Abstract
We investigated why a clinical meticillin-resistant Staphylococcus aureus (MRSA) isolate yielded false-negative results with some commercial PCR tests for MRSA detection. We found that an epidemic European CC1-MRSA-IV clone generally exhibits this behaviour. The failure of the assays was attributable to a large insertion in the orfX/SCCmec integration site. To ensure the reliability of molecular MRSA tests, it is vital to monitor emergence of new SCCmec types and junction sites.
Keywords: Staphylococci; Staphylococcus aureus; CC1-MRSA-IV; commercial PCR; Cepheid GenXpert; BD MAX, next generation sequencing
We investigated why a clinical meticillin-resistant Staphylococcus aureus (MRSA) isolate, collected in Austria in late 2019, yielded false-negative results with two widely used commercial orfX/SCCmec junction assays (Cepheid GeneXpert MRSA/SA BC, BD MAX Staph SR). The aim of this study was to investigate false-negative results with these two assays. Therefore, we tested and sequenced the index isolate and other isolates assigned by microarray to the same strain (i.e. the European CC1-MRSA-IV [1]).
Index case
A 62-year-old patient with metastasised cancer was admitted with suspected pneumonia to the Medical University Hospital of Graz, Austria. Blood cultures (BACTEC, Becton Dickinson, Heidelberg, Germany) became rapidly positive and Staphylococcus aureus was identified by in situ hybridisation (PNA FISH, AdvanDx, Woburn, United States (US)). In order to identify meticillin-resistant S. aureus (MRSA), the blood culture was investigated using the GeneXpert MRSA/SA BC PCR (Cepheid, Sunnyvale, US). Simultaneously, rapid antimicrobial susceptibility testing (RAST) was performed [2,3]. The GeneXpert test was negative for MRSA but RAST revealed cefoxitin resistance after 6 h. Antimicrobial susceptibility testing (Vitek2, bioMérieux, Marcy-l’Étoile, France) confirmed meticillin resistance. BD MAX StaphSR (Becton Dickinson) yielded a negative MRSA result. Microarray-based characterisation (S. aureus Genotyping Kit 2.0, Abbott, Alere Technologies, Jena, Germany) detected mecA and assigned the isolate (Graz_511421–19) to clonal complex CC1-MRSA-IV. Although the antibiotic treatment was adapted, the patient died shortly after because of tumour progression.
Isolates
Ten CC1-MRSA-IV isolates originated from the Sfanta Parascheva Hospital, Iasi, in north-eastern Romania [4]. Four isolates originated from the Dresden University Hospital, Saxony, Germany. Nineteen isolates from Regensburg University Medical Centre in Bavaria, Germany and five isolates from other Bavarian hospitals were also included. All included isolates had been collected and preliminarily analysed as part of earlier collaborations. Four fully sequenced reference strains were used as controls (Table 1). Additional controls comprised four isolates from Dresden that belonged to local epidemic strains or to another CC1-MRSA strain (isolate Dresden-220663).
Table 1. MRSA strains and isolates investigated in the present study and their detection using commercially available orfX/SCCmec junction site assays (n = 47).
