Abstract
The plant hormone auxin is essential for plant growth and development. YUCCA proteins catalyse the rate-limiting step for endogenous auxin biosynthesis. In this study, we isolated 20 MdYUCCA genes from apple genome. MdYUCCA6a, MdYUCCA8a, and MdYUCCA10a were expressed in most organs and could support whole plant basal auxin synthesis. MdYUCCA4a, MdYUCCA10b, and MdYUCCA11a expression indicated roles for these genes in auxin biosynthesis in vegetative organs. MdYUCCA2b, MdYUCCA11b, and MdYUCCA11d were mainly expressed in flower organs. High temperature induced the expression of MdYUCCA4a, MdYUCCA6a, MdYUCCA8a, and MdYUCCA10a, and down-regulated the expression of MdYUCCA2b and MdYUCCA6b. Dual-luciferase assay indicated that MdPIF4 could trans-activate the MdYUCCA8a promoter. Overexpression of MdYUCCA8a increased IAA content, increased stem height, enhanced apical dominance, and led to silique malformation. These results provide a foundation for further investigation of the biological functions of apple MdYUCCAs.
Subject terms: Plant sciences, Plant development, Plant hormones
Introduction
The plant hormone auxin is essential for plant growth and development, and plays important roles in biological processes, such as organ formation and development and apical dominance1,2. Auxin synthesis mainly occurs in young tissues, such as developing leaves, cotyledons, flowers, and shoots and root apex. Indole-3-acetic acid (IAA), the main auxin in plants, is de novo synthesized via tryptophan (Trp)-independent and Trp-dependent pathways, with synthesis of IAA from Trp proceeding independently via distinct pathway intermediates: indole-3-pyruvic acid (IPA), indole-3-acetaldoxime, indole-3-acetamide and tryptamine (TAM). The indole-3-pyruvic acid pathway is the primary Trp-dependent pathway for auxin biosynthesis and is conserved in plants. However, the physiological and molecular mechanism of the proposed Trp-independent pathway is less known. In the indole-3-pyruvic acid pathway, at the first step, Trp is converted into indole-3-pyruvate by tryptophan aminotransferase (TAA), then, YUCCA (YUC) flavin-containing monooxygenases catalyse the conversion of IPA to IAA3,4. The second step, catalysed by YUCCA, is a rate-limiting step for the indole-3-pyruvic acid pathway. The YUCCA family genes play a significant role in auxin-mediated developmental processes and have received much attention since their discovery.
YUCCA proteins contain several conserved motifs, including FAD-binding site, GC-motif, FMO-identifying sequence motif, NADPH-binding site and ATG-containing motif. Among these motifs, the FAD-binding site and NADPH-binding site are essential for YUCCA enzymatic activities. Arabidopsis YUCCAs also share low sequence similarity with thioredoxin (Trx) reductases (TrxR) from bacteria and plants and possess thiol-reductase (TR) activity5.
The Arabidopsis YUCCA gene family includes 11 members. Knockdown of any single YUCCA gene did not result in obvious abnormal phenotypes, whereas triple or quadruple yucca mutants showed severe defects in various developmental processes, suggesting that spatially and temporally regulated auxin biosynthesis involving multiple YUCCA genes is essential for plant development2. In Arabidopsis, YUC1, YUC2, YUC4, and YUC6 are the main YUC proteins functioned in shoots. Double mutants, yuc1yuc4 and yuc2yuc6, all triple mutant combinations, and yuc1yuc2yuc4yuc6 quadruple mutants had severe defects in vascular patterning and flower development2. Microspore development in yuc2yuc6 double mutants was arrested before the first asymmetric mitotic division (PMI), and consequently, yuc2yuc6 failed to produce viable pollen6. YUC3, YUC5, YUC7, YUC8, and YUC9 were expressed during root development and yuc3yuc5yuc7yuc8yuc9 quintuple mutants showed short and agravitropic root patterns7. AtYUC10 and AtYUC11 have been suggested to have overlapping functions with AtYUC1 and AtYUC4 during embryogenesis1. Similarly, in maize, a monocot-specific YUCCA like gene mutant sparse inflorescence1, showed defects in the formation of branches, spikelets, florets, and floral organs8. In rice, the YUC loss-of-function mutant oscow1 demonstrated a reduced root-to-shoot ratio9. The petunia ortholog of Arabidopsis YUC1 gene FLOOZY (FZY) was expressed in young leaves and developing flowers, which indicated that it was required for the formation of leaf secondary veins and flower architecture10. In the root parasitic plant Phtheirospermum japonicum, expression of YUC3 at the epidermal cells near the contact site contributed to haustorium formation11. In peach, PpYUC11 showed obvious tissue-specific expression during the mature climacteric stage of the melting flesh fruit and was involved in peach flesh texture development12. Overexpression of YUCCA genes resulted in auxin overproduction phenotypes, such as elongated hypocotyl; enhanced apical dominance; tall, slender stems; leaf hyponasty; changes in leaf shape; increased lateral root numbers; abnormal flower architecture; and sterility7,13–15.
Some YUCCA genes were also involved in plant responses to abiotic stress. High temperature increased IAA level and led to hypocotyl elongation in Arabidopsis16. The basic helix-loop-helix transcription factor PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulated high temperature-induced hypocotyl elongation by activating YUC8 in Arabidopsis17,18. RNA-binding protein FCA (Flowering time control protein) tuned down the high-temperature-induced stem elongation by modifying YUC8 chromatin and dissociating PIF4 from the YUC8 promoter19. In contrast, in barley and Arabidopsis, high temperature suppression of YUC2 and YUC6 gene expression in developing anthers resulted in male sterility, and exogenous IAA rescued the defects20. This indicated that the YUC-mediated local auxin biosynthesis in the anther was essential for male fertility. Furthermore, a 24-nt heterochromatic siRNA locus in the promoter of YUC2 may contribute to epigenetic regulation of auxin homeostasis at ambient temperature21. Overexpression of Arabidopsis YUCCA6 delayed dark-induced and hormone-induced leaf senescence in Arabidopsis22. Overexpression of Arabidopsis YUCCA6 and YUCCA7 enhanced resistance to water deficit23,24. Auxin concentrations were highly elevated under shade condition, and the expression of YUC2, YUC5, YUC8, and YUC9 were up-regulated to enable shade avoidance induced by auxin increases25. Under aluminium stress, YUCCA regulated local auxin biosynthesis in the root apex transition zone, mediating root growth inhibition26.
Systematic identification of YUCCA genes has been performed in only a few plant species, such as Arabidopsis, strawberry, peach, poplar and cucumber2,12,15,27. Apple is a favourite fruit crop with high economic and nutritional value. In this study, we systematically identified 20 YUCCA family genes in apple genome, explored the expression of these MdYUCCAs in different tissues, and measured expression changes in response to high temperature. We also verified an interaction between the MdPIF4 protein and MdYUCCA8a promoter, and analysed the phenotype of MdYUCCA8a overexpression in Arabidopsis.
Results
Identification and classification of apple MdYUCCA genes
Using the Arabidopsis AtYUCCA BLAST homology search and conserved domain analysis, 20 MdYUCCA genes were identified in apple (Table 1). They were named based on their similarity to Arabidopsis YUCCA genes. Deduced amino acid lengths of these MdYUCCAs were between 338 and 563 aa. The pIs were between 8.56 and 9.88, and molecular weights were between 37.08 and 64.48 kD. These MdYUCCA genes were located on nine different chromosomes. Chromosome 10 had six MdYUCCAs. Chromosome 15 had five MdYUCCAs. Chromosome 8 contained two MdYUCCAs. Chromosome 1, 2, 3, 5, 13, and 16 each included one MdYUCCA. MdYUCCA11a, MdYUCCA11b, MdYUCCA11d, and MdYUCCA11e on Chromosome 10 appeared to result from tandem duplication.
Table 1.
Characteristics of the MdYUCCA gene family in apple.
| Gene name | GDDH13_1-1 | NCBI | GDR v3.01 | Location | Strand | Length (aa) | pI | MW |
|---|---|---|---|---|---|---|---|---|
| MdYUCCA2a | MD15G1184800 | XM_008339168.1 | Chr15:14580569..14584004 | − | 437 | 8.8 | 48.73 | |
| MdYUCCA2b | MD01G1067600 | XM_008388239.1 | Chr01:17093902..17096602 | − | 436 | 9.05 | 48.61 | |
| MdYUCCA2c | MD02G1046700 | Chr02:3719433..3722141 | − | 381 | 8.96 | 42.55 | ||
| MdYUCCA3a | MD16G1232600 | XM_008344329.1 | Chr16:23900387..23902351 | + | 426 | 9.08 | 47.69 | |
| MdYUCCA3b | MD13G1227000 | Chr13:22205775..22207631 | + | 338 | 8.86 | 37.08 | ||
| MdYUCCA4a | MD08G1139500 | XM_008368432.1 | Chr08:13470642..13472655 | − | 410 | 9.06 | 45.29 | |
| MdYUCCA4b | MD15G1117200 | XM_008395801.1 | Chr15:8337905..8340244 | − | 411 | 9.19 | 45.35 | |
| MdYUCCA6a | MD15G1098700 | XM_008395672.1 | Chr15:6833888..6838031 | − | 460 | 9.05 | 51.04 | |
| MdYUCCA6b | MD08G1119300 | XM_008379894.1 | Chr08:10869749..10873628 | − | 448 | 9.26 | 49.79 | |
| MdYUCCA8a | MD10G1172800 | XM_008367718.1 | MDP0000262982 | Chr10:26514039..26515625 | − | 423 | 8.72 | 47.42 |
| MdYUCCA8b | MD05G1184800 | Chr05:31306814..31308517 | − | 423 | 8.72 | 47.37 | ||
| MdYUCCA10a | MDP0000138851 | XM_008370693.1 | MDP0000138851 | Chr03:31907169..31927564a | + | 382 | 9.01 | 42.68 |
| MdYUCCA10b | MD15G1133800 | Chr15:9725184..9729059 | − | 563 | 9.88 | 64.48 | ||
| MdYUCCA10c | MD00G1056000 | XM_008370695.1 | MDP0000208234 | Chr00:10506121..10507905 | + | 381 | 8.96 | 42.50 |
| MdYUCCA11a | MD10G1010300 | XM_008366814.1 | MDP0000259265 | Chr10:1431009..1433012 | + | 383 | 8.78 | 43.01 |
| MdYUCCA11b | MD10G1010000 | XM_008380434.1 | MDP0000582079 | Chr10:1378296..1380232 | + | 380 | 8.56 | 41.98 |
| MdYUCCA11c | MD15G1274600 | MDP0000686821 | MDP0000686821 | Chr15:24088780..24090298 | − | 385 | 9.09 | 42.85 |
| MdYUCCA11d | MD10G1010200 | Chr10:1404169..1405726 | + | 365 | 8.96 | 40.67 | ||
| MdYUCCA11e | MD10G1010100 | Chr10:1386074..1387630 | + | 365 | 8.96 | 40.67 | ||
| MdYUCCA11f | MDP0000209681 | XM_008350515.1 | MDP0000209681 | Chr10:956025..957926a | − | 365 | 8.88 | 40.65 |
aChromosome location at the GDR v3.01 genome. The chromosome location information of the other MdYUCCAs was from the GDDH13_1-1 version genome.
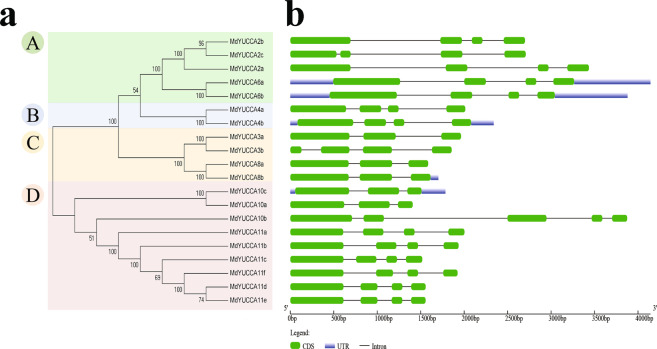
Phylogenetic analysis of MdYUCCA and exon distribution
MdYUCCAs were divided into four subfamilies based on their similarity. MdYUCCA2s and MdYUCCA6s were in the subfamily A. MdYUCCA4s were in the subfamily B. MdYUCCA3s and MdYUCCA8s were in the subfamily C. MdYUCCA10s and MdYUCCA11s were in the subfamily D (Fig. 1a). MdYUCCA10b contained five exons. MdYUCCA3a, MdYUCCA8a, MdYUCCA8b, MdYUCCA10a and MdYUCCA10c contained three exons. The other MdYUCCA genes contained four exons (Fig. 1b).
Figure 1.
Phylogenetic tree and gene structure of MdYUCCA. (a) Phylogenetic analysis of MdYUCCA, constructed with the neighbor-joining method in MEGA 7.0 software. A, B, C, and D represent different gene subfamilies. (b) Structure of MdYUCCA genes. Intron, exon and untranslation region (UTR) are represented by black lines, green boxes and light blue boxes respectively.
Conserved domain analysis revealed that most MdYUCCAs included five conserved motifs: FAD-binding domain, GC-motif, FMO-identifying sequence, NADPH domain, and ATG-containing motif (Fig. 2a). MdYUCCA2c lacked the FMO-identifying sequence and ATG-containing motif. MdYUCCA3b lacked the GC-motif (Fig. 2b).
Figure 2.
Conserved domains in MdYUCCA. (a) Conserved domains of MdYUCCA. (b) Alignment of conserved domains in MdYUCCA.
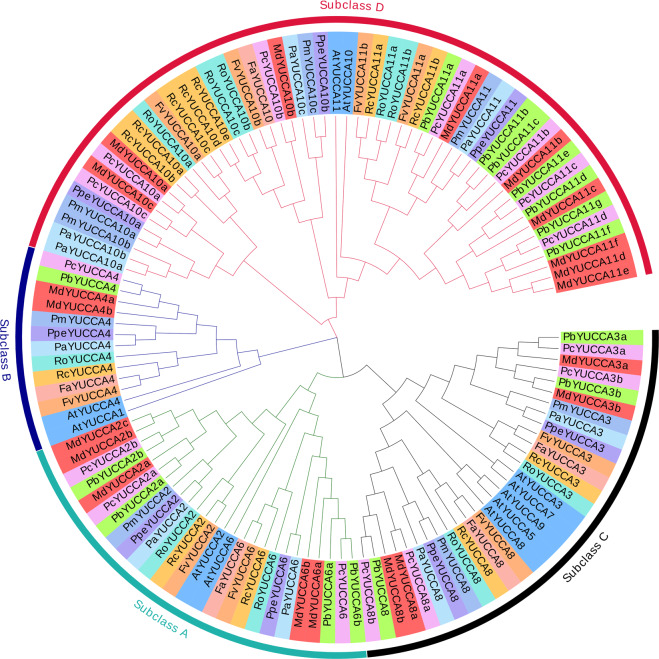
Evolutionary relationship of YUCCAs
To describe the evolutionary relationship of YUCCA genes in Rosaceae, MdYUCCAs were compared with YUCCA genes from Arabidopsis (as reference), peach, European pear, white pear, wood strawberry, strawberry, sweet cherry, rose, and rubus (Fig. 3). Rosaceae YUCCAs were divided into four subclasses based on their similarity. MdYUCCA2s and MdYUCCA6s were placed in subclass A. MdYUCCA4s comprised subclass B. MdYUCCA3s and MdYUCCA8s were included in subclass C. Nine MdYUCCA genes, including MdYUCCA10s and MdYUCCA11s comprised subclass D. There were 20 MdYUCCA genes identified in apple, more than in other studied Rosaceae species. Homologs of Arabidopsis YUCCA1, YUCCA5, YUCCA7, and YUCCA9 were not found in apple and other Rosaceae species. YUCCA10 and YUCCA11 were expanded in some Rosaceae species. For example, there were six and seven YUCCA11 homologs in apple and white pear, respectively.
Figure 3.
Phylogenetic tree of YUCCA from Rosaceae species. At: Arabidopsis thaliana; Md: Malus domestica; Fv: Fragaria vesca; Fa: Fragaria ananassa; Ppe: Prunus persica; Pb: Pyrus bretschneideri; Pc: Pyrus communis; Rc: Rosa chinensis; Pm: Prunus mume; Ro: Rubus occidentalis.
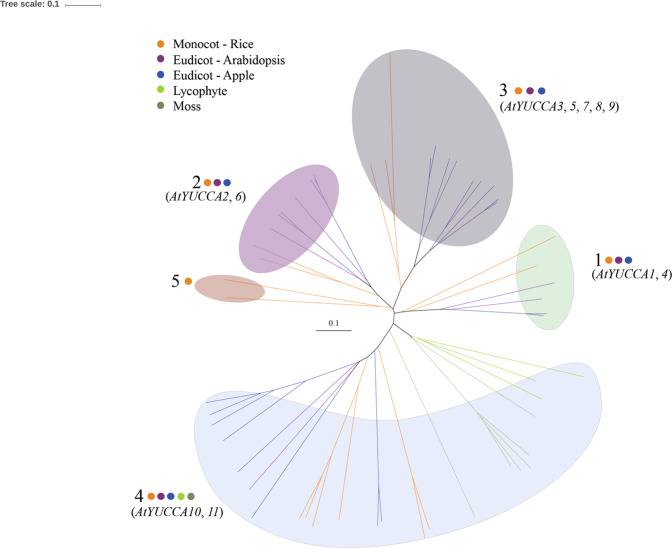
To explore the evolution of YUCCA genes, a phylogenetic tree was constructed with YUCCA genes from six species: a monocot, two eudicots, a lycophyte, a moss, and an algae. As shown in Fig. 4, 55 YUCCA proteins were classified into five subgroups based on clades and topology in the tree. The 20 apple YUCCAs, together with 11 AtYUCCAs and 13 OsYUCCAs, were distributed into subgroup 1, 2, 3, and 4. Two OsYUCCAs were placed in subgroup 5. YUCCAs from lycophyte and moss were only placed in subgroup 4, which included YUCCA10 and YUCCA11. There were no YUCCA homologs detected in the algae Chlamydomonas reinhardtii.
Figure 4.
Phylogenetic relationships of YUCCAs in vascular plants, moss, and algae. The bubbles indicate the five YUCCA subgroups. The dots symbolize the species to which the YUCCA proteins in each group belong (brown: rice [monocot]; purple: Arabidopsis thaliana [eudicot]; cyan-blue: apple [eudicot]; green: Selaginella moellendorffii [lycophyte]; and dark green: Physcomitrella patens [moss]).
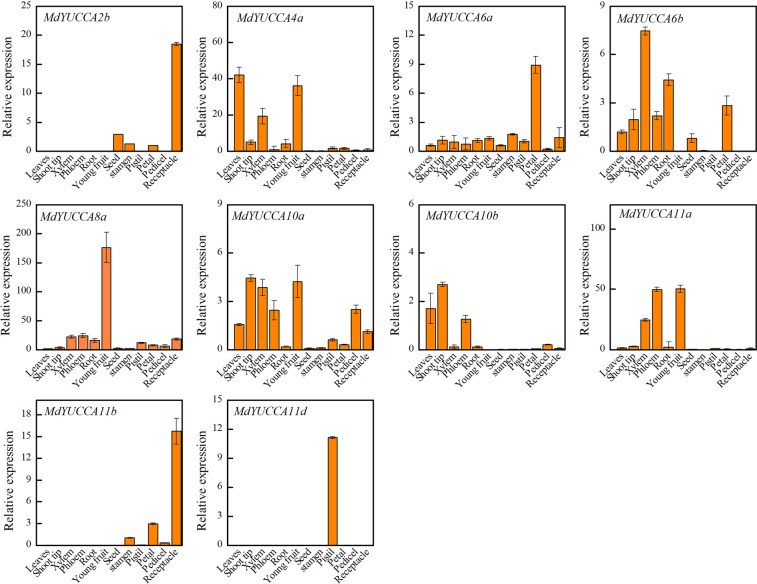
Expression pattern of MdYUCCA genes in different tissues
To explore MdYUCCA expression patterns in different tissues, real-time RT-PCR was used to measure transcript levels in the leaves, shoot tip, xylem, phloem, root, young fruit, seed, stamen, pistil, petal, pedicel, and receptacle of 4-year-old apple trees (Fig. 5). MdYUCCA2b was mainly expressed in the flower organ, with the highest expression in the receptacle. MdYUCCA4a was mainly expressed in the leaves, xylem, and young fruit. MdYUCCA6a was detected in all examined tissues and showed high expression in petal. MdYUCCA6b was highly expressed in xylem, root, and petal. MdYUCCA8a was highly expressed in young fruit. MdYUCCA10a was highly expressed in the shoot tip, young fruit, and xylem. MdYUCCA10b showed high expression in shoot tip. MdYUCCA11a was highly expressed in the phloem and young fruit. MdYUCCA11b showed high expression in receptacle. MdYUCCA11d was only expressed in the pistil.
Figure 5.
Expression of MdYUCCAs in different tissues. Gene expression of MdYUCCAs was measured in leaves, shoot tip, xylem, phloem, root, young fruit, seed, stamen, pistil, petal, pedicel, and receptacle. The MdACTIN gene and MdEF were used as the internal control to normalize the real-time PCR data. Error bars indicate SEs (standard errors) from 3 biological repetitions.
YUCCA enzyme inhibitor Yucasin decreased flower petal width
MdYUCCAs gene expression revealed that some MdYUCCAs were highly expressed in floral organs. Auxin plays important roles in floral organ development2,28. Yucasin is an effective competitive inhibitor of the YUCCA enzyme13. Thus, Yucasin was used to determine if inhibition of YUCCA activity affected flower organ development. The results showed that injecting Yucasin into dormant apple flower buds significantly decreased flower petal width (Fig. 6).
Figure 6.
YUCCA enzyme inhibitor Yucasin decreased flower petal width. (a) Control, the flower buds were injected with the sterile water. (b,c) Yucasin treatment. The flower buds were injected with 300 μL 100 μM Yucasin.
High temperature regulated the expression of MdYUCCAs
High temperature led to auxin-mediated hypocotyl elongation in Arabidopsis thaliana17,18. In apple, high temperature significantly increased MdYUCCA4a, MdYUCCA6a, MdYUCCA8a, and MdYUCCA10a expression, but down-regulated the expression of MdYUCCA2b and MdYUCCA6b. Expression of MdYUCCA8a was markedly induced by high temperature, with expression at 33 °C and 28 °C that was 5.0 and 1.8 times higher than at 23 °C, respectively (Fig. 7).
Figure 7.
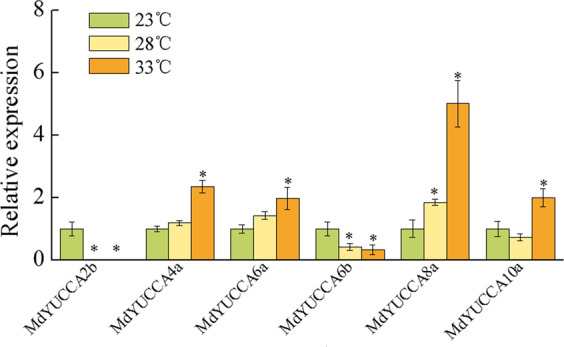
Effect of temperature on the expression of MdYUCCAs. The asterisks indicated statistical significance (p < 0.05) compared with the control (23 °C). Error bars indicate SEs from 3 biological repetitions.
Activity of MdYUCCA8a and MdYUCCA10a promoters in response to hormone treatment
We used a GUS reporter to measure MdYUCCA8a and MdYUCCA10a promoter activity in tobacco leaves under various hormone treatment (Fig. 8). The MdYUCCA8a promoter activity was induced 2.18, 2.36, and 2.00 times over that of mock treatment by gibberellic acid, methyl jasmonate, and brassinolide, respectively. Spermidine (a polyamine) and ethephon (a synthetic compound which decomposes into ethylene) significantly inhibited MdYUCCA8a promoter activity (Fig. 8a). The MdYUCCA10a promoter was induced 1.32 and 1.33 times over that of mock treatment by ABA and spermidine, respectively. Ethephon significantly inhibited MdYUCCA10a promoter activity (Fig. 8b).
Figure 8.
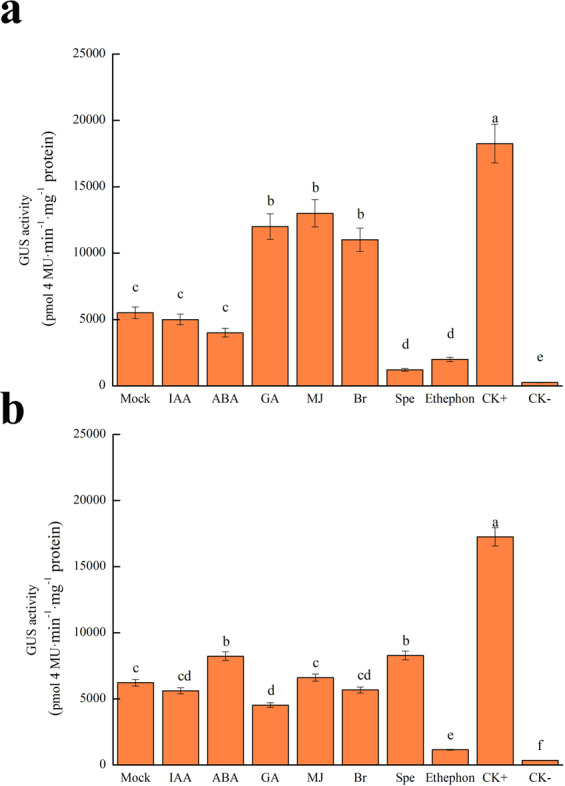
GUS activity of MdYUCCA8a and MdYUCCA10a promoters in response to hormone treatment. (a) GUS activity of transiently transforming the tobacco leaves by MdYUCCA8a promoter. (b) GUS activity of transiently transforming the tobacco leaves by MdYUCCA10a promoter. Mock: inoculation of MdYUCCApro-pCAMBIA1381-GUS bacterium solution and then spraying water, IAA: indole-3-acetic acids; ABA: abscisic acid; GA: gibberellin 3; MJ: methyl jasmonate; Br: brassinolide; Spe: spermidine; CK + : positive control, inoculation of 35S-pCAMBIA1381-GUS bacterium solution and then spraying water; CK−: negative control, inoculation of pCAMBIA1381-GUS bacterium solution and then spraying water. Error bars indicate SEs from three biological replications. Statistically significant differences are indicated by different lower case letters (p < 0.05, one-way ANOVA).
Interaction of transcription factor MdPIF4 with the MdYUCCA8a promoter
PIF4 binds to the YUCCA promoter and promotes YUCCA expression and auxin synthesis18. The dual luciferase transient expression system was used to determine if there was an interaction between MdPIF4 and the MdYUCCA8a promoter in tobacco leaves. A 2.8-fold enhancement of activity in the dual-luciferase assay indicated that MdPIF4 can trans-activate the MdYUCCA8a promoter (Fig. 9). Although the MdYUCCA10a promoter also contained G-box elements, we did not observe MdPIF4 activation of the MdYUCCA10a promoter.
Figure 9.
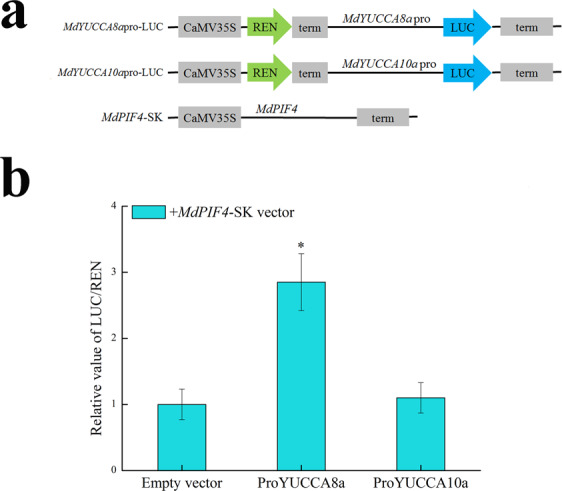
Dual luciferase transient expression assay to probe interaction of transcription factor MdPIF4 and MdYUCCA8a promoter in tobacco leaves. (a) Vector construction diagram. (b) The activity of firefly and renilla luciferase in tobacco leaves was measured 3 d after infiltration. Error bars show SEs of three independent experiments with at least four replicate reactions. The asterisks indicate statistical significance (p < 0.05).
The phenotype of plants overexpressing MdYUCCA8a
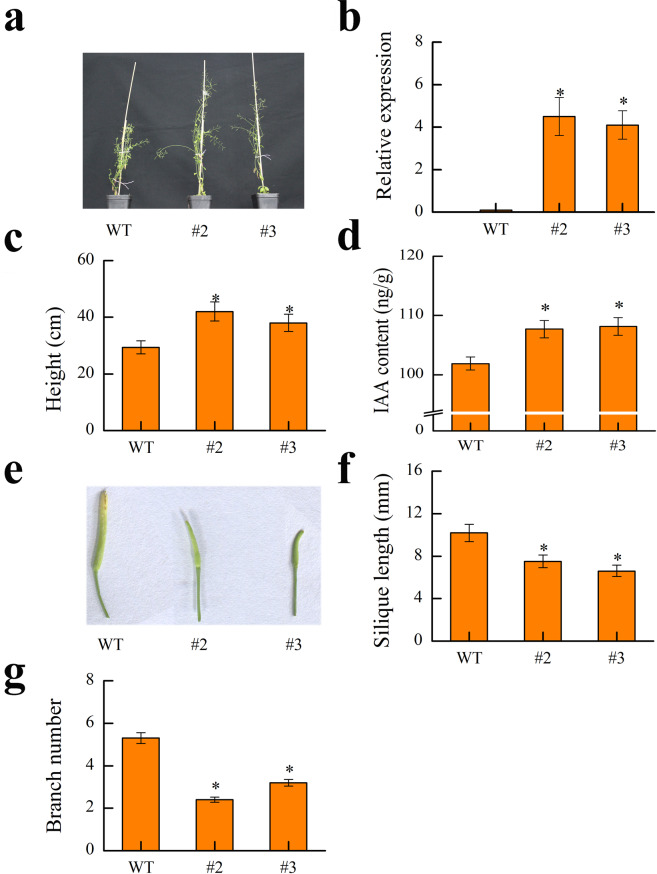
MdYUCCA8a was ectopically expressed in wild type Columbia ecotype Arabidopsis thaliana with a constitutive 35S strong promoter. Eight independent T3 generation transgenic lines were obtained, and line #2 and #3 were selected for phenotypic analysis (Fig. 10). Overexpression of MdYUCCA8a increased stem height, as the height of line #2 and #3 were 42% and 29% higher than that of wild type, respectively (Fig. 10a,c). The transgenic lines showed high IAA content (Fig. 10d). Overexpression of MdYUCCA8a led to silique malformation and the silique length was significantly reduced (Fig. 10e,f). Apical dominance significantly increased in the transgenic lines, as the number of lateral shoots in overexpression lines was significantly lower than in wild-type (Fig. 10g).
Figure 10.
Phenotype of MdYUCCA8a overexpression in Arabidopsis thaliana. (a) Photos of wild type (WT) and overexpressing MdYUCCA8a gene (#2 and #3) Arabidopsis thaliana. (b) qRT-PCR analysis of MdYUCCA8a expression in WT and transgenic lines. Arabidopsis thaliana AtACTIN2 was used as the reference gene. (c) Plant height. (d) IAA content. (e) Photos of siliques. (f) Siliques length. (g) Branch number. The asterisks indicate statistical significance (p < 0.05). Error bars indicate SEs from three replications.
Discussion
YUCCA is a rate-limiting enzyme in the indole-3-pyruvate (IPA) pathway and converts IPA into IAA. In this study, we identified 20 MdYUCCA genes in apple. Phylogenetic analysis shown MdYUCCA genes could be divided into four subfamilies. Homologs of Arabidopsis YUCCA1, YUCCA5, YUCCA7, and YUCCA9 were not found in Rosaceae species and cucumber15. These YUCCA genes may have been lost via chromosomal fragment deletion. YUCCA11 was expanded in apple and white pear, which contained six and seven YUCCA11 members, respectively. There has been a tandem duplication of MdYUCCA11s in Chromosome 10.
Auxin biosynthesis occurs both at local auxin response sites and at distant sites as a source for polar auxin transport. Auxin functions may be dependent on the location of auxin production. Plants use different YUCCA genes for auxin biosynthesis in different organs. In apple, MdYUCCA6a, MdYUCCA8a, and MdYUCCA10a were expressed in most organs and may be involved in whole plant basal auxin synthesis. MdYUCCA4a, MdYUCCA10b, and MdYUCCA11a were the main YUCCA genes for auxin biosynthesis in vegetative organs. MdYUCCA2b, MdYUCCA11b, and MdYUCCA11d may be responsible for auxin synthesis in flower organs. In other plants, the expression of certain YUCCA in specific tissues has indicated involvement in the development of specific organs. Peach PpYUCCA11 transcripts specifically accumulated during the late ripening stage in melting flesh peaches, suggesting involvement in fruit softening12. In cucumber, CsYUCCA11 uniquely accumulated in the opened male flower15. In apple, MdYUCCA11d appeared to be specifically expressed in the pistil, suggesting that it may be responsible for auxin synthesis there. YUCCA11 seem to have specific roles in flower and fruit development.
High temperature has been shown to induce elevated levels of endogenous free indole-3-acetic acid (IAA). In this study, high temperature induced the expression of MdYUCCA4a, MdYUCCA6a, MdYUCCA8a, and MdYUCCA10a. Dual-luciferase assay results showed that MdPIF4 could bind to G-box CACGTG DNA fragments in the promoter of MdYUCCA8a, but not MdYUCCA10a. This is in accordance with previous results in Arabidopsis that showed PIF4 directly binds to the YUCCA8 promoter, but not to other YUCCA promoters, with preferential binding of the CACGTG G-box motif18,29. PIF4-activated expression of YUCCA8 may be a conserved molecular mechanism that elevates endogenous IAA level under high temperature conditions. High temperature may regulate other YUCCA genes through indirect or PIF4-independent pathways. High temperature down-regulated the expression of MdYUCCA2b and MdYUCCA6b. In barley and Arabidopsis, high temperature conditions caused male sterility by repressing YUCCA2 and YUCCA6 expression, which decreased auxin levels in the developing anthers20. Thus, high temperature has varying up-regulation and down-regulation effects on different YUCCA genes in the different biological process.
Overexpression of MdYUCCA8a in Arabidopsis increased the free auxin level and produced expected auxin overproduction phenotypes, like taller plant height and enhanced apical dominance, but also induced silique malformation. In previous studies, YUCCA transgenic lines also showed reproductive organ defects. Overexpression of strawberry FaYUC1, wheat TaYUC10, and cucumber CsYUC11 in Arabidopsis resulted in defective pollen and sterility15,30,31. Overexpression of FvYUC6 in woodland strawberry also delayed flowering and led to severe male sterility32. Repression of YUCCA2 and YUCCA6 expression by high temperature stress decreased auxin content and caused male sterility in barley and Arabidopsis20. Hence, appropriate auxin content is necessary for reproductive organ development.
In conclusion, we have systematically identified 20 MdYUCCAs in apple. Gene expression results suggested that MdYUCCA6a, MdYUCCA8a, and MdYUCCA10a were expressed in most organs and could support whole plant basal auxin synthesis. MdYUCCA4a, MdYUCCA10b, and MdYUCCA11a were the main YUCCA genes for auxin biosynthesis in vegetative organs. MdYUCCA2b, MdYUCCA11b, and MdYUCCA11d could be mainly responsible for auxin synthesis in flower organs. High temperature induced expression of MdYUCCA4a, MdYUCCA6a, MdYUCCA8a, and MdYUCCA10a and down-regulated expression of MdYUCCA2b and MdYUCCA6b. MdPIF4 interacted with the MdYUCCA8a promoter. Overexpression of MdYUCCA8a increased IAA content, increased stem height, enhanced apical dominance, and led to silique malformation.
Methods
Identification of MdYUCCA genes in the apple genome
Arabidopsis AtYUCCA protein sequences retrieved from TAIR (The Arabidopsis Information Resource: http://www.arabidopsis.org/) were separately used as blastp queries against apple genome GDR v3.01 (downloaded from the Genome Database for Rosaceae, http://www.rosaceae.org), the protein sequence of Malus domestica from NCBI (https://www.ncbi.nlm.nih.gov), and the apple genome GDDH13_1-1 (https://iris.angers.inra.fr/gddh13/the-apple-genome-downloads.html) using a stand-alone version of BLAST (Basic Local Alignment Search Tool: http://blast.ncbi.nlm.nih.gov)33. Similar sequences with e-values <0.0001 were further inspected for FMO-like and NAD binding domains using the domain analysis programs Pfam (Protein family: http://pfam.sanger.ac.uk/)34 and SMART (Simple Modular Architecture Research Tool: http://smart.embl-heidelberg.de/)35 with default cutoff parameters. After combining the predicted proteins and eliminating redundancy, the remaining peptide sequences were considered MdYUCCA candidates. Redundant MdYUCCA genes isolated from different apple reference genomes were identified by multiple sequence alignment and from their genome location. ExPASY (http://www.expasy.org/tools/) was used to predict the isoelectric point (pI) and molecular weight (MW) of each MdYUCCA. Exon/intron structure was determined with alignments of coding and genomic sequences and depicted with the online tool GSDS (http://gsds.cbi.pku.edu.cn/).
Multiple sequence alignment, phylogenetic analysis, and classification of apple MdYUCCA genes
To establish the evolutionary relationship of MdYUCCAs to other Rosaceae YUCCA proteins, MdYUCCA protein sequences were compared with YUCCA sequences from Arabidopsis thaliana (as a reference), peach (Prunus persica L. Batsch), European pear (Pyrus communis), white pear (Pyrus bretschneideri Rehd.), woodland strawberry (Fragaria vesca), strawberry (Fragaria ananassa), sweet cherry (Prunus avium), rosa (Rosa chinensis), black raspberry (Rubus occidentalis), and mei (Prunus Mume).
To investigate YUCCA evolution in species ranging from algae to land plants, a phylogenetic tree was constructed with YUCCA proteins from six species (monocot-Oryza sativa, eudicots-apple and Arabidopsis, lycophyte-Selaginella moellendorffii, moss-Physcomitrella patens, and algae-Chlamydomonas reinhardtii). The phylogenetic tree was constructed using MEGA 7.0 with the neighbor-joining (NJ) method, 1000 bootstrap replicates, and complete delete parameters.
Plant materials and treatment
Leaves, shoot tip, xylem, phloem, root, young fruit, seed, stamen, pistil, petal, pedicel, and receptacle were sampled from 4-year-old Fuji apple trees. Trees were located in experimental plots of the Yangling Subsidiary Center at the National Apple Improvement Center, Yangling, China (34.31°N, 108.04°E), and were trained and managed with standard horticultural practices.
For the temperature treatment, 1-year-old apple trees were grown in the growth chambers for 4 weeks under 12 h/12 h (light/dark) daily cycle at 22 °C, 28 °C, or 33 °C during the daytime, and at 18 °C at night. Shoot tips were harvested for RNA extraction and gene expression analysis.
For YUCCA inhibition experiments, 300 μL of 100 μM Yucasin, a YUCCA enzyme inhibitor (cat. 573760, Sigma, USA), were injected into dormant Fuji flower buds in mid-November with an injection syringe. The control was injected with sterile water. Apple flower phenotype was assessed at the full-bloom stage of the second year.
RNA extraction, cDNA synthesis, and quantitative RT-PCR gene expression analysis
Total RNA was extracted from frozen samples according to the cetyltrimethylammonium bromide (CTAB) method36. The first-strand cDNA was synthesized with 1 µg of total RNA in a volume of 20 µL using reverse transcriptase. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was used to analyze the gene expression. PCR amplification conditions was as follows: 95 °C for 5 min for initial denaturation, then 45 cycles of 94 °C for 20 s, 60 °C for 20 s (determined by the primer), and 72 °C for 10 s. Fluorescence was measured at the end of each cycle. Primer sequences for the quantification of transcripts by RT-PCR are listed in Supplemental Table S1. MdActin and MdEF were used as internal references for apple gene expression. Gene relative expression was calculated using the comparative 2−∆∆CT method37.
The dual-luciferase assay
The promoter sequences of MdYUCCA8a and MdYUCCA10a were ligated into pGreenII_0800-5_LUC vector. The CDS sequence of transcription factor MdPIF4 was ligated into pGreenII_0029_62-SK vector. The recombinant plasmids were transformed into Agrobacterium GV3101. Dual-luciferase transient expression in tobacco leaves was performed in accordance with Hellens’ method38. Primer information is listed in Supplemental Table S1.
Promoter activity assay
About 2000 bp promoter fragments from MdYUCCA8a and MdYUCCA10a were each cloned from Fuji genomic DNA. The promoter fragments were ligated into the pCAMBIA1381-GUS plant transformation vector with a ClonExpress II kit (Vazyme Biotech Co., Ltd, China). Primer information is listed in Supplemental Table S1. The recombinant plasmids were transformed into Agrobacterium strain LBA4404 and then transformed into leaf epidermal cells of 4-week-old tobacco (Nicotiana Benthamiana) plants by syringe infiltration. Next, transfected leaves were sprayed with different hormones respectively, including 100 μM indole-3-acetic acids, 100 μM ABA, 100 μΜ GA3, 100 μΜ methyl jasmonate, 0.416 μΜ brassinolide, 100 μΜ spermidine, 3.46 mM ethephon. The GUS activity assay was performed as described by Jefferson et al.39.
Construction of MdYUCCA8a overexpression vector and Arabidopsis transformation
The full CDS of MdYUCCA8a was cloned from Fuji apple and ligated into pCAMBIA1301 vector with a ClonExpress II kit (Vazyme Biotech Co., Ltd, China). Primer information is listed in Supplemental Table S1. Recombinant plasmids were transformed into Agrobacterium strain LBA4404. MdYUCCA8a was introduced into the wild type (WT) Columbia ecotype Arabidopsis (Col-0) using the floral dip method40. Seeds from positive transgenic plants were harvested individually. T3 generation homozygous transgenic lines were used for further investigation. Four weeks old rosette leaves were used to measure IAA and for RNA extraction and gene expression analysis. AtActin2 gene was used as internal references. The IAA was extracted using solid-phase extraction methods, quantifed by the High Pressure Liquid Chromatography (Waters, USA) and enzyme-linked immunosorbent assay41–43.
Supplementary information
Acknowledgements
This research was supported by the Science and Technology Innovative Engineering Project in Shaanxi Province of China (2016KTZDNY01-04), National Apple Industry Technology System of Agriculture Ministry of China (CARS-27), Youth Program of National Natural Science Foundation of China (31801844), and Yan’an Virus-Free Apple Nursery Technology Innovation and Extention Demonstration (2019TSLNY02-04). We thank Liwen Bianji (www.liwenbianji.cn) for the English text of a draft of this manuscript.
Author contributions
C.S., D.Z., N.A., G.L., and M.H. participate in the experimental design. C.S., L.Z., Y.S., X.Z., Ji.M., Y.M., H.W., Y.Z., X.L., M.Q., J.Z., C.Z., L.X., and Ju.M. participate in material sampling, field measurements and the laboratory data measurement. C.S., D.Z., N.A., and M.H. participate in the paper writing and discussion. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chunhui Song and Dong Zhang.
Supplementary information
is available for this paper at 10.1038/s41598-020-66483-y.
References
- 1.Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng, Y., Dai, X. & Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes & Development20, 1790–1799, 10.1101/gad.1415106 (2006). [DOI] [PMC free article] [PubMed]
- 3.Hofmann NR. YUC and TAA1/TAR proteins function in the same pathway for auxin biosynthesis. Plant Cell. 2011;23:3869. doi: 10.1105/tpc.111.231112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stepanova AN, et al. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 2011;23:3961–3973. doi: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cha J, et al. A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nature communications. 2015;6:8041–8041. doi: 10.1038/ncomms9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao X, et al. Auxin production in diploid microsporocytes is necessary and sufficient for early stages of pollen development. Plos Genetics. 2018;14:e1007397. doi: 10.1371/journal.pgen.1007397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Q, et al. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant and Cell Physiology. 2014;55:1072–1079. doi: 10.1093/pcp/pcu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallavotti A, et al. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15196–15201. doi: 10.1073/pnas.0805596105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo Y, et al. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Molecular Biology. 2007;65:125–136. doi: 10.1007/s11103-007-9203-6. [DOI] [PubMed] [Google Scholar]
- 10.Tobenasantamaria R, et al. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes & Development. 2002;16:753–763. doi: 10.1101/gad.219502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishida JK, et al. Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates haustorium development in the parasitic plant Phtheirospermum japonicum. Plant Cell. 2016;28:1795–1814. doi: 10.1105/tpc.16.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan L, et al. PpYUC11, a strong candidate gene for the stony hard phenotype in peach (Prunus persica L. Batsch), participates in IAA biosynthesis during fruit ripening. Journal of Experimental Botany. 2015;66:7031–7044. doi: 10.1093/jxb/erv400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimura T, et al. Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant Journal. 2014;77:352–366. doi: 10.1111/tpj.12399. [DOI] [PubMed] [Google Scholar]
- 14.Kim JI, et al. yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiology. 2007;145:722–735. doi: 10.1104/pp.107.104935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan S, et al. Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Scientific Reports. 2016;6:20760. doi: 10.1038/srep20760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koini MA, et al. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Current Biology. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 18.Sun, J., Qi, L., Li, Y., Chu, J. & Li, C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLOS Genetics8 (2012). [DOI] [PMC free article] [PubMed]
- 19.Lee, H. et al. FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nature communications5, 10.1038/ncomms6473 (2014). [DOI] [PubMed]
- 20.Sakata T, et al. Auxins reverse plant male sterility caused by high temperatures. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8569–8574. doi: 10.1073/pnas.1000869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gyula, P. et al. Ambient temperature regulates the expression of a small set of sRNAs influencing plant development through NF-YA2 and YUC2. Plant, cell & environment, 10.1111/pce.13355 (2018). [DOI] [PubMed]
- 22.Kim JI, et al. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. Journal of Experimental Botany. 2011;62:3981–3992. doi: 10.1093/jxb/err094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JI, et al. Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Molecular Plant. 2013;6:337–349. doi: 10.1093/mp/sss100. [DOI] [PubMed] [Google Scholar]
- 24.Lee M, et al. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta. 2012;235:923–938. doi: 10.1007/s00425-011-1552-3. [DOI] [PubMed] [Google Scholar]
- 25.Mullermoule, P. et al. YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance. PeerJ4 (2016). [DOI] [PMC free article] [PubMed]
- 26.Liu, G. et al. Local transcriptional control of YUCCA regulates auxin promoted root-growth inhibition in response to aluminium stress in Arabidopsis. Plos Genetics12, e1006360, 10.1371/journal.pgen.1006360 (2016). [DOI] [PMC free article] [PubMed]
- 27.Ye X, Kang B, Osburn LD, Li Y, Zong-Ming C. Identification of the flavin-dependent monooxygenase-encoding YUCCA gene family in Populus trichocarpa and their expression in vegetative tissues and in response to hormone and environmental stresses. Plant Cell Tissue and Organ Culture. 2009;97:271–283. doi: 10.1007/s11240-009-9526-x. [DOI] [Google Scholar]
- 28.Cheng Y, Zhao Y. A role for auxin in flower development. Journal of Integrative Plant Biology. 2007;49:99–104. doi: 10.1111/j.1744-7909.2006.00412.x. [DOI] [Google Scholar]
- 29.Oh E, Zhu J, Wang Z. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nature Cell Biology. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li N, et al. Isolation and characterization of three TaYUC10 genes from wheat. Gene. 2014;546:187–194. doi: 10.1016/j.gene.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 31.Liu H, et al. Isolation and characterization of two YUCCA flavin monooxygenase genes from cultivated strawberry (Fragaria x ananassa Duch.) Plant Cell Reports. 2012;31:1425–1435. doi: 10.1007/s00299-012-1258-4. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, et al. Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. Journal of Integrative Plant Biology. 2014;56:350–363. doi: 10.1111/jipb.12150. [DOI] [PubMed] [Google Scholar]
- 33.Mount, D.W. Using the Basic Local Alignment Search Tool (BLAST). CSH Protoc (2007). [DOI] [PubMed]
- 34.Finn RD, et al. Pfam: the protein families database. Nucleic acids research. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic acids research. 2012;40:D302–305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang SJ, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- 37.Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods25, 402–408, 10.1006/meth.2001.1262 (2001). [DOI] [PubMed]
- 38.Hellens RP, et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jefferson RA. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Molecular Biology Reporter. 1987;5:387–405. doi: 10.1007/bf02667740. [DOI] [Google Scholar]
- 40.Zhang X, Henriques R, Lin S, Niu Q, Chua N. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocols. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 41.Dobrev PI, Havlicek L, Vagner M, Malbeck J, Kaminek M. Purification and determination of plant hormones auxin and abscisic acid using solid phase extraction and two-dimensional high performance liquid chromatography. Journal of Chromatography A. 2005;1075:159–166. doi: 10.1016/j.chroma.2005.02.091. [DOI] [PubMed] [Google Scholar]
- 42.Dobrev PI, Kaminek M. Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. Journal of Chromatography A. 2002;950:21–29. doi: 10.1016/S0021-9673(02)00024-9. [DOI] [PubMed] [Google Scholar]
- 43.Maldiney R, et al. A biotin-avidin-based enzyme immunoassay to quantify three phytohormones: auxin, abscisic acid and zeatin-riboside. Journal of Immunological Methods. 1986;90:151–158. doi: 10.1016/0022-1759(86)90070-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.