Short abstract
The National COVID-19 Chest Imaging Database (NCCID) is a repository of chest radiographs, CT and MRI images and clinical data from COVID-19 patients across the UK, to support research and development of AI technology and give insight into COVID-19 disease https://bit.ly/3eQeuha
Introduction
Since the emergence of the novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), in Wuhan, China in 2019 [1], the resulting coronavirus disease 2019 (COVID-19) disease has rapidly transitioned into a global pandemic with over 31 000 deaths and 215 000 infections in the UK as of 10 May 2020 [2]. Confirmation of SARS-CoV-2 infection requires reverse transcriptase PCR (RT-PCR) testing [3]. Chest radiographic and/or computed tomography (CT) imaging is also central to diagnosis and management [4].
The scale of the COVID-19 pandemic has resulted in the acquisition of huge volumes of imaging data. Traditionally, research using imaging data constituted collation of data within single hospitals or groups of hospitals at most. Endeavours on a local scale have the constraint that not all patient subgroups or disease manifestations might be captured in the collected data. It has long been recognised that there is an acute need to curate larger, more comprehensive datasets to better understand a disease. COVID-19 has arrived in an era where advances in computational power, aligned with an increased availability of big data and the development of self-learning neural networks, have begun to redefine research in medicine. In recent years, computer algorithms trained on imaging data, widely available on the internet, have been adapted to the task of medical image analysis [5, 6]. For computer algorithms to be successfully applied to medical image analysis, it is imperative that they train on large volumes and representative examples of imaging data. These are typically orders of magnitude larger than traditional imaging research datasets, and beyond the capacity of traditional research e-infrastructure.
The National Health Service (NHS) in the UK has long sought national repositories of linked clinical and imaging data, which are essential for applications of artificial intelligence (AI) computing systems in healthcare. Historically, logistical barriers to this have seemed insurmountable and progress has been notoriously slow. Yet, one aspect of the COVID-19 response has been the issue of a notice under Regulation 3(4) of the Health Service Control of Patient Information Regulations 2002 in the UK, which has temporarily eased data sharing restrictions to facilitate COVID-19-specific public health research and scientific collaboration over the course of the emergency.
The British Society of Thoracic Imaging research network began a multicentred COVID-19 imaging study [7], which grew into a partnership with NHSX to create the National COVID-19 Chest Imaging Database (NCCID; https://nhsx.github.io/covid-chest-imaging-database). NCCID has put in place mechanisms to collate all chest imaging and pre-specified clinical data from every UK hospital where patients undergo a RT-PCR test for COVID-19. This will include all RT-PCR-positive patients and a representative sample of RT-PCR-negative patients. The study aims to identify information in COVID-19 imaging that may be inconspicuous to the human eye, but which is extractable by computer algorithms. Such buried information may allow the early identification of patients at risk of deterioration, thereby anticipating future intensive care needs. AI algorithms may also demonstrate how comprehensive characterisation of COVID-19 may improve care/outcomes.
NCCID data and image transfer solutions are robust and secure, including those adapted from techniques tried and tested on numerous research studies involving large-scale medical image collection [8]. To maximise efficient resource utilisation in busy hospitals during the course of the pandemic, we are linking our imaging data to the International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC) UK Clinical Characterisation Protocol (https://isaric.tghn.org/UK-CCP), and aim to link to the Intensive Care National Audit and Research Centre (ICNARC; www.icnarc.org). ISARIC investigators are collating clinical information and biological samples for COVID-19 cases of all ages admitted to hospitals, while ICNARC collates detailed data from adults in the intensive care setting. The study has also been supported by Health Data Research UK as part of its UK response to COVID-19.
An endeavour of this scale, rarely attempted before in the NHS, can only succeed if the radiology and scientific community contribute their time, effort and available data. ISARIC has open source processes and already has established data and material sharing from UK COVID-19 cases [9]. The NCCID initiative will create an open, well-governed database for researchers from academia and industry to add to COVID-19 knowledge collectively. Public, patient and professional trust is vital to NCCID and COVID-19 in general, and we consider accessibility and feedback on data uses as central to good governance. The NCCID data access committee aim to link to researchers who are asking related scientific questions, using complementary methodologies. NCCID will also set aside a portion of its data to enable validation of AI models. This will maintain the highest standards of governance throughout the emergency and allow technology developers to validate their algorithms promptly.
