Abstract
Genome-scale metabolic models (GEMs) are valuable tools to study metabolism and provide a scaffold for the integrative analysis of omics data. Researchers have developed increasingly comprehensive human GEMs, but the disconnect among different model sources and versions impedes further progress. We therefore integrated and extensively curated the most recent human metabolic models to construct a consensus GEM, Human1. We demonstrated the versatility of Human1 through the generation and analysis of cell- and tissue-specific models using transcriptomic, proteomic, and kinetic data. We also present an accompanying web portal, Metabolic Atlas (https://www.metabolicatlas.org/), which facilitates further exploration and visualization of Human1 content. Human1 was created using a version-controlled, open-source model development framework to enable community-driven curation and refinement. This framework allows Human1 to be an evolving shared resource for future studies of human health and disease.
Introduction
Human metabolism is an integral part of cellular function, and many health conditions such as obesity, diabetes, hypertension, heart disease, and cancer (1, 2) are associated with abnormal metabolic states. Several of these conditions can be diagnosed by screening for metabolite biomarkers in a patient’s blood or urine (3), and recent studies have explored targeting metabolic processes for disease treatment (4, 5).
Despite the importance of metabolism and advances allowing for simultaneous measurement of thousands of metabolites (6), understanding metabolism in a holistic manner in human cells remains challenging. One reason for this difficulty is that the defining feature of metabolism is not the concentrations of biomolecules themselves (such as metabolites, mRNA, or proteins), but metabolic fluxes through reactions, for which concentrations can only be used as indirect proxies for biological activity (7). This challenge has been addressed by building genome-scale metabolic models (GEMs), which have been used for instance in industrial applications involving Saccharomyces cerevisiae and Escherichia coli to understand metabolism, engineer new cellular objectives (such as biofuel production), and increase product yield (8, 9).
Over the past 15 years, researchers have devoted a concerted effort to develop and improve such GEMs for human metabolism. This effort began in earnest with the development of Recon1 (10) and the Edinburgh Human Metabolic Network (EHMN) (11), which served as the starting point for two parallel model series: the Recon series (Recon1, 2, and 3D) (10, 12, 13) and the Human Metabolic Reaction series (HMR1 and 2) (14, 15). These two model lineages incorporate heavily from each other during updates (Fig. S1) and have been used to investigate diseases that include dysbiosis, diabetes, fatty liver disease, and cancer (16–19). Nevertheless, several challenges remain in the development of a human GEM, including the use of non-standard identifiers for genes, metabolites, and reactions; duplication of model components; propagation of errors from previous model iterations; effort divided among multiple model lineages; and model updates that are delayed, non-transparent, and difficult to coordinate among the scientific community.
Here, we present Human1, the first version of a unified human GEM lineage (Human-GEM), and Metabolic Atlas, its companion web portal. Human-GEM was developed by integrating and extensively curating the Recon and HMR model lineages. The entire development process was conducted systematically in a version-controlled Git repository to make all past and future changes publicly accessible, and to facilitate collaboration with the larger research community. We demonstrate the versatility and predictive accuracy of Human1 through an integrative analysis of transcriptomic data from 33 tumors and 53 healthy tissues; a gene-essentiality investigation involving over 620 different cell types; and the prediction of nutrient exchange and growth rates of NCI-60 cell lines using an enzyme-constrained GEMs (ecGEMs) derived from Human1.
Results
Human1 generation and curation
Our primary focus was to establish a systematically curated model of human metabolism that accurately represents the underlying biology. We therefore leveraged the collective knowledge contained within existing human GEMs by integrating their information into a single resource. Components and information from HMR2, iHsa (20), and Recon3D were integrated and reconciled to yield a unified GEM consisting of 13,417 reactions, 10,138 metabolites (4,164 unique), and 3,625 genes (Fig. 1, Table S1).
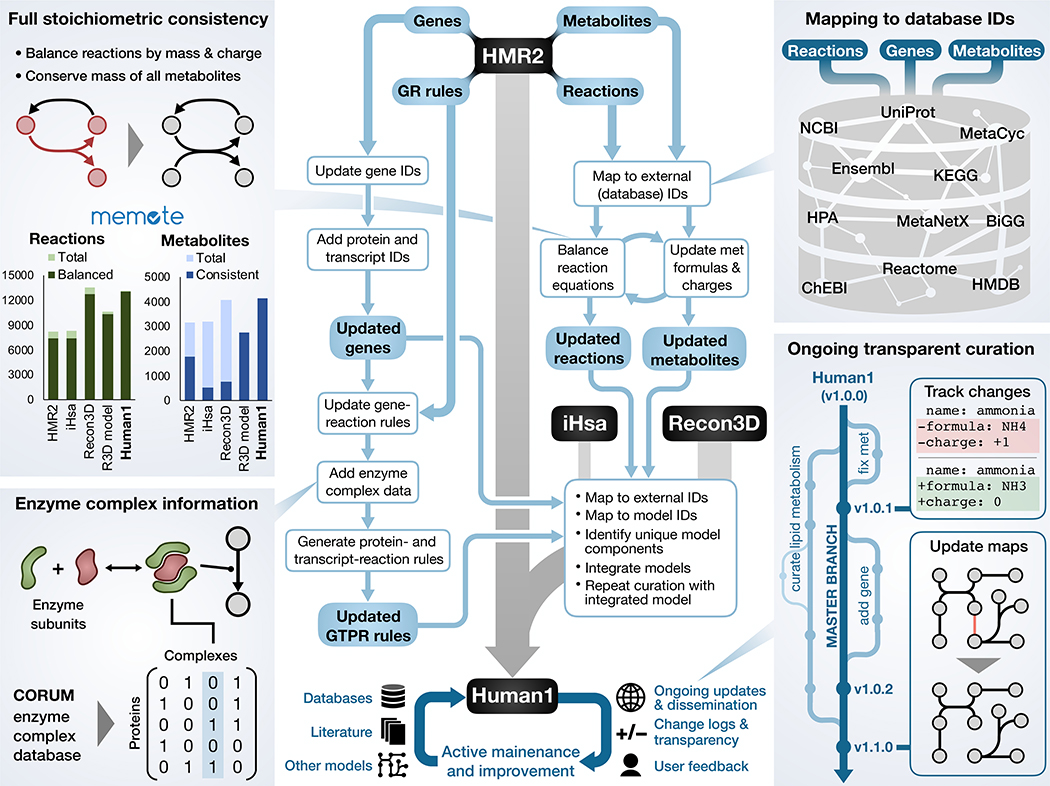
Fig. 1. Overview of Human1 generation and curation.
A simplified illustration of the key steps involved in the generation of Human1 from HMR2, Recon3D, and iHsa. The bottom of the diagram represents the ongoing open-source curation of Human1 using input from databases, literature, other models, and the scientific community. The four side panels provide further detail into selected Human1 features: extensive reaction mass and charge balancing to achieve 100% stoichiometric consistency; incorporation of new enzyme complex information; mapping model components to standard database identifiers; version-controlled and open-source model curation framework. In the bar graphs in the upper left panel, “Balanced” reactions represent the number of mass-balanced reactions, “Consistent” metabolites are the number of stoichiometrically consistent metabolites, and “R3D model” is the model version of Recon3D.
Curation of the integrated model to generate Human1 involved the removal of 8,185 duplicated reactions and 3,215 duplicated metabolites; revision of 2,016 metabolite formulas; re-balancing of 3,226 reaction equations; correction of reversibility for 83 reactions; and the inactivation or removal of 576 reactions that were inconsistent (violated mass or energy conservation) or deemed unnecessary (Tables S1–S3). We also constructed a new generic human biomass reaction based on various tissue and cell composition data sources to facilitate flux simulations and other analyses relying on a such a reaction (Data files S1 and S2). All model changes were documented to provide justification and to ensure reproducibility. Furthermore, to ensure that these changes remained consistent with previous human GEM simulation studies, we repeated the infant growth simulation presented by Nilsson et al. (21) and found excellent agreement between their HMR2-based results and our Human1-based simulations (Fig. S2).