| Isolate | Strain affiliation (according to microarray) | SCCmec element | Origin | BD MAX results | GeneXpert results |
|---|---|---|---|---|---|
| Graz_511421-19 (index case) | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Austria, 2019 | Negative (G) | Negative (2 X BC; G) |
| Dresden-94757 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Saxony, 2010 | Negative (D, G) | N/A |
| Dresden-94758 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Saxony, 2014 | Negative (D, G) | N/A |
| Dresden-94759 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Saxony, 2009 | Negative (D, G) | N/A |
| Dresden-94760 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Saxony, 2010 | Negative (D, G) | N/A |
| Iasi-95033 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Romania, 2009 | Negative (D, G) | N/A |
| Iasi-95034 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Romania, 2009 | Negative (D)/ambiguous (G)a | N/A |
| Iasi-95035 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Romania, 2009 | Negative (D)/ambiguous (G)a | N/A |
| Iasi-95037 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Romania, 2009 | Negative (D, G) | N/A |
| Iasi-95038 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Romania, 2009 | Negative (D, G) | N/A |
| Iasi-95039 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Romania, 2009 | Negative (D)/ambiguous (G)a | N/A |
| Iasi-95040 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Romania, 2009 | Negative (D, G) | N/A |
| Iasi-95041 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Romania, 2009 | Negative (D, G) | N/A |
| Iasi-174752 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Romania, 2010 | Negative (D, G) | N/A |
| Iasi-176047 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Romania, 2009 | Negative (D, G) | N/A |
| Bavaria-0643 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2018 | N/A | Positive (SSTI; R) |
| Bavaria-0824 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2015 | N/A | Positive (SSTI; R) |
| Bavaria-1185 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2018 | N/A | Positive (SSTI; R) |
| Bavaria-1274 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2014 | N/A | Positive (SSTI; R) |
| Bavaria-1537 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2013 | N/A | Positive (SSTI; R) |
| Bavaria-1780 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2013 | N/A | Positive (SSTI; R) |
| Bavaria-1962 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria | N/A | Positive (SSTI; R) |
| Bavaria-2102 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2019 | N/A | Positive (SSTI; R) |
| Bavaria-2220 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria | N/A | Positive (SSTI; R) |
| Bavaria-2312 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria | N/A | Positive (SSTI; R) |
| Bavaria-2360 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria | N/A | Positive (SSTI; R) |
| Bavaria-2391 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2018 | N/A | Positive (SSTI; R) |
| Bavaria-2483 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2019 | N/A | Positive (SSTI; R) |
| Bavaria-2535 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2019 | N/A | Positive (SSTI; R) |
| Bavaria-2584 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2019 | N/A | Positive (SSTI; R) |
| Bavaria-2585 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2019 | N/A | Positive (SSTI; R) |
| Bavaria-2588 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2019 | N/A | Positive (SSTI; R) |
| Bavaria-2596 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2019 | N/A | Positive (SSTI; R) |
| Bavaria-2618 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2012 | N/A | Positive (SSTI; R) |
| Bavaria-3012 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2011 | N/A | Positive (SSTI; R) |
| Bavaria-3254 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2010 | N/A | Positive (SSTI; R) |
| Bavaria-3702 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2019 | N/A | Positive (SSTI; R) |
| Bavaria-3741 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2019 | N/A | Positive (SSTI; R) |
| Bavaria-3784 | CC1-MRSA-IV (PVL-neg, aphA3/sat-pos) | SCCmec IVa with insertion | Bavaria, 2019 | N/A | Positive (SSTI; R) |
| Dresden-220663 | CC1-MRSA-IV, (PVL-neg, aphA3/sat-neg) | SCCmec IVa (as in reference strain MW2) | Saxony, 2007 | Positive (D) | N/A |
| Dresden-124288 | CC22-MRSA-IV (Barnim/UK EMRSA-15) | SCCmec IVh/j | Saxony | Positive (D) | N/A |
| Dresden-124289 | CC22-MRSA-IV (Barnim/UK EMRSA-15) | SCCmec IVh/j | Saxony | Positive (D) | N/A |
| Dresden-124281 | CC45-MRSA-IV (Berlin EMRSA) | SCCmec IVa | Saxony | Positive (D) | N/A |
| MU50 | CC5-MRSA-II (New York/Japan clone) | SCCmec II | Japan (sequenced reference strain) | Positive (D) | N/A |
| MW2 | CC1-MRSA-IV (PVL-pos US400) | SCCmec IVa | United States (sequenced reference strain) | Positive (D) | N/A |
| N315 | CC5-MRSA-II (New York/Japan clone) | SCCmec II | Japan (sequenced reference strain) | Positive (D) | N/A |
| US300-FPR3757 | CC8-MRSA-[Iva-pos ACME1] (PVL-pos ), US300 | SCC [mec IVa + ACME1 + Cu] | US (GenBank CP000255.1) | Positive (D) | N/A |
BC: GeneXpert MRSA/SA BC; CC: clonal complex; MRSA: meticillin-resistant Staphylococcus aureus; EMRSA: epidemic strain of MRSA; N/A: not available; PVL: Panton–Valentine leukocidin; SCCmec: staphylococcal cassette chromosome mec; SSTI: GeneXpert MRSA/SA SSTI.
