Abstract
The global pandemic caused by the newly described severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused worldwide suffering and death of unimaginable magnitude from coronavirus disease 2019 (COVID-19). The virus is transmitted through aerosol droplets, and causes severe acute respiratory syndrome. SARS-CoV-2 uses the receptor-binding domain of its spike protein S1 to attach to the host angiotensin-converting enzyme 2 receptor in lung and airway cells. Binding requires the help of another host protein, transmembrane protease serine S1 member 2. Several factors likely contribute to the efficient transmission of SARS-CoV-2. The receptor-binding domain of SARS-CoV-2 has a 10- to 20-fold higher receptor-binding capacity compared with previous pandemic coronaviruses. In addition, because asymptomatic persons infected with SARS-CoV-2 have high viral loads in their nasal secretions, they can silently and efficiently spread the disease. PCR-based tests have emerged as the criterion standard for the diagnosis of infection. Caution must be exercised in interpreting antibody-based tests because they have not yet been validated, and may give a false sense of security of being “immune” to SARS-CoV-2. We discuss how the development of some symptoms in allergic rhinitis can serve as clues for new-onset COVID-19. There are mixed reports that asthma is a risk factor for severe COVID-19, possibly due to differences in asthma endotypes. The rapid spread of COVID-19 has focused the efforts of scientists on repurposing existing Food and Drug Administration–approved drugs that inhibit viral entry, endocytosis, genome assembly, translation, and replication. Numerous clinical trials have been launched to identify effective treatments for COVID-19. Initial data from a placebo-controlled study suggest faster time to recovery in patients on remdesivir; it is now being evaluated in additional controlled studies. As discussed in this review, till effective vaccines and treatments emerge, it is important to understand the scientific rationale of pandemic-mitigation strategies such as wearing facemasks and social distancing, and implement them.
Key words: ACE2, asthma, allergic rhinitis, COVID-19, severe acute respiratory syndrome coronavirus 2, receptor-binding domain, TMPRSS2
Abbreviations used: ACE2, Angiotensin-converting enzyme 2; ARDS, Acute respiratory distress syndrome; CDC, Centers for Disease Control and Prevention; CoV, Coronavirus; COVID-19, Coronavirus disease 2019; FDA, Food and Drug Administration; IDO, Indoleamine 2,3-dioxygenase; JAK, Janus kinase; MERS, Middle East respiratory syndrome; MERS-CoV, Middle East respiratory syndrome coronavirus; MMWR, Morbidity and Mortality Weekly Report; MX1, MX dynamin-like GTPase 1; NIH, National Institutes of Health; RBD, Receptor-binding domain; RSV, Respiratory syncytial virus; RV, Rhinovirus; SARS, Severe acute respiratory syndrome; SARS-CoV, Severe acute respiratory syndrome coronavirus; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; S protein, Spike glycoprotein; TMPRSS2, Transmembrane protease serine S1 member 2; WHO, World Health Organization
In December 2019, a distinct coronavirus (CoV), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was identified as the cause of an outbreak of severe acute respiratory syndrome (SARS) associated with atypical pneumonia (coronavirus disease 2019 [COVID-19]).1 , 2 The index cases had visited or worked in the Huanan Wholesale Seafood Market in Wuhan, China.1 , 2 COVID-19 spread rapidly to mainland China. Outbreaks were subsequently reported in cruise ships such as Diamond Princess, where it infected 712 (19%) of the 3700 passengers and crew.3 In January 2020, SARS-CoV-2 spread to Europe,4 with most confirmed cases reported from Italy, Spain, Germany, France, and the United Kingdom. In the United States, the first case was detected in Washington on January 19, 2020,5 and had a travel history to Wuhan. Genome sequences of SARS-CoV-2 were uploaded from around the globe into the Global Initiative on Sharing All Influenza Data.6 Genome epidemiologists performed big data analysis of the Global Initiative on Sharing All Influenza Data, and suggested a pattern of spread of the virus from Wuhan to Europe, then the United States and the rest of the world.7 They determined that COVID-19 spread coast to coast across the United States.8 On March 11, 2020, the World Health Organization (WHO) declared COVID-19 as a global pandemic. As of June 26, there have been over 2,422,312 confirmed cases in the US and 9.635 million cases worldwide that have contributed to more than 124,415 deaths in the US and 489,922 deaths worldwide (Table I ).3 , 9, 10, 11, 12 The WHO estimates that COVID-19 is fatal in about 3.4% of reported cases.13 The number of people infected and its associated death toll make the COVID-19 pandemic one of the worst pandemics in recent history, and certainly worse than previous CoV pandemics—SARS and Middle East respiratory syndrome (MERS).14 , 15
Table I.
Confirmed COVID-19 cases and death, and ICU bed availability by country
| Country | Total confirmed cases | Total deaths | Total ICU beds per 100,000 capita |
|---|---|---|---|
| World | 9,635,935 | 489,922 | NA |
| United States | 2,422,312 | 124,415 | 34.7 |
| New York | 389,085 | 24,766 | NA |
| Spain | 247,486 | 28,330 | 9.7 |
| Italy | 239,706 | 34,678 | 12.5 |
| France | 197,885 | 29,775 | 11.6 |
| Germany | 193,790 | 8,962 | 29.2 |
| United Kingdom | 309,456 | 43,314 | 6.6 |
Rarely in human history have hospitals, clinicians, epidemiologists, scientists, and pharmaceutical companies worked so rapidly toward a common goal like we are seeing today—to fight the COVID-19 pandemic. This vast scientific and clinical effort has generated a wealth of information at an unbelievable pace. We navigated through this scientific literature, and here we summarize the major developments in this rapidly changing field. We examine the scientific basis and big data aspects of the spread of SARS-CoV-2 and transmission-mitigation strategies such as social distancing and wearing facemasks. We discuss how the development of some symptoms in people suffering from allergic rhinitis can serve as a clue for new-onset COVID-19. We examine how patients with asthma can be at a higher risk for severe COVID-19. We review the molecular pathogenesis of COVID-19, and examine how this knowledge has been critical in providing the scientific rationale for identifying novel and Food and Drug Administration (FDA)-approved repurposed therapeutic targets.
Distinguishing mild COVID-19 symptoms from those seen in allergic diseases
The median incubation period for COVID-19 has been estimated to be 4 to 5 days,16 , 17 and 98% of the subjects develop the symptoms within 12 days of infection.17 The clinical presentation and current recommendations in the management of COVID-19 are described in considerable detail on the American College of Physicians18 and National Institutes of Health (NIH)19 Web sites. There are some differences in symptoms observed in SARS-CoV-2–infected individuals from those observed in seasonal allergies. SARS-CoV-2–infected individuals usually develop symptoms such as dry cough, sore throat, nasal congestion, shortness of breath, myalgia, fatigue, fever,16 , 20, 21, 22, 23, 24 and rarely (about 1%) conjunctival congestion,16 and most recover spontaneously. In contrast, seasonal allergies almost universally present with a seasonally reproducible constellation of allergic rhinitis symptoms consisting of runny itchy nose, itchy eyes, sneezing, postnasal drip, and conjunctival congestion.25, 26, 27, 28, 29 From an allergist’s perspective, a shift from these allergic rhinitis symptoms to those observed in COVID-19 with fever, cough, and shortness of breath (Table II )16 , 22 , 29 may suggest the possibility of new-onset COVID-19 in allergic individuals. Taste or olfactory dysfunctions such as anosmia and dysgeusia can occur in about 35% to 90% of patients who reported olfactory and gustatory dysfunction,30 , 31 which is similar to higher to that seen in allergic rhinitis (taste dysfunction 20%,32 , 33 olfactory dysfunction 20%-40%33 , 34). Wheezing, a common feature of asthma exacerbation, rarely occurs in patients hospitalized with COVID-19.35, 36, 37, 38, 39 However, both asthma and COVID-19 are often associated with cough and shortness of breath,16 , 20, 21, 22, 23, 24 , 40 and testing for SARS-CoV-2 may be required to exclude the possibility of new-onset COVID-19 in individuals with asthma. The mechanisms underlying the lack of a strong association between asthma and COVID-19 are discussed in greater detail later in this article. Because about a fifth of hospitalized patients with COVID-19 develop cutaneous manifestations such as erythematous rash, urticaria, and chickenpox-like vesicles,41 if a patient with recurrent urticaria has new-onset urticarial rash together with fever, cough, or shortness of breath, it may suggest new-onset COVID-19. SARS-CoV-2 infection can induce severe Kawasaki-like disease,42 , 43 a multisystem vasculitis characterized by persistent fever, conjunctival injection, exanthema, lymphadenopathy, inflammation of the tongue and pharyngeal mucosa, and edema in peripheral extremities.44 Because its diagnosis is established by the presence of 5 or 6 principal symptoms, Kawasaki disease is not difficult to distinguish from allergic conjunctivitis or skin eruption. Because Kawasaki disease is a risk factor for subsequent allergic diseases,45 , 46 children who develop this disease during SARS-CoV-2 infection should be followed longitudinally for the development of allergic diseases.
Table II.
Prevalence of clinical symptoms of COVID-19 and AR
Clinical features of severe COVID-19
One of the reasons COVID-19 has developed into such a feared pandemic is that a subset of SARS-CoV-2– infected persons develop severe life-threatening complications such as pulmonary edema, severe pneumonia, and acute respiratory distress syndrome (ARDS), heart failure and other organ failures, and septic shock.15 , 20, 21, 22 A comparison of clinical features of PCR-confirmed patients with COVID-19 hospitalized in China versus those hospitalized in New York suggests that the patients in New York had 4- to 5-fold higher gastrointestinal symptoms such as nausea, vomiting, and diarrhea, and a higher incidence of shortness of breath (Table III ).16 , 23 These differences could reflect a more severe cohort of patients being included in the report from New York, or suggest racial or other differences.
Table III.
Prevalence of clinical symptoms of COVID-19 reported from China and the United States (New York)
| Symptom | China∗ |
New York† |
||
|---|---|---|---|---|
| Nonsevere | Severe | Noninvasive MV | Invasive MV | |
| Sex: male (%) | 58.2 | 57.8 | 55.5 | 70.8 |
| Median age (y) | 45 | 52 | 61.5 | 64.5 |
| Cough (%) | 67.3 | 70.5 | 77.6 | 83.1 |
| Fever (%) | 89.8 | 91.4 | 77.2 | 76.9 |
| Shortness of breath (dyspnea) (%) | 15.1 | 37.6 | 51.7 | 66.2 |
| Myalgia, arthralgia, and/or fatigue (%) | 14.5 | 17.3 | 28.9 | 23.8 |
| Diarrhea (%) | 3.5 | 5.8 | 25.1 | 20.8 |
| Nausea and/or vomiting (%) | 4.6 | 6.9 | 20.2 | 16.9 |
Risk factors for severe or fatal COVID-19 include age above 60 years, presence of comorbid conditions such as diabetes mellitus, hypertension, chronic obstructive lung disease, asthma, coronary artery disease, cerebrovascular disease, chronic renal disease, history of cigarette smoking, obesity, high Sequential Organ Failure Assessment score, and d-dimer level more than 1 μg/mL.16 , 20 , 37 , 47 The presence of shortness of breath as an early symptom is associated with more severe COVID-19.16 Although myocardial injury from SARS-CoV-2 occurs rarely in about 5% of patients hospitalized for COVID-19, its presence has been identified as a risk factor for death.48 , 49 The availability of intensive care unit beds to manage patients with severe COVID-19 can be a problem50 , 51 because the number of available intensive care unit beds varies widely in the United States11 or other countries12 (Table I).