The quality of Human1 was evaluated using Memote, a community-maintained framework for assessing GEMs with a standardized set of tests and metrics (22). In terms of consistency, Human1 exhibited excellent performance with 100% stoichiometric consistency, 99.4% mass-balanced reactions, and 98.2% charge-balanced reactions (Fig. S3). This is a considerable improvement over the most recent GEM, Recon3D, which had 19.8% stoichiometric consistency and 94.2% mass- and 95.8% charge-balanced reactions. Although the “model” version of Recon3D is fully stoichiometrically consistent and has a similar charge balance percentage (98.7%) as Human1, it has a lower percentage of mass balanced reactions (97.3%) and contains 20% fewer total reactions and 33% fewer metabolites compared to Human1. The average Memote annotation score for metabolites, reactions, genes, and SBO (systems biology ontology) terms in Human1 was 66%; although this is a substantial improvement over previous models (46% for HMR2 and 25% for Recon3D), it indicates an area requiring further attention. We also used Memote to evaluate all 27 Human-GEM releases (versions) preceding Human1 to resolve the effect of different curation processes on the various quality metrics (Fig. S4A–C).
A major advantage of GEMs is their ability to integrate different molecular datatypes to enable the interpretation of such data within the context of metabolism (23). We prioritized the curation and enhancement of gene-reaction associations for Human1 because such associations serve as an important link for the integration of multi-omics data. To this end, gene-reaction associations from HMR2, Recon3D, and iHsa were combined and integrated with enzyme complex information from Recon3D, iHsa, and the comprehensive resource of mammalian protein complexes database (CORUM (24)) to obtain gene-reaction rules for Human1. We also made available the transcript- and protein-reaction rules to facilitate direct integration of protein- or transcript-level data into the model, respectively (25). Furthermore, a key contribution of Recon3D was the association of protein structure information (such as 3D structure data) in a GEM-PRO data frame (13). We therefore regenerated the GEM-PRO data frame for Human1 to ensure that this same detailed protein information is also available for Human1.
An obstacle with existing human GEMs is their insufficient use of standard identifiers (such as KEGG, MetaCyc, and ChEBI) for many metabolites and reactions, thus impeding the retrieval of associated information from databases or the comparison of different models. To address this issue, we combined the available reaction and metabolite formulas, names, and identifiers in a semi-automated curation process using the MetaNetX reference database (26) to map 88.1% of reactions and 92.4% of metabolites to at least one standard identifier in Human1.
Other challenges facing human models are the ineffective communication and dissemination of their construction or revision. Traditionally, GEMs have been provided as a static object accompanying a publication, and thus errors can remain without correction for years. Based on the approach applied for the Yeast8 GEM (27), we developed Human1 using a Git repository hosted on GitHub to establish a more systematic and community-driven development process. This configuration enables version control and tracking of all changes made to the model since its inception, accompanied by documentation such as commit messages and log files. The use of a public repository allows users to view or download the curation history of Human1 and submit issues to suggest changes or highlight errors. Thus, new knowledge can be efficiently integrated in future updates of the model using a community wide effort.
Collectively, these improvements yield a standardized model enabling simple and accurate integration with different databases or omics datasets. We observed that the implementation of Human1 in a version-controlled framework such as Git is necessary to address many of the reproducibility and transparency concerns associated with computational research (28, 29).
Metabolic Atlas
In parallel with the development of Human1, we developed Metabolic Atlas (https://www.metabolicatlas.org/), an online platform that enables interactive exploration of cell metabolism and convenient integration of -omics data. Metabolic Atlas is an open-source re-implementation and complete re-design of its predecessor, the Human Metabolic Atlas (30).
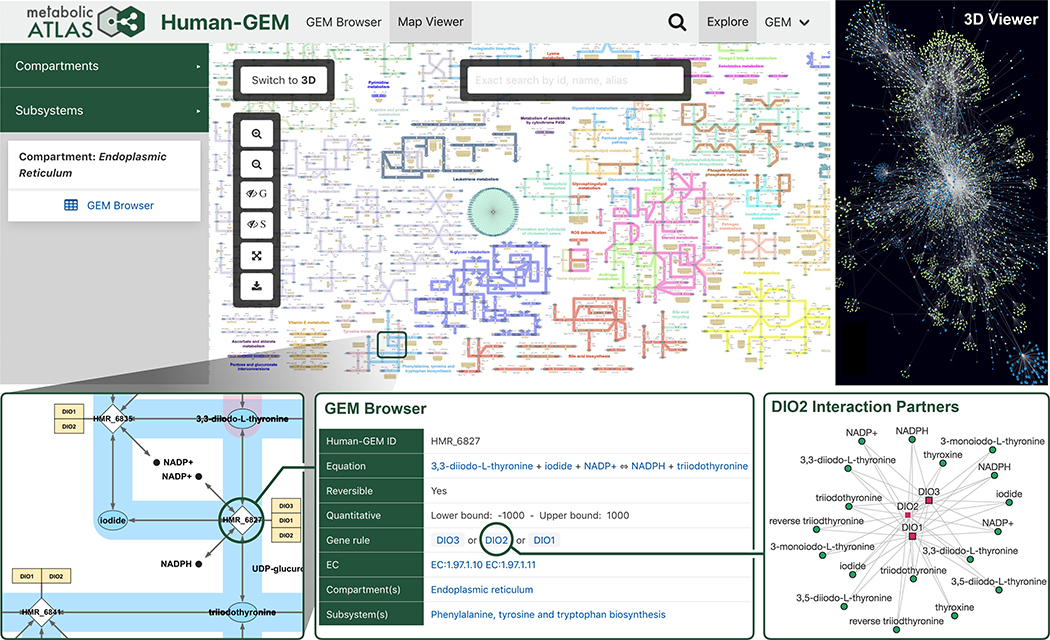
Metabolic Atlas enables visualization of the complex metabolic network and interconnects model components (Fig. 2). It contains interactive 2D maps at compartment and subsystem levels, allowing the use of smaller, more focused maps that pertain to metabolic areas of interest. The manually curated 2D maps cover 6,793 non-transport/non-exchange reactions (90%), 4,027 metabolites (97%), and 3,316 genes (91%) present in Human1. These maps are integrated with transcriptomic data from the Human Protein Atlas (HPA (31)), upon which gene expression levels from 37 different tissue types can be overlaid. Users can also upload their own transcriptomic data to be visualized on the maps, and an expression comparison feature allows the overlaying of expression fold-changes between two samples (such as different HPA tissues and/or user-uploaded data).
Fig. 2. Highlighted features provided by the Metabolic Atlas web portal.
A collection of screen captures from Metabolic Atlas, illustrating key features such as 2D and 3D metabolic network maps. A zoomed inset shows a subset of the endoplasmic reticulum compartment map, from which further information on components such as reactions, enzymes, or metabolites can be accessed in the GEM Browser. Interaction partner graphs are dynamically generated for any given enzyme or metabolite in Human1, which show the connectivity to other metabolites and enzymes based on their associated reactions.
Selection of a component (gene, reaction, metabolite, subsystem) on any Metabolic Atlas map provides a descriptive summary on the sidebar, which includes a link to its complete information page with further details and links to external databases. Moreover, automatically generated 3D maps are available, which cover 100% of the Human1 network. In addition to maps, Metabolic Atlas dynamically generates graphs of interaction partners for any given enzyme or metabolite in Human1, which show the connectivity to other metabolites and enzymes based on their associated reactions. These graphs can be expanded to include more distant interaction partners and are also integrated with HPA transcriptomic data.
Metabolic Atlas continues to serve as a repository for an increasing number of GEMs (over 350), ranging from those of individual human tissues and tumors to S. cerevisiae and other model organisms for fungi or bacteria. These models are summarized in a searchable table including information such as organism name; condition; year of publication; and number of reactions, metabolites, and enzymes. Furthermore, the content of Human1 can be accessed programmatically using the API to retrieve, for example, all information associated with a given metabolite.
Metabolic Atlas provides a valuable resource and intuitive tool that complements the functionality of the Human1 model for studying metabolism. The coupling of Human1 and Metabolic Atlas enables valuable infrastructural support for future research in human health and disease.