D, G, and R indicate that assays were performed in Dresden, Graz and Regensburg, respectively.
a Ambiguous: weak signal observed at cycle threshold Ct > 35.
Commercial MRSA assays
Test results are provided in Table 1. The index isolate tested negative using the BD MAX Staph SR assay (Lot 9303156). Isolates from Iasi and Dresden and controls were tested twice, in Graz and Dresden, using this assay (Graz, Lot 9303156; Dresden, Lot K55928980720210312). It failed to identify these 15 isolates although controls handled in parallel were correctly identified.
Testing of the index isolate with GeneXpert MRSA/SA BC (Lots 1000148707 and 1000179462) yielded negative results, too. Further investigations on this assay were not possible because that laboratory became involved in diagnostics for the coronavirus (Covid-19) pandemic.
All Bavarian isolates were tested using Cepheid GeneXpert MRSA/SA SSTI (Lot 1000180532) that gave correct, positive results.
Genotyping by microarray and sequencing
All isolates were genotyped using the S. aureus Genotyping Kit 2.0, a microarray covering 333 different target sequences corresponding to ca 170 different genes. Target genes, assay protocols and sequences of probes and primers have been published previously [5]. Isolates were assigned to clonal complexes, strains and SCCmec types based on microarray data as described [5].
All isolates underwent whole-genome sequencing. DNA was extracted as for array experiments. Its quality was assessed as previously described [6]. The Nextera DNA Flex Library Preparation Kit (Illumina, Eindhoven, the Netherlands) was used and libraries underwent paired-end sequencing using the 500-cycle MiSeq Reagent Kit v2 (Illumina). Libraries were scaled to exhibit at least 50-fold coverage. Sequencing run quality was assured following cluster density and Q30 assessment. Raw sequence reads were trimmed using fastp 0.19.11 [7] and assembled using SPAdes v3.9.1 [8]. Contigs under 1,000 bp were removed.
The sequence of the SCCmec element one representative isolate, Iasi-95037, was deposited in GenBank (accession number: MT380478).
Description of the strain and its SCCmec element
Microarray profiling and genome sequencing showed that the index isolate belonged to a CC1-MRSA-IV clone previously described as ‘European CC1-MRSA-IV’ that may have emerged in south-eastern Europe [1,4,9]. A putative, meticillin-susceptible ancestor is common in Romania where this MRSA clone frequently observed already several years ago [1,4]. A high prevalence or outbreaks have been reported from Ireland [1], Italy [10] and Germany (North Rhine-Westphalia and Bavaria) [1,11]. In Regensburg, retrospective microarray-based typing of 3,067 isolates revealed that the occurrence of the European CC1-MRSA-IV clone increased from < 1% of typed MRSA between 2010 and 2013 to 9.4% in 2019. In Dresden, this strain has only sporadically been observed, accounting for seven in 1,758 isolates genotyped since 2000 ([1,12] and data not shown). Microarray genotyping data indicated that this clone was also recovered from horses and wild birds in Austria [13,14] and from livestock in Italy [15].
Isolates of this clone typically exhibit sequence type (ST)1 (1–1-1–1-1–1-1) or ST4110 (1–1-1–1-1–1-558) and spa types t127 (07–23–21–16–34–33–13), t386 (07–23–13) or t13790 (07–23–21–16–34–33–34–34–33–34). Isolates usually carry ermC (erythromycin/clindamycin resistance), tetK (tetracycline resistance), aphA3 (kana-/neomycin resistance), aadE (streptomycin resistance) and sat (streptothricin resistance). Some isolates harbour aacA-aphD (gentamicin resistance). Isolates from Ireland frequently exhibit resistance to mupirocin, chlorhexidine and quaternary ammonium compounds because of plasmid-borne iles2/mupR and qacA [1]. Fusidic acid resistance has not yet been detected in this clone, in contrast to other CC1-MRSA of from the Middle East or the southern hemisphere.
