Highlights
-
•
PDCoV infection fails to induce IFN-λ1 production in IPI-2I cells.
-
•
PDCoV infection suppresses Sendai virus-induced IFN-λ1 production.
-
•
PDCoV infection inhibits the nuclear translocation of IRF1.
-
•
PDCoV infection results in reduction of the number of peroxisomes.
-
•
MAVS is the possible target for PDCoV to inhibit IFN-λ1 production.
Keywords: Porcine deltacoronavirus, Type III interferon, Immunosuppression
Abstract
Porcine deltacoronavirus (PDCoV) is a novel swine enteropathogenic coronavirus that causes watery diarrhea, vomiting and mortality in nursing piglets. Type III interferons (IFN-λs) are the major antiviral cytokines in intestinal epithelial cells, the target cells in vivo for PDCoV. In this study, we found that PDCoV infection remarkably inhibited Sendai virus-induced IFN-λ1 production by suppressing transcription factors IRF and NF-κB in IPI-2I cells, a line of porcine intestinal mucosal epithelial cells. We also confirmed that PDCoV infection impeded the activation of IFN-λ1 promoter stimulated by RIG-I, MDA5 and MAVS, but not by TBK1 and IRF1. Although the expression levels of IRF1 and MAVS were not changed, PDCoV infection resulted in reduction of the number of peroxisomes, the platform for MAVS to activate IRF1, and subsequent type III IFN production. Taken together, our study demonstrates that PDCoV suppresses type III IFN responses to circumvent the host’s antiviral immunity.
1. Introduction
Porcine deltacoronavirus (PDCoV) is an emerging swine enteric coronavirus (CoV) that belongs to the newly identified genus Deltacoronavirus, within the family Coronaviridae, in the order Nidovirales (Ma et al., 2015; Wang et al., 2016; Woo et al., 2012). The full-length genome of PDCoV is approximately 25.4 kb, encoding 15 mature nonstructural proteins (nsp2−16), four structural proteins (spike (S) protein, envelope (E) protein, membrane (M) protein, nucleocapsid (N) protein) and three accessory proteins (NS6, NS7, NS7a) (Fang et al., 2017, 2016; Wang et al., 2019). PDCoV infection causes enteric disease of piglets characterized by severe atrophic enteritis, diarrhea, vomiting and dehydration (Chen et al., 2015; Jung et al., 2016; Zhang, 2016). PDCoV was first discovered in 2012 in Hong Kong (Woo et al., 2012). The first outbreak of PDCoV was reported in 2014 in the United States and rapidly spread to at least 20 states, resulting in significant economic losses (Hu et al., 2015; Marthaler et al., 2014; Wang et al., 2014). Thereafter, many countries reported the emergence of PDCoV, including China, Canada, Japan, South Korea, Thailand, Lao People’s Democratic Republic and Vietnam (Dong et al., 2015; Janetanakit et al., 2016; Jang et al., 2017; Lee et al., 2016; Lorsirigool et al., 2016; Saeng-Chuto et al., 2017; Song et al., 2015). Recent studies reported that calves, chickens and turkey poults are also susceptible to PDCoV (Boley et al., 2020; Jung et al., 2017; Liang et al., 2019), and that PDCoV possesses the potential to infect humans (Li et al., 2018; Wang et al., 2018), posing a significant threat to human and animal health.
Interferons (IFNs) are key components of the host’s antiviral innate immunity. To date, three different types of IFNs, type I IFN (IFN-α, IFN-β, IFN-ε, IFN-κ and IFN-ω), type II IFN (IFN-γ) and type III IFN (IFN-λ), have been discovered (Kotenko et al., 2003). Relative to the well-known type I and type II IFNs, type III IFNs were recently discovered as a distinct class of antiviral cytokines. Similar to type I IFN, type III IFN is a multigene family consisting of four members in humans (IFN-λ1, IFN-λ2, IFN-λ3 and IFN-λ4), two in mice (IFN-λ2 and IFN-λ3) and three in swine (IFN-λ1, IFN-λ3 and IFN-λ4) (Kotenko et al., 2019; Zanoni et al., 2017). Type III IFNs have many similar, but some different, induction processes to type I IFNs (Lazear et al., 2019; Onoguchi et al., 2007). In virus-infected cells, pathogen-associated molecular patterns (PAMPs), such as certain viral RNA replication intermediates or leader RNAs, can be recognized by host pattern-recognition receptors (PRRs), such as retinoic acid-induced gene I (RIG-I) or melanoma differentiation gene 5 (MDA5). After recognition of PAMPs, RIG-I and/or MDA5 interact with the mitochondrial antiviral-signaling (MAVS) protein, leading to the activation of transcription factors, which include interferon regulatory factors (IRFs) and NF-κB. The activated IRFs and NF-κB are translocated to the nucleus and induce the production of type I and type III IFNs. Among these IRFs, IRF3 and IRF7 are mainly involved in the induction of type I IFN, while IRF1 appears to play a more important role than IRF3 and IRF7 in the induction of type III IFN (Odendall et al., 2014). Another main difference between the induction of type I and type III IFN systems is that mitochondrial MAVS induces activation of type I IFNs, while peroxisomal MAVS is responsible for the type III IFN response (Lee and Baldridge, 2017).
Previous studies have reported that PDCoV infection inhibits the type I IFN response to evade the host’s antiviral immune responses (Fang et al., 2018; Likai et al., 2019; Liu et al., 2019; Luo et al., 2016; Zhu et al., 2017a, b). However, the small intestines, particularly the jejunum and ileum, are the targets of PDCoV infection in vivo (Chen et al., 2015; Jung et al., 2016; Wang et al., 2016). It is not clear whether PDCoV inhibits the type III IFN response. In this study, we demonstrated that PDCoV infection remarkably suppressed Sendai virus (SeV)-induced IFN-λ1 production. Mechanistically, PDCoV decreases the number of peroxisomes and inhibits IRF1 nuclear translocation, impairing the induction of IFN-λ1.