Generation and comparison of healthy tissue- and tumor-specific models
To demonstrate the utility of Human1, we explored metabolic patterns across healthy tissues and primary cancers arising within those tissues. We performed GEM contextualization to construct tissue- and cancer-specific models because Human1 contains reactions across many human cell types and is thus not representative of any individual tissue or tumor type. The contextualization was performed using tINIT (32) based on gene expression levels from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression database (GTEx (33)) to construct 53 healthy tissue metabolic models and 33 cancer metabolic models.
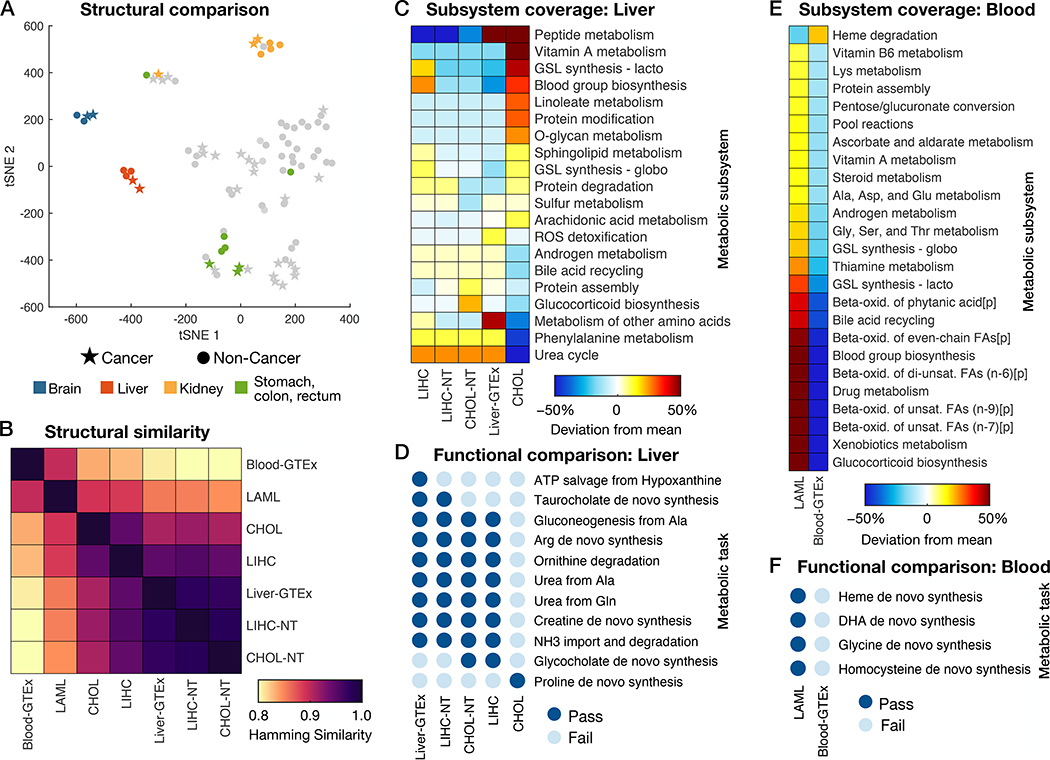
We first investigated the global similarity in the structure of the metabolic models by comparing which reactions were included in each model. We visualized relationships across the reaction structures of the 86 models using a 2D t-distributed stochastic neighbor embedding (tSNE) projection, which showed that each cancer type’s metabolic signature is more similar to the metabolism of its tissue of origin than to that of other cancer types (Fig. 3A and Fig. S5). This phenomenon has also been observed when comparing gene expression data among different tissue and cancer types (34). Several tissues and their associated tumors had markedly different metabolic capabilities than the other tissue models; these included the brain, liver, kidney, and tissues in the digestive system (stomach, colon, and rectum). This result highlights the role of these tissues as “metabolic specialists” as opposed to other human tissues.
Fig. 3. Structural and functional comparison of cancer- and healthy tissue-specific GEMs.
(A) Visualization of differences in models’ reaction content using a tSNE projection to two dimensions based on the Hamming similarity. See Fig. S5 for individual point labels. (B) Heatmap showing pairwise comparisons of reaction content between GEMs specific to healthy liver (CHOL-NT, LIHC-NT, and Liver-GTEx), blood, and their corresponding cancers (CHOL, LIHC, and LAML). (C) Relative subsystem coverage (number of reactions present in a model that are associated with the given subsystem) compared among GEMs of liver and liver tumors. Only subsystems with at least a 10% deviation from mean subsystem coverage among the models are shown. (D) Summary of metabolic task performance by the healthy and cancerous liver models, showing only the tasks that differed in at least one of the models. (E) Comparison of relative subsystem coverage between LAML- and blood-specific GEMs, showing only subsystems with at least a 10% deviation between the two models. (F) A summary of the five metabolic tasks that could be completed by the LAML GEM but failed in the healthy blood GEM. ROS, reactive oxygen species; GSL, glycosphingolipid; FA, fatty acid; [p], peroxisomal compartment; DHA, docosahexaenoic acid.
We next focused on the GEMs of liver, liver cancer, blood, and blood cancer. A more detailed reaction structure comparison showed that liver and blood models (and their associated tumors) have distinct metabolic reaction structures, and that, within liver models, cholangiocarcinoma (CHOL) was more distinct from healthy liver tissue whereas hepatocellular carcinoma (LIHC) laid between the two states (Fig. 3B).
To further explore these differences, we investigated the metabolic subsystem coverage and functional differences between liver tissue and liver cancers. We found a distinct loss of metabolic functions in the CHOL GEM, including a deficiency in metabolic reactions associated with the urea cycle, bile acid recycling, metabolism of other amino acids, phenylalanine metabolism, and glucocorticoid biosynthesis (Fig. 3C), leading to a loss of function in urea production, ornithine degradation, arginine and creatine synthesis, ammonia import and degradation, and other metabolic tasks (Fig. 3D). The exception was proline de novo synthesis, which was the only metabolic task active in CHOL that was inactive in the other liver-related GEMs. This was supported at the mRNA level (visualized using Metabolic Atlas in Fig. S6) and reflects previous studies that have shown increased proline synthesis and decreased proline degradation in other cancers in response to signaling through c-MYC and PI3K oncogenes, where the disruption of such metabolic activity constitutes a potential therapeutic strategy (35, 36). These and other approaches targeting metabolic functions such as ammonia buildup may constitute beneficial areas of research for developing CHOL treatments, which currently suffers from a lack of targeted therapies (37).
The construction of healthy and cancer-specific GEMs allowed us to compare cancer metabolism to healthy metabolism in systems for which paired normal tissue was not collected along with cancer tissue. An example is the comparison of the metabolism of acute myeloid leukemia (LAML) to that of healthy blood. The LAML GEM was characterized by a large increase in metabolic function over healthy blood (Figs. 3E and 3F), including processes such as glucocorticoid biosynthesis, fatty acid oxidation (Fig. S7), glycosphingolipid synthesis, and amino acid metabolism. This observation is consistent with previous studies showing that LAML relies on elevated fatty acid oxidation (38) and exhibits increased glycosphingolipid biosynthesis (39) which is associated with resistance to chemotherapeutics (40).
The large gain of metabolic function in LAML provides a rich number of pathways to target, such as heme biosynthesis, which constitutes a potential target for the treatment of LAML (41, 42). Moreover, reduced coverage of a metabolic pathway in the disease-state GEM may indicate a less robust metabolic function that is more susceptible to therapeutic disruption. For example, the LAML GEM contained fewer reactions in the heme degradation subsystem compared to that of healthy blood, suggesting that targeting such activity could prove beneficial for treating LAML. Supporting this observation, inhibition of oxidative heme degradation has been demonstrated to be a promising treatment for myeloid leukemia (43).
Prediction of metabolic task-essential genes in human cell lines
Following the construction and analysis of context-specific GEMs derived from Human1, we performed additional analyses to validate the network topologies of such models. Gene-reaction associations encoded within GEMs enable predictions of how gene perturbations (such as deletions) impact metabolic functionality. A common approach involves the prediction of essential genes by determining which genes, when deleted in silico, sufficiently reduce or eliminate the function of a specified objective reaction, such as biomass production (44). This predicted set of essential genes can then be compared with experimental gene essentiality measurements to quantitatively evaluate model performance.
