ABSRACT
Background
Several studies have investigated the associations between the podocalyxin-like protein (PODXL) expression quantity or locations and cancers survival, but the results were far from conclusive. Therefore, we proceeded a meta-analysis on PODXL in various human cancers to find its prognostic value and followed confirmation using the TCGA datasets.
Methods
We performed a systematic search, and 18 citations, including 5705 patients were pooled in meta-analysis. The results were verified with TCGA datasets.
Results
Total eligible studies comprised 5705 patients with 10 types of cancer. And the result indicated that PODXL high-expression or membrane-expression were significantly related to poor overall survival (OS). However, subgroup analysis showed a significant association between high expressed PODXL and poor OS in the colorectal cancer, pancreatic cancer, urothelial bladder cancer, renal cell carcinoma and glioblastoma multiforme. Then, we validated the inference using TCGA datasets, and the consistent results were demonstrated in patients with pancreatic cancer, glioblastoma multiforme, gastric cancer, esophageal cancer and lung adenocarcinoma.
Conclusion
The result of meta-analysis showed that high expressed PODXL was significantly linked with poor OS in pancreatic cancer and glioblastoma multiforme, but not in gastric cancer, esophageal cancer or lung adenocarcinoma. And the membrane expression of PODXL might also associate with poor OS. PODXL may act as tumor promotor and may serve as a potential target for antitumor therapy.
Keywords: Cancer, Meta-analysis, Podocalyxin-like protein, Prognosis, TCGA
Background
Nowadays, noncommunicable diseases (NCDs) account for the majority of global deaths, and cancer predicts to be the leading cause. According to the latest global cancer statistics, 18.1 million new cancer diagnoses and 9.6 million deaths are expected in 2018 [1].
Podocalyxin-like protein (PODXL) is a highly glycosylated type I transmembrane protein associated with CD34 [2–4]. PODXL expression has been reported in the cytoplasm of some tumor cells, in some cases protruding toward the cell membrane, but not in the nucleus [5]. PODXL is encoded on chromosome 7q32-q33, and highly expressed by glomerular podocytes, vascular endothelium, hematopoietic cells and breast epithelial cells [6–8], which involved in many physiologic processes, such as hematopoiesis [9], leucocyte-endothelial cell interaction [10], regulating vascular permeability [11] and neural development [12].
The clinical significance of PODXL in the progression of various cancers has been studied, and it was found as a stem cell marker in the testicular cancer at the first time [3]. The later findings proved that, PODXL associates with advanced tumor phenotype in some cancers, including breast cancer [1, 13], colorectal cancer [5, 14–16], esophageal cancer [17], gastric cancer [17–19], glioblastoma multiforme [20], lung adenocarcinoma [21], oral squamous cell carcinoma [4, 22], ovarian cancer [23], pancreatic cancer [24–27], prostate cancer [28, 29], renal cell carcinoma [30], urothelial bladder cancer [31], and so on.
In addition, the prognostic role of PODXL protein expression had been analyzed with systematic review and meta-analysis in 2017 [32]. But as new researches emerged, we performed a new meta-analysis at pooling data, in order to estimate the potential prognostic value of PODXL in deep. We explored the relationship between the expression level or site of PODXL and prognosis of multiple cancers. And the validation with the Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) datasets even had been added for further analysis.
Methods
Publication search
Our meta-analysis followed the guidance of the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) [33]. We performed a systematic search of the PubMed, Web of Science, Embase and Cochrane Library database from January 1, 2000 to October 31, 2018, using both MeSH search for keywords and full text. Our search terms were: (“cancer” OR “tumor” OR “neoplasm” OR “carcinoma”) AND (“Podocalyxin like protein” OR “Podocalyxin” OR “PODXL”) AND (“prognosis” OR “prognostic” OR “outcome”). Additionally, the references and other related researches were reviewed to find more potential articles.
Inclusion and exclusion criteria
The eligible articles selection process was done by two authors (Siying He and Menglan Li). The inclusion criteria were as followed: (1) involved the correlation between the expression of PODXL and survival data of cancer patients; (2) provided the relevant clinicopathological parameters; (3) the number of patients involved in the studies should be more than 50.
The exclusion criteria were as followed: (1) studies that not based on human; (2) insufficient Hazard ratios (HRs) or other data; (3) repetitive patients; (4) reviews, case reports or a meta-analysis.
Data collection and quality detection
Two researchers evaluated and collected data from these eligible articles with a predefined standard independently. The following information was recorded: (1) first author’s name; (2) publication year; (3) countries; (4) types of cancers; (5) number of patients; (6) detection methods; (7) cut-off criteria; (8) clinical parameters; (9) data about overall survival (OS), disease-free survival (DFS) or cancer-specific survival (CSS). The Engauge Digitizer 4.1 software was used to extract data from Kaplan-Meier (K-M) plot, when there was no HRs and its 95% confidence inter (CIs) offered directly [34]. In addition, the included studies should be evaluated with the Newcastle-Ottawa Scale (NOS) [35].
Data collection and analysis in TCGA
Data for the expression of PODXL and clinicopathological parameters in TCGA were recorded from the Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn) [36] and the UALCAN (http://ualcan.path.uab.edu) [37]. There were 31 types of cancer, including 9040 subjects which had both PODXL expression and cancer survival data. In order to make the K-M survival analysis and generated overall survival plots, the expression levels of PODXL were divided into low/median and high expression group according to the TPM value. The difference between two groups was conducted by Log-rank test.
Mechanism prediction of PODXL
We used the STRING database (http://string-db.org/) [38], online common software, for finding PODXL-related genes and providing a critical assessment and integration of protein-protein interactions (PPI) of PODXL and PODXL-related genes. And these PODXL-related genes were performed functional enrichment analysis by using DAVID database (http://david.abcc.ncifcrf.gov/), which means a common bioinformatics database for annotation, visualization and integrated discovery [39].
Statistical analysis
Our meta-analysis was based on the Stata12.0 software (Stata Corporation, College Station, TX, United States). The prognostic value of PODXL on OS, DFS and CSS was calculated by pooled HRs with 95% CIs. On the other hand, odds ratios (ORs) with corresponding 95% CIs were used to assess the relation between PODXL and clinicopathological features. Chi square-based Cochran Q test and I2 test were used to determine the heterogeneity among these eligible articles. I2 > 50% or P-value < 0.05 was considered as significant heterogeneity, and a random-effect model would be adopted; otherwise, a fix-effect model would be chose. The effect of covariates have been evaluated with regression analysis. The sources of heterogeneity could be dissect with subgroup analysis. In addition, the sensitivity and publication bias were performed. P < 0.05 was considered statistically significant with two-sided.
