Abstract
Treatment modalities in esophageal squamous cell carcinoma (ESCC) depend largely on lymph node metastasis (LNM) status. With sub-optimal detection sensitivity of existing imaging techniques, we propose a methylation signature which identifies patients with LNM with greater accuracy. This would allow precise stratification of high-risk patients requiring more aggressive treatment from low-risk ESCC patients who can forego radical surgery. An unbiased genome-wide methylation signature for LNM detection was established from an initial in-silico discovery phase. The signature was tested in independent clinical cohorts comprising 249 ESCC patients. The prognostic potential of the methylation signature was compared to clinical variables including LNM status. A 10-probe LNM associated signature (LNAS) was developed using stringent bioinformatics analyses. The area under the curve values for LNAS risk scores were 0.81 and 0.88 in training and validation cohorts respectively in association with lymphatic vessel invasion and tumor stage. High LNAS risk-score was also associated with worse overall survival [HR (95% CI) 3 (1.8 to 4.8), p < 0.0001 training and 3.9 (1.5 to 10.2), p = 0.001 validation cohort]. In conclusion, our novel methylation signature is a powerful biomarker that identifies LNM status robustly and is also associated with worse prognosis in ESCC patients.
Keywords: Epigenetic panel, biomarker, prognostic
Introduction
Esophageal cancer is the eighth most common cancer with the sixth highest mortality rate worldwide 1, 2. Among the two major histological subtypes of esophageal cancer, esophageal squamous cell carcinoma (ESCC) accounts for ~80% of all patients 3. Although ESCC is more frequently seen in developing countries, there has been a dramatic increase in its incidence rates even in the western world over the past few decades, with 16,940 new cases and 15,690 deaths annually in United States alone 4, 5. One of the major clinical concerns with ESCC is the dismal overall 5-year survival rates associated with this malignancy in spite of advances in its management and treatment 5.
Among various clinical risk factors associated with ESCC pathogenesis, lymph node metastasis (LNM) remains the most significant contributor to poor prognosis; with overall 5-year survival rates post-surgery dropping from 70–92% to 18–47% in patients with LNM 1. ESCC patients with LNM often reveal a more aggressive disease behavior with a higher tendency for loco-regional and distant recurrence, leading to worse prognosis 6. Surgical decision-making for performing less invasive endoscopic tumor resection over radical esophagectomy or determining extent of lymphadenectomy depends largely on whether the tumor is associated with LNM or not 7–9. Hence accurate identification of LNM status plays a crucial role in determining treatment strategies as well as prognostic outcomes 10, 11.
Current LNM detection strategies fall short of being a gold standard modality for multiple reasons. Non-invasive conventional imaging techniques have been found to be inaccurate in approximately 40% of patients where they are unable to detect micro-metastases, which often leads to under-staging and subsequent inadequate treatment 12. There have also been few recent reports wherein false positive cases of inflammatory lymphadenopathy were misdiagnosed as LNM positive [LNM(+)] by positron emission tomography/computed tomography (PET/CT) scans 13. In view of these clinical concerns, there is a dire need for development of molecular biomarkers that can facilitate LNM detection and allow clinicians in reaching a more informed decision-making for performing radical surgeries and improving patients’ quality of life.
Epigenetic alterations are recognized as key contributors to cancer initiation and progression 14. Among these, DNA methylation is one of the most extensively studied epigenetic modifications, even within a clinical context, essentially because methylation changes are dynamic yet stable, are disease-specific, and can be quantitatively measured in clinical specimens without relying upon the availability of an endogenous normalization control 15–19. There are candidate methylation markers reported in the literature that can distinguish normal tissues from esophageal cancers and even differentiate both histologic subtypes of esophageal cancers, namely squamous and adenocarcinoma 20, 21; however, such individual methylated genes or a signature for the identification of LNM remain limited 17, 22, 23. Recent studies indicate that a biomarker signature consisting of multiple targets offers superior performance compared to individual candidates as it integrates the effect of multiple genes and accordingly improves the predictive and prognostic accuracy for a given disease state 24, 25. Keeping this in mind, we systematically developed and established a novel methylation signature with well-defined risk scores for identifying LNM (+) ESCC patients which offers a distinct superiority and potential clinical application.
Materials and methods
Patient cohorts and sample selection
A total of 261 ESCC patients who underwent surgical resection were included in this study from the Nagoya University Hospital, Japan, between February 2001 and February 2015; and from the National Cancer Center Hospital, Japan, between January 2004 and January 2006. The tumor stage was evaluated according to American Joint Committee on Cancer (AJCC) Tumor-Node-Metastasis (TNM) grading system 7th edition26 and clinicopathological profiles of the patients were analyzed according to the classification of esophageal cancer proposed by the Japanese Society of Esophageal Diseases 27, 28. The LNM status was determined from histopathologic examination of resected LNs. The patient cohort from the Nagoya University Hospital (N = 219) were considered as the training cohort, while the patients from the National Cancer Center Hospital (N=42) comprised of the validation cohort. In the training cohort, out of 219 patients, 16 cases were treated with neo-adjuvant chemoradiotherapy (7.1%), 90 cases treated with neo-adjuvant chemotherapy (41.1%). Written informed consent was obtained from all patients and the study was approved by the institutional review boards of all participating institutions.
The biomarker discovery analysis
For the comprehensive biomarker discovery step, we first performed in-silico analysis on The Cancer Genome Atlas (TCGA) data which included normalized methylation profiling results (Infinium HumanMethylation450) and clinical data from esophageal cancer patients, which was downloaded from the UCSC Cancer Browser portal in February 2017 (https://genome-cancer.ucsc.edu/). The methylation and clinical data was available for 186 esophageal cancer patients from which information on 86 ESCC cases was extracted. TCGA dataset comprised of 34 lymph node metastasis positive (LNM (+)) and 52 lymph node metastasis negative (LNM (−)) patients. The normalized beta-values for methylation ranged from −0.5 to 0.5.