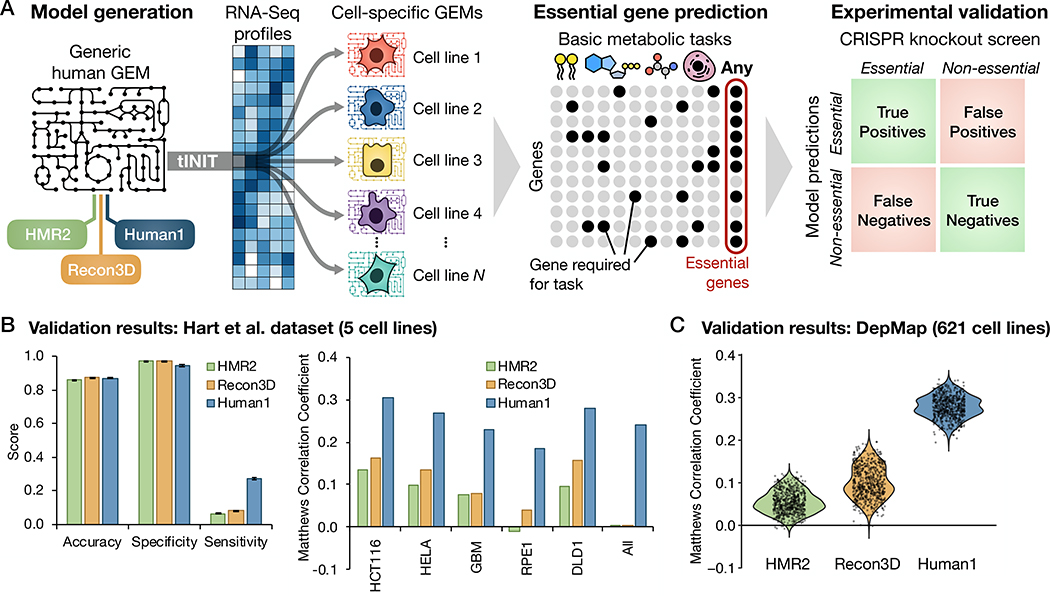
Genome-wide knockout screens have provided gene essentiality data to validate microbial GEMs, but such data has been unavailable for human cells due to challenges in genetically engineering these cells. Because the development of CRISPR technologies has enabled high-throughput genome-wide knockout screens in human cell lines, we leveraged this new data source to evaluate Human1 gene essentiality predictions. We retrieved gene essentiality data from a CRISPR knockout screen performed in five different human cell types: GBM, a patient-derived glioblastoma cell line; RPE1, retinal epithelial cells; HCT116 and DLD1, colorectal carcinoma cell lines; and HeLa, a cervical cancer cell line (45). Five cell-line-specific GEMs were constructed from Human1 using tINIT and their respective gene expression (RNA-Seq) profiles (45), and in silico gene deletions were performed on each GEM (Fig. 4A). Rather than focusing solely on growth, essential genes were defined as those which, upon deletion, impaired any of the 57 basic metabolic tasks (including biomass production) that are required for human cell viability (32) (Data file S3). This more general definition of gene essentiality reduces the extent to which predictions depend on the formulation of the biomass reaction and was hypothesized to increase sensitivity of the predictions by accounting for more functions of the metabolic network. We repeated this process using HMR2 and Recon3D as the template GEMs to enable comparison of Human1 performance with previous human model iterations.
Fig. 4. Predicted gene essentiality among different cell lines and human GEMs.
(A) Schematic illustration of the generation of cell-line-specific GEMs from HMR2, Recon3D, and Human1, and subsequent prediction of gene essentiality based on the GEMs’ ability to perform basic metabolic tasks. Genes predicted to be essential by the GEMs were compared to experimental measures of gene essentiality (45, 49) obtained from CRISPR knockout screens. (B) Comparison of gene essentiality predictions among the three reference GEMs and their 5 derivative cell line models with CRISPR screen results from Hart et al. (45). Left: Average accuracy, specificity, and sensitivity of predictions across the 5 cell lines for each reference GEM, with error bars representing the standard error of the mean. Right: Comparison of the Matthews Correlation Coefficient (MCC) of the predictions for each of the reference GEMs and cell lines. The “All” category indicates genes found to be essential in all 5 cell lines. (C) Comparison of gene essentiality predictions among the three reference GEMs and their 621 derivative cell line models with CRISPR screen results from the DepMap database (49).
We compared model-predicted essential genes for each individual cell line (as well as those essential in all five cell lines) to the set of essential genes identified in the corresponding CRISPR screen. The results were organized as confusion matrices quantifying the number of true and false positives and negatives (Fig. 4A), which were then used to evaluate prediction performance using several metrics (Fig. 4B). The general robustness of cells toward perturbations such as single gene knockouts (46) yields a much smaller number of essential genes than non-essential genes, resulting in highly imbalanced group sizes. Accuracy is therefore an inappropriate metric for assessing the quality of gene essentiality predictions. For example, although all reference models (HMR2, Recon3D, and Human1) achieved similarly high accuracy across all cell types (mean accuracy 86–88%), the same degree of accuracy is achieved if all genes are simply predicted as non-essential. This feature is reflected in the high specificity but low sensitivity exhibited by all three reference models. A more balanced prediction metric, the Matthews Correlation Coefficient (MCC) (47), was therefore calculated and compared among the different reference and cell-specific GEMs. Although the MCC values were relatively low overall, they showed a substantial increase (over 2.5-fold) in prediction quality for Human1-derived GEMs compared to HMR2- and Recon3D-derived models. Moreover, a hypergeometric test for enrichment of true positives in each model’s set of predicted essential genes showed significant enrichment for predictions from all Human1-derived GEMs (all p < 10−20), whereas HMR2- and Recon3D-derived GEMs performed no better than random (p > 0.05) in predicting essential genes for the RPE1 cell line and/or those common to all five cell lines (Fig. S8).
To further verify the improvement in Human1 gene essentiality predictions, we repeated the same pipeline (Fig. 4A) using RNA-Seq profiles and CRISPR knockout screen data for 621 human cell lines retrieved from the DepMap database (48, 49). The prediction performance of these 1,863 cell-specific GEMs (621 models derived from each of the 3 reference GEMs) was again evaluated using several different metrics (Fig. S9, A to D), including MCC (Fig. 4C). The analysis further confirmed the improvement in the performance of Human1, which exhibited a 2.8-fold mean increase in MCC over Recon3D. Because the CRISPR knockout screen scored genes on a continuous scale, it required the use of a threshold to categorize genes as essential or non-essential. We therefore repeated the analysis with a range of threshold values to confirm that our results were insensitive to this parameter (Fig. S10). To ensure that the selection of metabolic tasks was not biasing the results, we repeated the analysis using only biomass production to define gene essentiality. Although the relative performance between the three reference models was not affected, the results demonstrated an increased sensitivity in all GEMs’ predictions when using metabolic tasks instead of only biomass to define gene essentiality (Fig. S11, A and B).
Collectively these results demonstrated a marked improvement in Human1 over previous GEMs. However, the large number and diversity of curations involved in the development of Human1 make it difficult to resolve which changes contributed to the improved gene essentiality predictions. We therefore repeated the gene essentiality analysis pipeline (Fig. 4A) and comparison with the 5 cell-lines from the Hart 2015 dataset (45) for all 27 versions preceding the current release of Human1 (v1.3.0). Although the largest increases in performance were the result of updates to model genes or gene-reaction rules (based on database information, other GEMs, or the literature), other curations such as mass-balancing reactions and correcting reversibility of reactions associated with the electron transport chain also contributed to increases in Human1 predictive performance (Table S3 and Fig. S4D).