Results
Search results and research characteristics
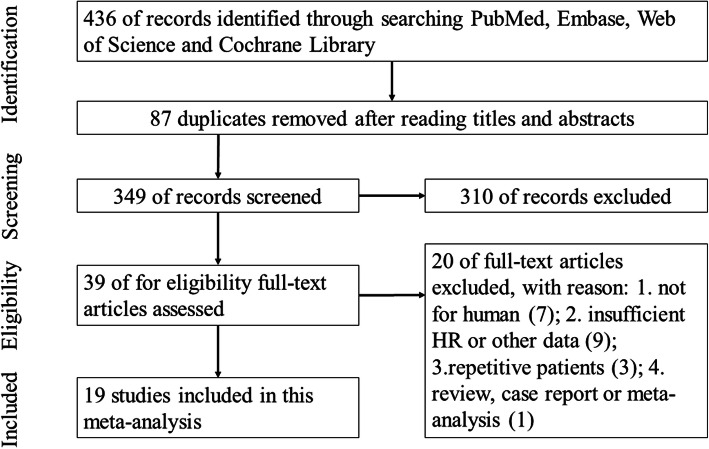
In total, 436 records were identified and 87 duplicates were excluded. 39 articles remained after scanning the titles and abstracts, and among the 39 studies, 7 were excluded for not for human, 9 were excluded for insufficient HRs or other data, 3 were excluded because the included patients were repetitive in other studies, and 1 meta-analysis was excluded, and the flow diagram was shown in Fig.1. Finally, 18 eligible studies were include in this meta-analysis [1, 5, 13–21, 23–27, 30, 31].. These eligible researches contained 5705 patients, involved 10 types of cancers, including the breast cancer (n = 2), renal cell carcinoma (n = 1), colorectal cancer (n = 4), ovarian cancer (n = 1), glioblastoma multiforme (n = 1), urothelial bladder cancer (n = 2), pancreatic adenocarcinoma (n = 4), esophageal cancer (n = 1), gastric cancer (n = 3) and lung adenocarcinoma (n = 1). In these studies, PODXL expression levels were evaluated by immunohistochemistry (IHC). The characteristics of the eligible articles were listed in Table 1.
Fig. 1.
Flow diagram of study selection
Table 1.
Characteristics of eligible studies in this meta-analysis
| Author | Year | Country | No. of Patient | Tumor type | Method | Cut-off | Outcome | Analysis | Antibody | NOS |
|---|---|---|---|---|---|---|---|---|---|---|
| Somasiri | 2004 | Canada | 272 | Breast cancer | IHC | IHC ≥ 50% | CSS | K-M Curve | M | 7 |
| Hsu | 2010 | Taiwan | 303 | Renal cell carcinoma | IHC | IHC score ≥ 1 | OS, CSS, MFS | Multivariate | P | 8 |
| Larsson | 2011 | Sweden | 626 | Colorectal cancer | IHC | IHC score ≥ 3 | OS, CSS | Multivariate | P | 8 |
| Cipollone | 2012 | Canada | 479 | Ovarian cancer | IHC | IHC score ≥ 1 | DFS | K-M Curve | M | 8 |
| Larsson | 2012 | Sweden | 607 | Colorectal cancer | IHC | IHC score ≥ 3 | OS, DFS, TTR | Multivariate | P | 9 |
| Binder | 2013 | America | 181 | Glioblastoma multiforme | IHC | NA | OS | Multivariate | NA | 7 |
| Boman | 2013 | Sweden | 100 | Urothelial bladder cancer | IHC | IHC score ≥ 3 | OS | Multivariate | M/P | 7 |
| Boman | 2013 | Sweden | 343 | Urothelial bladder cancer | IHC | IHC score ≥ 3 | OS, CSS, PFS | Multivariate | M/P | 8 |
| Forse | 2013 | Canada | 698 | Breast cancer | IHC | IHC score ≥ 3 | DFS | Multivariate | P | 9 |
| Kaprio | 2014 | Finland | 840 | Colorectal cancer | IHC | IHC score ≥ 3 | CSS | K-M Curve | M/P | 9 |
| Heby | 2015 | Sweden | 175 | Pancreatic and periampullary adenocarcinoma | IHC | IHC score ≥ 2 | OS, DFS | Multivariate | P | 7 |
| Laitinen | 2015 | Finland | 337 | Gastric cancer | IHC | IHC score ≥ 1 | CSS | Multivariate | M/P | 8 |
| Saukkonen | 2015 | Finland | 189 | Pancreatic ductal adenocarcinoma | IHC | IHC score ≥ 3 | CSS | Multivariate | M/P | 7 |
| Borg | 2016 | Sweden | 106 | Esophageal cancer | IHC | IHC score ≥ 1 | OS, TTR | K-M Curve | P | 7 |
| Borg | 2016 | Sweden | 65 | Gastric cancer | IHC | IHC score ≥ 1 | OS, TTR | K-M Curve | p | 7 |
| Chijiiwa | 2016 | Japan | 70 | Pancreatic cancer | IHC | IHC score ≥ 4 | OS, DFS | K-M Curve | M | 7 |
| Taniuchi | 2016 | Japan | 102 | Pancreatic cancer | IHC | IHC score ≥ 3 | OS | Multivariate | P | 7 |
| Kusumoto | 2017 | Japan | 114 | Lung adenocarcinoma | IHC | IHC score ≥ 1 | OS, DFS, CSS | K-M Curve | NA | 8 |
| Yuan | 2018 | China | 87 | Colorectal cancer | IHC | IHC score ≥ 3 | OS | Multivariate | M | 7 |
| Zhang | 2018 | China | 54 | Gastric cancer | IHC | IHC score ≥ 1 | OS, DFS | Multivariate | NA | 7 |
IHC Immunohistochemistry, NA Not Available, OS Overall Survival, DFS Disease-free Survival, CSS Cancer-specific Survival, NOS Newcastle-Ottawa Scale
Meta-analysis of PODXL expression levels and locations on OS/ DFS/ CSS
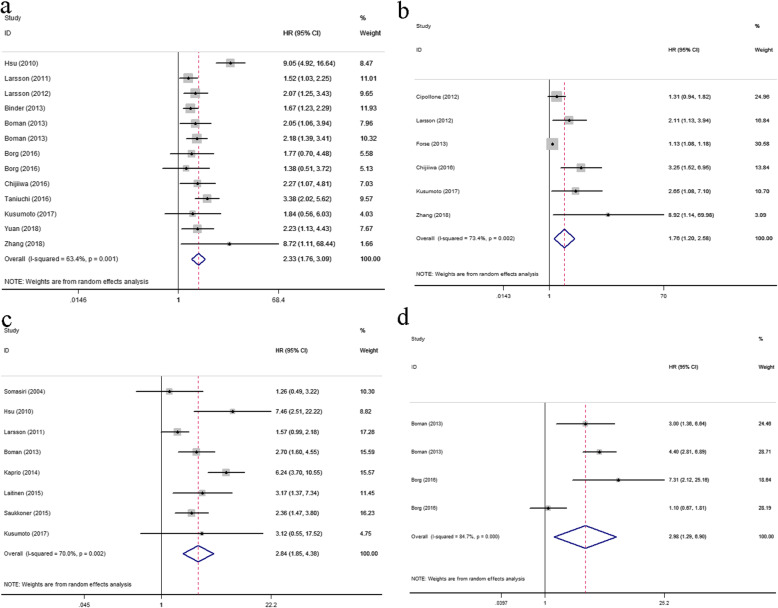
A total of 11 eligible studies, including 13 cohorts and 2272 patients, were recruited to evaluate the expression level of PODXL on OS. The pooled HR and 95% CI indicated that high-expressed PODXL was significantly related to poor OS in patients with various cancers (HR = 2.33, 95% CI = 1.76–3.09, P < 0.0001) with a significant heterogeneity across these studies (I2 = 63.4%, P = 0.001) (Fig.2a). In addition, there were 6 studies performed the relationships between PODXL expression levels and DFS, and 8 studies investigated the associations between PODXL expression levels and CSS respectively. Heterogeneity test indicated both the DFS (I2 = 73.4%, P = 0.002) and CSS (I2 = 70.0%, P = 0.002) should be analyzed using the random-effect model. Finally, the results indicated the association between the high expressed PODXL and the shorter DFS (HR = 1.76, 95% CI =1.20–2.58, P = 0.004) or the shorter CSS (HR = 2.84, 95% CI = 1.85–4.38, P < 0.0001) (Fig.2b-c). On the other hand, among these eligible 18 papers, 5 studies involved the expression locations of PODXL and the prognosis of cancers, and only 2 studies, including 4 cohorts, showed the association between membrane expressed PODXL and poor OS (HR = 2.98, 95% CI =1.29–6.90, P = 0.011), also by using the random-effect model (I2 = 84.7%, P < 0.0001) (Fig.2d).