2. Materials and methods
2.1. Cells, viruses and reagents
LLC-PK1 cells (porcine kidney cells) were maintained in modified Eagle's medium (MEM) with 10 % heat-inactivated fetal bovine serum (FBS) at 37 °C with 5 % CO2 in a humidified incubator. IPI-2I cells (porcine intestinal mucosal epithelial cells) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % heat-inactivated FBS. PDCoV strain CHN-HN-2014 used in this study was isolated from a piglet with severe diarrhea in 2014 in China (Dong et al., 2016). SeV was obtained from the Centre of Virus Resource and Information, Wuhan Institute of Virology, Chinese Academy of Sciences. Rabbit anti-IRF1 polyclonal antibody (Bioss Antibodies, bs-21318R) and rabbit anti-PMP70 polyclonal antibody (Novus Biologicals, NBP187258) were used for immunofluorescence assays (IFAs) and western blotting. Alexa Fluor 488-conjugated donkey anti-mouse and 594-conjugated donkey anti-rabbit antibodies were purchased from Abbkine. Monoclonal antibodies against Flag and β-actin were purchased from MBL. Mouse monoclonal antibody against PDCoV N protein was created in-house as described previously (Zhu et al., 2018).
2.2. Plasmids
The luciferase reporter plasmids p-55λ1-(-225/-36)-Luc, p-55λ1mut.IRF-Luc, p-55λ1mut.NF-κB-Luc and p-55λ1mut.IRF/mut.NF-κB-Luc were kindly provided by Dr. Takashi Fujita (Kyoto University, Kyoto, Japan) (Onoguchi et al., 2007). The eukaryotic expression plasmids encoding Flag-tagged RIG-I, MDA5, MAVS, IRF1, IRF3 or IRF7 have been described previously (Wang et al., 2011).
2.3. RNA extraction and quantitative real-time PCR
Cells were washed twice with phosphate-buffered saline (PBS) before RNA isolation. Total cellular RNA was isolated using TRIzol reagent (OMEGA) following the manufacturer's protocol. Cellular RNA was then reverse transcribed into cDNA using a transcriptor first strand cDNA synthesis kit (Roche). The cDNA was subjected to quantitative PCR using the SYBR green PCR assay system (Applied Biosystems) at least in triplicate. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was chosen as an internal control for each experiment.
2.4. Indirect immunofluorescence assay (IFA)
IPI-2I cells in 24-well plates were mock-infected or infected with PDCoV at a multiplicity of infection (MOI) of 0.5. At 24 h post-infection (hpi), cells were fixed with 4 % paraformaldehyde for 15 min and permeabilized using 100 % methanol at room temperature for 10 min. After three washes with PBS, the cells were blocked with PBS containing 5 % bovine serum albumin for 1 h and then incubated with primary antibody for 1 h. The cells were then treated with secondary antibodies for 1 h at 37 °C in the dark, followed by treatment with 4ʹ, 6-diamidino-2-phenylindole (DAPI) for 15 min. After three washes with PBST, fluorescent images were visualized and examined using a confocal laser scanning microscope (Olympus IX73).
2.5. Luciferase reporter assay
To examine activation of the IFN-λ1 promoter or its mutants during PDCoV infection, IPI-2I or LLC-PK1 cells were grown in 24-well plates and transfected with 0.1 μg/well reporter plasmid (p-55λ1-(-225/-36)-Luc, p-55λ1mut.IRF-Luc, p-55λ1mut.NF-κB-Luc or p-55λ1mut.IRF/mut.NF-κB-Luc) and 0.02 μg/well pRL-TK plasmid. At 24 h after transfection, cells were infected with PDCoV for 12 h followed by stimulation with SeV. After 12 h, cells were collected and subjected to luciferase activity detection. To determine at which step the PDCoV displays its inhibitory activities, IPI-2I cells were transfected for 24 h with luciferase reporter plasmid, an expression plasmid (RIG-I, MDA5, MAVS, IRF1) and pRL-TK at a ratio of 1:4:0.2. At 24 h post-transfection, cells were infected with PDCoV (MOI = 0.5). At 24 hpi, the cell lysates were harvested to measure the luciferase activities. The luciferase activities of firefly and renilla were determined with the Dual-Luciferase reporter assay system according to the manufacturer’s protocol (Promega). Transfections were performed using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen).
2.6. Western blot analysis
The infected or transfected IPI-2I cells were lysed at the indicated time points in RIPA lysis buffer [50 mM Tris−HCl (pH 7.4), 50 mM NaCl, 2 mM EDTA.2Na, 10 % glycerin, 0.1 % SDS, 1 % NP-40] supplemented with a protease inhibitor cocktail. The lysates were separated by 12 % SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5 % nonfat dry milk in TBST for 1 h and incubated with primary antibody at room temperature for 3 h. After washing three times with TBST, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody at room temperature for 1 h. After washing three times, the proteins were visualized by enhanced chemiluminescence (ECL) reagents (Bio-Rad). The endogenous expressions of IRF1 and MAVS proteins were analyzed with the indicated antibodies. The expressions of PDCoV N protein, Flag-tagged protein and β-actin were detected with monoclonal antibodies against N protein, Flag, and β-actin, respectively.
2.7. Statistical analysis
Data are shown as the mean ± standard deviation of three independent experiments. The results were analyzed for significance by a Student’s t test using GraphPad Prism 6 software. Differences between groups were considered statistically significant when P < 0.05.