An enzyme-constrained human model
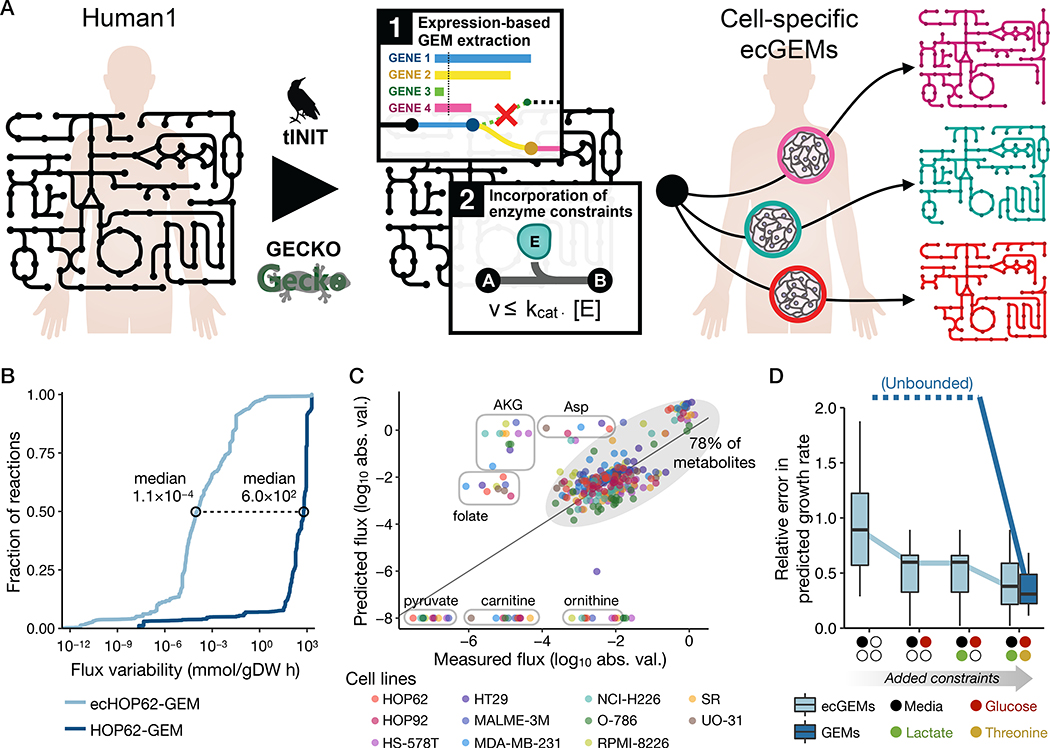
Human GEMs are often poorly constrained due to the limited availability of measured flux data, as well as the reliance of human cells on essential amino acids and vitamins as nutrients in addition to a dominant carbon source such as glucose (50). The GECKO (enhancement of a Genome-scale model with Enzymatic Constraints using Kinetic and Omics data) modeling framework was developed to integrate enzyme abundance and kinetic data into GEMs to constrain the flux space to a more meaningful region without requiring extensive nutrient exchange data (51). We therefore applied the GECKO framework to Human1-derived GEMs to generate enzyme constrained ecGEMs. GECKO implements enzyme constraints by incorporating the enzymes into their catalyzed reactions as pseudo-metabolites with a stoichiometric coefficient inversely proportional to their turnover rate (kcat). The explicit incorporation of enzymes allows the use of absolute proteomics datasets as constraints for each protein. If protein measurements are not available, the total protein content can be used as a global constraint for an additional pseudo-metabolite (protein pool) from which all enzymes are drawn.
To evaluate the improvement in flux predictions for enzyme-constrained GEMs derived from Human1, we used 11 NCI-60 cell-line-specific GEMs generated during the gene essentiality analysis (part of the DepMap dataset) for which reliable nutrient exchange rate data (52, 53) were available. Other NCI-60 cell lines were excluded as their metabolite exchange data was deemed unreliable due to early depletion of one or more nutrients (53, 54). Enzyme constraints were incorporated into each of these cell-specific GEMs using the GECKO framework, yielding 11 cell-specific ecGEMs (Fig. 5A).
Fig. 5. Generation and analysis of human ecGEMs.
(A) Graphical representation of the pipeline used to construct NCI-60 cell-line specific ecGEMs from Human1. (B) Cumulative distribution of flux variability among reactions in HOP62-GEM and ecHOP62-GEM. Only the ~3,200 reactions that carried a flux of >10−8 mmol/gDW h when optimizing biomass production in HOP62-GEM were included in the plot. Distributions for all 11 cell lines are shown in Fig. S12. (C) Comparison of predicted with measured exchange fluxes (log10-transformed absolute flux values) for the 11 cell-specific ecGEMs, where only the set of metabolites present in the growth medium (Ham’s medium) was specified. Different colored markers represent the different cell lines. Metabolites whose fluxes were systematically under- or over-predicted among the different models are labeled in circles, whereas the other ~78% lie within the shaded oval. Note that metabolites along the bottom of the plot have a predicted flux of zero but are shown here as having the absolute minimum measured value to avoid logarithm of zero. (D) Boxplots showing the relative error in predicted growth rate for the 11 cell-specific ecGEMs and non-ecGEMs. “Unbounded” indicates that the solutions are effectively unbounded and therefore have unquantifiable (infinite) error. Colored markers on the x-axis denote the exchange constraints that were cumulatively added to the models when making predictions. “Media” indicates that only the metabolites present in the growth medium were specified, without constraining their exchange rates. “Glucose”, “Lactate”, and “Threonine” indicate that the exchange flux for those metabolites in the model were constrained to the measured value.
After generating the cell-specific ecGEMs, we sought to evaluate the impact of the enzyme constraints on the accessible (feasible) flux space. An approach often used to assess the feasible flux range for all reactions in a model is flux variability analysis (FVA) (55). We conducted FVA on each of the 11 cell-specific ecGEMs and compared the flux variabilities with the corresponding non-ecGEMs. The analysis revealed a substantial reduction in solution space, where the median decrease in flux variability across the 11 cell line models ranged from 3.5 to 7 orders of magnitude (Fig. 5B, Fig. S12, Data file S4).
The integration of enzyme constraints substantially reduced the available flux space of Human1 but did not guarantee that this space was more accurate or biologically meaningful. We therefore sought to validate the ecGEMs by comparing predicted exchange fluxes with measured fluxes for 26 different metabolites, as well as comparing growth rates (Data file S5) (52). Fluxes were simulated by maximizing biomass production while specifying only which metabolites were present in the medium (Ham’s medium)—no uptake or excretion rates were provided. Under these conditions, exchange fluxes cannot be predicted by non-ecGEMs because the solution is unbounded (the maximum growth rate is effectively infinite). However, all ecGEMs were able to predict finite exchange fluxes for each of the 26 metabolites as well as growth rates, where most (~78%) were in reasonably good agreement with experimental measurements (Fig. 5C). The largest disagreements involved the overprediction of fluxes for folate, alpha ketoglutarate (AKG), and aspartate, and an underprediction for pyruvate, carnitine, and ornithine.
To further explore the improvement in flux predictions upon incorporating enzyme constraints into Human1-derived GEMs, we investigated the effect of specifying one or more metabolite exchange rates in addition to the media composition. Comparison of predicted to measured growth rates for the 11 cell lines revealed that non-ecGEMs could only achieve bounded solutions with errors comparable to their enzyme-constrained counterparts if the exchange rates of glucose, lactate, and at least one essential amino acid (threonine, in this case) were specified (Fig. 5D). These results also highlight an important feature of the enzyme-constraint framework: the greatest advantages and improvement in flux predictions are achieved when experimental exchange rates are limited or unavailable, which is most often the case when modeling human systems. However, when such flux measurements are available, the potential improvement offered by enzyme constraints becomes limited, as illustrated in the most constrained simulation in Fig. 5D.
The ability to estimate metabolic fluxes and growth rates with reasonable accuracy through the integration of enzyme constraints with Human1 represents a substantial development in human metabolic modeling. Whereas previous applications of human GEMs have largely been restricted to network-based analyses, the enzyme constraint formulation enables simulation-based approaches in the absence of metabolite exchange information.
Discussion
We developed Human1, a systematically curated and version-controlled human genome-scale metabolic model. Human1 is the unification of the parallel HMR and Recon human GEM lineages and effectively represents HMR3 and Recon4 with the aim of consolidating scientific efforts into a more efficient and coordinated approach to modeling human metabolism. We used Human1 to compare metabolic network structure and function across different healthy tissue and tumor types and demonstrated improved reliability of gene essentiality predictions for human cells; Human1 furthermore enables accurate simulation of cell growth and metabolite exchange rates given limited flux information.