The clone is PVL-negative and lacks the splE protease gene. It only rarely carries enterotoxin genes sek/seq in addition to seh that is ubiquitously present in CC1. Its relationship to other CC1-MRSA clones has been discussed previously in detail [1].
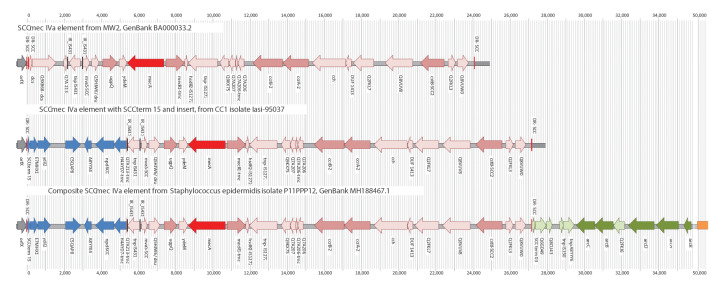
This CC1-MRSA clone has an SCCmec IVa element which is essentially identical in all isolates and in a previous Irish sequence (GenBank RBVO00000000.1) [1]. In contrast to MW2 (BA000033.2), it harbours an insertion of ca 5,350 nt, adjacent to orfX (Table 2, Figure). The insertion affects the orfX/SCCmec junction that is targeted by molecular tests for the detection of MRSA. It starts with a SCC terminal sequence alternate to dcs (‘SCCterm 15’) and encodes six hypothetical proteins (E7MHX1, ydiL2, C5QAP8, A8YYX4, npd and H4AYD7; RBVO000005.1: 280,690–286,024). This insertion replaces dcs/Q9XB68-dcs and removes most (212 of 240 nt) of a gene encoding hypothetical protein Q7A213.
Table 2. Genes in the variant SCCmec IVa element of the European CC1-MRSA-IV strain .
| Gene ID | Definition of gene product and comments (see also annotation of MH188467.1) | Orientation | Locus tag in MW2 (BA000033.2) | Nucleotide positions in GenBank RBVO | Nucleotide positions in GenBank MT380478 (Iasi-95037) |
|---|---|---|---|---|---|
| orfX | 23S rRNA methyltransferase | Forward | MW0024 |
RBVO01000005.1; nt 280,209–280,689 |
N/A |
| sRNA6 | Antisense RNA associated with orfX | Reverse | N/A |
RBVO01000005.1; nt 280,389–280,673 |
N/A |
| DR_SCC | Direct repeat of SCC, to 19 nt of the 3' end of the coding sequence of orfX | N/A | N/A |
RBVO01000005.1; nt 280,670–280,689 |
nt 1–19 |
| sccterm15 | SCC-terminal sequence adjacent to orfX, and alternate to dcs, see Discussion | N/A | Not present |
RBVO01000005.1; nt 280,689–280,912 |
nt 20–242 |
| E7MHX1 | Transcription regulator | Forward | Not present |
RBVO01000005.1; nt 280,912–281,239 |
nt 243–569 |
| ydiL2 | Hypothetical protein/putative membrane peptidase, associated with SCC elements | Forward | Not present |
RBVO01000005.1; nt 281,275–282,109 |
nt 606–1,439 |
| C5QAP8-M299 | Hypothetical protein | Forward | Not present |
RBVO01000005.1; nt 282,794–283,568 |
nt 2,125–2,898 |
| A8YYX4 | Hypothetical protein | Reverse | Not present |
RBVO01000005.1; nt 283,805–284,144 |
nt 3,136–3,474 |
| npd-SCC | Enoyl-[acyl-carrier-protein] reductase-like protein | Reverse | Not present |
RBVO01000005.1; nt 284,329–285,400 |
nt 3,660–4,730 |
| H4AYD7-trunc | Transcriptional regulator, LysR family | Truncated | Not present |
RBVO01000005.1; nt 285,412–286,024 |
nt 4,743–5,354 |
| Q7A213-trunc | Putative protein; it comprises the inverted repeat of IS431. In MW2 it is not truncated and comprises 240 nt | Truncated | MW0026 |
RBVO01000005.1; nt 286,024–286,052 |
nt 5,355–5,382 |
| IR_IS431 | Inverted repeat of IS431 | Truncated | N/A |
RBVO01000005.1; nt 286,024–286,040 |
nt 5,355–5,370 |
| tnpIS431 | Transposase for IS431 | Reverse | MW0027 |