Fig. 2.
Forest plot of studies evaluating HRs of PODXL expression and the prognosis of cancer patients. a High expressed PODXL and the OS; b high expressed PODXL and the DFS; c high expressed PODXL and the CSS; d membrane expressed PODXL and the OS
Subgroup analysis for OS
In order to find the source of heterogeneity, the subgroup analysis of OS was performed, and all of the 2272 patients were classified based on cancer types, analysis types, antibody types, ethnicities and sample sizes (Table 2). Single study which assessed the relationship between the expression and OS in renal cell carcinoma, glioblastoma multiforme, esophageal cancers and lung adenocarcinoma were defined as “other cancers” in the other cancers subgroup. Subgroup analysis showed that, high expressed PODXL were linked with poor OS in colorectal cancer (HR = 1.79, 95% CI = 1.35–2.37, P < 0.0001), pancreatic cancer (HR = 2.98, 95% CI = 1.95–4.55, P < 0.0001), urothelial bladder cancer (HR = 2.14, 95% CI = 1.48–3.10) and other cancers (HR = 2.60, 95% CI = 1.45–4.66, P = 0.001), but not in patients with the gastric cancer (HR = 2.76, 95% CI = 0.45–15.84, P = 0.256). In conclusion, high expressed level of PODXL was associated with poor OS in 6 types of cancers.
Table 2.
Subgroup analysis of pooled HR for OS
| Categories | No. of studies | No. of patients | Pooled HR (95%CI) | Heterogeneity | ||
|---|---|---|---|---|---|---|
| Fix/Random | P-value | I2 (%) | P-value | |||
| OS | 13 | 2272 | 2.33 (1.76, 3.09) | 0 | 63.4 | 0.001 |
| Cancer type | 8 | |||||
| Colorectal cancer | 3 | 834 | 1.79 (1.35, 2.37) | 0 | 0 | 0.499 |
| Pancreatic cancer | 2 | 172 | 2.98 (1.95, 4.55) | 0 | 0 | 0.391 |
| Gastric cancer | 2 | 119 | 2.76 (0.48, 15.84) | 0.256 | 59.9 | 0.114 |
| Urothelial bladder cancer | 2 | 443 | 2.14 (1.48, 3.10) | 0 | 0 | 0.880 |
| Other cancers | 4 | 704 | 2.60 (1.45, 4.66) | 0.001 | 83.3 | 0 |
| Analysis | ||||||
| K-M curve | 4 | 355 | 1.85 (1.17, 2.95) | 0.009 | 0 | 0.89 |
| Multivariate | 9 | 2017 | 2.59 (1.77, 3.80) | 0 | 74.7 | 0 |
| Antibody type | ||||||
| Monoclonal antibody | 2 | 157 | 2.25 (1.36, 3.73) | 0.002 | 0 | 0.975 |
| Polyclonal antibody | 6 | 1672 | 2.55 (1.45, 4.50) | 0.001 | 0 | 81.6 |
| M + P | 2 | 443 | 2.14 (1.48, 3.10) | 0 | 0 | 0.880 |
| Ethnicity | ||||||
| European | 6 | 1361 | 1.84 (1.47, 2.30) | 0 | 0 | 0.834 |
| Asian | 6 | 730 | 3.49 (2.02, 6.02) | 0 | 64.5 | 0.015 |
| North American | 1 | 181 | 1.67 (1.23, 2.29) | – | – | |
| Sample size | ||||||
| < 150 | 8 | 628 | 2.46 (1.81, 3.33) | 0 | 0 | 0.536 |
| ≥150 | 5 | 1644 | 2.36 (1.53, 3.65) | 0 | 81.3 | 0 |
OS overall survival, HR hazard ratio
And regarding the analysis type, we also found that the high expression of PODXL was significantly associated with the much shorter OS, when the studies were assessed with K-M curve. In the subgroups based on ethnicities, antibody types and sample sizes, we also found that, the relation between high expression level of PODXL and poor OS, except for patients from Asia or the sample size ≥150.
PODXL overexpression and relative clinical parameters
In order to obtain more clinical values of PODXL, we investigated the associations between PODXL expression levels and clinical parameters in several cancers (Table 3). From these results, we found that the expression level of PODXL was related with the TNM stage (HR = 1.63, 95% CI = 1.19–2.23, P = 0.002, fixed-effects), tumor grade (HR = 4.29, 95% CI = 1.84–9.99, P = 0.001, random-effects), differentiation (HR = 2.84, 95% CI = 1.82–4.42, P < 0.0001, fixed-effects), distant metastasis (HR = 5.46, 95% CI = 2.55–11.66, P < 0.0001, fixed-effects), lymph node metastasis (HR = 1.51, 95% CI = 1.03–2.22, P = 0.034, fixed-effects), neural invasion (HR = 2.43, 95% CI = 1.02–5.79, P = 0 .45, fixed-effects) and vascular invasion (HR = 2.27, 95% CI = 1.56–3.30, P < 0.0001, fixed-effects) significantly. Whereas, there was no significant correlations between PODXL expression and age (HR = 0.88, 95% CI = 0.71–1.10, P = 0.269, fixed-effects), gender (HR = 1.04, 95% CI = 0.82–1.32, P = 0.749, fix-effects) and tumor size (HR = 0.90, 95% CI = 0.61–1.34, P = 0.614, fixed-effects). As a result, these correlations indicated that the high expressed PODXL was associated with the advanced biological behavior in various cancers. No covariate analyzed in this study had a statistically significant effect on degree of tumor malignancy and survival.
Table 3.