3. Results
3.1. PDCoV infection blocks SeV-induced IFN-λ1 production
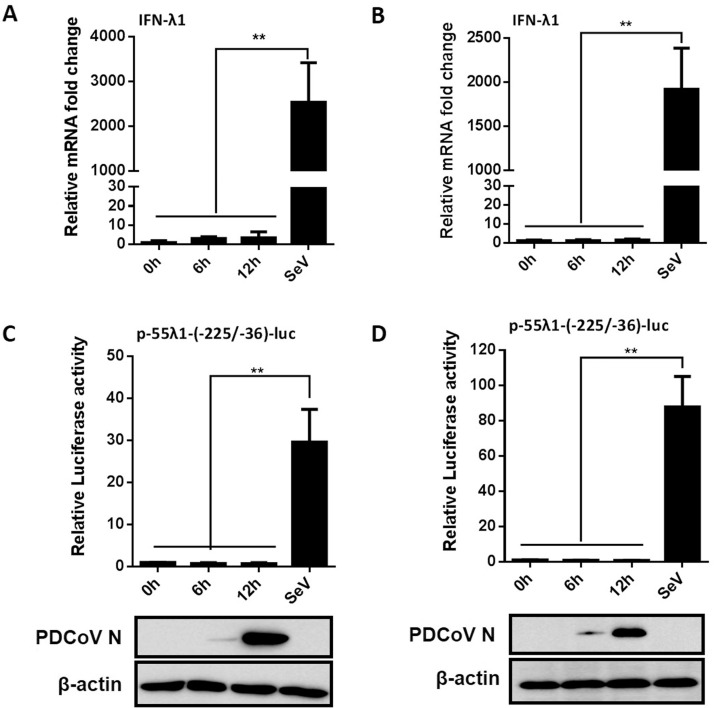
To investigate whether PDCoV infection inhibits the production of IFN-λ1, we first detected the mRNA expression of IFN-λ1 in PDCoV-infected cells. To this end, IPI-2I or LLC-PK1 cells were infected with PDCoV at a MOI of 0.5 and cells were collected at 0, 6 and 12 hpi for real-time RT-PCR. As shown in Fig. 1 A and 1B, there was no significant difference in the expression of IFN-λ1 mRNA in PDCoV-infected cells compared with the mock-infected cells, whereas the amount of IFN-λ1 mRNA in SeV-infected cells was significantly increased in the two cell types, indicating that PDCoV infection fails to induce the production of IFN-λ1.
Fig. 1.
PDCoV infection does not induce IFN-λ1 production. (A, B) IPI-2I cells (A) or LLC-PK1 cells (B) were infected with PDCoV at a MOI of 0.5 or 0.05, respectively. Cells were collected at 0, 6 and 12 h post-infection (hpi) to determine the mRNA expression of IFN-λ1 by RT-qPCR. Mock-infected cells stimulated with SeV for 12 h were used as a positive control. (C, D) IPI-2I cells (C) or LLC-PK1 cells (D) were co-transfected with p-55λ1-(-225/-36)-Luc and pRL-TK, followed by PDCoV infection. At 0, 6 and 12 hpi, cells were lysed for dual-luciferase reporter assays. Mock-infected cells stimulated with SeV for 12 h were used as a positive control. PDCoV infection was verified by western blot with anti-PDCoV N protein antibody. β-actin was detected by western blot analysis and used as a control for sample loading. All data are presented as the means ± SD of three independent experiments (**p < 0.01).
To examine whether PDCoV infection affects the activation of IFN-λ1 promoter, IPI-2I or LLC-PK1 cells were co-transfected with the luciferase reporter plasmid p-55λ1-(-225/-36)-Luc and the internal control plasmid pRL-TK. At 24 h after transfection, the cells were infected with PDCoV (MOI = 0.5), then collected at different time points for detecting IFN-λ1 promoter-driven luciferase activity. The infection of PDCoV was confirmed by western blot with monoclonal antibody against PDCoV N protein (Fig. 1C and D). Consistent with the results obtained by real-time RT-PCR, the IFN-λ1 promoter-driven luciferase activity was barely detectable in PDCoV-infected cells (Fig. 1C and D). As expected, SeV stimulation significantly activated IFN-λ1 promoter activity. Thus, PDCoV infection also failed to activate IFN-λ1 promoter activity.
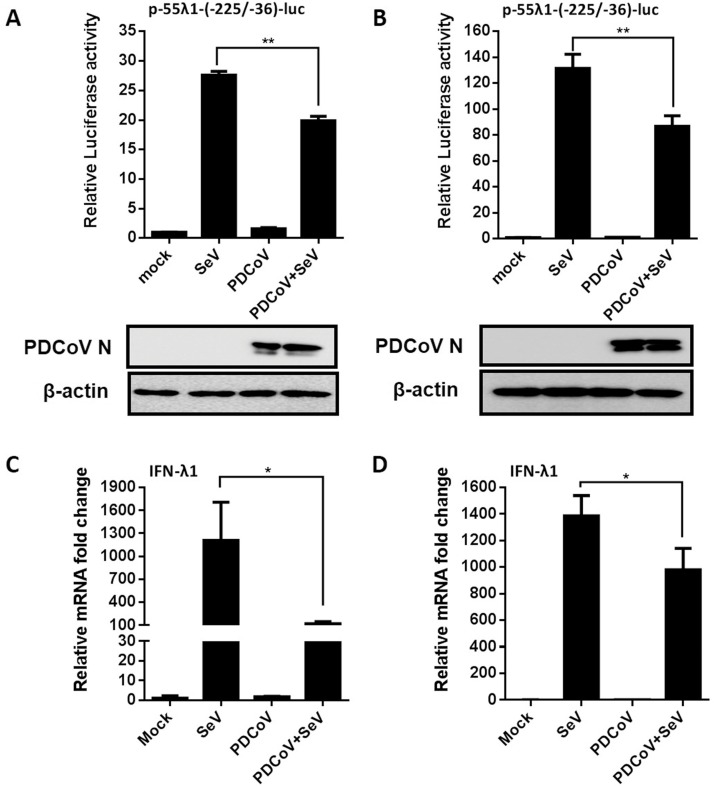
Since PDCoV infection did not induce IFN-λ1 production, we further examined whether PDCoV inhibits SeV-induced IFN-λ1 promoter activity. IPI-2I or LLC-PK1 cells were co-transfected with p-55λ1-(-225/-36)-Luc and pRL-TK, followed by infection with PDCoV. At 12 hpi, the cells were mock-stimulated or stimulated with SeV. As shown in Fig. 2 , SeV stimulation activated IFN-λ1 promoter activity in cells without PDCoV infection; however, SeV-induced IFN-λ1 promoter activation was significantly inhibited after PDCoV infection in both IPI-2I (Fig. 2A) and LLC-PK1 cells (Fig. 2B). We also examined the mRNA expression of IFN-λ1 in PDCoV-infected, SeV-stimulated cells by RT-qPCR. As expected, SeV stimulation significantly upregulated the expression of IFN-λ1 mRNA; however, PDCoV infection significantly inhibited SeV-induced IFN-λ1 mRNA expression (Fig. 2C and D).
Fig. 2.