RBVO01000005.1; nt 286,083–end of contig (nt 286,184) (partial) |
nt 5,414–6,088 |
| Teg143 | Trans-encoded RNA associated with tnpIS431 | Forward | N/A |
RBVO01000003.1; nt 203–237 |
nt 6,119–6,152 |
| IR_IS431 | Inverted repeat of IS431 | Truncated | N/A |
RBVO01000003.1; nt 213–229 |
nt 6,129–6,144 |
| mvaS-SCC | Truncated HMG-CoA synthase | Forward | MW0028 |
RBVO01000003.1; nt 245–598 |
nt 6,161–6,513 |
| Q5HJW6 | Hypothetical protein | Forward | N/A |
RBVO01000003.1; nt 695–1,046 |
nt 6,611–6,841 |
| dru | SCC direct repeat units | Truncated | N/A |
RBVO01000003.1; nt 835–1,273 |
nt 6,751–7,148 |
| ugpQ | Glycerophosphoryl diester phosphodiesterase-like protein | Forward | MW0029 |
RBVO01000003.1; nt 1,474–2,218 |
nt 7,350–8,093 |
| ydeM | Acyl dehydratase MaoC | Forward | MW0030 |
RBVO01000003.1; nt 2,314–2,743 |
nt 8,190–8,618 |
| mecA | Encodes penicillin binding protein 2 prime, defining MRSA | Reverse | MW0031 |
RBVO01000003.1; nt 2,812–4,795 |
nt 8,664–10,670 |
| mecR1-trunc | Meticillin resistance operon repressor 1, signal transducer protein, truncated in SCCmec IV | Truncated | MW0032 |
RBVO01000003.1; nt 4,894–5,862 |
nt 10,770–10,816 |
| hsdR2-IS1272 | Type I site-specific deoxyribonuclease restriction subunit | Truncated | MW0033 |
RBVO01000003.1; nt 5,869–6,103 |
nt 11,745–11,978 |
| tnpIS1272 | Transposase | Reverse | MW0034 |
RBVO01000003.1; nt 6,103–7,627 |
nt 11,979–13,502 |
| Q9KX75 | Hypothetical protein | Reverse | MW0035 |
RBVO01000003.1; nt 7,762–8,269 |
nt 13,638–14,144 |
| Q7A207 | Hypothetical protein | Reverse | MW0036 |
RBVO01000003.1; nt 8,283–8,595 |
nt 14,159–14,470 |
| Q7A206-trunc | Hypothetical protein, truncated | Truncated | N/A |
RBVO01000003.1; nt 8,596–8,683 |
nt 14,472–14,558 |
| Q7A206 | Hypothetical protein | Reverse | MW0037 |
RBVO01000003.1; nt 8,681–9,032 |
nt 14,557–14,907 |
| UTR_ccrB-2 | Highly conserved 3'-untranslated region of ccrB | N/A | N/A |
RBVO01000003.1; nt 9,032–9,553 |
nt 14,908–15,428 |
| ccrB-2 | Cassette chromosome recombinase B2 | Reverse | MW0038 |
RBVO01000003.1; nt 9,553–11,182 |
nt 15,429–17,057 |
| ccrA-2 | Cassette chromosome recombinase A2 | Reverse | MW0039 |
RBVO01000003.1; nt 11,203–12,553 |
nt 17,079–18,428 |
| cch-2 | Hypothetical protein/cassette chromosome helicase | Reverse | MW0040 |
RBVO01000003.1; nt 12,786–14,574 |
nt 18,662–20,449 |
| DUF1413 | Hypothetical protein, associated with cch | Reverse | MW0041 |
RBVO01000003.1; nt 14,573–14,864 |
nt 20,449–20,739 |
| Q2FKL7 | Putative membrane protein | Forward | MW0042 |
RBVO01000003.1; nt 15,002–16,052 |
nt 20,878–21,927 |
| Q8VUV8 | Putative transcriptional regulator | Forward | MW0043 |
RBVO01000003.1; nt 16,504–17,995 |
nt 22,380–23,870 |
| cstB-SCC2 | Includes a putative beta-lactamase; marker for SCCmec IVa | Truncated | MW0045 |
RBVO01000003.1; nt 18,367–19,685 |
nt 24,243–25,560 |
| Q2FKL3 | HNH endonuclease family protein | Forward | MW0046 |
RBVO01000003.1; nt 19,875–20,247 |
nt 25,751–26,122 |