The value of the rigorous curation process that was applied to Human1 is exemplified in part by the improved performance in gene essentiality predictions compared to other human GEMs (Figs. 4 B and C). Such improvements can be attributed to the integration of enzyme complex information from multiple models and databases into Human1 followed by careful curation of gene-reaction associations. The development of Human1 extended beyond gene-reaction associations and gene essentiality analyses, including an extensive mass and energy balancing process yielding a 100% stoichiometrically consistent GEM with over 99% mass-balanced reactions. Furthermore, the quantification of these metrics over the curation process (Fig. S4, A to D) enabled us to link various operations to changes in model performance or quality. This can help others identify where to focus efforts when applying this procedure to another organism or system, particularly if they are interested in improving one or two specific metrics.
An important feature of GEM-based analyses is that GEMs allow for simulation of flux through a metabolic network, enabling prediction of growth rates and intracellular reaction fluxes. Traditional simulations of human GEMs involve specifying external parameters (such as metabolite uptake rates) and internal parameters (such as specific growth rate and internal flux splits) to capture metabolic phenotypes, particularly in cancer (52). Measurements to determine these parameters in vivo are challenging or currently impossible, resulting in poorly constrained flux predictions and hindering the ability of GEMs to describe human metabolism where it matters most—within humans. In this work, we presented the construction and analysis of human ecGEMs which integrate enzyme kinetics and optionally proteomic data to allow physiologically meaningful flux simulations given little or no metabolite exchange information (51). This formalism enables flux simulations by specifying internal model constraints using more readily available -omics data rather than defining external model constraints based on metabolite exchange rates, greatly expanding the application potential of Human1, particularly for modeling metabolism of tissues and tumors in vivo.
As a complement to Human1, we developed the Metabolic Atlas web portal. This portal supplements and enriches the features of Human1 by providing users with deeper information on model components (for example, listing all reactions involving a given metabolite) and links to external databases (such as HPA, Ensembl, and MetaNetX). Metabolic Atlas also offers interactive compartment and subsystem maps to visualize and navigate Human1 content. By presenting the content in a more visual and connected format, Metabolic Atlas unlocks the information and potential of Human1 for those who are unfamiliar with GEMs but are interested in human metabolism.
Although GEMs provide versatile tools for the exploration of metabolism, their value is contingent upon their quality. Researchers rely on GEMs to be meticulously curated and frequently updated to ensure that they are consistent with current knowledge. Furthermore, this process should be done in a manner that is open, systematic, and reproducible. We therefore constructed Human1 in a version-controlled GitHub repository (https://github.com/SysBioChalmers/Human-GEM), where its latest iteration (v1.3.0 at the time of writing) and complete history are publicly available. This formulation allows the implementation of improvements and repairs to the model on the order of days to weeks, rather than several months to years as is the case with traditional GEM releases. We expect this or analogous approaches to become common practice in GEM development, as the rapid progress of the field requires a model development framework that can keep pace while maintaining transparency and reproducibility.
Supplementary Material
Materials and Methods
Fig. S1. The evolution of generic human GEMs.
Fig. S2. Replication of infant growth simulation using Human1.
Fig. S3. Memote report screenshot for Human1.
Fig. S4. Human1 quality and performance over the curation process.
Fig. S5. Labeled 2D tSNE projection of tissue- and tumor-specific GEM reaction content comparison based on Hamming similarity.
Fig. S6. Visualization of altered proline metabolism in cholangiocarcinoma (CHOL) using Metabolic Atlas.
Fig. S7. Visualization of increased expression in fatty acid beta oxidation subsystems for acute myeloid leukemia (LAML) using Metabolic Atlas.
Fig. S8. Enrichment of true positives in model-predicted essential genes.
Fig. S9. Comparison of gene essentiality predictions among the three reference GEMs and their 621 derivative cell line models with CRISPR knockout screen results from the DepMap database.
Fig. S10. Impact of gene essentiality threshold on DepMap gene essentiality analysis results.
Fig. S11. Gene essentiality predictions when considering only biomass production compared to considering the activity of 57 different metabolic tasks.
Fig. S12. Effect of enzyme constraints on GEM flux variability.
Table S1. Comparison of generic human GEM statistics.
Table S2. Issue-guided model curation workflow implemented on the Human-GEM GitHub repository.
Table S3. Summary of model changes associated with each version of Human-GEM.
Data file S1. Composition of the generic human cell biomass reaction
Data file S2. Average fatty acid composition for the curation of lipid metabolism
Data file S3. Metabolic tasks required for cellular viability
Data file S4. Flux variability analysis of ecHOP62-GEM
Data file S5. NCI-60 cell line experimental exchange fluxes
Acknowledgments
We thank Shan Huang and Jorrit Boekel for their contributions to the metabolicatlas.org web portal. The results published here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Funding: Research reported in this publication was supported by funding from the Knut and Alice Wallenberg Foundation, the National Cancer Institute of the National Institutes of Health under award number F32CA220848 (J.L.R.), the Novo Nordisk Foundation (grant no. NNF10CC1016517), and the European Union’s Horizon 2020 research and innovation program under grant agreement no. 720824 (I.D.).
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Human1 and its associated functions, scripts, and data are available on GitHub (https://github.com/SysBioChalmers/Human-GEM), and Metabolic Atlas is available at www.metabolicatlas.org. The Human1 GEM-PRO, as well as all cell-specific GEMs and enzyme-constrained ecGEMs described in this study, and the datasets and scripts necessary to reproduce the models, analyses, and figures are available on Zenodo (https://doi.org/10.5281/zenodo.3577466). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.DeBerardinis RJ, Thompson CB, Cellular metabolism and disease: what do metabolic outliers teach us? Cell 148, 1132–1144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghesquiere B, Wong BW, Kuchnio A, Carmeliet P, Metabolism of stromal and immune cells in health and disease. Nature 511, 167–176 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Emwas AHM, Salek RM, Griffin JL, Merzaban J, NMR-based metabolomics in human disease diagnosis: applications, limitations, and recommendations. Metabolomics 9, 1048–1072 (2013). [Google Scholar]
- 4.Day EA, Ford RJ, Steinberg GR, AMPK as a Therapeutic Target for Treating Metabolic Diseases. Trends in Endocrinology and Metabolism 28, 545–560 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Dey P, Baddour J, Muller F, Wu CC, Wang H, Liao WT, Lan Z, Chen A, Gutschner T, Kang Y, Fleming J, Satani N, Zhao D, Achreja A, Yang L, Lee J, Chang E, Genovese G, Viale A, Ying H, Draetta G, Maitra A, Wang YA, Nagrath D, DePinho RA, Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature 542, 119–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson CH, Ivanisevic J, Siuzdak G, Metabolomics: beyond biomarkers and towards mechanisms. Nature Reviews Molecular Cell Biology 17, 451–459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauer U, Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol 2, 62 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milne CB, Kim PJ, Eddy JA, Price ND, Accomplishments in genome-scale in silico modeling for industrial and medical biotechnology. Biotechnol J 4, 1653–1670 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oberhardt MA, Palsson BØ, Papin JA, Applications of genome-scale metabolic reconstructions. Mol Syst Biol 5, 320 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BØ, Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci U S A 104, 1777–1782 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma H, Sorokin A, Mazein A, Selkov A, Selkov E, Demin