Diagnostic tests for detecting and monitoring COVID-19 and immune response
The criterion standard test for COVID-19 is a RT-PCR– based test.52 Nasopharyngeal, oropharyngeal, middle turbinate, anterior nares specimens or swabs collected by health care professionals, and other Centers for Disease Control and Prevention (CDC)-recommended specimens are placed into a virus preservation solution,53 lysed to extract SARS-CoV-2 genes N, E, S, and RNA-dependent RNA polymerase, and amplified by real-time RT-PCR.54 COVID-19 point-of-care testing involves qualitative detection of nucleic acid from SARS-CoV-2 in nasopharyngeal swab and/or nasal wash/aspirate specimens. Some in-home testing kits for COVID-19 are now FDA-approved.55 Virus isolation and culture is not recommended as a routine diagnostic procedure. Seroconversion occurs in 7 days in 50% and 14 days in all patients.56 Even though many antibody-based tests have flooded the market, it is important to remember that they have not yet been validated, and “positive” test results may give a false sense of security of being “immune” to SARS-CoV-2.57 , 58
Scientific basis of using social distancing, quarantine, and facemasks to reduce spread of SARS-CoV-2
SARS-CoV-2 infection is transmitted through aerosol and droplets during coughing.59 Virus-laden small (<5 μm) aerosolized droplets can remain in the air and travel long distances (>1 m, and sometimes even 4 m),60 , 61 thus providing a scientific rationale for the CDC guidelines of social distancing of 6 ft (about 2 m). These droplets can spread and deposit on surfaces, where the virus remains viable for a few days.62 SARS-CoV-2 is more stable on plastic and stainless steel than on copper and cardboard, and viable virus can be detected up to 72 hours after application to the former surfaces.63 SARS-CoV-2 remains viable in aerosols for 3 hours, which is similar to that for SARS-CoV.63 The soles of shoes of medical staff can serve as carriers of SARS-CoV-2.61 Shedding of SARS-CoV-2 is high even before the onset of symptom,64 during the first week of symptoms, and continues till the end of symptoms.56 In one study, fecal samples were positive for SARS-CoV-2 in 52% of hospitalized patients with gastrointestinal symptoms and 39% of the subjects without gastrointestinal symptoms.65 However, the main mode of transmission appears to be through aerosol, droplets, and contact with surfaces that have deposits of the active virus.56 , 63 , 66
A big data study of infections in China estimated that 86% of all COVID-19 infections were undocumented.67 Their modeling studies estimated that because of their greater numbers, undocumented infections were the source for about 80% of infections, and facilitated the rapid dissemination of SARS-CoV2.67 However, because this report analyzed data from only the first few weeks of January, when local authorities were overwhelmed and underreporting cases, and did not include the data from the huge surge of cases in February, the 86% estimate should be treated with caution. During the previous SARS-CoV pandemic, the importance of social distancing, isolation of patients, contact tracing, and quarantine of exposed persons were identified as effective measures of mitigating the transmission of the virus.68 These measures were also effective in mitigating the human-to-human transmission of COVID-19 in China.69 Modeling studies suggest that the travel quarantine of Wuhan delayed the overall epidemic progression by 3 to 5 days in mainland China, but had a more marked effect at the international scale, where case importations were reduced by nearly 80% until mid-February.70 Recent studies have similarly shown the utility of facemask in mitigating transmission of COVID-19.71, 72, 73 However, some data suggest that surgical or cotton masks may not be enough to filter SARS-CoV-2.74 For these reasons, other CDC guidelines such as washing hands, not touching the face, and social distancing should also be followed to reduce the spread of this virus.
CoVs that have caused human diseases
CoVs are positive single-strand enveloped RNA viruses that belong to the family Coronoviridae. These viruses are characterized by club-like spikes that project from their surface, a large RNA genome, and a unique replication strategy. Before SARS-CoV-2 appeared, 6 human CoVs have been known to have contributed to human diseases: alpha CoVs HCoV-229E and HCoV-NL63, and beta CoVs HCoV-OC43, HCoV-HKU1, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV).75 The seasonal CoVs HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1 usually cause mild upper respiratory tract illness.76, 77, 78 However, pandemic CoVs SARS-CoV, SARS-CoV-2, and MERS-CoV behave differently, and have caused staggering illness and death. The lower respiratory tract symptoms such as severe acute respiratory illness, shortness of breath, and chest computed tomography findings of SARS-CoV2 infection are similar to symptoms of SARS-CoV and MERS-CoV infections.21 Similar to SARS and MERS,79 , 80 older age is a risk factor for adverse clinical outcomes in SARS-CoV-2. Although the 3.4% case-fatality rate of SARS-CoV213 appears to be lower than that reported for SARS (10%) or MERS (34%),81 the number of people who tested positive for SARS-CoV-2 (about 9.64 million) is many times greater than the number of people who tested positive for SARS-CoV (about 8500) or MERS-CoV (about 2500).82 Thus, the overall health effects of COVID-19 have greatly exceeded those observed in previous CoV pandemics.
Genome similarities, reservoir, and intermediate mammalian host of pandemic CoVs
The genome size of SARS-CoV-2 (29.9 kb) is similar to the genome size of SARS-CoV (27.9 kb) and MERS-CoV (30.1 kb).83, 84, 85 SARS-CoV-2 and SARS-CoV have about 80% genome sequence similarity.1 , 86 The SARS-CoV-2 and bat SARS-CoV-like CoVs share approximately 96% sequence similarities.86 Likewise, CoVs in Malayan pangolins (Manis javanica) have a high degree of similarity in all 6 residues of the receptor-binding domain (RBD) site of SARS-CoV-2.87 , 88 Because scientific data suggest that civets and camels served as reservoirs for maintenance of SARS-CoV and MERS-CoV,89 it has been proposed that bat CoV could have been transmitted to humans through pangolin reservoir to cause COVID-19.88
Receptors and lung cells that bind SARS-CoV-2
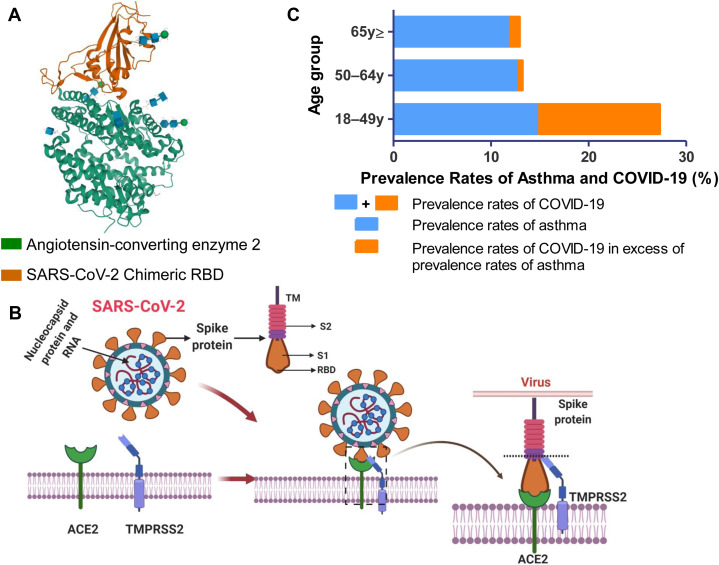
Angiotensin-converting enzyme 2 (ACE2) is a well-defined receptor for SARS-CoV.90 This receptor is expressed in most human respiratory cells,91 , 92 explaining its propensity to replicate in these cells. Like SARS-CoV, SARS-CoV-2 binds to the respiratory mucosa through the same ACE2 receptor.93 The spike glycoprotein (S protein) on SARS-CoV-2 plays a critical role in binding host ACE2 receptor and in membrane fusion.94 , 95 Structural studies have elucidated the conformational aspects of the interaction of the RBD of S protein with ACE2 (Fig 1 , A).37 , 94, 95, 96, 97 This binding induces conformational changes in amino acids that help create salt bridges, increase van der Waals interactions, and facilitate binding with ACE2 with much greater affinity than SARS-CoV.98 The S protein contains subunit S1 with the RBD that binds ACE2, the membrane-fusion subunit S2, the transmembrane anchor, and the intracellular tail (Fig 1, B). Attachment of the RBD of S1 to host ACE2 receptor requires the help of the cellular transmembrane protease serine S1 member 2 (TMPRSS2)99 to cleave S2 protein from S1, and help in membrane fusion100 , 101 (Fig 1, B). Some unique features of the S1 protein of SARS-CoV-214 , 56 account for its 10- to 20-fold higher receptor-binding capacity compared with SARS-CoV and MERS-CoV.99 Structural analysis also suggests that some variations of ACE2 can strengthen the interactions between the RBD of SARS-CoV-2 and ACE2.98 A neutralizing antibody CR3022 that recognizes the conserved epitope RBD of SARS-CoV102 also targets the RBD of SARS-CoV-2103 only when 2 RBDs on the trimeric S protein are changed to the “up” position conformationally.104 A careful study of this conformationally dependent interaction of this neutralizing antibody with RBD of SARS-CoV-2 may provide critical information required for developing additional high-potency neutralizing antibodies.
Fig 1.
A, Structure of RBD of spike protein S1 of SARS-CoV-2 bound to ACE2. Structure of ACE2 bound to the RBD of the S1 spike protein of SARS-CoV-2.94, 95, 96 The chimeric RBD is in orange, and human ACE2 is in green. The figure was created with Research Collaboratory for Structural Bioinformatics Protein Data Bank (https://www.rcsb.org/). RBD, RBD of S1 spike protein of SARS-CoV-2. B, Cartoon showing how SARS-CoV-2 binds to the lung epithelial cells. SARS-CoV-2 has a spike protein with transmembrane (TM), S1 and S2 part. S1 part has an RBD. The virion uses the spike protein S1 to attach with RBD of the host ACE2 receptor on the cell membrane with the help of the cellular TMPRSS2. Following attachment of S1 to ACE2, the host serine protease TMPRSS2 cleaves the S2 protein from S1, and plays a role in membrane fusion of CoVs. The figure was created using BioRender (https://biorender.com/). C, The prevalence of asthma in patients hospitalized for COVID-19 in United States. Data were extracted from April 8, 2020, MMWR report37 and Centers for Disease Control and Prevention.97 The total length of each bar represents the prevalence rates of COVID-19 in each age group. The length of the blue part of this bar is the expected prevalence rate of asthma in each age group. The orange part represents the prevalence rate of COVID-19 in excess of the expected prevalence rate of asthma in each age group.