| Q8VUW0 | Putative membrane protein | Forward | MW0047 |
RBVO01000003.1; nt 20,375–20,996 |
nt 26,251–26,871 |
| DR_SCC | Direct repeat of SCC | Truncated | N/A |
RBVO01000003.1; nt 21,300–21,319 |
nt 27,176–27,194 |
MRSA: meticillin-resistant Staphylococcus aureus; N/A: not available; nt: nucleotide position; SCC: staphylococcal cassette chromosome.
Figure.
SCCmec elements in the CC1 reference sequence MW2, the European CC1-MRSA-IV isolate Iasi-95037and the Staphylococcus epidermidis isolate P11PPP12
GenBank accession numbers: MW2: BA000033.23; Iasi-95037: MT380478; P11PPP12: MH188467.1.
SCCterm 15 is present in at least three other MRSA strains. In CC152-MRSA-XIII, it is close to orfX (MG674089, CP024998), possibly affecting MRSA PCRs. In two other strains, it is situated within complex SCCmec elements [16]. In a Danish CC8-MRSA strain (HM030720.1), the same insert as in the European CC1-MRSA is localised between an ACME-II and an SCCmec IVa element [17,18]. A Saudi Arabian CC22-MRSA strain (HF569105.1) harbours SCCterm 15, E7MHX1, ydiL2, IR_IS431 and tnpIS431, localised between a copper resistance element and a composite ACME-II/ SCCmec IVh/j element [19].
An identical 5,350 nt cluster is present in Staphylococcus epidermidis P11PPP12 (MH188467.1). Beyond that, the entire SCCmec IV element in P11PPP12 is identical to the one in the CC1-MRSA strain. Significantly, the site of recombination cutting short Q7A213 is conserved in both strains (position 5,818/5,819 in the Supplement). Thus, it is likely that the entire SCCmec IVa cassette including the insert was transferred between ancestors of the two strains, i.e. across the species. However, P11PPP12 also harbours an ACME-II/heavy metal resistance element downstream of SCCmec, which is absent from the European CC1-MRSA-IV. Therefore, it must have been lost during or after transfer of the SCCmec element, or it was acquired later by S. epidermidis.
Discussion
The study demonstrates that a CC1-MRSA-IV epidemic strain in Europe can yield false-negative results with common MRSA assays (GenXpert MRSA/SA BC, BD MAX Staph SR). Interestingly, GeneXpert MRSA/SA SSTI yielded correct results, indicating that the different tests utilise different primers. The absence of dcs and coverage of SCCterm 15 appear to be the reason for the discrepancy.
False-negative results of PCRs targeting the orfX/SCCmec junction site are concerning. Molecular assays are used to predict MRSA in positive blood cultures and to change therapy accordingly. The use of molecular assays is beneficial for a vast majority of patients because a result is available quickly. However, these assays can only detect target sequences that were available and considered at the time the primers were designed, and false-negative results have the potential to harm the patient by delaying effective therapy. Conventional antibiotic susceptibility tests are slower but are not constrained by the choice of primers or by the presence of unknown genotypes.