NCCID data
Imaging data
Chest radiographs
The UK prioritised the use of chest radiographs in the clinical work-up of patients suspected of COVID-19 [10]. The choice of chest radiographs as the primary diagnostic imaging test was pragmatic, given the reported limited sensitivity of CT imaging in COVID-19 diagnosis [11]. The choice also reflected concerns regarding seeding of infection via CT scanners contaminated with virus particles, as well as the limited numbers of CT scanners serving the UK population when compared with other European countries [12]. Accordingly, a large proportion of imaging in NCCID will consist of chest radiographs acquired at initial presentation of the patient to hospital and throughout the patient's hospital stay (figure 1). NCCID will collect chest radiographs from all RT-PCR-positive COVID-19 patients in hospitals throughout the UK. In addition, a number of chest radiographs from RT-PCR-negative patients will be collected from sites as a representative control population.
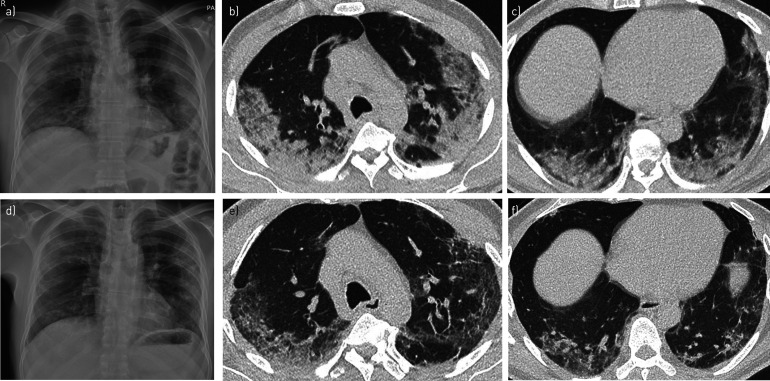
FIGURE 1.
Imaging from a 61-year-old man presenting with a 7-day history of feeling unwell and increasing breathlessness. a) The chest radiograph on admission demonstrated patchy peripheral consolidation in the middle and lower zones. Axial CT images of the b) upper and c) lower lung zones on day 6 of admission confirmed multifocal peripheral consolidation, in keeping with the diagnosis of COVID-19. d) A repeat chest radiograph taken 3.5 weeks later showed marked resolution of the consolidation, confirmed on repeat CT imaging of e) upper and f) lower lung zones, although a degree of architectural distortion was then evident within the lungs. Artificial intelligence tools, when applied to imaging and corresponding clinical data at scale, can allow the quantitation of patterns of disease progression in COVID-19. Unsupervised deep learning methods could help identify discrete COVID-19 patient phenotypes that manifest differing disease courses. Image analysis may also allow prediction of those patients that are likely to manifest chronic multisystem sequelae following their acute infection, which may inform future resource prioritisation in healthcare systems.
CT chest imaging
NCCID will collect all chest CT imaging in RT-PCR-positive COVID-19 patients. This will include non-contrast-enhanced chest CT imaging, CT pulmonary angiograms and CT coronary angiograms.
Previous chest imaging
For all RT-PCR-positive COVID-19 patients, NCCID will acquire all chest imaging performed in the previous 3 years. For RT-PCR-negative patients, NCCID will acquire any chest imaging performed in the previous 4 weeks.
Clinical data
Clinical data collected from RT-PCR-positive COVID-19 patients will include demographic information, patient comorbidity, smoking and medication histories, clinical observations, admission blood test results, and outcomes including time of intensive care unit admission and survival. The imaging will link to other clinical databases such as ISARIC to allow analyses against more detailed clinical information.
Data access
We anticipate that, in May 2020, NCCID data will be released to interested academic and commercial groups. Given that NCCID will be of particular interest to AI researchers, a portion of the data will be segregated to allow rigorous and independent validation of AI models. Data access will be initiated through online applications, to be filled in and submitted according to the instructions on the NCCID website. Applications will then be assessed by a central data access committee comprising scientific advisors, technology advisors, information-governance advisors, patient/ethics advisors and system advisors, to evaluate the positive impact on the NHS overall. Successful applicants can access the data on a cloud-based Amazon S3 bucket, and transfer it onto their computing infrastructure, which are required to fulfil high standards of IT security.
Discussion
In these challenging times, research efforts necessary to better understand COVID-19 cannot afford to be fragmented and uncoordinated. Accelerating insights and discovery necessitates leveraging economies of scale and resource amongst researchers, institutions and companies. We believe NCCID will quickly improve COVID-19 understanding and patient care and build on early insights into COVID-19 [13–15].
The scale of the current pandemic requires the best minds, data, algorithms and research programmes to come together quickly. We are fortunate in the UK to have an abundance of internationally recognised researchers working on COVID-19. We hope that NCCID provides a common UK resource to fuel international efforts in tackling COVID-19 and allows better preparedness for similar future needs.