The cells in the lungs and airways that are likely to be infected by SARS-CoV-2 have been investigated by single-nuclei and single-cell RNAseq analysis of human lung tissues. ACE2 and TMPRSS2 are expressed in transient secretory cells in the segmental bronchial branches and cells derived from lung tissues.105 Because SARS-CoV-2 has a furin-cleavage site in its S protein, a feature missing in SARS-CoV, it can use the serine endoprotease furin in host cells to streamline its internalization.94 , 105 The binding of SARS-CoV-2 to ACE2 increases the expression of ACE2, which further damages the alveolar cells. After fusion with the host cell, the viral genome RNA is released into the cytoplasm. The uncoated RNA translates the replicase-transcriptase polyproteins pp1a and pp1ab encoded in open-reading frame 1a and 1ab located at the 5’-terminus of the genome, and the replication-transcription complex106 replicates RNA for assembly and virus release.107, 108, 109
Respiratory viruses, CoVs, SARS-CoV-2, and asthma
Many respiratory viruses have been associated with asthma exacerbations, including respiratory syncytial virus (RSV), rhinoviruses (RVs), influenza virus, CoV, enterovirus, parainfluenza, adenovirus, bocavirus, and metapneumovirus.77 , 110, 111, 112, 113, 114, 115, 116 Atopy and asthma are risk factors for lower respiratory tract infection, more severe virus-induced wheezing, and asthma exacerbation.117, 118, 119 The contribution of RV or RSV to the initiation of asthma and asthma exacerbation has been investigated for many years. Positive family history of asthma, history of atopy, and wheezing are risk factors for RSV-induced lower respiratory tract infection,118 and hospitalizations due to RV infection.119 RSV-induced bronchiolitis is the most common cough, wheezing, and respiratory distress, and requires hospitalization in infants.120 Many prospective long-term follow-up studies demonstrated that the history of wheezing illnesses caused by RV or RSV infections is a predictor of the subsequent development of asthma.113 , 121, 122, 123, 124, 125 From these studies, 2 hypotheses have emerged; RSV- or RV-induced wheezing initiates the development of asthma, or, these viruses trigger wheezing and exacerbation of asthma. The validity of these 2 schools of thought has been debated for decades.
Like other respiratory viruses that infect the airway epithelial cells and pneumocytes through their receptors and induce asthma exacerbations,110 , 111 , 126, 127, 128, 129, 130, 131 seasonal human CoVs HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1 can also cause common cold and induce asthma exacerbations.110 , 131, 132, 133 Inoculation of SARS-CoV-2 or MERS-CoV pandemic CoVs into cynomolgus macaques infects airway cells with subtle differences.134 SARS-CoV-2 infects type I and II pneumocytes in ciliated airway mucosal epithelial cells and damages alveolar cells, whereas MERS-CoV infects predominantly type II pneumocytes, and causes less lung damage.134 Investigators have proposed a hypothesis to explain the mechanism by which respiratory viruses trigger asthma exacerbations—patients with asthma have an attenuated IFN-I and IFN-III response to these infections, and the resultant unopposed TH2 responses contribute to asthma exacerbation.135, 136, 137 Because infection of lung and airway cells with SARS-CoV-2 induces an attenuated IFN-I and INF-III signature138 , 139 similar to that observed in patients with asthma, COVID-19 would be expected to frequently trigger asthma exacerbations. Furthermore, the high levels of proinflammatory cytokines and their receptors such as CXCL1, CXCL2, CXCL8, CXCL17, CCL2, CCL3, CCL4, CCR1, CXCR2, IL5RA, IL-6, IL-1β, and IL-1R2 observed in the bronchoalveolar lavage and lungs of patients with COVID-19138 , 139 also suggests that SARS-CoV-2 infection should frequently induce asthma exacerbations. Yet, quite surprisingly, in the April 8, 2020, Morbidity and Mortality Weekly Report (MMWR)37 report of 1482 patients hospitalized for COVID-19 in the United States in March 2020, it was mentioned that wheezing was present in only about 7% of the 178 patients in whom data were available on underlying conditions, which is less than the prevalence rate of about 10% of asthma in the general population.140 These reports suggest that SARS-CoV-2 rarely induces asthma exacerbations during hospitalization for COVID-19.37 , 140
Could host immune-response factors contribute to the reduced ability of SARS-CoV-2–infected airway and lung cells from inducing asthma exacerbations? SARS-CoV-2 entry–associated genes ACE2 and TMPRSS2 are highly expressed in human nasal and lower airway epithelial cells, and cells that express these genes often coexpress innate immune genes such as indoleamine 2,3-dioxygenase (IDO) 1 and MX dynamin-like GTPase 1 (MX1).141 IDO is a rate-limiting enzyme in tryptophan catabolism that increases the synthesis of tryptophan metabolites such as kynurenine, 3-hydroxykynurenine, and xanthurenic acid that induce immune tolerance and suppress experimental allergic inflammation.142 , 143 Thus, increased expression of IDO in ACE2 and TMPRSS2-expressing cells could reduce asthma exacerbations in COVID-19. MX1 is an IFN-inducible GTPase with antiviral activities against a broad range of RNA viruses.144 The data from the Childhood Origins of Asthma study and Copenhagen Prospective Study on Asthma in Childhood suggest that polymorphisms of the MX1 gene are associated with asthma exacerbations.145 Thus, similar to IDO, increased expression of MX1 in ACE2 and TMPRSS2-expressing cells could reduce asthma exacerbations in COVID-19. IL-33, an alarmin that normally resides dormant exclusively in the nucleus, becomes a proallergic cytokine when it is secreted extracellularly and induces allergic inflammation and contributes to asthma exacerbation.146, 147, 148 Infection with respiratory viruses that trigger asthma exacerbation, such as influenza and rhinovirus, induce IL-33 secretion from bronchial epithelial cells and alveolar cells in the airways.146 , 148 , 149 Yet, our search of databases such as PubMed and Google Scholar did not find peer-reviewed publications that show that SARS-CoV-2 induces IL-33 secretion in the airways. Taken together, the unique host immune response to SARS-CoV-2 could explain why it does not frequently trigger asthma exacerbations.
If SARS-CoV-2 does not induce asthma exacerbations, can asthma be a risk factor for severe COVID-19 infection as suggested on the CDC Web site150? The comorbid diseases data in the April 8, 2020, MMWR report show that in 18- to 49-year-old patients hospitalized for COVID-19, 27% had a history of asthma.37 The COVID-19 task force of the American Academy of Allergy, Asthma & Immunology suggests that given the 10% prevalence of asthma in the United States, the higher 27% of patients with COVID-19 who were hospitalized in this age group suggests that they may be at increased risk of hospitalization due to COVID-19 (Fig 1, C).37 , 151 The same MMWR report suggests that African Americans have a disproportionally higher hospitalization for COVID-19, accounting for 33% of the US hospitalizations,37 just as they have a higher propensity for severe asthma.152, 153, 154 In contrast to these MMWR data, a retrospective analysis of 140 hospitalized patients with COVID-19 in China with confirmed results of SARS-CoV-2 viral infection reported that none of them had asthma.35 Another study performed in China involving 548 patients with COVID-19 revealed that 0.9% of patients with COVID-19 had asthma, which is lower than the prevalence rate of asthma in the general population in China.36 The discrepancy between the US MMWR report and these studies from China could reflect racial differences in the role of asthma in the severity of COVID-19.35 , 37 , 151 However, in a very large study of 5700 patients in New York City hospitalized for COVID-19, only 9% of the patients had underlying asthma,155 similar to the prevalence rates of asthma in the general population. That presence of asthma does not contribute to the severity of COVID-19 is also evident from another study performed in New York City that showed no increase in asthma in invasive compared with noninvasive mechanically ventilated patients with COVID-19.23
What could be the explanation for the paucity of patients with asthma in patients with COVID-19 in studies performed in China?35 , 36 Because the expression of airway levels of ACE2 is lower in atopic subjects compared with nonatopic subjects,156 TH2-high endotype of asthma may be at a lower risk for severe COVID-19 because their airways would have fewer receptors for entry of SARS-CoV-2. Likewise, because exposure of the airways of allergic patients with asthma to environmental allergens reduces ACE2 expression levels,156 seasonal exposure to aeroallergens may protect them from COVID-19. As discussed later, ciclesonide and formoterol are commonly used inhalers in asthma, and their antiviral properties could protect patients with asthma from COVID-19.
On the basis of MMWR data,37 if one assumes that asthma is a risk factor for severe COVID-19 particularly in the 18- to 49-year age group (Fig 1, C), what could be the mechanisms of this increased propensity toward severity of COVID-19? Because obesity is a known risk factor for severe COVID-19,37 obesity-related endotype of asthma157 , 158 could be a higher risk for severe COVID-19. The expression of ACE2 and TMPRSS2 in sputum cells is higher in males, in African Americans, and in patients with asthma with a history of diabetes, all risk factors for severe COVID-19.159 Persons with ACE D/D genotypes have higher immunoreactive ACE concentration in serum and a higher risk of asthma than those with other genotypes.160, 161, 162 Because SARS-CoV-2 uses ACE2 to infect cells, future studies should evaluate whether the ACE D/D genotype is a risk factor for COVID-19. TMPRSS2 is expressed in human airway epithelium163 and thought to contribute to the severity of SARS-CoV and MERS-CoV lung infection.164 Because subjects with atopic asthma have higher nasal levels of TMPRSS2 compared with healthy volunteers,165 these increased levels could be a risk factor for the severity of COVID-19 in asthma. Treatment of mice with ACE2 activator or angiotensin (1-7) reduces airway inflammation in experimental asthma.166 , 167 In addition, loss of ACE2 in the animal model study has been shown to aggravate severe acute lung injury, and the administration of recombinant human ACE2 alleviates lung injury.168 Taken together, these studies suggest that signaling through ACE2 provides protection against both allergic airway inflammation and acute lung injury. Because attachment of S1 part of the S protein to ACE2 can stimulate splicing of ACE2 by TMPRSS2,169 it is possible that this spliced ACE2 is less effective in providing protection against acute lung injury and asthma. Future studies will have to determine whether the administration of human recombinant ACE2 could be a treatment option for COVID-19 in patients with asthma.