Molecular assays are also used to guide infection control. False-negative tests may result in lapses facilitating further MRSA transmission. Another, less obvious consequence might be a shift in the clonal structure of MRSA populations. When molecular assays exert a selective pressure favouring a false-negative strain, PCR-positive strains might get ‘penalised’ with subsequent interventions, hindering proliferation and transmission. This could lead to an increasing prevalence of the false-negative strain and to more failures in therapy and infection control.
It is crucial to monitor the emergence of new SCCmec junction sites in S. aureus and in coagulase-negative staphylococci, as mobile SCCmec elements can readily be transmitted between different strains and species, as was the case in the strain described here. Such unknown genotypes represent a problem for established molecular assays. As illustrated here, updating existing tests and platforms to evolving genotypes of the target organisms is important for individual and public health.
The containment of the CC1 strain must rely on conventional susceptibility tests, culture-based screening using selective growth media or updated molecular tests. We propose screening of medical or nursing staff recruited from epidemic regions, not only in hospitals but also in other care facilities, as well as patients with travel histories to these regions.
Acknowledgements
DC and ME wish to acknowledge the support of the staff of the Irish National MRSA Reference Laboratory at St. James’s Hospital, Dublin, Ireland.
Supplementary Data
Conflict of interest: None declared.
Authors’ contributions: Stefan Monecke: Conception and design of the study, data visualisation, analysis and interpretation of data, drafting of the manuscript. Elisabeth König: Collection of strains and data generation, analysis and interpretation of data. Megan R. Earls: Data generation, analysis and interpretation of data; drafting of the manuscript. Eva Leitner: Conception and design of the study, data generation, analysis and interpretation of data, drafting of the manuscript. Elke Müller: Data generation, analysis and interpretation of data. Gabriel Wagner: Data generation, analysis and interpretation of data. David Poitz: Data generation, analysis and interpretation of data. Lutz Jatzwauk: Data generation, analysis and interpretation of data. Teodora Vremerǎ: Collection of strains and data generation. Olivia S. Dorneanu: Collection of strains and data generation. Alexandra Simbeck: Collection of strains and data generation, analysis and interpretation of data. Andreas Ambrosch: Data generation, analysis and interpretation of data. Ines Zollner-Schwetz: Data generation, analysis and interpretation of data. Robert Krause: Data generation, analysis and interpretation of data
Werner Ruppitsch: Data generation, analysis and interpretation of data. Wulf Schneider: Data generation, analysis and interpretation of data. David C. Coleman: Analysis and interpretation of data; drafting of the manuscript. Ivo Steinmetz: Analysis and interpretation of data; drafting of the manuscript. Ralf Ehricht: Conception and design of the study, analysis and interpretation of data; drafting of the manuscript,
References
- 1. Earls MR, Shore AC, Brennan GI, Simbeck A, Schneider-Brachert W, Vremerǎ T, et al. A novel multidrug-resistant PVL-negative CC1-MRSA-IV clone emerging in Ireland and Germany likely originated in South-Eastern Europe. Infect Genet Evol. 2019;69:117-26. 10.1016/j.meegid.2019.01.021 [DOI] [PubMed] [Google Scholar]
- 2.The European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST rapid antimicrobial susceptibility testing (RAST) directly from positive blood culture bottles. Version 1.1. Växjö: EUCAST; 2019. Available from: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/RAST/EUCAST_RAST_methodology_v1.1_Final.pdf
- 3.The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Zone diameter breakpoints for rapid antimicrobial susceptibility testing (RAST) directly from blood culture bottles. Version 1.1. Växjö: EUCAST; 2019. Available from: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/RAST/EUCAST_RAST_Breakpoint_Table_v_1.1_final_PDF.pdf