PDCoV infection inhibits SeV-induced IFN-λ1 production. (A, B) IPI-2I cells (A) or LLC-PK1 cells (B) were co-transfected with p-55λ1-(-225/-36)-Luc and pRL-TK, followed by PDCoV infection. At 12 hpi, the cells were stimulated with SeV for 12 h. Cell lysates were prepared for dual-luciferase reporter assays. PDCoV infection was verified by western blot with anti-PDCoV N protein antibody. β-actin served as a protein loading control. (C, D) IPI-2I cells (C) or LLC-PK1 cells (D) were infected with PDCoV at a MOI of 0.5 or 0.05, respectively, and then stimulated with SeV for 12 h. The total cellular RNAs were extracted to determine the IFN-λ mRNA levels by RT-qPCR. All data are presented as the means ± SD of three independent experiments (*p < 0.05 and **p < 0.01).
3.2. PDCoV infection inhibits the activation of IRF and NF-κB
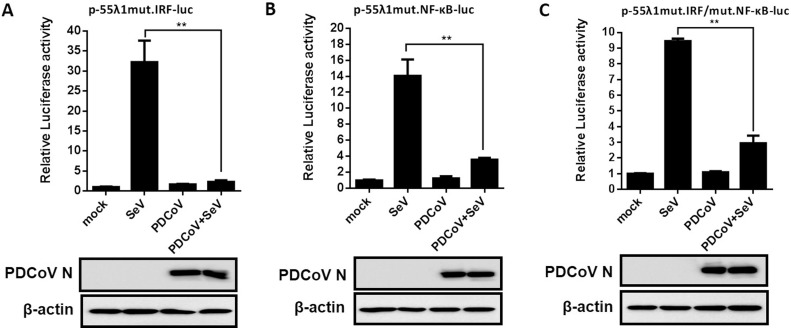
IRF and NF-κB are critical transcription factors for the production of IFN-λs (Onoguchi et al., 2007). To investigate whether PDCoV prevents the production of IFN-λ1 by inhibiting the activation of NF-κB and IRFs, LLC-PK1 cells were co-transfected with the internal control plasmid pRL-TK and the luciferase reporter plasmids p-55λ1mut.IRF-Luc, p-55λ1mut.NF-κB-Luc or p-55λ1mut.IRF/mut.NF-κB-Luc, which contain the binding-site mutation for IRF, NF-κB, and both IRF and NF-κB, respectively. At 24 h after co-transfection, cells were mock-infected or infected with PDCoV at a MOI of 0.01, then stimulated, or not, with SeV. The results showed that PDCoV infection failed to activate the promoter activity of these luciferase reporter plasmids (Fig. 3 A–C). The SeV-induced promoter activity of these luciferase reporter plasmids was also significantly impaired by PDCoV infection (Fig. 3A–C). These results indicated that PDCoV prevented the production of IFN-λ by blocking activation of the transcription factors NF-κB and IRF.
Fig. 3.
PDCoV infection prevents the production of IFN-λ1 by blocking activation of NF-κB and IRF. LLC-PK1 cells were transfected with the luciferase reporter plasmids p-55λ1mut.IRF-Luc (A), p-55λ1mut.NF-κB-Luc (B) or p-55λ1mut.IRF/mut.NF-κB-Luc (C) together with pRL-TK, and then mock-infected or infected with PDCoV at a MOI of 0.05. At 12 hpi, the cells were mock-infected or infected with SeV for 12 h. Cell lysates were prepared for dual-luciferase reporter assays. All data are presented as the means ± SD of three independent experiments (**p < 0.01). Western blot analysis with anti-PDCoV N protein antibody showed that PDCoV can normally infect cells, and β-actin was used as a control for sample loading.
3.3. PDCoV infection suppresses the IFN-λ1 promoter activity induced by RIG-I, MDA5 and MAVS
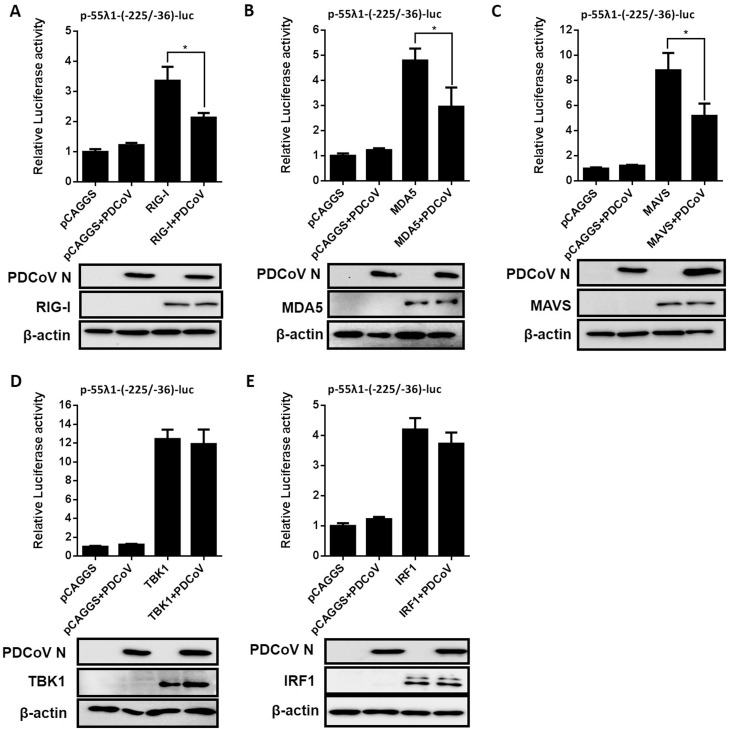
Previous studies have shown that PDCoV interrupts the RIG-I-like receptor (RLR)-mediated IFN signaling pathway (Fang et al., 2018; Luo et al., 2016), and the RLR-mediated signaling is involved in IFN-λs production (Onoguchi et al., 2007). To investigate at which step PDCoV displays its inhibitory activity on IFN-λ1 production, IPI-2I cells were co-transfected with p-55λ1-(-225/-36)-Luc and a plasmid expressing Flag-tagged RIG-I, MDA5, MAVS, TBK1 or IRF1, together with the pRL-TK internal control. At 24 h after transfection, cells were infected with PDCoV at a MOI of 0.5. The firefly and renilla luciferase activities of all samples were determined at 24 hpi. As shown in Fig. 4 , PDCoV infection reduced activation of the IFN-λ1 promoter activity mediated by RIG-I, MDA5 and MAVS, but had no obvious effects on activation induced by TBK1 and IRF1. The expressions of Flag-tagged RIG-I, MDA5, MAVS, TBK1 or IRF1 were confirmed by western blots with an anti-Flag antibody. These results indicated that PDCoV inhibits the IFN-λ1 production pathway by targeting MAVS or its associated molecules, but not TBK1 or its downstream molecules among the RLR signaling pathway.