Scientific strategies behind major clinical trials for COVID-19
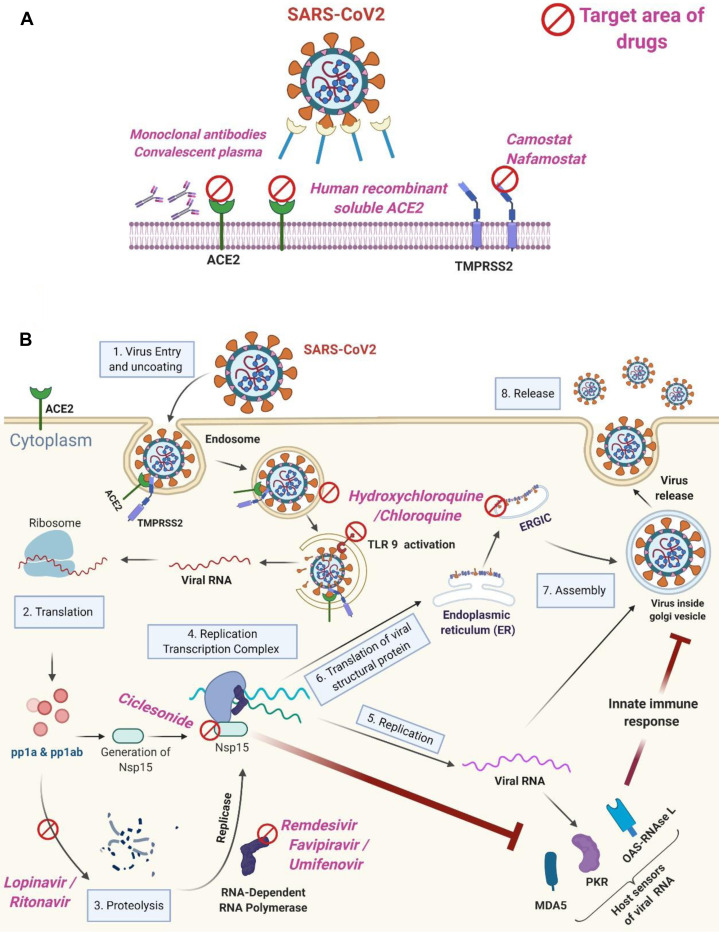
Many clinical trials such as seen in NIH ClinicalTrials.gov 170 or EU Clinical Trials Register171 are being performed worldwide for COVID-19 (Table IV ),172, 173, 174, 175, 176 but as of today there are no proven effective treatments. On March 20, 2020, WHO announced the launch of SOLIDARITY,177 an unprecedented, coordinated push to collect robust scientific data rapidly during a pandemic.178 Properly designed large clinical trials are required for assessing drugs in each category of the following mechanistic categories (Fig 2 ), and are being performed.
Table IV.
Clinical trials completed or are being performed worldwide for COVID-19 (listed in clinicaltrials.gov)
| Intervention | Category | Suggested mechanism of action | Design of trial | Status | Key outcome | Reference or ID |
|---|---|---|---|---|---|---|
| Lopinavir-ritonavir | Anti-HIV drug | Inhibition of protease | Open-label, randomized, and controlled trial | Completed | No benefit on the hospitalized adult patients with severe COVID-19 | 172 |
| Favipiravir vs umifenovir | Anti-influenza virus drug | Inhibition of viral RNA polymerase | Open-label randomized | Completed | Preferred clinical outcome in the favipiravir group than in the umifenovir group | 173 |
| Chloroquine | Immunosuppressive drug and antiparasite drug | Inhibition of virus entry | Clinical study | NA | Beneficial effect, but details have not been published | 174 |
| Hydroxychloroquine- azithromycin | Antimalarial drug, antibiotics | Inhibition of virus entry | Open-label nonrandomized | Completed | Combination drug reduced viral load in nasopharyngeal swabs | 175 |
| Hydroxychloroquine vs azithromycin | Antimalarial drug, antibiotics | Inhibition of virus entry | Open-label randomized | Recruiting Phase 2 |
NA |
clinicaltrials.gov (NCT04329832) |
| Lopinavir/ritonavir, ribavirin and IFN-β combination | Antivirus drug | Prodrug metabolized into nucleoside analogs that blocks and caps viral RNA | Open-label randomized | Completed | Preferred clinical outcome in the triple antiviral therapy group than in the lopinavir-ritonavir group | 176 |
| IFN-A2B | Antivirus drug | Activate multiple immunomodulatory and antiviral proteins | Open-label randomized, blank-controlled | Not yet recruiting Early phase 1 |
NA |
clinicaltrials.gov (NCT04293887) |
| Remdesivir | Antiebola drug | Inhibition of viral RNA polymerase | Open-label, randomized | Recruiting Phase 3 |
NA |
clinicaltrials.gov (NCT04292899) |
| Tocilizumab | Anti–IL-6 receptor antibody | Anti-inflammation | Open-label, single-group assignment | Recruiting Phase 2 |
NA |
clinicaltrials.gov (NCT04317092) |
| Ciclesonide vs ciclesonide plus hydroxychloroquine, vs no intervention | Inhaled corticosteroids | Anti-inflammation | Open-label randomized | Not yet recruiting Phase 2 | NA |
clinicaltrials.gov (NCT04330586) |
| Camostat mesilate | Antiproteinuric drug | Serine protease inhibitors | Randomized placebo-controlled | Recruiting Phases 1 and 2 |
NA |
clinicaltrials.gov (NCT04321096) |
| Recombinant human ACE2 | Monocarboxypeptidase that leads to degradation of angiotensin II | Antihypertensive | Double-blind randomized | Not yet recruiting Phase 2 | NA |
clinicaltrials.gov (NCT04335136) |
NA, Not applicable/available.
Fig 2.
Treatment strategies for COVID-19. A, Drugs that are designed to block entry of SARS-CoV into the cells. B, Drugs that act at different steps of virus replication inside the cell. The figure shows 8 steps from viral entry to virus release in airway epithelial cells. After fusion to the host cell, the viral genome RNA is released into the cytoplasm. The uncoated RNA translates pp1a and pp1ab polyproteins, and the replication-transcription complex replicates RNA for assembly and virus release. The figure was created using BioRender (https://biorender.com/). ERGIC, Reticulum-Golgi intermediate compartment; MDA5, melanoma differentiation-associated protein 5; NSP, nonstructural protein; OAS, 2'-5' oligoadenylate synthetase; PKR, protein kinase R; pp1a, polyprotein1a; pp1ab, polyprotein1ab; TLR9, Toll-like receptor 9.
Inhibiting viral entry: Decoy receptor, mAbs, convalescent plasma, camostat
Some data suggest that this strategy may be effective in COVID-19 (Fig 2, A). Acting as a decoy, human recombinant soluble ACE2 treatments dramatically inhibited the growth of SARS-CoV-2–infected Vero cells by more than 1000-fold,179 and suppressed SARS-CoV-2 infection with engineered human blood vessel and kidney organoids.179 These results suggest that human recombinant soluble ACE2 could block early stages of SARS-CoV-2 infections, and is a potential drug for use in COVID-19. In another study, transfusion of convalescent plasma containing neutralizing antibodies collected from the donors who had recovered from SARS-CoV-2 infection to 5 patients with COVID-19 and ARDS receiving mechanical ventilation improved their clinical status.180 Likewise, the administration of convalescent plasma had a beneficial effect on 10 patients with severe COVID-19.181 Human neutralizing mAbs from convalescent patients with COVID-19, B38 and H4, inhibit the binding of SARS-CoV-2 S protein RBD to ACE2182 (Fig 2, A). These antibodies reduced virus titers in the lungs and ameliorated the lung inflammation in an animal model developed to test the efficacy of drugs—human ACE transgenic mice infected with SARS-CoV-2.182 Treatment with serine protease inhibitor camostat inhibited entry of SARS-S and SARS-2-S protein into primary human lung cells.100 Likewise, nafamostat inhibits membrane fusion of S protein of MERS-CoV183 and SARS-2-S.184 Well-designed clinical trials are required to assess the role of human recombinant soluble ACE2, mAbs, convalescent sera, and camostat in COVID-19.
Inhibiting endocytosis and initial assembly of the virus genome
Chloroquine and hydroxychloroquine may inhibit SARS-CoV-2 by inhibiting pH-dependent viral fusion/replication and prevention of viral envelope glycoprotein as well as host receptor protein glycosylation, and virion assembly in endoplasmic reticulum-Golgi intermediate compartment–like structures185 (Fig 2, B). In addition to its suppressive effects on viral replication, hydroxychloroquine inhibits Toll-like receptor 7/9–dependent inflammatory responses.186 In a small study, azithromycin added to hydroxychloroquine was significantly more efficient for virus elimination than hydroxychloroquine alone or either drug given orally.175 The low cost and vast availability of chloroquine and hydroxychloroquine is one of the reasons it is being evaluated as part of SOLIDARITY trials. However, a parallel, double-masked, randomized clinical trial in hospitalized patients with COVID-19 revealed that the mortality rate until day 13 was higher in the high-dosage chloroquine diphosphate group than in the low-dosage group.187 The high-dosage group of chloroquine showed more often increased QTc interval than the low-dosage group.187 Likewise, another report suggests that neither hydroxychloroquine alone nor in combination with azithromycin had beneficial clinical effects in hospitalized patients with COVID-19.188 , 189 Furthermore, the administration of hydroxychloroquine may contribute to increased mortality180 and prolongation of QTc interval in electrocardiogram.19 These studies suggest that chloroquine and hydroxychloroquine may not have significant efficacy in COVID-19, and their excessive use may contribute to electrocardiogram changes and even death.
Inhibiting translation of the viral genome and replication of virus
Lopinavir-ritonavir is a boosted protease inhibitor used for treating HIV type 1 infection that had favorable effects on SARS190 and MERS191 in small studies. However, a randomized trial of lopinavir-ritonavir treatment demonstrated no beneficial effect in hospitalized adult patients with severe COVID-19.184 Remdesivir is an adenosine analog that incorporates into nascent viral RNA chains, and causes its early termination192 (Fig 2, B). In a case report, treatment with intravenous remdesivir was initiated on the evening of day 7 in a patient hospitalized for severe COVID-19, and on hospital day 8, the patient’s clinical condition improved.5 The administration of remdesivir on a compassionate-use basis to patients hospitalized with COVID-19 showed a beneficial effect in about 70% of patients.193 The preliminary results from the Adaptive COVID-19 Treatment Trial indicate that patients who received remdesivir had a 31% faster time (11 days) to recovery than those who received placebo (15 days; P < .001).194 However, a randomized, double-blind, placebo-controlled, multicenter trial from China demonstrated that remedesivir has no beneficial impact on hospitalized patients with COVID-19 compared with the placebo group.195 A study comparing the effect of favipiravir and umifenovir in the patients with moderate COVID-19 showed statistical superiority of favipiravir over umifenovir.173
Inhibition of the cytokine storm
SARS-CoV-2 triggers a cytokine storm with secretion of IL-6 and other proinflammatory cytokines that has been suggested as one of the mechanisms for organ damage and ARDS.21 , 196 , 197 A meta-analysis of 6 studies suggested that the mean IL-6 concentrations were about 3-fold higher in patients with complicated COVID-19 compared with uncomplicated disease.198 IL-6 binds to the IL-6 receptor on the cell surface and Janus kinase (JAK) is phosphorylated and the subsequent inflammatory cascade initiates.199 Inhibitor for IL-6 signaling and JAK1/2 are being evaluated to suppress the cytokine storm in COVID-19.197 Compared with patients with COVID-19 without ARDS, patients with COVID-19 with ARDS had increased level of IL-6 in serum, and the serum levels of IL-6 in patients with COVID-19 with ARDS who died were higher compared with levels in patients with COVID-19 with ARDS who survived.47 Thus, suppression of the IL-6 signaling pathway could be a therapeutic strategy against COVID-19. The FDA-approved drug tocilizumab is prescribed by rheumatologists, and has been administered to patients with COVID-19. A systematic review and meta-analysis of IL-6 and COVID-19 revealed that serum level of IL-6 was elevated among patients with COVID-19 with adverse clinical outcomes, and the administration of humanized monoclonal anti–IL-6 receptor antibody tocilizumab to these patients was efficacious and safe.198 Baricitinib is an oral JAK1/JAK2 inhibitor that binds to AP2-associated protein kinase 1,200 and has been used for the treatment of rheumatoid arthritis. Because AP2-associated protein kinase 1 is a regulator of endocytosis,201 baricitinib may inhibit SARS-CoV-2 replication and suppress IFN-controlled gene expression.202 A recent small study suggested that baricitinib therapy combined with lopinavir-ritonavir in moderate COVID-19 pneumonia is clinically more effective than the control treatment (lopinavir-ritonavir plus hydroxychloroquine) (intensive care unit transfer 0% vs 33%, discharge at week 2 8% vs 58%, respectively).203 Additional well-designed large-scale clinical trials are being performed to assess the role of tocilizumab and baricitinib in COVID-19.