O, Goryanin I, The Edinburgh human metabolic network reconstruction and its functional analysis. Mol Syst Biol 3, 135 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiele I, Swainston N, Fleming RMT, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, Thorleifsson SG, Agren R, Bolling C, Bordel S, Chavali AK, Dobson P, Dunn WB, Endler L, Hala D, Hucka M, Hull D, Jameson D, Jamshidi N, Jonsson JJ, Juty N, Keating S, Nookaew I, Le Novere N, Malys N, Mazein A, Papin JA, Price ND, Selkov E, Sigurdsson MI, Simeonidis E, Sonnenschein N, Smallbone K, Sorokin A, van Beek JHGM, Weichart D, Goryanin I, Nielsen J, Westerhoff HV, Kell DB, Mendes P, Palsson BØ, A community-driven global reconstruction of human metabolism. Nature Biotechnology 31, 419–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunk E, Sahoo S, Zielinski DC, Altunkaya A, Drager A, Mih N, Gatto F, Nilsson A, Preciat Gonzalez GA, Aurich MK, Prlic A, Sastry A, Danielsdottir AD, Heinken A, Noronha A, Rose PW, Burley SK, Fleming RMT, Nielsen J, Thiele I, Palsson BO, Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat Biotechnol 36, 272–281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mardinoglu A, Agren R, Kampf C, Asplund A, Nookaew I, Jacobson P, Walley AJ, Froguel P, Carlsson LM, Uhlen M, Nielsen J, Integration of clinical data with a genome-scale metabolic model of the human adipocyte. Mol Syst Biol 9, 649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mardinoglu A, Agren R, Kampf C, Asplund A, Uhlen M, Nielsen J, Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat Commun 5, 3083 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Väremo L, Nookaew I, Nielsen J, Novel insights into obesity and diabetes through genome-scale metabolic modeling. Frontiers in Physiology 4, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mardinoglu A, Shoaie S, Bergentall M, Ghaffari P, Zhang C, Larsson E, Backhed F, Nielsen J, The gut microbiota modulates host amino acid and glutathione metabolism in mice. Mol Syst Biol 11, 834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bidkhori G, Benfeitas R, Klevstig M, Zhang C, Nielsen J, Uhlen M, Boren J, Mardinoglu A, Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes. Proc Natl Acad Sci U S A 115, E11874–E11883 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Zhang C, Liu Z, Klevstig M, Mukhopadhyay B, Bergentall M, Cinar R, Stahlman M, Sikanic N, Park JK, Deshmukh S, Harzandi AM, Kuijpers T, Grotli M, Elsasser SJ, Piening BD, Snyder M, Smith U, Nielsen J, Backhed F, Kunos G, Uhlen M, Boren J, Mardinoglu A, Network analyses identify liver-specific targets for treating liver diseases. Mol Syst Biol 13, 938 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blais EM, Rawls KD, Dougherty BV, Li ZI, Kolling GL, Ye P, Wallqvist A, Papin JA, Reconciled rat and human metabolic networks for comparative toxicogenomics and biomarker predictions. Nature Communications 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilsson A, Mardinoglu A, Nielsen J, Predicting growth of the healthy infant using a genome scale metabolic model. Npj Systems Biology and Applications 3, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieven C, Beber ME, Olivier BG, Bergmann FT, Ataman M, Babaei P, Bartell JA, Blank LM, Chauhan S, Correia K, Diener C, Drager A, Ebert BE, Edirisinghe JN, Faria JP, Feist AM, Fengos G, Fleming RMT, Garcia-Jimenez B, Hatzimanikatis V, van Helvoirt W, Henry CS, Hermjakob H, Herrgard MJ, Kaafarani A, Kim HU, King Z, Klamt S, Klipp E, Koehorst JJ, Konig M, Lakshmanan M, Lee DY, Lee SY, Lee S, Lewis NE, Liu F, Ma H, Machado D, Mahadevan R, Maia P, Mardinoglu A, Medlock GL, Monk JM, Nielsen J, Nielsen LK, Nogales J, Nookaew I, Palsson BO, Papin JA, Patil KR, Poolman M, Price ND, Resendis-Antonio O, Richelle A, Rocha I, Sanchez BJ, Schaap PJ, Malik Sheriff RS, Shoaie S, Sonnenschein N, Teusink B, Vilaca P, Vik JO, Wodke JAH, Xavier JC, Yuan Q, Zakhartsev M, Zhang C, MEMOTE for standardized genome-scale metabolic model testing. Nat. Biotechnol, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bordbar A, Monk JM, King ZA, Palsson BO, Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 15, 107–120 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Giurgiu M, Reinhard J, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Ruepp A, CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 47, D559–D563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryu JY, Kim HU, Lee SY, Framework and resource for more than 11,000 gene-transcript-protein-reaction associations in human metabolism. Proceedings of the National Academy of Sciences of the United States of America 114, E9740–E9749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moretti S, Martin O, Tran TV, Bridge A, Morgat A, Pagni M, MetaNetX/MNXref - reconciliation of metabolites and biochemical reactions to bring together genome-scale metabolic networks. Nucleic Acids Research 44, D523–D526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu HZ, Li FR, Sanchez BJ, Zhu ZM, Li G, Domenzain I, Marcisauskas S, Anton PM, Lappa D, Lieven C, Beber ME, Sonnenschein N, Kerkhoven EJ, Nielsen J, A consensus S cerevisiae metabolic model Yeast8 and its ecosystem for comprehensively probing cellular metabolism. Nat. Commun. 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stodden V, McNutt M, Bailey DH, Deelman E, Gil Y, Hanson B, Heroux MA, Ioannidis JPA, Taufer M, Enhancing reproducibility for computational methods. Science 354, 1240–1241 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Marcisauskas S, Sanchez BJ, Domenzain I, Hermansson D, Agren R, Nielsen J, Kerkhoven EJ, RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLoS Comput. Biol. 14, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pornputtapong N, Nookaew I, Nielsen J, Human metabolic atlas: an online resource for human metabolism. Database (Oxford) 2015, bav068 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F, Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Agren R, Mardinoglu A, Asplund A, Kampf C, Uhlen M, Nielsen J, Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol. Syst. Biol. 10, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET, Guan P, Korzeniewski GE, Lockhart NC, Rabiner CA, Rao AK, Robinson KL, Roche NV, Sawyer SJ, Segre AV, Shive CE, Smith AM, Sobin LH, Undale AH, Valentino KM, Vaught J, Young TR, Moore HM, Consortium G, A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreservation and Biobanking 13, 311–319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J, Locasale JW, Bielas JH, O’Sullivan J, Sheahan K, Cantley LC, Vander Heiden MG, Vitkup D, Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat. Biotechnol. 31, 522–529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, Phang JM, Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc. Natl. Acad. Sci. U. S. A. 109, 8983–8988 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanner JJ, Fendt SM, Becker DF, The Proline Cycle As a Potential Cancer Therapy Target. Biochemistry 57, 3433–3444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simile MM, Bagella P, Vidili G, Spanu A, Manetti R, Seddaiu MA, Babudieri S, Madeddu G, Serra PA, Altana M, Paliogiannis P, Targeted Therapies in Cholangiocarcinoma: Emerging Evidence from Clinical Trials. Medicina (Kaunas) 55, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maher M, Diesch J, Casquero R, Buschbeck M, Epigenetic-Transcriptional Regulation of Fatty Acid Metabolism and Its Alterations in Leukaemia. Front Genet 9, 405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Wen L, Ma X, Chen Z, Yu Y, Zhu J, Wang Y, Liu Z, Liu H, Wu D, Zhou D, Li Y, High expression of lactotriaosylceramide, a differentiation-associated glycosphingolipid, in the bone marrow of acute myeloid leukemia patients. Glycobiology 22, 930–938 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gouaze-Andersson V, Cabot MC, Glycosphingolipids and drug resistance. Biochim. Biophys. Acta 1758, 2096–2103 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Lin KH, Xie A, Rutter JC, Ahn YR, Lloyd-Cowden JM, Nichols AG, Soderquist RS, Koves TR, Muoio DM, MacIver NJ, Lamba JK, Pardee TS, McCall CM, Rizzieri DA, Wood KC, Systematic Dissection of the Metabolic-Apoptotic Interface in AML Reveals Heme Biosynthesis to Be a Regulator of Drug Sensitivity. Cell Metab. 29, 1217–1231 e1217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuda Y, Wang Y, Lian SL, Lynch J, Nagai S, Fanshawe B, Kandilci A, Janke LJ, Neale G, Fan YP, Sorrentino BP, Roussel MF, Grosveld G, Schuetz JD, Upregulated heme biosynthesis, an exploitable vulnerability in MYCN-driven leukemogenesis. Jci Insight 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salerno L, Romeo G, Modica MN, Amata E, Sorrenti V, Barbagallo I, Pittala V, Heme oxygenase-1: A new druggable target in the management of chronic and acute myeloid leukemia. Eur. J. Med. Chem. 142, 163–178 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Joyce AR, Palsson BO, Predicting gene essentiality using genome-scale in silico models. Methods Mol. Biol. 416, 433–457 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, Mero P, Dirks P, Sidhu S, Roth FP, Rissland OS, Durocher D, Angers S, Moffat J, High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163, 1515–1526 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Masel J, Siegal ML, Robustness: mechanisms and consequences. Trends Genet. 25, 395–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matthews BW, Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta 405, 442–451 (1975). [DOI] [PubMed] [Google Scholar]