Fig. 4.
The effect of PDCoV infection on IFN-λ1 induced by molecules in the RIG-I signaling pathway. IPI-2I cells were transfected with a plasmid expressing RIG-I (A), MDA5 (B), MAVS (C), TBK1 (D), IRF1 (E) or an empty vector, along with p-55λ1-(-225/-36)-Luc and pRL-TK, then the cells were mock-infected or infected with PDCoV at a MOI of 0.5 for 24 h. Cell lysates were prepared for dual-luciferase reporter assays. All data are presented as the means ± SD of three independent experiments (*p < 0.05). The expressions of Flag-tagged RIG-I, MDA5, MAVS, TBK1 or IRF1 were confirmed by western bot with anti-Flag antibody. Antibodies against N protein was used to confirm the infection of PDCoV. β-actin was detected by western blot and used as a control for sample loading.
3.4. PDCoV infection decreases the number of peroxisomes and inhibits IRF1 nuclear translocation
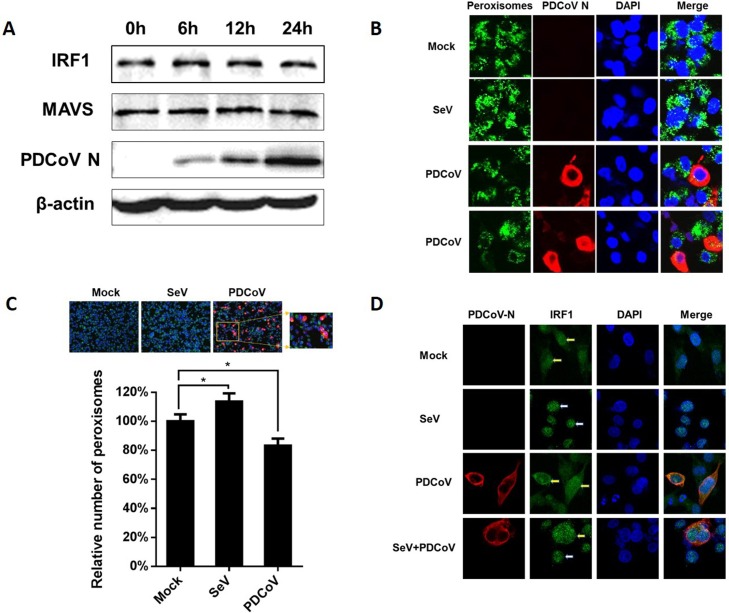
MAVS is the critical adaptor and IRF1 is the most important transcription factor for IFN-λs production, and previous studies showed that some viruses antagonize IFN-λs production by degrading MAVS or IRF1 (Ding and Robek, 2014; Ding et al., 2018). Thus, we further examined the expression of MAVS and IRF1 after PDCoV infection. The results showed that the protein levels of MAVS and IRF1 were not altered at different time points after PDCoV infection (Fig. 5 A).
Fig. 5.
PDCoV infection decreases the number of peroxisomes and inhibits IRF1 nuclear translocation. (A) IPI-2I cells were infected with PDCoV at a MOI of 0.5. At the indicated time points after infection, cells were collected and subjected to western blotting with antibodies against IRF1, MAVS and PDCoV N protein. β-actin was used as a control for sample loading. (B) IPI-2I cells were infected with PDCoV for 24 h or stimulated with SeV for 12 h as a positive control, and then fixed for indirect immunofluorescence assays with antibodies against PMP70 (peroxisome marker) and PDCoV N protein. Peroxisomes and PDCoV N protein were visualized by Alexa Fluor 488-conjugated donkey anti-mouse (green) and 594-conjugated donkey anti-rabbit (red) antibodies, respectively. Cellular nuclei were counterstained with 1 μg/mL of DAPI. Fluorescence was observed under a Fluoview ver. 3.1 confocal fluorescence microscope (Olympus) and representative images are shown. (C) IPI-2I cells were mock-infected or infected with PDCoV (MOI = 0.5) for 24 h or stimulated with SeV for 12 h as a positive control. The cells were fixed and the number of peroxisomes in PDCoV-infected cells, mock-infected cells or cells stimulated with SeV was quantified. The data are presented as the means ± SD of three independent experiments (*p < 0.05). (D) IPI-2I cells were infected with PDCoV (MOI = 0.5) for 12 h followed by stimulation with SeV for 12 h. Cells were fixed and incubated with anti-PDCoV N protein antibody and anti-IRF1 antibody for 1 h. IRF1 and PDCoV N were visualized by Alexa Fluor 488-conjugated (green) donkey anti-mouse and 594-conjugated (red) donkey anti-rabbit antibodies. Cellular nuclei were counterstained with DAPI. White arrows indicate the nucleus-localized IRF1 and yellow arrows indicate the cytoplasm-localized IRF1 (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Peroxisome is the innate antiviral signaling platform for the type III IFN signaling pathway (Ding and Robek, 2014). Peroxisome-localized MAVS is associated with the production of IFN-λs (Dixit et al., 2010). To investigate whether PDCoV infection affects the peroxisome-localization of MAVS, IPI-2I cells were infected with PDCoV for 24 h. Peroxisomal membrane protein 70 (PMP70) was used as a marker for peroxisomes to aid their enumeration. As shown in Fig. 5B, the number of peroxisomes was significantly reduced in PDCoV-infected cells compared with the mock-infected cells. We also calculated the average number of peroxisomes in mock-infected, PDCoV-infected cells and cells stimulated with SeV. The results showed that about 20 % reduction of peroxisomes in PDCoV-infected cells, while the number of peroxisomes in cells stimulated with SeV increased slightly (Fig. 5C).