Inhaled ciclesonide steroid and long-acting β-adrenergic receptor formoterol
Ciclesonide is prescribed by allergists as an intranasal or inhaled corticosteroid for treating allergic rhinitis and asthma, but recent studies suggest that it has antiviral properties (Fig 2, B). In one study, 48 FDA-approved drugs were screened for their antiviral properties against SARS-CoV2 using Vero cells.204 From this screening, ciclesonide was identified as one of the very few drugs that had significant antiviral properties against SARS-CoV-2, but had no toxicity.204 Likewise, in another study that involved screening of FDA-approved drugs for their antiviral properties, ciclesonide demonstrated antiviral effects against MERS-CoV.205 Nonstructural protein 15 produced by CoVs impair the ability of retinoic acid–inducible gene–I–like receptors such as retinoic acid–inducible gene–I and melanoma differentiation–associated protein (MDA-5) to detect viral RNA in the cytosol, thereby facilitating replication of the virus in host macrophages.206 Ciclesonide targets nonstructural protein 15 (Fig 2, B), thereby facilitating retinoic acid–inducible gene–I and MDA-5–mediated inhibition of MERS-CoV and SARS-CoV-2 replication.206 , 207 Lung imaging studies have shown that the small particle size of ciclesonide (1 μm) facilitates widespread lung deposition, including small airways.208 , 209 Thus, inhaled ciclesonide should be able to penetrate deep into the lungs and suppress SARS-CoV-2 infection. Indeed, inhaled ciclesonide clinically improved 3 patients with pneumonia caused by SARS-CoV-2 who required oxygen support.210 Likewise, formoterol, a long-acting selective β-adrenergic receptor agonist that is often prescribed by allergists as a combination drug for treatment of patients with persistent asthma, can suppress replication of HCoV-229E.211 Thus, well-designed large-scale clinical trials are required to assess the role of intranasal/nebulized ciclesonide and inhaled beta-adrenergic receptor agonists in treatment or prevention of COVID-19.
Structure-assisted drug screening for compounds that inhibit SARS-CoV-2 main protease activity
The SARS-CoV-2 main protease mediates viral replication and transcription.212 A study using structure-assisted drug design, virtual drug screening, and high-throughput screening identified 6 compounds, disulfiram, carmofur, ebselen, shikonin, tideglusib, and PX-12, that inhibited main protease activity.212 Additional studies will be required to assess the clinical efficacy of these compounds in COVID-19.
Vaccines to prevent COVID-19
The NIH launched a clinical trial of investigational vaccine for NIH-funded candidate mRNA vaccines for COVID-19 on March 16, 2020.213 , 214 Several clinical trials are in progress, and the WHO announced and updated a DRAFT landscape of COVID-19 candidate vaccines.215 As of today, there is no clinically available vaccine against SARS-CoV-2.
Conclusions
COVID-19 has become a feared pandemic because it has infected more than 200-fold greater number of people in the population than SARS-CoV or MERS-CoV pandemic, has spread at an unbelievable pace, and caused severe life-threatening complications in a significant subset of these infected persons. Here, we reviewed the molecular pathogenesis of COVID-19, and examined how this knowledge has been critical in providing the scientific rationale for identifying novel and FDA-approved repurposed therapeutic targets. From an allergists’ perspective, we discussed how the development of some symptoms in allergic rhinitis may serve as a clue for new-onset COVID-19 in subjects with allergy, and examined how asthma could be a risk factor for severe COVID-19. Till effective vaccines or treatments emerge, it is important to understand the scientific rationale discussed in this article that underlie pandemic-mitigation strategies such as wearing facemasks and social distancing. The knowledge gained from this review will give the readers a broad-based knowledge required to understand and correctly interpret current and future publications and developments in this rapidly changing field of COVID-19 pandemic.
Footnotes
This research was supported by the National Heart, Lung, and Blood Institute (grant nos. 5R01HL145477-02 and 3R01HL145477-01S1) and the Department of Defense (grant no. PR171425 W81XWH-18-1-0743).
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johns Hopkins University Coronavirus COVID-19 global cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU) https://coronavirus.jhu.edu/map.html Available at:
- 4.Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S., et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Global Initiative on Sharing All Influenza Data. https://www.gisaid.org/ Available at: Accessed April 9, 2020. [DOI] [PMC free article] [PubMed]
- 7.Genomic epidemiology of novel coronavirus—global subsampling. https://nextstrain.org/ncov/global Available at: Accessed April 9, 2020.
- 8.Fauver J.R., Petrone M.E., Hodcroft E.B., Shioda K., Ehrlich H.Y., Watts A.G., et al. Coast-to-coast spread of SARS-CoV-2 in the United States revealed by genomic epidemiology. medRxiv. 2020 Mar 26 doi: 10.1101/2020.03.25.20043828. 2020.03.25.20043828. Preprint. [DOI] [Google Scholar]
- 9.New York State Department of Health NYSDOH COVID-19 tracker. https://covid19tracker.health.ny.gov/views/NYS-COVID19-Tracker/NYSDOHCOVID-19Tracker-Map?%3Aembed=yes&%3Atoolbar=no&%3Atabs=n Available at: Accessed June 23, 2020.
- 10.New York State Department of Health NYSDOH COVID-19 tracker fatalities. https://covid19tracker.health.ny.gov/views/NYS-COVID19-Tracker/NYSDOHCOVID-19Tracker-Fatalities?%3Aembed=yes&%3Atoolbar=no&%3Atabs=n Available at: Accessed June 23, 2020.
- 11.Wallace D.J., Angus D.C., Seymour C.W., Barnato A.E., Kahn J.M. Critical care bed growth in the United States: a comparison of regional and national trends. Am J Respir Crit Care Med. 2015;191:410–416. doi: 10.1164/rccm.201409-1746OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes A., Ferdinande P., Flaatten H., Guidet B., Metnitz P.G., Moreno R.P. The variability of critical care bed numbers in Europe. Intensive Care Med. 2012;38:1647–1653. doi: 10.1007/s00134-012-2627-8. [DOI] [PubMed] [Google Scholar]
- 13.Why daily death tolls have become unusually important in understanding the coronavirus pandemic. Nature. https://www.nature.com/articles/d41586-020-01008-1 Available at: Accessed April 10, 2020. [DOI] [PubMed]
- 14.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 15.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 16.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.COVID-19: An ACP physician’s guide + resources. https://assets.acponline.org/coronavirus/scormcontent/?_ga=2.54152232.1605318961.1586281793-1274806679.1583854789&_gac=1.50270548.1586284027.Cj0KCQjwybD0BRDyARIsACyS8muepPoXZr0uLbZ8blIWVbi-aF5caFh-XrzlV8-CSJ2mvMxLLOdE64YaAojHEALw_wcB#/ Last Updated April 8, 2020. Available at: Accessed April 9, 2020.
- 19.National Institutes of Health COVID-19 treatment guidelines. https://covid19treatmentguidelines.nih.gov/introduction/ Available at: Accessed April 19, 2020. [PubMed]
- 20.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goyal P., Choi J.J., Pinheiro L.C., Schenck E.J., Chen R., Jabri A., et al. Clinical characteristics of Covid-19 in New York City. N Engl J Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chow E.J., Schwartz N.G., Tobolowsky F.A., Zacks R.L.T., Huntington-Frazier M., Reddy S.C., et al. Symptom screening at illness onset of health care personnel with SARS-CoV-2 infection in King County, Washington. JAMA. 2020;323:2087–2089. doi: 10.1001/jama.2020.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juniper E.F., Guyatt G.H., Dolovich J. Assessment of quality of life in adolescents with allergic rhinoconjunctivitis: development and testing of a questionnaire for clinical trials. J Allergy Clin Immunol. 1994;93:413–423. doi: 10.1016/0091-6749(94)90349-2. [DOI] [PubMed] [Google Scholar]
- 26.Skoner D.P. Allergic rhinitis: definition, epidemiology, pathophysiology, detection, and diagnosis. J Allergy Clin Immunol. 2001;108:S2–S8. doi: 10.1067/mai.2001.115569. [DOI] [PubMed] [Google Scholar]
- 27.Price D., Scadding G., Ryan D., Bachert C., Canonica G.W., Mullol J., et al. The hidden burden of adult allergic rhinitis: UK healthcare resource utilisation survey. Clin Transl Allergy. 2015;5:39. doi: 10.1186/s13601-015-0083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Canonica G.W., Mullol J., Pradalier A., Didier A. Patient perceptions of allergic rhinitis and quality of life: findings from a survey conducted in Europe and the United States. World Allergy Organ J. 2008;1:138–144. doi: 10.1097/WOX.0b013e3181865faf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schatz M. A survey of the burden of allergic rhinitis in the USA. Allergy. 2007;62:9–16. doi: 10.1111/j.1398-9995.2007.01548.x. [DOI] [PubMed] [Google Scholar]
- 30.Giacomelli A, Pezzati L, Conti F, Bernacchia D, Siano M, Oreni L, et al. Self-reported olfactory and taste disorders in SARS-CoV-2 patients: a cross-sectional study [published online ahead of print March 26, 2020]. Clin Infect Dis. https://doi.org/10.1093/cid/ciaa330. [DOI] [PMC free article] [PubMed]
- 31.Lechien J.R., Chiesa-Estomba C.M., De Siati D.R., Horoi M., Le Bon S.D., Rodriguez A., et al. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur Arch Otorhinolaryngol. 2020:1–11. doi: 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowart B.J., Flynn-Rodden K., McGeady S.J., Lowry L.D. Hyposmia in allergic rhinitis. J Allergy Clin Immunol. 1993;91:747–751. doi: 10.1016/0091-6749(93)90194-k. [DOI] [PubMed] [Google Scholar]
- 33.Rydzewski B., Pruszewicz A., Sulkowski W.J. Assessment of smell and taste in patients with allergic rhinitis. Acta Otolaryngol. 2000;120:323–326. doi: 10.1080/000164800750001189. [DOI] [PubMed] [Google Scholar]
- 34.Stuck B.A., Hummel T. Olfaction in allergic rhinitis: a systematic review. J Allergy Clin Immunol. 2015;136:1460–1470. doi: 10.1016/j.jaci.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, Yan YQ, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China [published online ahead of print February 19, 2020]. Allergy. https://doi.org/10.1111/all.14238. [DOI] [PubMed]
- 36.Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan [published online ahead of print April 12, 2020]. J Allergy Clin Immunol. https://doi.org/10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed]