Sample preparation
DNA was extracted from fresh frozen primary tissues using AllPrep DNA/RNA/miRNA Universal (Qiagen, Hilden, Germany) as per manufacturer’s instructions. Following DNA quantification using Nanodrop system (ThermoFisher Scientific, Massachusetts, USA), 500 ng of genomic DNA was bisulfite converted with EZ-DNA methylation Gold-Kit (Zymo, Irvine, CA, USA).
Bisulfite PCR and quantitative pyrosequencing
Primers for bisulfite-specific PCR (BSPCR) were designed using the PyroMark Assay Design Software 2.0 (Qiagen) and with amplicon size ranging from 120– 250 base pairs (Supplementary Table 1. Bisulfite converted DNA was amplified by BSPCR using PyroMark PCR Mastermix (Qiagen). Briefly, 10ng of bisulfite converted DNA was mixed with the HotStarTaq Master Mix (Qiagen) and bisulfite specific primers under the following conditions: 95 °C for 10 min, 45 cycles of 94 °C for 30 s, 56 °C for 30 s and 72 °C for 30 s, and an elongation step of 72 °C for 10 min. The amplified products were run on 2% agarose gels to check the specificity of primers. Pyrosequencing was performed on Pyromark Q48 Autoprep using Q48 advanced CpG Reagents (Qiagen). Concisely, 10 μl of BSPCR product was added to 3 μl of magnetic beads and 2 μl of 4 μM sequencing primer on the Pyromark Q48 discs as per manufacturer’s instructions. Output data were analyzed using PyroMark Q48 Autoprep Software (Qiagen), which calculates the CpG methylation value as the percentage (methylated cytosine/ [methylated cytosine+ unmethylated cytosine]) for each CpG site, allowing quantitative comparisons. Controls to assess proper bisulfite conversion of the DNA were included in each assay to ensure the fidelity of the measurements. Analysis was performed on the average methylation of all CpGs of a particular gene taken together.
Statistical analysis
Mann Whitney U test was carried out to compare methylation levels between LNM (+) and LNM (−) samples in TCGA dataset followed by Benjamini Hochberg’s multiple testing correction. Mann Whitney U test was performed in R using Wilcoxson’s signed rank test with (paired =FALSE) setting. A p-value of less than 0.05 was considered significant. All the above-mentioned analyses along with the heatmap construction were performed in R version 3.3.1 (Vienna, Austria). Criteria including at least 20% difference in methylation between two groups and at least 50% of probes in a CpG island being methylated were imposed to identify top methylated candidates. For both the training and validation cohorts, receiver operating characteristic (ROC) curves, area under the curve (AUC) and binary logistic regression analyses were performed using IBM SPSS version 23 (IBM, Armonk, NY, USA). Sensitivity, specificity, positive predictive values (PPV), negative predictive values (NPV), false discovery rate (FDR=1-PPV) and false omission rate (FOR=1-NPV were calculated using the median cut-off of the risk scores in both the cohorts. The Kaplan–Meier method was used to analyze the correlation between methylation signature and patient survival, and the log-rank test for comparing survival differences between groups using the MedCalc Statistical Software version 16.4.3 (MedCalc Software bvba, Ostend, Belgium; https://www.medcalc.org; 2016). The hazard ratio of the signature as well as the other clinical variables was calculated with Cox proportionality hazard model. The regression model and median cut-off derived from the methylation signature in the training cohort was applied to the independent validation cohort for calculating the AUC for LNM detection. Univariate binary logistic regression analysis was performed on age, sex, T-stage, differentiation, lymphatic vessel invasion (LVI), venous invasion, and LNM status along with the proposed LNM associated signature (LNAS). Only the significant variables in the univariate model were used to perform the multivariate logistic regression analysis.
Results
Genome wide methylation profiling for a methylation signature to detect LNM in ESCC patients
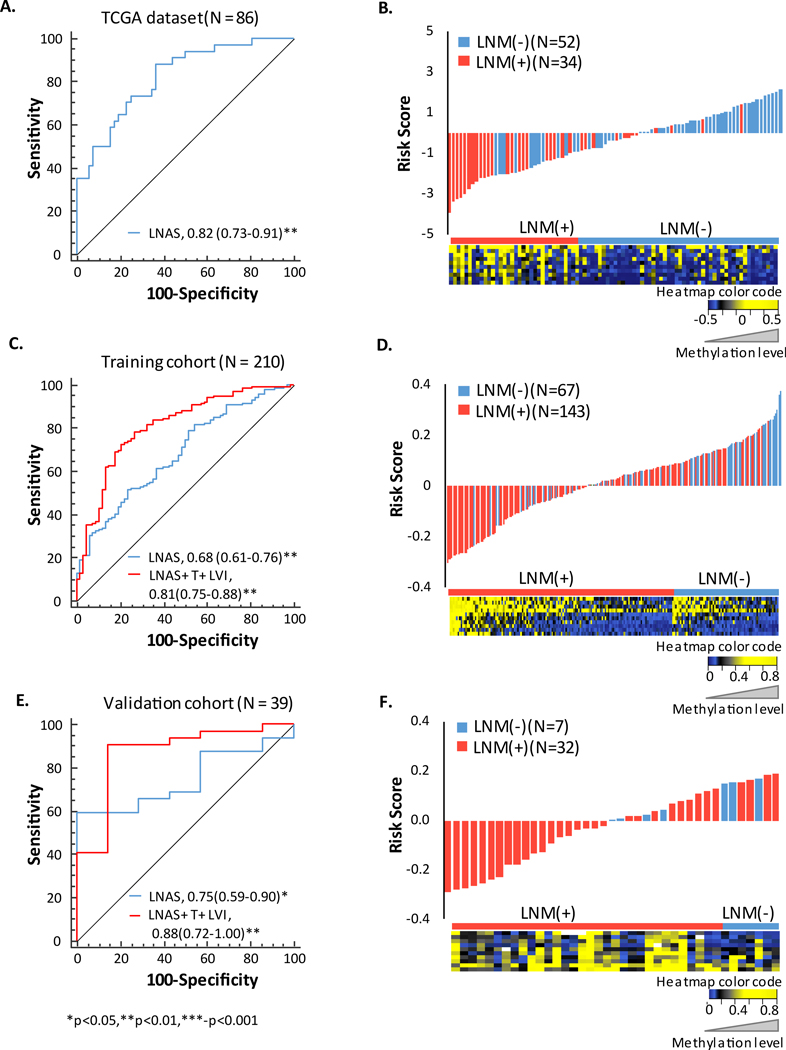
Eighty-six ESCC patients from in-silico TCGA dataset were analyzed in the discovery step to identify the most differentially methylated CpG islands between LNM (+) and LNM (−) patients (Figure 1A). At the outset, we selected 396,061 probes with data availability in at least 50% of the cases. After comparing the median normalized beta values of the LNM (+) and LNM (−) groups followed by multiple testing correction, 22,378 probes were found to be significant (Figure 1B-volcano plot). Using stringent criterion of at least 20% hyper or hypo methylation in LNM (+) vs. LNM (−) patients, we identified 22 hyper-methylated and 1 hypo-methylated probe (depicted by red and blue dots respectively in Figure 1B). Spearman’s rank