Shareable PDF
Supplementary Material
Footnotes
Conflict of interest: J. Jacob reports fees from Boehringer Ingelheim and Roche unrelated to the current submission and is supported by a Clinical Research Career Development Fellowship 209553/Z/17/Z from the Wellcome Trust, and by the NIHR Biomedical Research Centre at University College London.
Conflict of interest: D. Alexander has nothing to disclose.
Conflict of interest: J.K. Baillie reports grants from DHSC National Institute of Health Research UK, Medical Research Council UK, Wellcome Trust, Fiona Elizabeth Agnew Trust, Intensive Care Society and Chief Scientist Office, during the conduct of the study.
Conflict of interest: R. Berka is an Associate at Faculty; Faculty has a paid partnership with NHSX to build its new AI lab to help drive digital transformation and the use of AI in the NHS. This contract has been extended for setting up the platform environment that manages the data storage for the National COVID-19 Chest Imaging Database (NCCID); the platform is being made available by Faculty at zero licence cost. In addition, Faculty is contracted to support NHS England and Improvement as well as NHSX with its data response strategy to COVID-19, which includes developing dashboards, models and simulations to provide information to central government decision-makers.
Conflict of interest: O. Bertolli is an Associate at Faculty; Faculty has a paid partnership with NHSX to build its new AI lab to help drive digital transformation and the use of AI in the NHS. This contract has been extended for setting up the platform environment that manages the data storage for the National COVID-19 Chest Imaging Database (NCCID); the platform is being made available by Faculty at zero licence cost. In addition, Faculty is contracted to support NHS England and Improvement as well as NHSX with its data response strategy to COVID-19, which includes developing dashboards, models and simulations to provide information to central government decision-makers.
Conflict of interest: J. Blackwood has nothing to disclose.
Conflict of interest: I. Buchan reports personal fees from and acting as advisor for AstraZeneca, grants from NIHR, and is a former employee of Microsoft, outside the submitted work.
Conflict of interest: C. Bloomfield reports that NCIMI is funded through support from the Industry Strategy Challenge Fund, by Innovate UK grant 104688.
Conflict of interest: D. Cushnan has nothing to disclose.
Conflict of interest: A. Docherty has nothing to disclose.
Conflict of interest: A. Edey has nothing to disclose.
Conflict of interest: A. Favaro is a Lead Data Scientist at Faculty; Faculty has a paid partnership with NHSX to build its new AI lab to help drive digital transformation and the use of AI in the NHS. This contract has been extended for setting up the platform environment that manages the data storage for the National COVID-19 Chest Imaging Database (NCCID); the platform is being made available by Faculty at zero licence cost. In addition, Faculty is contracted to support NHS England and Improvement as well as NHSX with its data response strategy to COVID-19, which includes developing dashboards, models and simulations to provide information to central government decision-makers.
Conflict of interest: F. Gleeson has nothing to disclose.
Conflict of interest: M. Halling-Brown is an advisor to Google Health on the MAMMOTH project, and has a Visiting Professorship with University of Surrey as part of a research collaboration with Transpara.
Conflict of interest: S. Hare has nothing to disclose.
Conflict of interest: E. Jefferson reports grants from Medical Research Council (MRC), Health Data Research (HDR) UK, National Institute of Health Research (NIHR), Chief Scientist Office (CSO), Engineering and Physical Sciences Research Council (EPSRC), Health Foundation, Data Lab, Scottish Government, NHS Fife Health Board and EU Horizon 2020, during the conduct of the study.
Conflict of interest: A. Johnstone has nothing to disclose.
Conflict of interest: M. Kirby is a Principal at Faculty; Faculty has a paid partnership with NHSX to build its new AI lab to help drive digital transformation and the use of AI in the NHS. This contract has been extended for setting up the platform environment that manages the data storage for the National COVID-19 Chest Imaging Database (NCCID); the platform is being made available by Faculty at zero licence cost. In addition, Faculty is contracted to support NHS England and Improvement as well as NHSX with its data response strategy to COVID-19, which includes developing dashboards, models and simulations to provide information to central government decision-makers.
Conflict of interest: R. McStay has nothing to disclose.
Conflict of interest: A. Nair reports salary reimbursement from Biomedical Research Centre UCL, and is medical advisor to Aidence BV, outside the submitted work.
Conflict of interest: P.J.M. Openshaw reports personal fees for consultancy work with Janssen, J and J and Sanofi; grants from MRC, European Union, NIHR Biomedical Research Centre and Wellcome Trust, collaborative grants with GSK, personal fees from the European Respiratory Society and an NIHR Senior Investigator Award outside the submitted work; in addition, P.J.M. Openshaw was President of the British Society for Immunology; this was an unpaid appointment but travel and accommodation at some meetings was provided by the Society.
Conflict of interest: G. Parker is director and shareholder of Bioxydyn Limited, outside the submitted work.
Conflict of interest: G. Reilly has nothing to disclose.
Conflict of interest: G. Robinson has nothing to disclose.
Conflict of interest: G. Roditi has nothing to disclose.
Conflict of interest: J.C.L. Rodrigues has nothing to disclose.
Conflict of interest: N. Sebire has nothing to disclose.
Conflict of interest: M.G. Semple reports grants from DHSC National Institute of Health Research UK, Medical Research Council UK, Health Protection Research Unit in Emerging and Zoonotic Infections, and University of Liverpool, during the conduct of the study; and is minority owner of Integrum Scientific LLC, Greensboro, NC, USA, outside the submitted work.
Conflict of interest: C. Sudlow has nothing to disclose.
Conflict of interest: N. Woznitza reports grants from Cancer Research UK and Roy Castle Lung Cancer Foundation, personal fees from InHealth, outside the submitted work.
Conflict of interest: I. Joshi has nothing to disclose.
References
- 1.Huang C, Wang Y, Li X, et al. . Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395: 497–506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johns Hopkins University Center for Systems Science and Engineering COVID-19 Dashboard: World Map. https://coronavirus.jhu.edu/map.html [DOI] [PMC free article] [PubMed]
- 3.Guan WJ, Chen RC, Zhong NS. Strategies for the prevention and management of coronavirus disease 2019. Eur Respir J 2020; 55: 2000597. doi: 10.1183/13993003.00597-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues JCL, Hare SS, Edey A, et al. . An update on COVID-19 for the radiologist – a British Society of Thoracic Imaging statement. Clin Radiol 2020; 75: 323–325. doi: 10.1016/j.crad.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Fauw J, Ledsam JR, Romera-Paredes B, et al. . Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med 2018; 24: 1342–1350. doi: 10.1038/s41591-018-0107-6 [DOI] [PubMed] [Google Scholar]
- 6.Esteva A, Kuprel B, Novoa RA, et al. . Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542: 115–118. doi: 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hare SS, Rodrigues JCL, Jacob J, et al. . A UK-wide British Society of Thoracic Imaging COVID-19 imaging repository and database: design, rationale and implications for education and research. Clin Radiol 2020; 75: 326–328. doi: 10.1016/j.crad.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halling-Brown MD, Warren LM, Ward D, et al. . OPTIMAM Mammography Image Database: a large scale resource of mammography images and clinical data. arXiv 2020; preprint [https://arxiv.org/abs/2004.04742]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunning JW, Merson L, Rohde GGU, et al. . Open source clinical science for emerging infections. Lancet Infect Dis 2014; 14: 8–9. doi: 10.1016/S1473-3099(13)70327-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nair A, Rodrigues JCL, Hare S, et al. . A British Society of Thoracic Imaging statement: considerations in designing local imaging diagnostic algorithms for the COVID-19 pandemic. Clin Radiol 2020; 75: 329–334. doi: 10.1016/j.crad.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernheim A, Mei X, Huang M, et al. . Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology; 295: 200463. doi: 10.1148/radiol.2020200463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.EUROSTAT Healthcare resource statistics – technical resources and medical technology. https://ec.europa.eu/eurostat/statistics-explained/index.php/Healthcare_resource_statistics_-_technical_resources_and_medical_technology Date last updated: 28 November 2019.
- 13.Yang S, Shi Y, Lu H, et al. . Clinical and CT features of early stage patients with COVID-19: a retrospective analysis of imported cases in Shanghai, China. Eur Respir J 2020; 55: 2000407. doi: 10.1183/13993003.00407-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S, Li H, Huang S, et al. . High-resolution computed tomography features of 17 cases of coronavirus disease 2019 in Sichuan province, China. Eur Respir J 2020; 55: 2000334. doi: 10.1183/13993003.00334-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Gao YH, Lou LL, et al. . The clinical dynamics of 18 cases of COVID-19 outside of Wuhan, China. Eur Respir J 2020; 55: 2000398. doi: 10.1183/13993003.00398-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This one-page PDF can be shared freely online.
Shareable PDF ERJ-01809-2020.Shareable (478.2KB, pdf)