- 37.Centers for Disease Control and Prevention Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019—COVID-NET, 14 states, March 1–30, 2020. https://www.cdc.gov/mmwr/volumes/69/wr/pdfs/mm6915e3-H.pdf Available at: Accessed April 10, 2020. [DOI] [PMC free article] [PubMed]
- 38.Zimmermann P., Curtis N. COVID-19 in children, pregnancy and neonates: a review of epidemiologic and clinical features. Pediatr Infect Dis J. 2020;39:469–477. doi: 10.1097/INF.0000000000002700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Codispoti CD, Bandi S, Patel P, Mahdavinia M. Clinical course of asthma in 4 cases of COVID-19 infection [published online ahead of print May 11, 2020]. Ann Allergy Asthma Immunol. https://doi.org/10.1016/j.anai.2020.05.009. [DOI] [PMC free article] [PubMed]
- 40.Krishnan J.A., Lemanske R.F., Jr., Canino G.J., Elward K.S., Kattan M., Matsui E.C., et al. Asthma outcomes: symptoms. J Allergy Clin Immunol. 2012;129:S124–S135. doi: 10.1016/j.jaci.2011.12.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Recalcati S. Cutaneous manifestations in COVID-19: a first perspective [published online ahead of print March 2020]. J Eur Acad Dermatol Venereol. https://doi.org/10.1111/jdv.16387. [DOI] [PubMed]
- 42.Viner RM, Whittaker E. Kawasaki-like disease: emerging complication during the COVID-19 pandemic [published online ahead of print May 13, 2020]. Lancet. 10.1016/S0140-6736(20)31129-6. [DOI] [PMC free article] [PubMed]
- 43.Verdoni L, Mazza A, Gervasoni A, Martelli L, Ruggeri M, Ciuffreda M, et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study [published online ahead of print May 13, 2020]. Lancet. 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed]
- 44.Kawasaki T. Kawasaki disease. Proc Jpn Acad Ser B Phys Biol Sci. 2006;82:59–71. doi: 10.2183/pjab.82.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuo H.C., Chang W.C., Yang K.D., Yu H.R., Wang C.L., Ho S.C., et al. Kawasaki disease and subsequent risk of allergic diseases: a population-based matched cohort study. BMC Pediatr. 2013;13:38. doi: 10.1186/1471-2431-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai Y.J., Lin C.H., Fu L.S., Fu Y.C., Lin M.C., Jan S.L. The association between Kawasaki disease and allergic diseases, from infancy to school age. Allergy Asthma Proc. 2013;34:467–472. doi: 10.2500/aap.2013.34.3697. [DOI] [PubMed] [Google Scholar]
- 47.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China [published online ahead of print March 13, 2020]. JAMA Intern Med. https://doi.org/10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed]
- 48.Ma K-L, Liu Z-H, Cao C-F, Liu M-K, Liao J, Zou J-B, et al. COVID-19 myocarditis and severity factors: an adult cohort study [published online ahead of print March 23, 2020]. medRxiv. 10.1101/2020.03.19.20034124. [DOI]
- 49.Zhang F, Yang D, Li J, Gao P, Chen T, Cheng Z, et al. Myocardial injury is associated with in-hospital mortality of confirmed or suspected COVID-19 in Wuhan, China: a single center retrospective cohort study [published online ahead of print March 24, 2020]. medRxiv. 10.1101/2020.03.21.20040121. [DOI]
- 50.Grasselli G, Pesenti A, Cecconi M. Critical care utilization for the COVID-19 outbreak in Lombardy, Italy: early experience and forecast during an emergency response [published online ahead of print March 13, 2020]. JAMA. https://doi.org/10.1001/jama.2020.4031. [DOI] [PubMed]
- 51.Carenzo L., Costantini E., Greco M., Barra F.L., Rendiniello V., Mainetti M., et al. Hospital surge capacity in a tertiary emergency referral centre during the COVID-19 outbreak in Italy. Anaesthesia. 2020;75:928–934. doi: 10.1111/anae.15072. [DOI] [PubMed] [Google Scholar]
- 52.Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 Available at:
- 53.Centers for Disease Control and Prevention Interim guidelines for collecting, handling, and testing clinical specimens from persons for coronavirus disease 2019 (COVID-19) https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html Available at: Accessed May 15, 2020.
- 54.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.U.S. Food and Drug Administration Coronavirus (COVID-19) update: FDA authorizes first test for patient at-home sample collection. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-test-patient-home-sample-collection Available at: Accessed April 21, 2020.
- 56.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 57.Will antibody tests for the coronavirus really change everything? Nature. https://www.nature.com/articles/d41586-020-01115-z Available at: Accessed April 21, 2020.
- 58.Infectious Diseases Society of America IDSA COVID-19 antibody testing primer. https://www.idsociety.org/globalassets/idsa/public-health/covid-19/idsa-covid-19-antibody-testing-primer.pdf?fbclid=IwAR190ri4PlJ8jo-dRqWWmOm2qvofnWjH0LSVlpR8Vb6EuAhQONBDLjhz1Zw Available at: Accessed April 23, 2020.
- 59.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu J., Gu J., Li K., Xu C., Su W., Lai Z., et al. COVID-19 outbreak associated with air conditioning in restaurant, Guangzhou, China, 2020. Emerg Infect Dis. 2020;26 doi: 10.3201/eid2607.200764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo Z.D., Wang Z.Y., Zhang S.F., Li X., Li L., Li C., et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg Infect Dis. 2020;26 doi: 10.3201/eid2607.200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 65.Lin L., Jiang X., Zhang Z., Huang S., Zhang Z., Fang Z., et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 66.Ong S.W.X., Tan Y.K., Chia P.Y., Lee T.H., Ng O.T., Wong M.S.Y., et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323:1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lipsitch M., Cohen T., Cooper B., Robins J.M., Ma S., James L., et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chinazzi M., Davis J.T., Ajelli M., Gioannini C., Litvinova M., Merler S., et al. The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science. 2020;368:395–400. doi: 10.1126/science.aba9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X, Zhang S. COVID-19: face masks and human-to-human transmission [published online ahead of print March 29, 2020]. Influenza Other Respir Viruses. https://doi.org/10.1111/irv.12740. [DOI] [PMC free article] [PubMed]
- 72.Abd-Elsayed A, Karri J. Utility of substandard face mask options for health care workers during the COVID-19 pandemic [published online ahead of print March 31, 2020]. Anesth Analg. https://doi.org/10.1213/ANE.0000000000004841. [DOI] [PMC free article] [PubMed]
- 73.Feng S., Shen C., Xia N., Song W., Fan M., Cowling B.J. Rational use of face masks in the COVID-19 pandemic. Lancet Respir Med. 2020;8:434–436. doi: 10.1016/S2213-2600(20)30134-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bae S, Kim MC, Kim JY, Cha HH, Lim JS, Jung J, et al. Effectiveness of surgical and cotton masks in blocking SARS-CoV-2: a controlled comparison in 4 patients [published online ahead of print April 6, 2020]. Ann Intern Med. https://doi.org/10.7326/M20-1342. [DOI] [PMC free article] [PubMed] [Retracted]
- 75.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vabret A., Dina J., Gouarin S., Petitjean J., Tripey V., Brouard J., et al. Human (non-severe acute respiratory syndrome) coronavirus infections in hospitalised children in France. J Paediatr Child Health. 2008;44:176–181. doi: 10.1111/j.1440-1754.2007.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lau S.K., Woo P.C., Yip C.C., Tse H., Tsoi H.W., Cheng V.C., et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. 2006;44:2063–2071. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gerna G., Campanini G., Rovida F., Percivalle E., Sarasini A., Marchi A., et al. Genetic variability of human coronavirus OC43-, 229E-, and NL63-like strains and their association with lower respiratory tract infections of hospitalized infants and immunocompromised patients. J Med Virol. 2006;78:938–949. doi: 10.1002/jmv.20645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chan J.W., Ng C.K., Chan Y.H., Mok T.Y., Lee S., Chu S.Y., et al. Short term outcome and risk factors for adverse clinical outcomes in adults with severe acute respiratory syndrome (SARS) Thorax. 2003;58:686–689. doi: 10.1136/thorax.58.8.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arabi Y.M., Balkhy H.H., Hayden F.G., Bouchama A., Luke T., Baillie J.K., et al. Middle East respiratory syndrome. N Engl J Med. 2017;376:584–594. doi: 10.1056/NEJMsr1408795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mahase E. Coronavirus covid-19 has killed more people than SARS and MERS combined, despite lower case fatality rate. BMJ. 2020;368:m641. doi: 10.1136/bmj.m641. [DOI] [PubMed] [Google Scholar]
- 82.National Institute of Allergy and Infectious Diseases COVID-19, MERS & SARS. https://www.niaid.nih.gov/diseases-conditions/covid-19 Available at: Accessed April 24, 2020.
- 83.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3 doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang T, Wu Q, Zhang Z. Pangolin homology associated with 2019-nCoV [published online ahead of print February 20, 2020]. bioRxiv. 10.1101/2020.02.19.950253. [DOI]
- 88.Lam TT, Shum MH, Zhu HC, Tong YG, Ni XB, Liao YS, et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins [published online ahead of print March 26, 2020]. Nature. https://doi.org/10.1038/s41586-020-2169-0. [DOI] [PubMed]
- 89.Sabir J.S., Lam T.T., Ahmed M.M., Li L., Shen Y., Abo-Aba S.E., et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 90.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hamming I., Cooper M.E., Haagmans B.L., Hooper N.M., Korstanje R., Osterhaus A.D., et al. The emerging role of ACE2 in physiology and disease. J Pathol. 2007;212:1–11. doi: 10.1002/path.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus: a first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng Z.J., Shan J. 2019 Novel coronavirus: where we are and what we know. Infection. 2020;48:155–163. doi: 10.1007/s15010-020-01401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Simmons G., Zmora P., Gierer S., Heurich A., Pohlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Centers for Disease Control and Prevention Summary health statistics tables for U.S. adults: National Health Interview Survey, 2018, tables A-2b, A-2c. https://ftp.cdc.gov/pub/Health_Statistics/NCHS/NHIS/SHS/2018_SHS_Table_A-2.pdf Available at: Accessed April 23, 2020.