Clinicopathological features of the enrolled studies with high expressed PODXL in patients with cancer
| Clinicopathological parameters | Studies | No. of patients | Risk of high PODXL OR (95% CI) |
Significant Z |
P-value | Heterogeneity I2 (%) |
P-value | Model |
|---|---|---|---|---|---|---|---|---|
| Age (< 65 vs ≥ 65) | 10 | 2905 | 0.88 (0.71, 1.10) | 1.11 | 0.269 | 42.6 | 0.084 | Fixed effects |
| Gender (male vs female) | 11 | 3081 | 1.04 (0.82, 1.32) | 0.32 | 0.749 | 0 | 0.835 | Fixed effects |
| Tumor size (< 5 cm vs ≥5 cm) | 5 | 1334 | 0.90 (0.61, 1.34) | 0.50 | 0.614 | 0 | 0.703 | Fixed effects |
| TNM stage (III-IV vs I-II) | 12 | 2417 | 1.63 (1.19, 2.23) | 3.04 | 0.002 | 13.1 | 0.319 | Fixed effects |
| Tumor grade (3–4 vs 1–2) | 6 | 2268 | 4.29 (1.84, 9.99) | 3.38 | 0.001 | 78.6 | 0 | Random effects |
|
Tumor differentiation (moderate/well vs poor) |
6 | 1429 | 2.84 (1.82, 4.42) | 4.62 | 0 | 0 | 0.559 | Fixed effects |
|
Distant metastasis (positive vs Negative) |
3 | 475 | 5.46 (2.55, 11.66) | 4.38 | 0 | 44.5 | 0.165 | Fixed effects |
|
Lymph node metastasis (positive vs negative) |
6 | 1574 | 1.51 (1.03, 2.22) | 2.11 | 0.034 | 0 | 0.614 | Fixed effects |
|
Neural invasion (positive vs negative) |
3 | 264 | 2.43 (1.02, 5.79) | 2.00 | 0.045 | 0 | 1.000 | Fixed effects |
|
Vascular invasion (positive or negative) |
6 | 1240 | 2.27 (1.56, 3.30) | 4.29 | 0 | 2.1 | 0.403 | Fixed effects |
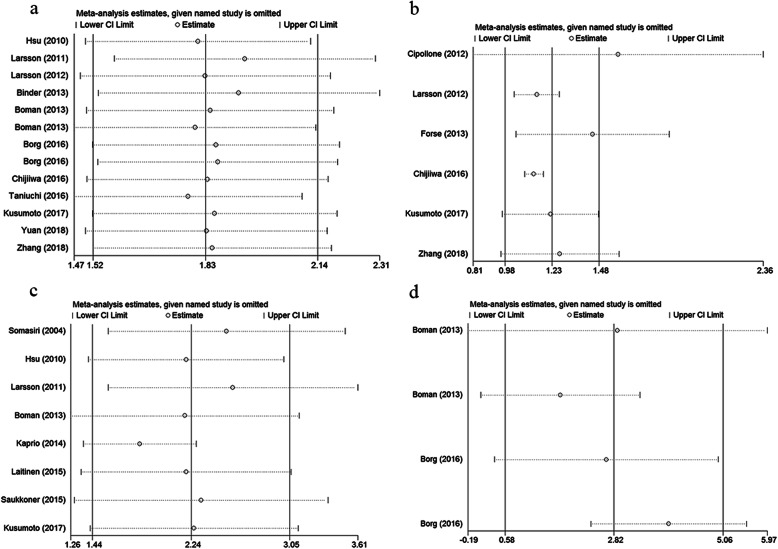
Sensitivity analysis and publication bias
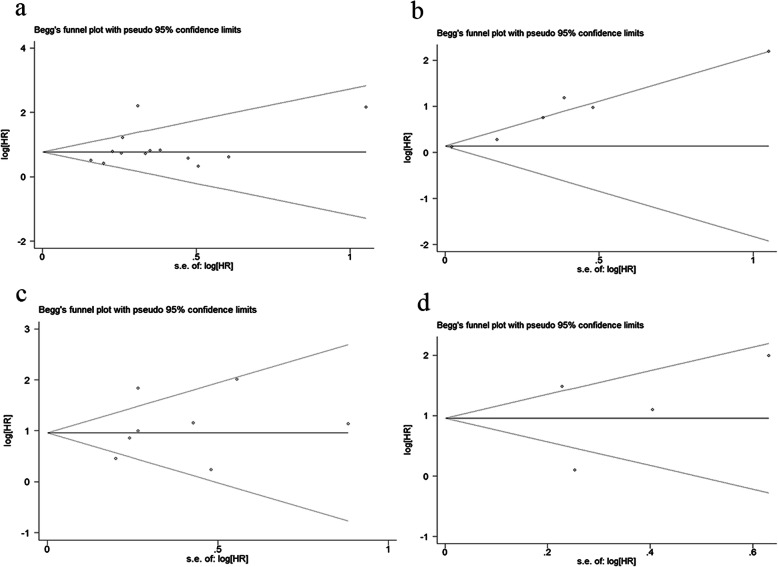
We performed sensitivity analysis to determine whether an individual study could affected the overall result. Results of association studies between PODXL expression and OS and CSS demonstrated that single study had no influence on the result of meta-analysis (Fig.3). Funnel plots and Begg’s test were performed and the results showed no publication bias existed in studies on associations between PODXL overexpression and OS (P = 0.502), DFS (P = 0.133) and CSS (P = 0.266). And no publication bias existed in our meta-analysis on associations between PODXL membrane expression and OS (P = 1.000) as well (Fig.4).
Fig. 3.
Sensitivity analysis of this meta-analysis. a OS of PODXL expression levels; b DFS of PODXL expression levels; c CSS of PODXL expression levels; d OS of PODXL expression locations
Fig. 4.
Begg’s funnel plots for the studies involved in the meta-analysis. a OS of PODXL expression levels; b DFS of PODXL expression levels; c CSS of PODXL expression levels; d OS of PODXL expression locations
The expression data of PODXL extracted from TCGA datasets
The differences of PODXL expression level between various tumor tissues and corresponding normal tissues were obtained with GEPIA, which was a common web-based tool that can provide a quick and customizable survey of function based on TCGA and GTEx data [36]. PODXL was detected in 23 types of cancers, and the result that the PODXL expression was significantly much higher than the corresponding normal tissues was found in 9 types of cancers, including the esophagus cancer, glioblastoma multiforme, acute myeloid leukemia, liver hepatocellular carcinoma, ovarian serous cystadenocarcinoma, pancreatic adenocarcinoma, rectum adenocarcinoma, stomach adenocarcinoma, testicular germ cell tumor (Table 4).
Table 4.
The difference of PODXL expression in cancers and corresponding normal tissues in TCGA datasets
| Types of cancer | TCGA dataset | No. of cancer tissues | No. of normal tissues | Log2(FC) | P value |
|---|---|---|---|---|---|
| Adenoid cystic carcinoma | ACC | 77 | 128 | − 1.068 | 1.10e-10 |
| Breast invasion carcinoma | BRCA | 1085 | 291 | −0.514 | 4.01e-16 |
| Cervical squamous cell carcinoma | CESC | 306 | 13 | −0.590 | 0.191 |
| Esophagus cancer | ESCA | 182 | 286 | 1.391 | 5.97e-22 |
| Glioblastoma multiforme | GBM | 163 | 207 | 0.866 | 1.12e-10 |
| Head and neck squamous cell carcinoma | HNSCC | 519 | 44 | 0.656 | 0.123 |
| Kidney chromophobe | KICH | 66 | 53 | −2.863 | 1.53e-10 |
| Kidney renal clear cell carcinoma | KIRC | 523 | 100 | −0.732 | 1.85e-9 |
| Kidney renal papillary cell carcinoma | KIRP | 286 | 60 | −4.247 | 1.31e-43 |
| Acute myeloid leukemia | LAML | 173 | 70 | 1.210 | 3.26e-2 |
| Liver hepatocellular carcinoma | LIHC | 369 | 160 | 1.508 | 8.20e-32 |
| Lung adenocarcinoma | LUAD | 483 | 347 | −2.064 | 6.27e-122 |
| Lung squamous cell carcinoma | LUSC | 486 | 338 | −2.832 | 3.86e-153 |
| Ovarian serous cystadenocarcinoma | OVSC | 426 | 88 | 1.449 | 3.96e-14 |
| Pancreatic adenocarcinoma | PAAD | 179 | 171 | 0.492 | 4.05e-5 |
| Prostate carcinoma | PRAD | 492 | 152 | −0.479 | 0.044 |
| Rectum adenocarcinoma | READ | 92 | 318 | 0.598 | 8.17e-8 |
| Skin cutaneous melanoma | SKCM | 461 | 558 | −0.636 | 3.83e-6 |
| Stomach adenocarcinoma | STAD | 408 | 211 | 1.597 | 1.64e-49 |
| Testicular germ cell tumor | TGCT | 137 | 165 | 2.750 | 3.93e-30 |
| Thyroid carcinoma | THCA | 512 | 337 | −0.796 | 6.95e-22 |
| Uterine corpus endometrial carcinoma | UCEC | 174 | 91 | −0.797 | 2.97e-5 |
| Uterine carcinosarcoma | UCS | 58 | 78 | −2.075 | 3.79e-12 |
Validation of prognostic correlation by TCGA datasets
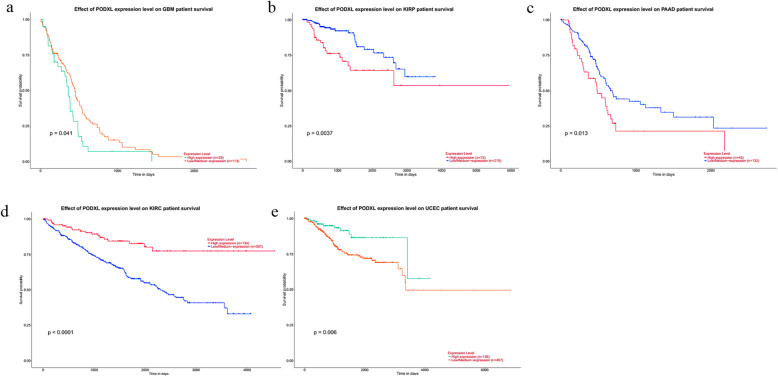
To validate the clinical prognosis indication value of PODXL, we explored TCGA datasets by using UALCAN, which was an interactive online tool that could analyze the expression data of genes in TCGA [37]. And among the 31 types of cancers, 9040 patients, the significant association between high expressed PODXL and poor OS was found in 3 types of cancers, including the glioblastoma multiforme, kidney renal papillary cell carcinoma and pancreatic adenocarcinoma (Table 5). But there were adverse results in kidney renal clear cell carcinoma and uterine corpus endometrial carcinoma, which showed a significant correlation between the low expressed PODXL and poor OS (Fig.5). The same results were also verified with KM Plotter, whose data sources were not completely consistent with TCGA datasets (Supplementary Fig. 1, SF.1).
Table 5.
The difference of overall survival in cancer patients with high PODXL expression vs low/median expression
| Cancer type | No. of cancer tissues | P value | ||
|---|---|---|---|---|
| High | Low/Median | Total | ||
| ACC | 20 | 59 | 79 | 0.37 |
| BLCA | 102 | 304 | 406 | 0.34 |
| BRCA | 272 | 809 | 1081 | 0.4 |
| CESC | 73 | 218 | 291 | 0.77 |
| CHOL | 9 | 27 | 36 | 0.57 |
| COAD | 69 | 210 | 279 | 0.32 |
| ESCA | 46 | 138 | 184 | 0.16 |
| GBM | 39 | 113 | 152 | 0.041 |
| HNSCC | 130 | 389 | 519 | 0.3 |
| KICH | 15 | 49 | 64 | 0.35 |
| KIRC | 134 | 397 | 531 | < 0.0001 |
| KIRP | 72 | 215 | 287 | 0.0037 |
| LAML | 43 | 120 | 163 | 0.64 |
| LIHC | 93 | 272 | 365 | 0.82 |
| LUAD | 125 | 377 | 502 | 0.37 |
| LUSC | 126 | 368 | 494 | 0.33 |
| DLBC | 12 | 35 | 47 | 0.21 |
| MESO | 22 | 63 | 85 | 0.23 |
| OVSC | 76 | 227 | 303 | 0.95 |
| PAAD | 45 | 132 | 177 | 0.013 |
| PCPG | 45 | 134 | 179 | 0.13 |
| PRAD | 125 | 372 | 497 | 0.92 |
| READ | 42 | 123 | 165 | 0.44 |
| SARC | 65 | 194 | 259 | 0.12 |
| SKCM | 115 | 344 | 459 | 0.22 |
| TGCT | 34 | 100 | 134 | 0.29 |
| THYM | 30 | 89 | 119 | 0.78 |
| THCA | 127 | 377 | 504 | 0.87 |
| UCS | 15 | 41 | 56 | 0.58 |
| UCEC | 136 | 407 | 543 | 0.006 |
| UVM | 20 | 60 | 80 | 0.36 |
ACC adrenocortical carcinoma, BLCA bladder urothelial carcinoma, BRCA breast invasion carcinoma, CESE cervical squamous cell carcinoma, CHOL cholangiocarcinoma, COAD colon adenocarcinoma, ESCA esophageal carcinoma, GBM glioblastoma multiforme, HNSCC head and neck squamous cell carcinoma, KICH kidney chromophobe, KIRC kidney renal clear cell carcinoma, KIRP kidney renal papillary cell carcinoma, LAML acute myeloid leukemia, LIHC liver hepatocellular carcinoma, LUAD lung adenocarcinoma, LUSC lung squamous cell carcinoma, DLBC lymphoid neoplasm diffuse large B-cell lymphoma, MESO mesothelioma, OVSC ovarian serous cystadenocarcinoma, PAAD pancreatic adenocarcinoma, PCPG pheochromocytoma and paraganglioma, PRAD prostate adenocarcinoma, READ rectum adenocarcinoma, SARC sarcoma, SKCM skin cutaneous melanoma, STAD stomach adenocarcinoma, TGCT testicular germ cell tumors, THYM thymoma, THCA thyroid carcinoma, UCS uterine carcinosarcoma, UCEC uterine corpus endometrial carcinoma, UVM uveal melanoma
Fig. 5.
Kaplan-Meier survival curves for cancer patients based on TCGA datasets. a glioblastoma multiforme; b kidney renal papillary cell carcinoma; c pancreatic adenocarcinoma; d kidney renal clear cell carcinoma; e uterine corpus endometrial carcinoma
A joint result of our meta-analysis and TCGA datasets validation identified the correlation between the expression level of PODXL and the glioblastoma multiforme, pancreatic adenocarcinoma, esophagus cancer, gastric cancer and lung adenocarcinoma.
PPI network construction and functional enrichment analysis
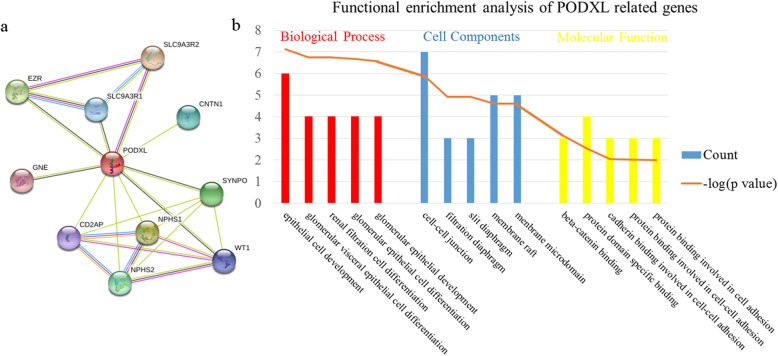
The PPI network of PODXL-related genes was obtained by using STRING, including 11 nodes and 23 edges (Fig.6a). The PODXL-related genes were collected for functional enrichment analysis (Fig.6b). The top GO terms, containing biological processes, cell components and molecular function, were selected based on the most significant. These PODXL-related genes were significantly enriched in cell development and differentiation, and played a significant role in cell-cell adhesion. These significant GO terms were matched with the pathogenesis of cancers, such as intercellular adhesion decrease, epithelial-mesenchymal transition (EMT), cell migration and invasion.