- 48.Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, Barretina J, Gelfand ET, Bielski CM, Li H, Hu K, Andreev-Drakhlin AY, Kim J, Hess JM, Haas BJ, Aguet F, Weir BA, Rothberg MV, Paolella BR, Lawrence MS, Akbani R, Lu Y, Tiv HL, Gokhale PC, de Weck A, Mansour AA, Oh C, Shih J, Hadi K, Rosen Y, Bistline J, Venkatesan K, Reddy A, Sonkin D, Liu M, Lehar J, Korn JM, Porter DA, Jones MD, Golji J, Caponigro G, Taylor JE, Dunning CM, Creech AL, Warren AC, McFarland JM, Zamanighomi M, Kauffmann A, Stransky N, Imielinski M, Maruvka YE, Cherniack AD, Tsherniak A, Vazquez F, Jaffe JD, Lane AA, Weinstock DM, Johannessen CM, Morrissey MP, Stegmeier F, Schlegel R, Hahn WC, Getz G, Mills GB, Boehm JS, Golub TR, Garraway LA, Sellers WR, Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, Goodale A, Lee Y, Ali LD, Jiang G, Lubonja R, Harrington WF, Strickland M, Wu T, Hawes DC, Zhivich VA, Wyatt MR, Kalani Z, Chang JJ, Okamoto M, Stegmaier K, Golub TR, Boehm JS, Vazquez F, Root DE, Hahn WC, Tsherniak A, Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cook DJ, Nielsen J, Genome-scale metabolic models applied to human health and disease. Wiley Interdisciplinary Reviews-Systems Biology and Medicine 9, (2017). [DOI] [PubMed] [Google Scholar]
- 51.Sanchez BJ, Zhang C, Nilsson A, Lahtvee PJ, Kerkhoven EJ, Nielsen J, Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 13, 935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zielinski DC, Jamshidi N, Corbett AJ, Bordbar A, Thomas A, Palsson BO, Systems biology analysis of drivers underlying hallmarks of cancer cell metabolism. Sci. Rep. 7, 41241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK, Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nilsson A, Haanstra JR, Teusink B, Nielsen J, Metabolite Depletion Affects Flux Profiling of Cell Lines. Trends in Biochemical Sciences 43, 395–397 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Mahadevan R, Schilling CH, The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 5, 264–276 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Kanehisa M, Goto S, KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caspi R, Billington R, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Midford PE, Ong Q, Ong WK, Paley S, Subhraveti P, Karp PD, The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Research 46, D633–D639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.King ZA, Lu J, Drager A, Miller P, Federowicz S, Lerman JA, Ebrahim A, Palsson BO, Lewis NE, BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Research 44, D515–D522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, To JK, Laird MR, Lavidas I, Liu Z, Loveland JE, Maurel T, McLaren W, Moore B, Mudge J, Murphy DN, Newman V, Nuhn M, Ogeh D, Ong CK, Parker A, Patricio M, Riat HS, Schuilenburg H, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Zadissa A, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Cunningham F, Yates A, Flicek P, Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heirendt L, Arreckx S, Pfau T, Mendoza SN, Richelle A, Heinken A, Haraldsdottir HS, Wachowiak J, Keating SM, Vlasov V, Magnusdottir S, Ng CY, Preciat G, Zagare A, Chan SHJ, Aurich MK, Clancy CM, Modamio J, Sauls JT, Noronha A, Bordbar A, Cousins B, El Assal DC, Valcarcel LV, Apaolaza I, Ghaderi S, Ahookhosh M, Ben Guebila M, Kostromins A, Sompairac N, Le HM, Ma D, Sun YK, Wang L, Yurkovich JT, Oliveira MAP, Vuong PT, El Assal LP, Kuperstein I, Zinovyev A, Hinton HS, Bryant WA, Artacho FJA, Planes FJ, Stalidzans E, Maass A, Vempala S, Hucka M, Saunders MA, Maranas CD, Lewis NE, Sauter T, Palsson BO, Thiele I, Fleming RMT, Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat. Protoc. 14, 639–702 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanchez BJ, Li F, Kerkhoven EJ, Nielsen J, SLIMEr: probing flexibility of lipid metabolism in yeast with an improved constraint-based modeling framework. BMC Syst. Biol. 13, 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shorten PR, Upreti GC, A mathematical model of fatty acid metabolism and VLDL assembly in human liver. Biochim Biophys Acta 1736, 94–108 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P, Molecular Biology of the Cell. (Garland Science, New York, NY, ed. 5th, 2008). [Google Scholar]
- 64.Wang DX, Eraslan B, Wieland T, Hallstrom B, Hopf T, Zolg DP, Zecha J, Asplund A, Li LH, Meng C, Frejno M, Schmidt T, Schnatbaum K, Wilhelm M, Ponten F, Uhlen M, Gagneur J, Hahne H, Kuster B, A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 15, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheikh K, Forster J, Nielsen LK, Modeling hybridoma cell metabolism using a generic genome-scale metabolic model of Mus musculus. Biotechnol Progr 21, 112–121 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Park JO, Rubin SA, Xu YF, Amador-Noguez D, Fan J, Shlomi T, Rabinowitz JD, Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat. Chem. Biol. 12, 482–489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mih N, Brunk E, Chen K, Catoiu E, Sastry A, Kavvas E, Monk JM, Zhang Z, Palsson BO, ssbio: a Python framework for structural systems biology. Bioinformatics 34, 2155–2157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Becker SA, Palsson BØ, Context-specific metabolic networks are consistent with experiments. PLoS Comput Biol 4, e1000082 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agren R, Bordel S, Mardinoglu A, Pornputtapong N, Nookaew I, Nielsen J, Reconstruction of Genome-Scale Active Metabolic Networks for 69 Human Cell Types and 16 Cancer Types Using INIT. Plos Computational Biology 8, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, Ceccarelli M, Bontempi G, Noushmehr H, TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Research 44, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.R Core Team. (R Foundation for Statistical Computing, Vienna, Austria, 2018). [Google Scholar]
- 72.Lewis NE, Hixson KK, Conrad TM, Lerman JA, Charusanti P, Polpitiya AD, Adkins JN, Schramm G, Purvine SO, Lopez-Ferrer D, Weitz KK, Eils R, Konig R, Smith RD, Palsson BO, Omic data from evolved E-coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol 6, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Fig. S1. The evolution of generic human GEMs.
Fig. S2. Replication of infant growth simulation using Human1.
Fig. S3. Memote report screenshot for Human1.
Fig. S4. Human1 quality and performance over the curation process.
Fig. S5. Labeled 2D tSNE projection of tissue- and tumor-specific GEM reaction content comparison based on Hamming similarity.
Fig. S6. Visualization of altered proline metabolism in cholangiocarcinoma (CHOL) using Metabolic Atlas.
Fig. S7. Visualization of increased expression in fatty acid beta oxidation subsystems for acute myeloid leukemia (LAML) using Metabolic Atlas.
Fig. S8. Enrichment of true positives in model-predicted essential genes.
Fig. S9. Comparison of gene essentiality predictions among the three reference GEMs and their 621 derivative cell line models with CRISPR knockout screen results from the DepMap database.
Fig. S10. Impact of gene essentiality threshold on DepMap gene essentiality analysis results.
Fig. S11. Gene essentiality predictions when considering only biomass production compared to considering the activity of 57 different metabolic tasks.
Fig. S12. Effect of enzyme constraints on GEM flux variability.
Table S1. Comparison of generic human GEM statistics.
Table S2. Issue-guided model curation workflow implemented on the Human-GEM GitHub repository.
Table S3. Summary of model changes associated with each version of Human-GEM.
Data file S1. Composition of the generic human cell biomass reaction
Data file S2. Average fatty acid composition for the curation of lipid metabolism
Data file S3. Metabolic tasks required for cellular viability
Data file S4. Flux variability analysis of ecHOP62-GEM
Data file S5. NCI-60 cell line experimental exchange fluxes