- 98.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., et al. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020 doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.de Wilde A.H., Snijder E.J., Kikkert M., van Hemert M.J. Host factors in coronavirus replication. Curr Top Microbiol Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hussain S., Pan J., Chen Y., Yang Y., Xu J., Peng Y., et al. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J Virol. 2005;79:5288–5295. doi: 10.1128/JVI.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Perrier A., Bonnin A., Desmarets L., Danneels A., Goffard A., Rouille Y., et al. The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J Biol Chem. 2019;294:14406–14421. doi: 10.1074/jbc.RA119.008964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wark P.A., Johnston S.L., Moric I., Simpson J.L., Hensley M.J., Gibson P.G. Neutrophil degranulation and cell lysis is associated with clinical severity in virus-induced asthma. Eur Respir J. 2002;19:68–75. doi: 10.1183/09031936.02.00226302. [DOI] [PubMed] [Google Scholar]
- 111.Khetsuriani N., Kazerouni N.N., Erdman D.D., Lu X., Redd S.C., Anderson L.J., et al. Prevalence of viral respiratory tract infections in children with asthma. J Allergy Clin Immunol. 2007;119:314–321. doi: 10.1016/j.jaci.2006.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chung J.Y., Han T.H., Kim S.W., Kim C.K., Hwang E.S. Detection of viruses identified recently in children with acute wheezing. J Med Virol. 2007;79:1238–1243. doi: 10.1002/jmv.20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kotaniemi-Syrjanen A., Vainionpaa R., Reijonen T.M., Waris M., Korhonen K., Korppi M. Rhinovirus-induced wheezing in infancy--the first sign of childhood asthma? J Allergy Clin Immunol. 2003;111:66–71. doi: 10.1067/mai.2003.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rakes G.P., Arruda E., Ingram J.M., Hoover G.E., Zambrano J.C., Hayden F.G., et al. Rhinovirus and respiratory syncytial virus in wheezing children requiring emergency care: IgE and eosinophil analyses. Am J Respir Crit Care Med. 1999;159:785–790. doi: 10.1164/ajrccm.159.3.9801052. [DOI] [PubMed] [Google Scholar]
- 115.Kloepfer K.M., Lee W.M., Pappas T.E., Kang T.J., Vrtis R.F., Evans M.D., et al. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J Allergy Clin Immunol. 2014;133:1301–1307. doi: 10.1016/j.jaci.2014.02.030. 1307. e1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zheng X.Y., Xu Y.J., Guan W.J., Lin L.F. Regional, age and respiratory-secretion-specific prevalence of respiratory viruses associated with asthma exacerbation: a literature review. Arch Virol. 2018;163:845–853. doi: 10.1007/s00705-017-3700-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Corne J.M., Marshall C., Smith S., Schreiber J., Sanderson G., Holgate S.T., et al. Frequency, severity, and duration of rhinovirus infections in asthmatic and non-asthmatic individuals: a longitudinal cohort study. Lancet. 2002;359:831–834. doi: 10.1016/S0140-6736(02)07953-9. [DOI] [PubMed] [Google Scholar]
- 118.Shi T., Balsells E., Wastnedge E., Singleton R., Rasmussen Z.A., Zar H.J., et al. Risk factors for respiratory syncytial virus associated with acute lower respiratory infection in children under five years: systematic review and meta-analysis. J Glob Health. 2015;5 doi: 10.7189/jogh.05.020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miller E.K., Lu X., Erdman D.D., Poehling K.A., Zhu Y., Griffin M.R., et al. Rhinovirus-associated hospitalizations in young children. J Infect Dis. 2007;195:773–781. doi: 10.1086/511821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meissner H.C. Viral bronchiolitis in children. N Engl J Med. 2016;374:1793–1794. doi: 10.1056/NEJMc1601509. [DOI] [PubMed] [Google Scholar]
- 121.Jackson D.J., Gangnon R.E., Evans M.D., Roberg K.A., Anderson E.L., Pappas T.E., et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178:667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lemanske R.F., Jr., Jackson D.J., Gangnon R.E., Evans M.D., Li Z., Shult P.A., et al. Rhinovirus illnesses during infancy predict subsequent childhood wheezing. J Allergy Clin Immunol. 2005;116:571–577. doi: 10.1016/j.jaci.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 123.Malmstrom K., Pitkaranta A., Carpen O., Pelkonen A., Malmberg L.P., Turpeinen M., et al. Human rhinovirus in bronchial epithelium of infants with recurrent respiratory symptoms. J Allergy Clin Immunol. 2006;118:591–596. doi: 10.1016/j.jaci.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 124.Stein R.T., Sherrill D., Morgan W.J., Holberg C.J., Halonen M., Taussig L.M., et al. Respiratory syncytial virus in early life and risk of wheeze and allergy by age 13 years. Lancet. 1999;354:541–545. doi: 10.1016/S0140-6736(98)10321-5. [DOI] [PubMed] [Google Scholar]
- 125.Sigurs N., Bjarnason R., Sigurbergsson F., Kjellman B. Respiratory syncytial virus bronchiolitis in infancy is an important risk factor for asthma and allergy at age 7. Am J Respir Crit Care Med. 2000;161:1501–1507. doi: 10.1164/ajrccm.161.5.9906076. [DOI] [PubMed] [Google Scholar]
- 126.Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., et al. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Broszeit F., Tzarum N., Zhu X., Nemanichvili N., Eggink D., Leenders T., et al. N-Glycolylneuraminic acid as a receptor for influenza A viruses. Cell Rep. 2019;27:3284–3294.e6. doi: 10.1016/j.celrep.2019.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Griggs T.F., Bochkov Y.A., Basnet S., Pasic T.R., Brockman-Schneider R.A., Palmenberg A.C., et al. Rhinovirus C targets ciliated airway epithelial cells. Respir Res. 2017;18:84. doi: 10.1186/s12931-017-0567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Weinheimer V.K., Becher A., Tonnies M., Holland G., Knepper J., Bauer T.T., et al. Influenza A viruses target type II pneumocytes in the human lung. J Infect Dis. 2012;206:1685–1694. doi: 10.1093/infdis/jis455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tan W.C., Xiang X., Qiu D., Ng T.P., Lam S.F., Hegele R.G. Epidemiology of respiratory viruses in patients hospitalized with near-fatal asthma, acute exacerbations of asthma, or chronic obstructive pulmonary disease. Am J Med. 2003;115:272–277. doi: 10.1016/s0002-9343(03)00353-x. [DOI] [PubMed] [Google Scholar]
- 131.Kistler A., Avila P.C., Rouskin S., Wang D., Ward T., Yagi S., et al. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Van Bever H.P., Chng S.Y., Goh D.Y. Childhood severe acute respiratory syndrome, coronavirus infections and asthma. Pediatr Allergy Immunol. 2004;15:206–209. doi: 10.1111/j.1399-3038.2004.00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chiu S.S., Chan K.H., Chu K.W., Kwan S.W., Guan Y., Poon L.L., et al. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin Infect Dis. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wark P.A., Johnston S.L., Bucchieri F., Powell R., Puddicombe S., Laza-Stanca V., et al. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med. 2005;201:937–947. doi: 10.1084/jem.20041901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhu J., Message S.D., Mallia P., Kebadze T., Contoli M., Ward C.K., et al. Bronchial mucosal IFN-alpha/beta and pattern recognition receptor expression in patients with experimental rhinovirus-induced asthma exacerbations. J Allergy Clin Immunol. 2019;143:114–125.e4. doi: 10.1016/j.jaci.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Papadopoulos N.G., Stanciu L.A., Papi A., Holgate S.T., Johnston S.L. A defective type 1 response to rhinovirus in atopic asthma. Thorax. 2002;57:328–332. doi: 10.1136/thorax.57.4.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Slejko J.F., Ghushchyan V.H., Sucher B., Globe D.R., Lin S.L., Globe G., et al. Asthma control in the United States, 2008-2010: indicators of poor asthma control. J Allergy Clin Immunol. 2014;133:1579–1587. doi: 10.1016/j.jaci.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 141.Sungnak W., Huang N., Becavin C., Berg M., Queen R., Litvinukova M., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hayashi T., Beck L., Rossetto C., Gong X., Takikawa O., Takabayashi K., et al. Inhibition of experimental asthma by indoleamine 2,3-dioxygenase. J Clin Invest. 2004;114:270–279. doi: 10.1172/JCI21275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Taher Y.A., Piavaux B.J., Gras R., van Esch B.C., Hofman G.A., Bloksma N., et al. Indoleamine 2,3-dioxygenase-dependent tryptophan metabolites contribute to tolerance induction during allergen immunotherapy in a mouse model. J Allergy Clin Immunol. 2008;121:983–991.e2. doi: 10.1016/j.jaci.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 144.Haller O., Staeheli P., Schwemmle M., Kochs G. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015;23:154–163. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 145.Loisel D.A., Du G., Ahluwalia T.S., Tisler C.J., Evans M.D., Myers R.A., et al. Genetic associations with viral respiratory illnesses and asthma control in children. Clin Exp Allergy. 2016;46:112–124. doi: 10.1111/cea.12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jackson D.J., Makrinioti H., Rana B.M., Shamji B.W., Trujillo-Torralbo M.B., Footitt J., et al. IL-33-dependent type 2 inflammation during rhinovirus-induced asthma exacerbations in vivo. Am J Respir Crit Care Med. 2014;190:1373–1382. doi: 10.1164/rccm.201406-1039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Allinne J., Scott G., Lim W.K., Birchard D., Erjefalt J.S., Sanden C., et al. IL-33 blockade affects mediators of persistence and exacerbation in a model of chronic airway inflammation. J Allergy Clin Immunol. 2019;144:1624–1637.e10. doi: 10.1016/j.jaci.2019.08.039. [DOI] [PubMed] [Google Scholar]
- 148.Ravanetti L., Dijkhuis A., Dekker T., Sabogal Pineros Y.S., Ravi A., Dierdorp B.S., et al. IL-33 drives influenza-induced asthma exacerbations by halting innate and adaptive antiviral immunity. J Allergy Clin Immunol. 2019;143:1355–1370.e16. doi: 10.1016/j.jaci.2018.08.051. [DOI] [PubMed] [Google Scholar]
- 149.Werder R.B., Zhang V., Lynch J.P., Snape N., Upham J.W., Spann K., et al. Chronic IL-33 expression predisposes to virus-induced asthma exacerbations by increasing type 2 inflammation and dampening antiviral immunity. J Allergy Clin Immunol. 2018;141:1607–1619.e9. doi: 10.1016/j.jaci.2017.07.051. [DOI] [PubMed] [Google Scholar]
- 150.Centers for Disease Control and Prevention Groups at higher risk for severe illness. https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/groups-at-higher-risk.html Available at:
- 151.American Academy of Allergy, Asthma and Immunology Messages from the COVID-19 Response Task Force. https://education.aaaai.org/task-force-messages_COVID-19#overlay-context= Available at: Accessed April 10, 2020.
