Domain Containing Protein Tyrosine Phosphatase 2 Modulates Ob Receptor SignalTtransduction. Proc Natl Acad Sci U S A. 1999;96:9677–9682. doi: 10.1073/pnas.96.17.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wakahara R, Kunimoto H, Tanino K, et al. Phospho‐ Ser727 of STAT3 Regulates STAT3 Activity by Enhancing Dephosphorylation of Phospho-Tyr705 Largely through TC45. Genes Cells. 2012;17:132–145. doi: 10.1111/j.1365-2443.2011.01575.x. [DOI] [PubMed] [Google Scholar]
- 29.Veeriah S, Brennan C, Meng S, et al. The Tyrosine Phosphatase PTPRD is a Tumor Suppressor that is Frequently Inactivated and Mutated in Glioblastoma and Other Human Cancers. Proc Natl Acad Sci U S A. 2009;106:9435–9440. doi: 10.1073/pnas.0900571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Guo A, Yu J, et al. Identification of STAT3 as a Substrate of Receptor Protein Tyrosine Phosphatase T. Proc Natl Acad Sci U S A. 2007;104:4060–4064. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen YW, Guo T, Shen L, et al. Receptor-Type Tyrosine-Protein Phosphatase Kappa Directly Targets STAT3 Activation for Tumor Suppression in Nasal NK/T-Cell Lymphoma. Blood. 2015;125:1589–1600. doi: 10.1182/blood-2014-07-588970. [DOI] [PubMed] [Google Scholar]
- 32.Alonso A, Sasin J, Bottini N, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 33.Bourdeau A, Dube N, Tremblay ML. Cytoplasmic protein tyrosine phosphatases, regulation and function: the roles of PTP1B and TC-PTP. Curr Opin Cell Biol. 2005;17:203–209. doi: 10.1016/j.ceb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Lee H, Morales LD, Slaga TJ, Kim DJ. Activation of T-cell protein-tyrosine phosphatase suppresses keratinocyte survival and proliferation following UVB irradiation. J Biol Chem. 2015;290:13–24. doi: 10.1074/jbc.M114.611681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavic K, Duan G, Köhn M. VHR/DUSP3 Phosphatase: Structure, Function and Regulation. FEBS J. 2015;282:1871–1890. doi: 10.1111/febs.13263. [DOI] [PubMed] [Google Scholar]
- 36.Vandereyken M, Jacques S, Van Overmeire E, et al. Dusp3 Deletion in Mice Promotes Experimental Lung Tumour Metastasis in a Macrophage Dependent Manner. PLoS One. 2017;12:e0185786. doi: 10.1371/journal.pone.0185786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh P, Dejager L, Amand M, et al. DUSP3 Genetic Deletion Confers M2-Like Macrophage-Dependent Tolerance to Septic Shock. J Immunol. 2015;194:4951–4962. doi: 10.4049/jimmunol.1402431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takaishi K, Komohara Y, Tashiro H, et al. Involvement of M2-Polarized Macrophages in the Ascites from Advanced Epithelial Ovarian Carcinoma in Tumor Progression via Stat3 Activation. Cancer Sci. 2010;101:2128–2136. doi: 10.1111/j.1349-7006.2010.01652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shiraishi D, Fujiwara Y, Komohara Y, Mizuta H, Takeya M. Glucagon-Like Peptide-1 (GLP-1) Induces M2 Polarization of Human Macrophages via STAT3 Activation. Biochem Biophys Res Commun. 2012;425:304–308. doi: 10.1016/j.bbrc.2012.07.086. [DOI] [PubMed] [Google Scholar]
- 40.Perovic V, Sumonja N, Gemovic B, Toska E, Roberts SG, Veljkovic N. TRI_Tool: a Web-Tool for Prediction of Protein-Protein Interactions in Human Transcriptional Regulation. Bioinformatics. 2017;33:289–291. doi: 10.1093/bioinformatics/btw590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.