coefficient (r2) was calculated between the 23 probes for determining the correlation between the probes. Hyper-methylated probes belonging to the same CpG island positively correlated with each other (r2 >0.6; depicted by darker shades of blue, Figure 1C) while the hypo-methylated probe negatively correlated with the other probes (depicted in yellow, Figure 1C). The heatmap shows the methylation values of these 23 probes in the LNM (+) and LNM (−) groups were hyper-methylated in the majority of the probes in LNM (+) group, while there were more hypo-methylation observed in the latter group (Figure 1D). The details of the 23 probes are summarized in Supplementary Table 2. As differential methylation is not an isolated event and is observed to be present in adjacent probes of a CpG island as well, we selected probes that belonged to CpG islands or differentially methylated regions (DMRs; in case of non-CpG probe) with more than half of probes with more than 10% differential methylation (Supplementary Figure 1). In the end, by using these stringent elimination criteria, we identified a panel of 10 probes representing regions that were hyper-methylated in ESCC patients with LNM (+) tumors.
Figure 1: Identification of methylation signature for lymph node metastasis detection in esophageal squamous cell carcinoma.
A) Flowchart illustrating the in-silico discovery, training and validation steps. B) 23 differentially methylated probes identified from TCGA data analysis. Red dots are probes with > 0.2 delta beta values and blue dot is the probe with < −0.2 delta beta value. C) Spearman correlation coefficient between the 23 CpG probes. D) Heatmap representing the methylation frequencies of the 23 probes in lymph node metastasis positive (LNM +) (N=34) and lymph node metastasis negative (LNM -) (N=52) samples.
Training and validation of a lymph node metastasis associated signature in two independent ESCC patient cohorts
To further train and validate the methylation signature in independent clinical cohorts, we designed and established optimal pyrosequencing assays and analyzed patient specimens in both the training (n = 219) and validation cohorts (n = 42). The final 10-gene methylation signature was referred to as lymph node metastasis associated signature (LNAS) henceforth, for convenience and consistency (Supplementary Tables 3 & 4). A very few cases (9 in the training cohort and 3 in the validation cohort) had missing LNM status information; and hence were excluded from subsequent analysis. The LNAS score was calculated by binary logistic regression analysis on methylation frequency values of all the 10 probes in the training cohort. The coefficients of the methylation signatures derived from the training cohort were applied to the independent validation cohort. In other words, risk score for each patient, in both cohorts, was based on the methylation frequency values for the 10 probes as follows: 1/ {1 + EXP [-(−0.015*cg01834022 + 0.031*cg20693607 – 0.003*cg22352818 – 0.019*cg24505892 + 0.005*cg13045134 + 0.034*cg25903779 + 0.002*cg21530266 + 0.009*cg04008703 – 0.015*cg04618333 + 0.01*cg08151857)]}. The demographic details of the cohorts based on their risk scores are summarized in Table 1. On plotting the risk scores, 88% of LNM (+) cases had a negative score while 62% of the LNM (−) cases had a positive score in in-silico dataset. Likewise, 56% of LNM (+) cases had a negative score while 63% of the LNM (−) cases had a positive score in the training cohort and 59% of LNM (+) cases had a negative score while 100% of the LNM (−) cases had a positive score in validation cohort (risk score distribution plots in Figures 2B, 2D and 2F). LNAS scores were used for ROC curve construction and an area under the curve (AUC) value of 0.68 was observed in the training cohort and 0.74 in the validation cohort (Figure 2C and 2E). The specificity of the signature in identifying LNM (+) cases was 81% in in-silico dataset, 70% in training cohort and 100% in validation cohort, while the sensitivity was 65% in in-silico and 53% in both the cohorts (Supplementary Table 5). As primary tumor size (T) and LVI were two important clinical variables that were associated with LNM in univariate logistic regression analysis in the training cohort, we combined these two factors along with the LNAS. This resulted in an improved AUC value of 0.81 in the training cohort and 0.88 in validation cohort, implying improved accuracy of the signature combined with clinical variables in identifying LNM in ESCC patients (Figures 2C and 2E; Supplementary Table 6). We also compared the AUCs of the LNAS in a subset of training cohort with respect to T1-T2 stages and neoadjuvant therapy, and observed similar AUCs in all the three scenarios (Supplementary Figure 2).
Table 1:
Baseline characteristics of the training and validation sets stratified with LNAS risk-score
| Training cohort (N=210) | Validation cohort (N=39) | |||||||
|---|---|---|---|---|---|---|---|---|
| Total | Low Risk | High Risk | p-value | Total | Low Risk | High Risk | p-value | |
| Sex | ||||||||
| Male | 161(77) | 81(77) | 80(76) | 0.87 | 35(90) | 17(90) | 18(90) | 1 |
| Female | 49(23) | 24(23) | 25(24) | 4(10) | 2(10) | 2(10) | ||
| Tumor Size | ||||||||
| <45mm | 99(47) | 44(42) | 55(52) | 0.14 | 18(46) | 9(47) | 9(45) | 0.88 |
| ≥45mm | 104(50) | 57(54) | 47(45) | 21(54) | 10(53) | 11(55) | ||
| Undefined | 7(3) | 4(4) | 3(3) | 0(0) | 0(0) | 0(0) | ||
| Differentiation | ||||||||
| Well | 22(10) | 11(11) | 11(11) | 0.91 | 17(44) | 6(32) | 11(55) | 0.65 |
| Moderate | 148(71) | 76(72) | 72(69) | 9(23) | 4(21) | 5(25) | ||
| Poor | 29(14) | 14(13) | 15(14) | 0.9 | 13(33) | 9(47) | 4(20) | 0.06 |
| Undefined | 11(5) | 4(4) | 7(7) | 0(0) | 0(0) | 0(0) | ||
| T category | ||||||||
| T1 | 44(21) | 22(21) | 22(21) | 0.69 | 3(8) | 3(16) | 0(0) | 0.27 |
| T2 | 33(16) | 15(14) | 18(17) | 3(8) | 2(10) | 1(5) | ||
| T3 | 119(57) | 63(60) | 56(53) | 0.73 | 32(82) | 14(74) | 18(90) | 0.06 |
| T4 | 12(6) | 4(4) | 8(8) | 0.3 | 1(2) | 0(0) | 1(5) | 0.04 |
| Undefined | 2(1) | 1(1) | 1(1) | 0(0) | 0(0) | 0(0) | ||
| Lymph Node metastasis | ||||||||
| Absent | 67(32) | 42(40) | 25(24) | 0.19 | 7(18) | 6(32) | 1(5) | 0.03 |
| Present | 143(68) | 63(60) | 80(76) | 32(82) | 13(68) | 19(95) | ||
| Distant Metastasis | ||||||||
| Present | 189(90) | 91(87) | 98(93) | 0.1 | 36(92) | 18(95) | 0(0) | 0.003 |
| Absent | 21(10) | 14(13) | 7(7) | 3(8) | 1(5) | 2(100) | ||
| TNM stage | ||||||||
| Stage I | 41(20) | 26(25) | 15(14) | 0.11 | 4(10) | 3(16) | 1(5) | 0.85 |
| Stage II | 45(21) | 21(20) | 24(23) | 5(13) | 4(21) | 1(5) | ||
| Stage III | 101(48) | 43(41) | 58(55) | 0.02 | 27(69) | 11(58) | 16(80) | 0.19 |
| Stage IV | 21(10) | 14(13) | 7(7) | 0.8 | 3(8) | 1(5) | 2(10) | 0.27 |
| Undefined | 2(1) | 1(1) | 1(1) | 0(0) | 0(0) | 0(0) | ||
| Lymphatic vessel Invasion | ||||||||
| Absent | 56(27) | 35(33) | 21(20) | 0.02 | 18(46) | 8(42) | 10(50) | 0.62 |
| Present | 151(72) | 68(65) | 83(79) | 21(54) | 11(58) | 10(50) | ||
| Undefined | 3(1) | 2(2) | 1(1) | 0(0) | 0(0) | 0(0) | ||
| Venous Invasion | ||||||||
| Absent | 128(61) | 61(58) | 67(64) | 0.39 | 15(38) | 9(47) | 6(30) | 0.26 |
| Present | 80(38) | 43(41) | 37(35) | 24(62) | 10(53) | 14(70) | ||
| Undefined | 2(1) | 1(1) | 1(1) | 0(0) | 0(0) | 0(0) | ||
Figure 2: AUC and associated risk scores of LNAS signature.
A) AUC of LNAS in TCGA. B) Risk score distribution plot along with heatmap of signature. C) AUC of LNAS and LNAS along with clinical variables T stage (T) and lymphatic vessel invasion (LVI) in training cohort. D) Risk score distribution plot in training cohort along with heatmap of signature. E) AUC of LNAS and LNAS along with clinical variables T and LVI in validation cohort. F) Waterfall plot of modified risk score in validation cohort along with heatmap of signature. Modified risk score was obtained from subtracting individual risk score from median value.
Risk assessment of clinicopathological variables for LNM in ESCC patients
Association between various clinicopathological factors including LNAS with LNM risk was calculated by univariate and multivariate logistic regression analysis. In univariate analysis, venous invasion [OR (95% CI) = 1.94 (1.04 to 3.62), p = 0.04], T stage [OR (95% CI) = 4.58 (2.47 to 8.48), p < 0.001] and LVI [OR (95% CI) = 6.51 (3.36 to 12.58), p < 0.001] significantly increased risk of LNM in the training cohort along with the LNAS [OR (95% CI) = 13.79 (4.13 to 46.04), p < 0.001] (Table 2). Even after adjustment for these significant variables, the LNAS remained as an independent risk factor for LNM [OR (95% CI) = 11.83 (3.02 to 46.40), p < 0.001] in multivariate analysis.
Table 2:
Multivariate logistic regression of clinicopathological factors and 10 gene methylation signature in the training cohort
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Characteristics | OR | 95%CI | p Value | OR | 95%CI | p Value |
| Age (< 66 vs ≥ 66) | 0.63 | 0.34–1.1 | 0.1 | |||
| Sex (male vs female) | 1.26 | 0.62–2.5 | 0.52 | |||
| Pathology (well vs moderate + poor) | 1.28 | 0.83–1.95 | 0.25 | |||
| T stage(T1+T2 vs T3+T4) | 4.58 | 2.47–8.48 | <0.01 | 3.88 | 1.91–7.85 | <0.01 |
| Venous Invasion (present or absent) | 1.94 | 1.04–3.62 | 0.04 | 1.04 | 0.48–2.26 | 0.91 |
| Lymphatic Vessel Invasion(present or absent) | 6.51 | 3.36–12.58 | <0.01 | 3.52 | 1.63–7.60 | 0.01 |
| LNAS (risk score < 0.67 or ≥ 0.67) | 13.79 | 4.13–46.04 | <0.01 | 11.83 | 3.02–46.40 | <0.01 |
Bold values indicate statistically significant values
In the validation cohort, LVI [OR (95% CI) = 9.62 (1.04 to 88.65), p = 0.04] was the only significant clinical variable in univariate analysis and LNAS again remained as a significant risk factor for LNM [OR (95% CI) = 226 (1.44 to >1000), p = 0.034] (Table3).
Table 3:
Multivariate logistic regression of clinicopathological factors and 10 gene methylation signature in the validation cohort
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Characteristics | OR | 95% CI | p Value | OR | 95% CI | p Value |
| Sex (male vs female) | 0.62 | 0.05–7.03 | 0.7 | |||
| Pathology (well vs moderate + poor) | 0.67 | 0.11–3.99 | 0.65 | |||
| T stage (T1+T2 vs T3+T4) | 2.8 | 0.40–19.60 | 0.3 | |||
| Venous invasion (present or absent) | 5.5 | 0.90–33.34 | 0.06 | |||
| Lymphatic vessel invasion (present or absent) | 9.62 | 1.04–88.65 | 0.04 | 18.38 | 1.53–220.74 | 0.02 |
| LNAS (Risk score < 0.67 or ≥ 0.67) | 57.28 | 0.69–4692 | 0.07 | 226 | 1.44–35357 | 0.03 |
Bold values indicate statistically significant values
Association of LNAS signature with survival in the training and validation cohorts
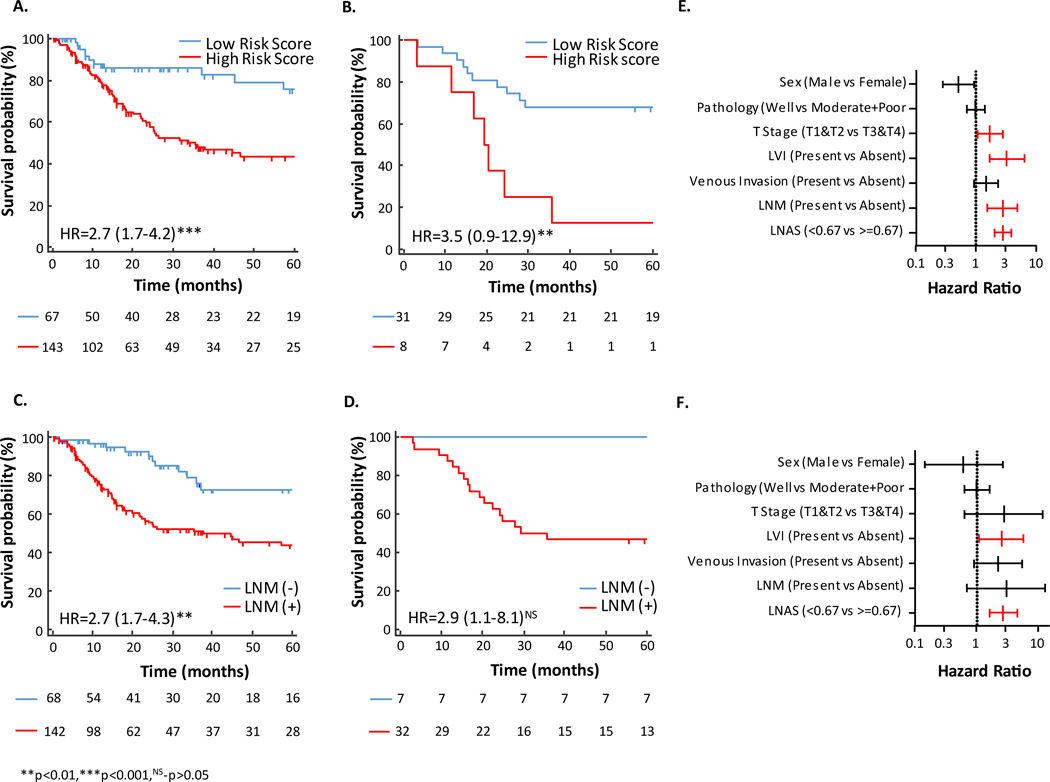
To determine the prognostic potential of LNAS and compare it to that of pathologically determined LNM status, survival analysis was performed for overall survival (OS) and disease-free survival (DFS). Patients in the high-risk group demonstrated shorter OS than those in the low-risk group in both the training and validation cohorts [HR (95% CI) 2.7 (1.7 to 4.2), p < 0.001 in training cohort, HR (95% CI) 3.5 (0.9 to 12.9), p = 0.003 in validation cohort] (Figures 3A & 3B). Positive LNM status was expectedly associated with worse OS in both cohorts. However, while the hazard ratio was significant and markedly similar to LNAS in the training cohort [HR (95%CI) 2.7 (1.7 to 4.3), p = 0.003], it did not reach statistical significance in the validation cohort [HR (95% CI) 2.9 (1.1 to 8.05), p = 0.11; Figures 3C & 3D). While DFS analysis for validation cohort could not be performed due to unavailability of recurrence data, higher LNAS risk score was also associated with worse DFS in the training cohort [HR (95% CI) 2.6 (1.6 to 4.3), p < 0.001; Supplementary Figure 3].
Figure 3: Kaplan Meier survival and Cox hazard ratio analysis of LNAS.
A) Kaplan Meier plot for 5 years OS (OS) associated with LNAS in training cohort. B) Kaplan Meier plot for 5 years OS associated with LNAS in validation cohort. C) Kaplan Meier plot for 5 years OS associated with lymph node metastasis in training cohort. D) Kaplan Meier plot for 5 years OS associated with lymph node metastasis in validation cohort. E) Forrest plot with hazard ratio of clinical variables and LNAS in training cohort. F) Forrest plot with hazard ratio of clinical variables and LNAS in validation cohort. Red bar indicate significantly higher hazard ratio. LVI-lymphatic vessel invasion, LNM-lymph node metastasis.
In multivariate analysis with other clinicopathological factors like presence of LVI and higher T stages were strongly associated with worse OS in both cohorts (Figures 3E & 3F).
Functional annotation of genes in LNAS
The annotation of the 10 probes constituting LNAS showed that three of these probes were located in the 5’ UTR of protein coding genes (cg01834022–EVC2, cg04008703–EVX1, cg0461833–PRAC2) while cg08151857 was located in the first exon of HOXB8. Two probes were at the 3’ UTR (cg20693607–EPHB3 and cg24505892–LBX1) while two of the remaining probes were located near long intergenic non-coding (LINC) RNA (cg22352818–LOC105373496 and cg21530266–LINC01391). Remaining probe cg13045134 was interestingly located in a gene desert marked by active H3K27Ac marks. Overall, we observed that three out of the ten methylated probes were associated with homeobox family of genes (EVX1, LBX1 and HOXB8).
Discussion
LNM is a major determinant of recurrence and prognosis in ESCC patients; hence identification of the LNM status is critical for deciding the course of treatment as well as extent of invasive surgery in this malignancy. However, in 20–40% of ESCC cases, LNM is misdiagnosed due to the limitations of current diagnostic methodologies 9, 12, 13, 29. This accentuates the need for molecular biomarkers that can detect these patients with increased accuracy. In this study we developed a methylation signature (lymph node metastasis associated signature or LNAS) that can identify lymph node metastasized cases of ESCC more accurately using an unbiased genome-wide discovery, followed by clinical validation with quantitative pyrosequencing in two independent clinical cohorts. This signature is of utmost clinical significance as it can help in risk stratification of patients that will directly impact treatment modality and patient care.
DNA methylation-based markers have emerged as clinically-relevant disease markers due to their stability and tissue-specificity 15–18. Previous studies have primarily focused on single candidates and emphasized more on their prognostic role in ESCC 30–32. Furthermore, these previous studies have additional shortcomings with respect to sample size analyzed and the inadequate representation of various stages of patients in the discovery and validation steps 17, 22, 23. To overcome these drawbacks, in this current study, we have systematically and comprehensively identified and developed a methylation signature by using stringent criteria during the biomarker discovery phase. Considering the regional “spread” of methylation, we also took into account the methylation status of adjacent probes in a CpG island or DMR while selecting our candidates, thereby reducing the false positive hits and maximizing signal to noise ratio 33. The LNAS had similar AUCs when evaluated within the early tumor stages (T1-T2), as well as within patients who had not received neoadjuvant chemo or chemoradiation therapy which underscores the stability of our model. Testing the performance of our risk scores in two independent cohorts in combination with T stage and LVI status revealed a 0.13 increase in the AUC in both cohorts. This is a significant finding with respect to the current treatment norms. With advancement in endoscopy, clinicians are now opting for endoscopic submucosal dissection (ESD) or endoscopic mucosal resection (EMR) methods, where they extract superficial lesions to evaluate tumor stage and assess the adequacy of resection 34. Our methylation signature-based risk scores can be easily measured from these specimens together with the determination of tumor stage and LVI status in the identification of LNM status more robustly. Through this study, we demonstrated that incorporating existing clinical variables along with our newly developed LNAS can enhance the detection accuracy of LNM in ESCC patients.
LNM is also the single most powerful predictor of poor prognosis in ESCC patients and it was interesting to find that LNAS was as strong a predictor of OS rates as pathologically determined LNM status, if not better. We propose that if this signature can be measured in the endoscopically resected/dissected specimens, clinicians can evaluate the 5-year overall and recurrence survival rates, as well help develop personalized treatment regimens for these patients.
We observed a few genes of homeobox family in our signature. LBX1 or ladybird homeobox 1 gene has been reported to be hyper methylated in prostate and lung cancer 35, 36 and is known to play a role in epithelial-mesenchymal transition by regulating genes like ZEB1, ZEB2 and TGFB2 37. EVX1 or even skipped homeobox-1 hyper methylation is also associated with prostate and lung cancer progression while a recent report by Mallak et al demonstrated reduced expression of EVX1 to be associated with ESCC aggressiveness by being a target gene in BMP signaling pathway 38–40. Lastly, HOXB8 was the other homeobox candidate and it is not surprising as HOX A and B gene clusters are frequently hyper methylated and silenced in cancer types with poor prognosis 41–43.
We would like to acknowledge few of the limitations of our present study. The validation cohort in our study was relatively modest in size and lacked some clinical data including recurrence free survival information. Also, there was a difference in the proportion of lymph node metastasis cases (32% in a larger training cohort and 17% in the smaller validation cohort). Hence future prospective studies will be necessary to confirm our findings. We did not have access to such clinical specimens, but the true potential and clinical utility of LNAS should to be evaluated using endoscopic submucosal dissected or mucosal resected patient samples in future studies.
To conclude, in the recent times, radiation and chemotherapy have been incorporated in multimodal treatment but surgical resection remains a standard procedure for resectable esophageal cancer. In this regard, outcome of surgery is largely dependent on extent of lymphadenectomy. Even in early stage esophageal cancer, endoscopic tissue removal is appropriate only in the absence of LNM. Use of our molecular signature with selected clinical features can increase the accuracy of LNM detection as well as predict prognosis in ESCC patients that can improve overall patient treatment and outcomes.
Supplementary Material
Supplementary Figure 1: A representation of selection criterion for probes based on methylation of adjacent CpG sites in a CpG island. Each figure represents a CpG island where each bar is a probe present in 450K array. The bars in orange are the ones significant in our study through Mann Whitney U test. The probes whose 50% of adjacent CpG sites were also at least 10% methylated were selected for signature construction.
Supplementary Figure 2: Comparative ROC curve of a)T1-T2 patients (N=75) b) Patients with neoadjuvant therapy (N=102) c) patients without neoadjuvant therapy in training cohort (N=98)
Supplementary Figure 3: Kaplan Meier plot for 5 year’s disease free survival in training cohort.
Novelty and impact.
Lymph node metastasis (LNM) is the single most significant determinant of disease recurrence and prognosis in esophageal squamous cell carcinoma (ESCC); however, currently there is a paucity of reliable molecular signatures that can identify LNM is ESCC patients. We developed a comprehensive methylation signature that can distinguish LNM cases with very high specificity in independent training and validation cohorts, which allows prediction of prognosis in ESCC patients.
Acknowledgments
Grant support: This work was supported by the CA72851, CA181572, CA184792, CA187956 and CA202797 grants from the National Cancer Institute, National Institute of Health; RP140784 from the Cancer Prevention Research Institute of Texas; grants from the Sammons Cancer Center and Baylor Foundation, as well as funds from the Baylor Scott & White Research Institute, Dallas, TX, USA awarded to AG.
Abbreviations:
- ESCC
Esophageal squamous cell carcinoma
- LNM
lymph node metastasis
- LNAS
Lymph node metastasis associated signature
- HR
Hazard ratio
- OR
Odds Ratio
- CI
Confidence interval
Footnotes
Disclosure
Authors have no conflict of interest to disclose.
References
- 1.Kayani B, Zacharakis E, Ahmed K, Hanna GB. Lymph node metastases and prognosis in oesophageal carcinoma--a systematic review. Eur J Surg Oncol 2011;37: 747–53. [DOI] [PubMed] [Google Scholar]
- 2.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet 2013;381: 400–12. [DOI] [PubMed] [Google Scholar]
- 3.Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent Advances From Basic and Clinical Studies of Esophageal Squamous Cell Carcinoma. Gastroenterology 2015;149: 1700–15. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67: 7–30. [DOI] [PubMed] [Google Scholar]
- 5.Huang FL, Yu SJ. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J Surg 2016. [DOI] [PubMed] [Google Scholar]
- 6.Herrera LJ. Extent of lymphadenectomy in esophageal cancer: how many lymph nodes is enough? Ann Surg Oncol 2010;17: 676–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gockel I, Sgourakis G, Lyros O, Hansen T, Lang H. Dissection of lymph node metastases in esophageal cancer. Expert Rev Anticancer Ther 2011;11: 571–8. [DOI] [PubMed] [Google Scholar]
- 8.Cho JW, Choi SC, Jang JY, Shin SK, Choi KD, Lee JH, Kim SG, Sung JK, Jeon SW, Choi IJ, Kim GH, Jee SR, et al. Lymph Node Metastases in Esophageal Carcinoma: An Endoscopist’s View. Clin Endosc 2014;47: 523–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakajima Y, Nagai K, Miyake S, Ohashi K, Kawano T, Iwai T. Evaluation of an indicator for lymph node metastasis of esophageal squamous cell carcinoma invading the submucosal layer. Jpn J Cancer Res 2002;93: 305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakegawa T Forty years’ experience in surgical treatment for esophageal cancer. Int J Clin Oncol 2003;8: 277–88. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S Management of complications of radical esophagectomy. Indian J Surg Oncol 2013;4: 105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kassis ES, Nguyen N, Shriver SP, Siegfried JM, Schauer PR, Luketich JD. Detection of occult lymph node metastases in esophageal cancer by minimally invasive staging combined with molecular diagnostic techniques. JSLS 1998;2: 331–6. [PMC free article] [PubMed] [Google Scholar]
- 13.Yamatsuji T, Ishida N, Takaoka M, Hayashi J, Yoshida K, Shigemitsu K, Urakami A, Haisa M, Naomoto Y. False-Positive Cases of Fluorodeoxyglucose-Positron Emission Tomography/Computed Tomographic Scans in Metastasis of Esophageal Cancer. Clin Med Insights Case Rep 2017;10: 1179547617703402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007;128: 683–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005;352: 997–1003. [DOI] [PubMed] [Google Scholar]
- 16.Toyota M, Suzuki H, Yamashita T, Hirata K, Imai K, Tokino T, Shinomura Y. Cancer epigenomics: implications of DNA methylation in personalized cancer therapy. Cancer Sci 2009;100: 787–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyobu K, Yamashita S, Matsuda Y, Igaki H, Niwa T, Oka D, Kushima R, Osugi H, Lee S, Suehiro S, Ushijima T. Identification and validation of DNA methylation markers to predict lymph node metastasis of esophageal squamous cell carcinomas. Ann Surg Oncol 2011;18: 1185–94. [DOI] [PubMed] [Google Scholar]
- 18.Levenson VV. DNA methylation as a universal biomarker. Expert Rev Mol Diagn 2010;10: 481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010;70: 27–56. [DOI] [PubMed] [Google Scholar]
- 20.Hoshimoto S, Takeuchi H, Ono S, Sim MS, Huynh JL, Huang SK, Marzese DM, Kitagawa Y, Hoon DS. Genome-wide hypomethylation and specific tumor-related gene hypermethylation are associated with esophageal squamous cell carcinoma outcome. J Thorac Oncol 2015;10: 509–17. [DOI] [PubMed] [Google Scholar]
- 21.Fukuoka T, Hibi K, Nakao A. Aberrant methylation is frequently observed in advanced esophageal squamous cell carcinoma. Anticancer Res 2006;26: 3333–5. [PubMed] [Google Scholar]
- 22.Nagata H, Kozaki KI, Muramatsu T, Hiramoto H, Tanimoto K, Fujiwara N, Imoto S, Ichikawa D, Otsuji E, Miyano S, Kawano T, Inazawa J. Genome-wide screening of DNA methylation associated with lymph node metastasis in esophageal squamous cell carcinoma. Oncotarget 2017;8: 37740–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otto B, Koenig AM, Tolstonog GV, Jeschke A, Klaetschke K, Vashist YK, Wicklein D, Wagener C, Izbicki JR, Streichert T. Molecular changes in pre-metastatic lymph nodes of esophageal cancer patients. PLoS One 2014;9: e102552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kratz JR, He J, Van Den Eeden SK, Zhu ZH, Gao W, Pham PT, Mulvihill MS, Ziaei F, Zhang H, Su B, Zhi X, Quesenberry CP, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet 2012;379: 823–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei JH, Haddad A, Wu KJ, Zhao HW, Kapur P, Zhang ZL, Zhao LY, Chen ZH, Zhou YY, Zhou JC, Wang B, Yu YH, et al. A CpG-methylation-based assay to predict survival in clear cell renal cell carcinoma. Nat Commun 2015;6: 8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Network NCC. NCCN Clinical Practice Guidelines in Oncology. Esophageal and Esophagogastric Junction Cancers, 2017. [Google Scholar]
- 27.Japan Esophageal S Japanese Classification of Esophageal Cancer, 11th Edition: part II and III. Esophagus 2017;14: 37–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Japan Esophageal S Japanese Classification of Esophageal Cancer, 11th Edition: part I. Esophagus 2017;14: 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter M, Gibson R, Ruszkiewicz A, Thompson SK, Thierry B. Beyond conventional pathology: towards preoperative and intraoperative lymph node staging. Int J Cancer 2015;136: 743–51. [DOI] [PubMed] [Google Scholar]
- 30.Hou J, Liao LD, Xie YM, Zeng FM, Ji X, Chen B, Li LY, Zhu MX, Yang CX, Qing Z, Chen T, Xu XE, et al. DACT2 is a candidate tumor suppressor and prognostic marker in esophageal squamous cell carcinoma. Cancer Prev Res (Phila) 2013;6: 791–800. [DOI] [PubMed] [Google Scholar]
- 31.Cao B, Yang W, Jin Y, Zhang M, He T, Zhan Q, Herman JG, Zhong G, Guo M. Silencing NKD2 by Promoter Region Hypermethylation Promotes Esophageal Cancer Progression by Activating Wnt Signaling. J Thorac Oncol 2016;11: 1912–26. [DOI] [PubMed] [Google Scholar]
- 32.Deng J, Zhang J, Wang C, Wei Q, Zhou D, Zhao K. Methylation and expression of PTPN22 in esophageal squamous cell carcinoma. Oncotarget 2016;7: 64043–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehmann-Werman R, Neiman D, Zemmour H, Moss J, Magenheim J, Vaknin-Dembinsky A, Rubertsson S, Nellgard B, Blennow K, Zetterberg H, Spalding K, Haller MJ, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A 2016;113: E1826–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah PM, Gerdes H. Endoscopic options for early stage esophageal cancer. J Gastrointest Oncol 2015;6: 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pradhan MP, Desai A, Palakal MJ. Systems biology approach to stage-wise characterization of epigenetic genes in lung adenocarcinoma. BMC Syst Biol 2013;7: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Massie CE, Mills IG, Lynch AG. The importance of DNA methylation in prostate cancer development. J Steroid Biochem Mol Biol 2017;166: 1–15. [DOI] [PubMed] [Google Scholar]
- 37.Yu M, Smolen GA, Zhang J, Wittner B, Schott BJ, Brachtel E, Ramaswamy S, Maheswaran S, Haber DA. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes Dev 2009;23: 1737–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Truong M, Yang B, Wagner J, Kobayashi Y, Rajamanickam V, Brooks J, Jarrard DF. Even-skipped homeobox 1 is frequently hypermethylated in prostate cancer and predicts PSA recurrence. Br J Cancer 2012;107: 100–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heller G, Babinsky VN, Ziegler B, Weinzierl M, Noll C, Altenberger C, Mullauer L, Dekan G, Grin Y, Lang G, End-Pfutzenreuter A, Steiner I, et al. Genome-wide CpG island methylation analyses in non-small cell lung cancer patients. Carcinogenesis 2013;34: 513–21. [DOI] [PubMed] [Google Scholar]
- 40.Mallak AJ, Abbaszadegan MR, Khorasanizadeh PN, Forghanifard MM. Contribution of EVX1 in Aggressiveness of Esophageal Squamous Cell Carcinoma. Pathol Oncol Res 2016;22: 341–7. [DOI] [PubMed] [Google Scholar]
- 41.Strathdee G, Holyoake TL, Sim A, Parker A, Oscier DG, Melo JV, Meyer S, Eden T, Dickinson AM, Mountford JC, Jorgensen HG, Soutar R, et al. Inactivation of HOXA genes by hypermethylation in myeloid and lymphoid malignancy is frequent and associated with poor prognosis. Clin Cancer Res 2007;13: 5048–55. [DOI] [PubMed] [Google Scholar]
- 42.Aine M, Sjodahl G, Eriksson P, Veerla S, Lindgren D, Ringner M, Hoglund M. Integrative epigenomic analysis of differential DNA methylation in urothelial carcinoma. Genome Med 2015;7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu L, Frommel SC, Oakes CC, Simon R, Grupp K, Gerig CY, Bar D, Robinson MD, Baer C, Weiss M, Gu Z, Schapira M, et al. BAZ2A (TIP5) is involved in epigenetic alterations in prostate cancer and its overexpression predicts disease recurrence. Nat Genet 2015;47: 22–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: A representation of selection criterion for probes based on methylation of adjacent CpG sites in a CpG island. Each figure represents a CpG island where each bar is a probe present in 450K array. The bars in orange are the ones significant in our study through Mann Whitney U test. The probes whose 50% of adjacent CpG sites were also at least 10% methylated were selected for signature construction.
Supplementary Figure 2: Comparative ROC curve of a)T1-T2 patients (N=75) b) Patients with neoadjuvant therapy (N=102) c) patients without neoadjuvant therapy in training cohort (N=98)
Supplementary Figure 3: Kaplan Meier plot for 5 year’s disease free survival in training cohort.