- 152.Shanawani H. Health disparities and differences in asthma: concepts and controversies. Clin Chest Med. 2006;27:17–28. doi: 10.1016/j.ccm.2005.11.002. v. [DOI] [PubMed] [Google Scholar]
- 153.Gupta R.S., Carrion-Carire V., Weiss K.B. The widening black/white gap in asthma hospitalizations and mortality. J Allergy Clin Immunol. 2006;117:351–358. doi: 10.1016/j.jaci.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 154.El-Ekiaby A., Brianas L., Skowronski M.E., Coreno A.J., Galan G., Kaeberlein F.J., et al. Impact of race on the severity of acute episodes of asthma and adrenergic responsiveness. Am J Respir Crit Care Med. 2006;174:508–513. doi: 10.1164/rccm.200603-431OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jackson DJ, Busse WW, Bacharier LB, Kattan M, O’Connor GT, Wood RA, et al. Association of respiratory allergy, asthma, and expression of the SARS-CoV-2 receptor ACE2 [published online ahead of print April 22, 2020]. J Allergy Clin Immunol. https://doi.org/10.1016/j.jaci.2020.04.009. [DOI] [PMC free article] [PubMed]
- 157.Kuruvilla M.E., Lee F.E., Lee G.B. Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin Rev Allergy Immunol. 2019;56:219–233. doi: 10.1007/s12016-018-8712-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Peters U., Dixon A.E., Forno E. Obesity and asthma. J Allergy Clin Immunol. 2018;141:1169–1179. doi: 10.1016/j.jaci.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Peters MC, Sajuthi S, Deford P, Christenson S, Rios CL, Montgomery MT, et al. COVID-19 related genes in sputum cells in asthma: relationship to demographic features and corticosteroids [published online ahead of print April 29, 2020]. Am J Respir Crit Care Med. https://doi.org/10.1164/rccm.202003-0821OC. [DOI] [PMC free article] [PubMed]
- 160.Rigat B., Hubert C., Alhenc-Gelas F., Cambien F., Corvol P., Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ding Q.L., Sun S.F., Cao C., Deng Z.C. Association between angiotensin-converting enzyme I/D polymorphism and asthma risk: a meta-analysis involving 11,897 subjects. J Asthma. 2012;49:557–562. doi: 10.3109/02770903.2012.685540. [DOI] [PubMed] [Google Scholar]
- 162.Yao S., Shi A., Li J., Ma J., Lu L. Angiotensin-converting enzyme gene polymorphisms might be associated with childhood asthma in East Asia. J Asthma. 2017;54:476–478. doi: 10.1080/02770903.2016.1236943. [DOI] [PubMed] [Google Scholar]
- 163.Bottcher E., Matrosovich T., Beyerle M., Klenk H.D., Garten W., Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93 doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kesic M.J., Hernandez M., Jaspers I. Airway protease/antiprotease imbalance in atopic asthmatics contributes to increased influenza A virus cleavage and replication. Respir Res. 2012;13:82. doi: 10.1186/1465-9921-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dhawale V.S., Amara V.R., Karpe P.A., Malek V., Patel D., Tikoo K. Activation of angiotensin-converting enzyme 2 (ACE2) attenuates allergic airway inflammation in rat asthma model. Toxicol Appl Pharmacol. 2016;306:17–26. doi: 10.1016/j.taap.2016.06.026. [DOI] [PubMed] [Google Scholar]
- 167.Magalhaes G.S., Barroso L.C., Reis A.C., Rodrigues-Machado M.G., Gregorio J.F., Motta-Santos D., et al. Angiotensin-(1-7) promotes resolution of eosinophilic inflammation in an experimental model of asthma. Front Immunol. 2018;9:58. doi: 10.3389/fimmu.2018.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.ClinicalTrials.gov National Institutes of Health. https://clinicaltrials.gov/ct2/home Available at: Accessed April 23, 2020.
- 171.EU Clinical Trials Register EudraCT. https://www.clinicaltrialsregister.eu/ctr-search/search Available at: Accessed April 23, 2020.
- 172.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., et al. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen C, Huang J, Cheng Z, Wu J, Chen S, Zhang Y, et al. Favipiravir versus Arbidol for COVID-19: a randomized clinical trial [published online ahead of print April 15, 2020]. medRxiv. 10.1101/2020.03.17.20037432. [DOI]
- 174.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 175.Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial [published online ahead of print March 20, 2020]. Int J Antimicrob Agents. https://doi.org/10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed]
- 176.Hung I.F., Lung K.C., Tso E.Y., Liu R., Chung T.W., Chu M.Y., et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.World Health Organization “Solidarity” clinical trial for COVID-19 treatments. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments Available at:
- 178.Kupferschmidt K., Cohen J. Race to find COVID-19 treatments accelerates. Science. 2020;367:1412–1413. doi: 10.1126/science.367.6485.1412. [DOI] [PubMed] [Google Scholar]
- 179.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. CellPress. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, et al. The feasibility of convalescent plasma therapy in severe COVID-19 patients: a pilot study [published online ahead of print March 23, 2020]. medRxiv. 10.1101/2020.03.16.20036145. [DOI]
- 182.Wu Y, Wang F, Shen C, Peng W, Li D, Zhao C, et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2 [published online ahead of print May 13, 2020]. Science. https://doi.org/10.1126/science.abc2241. [DOI] [PMC free article] [PubMed]
- 183.Yamamoto M., Matsuyama S., Li X., Takeda M., Kawaguchi Y., Inoue J.I., et al. Identification of nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother. 2016;60:6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Identification of an existing Japanese pancreatitis drug, Nafamostat, which is expected to prevent the transmission of new coronavirus infection (COVID-19) https://www.u-tokyo.ac.jp/focus/en/articles/z0508_00083.html Available at: Accessed April 8, 2020.
- 185.Hu T.Y., Frieman M., Wolfram J. Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat Nanotechnol. 2020;15:247–249. doi: 10.1038/s41565-020-0674-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Torigoe M., Sakata K., Ishii A., Iwata S., Nakayamada S., Tanaka Y. Hydroxychloroquine efficiently suppresses inflammatory responses of human class-switched memory B cells via Toll-like receptor 9 inhibition. Clin Immunol. 2018;195:1–7. doi: 10.1016/j.clim.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 187.Borba M.G.S., Val F.F.A., Sampaio V.S., Alexandre M.A.A., Melo G.C., Brito M., et al. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: a randomized clinical trial. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PubMed] [Google Scholar]
- 188.Magagnoli J, Narendran S, Pereira F, Cummings T, Hardin JW, Sutton SS, et al. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with Covid-19 [published online ahead of print April 21, 2020]. medRxiv. https://doi.org/10.1101/2020.04.16.20065920. [DOI] [PMC free article] [PubMed]
- 189.Rosenberg ES, Dufort EM, Udo T, Wilberschied LA, Kumar J, Tesoriero J, et al. Association of treatment with hydroxychloroquine or azithromycin with in-hospital mortality in patients with COVID-19 in New York state [published online ahead of print May 11, 2020]. JAMA. https://doi.org/10.1001/jama.2020.8630. [DOI] [PMC free article] [PubMed]
- 190.Chu C.M., Cheng V.C., Hung I.F., Wong M.M., Chan K.H., Chan K.S., et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kim U.J., Won E.J., Kee S.J., Jung S.I., Jang H.C. Combination therapy with lopinavir/ritonavir, ribavirin and interferon-alpha for Middle East respiratory syndrome. Antivir Ther. 2016;21:455–459. doi: 10.3851/IMP3002. [DOI] [PubMed] [Google Scholar]
- 192.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.National Institutes of Health NIH clinical trial shows remdesivir accelerates recovery from advanced COVID-19. https://www.nih.gov/news-events/news-releases/nih-clinical-trial-shows-remdesivir-accelerates-recovery-advanced-covid-19 Available at: Accessed May 14, 2020.
- 195.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chan JF, Zhang AJ, Yuan S, Poon VK, Chan CC, Lee AC, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility [published online ahead of print March 26, 2020]. Clin Infect Dis. https://doi.org/10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed]
- 197.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Coomes EA, Haghbayan H. Interleukin-6 in COVID-19: a systematic review and meta-analysis [published online ahead of print April 3, 2020]. medRxiv. 10.1101/2020.03.30.20048058. [DOI] [PMC free article] [PubMed]
- 199.Schaper F., Rose-John S. Interleukin-6: biology, signaling and strategies of blockade. Cytokine Growth Factor Rev. 2015;26:475–487. doi: 10.1016/j.cytogfr.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 200.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Conner S.D., Schmid S.L. Identification of an adaptor-associated kinase, AAK1, as a regulator of clathrin-mediated endocytosis. J Cell Biol. 2002;156:921–929. doi: 10.1083/jcb.200108123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Favalli EG, Biggioggero M, Maioli G, Caporali R. Baricitinib for COVID-19: a suitable treatment? [published online ahead of print April 3, 2020]. Lancet Infect Dis. https://doi.org/10.1016/S1473-3099(20)30262-0. [DOI] [PMC free article] [PubMed]
- 203.Cantini F, Niccoli L, Matarrese D, Nicastri E, Stobbione P, Goletti D. Baricitinib therapy in COVID-19: a pilot study on safety and clinical impact [published online ahead of print April 23, 2020]. J Infect. https://doi.org/10.1016/j.jinf.2020.04.017. [DOI] [PMC free article] [PubMed]
- 204.Jeon S, Ko M, Lee J, Choi I, Byun SY, Park S, et al. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs [published online ahead of print March 4, 2020]. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed]
- 205.Ko M, Chang SY, Byun SY, Choi I, d’Alexandry d’Orengiani A-LPH, Shum D, et al. Screening of FDA-approved drugs using a MERS-CoV clinical isolate from South Korea identifies potential therapeutic options for COVID-19 [published online ahead of print March 19, 2020]. bioRxiv. 10.1101/2020.02.25.965582. [DOI] [PMC free article] [PubMed]
- 206.Deng X., Hackbart M., Mettelman R.C., O’Brien A., Mielech A.M., Yi G., et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Matsuyama S, Kawase M, Nao N, Shirato K, Ujike M, Kamitani W, et al. The inhaled corticosteroid ciclesonide blocks coronavirus RNA replication by targeting viral NSP15. bioRxiv [published online ahead of print March 12, 2020]. 10.1101/2020.03.11.987016. [DOI]
- 208.Leach C.L., Bethke T.D., Boudreau R.J., Hasselquist B.E., Drollmann A., Davidson P., et al. Two-dimensional and three-dimensional imaging show ciclesonide has high lung deposition and peripheral distribution: a nonrandomized study in healthy volunteers. J Aerosol Med. 2006;19:117–126. doi: 10.1089/jam.2006.19.117. [DOI] [PubMed] [Google Scholar]
- 209.Nave R., Mueller H. From inhaler to lung: clinical implications of the formulations of ciclesonide and other inhaled corticosteroids. Int J Gen Med. 2013;6:99–107. doi: 10.2147/IJGM.S39134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Iwabuchi K., Yoshie K., Kurakami Y., Takahashi K., Kato Y., Morishima T. 2020. COVID-19. Three cases improved with inhaled ciclesonide in the early to middle stages of pneumonia. Available at: http://www.kansensho.or.jp/modules/topics/index.php?content_id=31. Accessed June 25, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Yamaya M., Nishimura H., Deng X., Sugawara M., Watanabe O., Nomura K., et al. Inhibitory effects of glycopyrronium, formoterol, and budesonide on coronavirus HCoV-229E replication and cytokine production by primary cultures of human nasal and tracheal epithelial cells. Respir Investig. 2020;58:155–168. doi: 10.1016/j.resinv.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. Structure of M(pro) from COVID-19 virus and discovery of its inhibitors [published online ahead of print April 9, 2020]. Nature. https://doi.org/10.1038/s41586-020-2223-y.
- 213.National Institutes of Health NIH clinical trial of investigational vaccine for COVID-19 begins. https://www.nih.gov/news-events/news-releases/nih-clinical-trial-investigational-vaccine-covid-19-begins Available at:
- 214.Cohen J. Vaccine designers take first shots at COVID-19. Science. 2020;368:14–16. doi: 10.1126/science.368.6486.14. [DOI] [PubMed] [Google Scholar]
- 215.World Health Organization DRAFT landscape of COVID-19 candidate vaccines–20 March 2020. https://www.who.int/blueprint/priority-diseases/key-action/novel-coronavirus-landscape-ncov.pdf?ua=1 Available at: