Summary
De novo germline mutations in the RNA helicase DDX3X account for 1–3% of unexplained intellectual disability (ID) cases in females, and are associated with autism, brain malformations, and epilepsy. Yet, the developmental and molecular mechanisms by which DDX3X mutations impair brain function are unknown. Here, we use human and mouse genetics, and cell biological and biochemical approaches to elucidate mechanisms by which pathogenic DDX3X variants disrupt brain development. We report the largest clinical cohort to date with DDX3X mutations (n=107), demonstrating a striking correlation between recurrent dominant missense mutations, polymicrogyria, and the most severe clinical outcomes. We show that Ddx3x controls cortical development by regulating neuron generation. Severe DDX3X missense mutations profoundly disrupt RNA helicase activity, induce ectopic RNA-protein granules in neural progenitors and neurons, and impair translation. Together, these results uncover key mechanisms underlying DDX3X syndrome, and highlight aberrant RNA metabolism in the pathogenesis of neurodevelopmental disease.
Keywords: cortical development, corpus callosum agenesis, DDX3X, helicase, intellectual disability, polymicrogyria, autism, radial glial progenitor, RNA metabolism, stress granule
Graphical ABSTRACT
eTOC
Using human and mouse genetics this study identifies 107 mutations in DDX3X, demonstrating DDX3X is essential for cortical development. A striking correlation between the severity of clinical mutations and abnormal RNA metabolism highlights unappreciated mechanisms of DDX3X syndrome.
Introduction
Recent reports estimate that 1–3% of females with unexplained intellectual disability (ID) have de novo nonsense, frameshift, splice site or missense mutations in DDX3X (Scala et al., 2019; Snijders Blok et al., 2015; Wang et al., 2018). The approximately 70 reported individuals present with diverse neurologic phenotypes, including microcephaly, corpus callosum hypoplasia, ventricular enlargement, and epilepsy, which together define the DDX3X syndrome. De novo mutations in DDX3X have also been implicated in autism spectrum disorder (ASD) (Iossifov et al., 2014; RK et al., 2017; Ruzzo et al., 2019; Takata et al., 2018) and Toriello-Carey syndrome, a causally heterogeneous disorder characterized by severe ID, microcephaly, corpus callosum agenesis and various systemic (including cardiac) comorbidities (Dikow et al., 2017; Toriello et al., 2016). Additionally, DDX3X is mutated in several cancers and in some cases at identical residues as ID (Jiang et al., 2015; Jones et al., 2012; Pugh et al., 2012; Robinson et al., 2012). Early studies suggested that both DDX3X missense and nonsense mutations function in a haploinsufficient manner (Snijders Blok et al., 2015). However, this phenotype/genotype correlation may be modified with a larger, fully phenotyped patient cohort.
DDX3X encodes an RNA binding protein of the DEAD-box family (Sharma and Jankowsky, 2014). While broadly implicated in mRNA metabolism, DDX3X is best characterized as a translational regulator (Lai et al., 2008; Shih et al., 2008), particularly for mRNAs with highly structured 5′ untranslated regions (UTRs) (Calviello et al., 2019; Chen et al., 2018; Phung et al., 2019) and for repeat-associated non-AUG translation (Cheng et al., 2019; Linsalata et al., 2019). DDX3X is also a component of ribonucleoprotein granules composed of mRNA and protein (RNPs) (Hondele et al., 2019), including neuronal transport (Elvira et al., 2006; Kanai et al., 2004) and cytoplasmic stress granules (Markmiller et al., 2018). RNP granules are a pathological hallmark of many neurodegenerative diseases (Ramaswami et al., 2013), but it is unknown whether they are associated with DDX3X syndrome.
In animal models, Ddx3x is essential for cell viability and division, including in mouse blastocysts and Drosophila germline stem cells (Kotov et al., 2016; Li et al., 2014; Pek and Kai, 2011). Moreover, germline Ddx3x hemizygous male mouse embryos exhibit early lethality at embryonic day (E) 6.5, whereas epiblast-specific mutants are lethal at E11.5 with pronounced apoptosis, cell cycle defects, and aberrant neural tube closure. Notably, Ddx3x heterozygous female mice are viable (Chen et al., 2016). DDX3X is in a chromosomal region that can escape X inactivation though this is likely context specific (Carrel and Willard, 2005; Garieri et al., 2018).
DDX3X-associated brain malformations overwhelmingly affect the cerebral cortex. Embryonic cortical development occurs between E11.5 and E18.5 in mice, and gestational weeks (GW) 7 to 20 in humans. In mice, the main neural precursors are radial glial cells (RGCs), which divide in the ventricular zone (VZ) to self-renew or produce neurons and intermediate progenitors (IPs) (Taverna et al., 2014). IPs then generate neurons in the subventricular zone (SVZ). Deep and then upper layer excitatory neurons are generated sequentially, migrating into the cortical plate (CP) along the RGC basal process. Disorganized RGC basal processes, aberrant proliferation, and defective neuronal migration are hypothesized to cause polymicrogyria (PMG) (Jamuar and Walsh, 2015). Likewise, impaired neuron generation and survival underlie microcephaly. Moreover, ID- and ASD- associated genes are highly expressed in the human prenatal neocortex, implicating neurogenesis in their disease etiology (Polioudakis et al., 2019; Ruzzo et al., 2019). Although DDX3X function in cortical development has yet to be examined, it is essential for neurite outgrowth (Chen et al., 2016). These studies suggest that defects in neural progenitor differentiation and/or neuronal migration could underlie brain malformations associated with the DDX3X syndrome.
While DDX3X mutations have been linked to profound clinical deficits, the developmental and molecular mechanisms and genotype-phenotype correlations remain almost entirely unknown (Figure 1A). Here we show that deleterious DDX3X variants impair cerebral cortex development in mice and humans. We identify 107 individuals with DDX3X mutations, including 101 de novo, as well as 11 missense mutations which are associated with more severe clinical impairment, including PMG. Using mice, we demonstrate that Ddx3x depletion impairs neural progenitors thus reducing cortical neuron generation in vivo. Finally, we show that these aforementioned clinically severe DDX3X missense mutations exhibit reduced helicase activity, which is associated with aberrant RNP granule formation and perturbed translation. This work sheds light on fundamental developmental and cellular mechanisms that underlie the etiology of DDX3X syndrome.
Figure 1. Overview of study and predicted amino acid changes in DDX3X in our cohort.
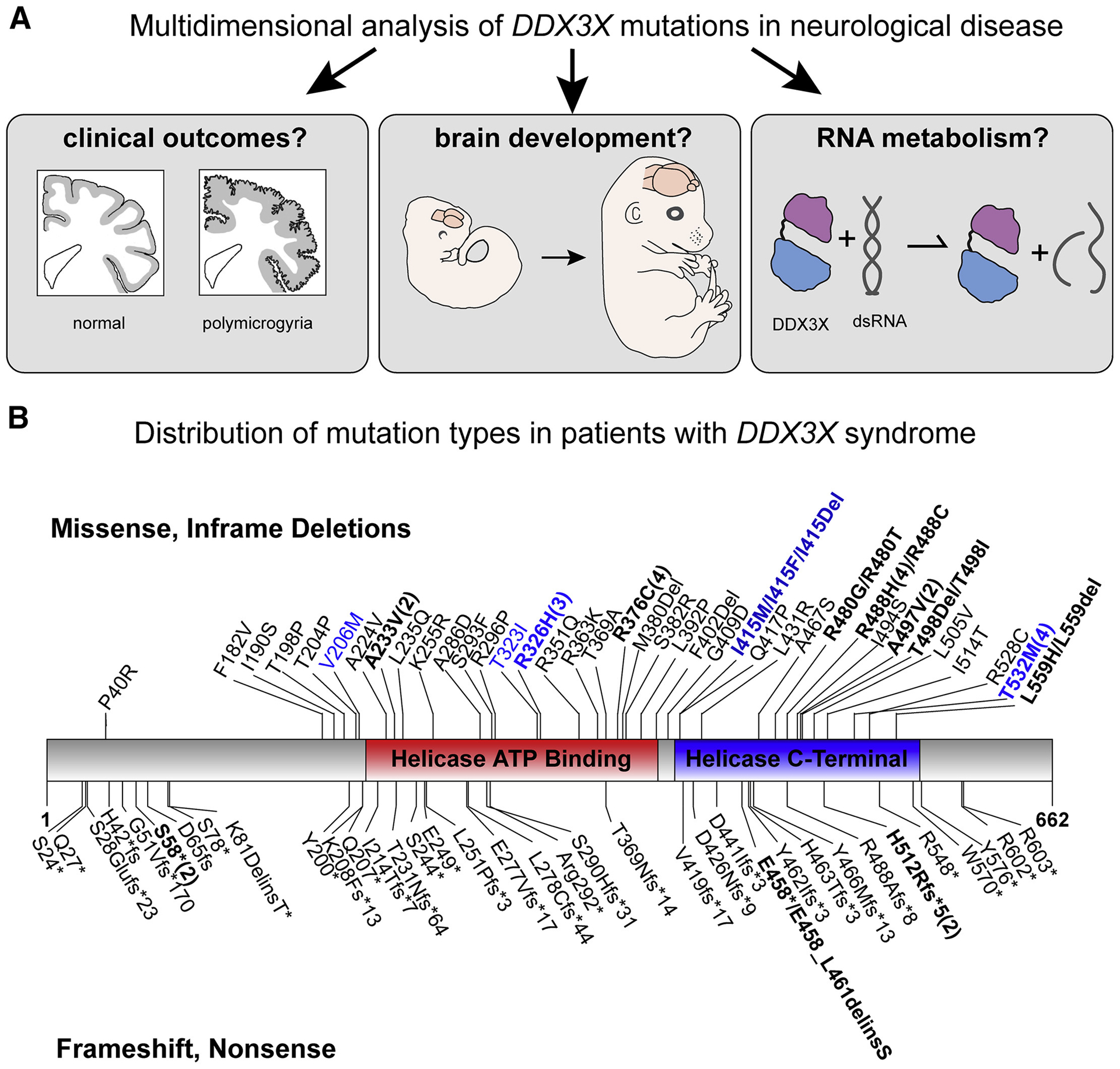
A, Overview of the three questions assessed in this study to understand the role of DDX3X de novo mutations in disease. B, Annotations include the following: Missense and in frame deletions (top); frameshift and nonsense mutations (bottom); Mutations found in patients with polymicrogyria (blue); Recurrent mutations (bolded and indicated by numbers in parentheses). Overall, mutations are enriched in the helicase domains at a rate higher than random chance (p=4.3*10−6 given 87 unique mutation positions including 12 splice sites). Not shown are the 12 splice site mutations (see Table S1).
Results
Identification of 107 individuals with DDX3X mutations and the associated clinical spectrum
To investigate the broad spectrum of clinical phenotypes associated with DDX3X mutations, we identified heterozygous de novo DDX3X mutations in 101 females, hemizygous, maternally inherited mutations in three males as in (Kellaris et al., 2018), and three female patients without parental testing (Table S1, N=107 total, 11 previously reported) (Figure 1B) (Snijders Blok et al., 2015).
To characterize clinical features, we obtained patients’ MRI scans (N=89) and medical records (N=106). 53 families completed the Vineland Adaptive Behavior Scales (VABS) a standardized measure of their child’s development (Sparrow, 2005). The Social Responsiveness Scale-II (SRS-II, N=49; (Constantino, 2013)) and the Social Communication Questionnaire (SQC, N=42; (Rutter M., 2003)) assessed risk for ASD and social impairment. The Child Behavior Checklist (CBC, N=49; (Achenbach, 2011)) assessed behavioral challenges (Figure S1). Of the 106 patients with clinical records, all had neurologic findings, the most common being ID followed by muscle tone abnormalities (91%, 85/93 patients; Table 1). Compared to neurotypical controls (100, <59, <15, 50), patients had a significant deviation in the mean score in all 3 exams (VABS: 56.4 (15), p<0.001; SRS-II 71.6 (12), p<0.001; SCQ 17.5 (7.6), p=0.03; CBC 58 (10), p<0.001). The SCQ scores suggest that 67% (28/42) of the cohort were above the “at risk” threshold for ASD and should be evaluated by a trained clinician. Of the three male individuals, two had VABS scores of 65 and 69, above average for our cohort but too small a sample size to assess statistically.
Table 1.
Clinical and Imaging Findings in Individuals with Mutations in DDX3X
| Non-PMG Individuals | PMG Individuals | Combined Total | |
|---|---|---|---|
| Neurologic | |||
| ID/DD | 95/95 (100%) | 11/11 (100%) | 106/106 (100%) |
| Nonverbal (in individuals above 5 years old) | 32/68 (47%) | 6/7 (86%) | 38/75 (51%) |
| Seizures | 15/83 (18%) | 2/10 (20%) | 17/93 (18%) |
| Microcephaly (≤3rd percentile) | 27/80 (34%) | 7/10 (70%) | 34/90 (38%) |
| Hypotonia | 52/82 (63%) | 2/11 (18%) | 54/93 (58%) |
| Hypertonia/Spasticity | 3/82 (4%) | 2/11 (18%) | 5/93 (5%) |
| Mixed Hypo and Hypertonia | 24/82 (29%) | 7/11 (64%) | 31/93 (33%) |
| Ophthalmologic | |||
| Coloboma | 2/82 (2%) | 2/10 (20%) | 4/92 (4%) |
| Strabismus | 22/82 (27%) | 3/10 (30%) | 25/92 (27%) |
| Congenital Cardiac Defects | 8/80 (10%) | 5/10 (50%) | 13/90 (14%) |
| Other Abnormalities | |||
| Precocious Puberty | 9/84 (11%) | 2/10 (20%) | 11/94 (12%) |
| Scoliosis | 12/84 (14%) | 3/10 (30%) | 15/94 (16%) |
| Non-PMG Individuals | PMG-spectrum Individuals | Combined Total | |
| Corpus Callosum Type | |||
| Complete ACC | 0/78 (0%) | 1/11 (9%) | 1/89 (1%) |
| Partial ACC | 1/78 (1%) | 3/11 (27%) | 4/89 (4%) |
| Diffusely thin | 13/78 (17%) | 1/11 (9%) | 14/89 (16%) |
| Thin posteriorly | 47/78 (60%) | 6/11 (55%) | 53/89 (60%) |
| Thick | 5/78 (7%) | 0/11 (0%) | 5/89 (6%) |
| Normal | 12/78 (15%) | 0/11 (0%) | 12/89 (13%) |
| Ventricles | |||
| Enlarged | 17/78 (22%) | 4/11 (36%) | 18/89 (24%) |
| Key-hole shaped temporal horns | 29/78 (37%) | 3/11 (27%) | 32/89 (36%) |
| Colpocephaly | 1/78 (1%) | 2/11 (18%) | 3/89 (3%) |
| Other | |||
| Small anterior commissure | 5/78 (6%) | 0/11 (0%) | 5/89 (6%) |
| Small pons | 6/78 (8%) | 5/11 (45%) | 11/89 (12%) |
| Small inferior vermis | 4/78 (5%) | 2/11 (18%) | 6/89 (7%) |
| Decreased white matter volume, (cortical) | 36/78 (46%) | 8/11 (72%) | 44/89 (49%) |
Bolded percentages indicate a p-value<0.05 and significant difference between a clinical feature found in Non-PMG and PMG individuals with the DDX3X mutation. Note: one patient had a low quality MRI scan and it could not be determined whether PMG was present or absent with the current data; therefore 106/107 patients are represented in the clinical data table.
The total numbers for each category may vary, reflecting the range of data available for each participant.
17 individuals (18%) had seizures and 34 (38%) had microcephaly (head circumference less than or equal to the 3rd percentile, Table 1). Cardiac malformations were observed (N=13 with 5 requiring surgical repair), linking DDX3X mutation to Toriello-Carey syndrome (Dikow et al., 2017). Three individuals presented with neuroblastoma, two incidentally, and all three individuals, after resection, were tumor-free at annual follow-ups. This observation is consistent with findings that DDX3X is mutated in several cancers (Jiang et al., 2015; Jones et al., 2012; Pugh et al., 2012; Robinson et al., 2012). These data suggest the full clinical spectrum of DDX3X syndrome includes involvement outside the CNS (Beal et al., 2019).
DDX3X mutations impair human brain development
To identify notable brain anatomic disruptions in our DDX3X cohort, we reviewed brain MRI scans from 89 patients (Figure 2 and Table 1). 77/89 (87%) had a corpus callosum malformation ranging from complete to more mild agenesis (Table 1, Figure 2A–C). 11/89 (12%) had PMG (presence of abnormally dense and small gyri in the cerebral cortex) (Figures 2D–G; 1B, blue), or dysgyria (abnormal gyrification patterns which may appear different on scans depending on age and myelination state of myelination) (Takanashi and Barkovich, 2003). Other noted features were globally diminished white matter volume (44/89, 49%) and lateral “key-hole-shaped” ventricles with characteristic enlargement in the temporal horns (32/89, 36%; Figure 2B1). This recognizable feature correlates closely with diminished size of the ventral aspect of the cingulum bundles (Nakata et al., 2009) (Figure 2A1–C1). Thus, in the vast majority of DDX3X patients, mutations result in a range of brain anatomy changes, consistent with disrupted brain development.
Figure 2. Common brain imaging findings in DDX3X cohort.
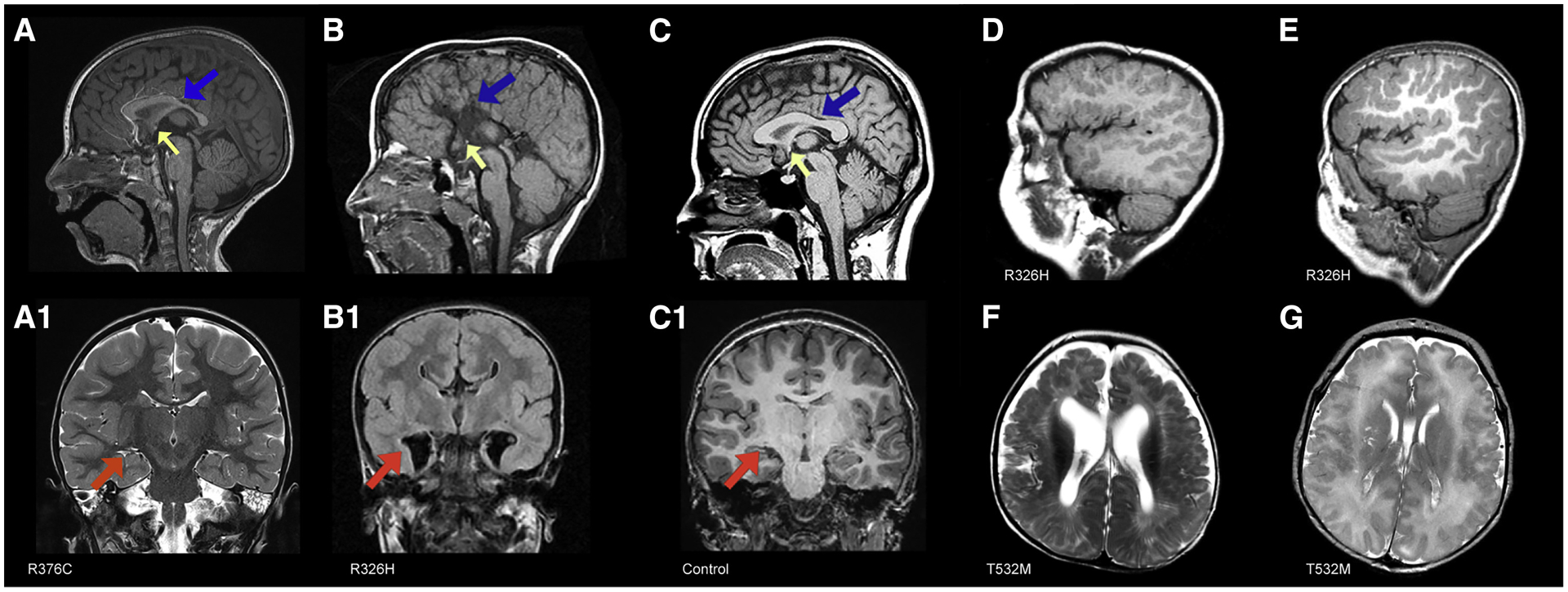
A, A1 Patient 2612–0 (R376C, mild clinical impairments) at 2 years of age. A, Sagittal image shows hypoplastic corpus callosum with a short, thinned posterior body (blue arrow) and a hypoplastic to absent splenium; small anterior commissure (yellow arrow); inferior genu and rostrum are absent. A1, Coronal image shows mildly diminished white matter volume, normal ventricle size, including temporal horns (red arrow). B, B1, Patient 1090–0 (R326H) at 7 months. B, Sagittal image shows complete agenesis of the corpus callosum and hippocampal commissure (blue arrow) and a small anterior commissure (yellow arrow). B1 Colpocephaly with enlarged keyhole-shaped temporal horns (red arrow). C, C1, Normal MRIs for comparison. C, Mid-sagittal T1-weighted image showing a normal sized corpus callosum (blue arrow) and anterior commissure (yellow arrow). C1, Coronal T1-weighted image shows normal cortical thickness and gyration, normal sized ventricle bodies and temporal horns (red arrow). Saggital images from patients 1090–0 (D) at 7 months and 3437–0 (E) at 4 years (both R326H) with extensive bilateral frontal PMG. Axial images from patients 3072–0 (F) at 6 months and 1954–0 (G) at 3 days of age, (both T532M) and bilateral perisylvian and frontal PMG and enlarged ventricles.
Location and type of mutation predicts imaging features and clinical outcomes
We assessed whether the location and type of DDX3X mutation found in patients correlated with their clinical impairment. 57 individuals had missense mutations or in-frame deletions, 38 had nonsense or frameshift mutations (LoF), and 12 had splice site mutations (Figure 1B, Table S1). Patients with missense mutations were significantly more likely to have severe phenotypes, such as PMG (p<0.001) compared to the cohort overall. In contrast, not a single patient with a frameshift or nonsense LoF mutation presented with PMG (p<0.001).
In the cohort, we identified 15 nucleotides that were repeatedly mutated (bolded in Figure 1B); and the probability of each individual loci having recurrent mutations, given the number of overall mutations, was low and statistically significant for each grouping. This includes six mutations at R488 (p<1×10−15, R488C, R488Afs, and four R488H), four at R376 (Arg to Cys, p<1×10−9), four at T532M (p<1×10−10), three at R326 (p<1×10−7), three at I415 (p<1×10−7), and two patients each with recurrent mutations at 10 additional loci (p<0.001).
Of the loci with recurrent mutations, qualitative analysis suggests they result in very similar phenotypes. For example, three of four patients with a T532M mutation had PMG or dysgyria (one patient had a low quality scan). Three patients with the same de novo mutation at R326H exhibited a similarly severe phenotype, notable for PMG, and severe developmental delay. Conversely, the four patients with the R376C mutation had no cortical malformations, thin but visible corpus callosal body and splenium; correspondingly with only mild cognitive challenges. Non-neurologic phenotypes also clustered; all three patients with I415 mutations had cardiac malformations (atrial septal defect, ASD; patent foramen ovale, PFO; patent ductus arteriosus, PDA; persistent pulmonary hypertension of the newborn, PPHN).
Of the 11 patients with PMG (Figure 1B, blue), ten had missense mutations while one had an in-frame single amino acid deletion. Nine mutations were at three recurrent loci (T532, I415, R326) demonstrating an association with PMG (p=0.01). Seven of ten (70%) individuals with PMG had microcephaly, whereas only 27 of 80 individuals (34%) without PMG had microcephaly (p<0.05), suggesting a mechanistic link between microcephaly and PMG in this cohort. In addition, patients with PMG were more delayed developmentally, with an average VABS of 43.8 vs. 57.5 in the non-PMG cohort (p<0.05). 5/13 patients with cardiac findings also had PMG, disproportionate to the number of PMG patients in the overall cohort (p<0.05). PMG patients were also more likely to have partial agenesis versus a thin corpus callosum (27% vs 1% p<0.05). These cumulative data link the severity of clinical features and mutation type, indicating that a subset of missense mutations function in a dominant manner.
Ddx3x is expressed in the developing mouse neocortex
These cortical and neurological findings suggest DDX3X plays a central role in development of the cerebral cortex. DDX3X is expressed across human fetal brain regions from post conceptual weeks (PCW) 11–22 (Miller et al., 2014), and in human fetal cortical progenitors and neurons (Nowakowski et al., 2017). Likewise, in the developing mouse neocortex, Ddx3x is highly expressed in progenitors and neurons (Ayoub et al., 2011; Molyneaux et al., 2015). DDX3X is highly conserved, with 98.6% amino acid identity between mice and humans. This suggests that mice are a suitable model for investigating DDX3X LoF impacts on cortical development (Figure 3A).
Figure 3. Ddx3x is expressed in progenitors and neurons in the embryonic mouse cortex and in vivo disruption alters neurogenesis.
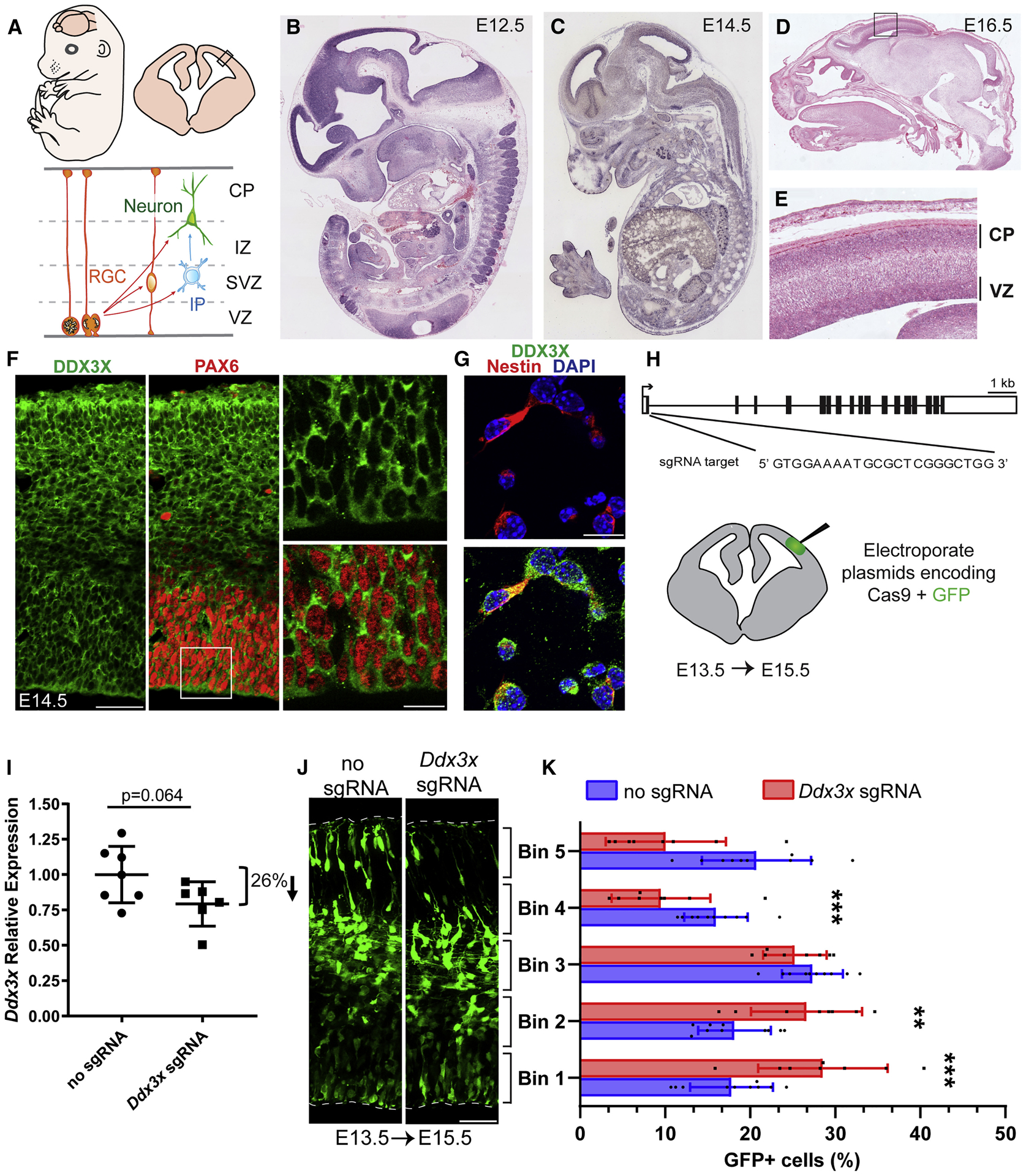
A, Top, cartoon representation of a mouse embryo and coronal cortical section. Bottom, cartoon representation of boxed region above depicting major embryonic cortical cell types examined in this study including radial glial progenitor cells (RGCs), intermediate progenitors (IPs), and neurons. The ventricular zone (VZ), subventricular zone (SVZ), intermediate zone (IZ), and cortical plate (CP) are indicated. B-E, Ddx3x In situ hybridization in sagittal sections of E12.5 (B), E14.5 (C), and E16.5 (D,E) embryos. Box in D is magnified in E. F, G, Immunofluorescence of E14.5 cortical sections (F) co-stained for DDX3X (green), the RGC marker PAX6 (red), and of E14.5 primary cells (G) co-stained for the RGC marker Nestin (red) and DAPI (blue). H, Schematic of mouse Ddx3x gene structure with exons (boxes) and introns (thin lines). Ddx3x sgRNA targets Exon 1 at the indicated sequence. Bottom, schematic of coronal section depicting electroporated region. I, Validation of Ddx3x mRNA knockdown in FACS purified GFP+ cells from E15.5 brains electroporated at E13.5. J, Representative E15.5 coronal sections, electroporated at E13.5, with GFP and either no sgRNA or Ddx3x sgRNA, immunostained for GFP (green). Dotted lines represent ventricular and pial surfaces, and brackets delineate equivalently sized bins. K, Quantification of distribution of GFP-positive cells with Bin1 at the ventricle and Bin5 at the pia. Scale bars: 500 μm and 50 μm (F) and 15 μm (G) and 50 μm (J). Error bars=SD.
We first assessed Ddx3x spatial and temporal expression in mouse cortical development by in situ hybridization. In E12.5 and E14.5 sagittal sections, Ddx3x mRNA is ubiquitous, but especially enriched in the developing cortex (Visel et al., 2004) (Figures 3B, C). Ddx3x expression was initially enriched in the VZ (Visel et al., 2004), and by E16.5 was also evident in the CP and SVZ (Figures 3B–E, S2A–D). By postnatal day (P) 2, Ddx3x mRNA was highly expressed throughout the hippocampus (CA1-CA3 and subiculum) and minimally in the corpus callosum (Figures S2D).
Consistent with its RNA expression, DDX3X protein was expressed in all embryonic cortical layers (Figure 3F). In E14.5 primary cells, DDX3X was evident in both Nestin-positive RGCs and TUJ1-positive neurons (Figure 3G, S2E, S2F). In P5 brains we detected DDX3X expression in neurons, astrocytes, oligodendrocytes, microglia and ependymal cells (Figures S2G–P). Further, DDX3X localized to puncta within the cytoplasm (Figures 3F, G, S2E, S2F), consistent with previous reports (Lai et al., 2008). Taken together with in situ and genomic expression data, this demonstrates DDX3X is expressed throughout cortical development in both neurons and progenitors.
In vivo depletion of Ddx3x alters corticogenesis
We next interrogated the requirement of Ddx3x in embryonic corticogenesis using CRISPR/Cas9 to deplete Ddx3x in vivo. We designed a short guide RNA (sgRNA) against Exon 1 of Ddx3x near the the start codon, in order to induce early frameshift LoF mutations (Figure 3H). 72 hour expression of Ddx3x sgRNA + Cas9 in Neuro2A (N2A) cells significantly reduced Ddx3x mRNA and protein levels, relative to control (Cas9 without sgRNA) (Figures S3A–C). These data confirm the efficacy of these sgRNAs in effectively depleting Ddx3x levels.
We next used in utero electroporation to deliver Ddx3x sgRNA and Cas9 along with GFP to the developing cortex, to deplete Ddx3x expression in both RGCs and their progeny (Saito, 2006) (Figure 3H). We targeted E14.5, when Ddx3x expression is robust and progenitors are neurogenic, and harvested brains at E17.5, allowing time for newborn neurons to migrate to the CP. We detected an average ~25% reduction in DDX3X immunostaining in GFP-positive cells expressing sgRNA compared to control (Figures S3D, E). To assess the impact of Ddx3x depletion in vivo, we quantified the distribution of GFP-positive cells within cortical bins (Figure S3F). While GFP-positive cells were evenly distributed in control cortices, Ddx3x sgRNA significantly altered their distribution resulting in 1.4-fold more cells in the VZ/SVZ (bins 1 and 2) and a 1.9-fold fewer cells in the CP (bins 4 and 5, Figure S3G).
We also independently assessed Ddx3x requirements using Ddx3x siRNAs validated in N2A cells (Figures S3H, I). We in utero electroporated either scrambled or Ddx3x siRNAs at E14.5 and harvested E17.5 brains. Ddx3x siRNA-mediated depletion caused more cells in VZ/SVZ and less in the CP compared to a scrambled control, thus phenocopying the CRISPR experiments (Figures S3J, K). Taken together, these findings demonstrate that a three-day transient depletion of Ddx3x alters distribution of newborn cells in the neocortex, suggesting a role for DDX3X during neurogenesis.
In vivo depletion of Ddx3x reduces neuronal differentiation
The aberrant cellular distribution following Ddx3x depletion could result from defective progenitor proliferation, cell death in the CP, and/or defects in neuronal migration. To investigate these possibilities, we employed CRISPR-mediated Ddx3x knockdown at E13.5 with analysis at E15.5 (Figure 3H). A two-day paradigm at these stages was used to pinpoint progenitor cellular defects. We used FACS-purified GFP+ cells from E15.5 electroporated brains and determined overall Ddx3x mRNA levels were ~26% reduced in gRNA relative to control (Figure 3I). This may underestimate the extent of DDX3X depletion within individual cells, given heterogeneity associated with CRISPR. Quantification of GFP-positive cell distribution at E15.5 (Figures 3J and K) showed significantly more cells in the VZ/SVZ and fewer in the CP. This phenotype is consistent with the three-day CRISPR and siRNA-mediated experiments and altogether, extends the developmental requirement of Ddx3x to include E13.5-E17.5.
We then assessed the impact of Ddx3x depletion upon progenitors and newborn neurons. We first quantified the fraction of GFP-positive cells which co-expressed either PAX6 or TBR2 transcription factors, to mark RGCs and IPs, respectively. Relative to control, Ddx3x depletion led to a significant 1.5-fold increase in RGCs and a 1.4-fold increase in IPs (Figures 4A–D). By measuring co-expression of GFP with NEUROD2, we quantified a significant 1.2-fold decrease in neurons (Figures 4E, F). NEUROD2 also has low expression in IPs and thus this may under-reflect the neuronal phenotype. Since reduced neurons were concomitant with increased progenitors, this suggests Ddx3x controls the normal balance of these cells. We noted equivalently low levels of apoptosis by cleaved-Caspase3 (CC3) staining in mutant and control brains, suggesting this imbalance is not due to selective neuronal death (Figure 4G, H). Together, these results indicate that Ddx3x is required for proper generation of neurons from RGCs and/or IPs.
Figure 4. Ddx3x is required for neuron generation in vivo.
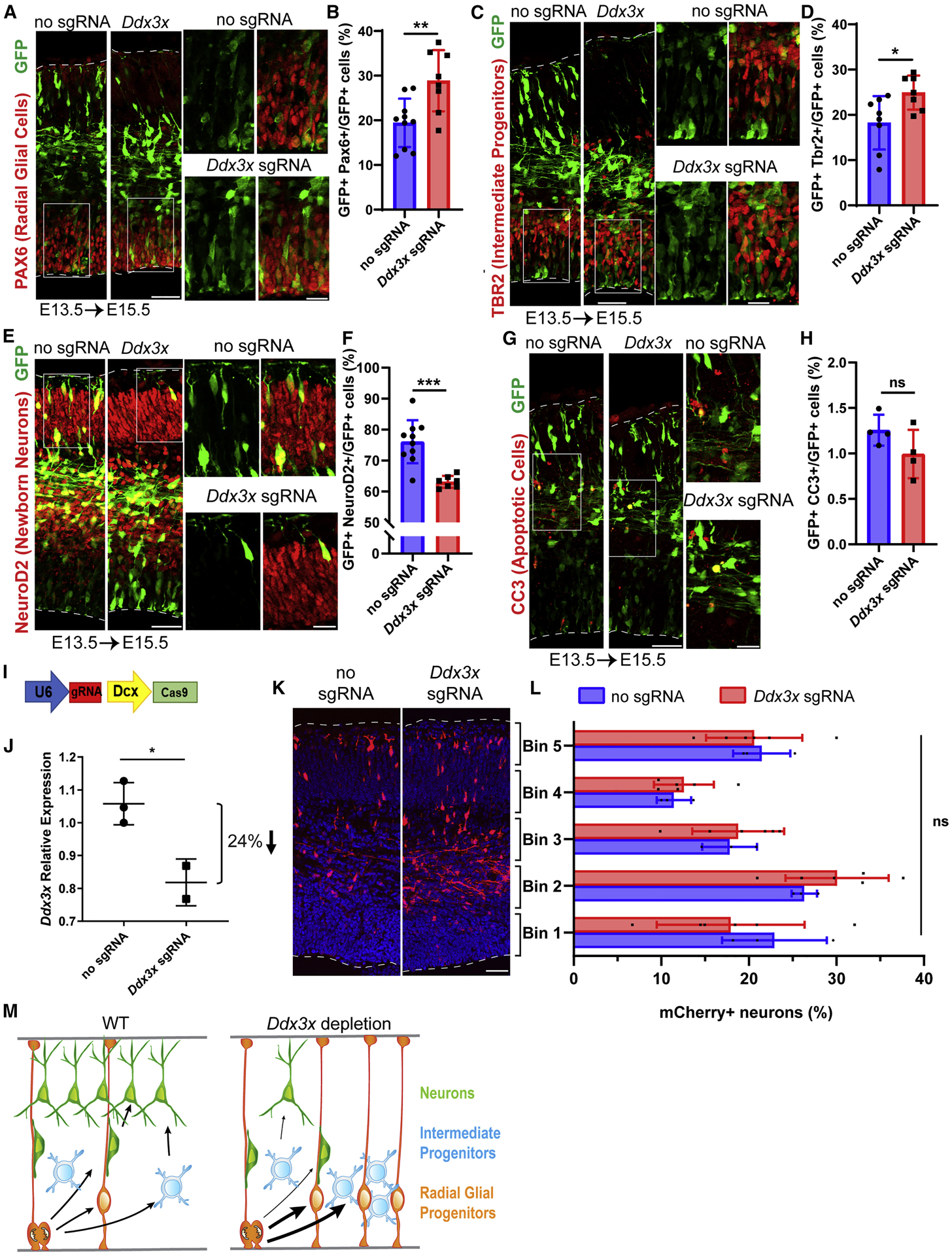
A, C, E, G Sections of E15.5 cortices, electroporated at E13.5, and co-stained with antibodies against GFP (green) and PAX6 (red) (A); TBR2 (red) (C), NeuroD2 (red) (E) and CC3 (red) (G). Boxed regions are shown at higher magnification on the right. B, D, F, H Quantification of percentage of GFP-positive cells expressing PAX6 (B), TBR2 (D), NEUROD2 (F), and CC3 (H). I, Schematic diagram of CRISPR/Cas9 targeting vector under control of neuron-specific Dcx promoter. J, Validation of Ddx3x mRNA knockdown in FACS purified mCherry+ cells from E17.5 brains electroporated at E14.5. K, Representative coronal sections of E17.5 brains coelectroporated at E14.5 with pDcx-mCherry and either pX330-Dcx-Cas9 (no sgRNA) or pX330Dcx-Cas9 plus Ddx3x sgRNA, stained with anti-RFP (red). Dotted lines represent ventricular and pial surfaces, and brackets on the right refer to equivalently sized bins. L, Quantitation of mCherry-positive cells distribution with Bin1 at the ventricle and Bin5 at the pia. M, Schematic model summarizing finding that Ddx3x LoF impairs neuron generation, associated with increased RGCs and IPs. Scale bars: 50 μm, low magnification, 15 μm, high magnification (A,C,E,G, K). Error bars=SD.
After neurons are generated in the VZ/SVZ, they migrate radially into the CP. As Ddx3x is also expressed in neurons, we monitored their migration following Ddx3x depletion. To assess Ddx3x autonomous requirements in neurons, we expressed both mCherry and Cas9 under control of the Dcx promoter, enabling Ddx3x depletion only in newborn neurons (Figure 4I)(Franco et al., 2011). In utero electroporations were performed at E14.5 and FACS isolated mCherry+ neurons at E17.5 showed ~24% reduction in Ddx3x mRNA levels in Ddx3x sgRNA relative to control (Figure 4J). This degree of knockdown is similar to that we observed with ubiquitous CRISPR (Figure 3I). Quantification of mCherry+ neuron distribution within cortical bins revealed slightly more neurons in proliferative zones, but this was statistically insignificant (Figures 4K, L). We also probed longer term Ddx3x requirements in neurons, by electroporating E14.5 brains and assessing neuronal distribution at P2. Again, the overall distribution in the Ddx3x sgRNA condition was subtly shifted towards the proliferative zones but was statistically insignificant (Figures S4A, B). Taken altogether, these results establish that during embryonic cortical development, DDX3X primarily functions in progenitors to promote neuronal differentiation (Figure 4M).
DDX3X has been reported to directly influence canonical Wnt signaling (Cruciat et al., 2013; Snijders Blok et al., 2015), which has roles in cortical progenitors and migrating neurons (Bocchi et al., 2017; Harrison-Uy and Pleasure, 2012). To investigate if Ddx3x depletion impacts Wnt signaling, we measured expression of Wnt targets in transfected N2A cells or electroporated GFP+ progenitors and neurons (FACs purified). Some but not all Wnt target transcripts were mis-expressed following Ddx3x knockdown (Figures S5A, B). Given the central role of Wnt in neurogenesis, these changes may also reflect differences in progenitor and neuron number.
To independently assess Wnt, we electroporated E14.5 brains with a canonical Wnt reporter along with scrambled or Ddx3x siRNAs (Figure S5C) (Ferrer-Vaquer et al., 2010). At E17.5, there was no significant difference in the fraction of Wnt reporter-positive cells between either condition (Figure S5D). Taken together, these data suggest there are both Ddx3xdependent and independent Wnt targets, and thus Wnt signaling may be indirectly controlled by Ddx3x.
Given canonical roles of DDX3X in translation, we measured global translation in N2A cells transfected with Ddx3x siRNAs (Figure S6A). Using FUNCAT (FlUorescent Non-Canonical Amino acid Tagging), we monitored the incorporation of a methionine analog, L-azidohomoalaine (AHA), into nascent peptides (Hinz et al., 2013). As controls, we used the translation elongation inhibitor, emetine, or omitted AHA. Notably, by either fluorescence or western analyses, translation levels were comparable between scrambled and Ddx3x knockdown conditions (Figures S6B–D). This finding is consistent with previous DDX3X LoF findings in immortalized cells (Calviello et al., 2019; Chen et al., 2018; Lai et al., 2008; Phung et al., 2019). These data strongly indicate that global translation is unaffected by Ddx3x depletion in neural cells, though it remains possible that DDX3X controls translation of individual targets during neurogenesis.
DDX3X missense mutations impair helicase activity that correlates with disease severity
The mouse studies establish essential requirements of Ddx3x in cortical development and indicate mechanisms by which DDX3X LoF may drive disease etiology. We next investigated the biochemical impact of DDX3X missense mutations, which comprise half of our clinical cohort and fall almost entirely within the two helicase domains. We hypothesized they may directly impair DDX3X helicase activity, which could correlate with disease severity. We therefore measured the ability of purified DDX3X helicase-domain mutants to unwind RNA duplexes (Figure 5A) (Floor et al., 2016b). We selected recurrent DDX3X missense mutations associated with (1) less severe clinical outcomes (R376C, I514T, A233V), (2) severe impairment (R475G), or (3) severe clinical impairment plus PMG (T323I, R326H, I415del, T532M). Duplex unwinding rate by all mutant proteins was lower than that of WT by varying degrees (Figure 5B). Notably, we found mild to moderate slowing in the rate of unwinding with mutations from less severely affected individuals (R376C, I514T, A233V), but a complete loss of unwinding activity in all four PMG-associated mutations. This indicates that the most severe DDX3X missense mutants lack biochemical activity, while less severe missense mutants retain some activity.
Figure 5. DDX3X missense mutants exhibit disrupted helicase activity.
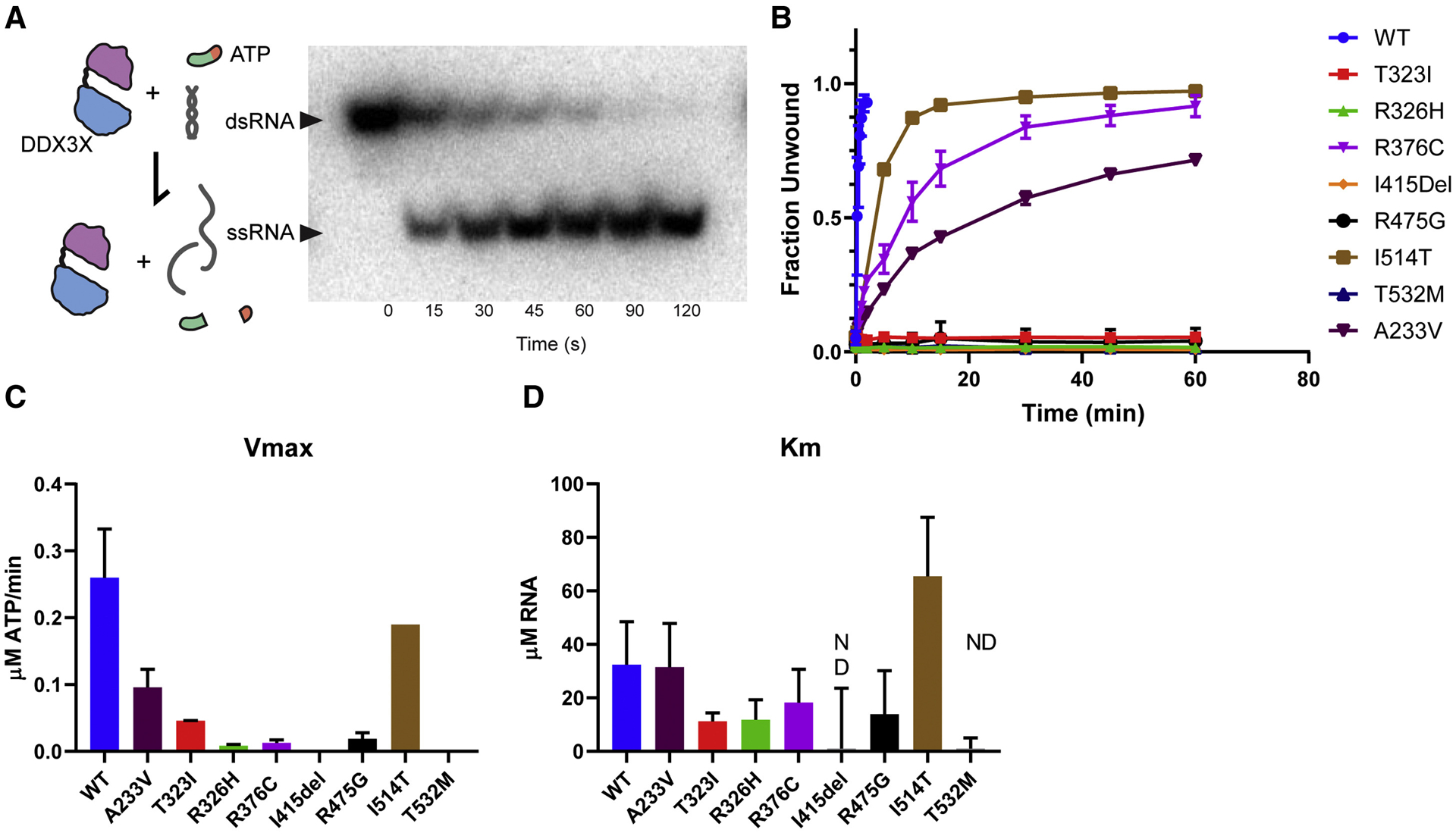
A, Left, Diagram of DDX3X activity tested in this assay. ATP hydrolysis is necessary for initial binding and release of RNA, but not for RNA unwinding. Right, Non-denaturing gel depicting time course of helicase assay in which amounts of dsRNA (not unwound by DDX3X) and ssRNA (unwinding by DDX3X) are measured. B, Unwinding assay for WT, A233V, T323I, R326H, R376C, I415del, R475G, I514T, and T532 DDX3X. C, D, Graphs depicting Vmax (C) and Km (D). Note, I415del and T532M had unwinding curves which did not vary within the range of tested RNA concentrations (0–40 uM) so Km was not determined (n.d.). The majority of mutants had lower Km (indicating higher affinity for RNA) than WT, with the exception of I514T. Error bars=SD.
To further define the biochemical defect in DDX3X missense mutants, we measured vmax and Km for RNA using ATP hydrolysis assays. RNA stimulates ATP hydrolysis by DEAD-box proteins, and thus, changing the RNA concentration modulates ATP hydrolysis kinetics (Lorsch and Herschlag, 1999). Overall vmax and Km were diminished in DDX3X mutants (Figures 5C, D), with stronger and more consistent effects for vmax. Taken together, these assays suggest that DDX3X missense mutations result in a range of biochemical activity loss, which correlates with clinical disease severity.
DDX3X missense mutations induce RNA-protein cytoplasmic aggregates
These biochemical experiments demonstrate that DDX3X missense mutations have strongly reduced helicase activity. This raises the question as to how impaired helicase activity affects RNA metabolism in progenitors and neurons. DDX3X is a component of RNP granules, including stress granules (Elvira et al., 2006; Kanai et al., 2004; Lai et al., 2008; Markmiller et al., 2018). DDX3X can also promote stress granule formation (Shih et al., 2012) and cancer-associated mutations do so potently (Valentin-Vega et al., 2016). We therefore postulated that impaired helicase activity of mutant DDX3X could be associated with aberrant formation of RNP granules. To test this hypothesis, we first sought to generate stable cells that endogenously express DDX3X missense mutations. However, we were only able to generate DDX3X missense mutations in approximately 30% of N2A cells at the population level, and this fraction steadily decreased over time. This suggests that some DDX3X missense mutations may be toxic or impair the rate of cell proliferation.
As an alternative approach, we investigated DDX3X missense mutations by exogenously expressing them, as done previously (Huang et al., 2019; Oh et al., 2016; Valentin-Vega et al., 2016). We investigated a clinically mild recurrent mutation (R376C), a clinically severe recurrent mutation (R326H), three previously reported mutations (R475G, R534H, P568L) (Snijders Blok et al., 2015), and a medulloblastoma-associated mutation shown to induce stress granules (G325E) (Valentin-Vega et al., 2016). These mutations were all located in the helicase/RNA binding domains, exhibited biochemical defects (Figure 5B) and had predicted functional impacts based on Polyphen-2 scoring (Table S1).
GFP-tagged human WT-DDX3X and DDX3X mutants were transiently expressed at equivalent low levels in N2A cells and DDX3X subcellular localization was monitored after 24 hours (Figures 6A, B, S6E). WT-DDX3X was primarily diffuse cytoplasmically with GFP-positive granules evident in ~7% of cells, whereas severe missense mutants and G325E formed granules in ~20% of cells. In contrast, the recurrent clinically mild R376C mutant did not significantly form DDX3X granules. Similar phenotypes were evident with transient expression of R475G DDX3X-GFP in E14.5 primary cortical cells. While WT-DDX3X was primarily diffuse and cytoplasmic, R475G induced granule formation 3-fold more frequently (Figure 6C, D), evident in both progenitors and neurons (Figure 6F). These data indicate that severe missense mutations disrupt DDX3X subcellular localization, which correlates with impaired DDX3X helicase activity and clinical severity.
Figure 6. DDX3X missense mutations induce ectopic RNP granules in neural progenitors.
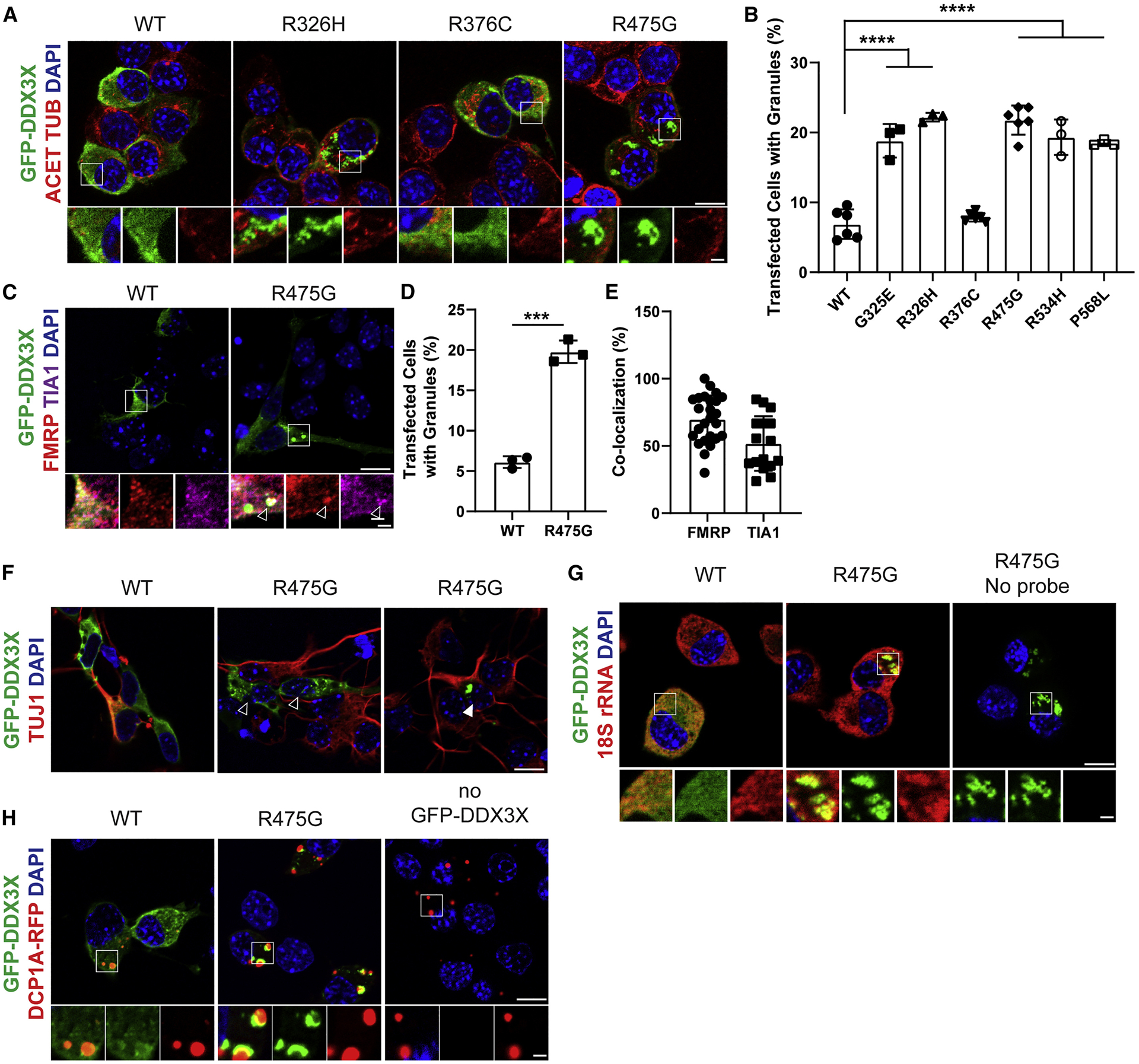
A, Images of N2A cells transfected for 24 hrs with WT or mutant DDX3X-GFP and immunostained for GFP (green), Acetylated-TUBULIN (red), and DAPI (blue). Below, high magnification images from boxed regions. B, Quantification of percentage of N2A cells containing WT or mutant DDX3X-GFP granules. C, Primary cortical cells transfected for 24 hrs with WT or R475G DDX3X-GFP (green) and co-stained for FMRP (red) and TIA1 (magenta), with granule co-localization granules (arrowhead). D, Quantification of percentage of primary cortical cells containing DDX3X-GFP granules. E, Quantification of DDX3X-granules co-localized with RNA-binding proteins TIA1 or FMRP. F, Primary cortical cells expressing either WT or R475G DDX3X-GFP (green) and stained with TUJ1 (red). Both TUJ1- progenitors (empty arrowhead) and TUJ1+ neurons (filled arrowhead) contain DDX3X granules. G, N2A cells transfected with DDX3X-GFP (green) and probed for 18S rRNA with smFISH probes (red). H, N2A cells transfected with DDX3X-GFP (green) and DCP1A-RFP (red) to mark P-bodies. Scale bars, 10 μm (low magnification) and 2 μm (high magnification) (A, C, F, G, H). Error bars=SD.
We then used both N2A cells and primary cortical cells to characterize DDX3X granules. Approximately 70% and 40% of DDX3X granules co-localized with the stress granule markers, FMRP and TIA1, respectively (Figures 6C, E) indicating they have some features of stress granules. DDX3X granules also co-localized with 18s rRNA indicating these are RNP granules (Figure 6G). Finally, we asked whether DDX3X granules are processing bodies (P-bodies), cytoplasmic RNPs involved in mRNA decay. Using the P-body marker DCP1A (Decapping MRNA1A), we found DCP1A-RFP and DDX3X-GFP granules were often adjacent but did not co-localize (Figure 6H), suggesting that DDX3X granules are not P-bodies. Together, these data indicate that DDX3X missense mutations induce formation of cytoplasmic RNP granules with some features of stress granules.
DDX3X missense mutations alter translation of select mRNA targets
We next sought to determine the impact of DDX3X missense mutations upon translation in primary cortical cells. First, we monitored global protein synthesis using a puromycylation assay, in which the tRNA analog puromycin is incorporated into nascent peptides (Schmidt et al., 2009). Primary cortical cells transfected with WT or R475G DDX3X-GFP, were pulsed with O-propragyl-puromycin and puromycin incorporation was detected (Figure S6F). Non-transfected and WT-DDX3X-expressing cells had diffuse and evenly distributed cytoplasmic puromycin signal, which was blocked by anisomycin, a translational inhibitor. A subset of R475G cells showed focal accumulation of puromycin in granules. These findings were also verified with FUNCAT assays, which detected some cells with AHA signal at granules (Figure S6G, arrowheads), as well as examples, even within the same cell, where this was not the case. These data suggest that DDX3X RNP granules are heterogenous and that, unlike canonical stress granules, they can contain either newly synthesized proteins and/or stalled polysomes (Graber et al., 2013).
We also quantified FUNCAT signal across all cells regardless of granule status. While aniosomycin abolished AHA incorporation, the R326H severe mutation and WT DDX3X showed similar translation levels (Figure S6G–H). This finding is consistent with previous reports and similar to Ddx3x depletion (Figure S6B–D) (Calviello et al., 2019; Lai et al., 2008). Hence, perturbing DDX3X either through depletion or missense mutations does not globally impair translation.
While global translation levels were unaffected in missense DDX3X-expressing cells, it remains possible that missense DDX3X impairs translation of specific RNAs. Indeed, DDX3X is implicated in translation of mRNAs which contain highly structured 5′ UTRs and/or high GC content (Calviello et al., 2019; Chen et al., 2018; Phung et al., 2019). We thus measured if DDX3X missense mutations impair translation of previously identified DDX3X-sensitive 5′ UTRs (ATF5, CCNE1, RPLP1, PRKRA and RAC1) or controls predicted to be insensitive to DDX3X LoF or depletion (SIKE1 and SRSF5) (Calviello et al., 2019; Oh et al., 2016). HEK 293T cells were transfected with DDX3X siRNA along with either DDX3X WT, R326H or R376C (rendered siRNA-resistant via synonymous mutations in the seed sequence) (Figure 7A). Transfections were optimized so that DDX3X variant and WT levels were similar and we performed in vitro translation assays with cell lysates. DDX3X missense mutations broadly decreased translation of DDX3X-sensitive reporters, with the R326H mutant showing a more severe impact than R376C (Figures 7B–C). Thus, the expression of DDX3X variants directly affects translation of a subset of mRNAs, in a manner consistent with the severity of clinical phenotype, unwinding activity in vitro, and cell biological phenotype (Figure 7D).
Figure 7. DDX3X missense mutations alter translation of select targets.
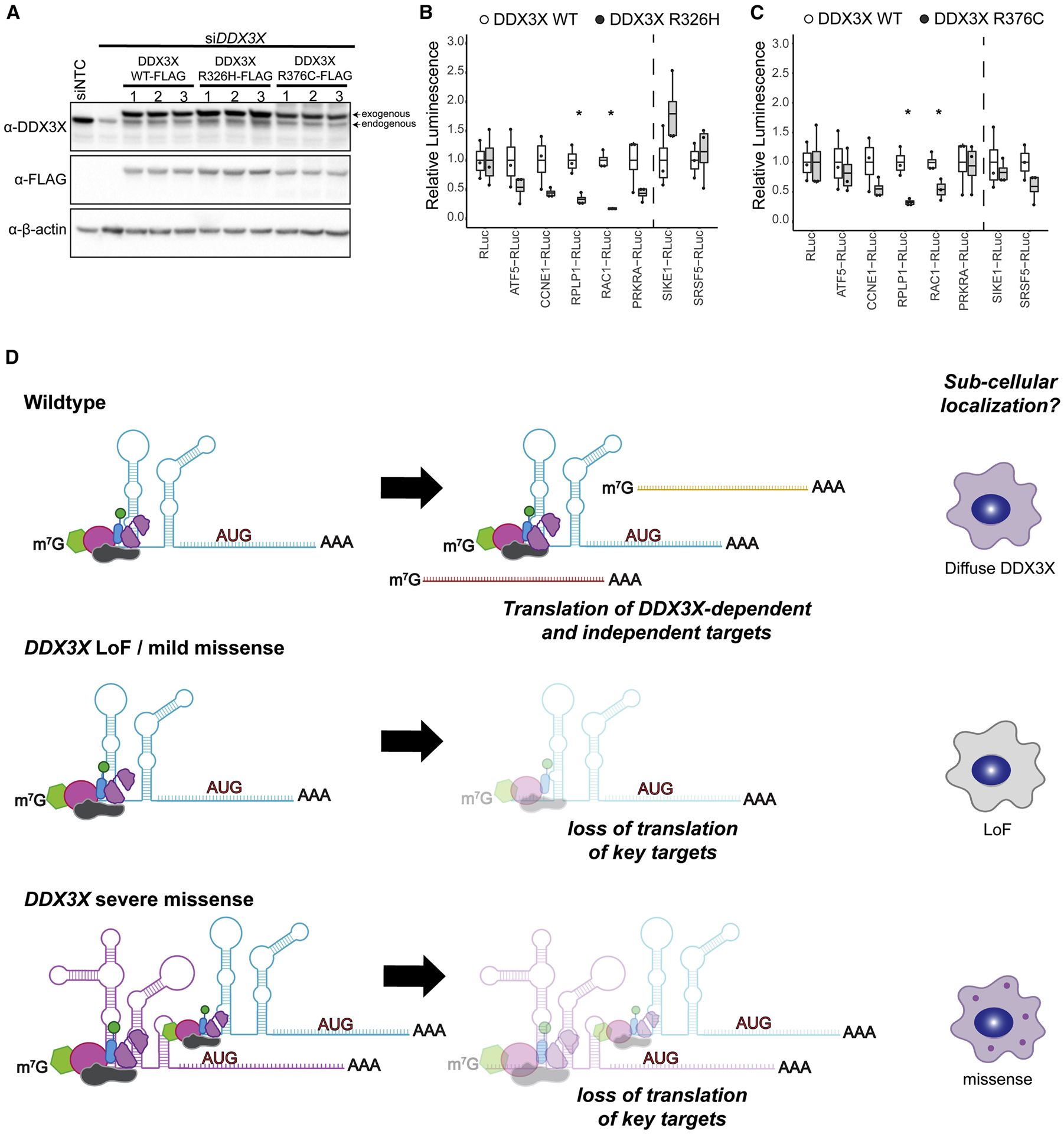
A, Western blots depicting DDX3X levels (WT, R326H, or R376C) with simultaneous knockdown of endogenous DDX3X. Endogenous and FLAG-tagged DDX3X is detected. B, C, Translation of in vitro transcribed reporter RNAs in lysates expressing either DDX3X WT, R326H (B) or R376C (C) Signal was normalized to WT. Predicted DDX3X sensitive reporters (ATF5, CCNE1, RPLP1, PRKRA and RAC1) show robust decreases in translation, while control transcripts (SIKE1, SRSF5) right of dotted line demonstrate either no change or modest increases. Box and whisker plot with bars indicating data range, box at upper and lower quartiles, and line at the median. D, Proposed model for mechanism of DDX3X mutants, based upon in vitro biochemical and cell biology studies. Mild missense or DDX3X LoF are impair translation of some targets but do not induce granule formation. Severe DDX3X missense mutations show impaired RNA release and helicase activity, and altered translation of DDX3X-dependent targets. This results in sequestration of RNAs and RNA binding proteins and formation of aberrant RNP granules. Created with BioRender.com.
Discussion
DDX3X is a frequently mutated gene in both ID and cancer, yet the mechanism by which DDX3X mutations impair brain development have not yet been defined. Here, we report the largest cohort of patients with mutations in DDX3X (n=107; 101 confirmed de novo) to date, highlighting corpus callosum abnormalities and PMG as two major brain anatomic phenotypes characteristic of DDX3X syndrome, with PMG strongly linked to missense mutations and significant clinical impairment. Further, using in vivo mouse studies, we demonstrate these phenotypes arise from developmental mechanisms, in which Ddx3x is essential for proper cortical development by controlling neuron generation. Finally, using biochemical and cell biological assays, we show that DDX3X missense mutations disrupt helicase activity, which is associated with altered RNP granule dynamics and translation of specific transcripts. The severity of these biochemical and cellular impairments correlates closely with clinical severity, supporting the hypothesis that a subset of missense mutations function in a dominant negative manner. Altogether, our study demonstrates cellular and molecular mechanisms by which DDX3X mutations induce severe cortical malformations in humans, and broadly implicates new pathogenic etiologies of neurodevelopmental disorders.
Location and type of mutation predict DDX3X clinical phenotype
In this study, we identify and characterize phenotypes for 107 patients, significantly extending our understanding of clinical outcomes associated with DDX3X mutations. Detailed analysis of our expanded cohort reveals that the same recurrent de novo mutations in unrelated individuals result in markedly similar phenotypes. Ten individuals with PMG have mutations at recurrent amino acids, R326, I415, and T532. The R326 mutation was also previously reported with PMG, underscoring the strong correlation between specific mutations and brain anatomy (Snijders Blok et al., 2015). We identified individuals with five new PMG mutations (V206M, T323I, I415F, I415del, T532M), who had a more severe cerebral anatomic phenotype, including complete or partial agenesis of the corpus callosum. The correlation with PMG is important as these individuals also had a more involved range of clinical deficits, including epilepsy, ASD, severe ID, and structural cardiovascular malformations than patients without PMG. Of the 11 individuals with PMG, ten had missense mutations and one had a three-nucleotide deletion. This striking association between PMG and missense mutations supports the hypothesis that certain missense mutations function in a dominant negative fashion. In comparison, the most common recurrent mutation, R376C, showed a mild clinical phenotype overall. In contrast, none of the 39 individuals with LoF mutations had PMG, possessing a milder spectrum of clinical phenotypes. This degree of genotype-phenotype association was not evident in the first reported cohort of DDX3X patients (Snijders Blok et al., 2015) or in subsequent cohorts (Wang et al., 2018), perhaps due to a smaller sample size.
Ddx3x loss-of-function in mice impairs neuronal generation
The malformations associated with DDX3X LoF, including microcephaly and corpus callosum abnormalities, are predicted to result from perturbed embryonic cortical development. Indeed, we show that DDX3X is required in RGCs to control neuron generation. Our findings reinforce the exquisite dosage sensitivity of the developing brain to DDX3X, as even a 25% reduction in DDX3X levels strongly perturbed neurogenesis. This degree of knockdown may model gene expression in female patients who are heterozygous for de novo DDX3X mutations and may be mosaic as DDX3X is X-linked. In the future, it will be critical to understand how Ddx3x loss impairs neuron generation and leads to more cycling progenitors. We speculate that DDX3X promotes neurogenic divisions and/or controls progenitor cell cycle duration. Consistent with the latter possibility, DDX3X is strongly implicated in cell cycle progression (Chen et al., 2016; Li et al., 2014; Pek and Kai, 2011). Notably, this role of DDX3X in regulating cell fate decisions may be broadly applicable in both physiological and stressed conditions (Samir et al., 2019).
Surprisingly, neuron-specific Ddx3x depletion over two different developmental windows (E14.5-E17.5 and E14.5-P2) did not significantly disrupt neuronal position, indicating that DDX3X is largely dispensable for radial migration of cortical neurons. However, DDX3X is highly expressed in cortical neurons suggesting it likely functions at postnatal stages, such as for synapse development and neuronal circuitry. In this light, future studies that utilize conditional in vivo mouse models to deplete Ddx3x post-natally will be invaluable.
These mouse studies suggest important mechanisms by which LoF mutations may drive disease pathology. Microcephaly, evident in approximately 40% of patients, is predicted to result from a generalized reduction of neurons. Corpus callosum defects, found in almost all the patients, may derive from aberrant neuronal generation (Fame et al., 2011). Altogether, these mouse studies establish critical requirements of DDX3X for neuron generation, giving important clues as to how DDX3X LoF impairs cortical development.
RNA metabolism is severely impaired by DDX3X missense mutants
Our data pinpoint defective RNA metabolism as an underlying mechanism by which DDX3X missense mutations impact progenitors and neurons. We show that some DDX3X missense mutants have reduced helicase activity and form aberrant RNP granules. Hence, we predict that missense mutants with impaired helicase activity act as dominant negatives, and by failing to release from RNA, mutant DDX3X may aberrantly sequester both RNA and RNA binding proteins within granules. Strikingly, DDX3X mutations associated with the most severe clinical outcomes produce the strongest cell biological and biochemical phenotypes. In contrast, mild mutants which maintain some helicase activity behave similar to LoF, and do not induce RNP granules. Altogether this provides a molecular explanation for severe clinical phenotypes associated with missense mutations compared to nonsense and frameshift (presumed LoF) mutations.
Based upon its canonical function and our findings, we posit that DDX3X controls neurogenesis by influencing translation. While global translation was unaffected by Ddx3x LoF or missense mutations, DDX3X missense mutants altered translation of specific targets. Hence, DDX3X may control neurogenesis by promoting translation of key targets, including those important for neuronal generation. It could do so by acting in concert with known translational regulators such as Nanos1 and Smaug2 which translationally repress a prol-proliferative program (Amadei et al., 2015), or with the eIF4E1/4ET complex, which translationally represses a pro-neurogenic program in neural progenitors (Yang et al., 2014). We also cannot exclude that DDX3X functions outside of translation (Soto-Rifo and Ohlmann, 2013). These questions should be addressed in the future with comprehensive genomic studies of both LoF and missense variants.
Our study is amongst the first to observe perturbed RNP granules in the context of neurodevelopmental disorders (Balak et al., 2019; Lessel et al., 2017), which to date have been primarily associated with neurodegenerative etiologies. Many RNA binding proteins, including helicases, are essential for cortical development and linked to neurodevelopmental disease (Lennox et al., 2018). Thus, going forward, it will be important to to consider widespread roles for aberrant RNP granules in cortical development and disease.
In sum, by thoroughly exploring the biology behind a larger cohort, including recurrent mutations, we show new genotype-phenotype correlations. Paired with our biochemical and cell biological studies, this suggests a dominant negative impact of mutant DDX3X in severely affected individuals. Our study reinforces the need for using mutation-based approaches to understand the role of DDX3X in neurodevelopment. This potential to ultimately predict disease severity and clinical outcomes (both mild and severe) from biochemical or cell biological “readouts” will be a benefit to families and clinicians.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Debra Silver (debra.silver@duke.edu).
All unique/stable reagents generated in this study are available from the Lead Contact either without restriction or in some cases with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Clinical Sample
This study includes data from 107 participants from a network of seven clinical sites. This cohort was recruited through collaboration with clinical geneticists and neurologists, clinical genetic laboratories, and through the DDX3X family support foundation (DDX3X.org). Data collection sites had study protocols approved by their Institutional Review Boards (IRB), and all enrolled subjects had informed consent provided by parent/guardian.
Mice
All animal use was approved by either the Division of Laboratory Animal Resources from Duke University School of Medicine or by the University of Queensland Animal Ethics committee in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes. C57BL/6J mouse embryos of either sex (Jackson Laboratory) were used for all experiments, except for coronal section in situ hybridizations (Figure 3 F–I) and postnatal immunofluorescence (Figure S2), which both used CD1 mice. Plug dates were defined as E0.5 on the morning the plug was identified.
Cell lines and primary cultures
N2A cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. N2A cells were transfected with Lipofectamine 2000 (Invitrogen). For the DDX3X granule formation assay, equal amounts of plasmid were transfected for each construct and cells were fixed 24 hours after transfection. Samples were blinded prior to quantification on the microscope. Primary cortical cultures were derived from E12.5 – E14.5 embryonic dorsal cortices, as previously described (Mao et al., 2015). Microdissected tissue was dissociated with 0.25% Trypsin-EDTA + DNase for 5 minute at 37°C bef ore trituration with a pipette. Cells were plated on poly-D-lysine coated coverslips in 12 well culture plates. Cells were grown in DMEM supplemented with B27, N2, N-acetyl-L-cystine, and bFGF. Primary cortical cells were transfected with the Amaxa Nucleofector in P3 solution.
METHOD DETAILS
Plasmids and subcloning
The pX330-U6-Chimeric_BB-CBh-hSpCas9 was a gift from Feng Zhang (AddGene plasmid # 42230) (Cong et al., 2013). Guide sequences were designed using an online program (http://crispr.mit.edu/) and cloned into the pX330 vector as described by the Zhang lab (http://www.addgene.org/crispr/zhang/). The following guide RNA was used: Ddx3x exon 1 5’-AGTGGAAAATGCGCTCGGGC-3’. The TCF/LEF-H2B:GFP plasmid was a gift from Anna-Katerina Hadjantonakis (AddGene plasmid # 32610 (Ferrer-Vaquer et al., 2010)). Dcp1a-RFP was a gift from Stacy Horner’s lab. pCAG-DDX3X was generated by amplifying full length human DDX3X from cDNA and subcloning into the pCAG-EX2 vector using NEBuilder HiFi DNA Assembly Kit. GFP or FLAG tags were added upstream of the DDX3X to generate fusion proteins. The NEBuilder kit was also used to engineer point mutations. The Dcx::mCherry plasmids were a kind gift of Santos Franco.
All biochemical experiments were performed using DDX3X from amino acid residues 132–607 (NP-001347.3) in a pHMGWA vector backbone containing a 5’ 6xHis-MBP tag, as described, (Floor et al., 2016b). Mutant clones were generated using Quikchange XL-II site directed mutagenesis kit (Agilent Cat Number: 200521) with the following primers:
R376C 5’-CACATCGTATGGCAAACGCCTTTCGGTGGCAT-3’
R326H 5’-CATATCCACCAGGTGACCCGGGGTAGC-3’
T323I 5’-CGACCCGGGATAGCAACCAGCAAGTGG-3’
A233V 5’-ATCGGCAACAGAAAGACAGCCGTCTTACCGC-3’
T532M 5’-GCCGACACGACCCATACGACCAATACGGTGCACA-3’
I514T 5’-CCAATGTCAAACACGTGACCAACTTTGATTTGCCGAG-3’
A497V 5’-CGCCACTGCCGTCACCACCAGAATCGG-3’
R475G 5’-CACGCTGGCTGCCATCACCGTGAATGC 3’
I415M 5’-ACCTTCTGCGTCATATTCTCGCTAGTGGAGCCA-3’
I415del 5’-GTTGGCTCCACTAGCGAGAATACGCAGAAGGT-3’
Purification of recombinant DDX3X WT and mutant
Plasmids encoding amino acid residues #132–607 were transformed into competent E.coli BL21-star cells, lysed by sonication in lysis buffer (0.5 M NaCl, 0.5% NP40, 10 mM Imidazole, 20 mM HEPES pH 7.5) and bound to nickel beads, as described (Floor et al., 2016b). Beads were washed with low salt (0.5M NaCl 20 mM Imidazole, 20 mM HEPES) and high salt wash buffers (1 M NaCl, 20 mM Imidazole, 20 mM HEPES pH 7.5) and eluted (0.5 M NaCl, 0.25 M imidazole, 100 mM Na2SO4, 9.25 mM NaH2PO4, 40.5 mM Na2HPO4). The 6xHis-MBP tag was cleaved overnight by tev protease at a 1:40 tev:protein ratio (w/w), and then dialyzed into ion exchange low salt buffer (200 mM NaCl, 20 mM HEPES ph7, 10% Glycerol, 0.5 mM TCEP). The sample was loaded onto a GE HiTrap heparin column (GE, 17-0406-01), eluted at 25% ion exchange high salt buffer (1 M NaCl, 20 mM HEPES pH 7, 10% Glycerol, 0.5 mM TCEP) and applied to Superdex 75 column (GE 17-0771-01) equilibrated in 500 mM NaCl, 10% Glycerol, 20 mM HEPES pH 7.5 and 0.5 mM TCEP. Peak fractions were collected, concentrated to ~50 μM, and then supplemented with 20% glycerol, diluted to 30 μM, and flash frozen in liquid nitrogen. Sample of the collected protein was run on a SDS-PAGE gel and stained with Coomassie to confirm protein purity and size.
Radiolabelling and formation of RNA Duplex
Two complementary RNA molecules were synthesized by IDT: “5’ duplex” (5’-AGCACCGUAAGAGC-3’) and “3’ overhang (5’GCGUCUUUACGGUGCUUAAAACAAAACAAAACAAAACAAAA-3’). 5’ duplex RNA was labeled with 32P using T4 PNK (NEB, M0201S) for one hour at which point stop buffer was added to terminate the reaction. Labeled RNA was run on a 15% DNA denaturing gel, imaged on a phosphorscreen, and then bands were excised and RNA eluted overnight in 600 μL elution buffer (300 mM NaOAc pH 5.2, 1 mM EDTA, 0.5% SDS). Eluted RNA was then precipitated using 750 μL of 100% ethanol and immersed in dry ice for 1 hour 30 minutes and then resuspended in 20 μL DEPC H2O to > 200k cpm. RNA duplex was created by annealing 3 μL of 100 μM 3’ overhang RNA to 2.5 μL 5’ duplex of duplex RNA, heated to 95°C for 1 minute and allowed to cool to 30°C before purifying on a 15% n ative gel run at 8 watts.
Duplex Unwinding Assays
This protocol was modified from Jankowski et al. (Jankowsky and Putnam, 2010). Radiolabeled, purified duplex RNA was diluted to ~333 cpm, and protein was diluted to 10 μM concentration. Reaction was run in 30 μL in reaction buffer (40 mM Tris-HCl pH 8, 5mM MgCl2, 0.1% IGEPAL, 20 mM DTT) with 3 μL of diluted RNA and 1 μL of 10 μM protein. 3 μL of 20 mM ATP:MgCl2 was added to initiate the reaction. At each time point 3 μL of reaction mix is removed and quenched in 3 μL of stop buffer (50 mM EDTA, 1% SDS, 0.1% Bromophenol blue, 0.1% Xylene cyanol, 20% glycerol). Reaction time points are then run on a 15% native polyacrylamide gel, at 5 watts for 30 minutes at 4°C. Gel is dried and ima ged on a phospho-screen and quantified using ImageQuant software.
ATP-ase activity Assay
The ATPase activity assay was performed with protein constructs previously purified and in the enzyme reaction buffer used the duplex unwinding assay reaction buffer, as described in Floor and Barkovich (Floor et al., 2016a). 100 μL of reaction master mix was made containing 3 μL of 10 μM DDX3X protein, 10 μL of 10x reaction buffer (400 mM Tris-HCl pH 8, 50mM MgCl2, 1% IGEPAL, 200 mM DTT) and 26.9 μL of DEPC H2O. 2 μL of single stranded RNA was added to each 7 μL to create a dilution series of 0 μM, 0.625 μM, 1.25 μM, 2.5 μM, 5 μM, 10 μM, 20 μM and 40 μM. The single-stranded RNA was transcribed and gel purified, with sequence GGAAUCUCGCUCAUGGUCUCUCUCUCUCUCUCUCUCUCUCU. Radioactive ATP-(32P) (Perkin Elmer) was diluted to 666 μCi/uL. 1 μL of ATP at varying concentrations was added to initiate each reaction. At each time point 1 μL were spotted and quenched on PEI cellulose thin layer chromatography plates. Chromotography plates were run in a buffer of 0.5 M LiCl and 0.5 M Formic acid. Plates were then exposed to a GE Phosphoscreen for 1.5 hours and then imaged on a GE Typhoon imager.
siRNAs
Mouse Ddx3x siRNAs and negative control siRNAs were obtained from Qiagen. siRNA experiments were performed by pooling 4 siRNAs with the following target sequences: 5’-CTGATAATAGTCTTTAAACAA-3’, 5’-TCCATAAATAATATAAGGAAA-3’, 5’-CTCAAAGTTAATGCAAGTAAA-3’, 5’-CACAGGTGTGATACAACTTAA-3’.
Western blot and qRT-PCR analysis.
N2A cells were transfected with either Cas9 with or without Ddx3x sgRNA or scrambled or Ddx3x siRNAs. Protein was harvested 72 hours after transfections in RIPA lysis buffer with protease inhibitors. Lysates were run on 4–20% mini-PROTEAN TGX precast gels. Gels were transferred to PVDF membranes, blocked with 5% milk/TBST, probed primary antibodies overnight at 4°C, and secondary HRP-conjugated anti bodies at room temperature for 1 hour. The following primary antibodies were used: mouse anti-DDX3X (Santa Cruz, sc-365768, 1:100) or mouse anti-Tubulin (Sigma, T6199, 1:1000). For Figure 7, rabbit polyclonal anti-DDX3 (custom made by Genemed Synthesis using peptide ENALGLDQQFAGLDLNSSDNQS (Lee et al. 2008), anti-actin HRP (Santa Cruz Biotechnology, sc-47778), anti-FLAG HRP (Sigma, A8592). Blots were developed with ECL and quantified by densitometry in ImageJ (Schneider et al., 2012). RNA was extracted from transfected N2A Cells using TriReagent (Sigma), followed by cDNA synthesis with iScript Reverse Transcriptase (BioRad). The primers used for qRT-PCR are as follows: Ddx3x forward 5’- TGGAAATAGTCGCTGGTGTG-3’ and reverse 5’- GGAGGACAGTTGTTGCCTGT-3’; Actb forward 5’- AGATCAAGATCATTGTCCT 3’ and reverse 5’ CCTGCTTGCTGATCCACATC 3’.
In utero electroporation
Plasmids were delivered to embryonic brains as previously described (Mao et al J Neuro 2015). Briefly, E13.5 or E14.5 embryos were injected with 1–1.5 uL of plasmid DNA mixed with Fast Green Dye. Plasmids were used at the following concentrations: pCAG-GFP (1.0 ug/uL), Dcx-mCherry (1.0 ug/uL), pX330 empty or pX330-Ddx3x Ex1 sgRNA (2.4 ug/uL), pX330 empty-Dcx-Cas9 empty or pX330-Ddx3x Ex1 sgRNA -Dcx-Cas9 (2 ug/uL), TCF/LEF-H2B:GFP (1.25 ug/uL), pCAG-mCherry (1.0 ug/uL). Scrambled or Ddx3x siRNAs were injected at 1 μM. Following injection, embryos were pulsed five times with 50 V for 50 ms. Embryonic brains were harvested 48–72 hours later or 7 days later (neuron-specific knockdown).
In situ Hybridization
In situ hybridization was performed as described in Moldrich et al, 2010. The Ddx3x riboprobe was generated in-house using the primers corresponding to those used in the Allen Developing Mouse Brain Atlas (Website: © 2015 Allen Institute for Brain Science. Allen Developing Mouse Brain Atlas [Internet]). Available from: http://developingmouse.brain-map.org): Forward primer: 5’ AAGGGAGCTCAAGGTCACAA 3’, Reverse primer: 5’ CCTGCTGCATAATTCTTCC 3’. Using mouse cortex cDNA, these primers were used to amplify a 908 base pair fragment. This fragment was purified and cloned into pGEM00ae-T Vector System (Promega). The plasmid was then linearized (SacII restriction enzyme, New England BioLabs), purified (PCR Clean up Kit, Qiagen), transcribed (Sp6 RNA Polymerase, New England BioLabs) and digoxigenin-labelled (DIG RNA labelling Mix, Roche) to generate the riboprobe. In situ hybridization against Ddx3x mRNA was performed on 20 μm cryostat sections for embryonic stages and 50 μm vibratome sections for postnatal brains in wildtype CD1 mice.
Immunofluorescence
Embryonic brains were fixed overnight in 4% PFA at 4°C, submerged in 30% sucrose overnight, and embedded in NEG-50. 20 μm frozen sections were generated on a cryotome and stored at −80. Sections were permeabilized with 0.25% TritonX-100, blocked with either 5% NGS/PBS or MOM block reagent (Vector Laboratories) for 1 hour at room temperature. Sections were incubated with primary antibodies overnight at 4°C, and secondary antibodies at room temperature for 30–60 minutes. Cultured cells were fixed for 15 minutes at room temperature with 4% PFA, permeabilized with 0.5% Triton X-100, blocked with 5% NGS, incubated with primary antibodies for 1 hour at room temperature and secondary antibodies for 15 minutes at room temperature. Images were captured using a Zeiss Axio Observer Z.1 equipped with an Apotome for optical sectioning. The following primary antibodies were used: mouse anti-DDX3 (Santa Cruz, sc-365768, 1:100), rabbit anti-DDX3X (Protein Tech, 11115–1-AP, 1:150), mouse anti-TUJ1 (Biolegend, 801202, 1:1000), mouse anti-NESTIN (BD Biosciences, BD401,1:100), rabbit anti-PAX6 (Millipore, AB2237, 1:1000), rabbit anti-TBR2 (Abcam, AB23345, 1:1000), rabbit anti-CC3 (Cell Signaling, 9661, 1:250), rabbit anti-NEUROD2 (Abcam, AB104430, 1:500), chicken anti-GFP (Abcam, Ab13970, 1:1000), rabbit anti-Laminin (Millipore, AB2034, 1:200), rabbit anti-acetylated Tubulin (Sigma, T7541, 1:500), rabbit anti-FMRP (Sigma, F1804,1:500), mouse anti-TIA1 (Abcam, AB2712, 1:100), mouse anti-puromycin (DSHB, PMY-2A4, 1:100), anti-RFP (Rockland, 600-401-379S, 1:500), guinea pig anti-NeuN (Merck, ABN90P, 1:1000), chicken anti-GFAP (Abcam, ab4674, 1:1000), goat anti-OLIG2 (Santa Cruz, sc19969, 1:200), goat anti-IBA1 (Abcam, ab5076, 1:1000), mouse anti-FOXJ1 (eBioscience, 149965–82, 1:1000), and rabbit anti-DDX3X (Sigma Aldrich, HPA001648, 1:500). Secondary antibodies used in embryonic brains and cell culture experiments were Alexa Fluor-conjugated (Thermo Fisher Scientific, 1:500). Those used in postnatal brains were Alexa Fluor-conjugated secondary antibodies (Thermo Fisher Scientific), biotinylated-conjugated secondary antibodies (Jackson Laboratories) used in conjunction with AlexaFluor 647-conjugated Streptavidin (Thermo Fisher Scientific) and CF dyes conjugated secondary antibodies (Biotium). Postnatal brains were collected and fixed as previously described (Moldrich et al., 2010 and Piper et al., 2011), then post-fixed with 4% paraformaldehyde (ProSciTech) for 2 to 4 days and stored in 1x Dulbecco’s phosphate buffered saline (Lonza) with 0. 1% sodium azide (Sigma Aldrich). Brains were sectioned on a vibratome (Leica) at 50 μm thickness in coronal orientation. Fluorescence immunohistochemistry was performed as previously described (Mao et al., 2016).
Puromycylation, Ribopuromycylation, and FISH
For puromycylation, cultured cells were pre-treated with 40 μM anisomycin or 0.5 mM sodium arsenite for 10 minutes, followed by a 20 minute incubation with 10 μM O-propargyl-puromycin (Life Technologies). Cells were rinsed with PBS, and fixed in 4% PFA for 15 minutes, and puromycin was conjugated to AlexaFluo594 with Click-It Technology according to the manufacturer’s protocol. For ribopuromycylation, cultured cells were pre-treated with 208 μM emetine for 15 minutes, then pulsed with 91 μM puromycin for 5 minutes in the presence of emetine. Anisomycin used at 40 μM as a competitive inhibitor of puromycin. Cells were extracted with cold polysome buffer (50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 25 mM KCl, 355 μM CHX, EDTA-free protease inhibitors, and 10 U/mL RNaseOut) containing 0.015% digitonin on ice for 2 minutes, rinsed with cold polysome buffer, then fixed with 4% PFA for 15 minutes at room temperature. Puromycin incorporation was detected using the 2A4 anti-PMY antibody (DSHB) at 1:100, and imaged with fluorescent microscopy. Stellaris single molecule FISH probes against 18S rRNA were purchased from BioSearch Technology. Cells were fixed with PFA for 10 minutes, permeabilized with 70% EtOH for 1–2 hours at 4°C, rinsed with wash buffer (10% formamide,2x SSC), hybridized with probe at 1:100 overnight at 37°C in hybridization buffer (100 mg/mL dextran sulfate, 10% formamide, 2x SSC), and rinsed again with buffer before mounting.
FUNCAT/AHA Click-it assays and analysis
48 hours post-transfection, N2A cells were washed 2X with DPBS and incubated with Methionine-free DMEM (Thermo, 21013024) for 1 hour. After 30 minutes, Anisomycin was added (20 μM) to some wells for the remaining 30 minutes. Afterwards, the methionine analog, AHA (100 μM; Thermo, C10102), was added to cells in Met-free media for 1 hour. The cells were then washed with DPBS. and fixed in 4% paraformaldehyde for 20 minutes at room temperature. The Click-it labeling was performed according to the manufacturer’s recommendations for both western- and fluorescent-based readouts. Fluorescent-based (Thermo, C10269): cells were fixed in 4% paraformaldehyde for 20 minutes at room temperature and washed 3X in DPBS. The cells were then permeabilized with 0.25% TritonX-100 for 10 minutes and the click-it assay was performed (4 μM 647-alkyne; Thermo, A10278).
For analysis, images were acquired on a 780 Zeiss confocal microscope and GFP+ cells were outlined and the fluorescence intensity in the 647 channel was measured in Fiji (Schindelin et al., 2012). Fluorescence intensity was normalized to no AHA controls. Western-based (Thermo, C10276): cells were lysed in 50 mM Tris pH 8, 0.1% SDS + protease inhibitors and then used for the click-it reaction (40 μM biotin-alkyne; Thermo, B10185).
In vitro transcription, capping, and 2′-O methylation of reporter RNAs
Annotated 5′ UTRs for selected transcripts were cloned upstream of Renilla Luciferase (RLuc) under the control of a T7 promoter, with 60 adenosine nucleotides downstream of the stop codon to mimic polyadenylation. Template was PCR amplified using Phusion polymerase from the plasmids using the following primers, and gel purified, as described (Floor and Doudna 2016).
pA60 txn rev: TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT CTG CAG; pA60 txn fwd: CGG CCA GTG AAT TCG AGC TCT AAT ACG ACT CAC TAT AGG. 100 uL in vitro transcription reactions were set up at room temperature with 1–5 micrograms of purified template, 7.5mM ACGU ribonucleotides, 30mM Tris-Cl pH 8.1, 125mM MgCl2, 0.01% Triton X-100, 2mM spermidine, 110mM DTT, T7 polymerase and 0.2 U/uL units of Superase-In RNase inhibitor (Thermo-Fisher Scientific). Transcription reactions were incubated in a PCR block at 37°C for 1 hour. 1 μL of 1 mg/mL pyrophosphatase (Roche) was added to each reaction, and the reactions were subsequently incubated in a PCR block at 37°C for 3 hours. 1 unit of RQ1 RNase-free DNase (Promega) was added to each reaction followed by further incubation for 30 minutes. RNA was precipitated by the addition of 200 μL 0.3M NaOAc pH 5.3, 15 ug GlycoBlue co-precipitant (Thermo-Fisher Scientific) and 750 μL 100% EtOH. Precipitated RNA was further purified over the RNA Clean & Concentrator-25 columns (Zymo Research). Glyoxal gel was run to assess the integrity of the RNA before subsequent capping and 2′ O-methylation.
20 μg of total RNA was used in a 40 μL capping reaction with 0.5mM GTP, 0.2mM sadenosylmethionine (SAM), 20 units of Vaccinia capping enzyme (New England Biolabs), 100 units of 2′-O-Me-transferase (New England Biolabs) and 25 units RNasin Plus RNase inhibitor (Promega). The reactions were incubated at 37 degrees C for 1 hour, followed by purification over the RNA Clean & Concentrator-25 columns (Zymo Research) and elution in DEPC H2O. Glyoxal gel was run to assess the integrity of the RNA before proceeding to in vitro translation reactions. In vitro translation was performed at 30 °C for 45 minutes, and substrate injection and signal integration (over 10 seconds) was performed in a GloMax Explorer plate reader (Promega).
Construction of si-resistant DDX3 mutant plasmids
Mutations in DDX3 were introduced by modifying the pCMV-DDX3X_WT-FLAG-Puromycin plasmid. Briefly, divergent overlapping primers containing the desired mutation were used to amplify the plasmid using the Kapa HiFi DNA Polymerase (Kapa Biosystems). The amplicon was gel extracted, any residual template plasmid was digested with DpnI (New England Biolabs), and the amplicon containing the desired mutation was circularized using the Gibson Assembly Master Mix (New England Biolabs). Presence of the desired mutation was confirmed by sanger sequencing individual isolates. Synonymous mutations to render the transfected DDX3X resistant to the siRNA are shown below:
DDX3X protein sequence: I Q M L A R D F L
DDX3X mRNA sequence: AUA CAG AUG CUG GCU CGU GAU UUC UUA
DDX3X siRNA sequence: G AUG CUG GCU CGU GAU UUC
Si-resistant DDX3X mRNA sequence: AUA CAA AUG TTA GCA AGA GAC UUU UUA
Si-resistant DDX3X protein sequence: I Q M L A R D F L
Transfection of siRNA and DDX3 mutant plasmids for in vitro translation
HEK293T cells in 150mM plates were transfected with 20 μL of siRNA (against DDX3 or a non-targeting control) using Lipofectamine 2000 (Thermo Fisher Scientific), following manufacturer’s instructions. After 24 hours, cells were further transfected with 10 μL of siRNA (against DDX3 or a non-targeting control) and 30 μg of plasmids expressing si-resistant mutants of DDX3 using Lipofectamine 2000 (Thermo Fisher Scientific), following manufacturer’s instructions. Cells were harvested for preparation of cellular extracts after 48 hours.
Preparation of cellular extracts for in vitro translation
150mm plates of HEK293T cells were trypsinized and pelleted at 1000g, 4°C. One cell-pellet volume of lysis buffer (10mM HEPES, pH 7.5, 10mM KOAc, 0.5mM MgOAc2, 5mM DTT, and 1 tablet miniComplete EDTA free protease inhibitor (Roche) per 10 mL) was added to the cell pellet and was incubated on ice for 45 minutes. The pellet was homogenized by trituration through a 26G needle attached to a 1 mL syringe 13–15 times. Efficiency of disruption was checked by trypan blue staining (>95% disruption target). The lysate was cleared by centrifugation at 14000g for 1 minute at 4°C, 2–5 μl was reserved for western blot analysis, and the remainder was aliquoted and flash frozen in liquid nitrogen.
In vitro translation assay
5 μL in vitro translation reactions were set up with 2.5 μL of lysate and 20 ng total RNA (0.84mM ATP, 0.21mM GTP, 21mM Creatine Phosphate, 0.009units/mL Creatine phosphokinase, 10mM HEPES pH 7.5, 2mM DTT, 2mM MgOAc, 100mM KOAc, 0.008mM amino acids, 0.25mM spermidine, 5 units RNasin Plus RNase inhibitor (Promega) as described (Lee et al. 2015). Reaction tubes were incubated at 37°C for 45 minutes, and expression of the reporter was measured using the Renilla Luciferase Assay System (Promega) on a GloMax Explorer plate reader (Promega).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical methods and sample size
Table S2 details the comparison, statistical test, p-value and sample size for each figure is provided. Data are presented as mean ± standard deviation. *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001
Data Reporting
No statistical methods were used to predetermine sample size for this study. Recruitment for this study was not blinded.
Data Collection Methods
The majority of mutations were identified through clinical whole exome sequencing either at referring and participating clinical centers or through the clinical genetic laboratories: GeneDx, Baylor, or BGI-Xome. Three patients (2839–0, 2839–3 and 2897–0) were ascertained as part of a larger study of genetic causes for Dandy-Walker malformation. Exome sequencing of 2839–0 and 2897–0 and their parents was performed as described (Van De Weghe et al., 2017); 2839–3 is the identical twin of 2839–0 and Sanger sequencing confirmed they shared the same mutation. Genetic mutation data were obtained for all 107 participants. All clinical features and findings were defined by the medical records obtained from participants or from the completion of standardized behavioral and development measures, including the Vineland Adaptive Behavior Scales, Second Edition (Vineland-II or VABS), Child Behavior Checklist (CBCL; (Achenbach, 2011)), Social Communication Questionnaire (SCQ) and Social Responsiveness Scale, Second Edition (SRS-2). 53 of 107 participants completed at least one neuropsychological questionnaire. Comparison of population means for behavioral scales were performed using a Wilcoxon test. One patient had a low-quality MRI scan and it could not be determined whether PMG was present or absent with the current data; therefore 106/107 patients are represented in the clinical data table.
MRI Review
This study includes MRI scans from 89 participants. 9 patients in the cohort did not have MRI scans in their records and 8 MRIs could not be obtained. All scans were initially reviewed locally by a radiologist for findings that, if present, were communicated to the participant and noted in the patient chart. DiCOM files of high quality MRI scans were then transmitted and neuroradiologic findings were noted in a standardized assessment of developmental features as previously utilized by our group (Hetts et al., 2006). All MRI findings were reviewed by a board certified pediatric neuroradiologist at UCSF blinded to genetic status as part of a larger more general review of potential agenesis of the corpus callosum cases.
Statistical analysis of clinical data
Comparisons of various frequencies between the groups were performed based on Fisher’s exact test. One-sample tests of frequencies of the observed mutations were performed using Binomial test. Comparisons of continuous measures were performed using Mann-Whitney U-Test.
Quantification and Binning Analysis
All mouse embryo experiments were blinded throughout processing, imaging, and analysis. For the binning analysis, 450 μm wide radial columns were broken down into 5 evenly spaced bins spanning from the ventricular to the pial surface. Each GFP+ cell was assigned to a bin to calculate the distribution. At least 3 sections were analyzed per embryo, with multiple embryos per condition (see figure legend for exact sample size).
DATA AND CODE AVAILABILITY
This study did not generate any unique code. Original/source data for figures in the paper are available from the corresponding authors on request.
Supplementary Material
Table S1 Comprehensive phenotypes for DDX3X patients, Related to Figure 1.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-DDX3X (western; 1:100) | Santa Cruz Biotechnology | Cat# sc-365768, RRID:AB_10844621 |
| mouse anti-Tubulin (western; 1:1000) | Sigma-Aldrich | Cat# T6199, RRID:AB_477583 |
| rabbit polyclonal anti-DDX3 (custom made using peptide ENALGLDQQFAGLDLNSSDNQS) | Genemed Synthesis (custom made) / Stephen Floor lab | N/A |
| rabbit anti-DDX3X (IF; 1:150) | Protein Tech | Cat# 11115–1-AP, RRID:AB_10896499 |
| rabbit anti-DDX3X (IF; 1:500) | Sigma-Aldrich | Cat# HPA001648, RRID:AB_1078635 |
| mouse anti-TUJ1 (IF; 1:1000) | Biolegend | Cat# 801202, RRID:AB_10063408 |
| mouse anti-NESTIN (IF; 1:100) | BD Biosciences | BD401 |
| rabbit anti-PAX6 (IF; 1:1000) | Millipore | Cat# AB2237, RRID:AB_1587367 |
| rabbit anti-TBR2 (IF; 1:1000) | Abcam | Cat# ab23345, RRID:AB_778267 |
| rabbit anti-CC3 (IF; 1:250) | Cell Signaling Technology | Cat# 9661, RRID:AB_2341188 |
| rabbit anti-NEUROD2 (IF; 1:500) | Abcam | Cat# ab104430, RRID:AB_10975628 |
| chicken anti-GFP (IF; 1:1000) | Abcam | Cat# ab13970, RRID:AB_300798 |
| rabbit anti-Laminin (IF; 1:200) | Millipore | Cat# AB2034, RRID:AB_91209 |
| rabbit anti-acetylated Tubulin (IF; 1:500) | Sigma-Aldrich | Cat# T7451, RRID:AB_609894 |
| rabbit anti-FMRP (IF; 1:500) | Sigma-Aldrich | Cat# F1804, RRID:AB_262044 |
| mouse anti-TIA1 (IF; 1:100) | Abcam | Cat# ab2712, RRID:AB_2201439 |
| mouse anti-puromycin (IF; 1:100) | DSHB | Cat# PMY-2A4, RRID:AB_2619605 |
| anti-RFP (IF; 1:500) | Rockland | Cat# 600-401-379S, RRID:AB_11182807 |
| guinea pig anti-NeuN (IF; 1:1000) | Merck | Cat# ABN90P, RRID:AB_2341095 |
| chicken anti-GFAP (IF; 1:1000) | Abcam | Cat# ab4674, RRID:AB_304558 |
| goat anti-OLIG2 (IF; 1:200) | Santa Cruz Biotechnology | Cat# sc-19969, RRID:AB_2236477 |
| goat anti-IBA1 (IF; 1:1000) | Abcam | Cat# ab5076, RRID:AB_2224402 |
| mouse anti-FOXJ1 (IF; 1:1000) | Thermo Fisher Scientific / eBioscience | Cat# 14-9965-82, RRID:AB_1548835 |
| Bacterial and Virus Strains | ||
| NEB 5-alpha Competent E. coli (High Efficiency) | NEB | C2987H |
| One Shot™ BL21 Star™ (DE3) Chemically Competent E. coli | Thermo Fisher Scientific | C601003 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| DDX3X from amino acid residues 132–607 (NP-001347.3) | Stephen Floor Lab | N/A |
| Anisomycin | Sigma-Aldrich | A9789–5MG |
| Puromycin | Sigma-Aldrich | P8833–10MG |
| Emetine | Sigma-Aldrich | E2375–500MG |
| biotin-alkyne | Thermo Fisher Scientific | B10185 |
| Alexa647-alkyne | Thermo Fisher Scientific | A10278 |
| AHA | Thermo Fisher Scientific | C10102 |
| Critical Commercial Assays | ||
| Methionine-free DMEM | Thermo Fisher Scientific | 21013024 |
| Fluorescence-based click-it | Thermo Fisher Scientific | C10269 |
| Western-based click-it | Thermo Fisher Scientific | C10276 |
| NEBuilder HiFi DNA Assembly Kit | NEB | E5520S |
| Renilla Luciferase Assay System | Promega | E2810 |
| Click-it Plus OPP 594 (Puromycin Click Kit) | Thermo Fisher Scientific | C10457 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Neuro2A (male) | ATCC | CCL131 |
| HEK293T (female) | ATCC | CRL-11268 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J mice (male and female) | The Jackson Laboratory | 000664 |
| CD1 mice (male and female; Figure 3B–E and Figure S2) | Animal Resource Centre | Arc:Arc(S) |
| Oligonucleotides | ||
| Formation of RNA duplex for unwinding assays: “5’ duplex” (5’-AGCACCGUAAGAGC-3’) and “3’ overhang (5’GCGUCUUUACGGUGCUUAAAACAAAACAAAACAAAACAAAA-3’) | Synthesized at IDT | |
| Transcribed RNA for ATPase assay: GGAAUCUCGCUCAUGGUCUCUCUCUCUCUCUCUCUCUCUCU | Stephen Floor lab | |
| Mouse Ddx3x siRNA pool: 5’-CTGATAATAGTCTTTAAACAA-3’, 5’-TCCATAAATAATATAAGGAAA-3’, 5’-CTCAAAGTTAATGCAAGTAAA-3’, 5’-CACAGGTGTGATACAACTTAA-3’ |
Qiagen | GS13205 |
| Negative control siRNA | Qiagen | 1022076 |
| Primers for RT-qPCR: mouse Ddx3x forward 5’- TGGAAATAGTCGCTGGTGTG-3’ and reverse 5’- GGAGGACAGTTGTTGCCTGT-3’; mouse Actb forward 5’- AGATCAAGATCATTGTCCT 3’ and reverse 5’ CCTGCTTGCTGATCCACATC 3’ | This paper (Debra Silver Lab) | |
| Primers for In Situ Hybridization (mouse): Forward primer: 5’ AAGGGAGCTCAAGGTCACAA 3’, Reverse primer: 5’ CCTGCTGCATAATTCTTCC 3’ | Allen Developing Mouse Brain Atlas | |
| Recombinant DNA | ||
| pX330-U6-Chimeric_BB-CBh-hSpCas9 | Feng Zhang Lab | AddGene plasmid # 42230; RRID:Addgene_42230 |
| Ddx3x exon 1 sgRNA (5’-AGTGGAAAATGCGCTCGGGC-3’) | This paper (Debra Silver Lab) | http://crispr.mit.edu/ |
| TCF/LEF-H2B:GFP plasmid | Anna-Katerina Hadjantonakis Lab | AddGene plasmid # 32610; RRID:Addgene_32610 |
| Dcx::mCherry plasmid | Santos Franco Lab | |
| pCAG-GFP-DDX3X | This paper (Debra Silver Lab) | pCAG-EX2 |
| pHM-GWA-6xHis-MBP-DDX3X (protein purification) | Stephen Floor Lab | pHM-GWA |
| pCMV-DDX3X_WT-FLAG-Puromycin | Stephen Floor Lab | pCMV |
| Software and Algorithms | ||
| Fiji | Schindelin, J, et al., 2012 | https://imagej.net/Fiji |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Other | ||
| Vineland Adaptive Behavior Scales, Second Edition (Vineland-II) | Sparrow, SS. et al., 2006 | N/A |
| Child Behavior Checklist (CBCL) | Achenbach, T.M., 2011 | N/A |
| Social Communication Questionnaire (SCQ) | Rutter, M., et al., 2003 | N/A |
| Social Responsiveness Scale, Second Edition (SRS-2) | Constantino, JN., 2013 | N/A |
Highlights.
Discovery of 107 mutations in the RNA helicase DDX3X causing cortical malformations
Clinical severity is linked to reduced helicase activity and RNA-protein granules
Ddx3x is required in neural progenitors to produce cortical neurons during development
Severe missense mutations cause polymicrogyria and impaired translation of targets
Acknowledgements
We thank the patients, their families, and referring physicians for their important contributions, and members of the Sherr and Silver labs for helpful discussions. This work was supported by the Holland-Trice Foundation, NIH R01NS083897, NIH R21NS098176, NIH R01NS110388 (DLS); NIH F31NS0933762 (ALL); Regeneration Next Initiative Postdoctoral Fellowship, NIH F32NS112566 (MLH); NIH R25 (BLJK); NIH 1R01NS058721 (EHS, WBD, and LJR); NIH 5R01NS050375 (WBD); the DDX3X Foundation (EHS), National Health and Medical Research Council, Australia project grant GNT1126153 (LJR and EHS); GNT1120615 Principal Research Fellowship from the NHMRC (LJR); UCSF Program for Breakthrough Biomedical Research, funded in part by the Sandler Foundation (SNF); California Tobacco-Related Disease Research Grants Program 27KT-0003, NIH DP2GM132932 (SNF); Dandy-Walker Alliance (KAA and WBD); the HUGODIMS consortium RC14_0107, funded in part by the French Ministry of Health and the Health Regional Agency from Poitou-Charentes, Frédérique Allaire from the Health Regional Agency of Poitou-Charentes (SK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors confirm there are no competing interests.
References
- Achenbach TM (2011). Child Behavior Checklist. In Encyclopedia of Clinical Neuropsychology, Kreutzer JS, DeLuca J, and Caplan B, eds. (New York, NY: Springer New York; ), pp. 546–552. [Google Scholar]
- Amadei G, Zander MA, Yang G, Dumelie JG, Vessey JP, Lipshitz HD, Smibert CA, Kaplan DR, and Miller FD (2015). A Smaug2-Based Translational Repression Complex Determines the Balance between Precursor Maintenance versus Differentiation during Mammalian Neurogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience 35, 15666–15681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub AE, Oh S, Xie Y, Leng J, Cotney J, Dominguez MH, Noonan JP, and Rakic P (2011). Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc Natl Acad Sci U S A 108, 14950–14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balak C, Benard M, Schaefer E, Iqbal S, Ramsey K, Ernoult-Lange M, Mattioli F, Llaci L, Geoffroy V, Courel M, et al. (2019). Rare De Novo Missense Variants in RNA Helicase DDX6 Cause Intellectual Disability and Dysmorphic Features and Lead to P-Body Defects and RNA Dysregulation. Am J Hum Genet 105, 509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal B, Hayes I, McGaughran J, Amor DJ, Miteff C, Jackson V, van Reyk O, Subramanian G, Hildebrand MS, Morgan AT, et al. (2019). Expansion of phenotype of DDX3X syndrome: six new cases. Clinical dysmorphology 28, 169–174. [DOI] [PubMed] [Google Scholar]
- Bocchi R, Egervari K, Carol-Perdiguer L, Viale B, Quairiaux C, De Roo M, Boitard M, Oskouie S, Salmon P, and Kiss JZ (2017). Perturbed Wnt signaling leads to neuronal migration delay, altered interhemispheric connections and impaired social behavior. Nature communications 8, 1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviello L, Venkataramanan S, Rogowski KJ, Wyler E, Tejura M, Thai B, Krol J, Filipowicz W, Landthaler M, and Floor SN (2019). DDX3 depletion selectively represses translation of structured mRNAs. bioRxiv, 589218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, and Willard HF (2005). X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404. [DOI] [PubMed] [Google Scholar]
- Chen CY, Chan CH, Chen CM, Tsai YS, Tsai TY, Wu Lee YH, and You LR (2016). Targeted inactivation of murine Ddx3x: essential roles of Ddx3x in placentation and embryogenesis. Hum Mol Genet 25, 2905–2922. [DOI] [PubMed] [Google Scholar]
- Chen HH, Yu HI, Yang MH, and Tarn WY (2018). DDX3 Activates CBC-eIF3-Mediated Translation of uORF-Containing Oncogenic mRNAs to Promote Metastasis in HNSCC. Cancer research 78, 4512–4523. [DOI] [PubMed] [Google Scholar]
- Cheng W, Wang S, Zhang Z, Morgens DW, Hayes LR, Lee S, Portz B, Xie Y, Nguyen BV, Haney MS, et al. (2019). CRISPR-Cas9 Screens Identify the RNA Helicase DDX3X as a Repressor of C9ORF72 (GGGGCC)n Repeat-Associated Non-AUG Translation. Neuron 104, 885–898 e888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN (2013). Social Responsiveness Scale. In Encyclopedia of Autism Spectrum Disorders, Volkmar FR, ed. (New York, NY: Springer New York; ), pp. 2919–2929. [Google Scholar]
- Cruciat CM, Dolde C, de Groot RE, Ohkawara B, Reinhard C, Korswagen HC, and Niehrs C (2013). RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-beta-catenin signaling. Science 339, 1436–1441. [DOI] [PubMed] [Google Scholar]
- Dikow N, Granzow M, Graul-Neumann LM, Karch S, Hinderhofer K, Paramasivam N, Behl LJ, Kaufmann L, Fischer C, Evers C, et al. (2017). DDX3X mutations in two girls with a phenotype overlapping Toriello-Carey syndrome. American journal of medical genetics Part A 173, 1369–1373. [DOI] [PubMed] [Google Scholar]
- Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sanchez-Carbente M, Servant F, Bell AW, Boismenu D, et al. (2006). Characterization of an RNA granule from developing brain. Mol Cell Proteomics 5, 635–651. [DOI] [PubMed] [Google Scholar]
- Fame RM, MacDonald JL, and Macklis JD (2011). Development, specification, and diversity of callosal projection neurons. Trends in neurosciences 34, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Vaquer A, Piliszek A, Tian G, Aho RJ, Dufort D, and Hadjantonakis AK (2010). A sensitive and bright single-cell resolution live imaging reporter of Wnt/ss-catenin signaling in the mouse. BMC developmental biology 10, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floor SN, Barkovich KJ, Condon KJ, Shokat KM, and Doudna JA (2016a). Analog sensitive chemical inhibition of the DEAD-box protein DDX3. Protein science : a publication of the Protein Society 25, 638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floor SN, Condon KJ, Sharma D, Jankowsky E, and Doudna JA (2016b). Autoinhibitory Interdomain Interactions and Subfamily-specific Extensions Redefine the Catalytic Core of the Human DEAD-box Protein DDX3. The Journal of biological chemistry 291, 2412–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, and Muller U (2011). Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron 69, 482–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garieri M, Stamoulis G, Blanc X, Falconnet E, Ribaux P, Borel C, Santoni F, and Antonarakis SE (2018). Extensive cellular heterogeneity of X inactivation revealed by single-cell allele-specific expression in human fibroblasts. Proceedings of the National Academy of Sciences of the United States of America 115, 13015–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber TE, Hebert-Seropian S, Khoutorsky A, David A, Yewdell JW, Lacaille JC, and Sossin WS (2013). Reactivation of stalled polyribosomes in synaptic plasticity. Proceedings of the National Academy of Sciences of the United States of America 110, 16205–16210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison-Uy SJ, and Pleasure SJ (2012). Wnt signaling and forebrain development. Cold Spring Harbor perspectives in biology 4, a008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetts SW, Sherr EH, Chao S, Gobuty S, and Barkovich AJ (2006). Anomalies of the corpus callosum: an MR analysis of the phenotypic spectrum of associated malformations. AJR American journal of roentgenology 187, 1343–1348. [DOI] [PubMed] [Google Scholar]
- Hinz FI, Dieterich DC, and Schuman EM (2013). Teaching old NCATs new tricks: using non-canonical amino acid tagging to study neuronal plasticity. Curr Opin Chem Biol 17, 738–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, Fontoura BMA, and Weis K (2019). DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Tailor J, Zhen Q, Gillmor AH, Miller ML, Weishaupt H, Chen J, Zheng T, Nash EK, McHenry LK, et al. (2019). Engineering Genetic Predisposition in Human Neuroepithelial Stem Cells Recapitulates Medulloblastoma Tumorigenesis. Cell Stem Cell 25, 433–446 e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. (2014). The contribution of de novo coding mutations to autism spectrum disorder. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamuar SS, and Walsh CA (2015). Genomic variants and variations in malformations of cortical development. Pediatric clinics of North America 62, 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, and Putnam A (2010). Duplex unwinding with DEAD-box proteins. Methods in molecular biology (Clifton, NJ: ) 587, 245–264. [DOI] [PubMed] [Google Scholar]
- Jiang L, Gu ZH, Yan ZX, Zhao X, Xie YY, Zhang ZG, Pan CM, Hu Y, Cai CP, Dong Y, et al. (2015). Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nature genetics 47, 1061–1066. [DOI] [PubMed] [Google Scholar]
- Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, et al. (2012). Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, and Hirokawa N (2004). Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525. [DOI] [PubMed] [Google Scholar]
- Kellaris G, Khan K, Baig SM, Tsai IC, Zamora FM, Ruggieri P, Natowicz MR, and Katsanis N (2018). A hypomorphic inherited pathogenic variant in DDX3X causes male intellectual disability with additional neurodevelopmental and neurodegenerative features. Hum Genomics 12, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotov AA, Olenkina OM, Kibanov MV, and Olenina LV (2016). RNA helicase Belle (DDX3) is essential for male germline stem cell maintenance and division in Drosophila. Biochimica et biophysica acta 1863, 1093–1105. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lee YH, and Tarn WY (2008). The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell 19, 3847–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox AL, Mao H, and Silver DL (2018). RNA on the brain: emerging layers of post-transcriptional regulation in cerebral cortex development. Wiley interdisciplinary reviews Developmental biology 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessel D, Schob C, Kury S, Reijnders MRF, Harel T, Eldomery MK, Coban-Akdemir Z, Denecke J, Edvardson S, Colin E, et al. (2017). De Novo Missense Mutations in DHX30 Impair Global Translation and Cause a Neurodevelopmental Disorder. Am J Hum Genet 101, 716–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhang P, Zhang C, Wang Y, Wan R, Yang Y, Guo X, Huo R, Lin M, Zhou Z, et al. (2014). DDX3X regulates cell survival and cell cycle during mouse early embryonic development. J Biomed Res 28, 282–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsalata AE, He F, Malik AM, Glineburg MR, Green KM, Natla S, Flores BN, Krans A, Archbold HC, Fedak SJ, et al. (2019). DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation. EMBO Rep 20, e47498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, and Herschlag D (1999). Kinetic dissection of fundamental processes of eukaryotic translation initiation in vitro. The EMBO journal 18, 6705–6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, McMahon JJ, Tsai YH, Wang Z, and Silver DL (2016). Haploinsufficiency for Core Exon Junction Complex Components Disrupts Embryonic Neurogenesis and Causes p53-Mediated Microcephaly. PLoS Genet 12, e1006282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Pilaz LJ, McMahon JJ, Golzio C, Wu D, Shi L, Katsanis N, and Silver DL (2015). Rbm8a haploinsufficiency disrupts embryonic cortical development resulting in microcephaly. J Neurosci 35, 7003–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, et al. (2018). Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 172, 590–604 e513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. (2014). Transcriptional landscape of the prenatal human brain. Nature 508, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Goff LA, Brettler AC, Chen HH, Hrvatin S, Rinn JL, and Arlotta P (2015). DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 85, 275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata Y, Barkovich AJ, Wahl M, Strominger Z, Jeremy RJ, Wakahiro M, Mukherjee P, and Sherr EH (2009). Diffusion abnormalities and reduced volume of the ventral cingulum bundle in agenesis of the corpus callosum: a 3T imaging study. AJNR American journal of neuroradiology 30, 1142–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Flynn RA, Floor SN, Purzner J, Martin L, Do BT, Schubert S, Vaka D, Morrissy S, Li Y, et al. (2016). Medulloblastoma-associated DDX3 variant selectively alters the translational response to stress. Oncotarget 7, 28169–28182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek JW, and Kai T (2011). DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proceedings of the National Academy of Sciences of the United States of America 108, 12007–12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung B, Ciesla M, Sanna A, Guzzi N, Beneventi G, Cao Thi Ngoc P, Lauss M, Cabrita R, Cordero E, Bosch A, et al. (2019). The X-Linked DDX3X RNA Helicase Dictates Translation Reprogramming and Metastasis in Melanoma. Cell reports 27, 3573–3586 e3577. [DOI] [PubMed] [Google Scholar]
- Polioudakis D, de la Torre-Ubieta L, Langerman J, Elkins AG, Shi X, Stein JL, Vuong CK, Nichterwitz S, Gevorgian M, Opland CK, et al. (2019). A Single-Cell Transcriptomic Atlas of Human Neocortical Development during Mid-gestation. Neuron 103, 785–801 e788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, et al. (2012). Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488, 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, and Parker R (2013). Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154, 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RK CY, Merico D, Bookman M, L.H. J, Thiruvahindrapuram B, Patel RV, Whitney J, Deflaux N, Bingham J, Wang Z, et al. (2017). Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nature neuroscience 20, 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. (2012). Novel mutations target distinct subgroups of medulloblastoma. Nature 488, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, B. ALC. (2003). Social Communication Questionnaire (Los Angeles, CA: Western Psychological Services; ). [Google Scholar]
- Ruzzo EK, Perez-Cano L, Jung JY, Wang LK, Kashef-Haghighi D, Hartl C, Singh C, Xu J, Hoekstra JN, Leventhal O, et al. (2019). Inherited and De Novo Genetic Risk for Autism Impacts Shared Networks. Cell 178, 850–866 e826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T (2006). In vivo electroporation in the embryonic mouse central nervous system. Nature protocols 1, 1552–1558. [DOI] [PubMed] [Google Scholar]
- Samir P, Kesavardhana S, Patmore DM, Gingras S, Malireddi RKS, Karki R, Guy CS, Briard B, Place DE, Bhattacharya A, et al. (2019). DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome. Nature 573, 590–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala M, Torella A, Severino M, Morana G, Castello R, Accogli A, Verrico A, Vari MS, Cappuccio G, Pinelli M, et al. (2019). Three de novo DDX3X variants associated with distinctive brain developmental abnormalities and brain tumor in intellectually disabled females. Eur J Hum Genet 27, 1254–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, and Pierre P (2009). SUnSET, a nonradioactive method to monitor protein synthesis. Nature methods 6, 275–277. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, and Jankowsky E (2014). The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit Rev Biochem Mol Biol 49, 343–360. [DOI] [PubMed] [Google Scholar]
- Shih JW, Tsai TY, Chao CH, and Wu Lee YH (2008). Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27, 700–714. [DOI] [PubMed] [Google Scholar]
- Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, and Wu Lee YH (2012). Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J 441, 119–129. [DOI] [PubMed] [Google Scholar]
- Snijders Blok L, Madsen E, Juusola J, Gilissen C, Baralle D, Reijnders MR, Venselaar H, Helsmoortel C, Cho MT, Hoischen A, et al. (2015). Mutations in DDX3X Are a Common Cause of Unexplained Intellectual Disability with Gender-Specific Effects on Wnt Signaling. American journal of human genetics 97, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Rifo R, and Ohlmann T (2013). The role of the DEAD-box RNA helicase DDX3 in mRNA metabolism. Wires Rna 4, 369–385. [DOI] [PubMed] [Google Scholar]
- Sparrow S, Cicchetti DV, & Balla D (2005). Vineland-II Adaptive Behavior Scales. (Circle Pines, MN: American Guidance Service; ). [Google Scholar]
- Takanashi J, and Barkovich AJ (2003). The changing MR imaging appearance of polymicrogyria: a consequence of myelination. AJNR American journal of neuroradiology 24, 788–793. [PMC free article] [PubMed] [Google Scholar]
- Takata A, Miyake N, Tsurusaki Y, Fukai R, Miyatake S, Koshimizu E, Kushima I, Okada T, Morikawa M, Uno Y, et al. (2018). Integrative Analyses of De Novo Mutations Provide Deeper Biological Insights into Autism Spectrum Disorder. Cell Rep 22, 734–747. [DOI] [PubMed] [Google Scholar]
- Taverna E, Gotz M, and Huttner WB (2014). The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annual review of cell and developmental biology 30, 465–502. [DOI] [PubMed] [Google Scholar]
- Toriello HV, Colley C, and Bamshad M (2016). Update on the Toriello-Carey syndrome. American journal of medical genetics Part A 170, 2551–2558. [DOI] [PubMed] [Google Scholar]
- Valentin-Vega YA, Wang YD, Parker M, Patmore DM, Kanagaraj A, Moore J, Rusch M, Finkelstein D, Ellison DW, Gilbertson RJ, et al. (2016). Cancer-associated DDX3X utations drive stress granule assembly and impair global translation. Sci Rep 6, 25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Weghe JC, Rusterholz TDS, Latour B, Grout ME, Aldinger KA, Shaheen R, Dempsey JC, Maddirevula S, Cheng YH, Phelps IG, et al. (2017). Mutations in ARMC9, which Encodes a Basal Body Protein, Cause Joubert Syndrome in Humans and Ciliopathy Phenotypes in Zebrafish. American journal of human genetics 101, 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Thaller C, and Eichele G (2004). GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic acids research 32, D552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Posey JE, Rosenfeld JA, Bacino CA, Scaglia F, Immken L, Harris JM, Hickey SE, Mosher TM, Slavotinek A, et al. (2018). Phenotypic expansion in DDX3X - a common cause of intellectual disability in females. Ann Clin Transl Neurol 5, 1277–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Smibert CA, Kaplan DR, and Miller FD (2014). An eIF4E1/4E-T complex determines the genesis of neurons from precursors by translationally repressing a proneurogenic transcription program. Neuron 84, 723–739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Comprehensive phenotypes for DDX3X patients, Related to Figure 1.
Data Availability Statement
This study did not generate any unique code. Original/source data for figures in the paper are available from the corresponding authors on request.