Previous studies showed that peroxisomal MAVS activates IRF1-mediated IFN-λs production (Ding and Robek, 2014; Dixit et al., 2010). Because PDCoV infection reduced the number of peroxisomes in cells and PDCoV infection does not affect IRF1 expression, we further investigated whether PDCoV infection affects the nuclear translocation of IRF1, an essential step for IRF1-mediated IFN-λs production. Thus, we analyzed the subcellular localization of IRF1 after PDCoV infection by indirect immunofluorescence assays (IFAs). As shown in Fig. 5D, in mock-infected cells, IRF1 was widely distributed in the cytoplasm (yellow arrows), and translocated to the nucleus (white arrows) following SeV stimulation, whereas, in PDCoV-infected cells, IRF1 remained in the cytoplasm (yellow arrows) even after SeV stimulation.
4. Discussion
In recent years, type III IFNs have received increasing attention because they play unique and non-redundant functions, especially in mucosal tissues, compared with type I IFNs (Galani et al., 2017). For example, type III IFNs have been demonstrated to be critical host factors that determine susceptibility to influenza virus infection in allergic nasal mucosa (Jeon et al., 2018), and mice lacking functional IFN-λ receptors (Ifnlr1˗/˗) no longer prevent influenza virus spread from the upper airways to the lungs (Klinkhammer et al., 2018). Similarly, mice lacking functional receptors for IFN-λ are impaired in the control of oral infection of rotavirus, and systemic treatment of suckling mice with IFN-λ represses rotavirus replication in the gut, whereas treatment with type I IFN had no effect (Pott et al., 2011). Another study also reported that IFN-λ preferentially inhibits the infection of porcine epidemic diarrhea virus (PEDV), an enteropathogenic coronavirus, in porcine intestinal epithelial cells compared with IFN-α (Li et al., 2017). Because type III IFNs have important immune functions in mucosal tissues, it is not surprising that many viruses take various strategies to antagonize type III IFN responses. For example, rotavirus VP3 targets MAVS for degradation to inhibit type III IFN expression in intestinal epithelial cells (Ding et al., 2018), and respiratory syncytial virus (RSV), a significant human pathogen that causes pneumonia and contributes to exacerbations of chronic lung diseases, activates epidermal growth factor receptor (EGFR) to suppress IRF1-dependent IFN-λ and antiviral defenses in airway epithelium (Kalinowski et al., 2018). In this study, we demonstrated that PDCoV infection can also antagonize the production of IFN-λ1 to evade host innate immune defenses. In addition to IFN-λ1, we also tested whether PDCoV infection affects the expression of IFN-λ3. We found that the basal expression level of IFN-λ3 mRNA was much lower than that of IFN-λ1 in both IPI-2I and LLC-PK1 cells. SeV stimulation significantly up-regulated the expression level of IFN-λ3 mRNA, however, the increase was also inhibited notably by PDCoV infection (data not shown). Considering the very low basal expression of IFN-λ3 in the tested cells, we did not further investigate the inhibition mechanism of PDCoV on IFN-λ3.
Different to type I IFN production, transcription factor IRF1 plays a vital role in inducing type III IFN (Odendall et al., 2014). Consistent with this concept, PDCoV infection inhibits the nuclear translocation of IRF1, a key step in IRF1 activation, to antagonize the production of IFN-λ1, although PDCoV infection does not regulate the expression of IRF1. In a recent study, Jiang et al. found that PDCoV infection can upregulate IRF1 mRNA expression at 24 h post-infection in PK-15 cells (Jiang et al., 2019). However, in the present study, we demonstrated that PDCoV infection does not affect the protein level of IRF1. These differences may be caused by different cell types because the infection dynamics of PDCoV in PK-15 cells is different from other permissive cell lines such as LLC-PK1 and IPI-2I (Jiang et al., 2019). In addition, we cannot rule out the possibility that PDCoV infection degrades IRF1 protein, although its mRNA expression is upregulated after PDCoV infection.
To identify the possible target(s) in the RLR pathway of PDCoV to inhibit IFN-λ1 production, we screened the components of the RLR pathway and found that PDCoV infection significantly impeded the activation of IFN-λ1 promoter stimulated by RIG-I, MDA5 and MAVS, but not by TBK1 and IRF1, suggesting that MAVS is the target of PDCoV to inhibit IFN-λ1 production. In mucosal tissues, peroxisomal MAVS is responsible for type III IFN responses, while mitochondrial MAVS induces type I IFN responses (Lee and Baldridge, 2017). Another study also showed that intestinal epithelial cells produce abundant type III IFNs by upregulation of the biogenesis of peroxisomes (Odendall et al., 2014). Peroxisomes in the intestinal epithelial cells are particularly crucial for the production of type III IFNs. Due to the lack of antibody against porcine MAVS and the poor cross-reactivity to porcine MAVS of antibody against human MAVS, we did not detect the number or localization of MAVS on peroxisomes by an indirect immunofluorescence assay following viral infection. As an alternative strategy, we detected the number of peroxisomes in the context of PDCoV infection. The results showed that the number of peroxisomes in PDCoV-infected cells was significantly reduced compared with mock-infected cells. These results further supported the conclusion that PDCoV inhibits the type III IFN response by targeting MAVS.
Previous studies from our laboratory and other groups have demonstrated that PDCoV infection inhibits the type I IFN response (Fang et al., 2018; Likai et al., 2019; Luo et al., 2016; Zhu et al., 2017a, b). In this study, we further showed that PDCoV infection can antagonize type III IFN production, suggesting that PDCoV possesses the ability to systematically suppress the host’s IFN responses. Although type III IFNs share a similar induction pathway to type I IFNs (Lazear et al., 2019), PDCoV appears to use a different strategy to antagonize the production of IFN-λs and IFN-α/β. Evidence for this is that PDCoV infection significantly suppresses the activation of IFN-β promoter stimulated by IRF3 or its upstream molecules (RIG-I, MDA5, MAVS, TBK1) in the RIG-I signaling pathway, but does not inhibit TBK1-mediated IFN-λ1 production. We also detected whether PDCoV infection affects IRF3 or IRF7-induced IFN-λ1 production, and found that PDCoV infection did not affect the production of IFN-λ1 induced by IRF3 or IRF7 (data not shown). Further comparing the anti-PDCoV activity of type III and I IFNs and dissecting the different mechanisms used by PDCoV to antagonize the production of type III and I IFNs, will help us to better understand the interaction between PDCoV and the host’s IFN system.
As a newly identified porcine enteric coronavirus, elucidation of the immune evasion strategy used by PDCoV is still at an early stage. Previous studies have shown that PEDV, another porcine enteropathogenic coronavirus, downregulates and evades IRF1-mediated type III IFN responses by reducing the number of peroxisomes, and several PEDV-encoded proteins, particularly nonstructural protein 1 (nsp1), are associated with the suppression of type III IFN activity (Zhang et al., 2018). PDCoV does not encode the orthologue nsp1 in PEDV (Ma et al., 2015). Whether other PDCoV-encoded proteins antagonize type III IFN production and the mechanisms involved, are issues that are currently under investigation in our laboratory.
5. Conclusions
In summary, this study is the first to demonstrate that PDCoV infection antagonizes IFN-λ1 production in IPI-2I cells. Mechanistically, PDCoV decreases the number of peroxisomes and inhibits IRF1 nuclear translocation, impairing the induction of IFN-λ1. Future studies to identify the virus-encoded protein(s) involved will help us to better understand the immune evasion mechanisms of this emerging porcine enteric coronavirus.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31730095, U1704231), the National Key R&D Program of China (2016YFD0500103), and the Special Project for Technology Innovation of Hubei Province (2017ABA138).
References
- Boley P.A., Alhamo M.A., Lossie G., Yadav K.K., Vasquez-Lee M., Saif L.J., Kenney S.P. Porcine deltacoronavirus infection and transmission in poultry, United States(1) Emerg Infect Dis. 2020;26:255–265. doi: 10.3201/eid2602.190346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P., Stafne M., Thomas J., Arruda P., Burrough E., Madson D., Brodie J., Magstadt D., Derscheid R., Welch M., Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Robek M.D. Peroxisomal MAVS activates IRF1-mediated IFN-lambda production. Nat. Immunol. 2014;15:700–701. doi: 10.1038/ni.2924. [DOI] [PubMed] [Google Scholar]
- Ding S., Zhu S., Ren L., Feng N., Song Y., Ge X., Li B., Flavell R.A., Greenberg H.B. Rotavirus VP3 targets MAVS for degradation to inhibit type III interferon expression in intestinal epithelial cells. Elife. 2018;7 doi: 10.7554/eLife.39494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E., Boulant S., Zhang Y., Lee A.S., Odendall C., Shum B., Hacohen N., Chen Z.J., Whelan S.P., Fransen M., Nibert M.L., Superti-Furga G., Kagan J.C. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Zeng S., Sun Q., Chen H., Xiao S. Porcine deltacoronavirus in Mainland China. Emerg Infect Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Yang H., Liu H., Du T., Fang P., Wang D., Chen H., Xiao S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet. Microbiol. 2016;196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Liu X., Hong Y., Wang Y., Dong N., Ma P., Bi J., Wang D., Xiao S. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology. 2016;499:170–177. doi: 10.1016/j.virol.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Hong Y., Liu X., Dong N., Ma P., Bi J., Wang D., Xiao S. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J. Gen. Virol. 2017;98:173–178. doi: 10.1099/jgv.0.000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Ren J., Hong Y., Liu X., Zhao Y., Wang D., Peng G., Xiao S. Porcine deltacoronavirus accessory protein NS6 antagonizes interferon Beta production by interfering with the binding of RIG-I/MDA5 to double-stranded RNA. J. Virol. 2018;92:e00712–18. doi: 10.1128/JVI.00712-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani I.E., Triantafyllia V., Eleminiadou E.E., Koltsida O., Stavropoulos A., Manioudaki M., Thanos D., Doyle S.E., Kotenko S.V., Thanopoulou K., Andreakos E. Interferon-lambda mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity. 2017;46:875–890. doi: 10.1016/j.immuni.2017.04.025. e6. [DOI] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Chepngeno J., Lu Z., Wang Q., Saif L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015;53:1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetanakit T., Lumyai M., Bunpapong N., Boonyapisitsopa S., Chaiyawong S., Nonthabenjawan N., Kesdaengsakonwut S., Amonsin A. Porcine Deltacoronavirus, Thailand, 2015. Emerg Infect Dis. 2016;22:757–759. doi: 10.3201/eid2204.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang G., Lee K.K., Kim S.H., Lee C. Prevalence, complete genome sequencing and phylogenetic analysis of porcine deltacoronavirus in South Korea, 2014-2016. Transbound. Emerg. Dis. 2017;64:1364–1370. doi: 10.1111/tbed.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y.J., Lim J.H., An S., Jo A., Han D.H., Won T.B., Kim D.Y., Rhee C.S., Kim H.J. Type III interferons are critical host factors that determine susceptibility to Influenza A viral infection in allergic nasal mucosa. Clin. Exp. Allergy. 2018;48:253–265. doi: 10.1111/cea.13082. [DOI] [PubMed] [Google Scholar]
- Jiang S., Li F., Li X., Wang L., Zhang L., Lu C., Zheng L., Yan M. Transcriptome analysis of PK-15 cells in innate immune response to porcine deltacoronavirus infection. PLoS One. 2019;14 doi: 10.1371/journal.pone.0223177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch. Virol. 2017;162:2357–2362. doi: 10.1007/s00705-017-3351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski A., Galen B.T., Ueki I.F., Sun Y., Mulenos A., Osafo-Addo A., Clark B., Joerns J., Liu W., Nadel J.A., Dela Cruz C.S., Koff J.L. Respiratory syncytial virus activates epidermal growth factor receptor to suppress interferon regulatory factor 1-dependent interferon-lambda and antiviral defense in airway epithelium. Mucosal Immunol. 2018;11:958–967. doi: 10.1038/mi.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkhammer J., Schnepf D., Ye L., Schwaderlapp M., Gad H.H., Hartmann R., Garcin D., Mahlakoiv T., Staeheli P. IFN-lambda prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife. 2018:7. doi: 10.7554/eLife.33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- Kotenko S.V., Rivera A., Parker D., Durbin J.E. Type III IFNs: beyond antiviral protection. Semin. Immunol. 2019;43 doi: 10.1016/j.smim.2019.101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H.M., Schoggins J.W., Diamond M.S. Shared and distinct functions of type I and type III interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Baldridge M.T. Interferon-lambda: a potent regulator of intestinal viral infections. Front. Immunol. 2017;8:749. doi: 10.3389/fimmu.2017.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Chung H.C., Nguyen V.G., Moon H.J., Kim H.K., Park S.J., Lee C.H., Lee G.E., Park B.K. Detection and phylogenetic analysis of porcine deltacoronavirus in korean swine farms, 2015. Transbound. Emerg. Dis. 2016;63:248–252. doi: 10.1111/tbed.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Fu F., Xue M., Chen W., Liu J., Shi H., Chen J., Bu Z., Feng L., Liu P. IFN-lambda preferably inhibits PEDV infection of porcine intestinal epithelial cells compared with IFN-alpha. Antiviral Res. 2017;140:76–82. doi: 10.1016/j.antiviral.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hulswit R.J.G., Kenney S.P., Widjaja I., Jung K., Alhamo M.A., van Dieren B., van Kuppeveld F.J.M., Saif L.J., Bosch B.J. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci USA. 2018;115:E5135–E5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Zhang H., Li B., Ding Q., Wang Y., Gao W., Guo D., Wei Z., Hu H. Susceptibility of chickens to porcine deltacoronavirus infection. Viruses. 2019;11:573. doi: 10.3390/v11060573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likai J., Shasha L., Wenxian Z., Jingjiao M., Jianhe S., Hengan W., Yaxian Y. Porcine Deltacoronavirus Nucleocapsid Protein Suppressed IFN-beta Production by Interfering Porcine RIG-I dsRNA-Binding and K63-Linked Polyubiquitination. Front. Immunol. 2019;10:1024. doi: 10.3389/fimmu.2019.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Fang P., Fang L., Hong Y., Zhu X., Wang D., Peng G., Xiao S. Porcine deltacoronavirus nsp15 antagonizes interferon-beta production independently of its endoribonuclease activity. Mol. Immunol. 2019;114:100–107. doi: 10.1016/j.molimm.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsirigool A., Saeng-Chuto K., Temeeyasen G., Madapong A., Tripipat T., Wegner M., Tuntituvanont A., Intrakamhaeng M., Nilubol D. The first detection and full-length genome sequence of porcine deltacoronavirus isolated in Lao PDR. Arch. Virol. 2016;161:2909–2911. doi: 10.1007/s00705-016-2983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Fang L., Dong N., Fang P., Ding Z., Wang D., Chen H., Xiao S. Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-beta production. Virology. 2016;495:10–17. doi: 10.1016/j.virol.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6 doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler D., Raymond L., Jiang Y., Collins J., Rossow K., Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg Infect Dis. 2014;20:1347–1350. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odendall C., Dixit E., Stavru F., Bierne H., Franz K.M., Durbin A.F., Boulant S., Gehrke L., Cossart P., Kagan J.C. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoguchi K., Yoneyama M., Takemura A., Akira S., Taniguchi T., Namiki H., Fujita T. Viral infections activate types I and III interferon genes through a common mechanism. J. Biol. Chem. 2007;282:7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- Pott J., Mahlakoiv T., Mordstein M., Duerr C.U., Michiels T., Stockinger S., Staeheli P., Hornef M.W. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeng-Chuto K., Lorsirigool A., Temeeyasen G., Vui D.T., Stott C.J., Madapong A., Tripipat T., Wegner M., Intrakamhaeng M., Chongcharoen W., Tantituvanont A., Kaewprommal P., Piriyapongsa J., Nilubol D. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and lao PDR in 2015. Transbound. Emerg. Dis. 2017;64:3–10. doi: 10.1111/tbed.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Zhou X., Peng Q., Chen Y., Zhang F., Huang T., Zhang T., Li A., Huang D., Wu Q., He H., Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in Swine in China: identification, prevalence and full-length genome sequence analysis. Transbound. Emerg. Dis. 2015;62:575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Liu L., Zhong H., Chen Q., Luo R., Liu X., Zhang Z., Chen H., Xiao S. Foot-and-mouth disease virus (FMDV) leader proteinase negatively regulates the porcine interferon-lambda1 pathway. Mol. Immunol. 2011;49:407–412. doi: 10.1016/j.molimm.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis. 2014;20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hayes J., Sarver C., Byrum B., Zhang Y. Porcine deltacoronavirus: histological lesions and genetic characterization. Arch. Virol. 2016;161:171–175. doi: 10.1007/s00705-015-2627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Liu Y., Ji C.M., Yang Y.L., Liang Q.Z., Zhao P., Xu L.D., Lei X.M., Luo W.T., Qin P., Zhou J., Huang Y.W. Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J. Virol. 2018;92:e00318–18. doi: 10.1128/JVI.00318-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Vlasova A.N., Kenney S.P., Saif L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019;34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni I., Granucci F., Broggi A. Interferon (IFN)-lambda takes the helm: immunomodulatory roles of type III IFNs. Front. Immunol. 2017;8:1661. doi: 10.3389/fimmu.2017.01661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 2016;226:71–84. doi: 10.1016/j.virusres.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ke H., Blikslager A., Fujita T., Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J. Virol. 2018;92:e01677–17. doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Fang L., Wang D., Yang Y., Chen J., Ye X., Foda M.F., Xiao S. Porcine deltacoronavirus nsp5 inhibits interferon-beta production through the cleavage of NEMO. Virology. 2017;502:33–38. doi: 10.1016/j.virol.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang D., Zhou J., Pan T., Chen J., Yang Y., Lv M., Ye X., Peng G., Fang L., Xiao S. Porcine deltacoronavirus nsp5 antagonizes type I interferon signaling by cleaving STAT2. J. Virol. 2017;91:e00003–17. doi: 10.1128/JVI.00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Liu S., Wang X., Luo Z., Shi Y., Wang D., Peng G., Chen H., Fang L., Xiao S. Contribution of porcine aminopeptidase N to porcine deltacoronavirus infection. Emerg. Microbes Infect. 2018;7:65. doi: 10.1038/s41426-018-0068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]