- 4. Monecke S, Müller E, Dorneanu OS, Vremeră T, Ehricht R. Molecular typing of MRSA and of clinical Staphylococcus aureus isolates from Iaşi, Romania. PLoS One. 2014;9(5):e97833. 10.1371/journal.pone.0097833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One. 2011;6(4):e17936. 10.1371/journal.pone.0017936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Earls MR, Coleman DC, Brennan GI, Fleming T, Monecke S, Slickers P, et al. Intra-hospital, inter-hospital and intercontinental spread of ST78 MRSA from two neonatal intensive care unit outbreaks established using whole-genome sequencing. Front Microbiol. 2018;9(1485):1485. 10.3389/fmicb.2018.01485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884-90. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455-77. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Earls MR, Kinnevey PM, Brennan GI, Lazaris A, Skally M, O’Connell B, et al. The recent emergence in hospitals of multidrug-resistant community-associated sequence type 1 and spa type t127 methicillin-resistant Staphylococcus aureus investigated by whole-genome sequencing: Implications for screening. PLoS One. 2017;12(4):e0175542. 10.1371/journal.pone.0175542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Manara S, Pasolli E, Dolce D, Ravenni N, Campana S, Armanini F, et al. Whole-genome epidemiology, characterisation, and phylogenetic reconstruction of Staphylococcus aureus strains in a paediatric hospital. Genome Med. 2018;10(1):82. 10.1186/s13073-018-0593-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scheithauer S, Trepels-Kottek S, Häfner H, Keller D, Ittel T, Wagner N, et al. Healthcare worker-related MRSA cluster in a German neonatology level III ICU: a true European story. Int J Hyg Environ Health. 2014;217(2-3):307-11. 10.1016/j.ijheh.2013.07.006 [DOI] [PubMed] [Google Scholar]
- 12. Monecke S, Jatzwauk L, Müller E, Nitschke H, Pfohl K, Slickers P, et al. Diversity of SCCmec elements in Staphylococcus aureus as observed in south-eastern Germany. PLoS One. 2016;11(9):e0162654. 10.1371/journal.pone.0162654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loncaric I, Künzel F, Licka T, Simhofer H, Spergser J, Rosengarten R. Identification and characterization of methicillin-resistant Staphylococcus aureus (MRSA) from Austrian companion animals and horses. Vet Microbiol. 2014;168(2-4):381-7. 10.1016/j.vetmic.2013.11.022 [DOI] [PubMed] [Google Scholar]
- 14. Loncaric I, Stalder GL, Mehinagic K, Rosengarten R, Hoelzl F, Knauer F, et al. Comparison of ESBL--and AmpC producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus (MRSA) isolated from migratory and resident population of rooks (Corvus frugilegus) in Austria. PLoS One. 2013;8(12):e84048. 10.1371/journal.pone.0084048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alba P, Feltrin F, Cordaro G, Porrero MC, Kraushaar B, Argudín MA, et al. Livestock-associated methicillin resistant and methicillin susceptible Staphylococcus aureus sequence type (CC)1 in european farmed animals: high genetic relatedness of isolates from Italian cattle herds and humans. PLoS One. 2015;10(8):e0137143. 10.1371/journal.pone.0137143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baig S, Johannesen TB, Overballe-Petersen S, Larsen J, Larsen AR, Stegger M. Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus. Infect Genet Evol. 2018;61:74-6. 10.1016/j.meegid.2018.03.013 [DOI] [PubMed] [Google Scholar]
- 17. Bartels MD, Boye K, Rohde SM, Larsen AR, Torfs H, Bouchy P, et al. A common variant of staphylococcal cassette chromosome mec type IVa in isolates from Copenhagen, Denmark, is not detected by the BD GeneOhm methicillin-resistant Staphylococcus aureus assay. J Clin Microbiol. 2009;47(5):1524-7. 10.1128/JCM.02153-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bartels MD, Hansen LH, Boye K, Sørensen SJ, Westh H. An unexpected location of the arginine catabolic mobile element (ACME) in a USA300-related MRSA strain. PLoS One. 2011;6(1):e16193. 10.1371/journal.pone.0016193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hill-Cawthorne GA, Hudson LO, El Ghany MFA, Piepenburg O, Nair M, Dodgson A, et al. Recombinations in staphylococcal cassette chromosome mec elements compromise the molecular detection of methicillin resistance in Staphylococcus aureus. PLoS One. 2014;9(6):e101419. 10.1371/journal.pone.0101419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.