Fig. 6.
Mechanism prediction of PODXL-related genes with bioinformatics. a The protein-protein interaction network of PODXL-related genes. The lines represented the interaction between the nodes. b The functional enrichment analysis of PODXL-related genes
Discussion
Recently, increasing evidences have suggested that PODXL was involved in multiple links in several process of tumor development, such as cell adhesion and morphology [40], lymphatic metastasis [41], tumor cells motility and invasiveness [26], tumor angiogenesis [42] and prognosis. Recent researches indicated that the expression level and location of PODXL could be a new biomarker to assess the prognosis of various types of cancers. However, a single study is limited by insufficient data and single experimental model, so that a meta-analysis of pooling studies is necessary to explore the potential clinical value of PODXL.
Among these published studies, there were 10 types of cancers, including 5705 patients. Our meta-analysis not only indicated that high expressed PODXL was associated with poor OS, DFS or CSS in patients with cancers, but also showed that membrane expression was correlated with poor OS as well. Clinicopathological features analysis showed that the overexpressed PODXL was linked with poor stage and differentiation, and high incidences of metastasis and invasion in cancers, which indicated that there might be a significant association between PODXL expression level and advanced features of cancer. Subgroup analysis showed that the association between overexpressed PODXL and poor OS in patients with cancers, was only significative in the glioblastoma multiforme, pancreatic cancer, renal cell carcinoma, colorectal cancer and urothelial bladder cancer, but not in the esophageal cancer, gastric cancer and lung adenocarcinoma. Then we used GEPIA and UALCAN to explore TCGA datasets, to compare the expression difference of PODXL among tumor tissues and correlated normal tissues, and the survival curves. Consistent results of meta-analysis and TCGA datasets validation were found in 5 types of cancers. Beside TGGA datasets, Oncomine was used to further verify the differences of PODXL expression level between various tumor tissues and corresponding normal tissues. And On the other hand, KM Plotter was used to validate the clinical prognosis indication value of PODXL. The results of these databases also supported the consequence of TCGA datasets.
The prognostic value of PODXL had been indicated by meta-analysis in 2017 [32], the conclusion put forward by Wang et al. was approximately consistent with our results. But we revisited and gathered relevant research for another meta-analysis, in order to further explore its clinical significance. Compared with the meta-analysis in 2017, our research contained more studies and patients, which reinforced the conclusion. In addition, both of the expression level and site of PODXL were found to be associated with prognosis of various cancers. And the results of meta-analysis were filtrated by validation with TCGA datasets, which made our conclusion seem more convincing.
Among the eligible 18 studies, there were only 2 researches mentioned the expression location of PODXL and prognosis of cancers, containing 4 cohorts. The studies showed a significant association between membrane expression of PODXL and poor OS, but the sensitivity analysis showed that this result is not credible. On the premise of appropriate number of included studies, samples that may introduce heterogeneity are moved, but the sensitivity is still high, so this result can only be used as a descriptive hypothesis, and need more included studies. As PODXL is a transmembrane glycoprotein, whose high expression level and membrane expression lead to cell motility increasing, and over-activated tumor cell migration ability promotes tumor progression. Combined with the existing results, the expression site of PODXL was a promising markers in predicting the prognosis of cancers.
Although, PODXL has been found to be highly expressed in various malignancies and was related to a more aggressive phenotype and poor prognosis, the exact mechanisms of which role did PODXL play in tumorigenesis remains unclear [43]. The gene functional enrichment analysis showed that PODXL was a fatal gene in cell development and differentiation, which played an important role in cell-cell adhesion. Some latest studies showed that PODXL promoted the gelsolin-actin interaction in cell protrusions to enhance the motility and invasiveness [26], and some showed that the PODXL-ezrin signaling axis could rearrange the dynamic cytoskeleton for transendothelial migration [44]. According to these reports, it could be deduced that high expressed PODXL promoted tumor progression by enhancing a series of cell changes such as EMT, cell migration and invasion. In addition, the result that membrane-expressed PODXL was associated with poor survival, further supported the deduction that PODXL promoted tumor progression by enhancing the motility and invasiveness of tumor cells. PODXL also took part in the NF-kB, PI3K/AKT, Hippo and MAPK/ERK signaling pathway, and facilitated tumor progression by increasing cell proliferation, migration and invasion as well as suppressing apoptosis [21, 45, 46].
PODXL was expected to be a novel therapeutic and monitoring biomarker in certain cancers, because the high expressed PODXL might be a potential indicator of poor prognosis of cancers. Overexpressed PODXL could be detected in peripheral blood and used as a non-invasive diagnostic biomarker for the detection of pancreatic cancer [47]. ATF3 could activate PODXL transcription, which suggested that ATF3 pathway might be beneficial for anticancer therapy [48]. High expression of miR-509-3-5p and miR-5100 inhibited the invasion and metastasis of gastric cancers and pancreatic cancers by directly targeting PODXL, functioning as a tumor suppressor [27, 41]. A core fucose-deficient monoclonal antibody (mAb) of PODXL might be a new antibody-based therapy method against PODXL high-expressed oral squamous cell carcinoma [49]. And patients with gastric or esophageal adenocarcinoma would have a much better prognosis after treating with neoadjuvant ± adjuvant fluoropyrimidine– and oxaliplantin-based chemotherapy, if the expression level of PODXL is high [50].
However, there are still some limitations. First of all, many unavoidable reasons, such as different types of cancers, the analysis methods, ethnicities and sample sizes might attribute to the heterogeneity. Secondly, we extracted the data of HRs and 95% CIs from the K-M plots when it could not be obtained from the paper directly, and this process might decrease the accuracy of results. Thirdly, the sensitivity analysis only showed that individual study had no influence on the association study between the high expressed PODXL and poor OS or CSS, that is to say, the results of the association between the membrane expressed PODXL and poor OS in cancers can only be seen as a descriptive hypothesis, might be induced by the insufficient studies or the small sample size. Fourthly, our meta-analysis seemed have no publication bias, but as the chance of negative results being published is very small, more studies are needed to verify the results of our meta-analysis.
Conclusion
PODXL is a significant clinical indicator for tumor prognosis and detection, the expression level and location in tumor tissues, and even the serum concentration of which could be associated significantly with tumor progression [47]. Our meta-analysis showed that PODXL plays a significant role in cancer progression, and high-expressed PODXL could be linked to aggressive biological phenotype and poor prognosis. Specifically, the high expressed PODXL was correlated with poor prognosis significantly in the glioblastoma multiforme and pancreatic cancer, but not in the esophageal adenocarcinoma, gastric cancer and lung adenocarcinoma.
Supplementary information
Additional file 2 SF.1 Kaplan-Meier survival curves for cancer patients from KM Plotter. (a) Pancreatic adenocarcinoma; (b) kidney renal papillary cell carcinoma; (c) kidney renal clear cell carcinoma; (d) uterine corpus endometrial carcinoma.
Acknowledgements
Not applicable.
Abbreviations
- CI
Confidence inter
- CSS
Cancer-specific survival
- DFS
Disease-free survival
- GEPIA
Gene Expression Profiling Interactive Analysis
- HR
Hazard ratio
- IHC
Immunohistochemistry
- K-M
Kaplan-Meier
- mAb
Monoclonal antibody
- NCD
Noncommunicable disease
- NOS
Newcastle-Ottawa Scale
- OR
Odds ratio
- OS
Overall survival
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analysis
- PODXL
Podocalyxin-like protein
- TCGA
The Cancer Genome Atlas
Authors’ contributions
MY and SH conceived the study. SH and ML searched the databases and extracted the data. WD and SH analyzed the data. SH and WD wrote the draft of the paper. FZ and MY reviewed the manuscript. All authors have read and approved the manuscript.
Funding
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this article and referenced articles are listed in the References section.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Siying He and Wenjie Du contributed equally to this work.
Contributor Information
Ming Yan, Email: yanming72@whu.edu.cn.
Fang Zheng, Email: zhengfang@whu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12885-020-07108-5.
References
- 1.Somasiri A, Nielsen JS, Makretsov N, McCoy ML, Prentice L, Gilks CB, et al. Overexpression of the anti-adhesin podocalyxin is an independent predictor of breast cancer progression. Cancer Res. 2004;64(15):5068–5073. doi: 10.1158/0008-5472.can-04-0240. [DOI] [PubMed] [Google Scholar]
- 2.McNagny KM, Pettersson I, Rossi F, Flamme I, Shevchenko A, Mann M, et al. Thrombomucin, a novel cell surface protein that defines thrombocytes and multipotent hematopoietic progenitors. J Cell Biol. 1997;138(6):1395–1407. doi: 10.1083/jcb.138.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schopperle WM, Kershaw DB, DeWolf WC. Human embryonal carcinoma tumor antigen, Gp200/GCTM-2, is podocalyxin. Biochem Biophys Res Commun. 2003;300(2):285–290. doi: 10.1016/S0006-291X(02)02844-9. [DOI] [PubMed] [Google Scholar]
- 4.Itai S, Yamada S, Kaneko MK, Sano M, Nakamura T, Yanaka M, et al. Podocalyxin is crucial for the growth of oral squamous cell carcinoma cell line HSC-2. Biochem Biophys Rep. 2018;15:93–96. doi: 10.1016/j.bbrep.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsson A, Johansson ME, Wangefjord S, Gaber A, Nodin B, Kucharzewska P, et al. Overexpression of podocalyxin-like protein is an independent factor of poor prognosis in colorectal cancer. Br J Cancer. 2011;105(5):666–672. doi: 10.1038/bjc.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kershaw DB, Beck SG, Wharram BL, Wiggins JE, Goyal M, Thomas PE, et al. Molecular cloning and characterization of human podocalyxin-like protein. Orthologous relationship to rabbit PCLP1 and rat podocalyxin. J Biol Chem. 1997;272(25):15708–15714. doi: 10.1074/jbc.272.25.15708. [DOI] [PubMed] [Google Scholar]
- 7.Li P, Karaczyn AA, McGlauflin R, Favreau-Lessard AJ, Jachimowicz E, Vary CP et al. Novel roles for podocalyxin in regulating stress myelopoiesis, Rap1a, and neutrophil migration. Exp Hematol. 2017;50:77–83.e6. doi:10.1016/j.exphem.2017.04.001. [DOI] [PMC free article] [PubMed]
- 8.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 9.Kerosuo L, Juvonen E, Alitalo R, Gylling M, Kerjaschki D, Miettinen A. Podocalyxin in human haematopoietic cells. Br J Haematol. 2004;124(6):809–818. doi: 10.1111/j.1365-2141.2004.04840.x. [DOI] [PubMed] [Google Scholar]
- 10.Porras G, Ayuso MS, Gonzalez-Manchon C. Leukocyte-endothelial cell interaction is enhanced in podocalyxin-deficient mice. Int J Biochem Cell Biol. 2018;99:72–79. doi: 10.1016/j.biocel.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Horrillo A, Porras G, Ayuso MS, Gonzalez-Manchon C. Loss of endothelial barrier integrity in mice with conditional ablation of podocalyxin (Podxl) in endothelial cells. Eur J Cell Biol. 2016;95(8):265–276. doi: 10.1016/j.ejcb.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Vitureira N, Andres R, Perez-Martinez E, Martinez A, Bribian A, Blasi J, et al. Podocalyxin is a novel polysialylated neural adhesion protein with multiple roles in neural development and synapse formation. PLoS One. 2010;5(8):e12003. doi: 10.1371/journal.pone.0012003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forse CL, Yilmaz YE, Pinnaduwage D, O'Malley FP, Mulligan AM, Bull SB, et al. Elevated expression of podocalyxin is associated with lymphatic invasion, basal-like phenotype, and clinical outcome in axillary lymph node-negative breast cancer. Breast Cancer Res Treat. 2013;137(3):709–719. doi: 10.1007/s10549-012-2392-y. [DOI] [PubMed] [Google Scholar]
- 14.Larsson A, Fridberg M, Gaber A, Nodin B, Leveen P, Jonsson G, et al. Validation of podocalyxin-like protein as a biomarker of poor prognosis in colorectal cancer. BMC Cancer. 2012;12:282. doi: 10.1186/1471-2407-12-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaprio T, Hagstrom J, Fermer C, Mustonen H, Bockelman C, Nilsson O, et al. A comparative study of two PODXL antibodies in 840 colorectal cancer patients. BMC Cancer. 2014;14:494. doi: 10.1186/1471-2407-14-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan D, Chen H, Wang S, Liu F, Cheng Y, Fang J. Identification of LEA, a podocalyxin-like glycoprotein, as a predictor for the progression of colorectal cancer. Cancer Med. 2018;7(10):5155–5166. doi: 10.1002/cam4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borg D, Hedner C, Nodin B, Larsson A, Johnsson A, Eberhard J, et al. Expression of podocalyxin-like protein is an independent prognostic biomarker in resected esophageal and gastric adenocarcinoma. BMC Clin Pathol. 2016;16:13. doi: 10.1186/s12907-016-0034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laitinen A, Bockelman C, Hagstrom J, Kokkola A, Fermer C, Nilsson O, et al. Podocalyxin as a prognostic marker in gastric Cancer. PLoS One. 2015;10(12):e0145079. doi: 10.1371/journal.pone.0145079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Zhu Z, Wu H, Yu Z, Rong Z, Luo Z, et al. PODXL, negatively regulated by KLF4, promotes the EMT and metastasis and serves as a novel prognostic indicator of gastric cancer. Gastric Cancer. 2018. 10.1007/s10120-018-0833-y. [DOI] [PMC free article] [PubMed]
- 20.Binder ZA, Siu IM, Eberhart CG, Ap Rhys C, Bai RY, Staedtke V, et al. Podocalyxin-like protein is expressed in glioblastoma multiforme stem-like cells and is associated with poor outcome. PLoS One. 2013;8(10):e75945. doi: 10.1371/journal.pone.0075945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusumoto H, Shintani Y, Kanzaki R, Kawamura T, Funaki S, Minami M, et al. Podocalyxin influences malignant potential by controlling epithelial-mesenchymal transition in lung adenocarcinoma. Cancer Sci. 2017;108(3):528–535. doi: 10.1111/cas.13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin CW, Sun MS, Wu HC. Podocalyxin-like 1 is associated with tumor aggressiveness and metastatic gene expression in human oral squamous cell carcinoma. Int J Oncol. 2014;45(2):710–718. doi: 10.3892/ijo.2014.2427. [DOI] [PubMed] [Google Scholar]
- 23.Cipollone JA, Graves ML, Kobel M, Kalloger SE, Poon T, Gilks CB, et al. The anti-adhesive mucin podocalyxin may help initiate the transperitoneal metastasis of high grade serous ovarian carcinoma. Clin Exp Metastasis. 2012;29:239–252. doi: 10.1007/s10585-011-9446-0). [DOI] [PubMed] [Google Scholar]
- 24.Saukkonen K, Hagstrom J, Mustonen H, Juuti A, Nordling S, Fermer C, et al. Podocalyxin is a marker of poor prognosis in pancreatic ductal adenocarcinoma. PLoS One. 2015;10(6):e0129012. doi: 10.1371/journal.pone.0129012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heby M, Elebro J, Nodin B, Jirstrom K, Eberhard J. Prognostic and predictive significance of podocalyxin-like protein expression in pancreatic and periampullary adenocarcinoma. BMC Clin Pathol. 2015;15:10. doi: 10.1186/s12907-015-0009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taniuchi K, Furihata M, Naganuma S, Dabanaka K, Hanazaki K, Saibara T. Podocalyxin-like protein, linked to poor prognosis of pancreatic cancers, promotes cell invasion by binding to gelsolin. Cancer Sci. 2016;107(10):1430–1442. doi: 10.1111/cas.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chijiiwa Y, Moriyama T, Ohuchida K, Nabae T, Ohtsuka T, Miyasaka Y, et al. Overexpression of microRNA-5100 decreases the aggressive phenotype of pancreatic cancer cells by targeting PODXL. Int J Oncol. 2016;48(4):1688–1700. doi: 10.3892/ijo.2016.3389. [DOI] [PubMed] [Google Scholar]
- 28.Casey G, Neville PJ, Liu X, Plummer SJ, Cicek MS, Krumroy LM, et al. Podocalyxin variants and risk of prostate cancer and tumor aggressiveness. Hum Mol Genet. 2006;15(5):735–741. doi: 10.1093/hmg/ddi487. [DOI] [PubMed] [Google Scholar]
- 29.Heath EI, Heilbrun LK, Smith D, Schopperle WM, Ju Y, Bolton S, et al. Overexpression of the Pluripotent Stem Cell Marker Podocalyxin in Prostate Cancer. Anticancer Res. 2018;38(11):6361–6366. doi: 10.21873/anticanres.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu YH, Lin WL, Hou YT, Pu YS, Shun CT, Chen CL, et al. Podocalyxin EBP50 ezrin molecular complex enhances the metastatic potential of renal cell carcinoma through recruiting Rac1 guanine nucleotide exchange factor ARHGEF7. Am J Pathol. 2010;176(6):3050–3061. doi: 10.2353/ajpath.2010.090539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boman K, Larsson AH, Segersten U, Kuteeva E, Johannesson H, Nodin B, et al. Membranous expression of podocalyxin-like protein is an independent factor of poor prognosis in urothelial bladder cancer. Br J Cancer. 2013;108(11):2321–2328. doi: 10.1038/bjc.2013.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Zhao Y, Qi R, Zhu X, Huang C, Cheng S, et al. Prognostic role of podocalyxin-like protein expression in various cancers: A systematic review and meta-analysis. Oncotarget. 2017;8(32):52457–52464. doi: 10.18632/oncotarget.14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D M, A l, J T, G AD, Group TP Preferred reporting items for systematic reviews and meta-analysis: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng X, Zhang Y, Kwong JS, Zhang C, Li S, Sun F, et al. The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: a systematic review. J Evid Based Med. 2015;8(1):2–10. doi: 10.1111/jebm.12141. [DOI] [PubMed] [Google Scholar]
- 36.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiaoli Jiao BTS, Huang DW, Stephens R, Baseler MW, Lane HC, Richard A. Lempicki. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Oxford J. 2012. [DOI] [PMC free article] [PubMed]
- 40.Nielsen JS, Graves ML, Chelliah S, Vogl AW, Roskelley CD, McNagny KM. The CD34-related molecule podocalyxin is a potent inducer of microvillus formation. PLoS One. 2007;2(2):e237. doi: 10.1371/journal.pone.0000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Zhu Z, Sheng J, Yu Z, Yao B, Huang K, et al. miR-509-3-5P inhibits the invasion and lymphatic metastasis by targeting PODXL and serves as a novel prognostic indicator for gastric cancer. Oncotarget. 2017;8(21):34867–34883. doi: 10.18632/oncotarget.16802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amo L, Tamayo-Orbegozo E, Maruri N, Eguizabal C, Zenarruzabeitia O, Rinon M, et al. Involvement of platelet-tumor cell interaction in immune evasion. Potential role of podocalyxin-like protein 1. Front Oncol. 2014;4:245. doi: 10.3389/fonc.2014.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsson AH, Lehn S, Wangefjord S, Karnevi E, Kuteeva E, Sundström M, et al. Significant association and synergistic adverse prognostic effect of podocalyxin-like protein and epidermal growth factor receptor expression in colorectal cancer. J Transl Med. 2016;14(1):128. doi: 10.1186/s12967-016-0882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fröse J, Chen MB, Hebron KE, Reinhardt F, Hajal C, Zijlstra A, et al. Epithelial-Mesenchymal transition induces Podocalyxin to promote extravasation via Ezrin signaling. Cell Rep. 2018;24(4):962–972. doi: 10.1016/j.celrep.2018.06.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee WY, Kuo CC, Lin BX, Cheng CH, Chen KC, Lin CW. Podocalyxin-Like Protein 1 Regulates TAZ Signaling and Stemness Properties in Colon Cancer. Int J Mol Sci. 2017;18(10). 10.3390/ijms18102047. [DOI] [PMC free article] [PubMed]
- 46.Zhi Q, Chen H, Liu F, Han Y, Wan D, Xu Z, et al. PODXL promotes gastric cancer progression through interacting with RUFY1 protein. Cancer Sci. 2018. 10.1111/cas.13864. [DOI] [PMC free article] [PubMed]
- 47.Taniuchi K, Tsuboi M, Sakaguchi M, Saibara T. Measurement of serum PODXL concentration for detection of pancreatic cancer. Onco Targets Ther. 2018;11:1433–1445. doi: 10.2147/ott.s155367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buganim Y, Madar S, Rais Y, Pomeraniec L, Harel E, Solomon H, et al. Transcriptional activity of ATF3 in the stromal compartment of tumors promotes cancer progression. Carcinogenesis. 2011;32(12):1749–1757. doi: 10.1093/carcin/bgr203. [DOI] [PubMed] [Google Scholar]
- 49.Itai S, Ohishi T, Kaneko MK, Yamada S, Abe S, Nakamura T, et al. Anti-podocalyxin antibody exerts antitumor effects via antibody-dependent cellular cytotoxicity in mouse xenograft models of oral squamous cell carcinoma. Oncotarget. 2018;9(32):22480–22497. doi: 10.18632/oncotarget.25132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borg D, Larsson AH, Hedner C, Nodin B, Johnsson A, Jirstrom K. Podocalyxin-like protein as a predictive biomarker for benefit of neoadjuvant chemotherapy in resectable gastric and esophageal adenocarcinoma. J Transl Med. 2018;16(1):290. doi: 10.1186/s12967-018-1668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 2 SF.1 Kaplan-Meier survival curves for cancer patients from KM Plotter. (a) Pancreatic adenocarcinoma; (b) kidney renal papillary cell carcinoma; (c) kidney renal clear cell carcinoma; (d) uterine corpus endometrial carcinoma.
Data Availability Statement
All data generated or analyzed during this study are included in this article and referenced articles are listed in the References section.